SUMMARY
Autism spectrum disorder (ASD) and intellectual disability (ID) are often comorbid, but the extent to which they share common genetic causes remains controversial. Here, we present two autosomal-recessive “founder” mutations in the CC2D1A gene causing fully penetrant cognitive phenotypes, including mild-to-severe ID, ASD, as well as seizures, suggesting shared developmental mechanisms. CC2D1A regulates multiple intracellular signaling pathways, and we found its strongest effect to be on the transcription factor nuclear factor κB (NF-κB). Cc2d1a gain and loss of function both increase activation of NF-κB, revealing a critical role of Cc2d1a in homeostatic control of intra-cellular signaling. Cc2d1a knockdown in neurons reduces dendritic complexity and increases NF-κB activity, and the effects of Cc2d1a depletion can be rescued by inhibiting NF-κB activity. Homeostatic regulation of neuronal signaling pathways provides a mechanism whereby common founder mutations could manifest diverse symptoms in different patients.
INTRODUCTION
Autism spectrum disorder (ASD) is a highly heritable neuropsychiatric condition, but its genetic architecture remains unclear. Although common genetic variants have been shown to confer risk for this disorder (Cross-Disorder Group of the Psychiatric Genomics Consortium et al., 2013), ASD is also caused by rare mutations of large effect, suggesting a genetic architecture partially similar to intellectual disability (ID) (Devlin and Scherer, 2012). Because ASD is comorbid with ID in 30%–50% of cases (Developmental Disabilities Monitoring Network Surveillance Year 2010 Principal Investigators, 2014), these disorders may not be genetically distinct. However, it is not clear how mutations in the same gene can have divergent consequences, ranging from severe ID to prominent social defects with preserved intellectual function.
Here, we report that null mutations in coiled-coil and C2 domain containing 1A (CC2D1A) consistently cause a variable spectrum of presentations including ID, ASD, and seizures even within the same family. CC2D1A encodes a multifunctional signaling scaffold that regulates multiple pathways involved in neuronal differentiation by linking transmembrane receptors and their downstream effectors, including protein kinase B (PKB/AKT) activators (Nakamura et al., 2008), and multiple effectors upstream of activation of the transcription factor nuclear factor κB (NF-κB) (Chang et al., 2011; Zhao et al., 2010).
We performed both gain- and loss-of-function experiments to define the role of Cc2d1a in neuronal differentiation and intracellular trafficking. Cc2d1a depletion in murine neurons leads to a striking reduction in dendritic complexity and dendritic spine number. By exploring the involvement of Cc2d1a in a variety of signaling pathways, we discovered that the NF-κB pathway was most strongly affected. Cc2d1a loss and gain of function both activated NF-κB in developing neurons, and restoring NF-κB activity during differentiation rescued dendritic complexity in Cc2d1a knockdown neurons. Our results suggest an important role of Cc2d1a in regulating NF-κB activity during brain development and that NF-κB activation may underlie some of the defects caused by Cc2d1a loss of function.
RESULTS AND DISCUSSION
Null Mutations in CC2D1A Cause ASD, ID, and Seizures
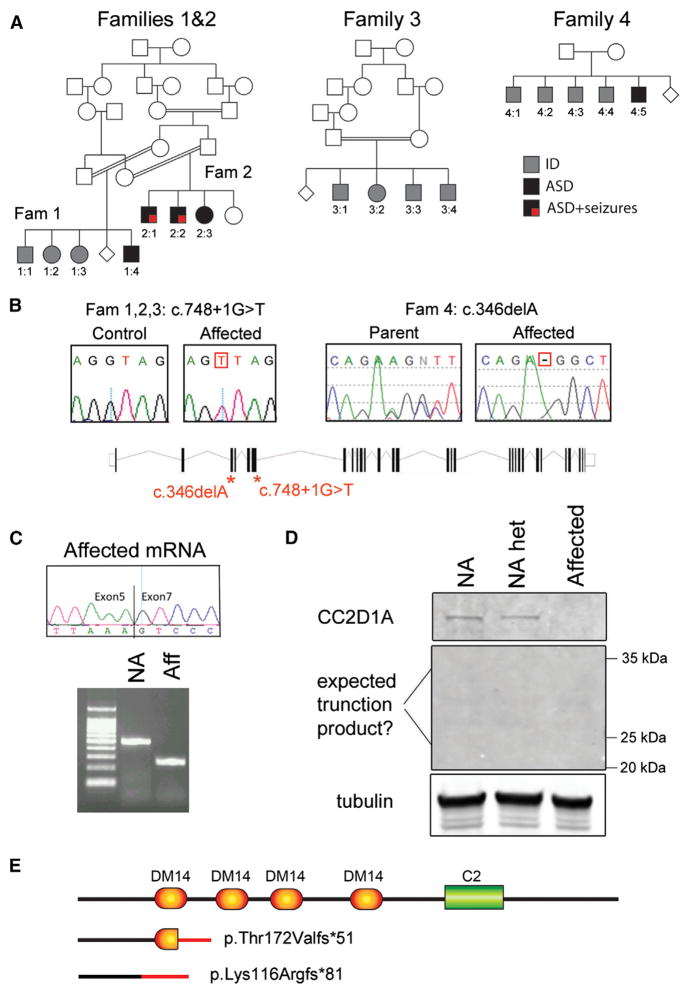
We report 4 families with a total of 16 individuals affected by a spectrum of cognitive and social impairments, including ASD, nonsyndromic ID (NSID), and seizures (Figure 1A). Families 1 and 2 represent two related consanguineous families from Saudi Arabia. In family 1, one male is affected by ASD and ID (individual 1:4), one male by cognitive problems and aggressive behavior (1:1), and two females by moderate-to-severe NSID (1:2 and 1:3). In family 2, two males (2:1 and 2:2) and one female (2:3) are affected by combinations of ASD, severe NSID, language impairment, and seizures (Table S1 for clinical information). The parents in family 3 are first cousins once removed, also from Saudi Arabia, and four siblings (three males and one female) are affected by severe NSID with language impairment (Table S1). Finally, five Pakistani males with variable presentation belonged to family 4: individuals 4:1, 4:2, and 4:4 had moderate NSID, individual 4:3 was more severely affected with language impairment and borderline autistic features, and individual 4:5 had moderate ASD/ID (Table S1). The parents reported being unrelated.
Figure 1. CC2D1A Null Mutations Cause Severe ID, ASD, and Seizures.
(A) Four families with 16 individuals affected with ID and ASD plus seizures. Affected individuals are represented by shaded symbols (squares indicate males; circles indicate females). Fam, family.
(B) All individuals in families 1, 2, and 3 carried a splicing mutation at the junction of exon and intron 6 of CC2D1A (see gene schematic). Individuals in family 4 carried a 1 bp deletion in exon 3 leading to a frameshift in the coding sequence.
(C) The splicing mutation causes complete skipping of exon 6 as shown by RT-PCR analysis of cDNA from patient cell lines. NA, not affected; aff, affected.
(D) The protein change p.T172Vfs*51 is predicted to generate a 223 amino acid truncated protein that could not be found by western blot in cell lines from an affected individual, a parent (NA het), and a normal control (NA).
(E) Schematic representation of the predicted protein fragments generated by the identified mutations. In all cases, mutations are predicted to generate a complete loss-of-function phenotype.
Genomic DNA from all family members was run on genome-wide SNP arrays, and genotyping data were analyzed under an autosomal recessive model. Linkage analysis was performed independently on the first three families, but all studies identified a common region on chromosome 19p13 with a LOD score of 3.4 for families 1 and 2 and of 3.8 for family 3. The locus had been previously linked to NSID in Israeli-Arab families, who carried a 3,589 bp deletion in CC2D1A generating an 85 kDa protein fragment (Basel-Vanagaite et al., 2006) making CC2D1A a strong positional candidate gene. The Saudi families did not carry the Israeli-Arab deletion (data not shown), but upon Sanger sequencing of CC2D1A, we identified a homozygous G > T transversion at the exon-intron junction of exon 6 (c.748+1 G > T), which segregated with the disease in the families (Figure 1B). The shared SNP haplotype observed in all affected individuals indicated that c.748+1 G > T is a founder mutation in the Saudi population shared by all three families and inherited from a distant common ancestor (Figure S1). A common region of homozygosity including CC2D1A was also identified in all affected individuals in family 4. Exome sequencing identified a 1 bp deletion in exon 3 (c.346 delA) leading to an early frameshift (p.Lys116Argfs*81) (Figure 1B). To determine whether additional mutations in a different gene could be present in families 1, 2, and 4 to explain the ASD phenotype, we reanalyzed the SNP genotyping to ascertain whether a second region with identical homozygous genotype was shared among the individuals with ASD, but no such region was present.
When CC2D1A mRNA derived from patient lymphoblasts from family 1 was analyzed via RT-PCR, it showed complete exclusion of exon 6 (Figure 1C). This skipping of exon 6 leads to a predicted frameshift at position 172 in the protein and an early stop codon at position 223 (p. Thr172Valfs*51). Western blot analysis of patient cell lines showed reduced CC2D1A expression in a heterozygous carrier and complete absence of CC2D1A in the patient, either in the full-length or truncated form (p.Thr172Valfs*51), showing that this mutation is null (Figure 1D). The frameshift mutation found in family 4 is predicted to lead to an N-terminal fragment and not to preserve any known domain, also leading to a complete loss of function (Figure 1E), indicating that the ASD/ID presentations in the patients reflect null mutations in CC2D1A.
Our findings show that CC2D1A loss of function causes an unexpectedly wide range of cognitive and social phenotypes, as well as seizures, in humans. Autosomal recessive mutations have been recently estimated to account for at least 3% of ASD cases (Lim et al., 2013), but many of these genes still remain unidentified. Although in many instances ASD caused by such recessive mutations represents a hypomorphic phenotype of a known syndrome (Yu et al., 2013), CC2D1A stands out as the only autosomal gene known to date whose complete loss of function of both alleles leads to ASD with no other syndromic presentations in males and females.
Loss of Cc2d1a Reduces Dendritic Complexity and Synapse Number in Neurons
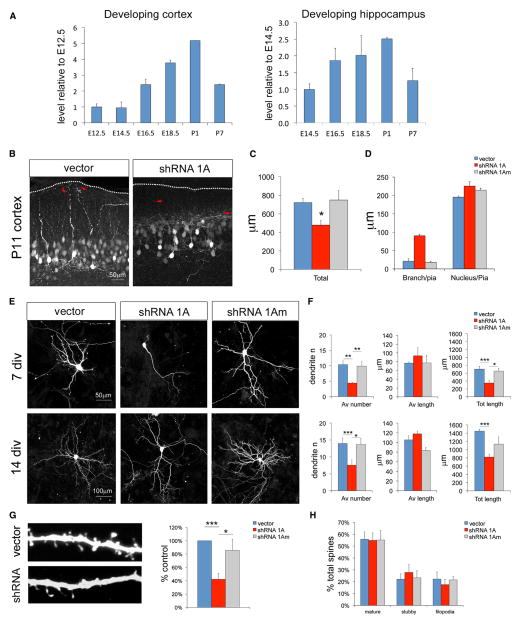
Cc2d1a has been shown to be expressed throughout the brain (Basel-Vanagaite et al., 2006), and we asked whether expression is dynamic during development. Using quantitative PCR (qPCR), we determined that Cc2d1a mRNA expression in the developing murine cortex and hippocampus shows an increase during the postnatal period corresponding to early stages of postmitotic neuronal differentiation, between embryonic day 18.5 (E18.5) and postnatal day 1 (P1) in the cortex and hippocampus (Figure 2A), suggesting possible roles in neuronal morphogenesis or synapse formation.
Figure 2. Cc2d1a Knockdown Reduces Dendritic Complexity in Hippocampal Neurons.
(A) Expression levels of Cc2d1a increase during early neuronal development in the mouse brain. qPCR analysis was performed in different tissues (cortex and hippocampus) at the ages indicated. Mean and SEM were calculated from three independent samples per time point.
(B) A simplified dendritic arbor was observed in cortical neurons at P11 following in utero electroporation of shRNA constructs at E15.5. Dendrites also appeared to be misoriented, terminating farther from the pial surface (red arrows). Scale bar, 50 μm.
(C) Total dendritic length was reduced by approximately 40% (three electroporated animals).
(D) Quantification of the disruption in dendrite orientation shown in (B). Although the nucleus was at the same distance from the pia, the apical dendritic arbor ended much farther from the surface.
(E and F) Hippocampal neurons at 7 and 14 DIV following Cc2d1a knockdown at 1 DIV also showed a reduction in total dendritic length caused by a reduction in process number. Average process length was not affected (n > 3 independent cultures). Scale bars, 50 μm (7 DIV) and 100 μm (14 DIV).
(G) The number of dendritic spines in hippocampal neurons (tested at 14 DIV) was reduced.
(H) The type of spine (mature, spine with a neck and head; stubby, spine with no neck; filopodia, elongated filamentous protrusion) was not changed in the experimental group, suggesting that spine maturation was not affected (n = 6 for G and H).
All graphs show mean ± SEM. *p < 0.5; **p < 0.01; ***p < 0.001.
Cc2d1a knockout mouse studies have only provided limited information on the role of this gene because both mouse lines developed so far lead to lethality immediately after birth despite the lack of gross abnormalities (Al-Tawashi et al., 2012; Zhao et al., 2011). Analysis of primary neurons obtained from these lines has yielded either a reduction (Al-Tawashi et al., 2012) or no change in the dendritic arbor and defects in synaptic maturation (Zhao et al., 2011). These findings suggest that compensatory mechanisms may be in place leading to phenotypic variability as observed in the patients. We used an RNAi approach to remove Cc2d1a during the perinatal period of increasing expression and study its effect on neuronal development. Multiple small hairpin RNA (shRNA) constructs targeted to different sites on the Cc2d1a mRNA reduced protein expression more than 85% (Figure S2A). An untargetable shRNA was designed by inserting five point mutations into one of the most effective shRNAs to control for possible nonspecific effects of the RNAi approach. All constructs included a GFP reporter to identify transfected cells and outline neuronal morphology.
We first knocked down Cc2d1a in neuronal progenitors of the developing mouse cortex via in utero electroporation of experimental and control plasmids at E15.5 when Cc2d1a expression begins to increase. Electroporated cells were analyzed at E18.5 and P11. At E18.5, Cc2d1a knockdown did not affect migration or leading process extension or orientation because cortices looked undistinguishable from control (Figure S2B). At P11, electroporated neurons were found in the correct location in layer 2/3, but their dendrites appeared abnormal: dendrites were jumbled and misoriented, and the overall dendritic arbor was smaller than in control conditions (Figure 2B). Dendritic arbor length measurements in isolated cells showed a reduction in the total dendritic length following Cc2d1a knockdown (Figure 2C; vector, 721.20 ± 45.84 μm; shRNA 1A, 477.61 ± 51.15 μm; shRNA 1Am, 747.90 ± 101.06 μm; p = 0.05). In addition, the apical dendrites were bent and did not extend toward the pial surface as in control cells terminating in the middle of the molecular layer (Figure 2D).
To better quantify this phenotype, Cc2d1a was knocked down in vitro in hippocampal cultures. shRNA plasmids were transfected into primary hippocampal neurons 18 hr after plating. Results from only one shRNA will be shown for simplicity, but multiple shRNAs produced identical results (Figure S2C). In neurons deficient in Cc2d1a, we observed abnormal neuronal morphology starting 4–5 days after plating, and by 7 days in vitro (DIV), there was a 50%–60% reduction in the number of dendritic processes (dendrite number, 43.7% ± 6.9% of control; p = 0.007; n = 3) and in the total dendritic arbor length (total dendrite length, 52.6% ± 14.9% of control; p = 0.02; n = 3) (Figures 2E and 2F). Reduced dendritic complexity was also evident at 14 DIV following Cc2d1a knockdown (dendrite number, 52.7% ± 4.9% of control, p = 0.03; total dendrite length, 56.6% ± 9.0% of control, p = 0.004, n = 4; Figures 2E and 2F), and dendritic spine density was significantly reduced (spine number per unit length, 42.4% ± 8.6% of control, p < 0.001, n = 6; Figure 2G). To determine whether there were differences in spine maturation or spine type, we counted the number of spines with a head and a neck (mature), without a neck (stubby) or filopodia, and found no differences in experimental or control conditions (Figure 2H).
We then analyzed both excitatory and inhibitory synapse density by determining colocalization of the AMPA receptor subunit 1 (GluA1) and the synaptic vesicle marker synapsin I, or the inhibitory synaptic scaffold gephyrin and the vesicular γ-aminobutyric acid (GABA) transporter (VGAT), respectively. No change in synaptic density (number of puncta per 10 μm of dendrite length) was observed in either excitatory or inhibitory contacts (Figures.S2D and S2E), indicating that loss of Cc2d1a may not affect synapse formation rate directly. Because synaptic density is maintained, the reduced dendritic arbor length would lead to a 50%–60% reduction in the total number of excitatory and inhibitory synapses for each neuron. These linked effects of Cc2d1a knockdown on dendritic arbor complexity, synaptic spine number, and synapse number show crucial roles in neuronal morphogenesis and circuit formation.
NF-κB Signaling Is Primarily Affected by Cc2d1a Gain of Function
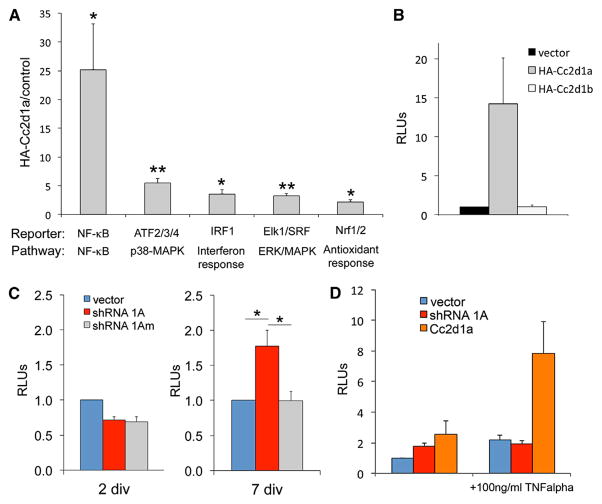
CC2D1A has been independently involved in multiple signaling pathways, including AKT (Nakamura et al., 2008), NF-κB (Chang et al., 2011; Zhao et al., 2010), RIG-I-like receptors (RLRs) (Chen et al., 2012), and protein kinase A (PKA) (Al-Tawashi et al., 2012). To determine the level of CC2D1A-mediated activation across different pathways and whether other pathways are regulated by CC2D1A, we employed a commercial array of 45 different luciferase reporters assaying a range of signaling pathways, including all known targets of CC2D1A (NF-κB, MAPK, CREB, and AKT) and many other promoters that had not been tested before (Figure S3A). Of all the promoters tested, Cc2d1a overexpression caused significant activation of NF-κB, activation transcription factors 2, 3, and 4 (ATF2/ATF3/ATF4), interferon response factor 1 (IRF1), Elk1/serum response factor (SRF), and nuclear respiration factors 1 and 2 (Nrf1/Nrf2) (Figure 3A). NF-κB showed by far the strongest response with 25.2- ± 7.9-fold increase compared to cells transfected with empty vector (p = 0.038). This was almost five times the activation of the closest reporter ATF2/ATF3/ATF4, which was activated 5.5- ± 0.8-fold (p = 0.005). IRF1, which responds to interferon, antiviral immunity downstream of RLRs (Dixit et al., 2010), and also shows close interactions with the NF-κB pathway, was elevated 3.6 ± 0.7 times (p = 0.025). All reporters activated have been involved in signaling crosstalk with NF-κB (Hoffmann and Baltimore, 2006), but it cannot be excluded that Cc2d1a is important for regulation of additional signaling pathways. Even within immune pathways upstream of NF-κB and interferon response activation, Cc2d1a appears to be quite promiscuous (Zhao et al., 2010), suggesting an important multifunctional nature.
Figure 3. Cc2d1a Overexpression Preferentially Activates NF-κB Signaling.
(A) NF-κB shows the highest activation among 45 pathways assayed using luciferase reporter arrays. Pathways showing significant activation are presented here, and remaining pathways are given in Figure S3 (n = 3).
(B) NF-κB activation is specific to Cc2d1a because expression of the homolog Cc2d1b does not activate NF-κB (n = 3). RLUs, relative luminescence units.
(C) Cc2d1a knockdown only elicits a response in NF-κB signaling at 7 DIV, but not at 2 DIV (n = 6).
(D) TNF-α treatment has different effects on neurons following Cc2d1a knockdown or over-expression. Cc2d1a knockdown prevents any further activation of NF-κB transcriptional activity, whereas treatment during overexpression causes enhanced NF-κB activation (n = 4).
All graphs show mean ± SEM. *p < 0.5; **p < 0.01.
We focused on NF-κB as a preferred target because its activation by Cc2d1a was disproportionately higher than other pathways, and NF-κB has been involved in the regulation of neuronal differentiation and survival (Mattson and Meffert, 2006). CC2D1A has been shown to act as a scaffold, coupling NF-κB activation with Toll-like receptor signaling in the endosome (Chang et al., 2011) and to control NF-κB via the canonical pathway (Zhao et al., 2010). The Cc2d1a homolog, Cc2d1b, has similar topology and structure (Figure S3B) and has been shown to share interactors with Cc2d1a (Martinelli et al., 2012; Usami et al., 2012), suggesting that these proteins may be redundant. However, Cc2d1b overexpression did not elicit an increase in NF-κB activation (Figure 3B), indicating that NF-κB activation is specific to Cc2d1a.
NF-κB Modulation Restores Dendritic Complexity following Cc2d1a Loss of Function
NF-κB is an important regulator of neurite outgrowth downstream of brain-derived neurotrophic factor (BDNF) and in concert with Notch (Bonini et al., 2011; Salama-Cohen et al., 2005), and NF-κB activation can increase or decrease dendritic complexity in ways that are pathway and context dependent (Gavaldá et al., 2009; Gutierrez et al., 2008). Therefore, we asked whether the dendritic phenotype observed following Cc2d1a loss of function in developing neurons could reflect dysregulation of NF-κB transcriptional activity. We tested NF-κB activity changes by cotransfecting luciferase reporter constructs in parallel with Cc2d1a knockdown. No change in NF-κB activity was observed following CC2D1A knockdown in human embryonic kidney 293 (HEK293) cells (Figures S4A and S4B), whereas Cc2d1a knockdown during hippocampal neuron differentiation had differential effects on NF-κB function. Early during differentiation at 2 DIV, we found no effect of Cc2d1a depletion on NF-κB activity, but NF-κB activation was moderately and significantly increased above control conditions during the peak of dendrite extension at 7 DIV in Cc2d1a knockdown neurons (luciferase activity, 1.78 ± 0.22 of control; n = 5; p = 0.014; Figure 3C). These data suggest that the regulation of NF-κB via Cc2d1a may vary in different contexts and that neurons may be more sensitive to Cc2d1a loss of function. Because our data are normalized to control conditions at 2 and 7 DIV, the known developmental increase in NF-κB activity during normal neuronal differentiation may not be immediately apparent (Boersma et al., 2011). In fact, NF-κB activity varies dynamically during development, and we speculate that the further increase in NF-κB-mediated transcription following Cc2d1a knockdown could reflect the dysregulation of this mechanism. The fly ortholog of Cc2d1a, lgd, displayed similar effects in gain- and loss-of-function experiments, where both manipulations led to increases in Notch signaling (Jaekel and Klein, 2006). In the mouse brain, Cc2d1a may similarly regulate a finely balanced NF-κB signaling mechanism, which is disrupted both by overexpression and knockdown.
To define whether the NF-κB activation following Cc2d1a overexpression and knockdown was mediated via the same mechanisms, we also challenged the neurons with tumor necrosis factor α (TNF-α). Treatment with 100 ng/ml of TNF-α elicited different responses in neurons following Cc2d1a overexpression and knockdown leading to a substantial increase in NF-κB activation following overexpression and to no effect upon knockdown (Figure 3D). The knockdown result is in concordance with studies in cell lines showing that removal of Cc2d1a prevents the cell from responding to stimuli (Chang et al., 2011; Zhao et al., 2010), whereas the increase following overexpression suggests that different mechanisms may be at play whereby stimulation is amplified by increased Cc2d1a expression. Cc2d1a overexpression has been shown to be involved in regulating NF-κB activation via the canonical activation pathway (Zhao et al., 2010), and it is possible that in the absence of Cc2d1a, noncanonical NF-κB activation can be triggered either because Cc2d1a is normally sequestering and trafficking specific effector proteins or tethering effectors to the canonical branch of the pathway.
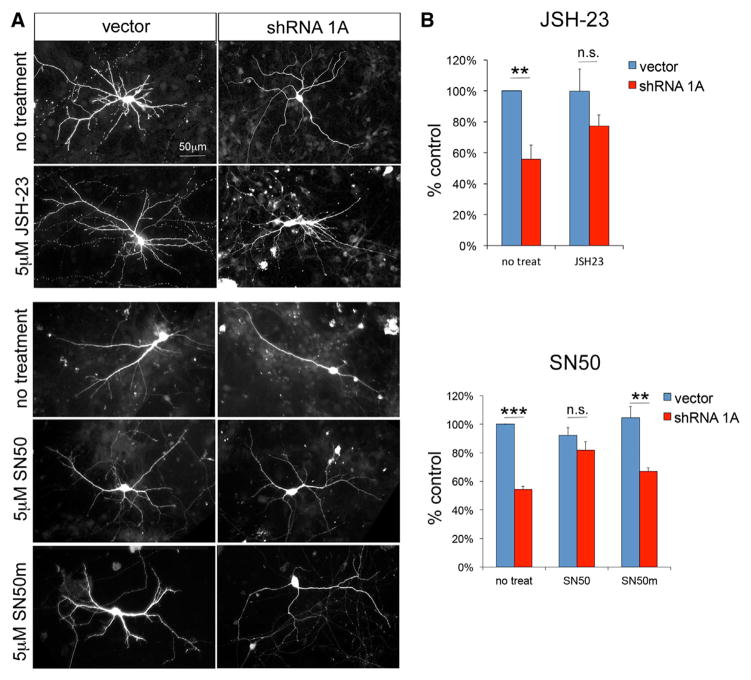
To test whether increased NF-κB activation could be responsible for the dendritic phenotype observed in neurons following Cc2d1a knockdown, we reduced NF-κB activity using two compounds targeting NF-κB nuclear translocation: SN50, a peptide inhibitor, and JSH-23, a small molecule inhibitor. Although SN50 has been widely used to modulate NF-κB activity, JSH-23, which has been shown to be very effective in reducing NF-κB activation following lipopolysaccharide treatment, is relatively untested (Shin et al., 2004). Therefore, before treating neuronal cultures with JSH-23, we tested its effectiveness in HEK293 cells and determined that it was able to block NF-κB activation following a variety of stimuli, such as TNF-α and Cc2d1a overexpression (Figure S4C). Because constitutive NF-κB activity increases during differentiation and is necessary for neuronal survival (Bhakar et al., 2002), blocking NF-κB with maximal concentrations of SN50 (20 μM) led to cell death within 12 hr (data not shown). In an attempt to only partially reduce NF-κB activity without completely blocking it, we lowered the concentration of both drugs below the half-maximal effective concentration (EC50) to try and restore normal signaling levels. We treated hippocampal neurons with 5 μM of SN50 and the inactive peptide SN50m or 5 μM of JSH-23 every 1.5 days from 4 DIV, when the dendritic phenotype first becomes evident, to 7 DIV. We found that treatment with SN50 and JSH-23, but not the inactive control peptide SN50m, rescued dendritic complexity in Cc2d1a RNAi-treated cells (Figures 4A and 4B; SN50 treatment: vector, 92.1% ± 5.5% of untreated control, and shRNA 1A, 81.9% ± 6.0% of untreated control, p = 0.26, n = 4; SN50m treatment: vector, 104.6% ± 7.8% of untreated control, and shRNA 1A, 66.8% ± 2.8% of untreated control, p = 0.004, n = 4; JSH-23 treatment: vector, 99.7% ± 9.1% of untreated control, and shRNA 1A, 77.4% ± 7.1%, p = 0.21, n = 4), indicating that loss of Cc2d1a impairs dendritic initiation via an NF-κB-dependent mechanism. NF-κB signaling is finely controlled in neurons, presumably because it is responsible for controlling morphogenesis, synapse formation, and plasticity but can also trigger cell death (Mattson and Meffert, 2006), and our results show that Cc2d1a levels must be also tightly regulated. Because of the essential role of NF-κB in neuronal survival, it is unclear whether the reduction in dendritic complexity following Cc2d1a knockdown is a reflection of reduced neuronal health or whether it is a specific effect of NF-κB on dendrite formation.
Figure 4. Treatment with NF-κB Inhibitors Rescues Dendritic Complexity following Cc2d1a Knockdown.
(A) In independent experiments, hippocampal neurons were treated with NF-κB inhibitor JSH-23 (top panels) or NF-κB inhibitor SN50 and the inactive control peptide SN50m (bottom panels). Scale bar, 50 μm.
(B) Dendrite number quantification shows that both inhibitors restore dendritic complexity in hippocampal cultures (n = 4 for each inhibitor). All graphs show mean ± SEM. **p < 0.01; ***p < 0.001; n.s., not significant.
CC2D1A’s cellular role appears to be very complex. It has been studied independently in endosomal trafficking in the fruit fly (Gallagher and Knoblich, 2006; Troost et al., 2012), membrane scission in HIV budding (Martinelli et al., 2012; Usami et al., 2012), intracellular signaling in mouse and human cells (Chen et al., 2012; Nakamura et al., 2009; Zhao et al., 2010), and transcriptional control of serotonin receptors (Ou et al., 2003), and it is still not known how these functions relate to each other. Because the Cc2d1 proteins have a unique structure and are the only genes in the genome with DM14 protein-binding domains, they may have a dedicated role in integrating different pathways, and Cc2d1a may be the only family member involved in NF-κB regulation.
The effects of CC2D1A on NF-κB function provide an intriguing potential explanation for the phenotypes of patients that share identical homozygous null mutations in CC2D1A because in different patients, loss of CC2D1A might manifest slightly differently, leading to distinct phenotypes. Although complete loss of NF-κB function is lethal in the mouse, deficits in regulating NF-κB signaling disrupt learning and memory in multiple mouse models (Meffert et al., 2003; O’Mahony et al., 2006; O’Riordan et al., 2006), and mutations in TRAPPC9, a binding protein for both NF-κB-inducing kinase and the IκB kinase, have been described in families with ID (Mir et al., 2009; Mochida et al., 2009; Philippe et al., 2009). Our findings on CC2D1A and findings on TRAPPC9 indicate that the NF-κB pathway deserves much stronger scrutiny in human genetic studies and that NF-κB signaling is a possible therapeutic target for cognitive disorders.
EXPERIMENTAL PROCEDURES
Patient Enrollment and Clinical Studies
This study was approved by the institutional review boards of Boston Children’s Hospital and of King Faisal Specialist Hospital and Research Center. All affected individuals were examined by a neurologist or a medical geneticist (M.A.S., M.G., Z.A.N., R.S., or F.S.A.) and received a comprehensive health assessment.
Linkage Analysis and DNA Sequencing
Linkage analysis using genotyping of genomic DNA using SNP arrays or micro-satellites and sequencing of CC2D1A (NM_017721)-coding region was performed using standard methods as detailed in the Supplemental Experimental Procedures.
Patient Cell Lines and Mutated Protein Detection
Transformed lymphoblastoid cell lines were established from the peripheral blood of affected and unaffected individuals at the Partners Center for Personalized Genetic Medicine (Cambridge). Cell line maintenance, lysate preparation, and western blotting are detailed in the Supplemental Experimental Procedures. The presence of CC2D1A was assessed using a mouse polyclonal antibody raised against the full-length human CC2D1A protein (Abcam).
qPCR
All animal care and use were in accordance with institutional guidance and approved by the Institutional Animal Care and Use Committee of Boston Children’s Hospital. cDNA samples from mouse brain tissue were dissected at the times indicated from NIH Swiss dams. mRNA was extracted using ReliaPrep RNA Miniprep System (Promega), and cDNA was synthesized using iScript Reverse Transcription Supermix (Bio-Rad). qPCR was performed using SYBR green reagents (SsoFast EvaGreen Supermix from Bio-Rad) on a Bio-Rad platform (CFX384). Expression was normalized to β-actin (Actb); relative expression levels are indicated as ratio to the cDNA from the earliest time available.
In Utero Electroporation
In utero electroporations were performed as previously described by Olson et al. (2006). shRNA description and validation are available in the Supplemental Experimental Procedures. As many as ten embryos were electroporated per dam and were then allowed to develop after surgery for defined periods. Pups were transcardially perfused, and brains were then sectioned to 150–300 μm sections on a vibratome (Leica), and sections were immunostained for GFP to visualize electroporated cells. 3D reconstruction of electroporated neurons was performed by generating z stacks on a Zeiss LSM510 confocal microscope. Dendrite length was measured on 3D reconstructed cells using the Filament function of Imaris software (Bitplane).
Neuronal Cell Culture and Analysis
Hippocampal cultures were prepared as described in the Supplemental Experimental Procedures, fixed with 4% paraformaldehyde at the time points indicated, and transfected neurons were immunostained for GFP to reveal dendrite morphology. A detailed description of dendritic and synaptic analysis is available in the Supplemental Experimental Procedures. Briefly, dendrite length and number and spine number were measured for at least ten cells per condition. Spine number was normalized by dendrite length to determine density. For synaptic density analysis, cells were stained for excitatory (GluA1, synapsin I) or inhibitory (gephyrin, VGAT) markers, and puncta colocalization was quantified on GFP-positive dendrites.
45-Pathway Array and Luciferase Assays
N-terminal HA-tagged Cc2d1a was generated by cloning full-length Cc2d1a in frame into pCMV-HA vector from Clontech Laboratories. Cignal Finder 45-Pathway Reporter arrays (SABiosciences) were transfected with HA-Cc2d1a or control vector following the protocol from the manufacturer, and firefly and Renilla luciferase activity was assayed 24 hr after transfection using Dual-Luciferase Reporter Assay System (Promega) in a Victor2 plate reader (PerkinElmer). Follow-up experiments testing NF-κB activity in HEK293 cells and neuronal cultures were performed as described in the Supplemental Experimental Procedures.
SN50 and JSH-23 Treatments
Primary hippocampal neuronal cultures were transfected using shRNA or control vectors as in previous experiments. At 4 DIV, cells were treated with 5 μM of SN50, SN50m (both from Calbiochem), or JSH-23 (Sigma-Aldrich) or fresh medium. Treatments to replenish fresh drugs were repeated after 1.5 days by a complete medium change, and cells were fixed at 7 DIV using 4% paraformaldehyde. Dendrite analysis was performed as described above.
Supplementary Material
Acknowledgments
The authors would like to thank the families for their constant availability and collaboration, Dilenny Gonzalez and Daniel Rakiec for technical help, Adria Bodell for help with patient recruitment, Tom Maynard at the George Washington University for help with qPCR, and the Genotyping and Sequencing Core Facility at KFSHRC for help with analysis of families 1 and 2. They are also grateful to Jenny Yang, Tim Yu, and Adam Oaks for helpful discussion. Research was supported by grants from the NIH (R01MH083565 and R01NS032457 to C.A.W. and R00HD067379 to M.C.M.) the Simons Foundation (to C.A.W.), and KACST (Grant 13-BIO1113-20 to F.S.A.). M.C.M. was also supported by grants from the Centro per la Comunicazione e Ricerca del Collegio Ghislieri, the Hearst Fund, and the Manton Center for Orphan Disease Research. M.A.S. was supported by the Deanship of Scientific Research, King Saud University, Riyadh, Saudi Arabia through the research group project number RGP-VPP-301. A.C. was supported by a fellowship from the Italian National Research Council and the Giovanni Armenise-Harvard Foundation. C.A.W. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, four figures, and one table and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2014.06.039.
AUTHOR CONTRIBUTIONS
M.C.M., L.X., M.A.S., F.S.A., and C.A.W. designed the study. M.C.M., L.X., R.S., Q.J., S.L., R.S.H., S.B.L., D.E.T., S.D.C., V.M., D.J.T., A.C., D.G., and D.S. designed and performed experiments and analyzed data. M.G., M.C., Z.A.N., S.R., M.A.S., and F.S.A. examined patients and provided materials. M.C.M., M.A.S., F.S.A., and C.A.W. wrote the manuscript.
References
- Al-Tawashi A, Jung SY, Liu D, Su B, Qin J. Protein implicated in nonsyndromic mental retardation regulates protein kinase A (PKA) activity. J Biol Chem. 2012;287:14644–14658. doi: 10.1074/jbc.M111.261875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basel-Vanagaite L, Attia R, Yahav M, Ferland RJ, Anteki L, Walsh CA, Olender T, Straussberg R, Magal N, Taub E, et al. The CC2D1A, a member of a new gene family with C2 domains, is involved in autosomal recessive non-syndromic mental retardation. J Med Genet. 2006;43:203–210. doi: 10.1136/jmg.2005.035709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhakar AL, Tannis LL, Zeindler C, Russo MP, Jobin C, Park DS, MacPherson S, Barker PA. Constitutive nuclear factor-kappa B activity is required for central neuron survival. J Neurosci. 2002;22:8466–8475. doi: 10.1523/JNEUROSCI.22-19-08466.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boersma MC, Dresselhaus EC, De Biase LM, Mihalas AB, Bergles DE, Meffert MK. A requirement for nuclear factor-kappaB in developmental and plasticity-associated synaptogenesis. J Neurosci. 2011;31:5414–5425. doi: 10.1523/JNEUROSCI.2456-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonini SA, Ferrari-Toninelli G, Uberti D, Montinaro M, Buizza L, Lanni C, Grilli M, Memo M. Nuclear factor κB-dependent neurite remodeling is mediated by Notch pathway. J Neurosci. 2011;31:11697–11705. doi: 10.1523/JNEUROSCI.1113-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CH, Lai LC, Cheng HC, Chen KR, Syue YZ, Lu HC, Lin WY, Chen SH, Huang HS, Shiau AL, et al. TBK1-associated protein in endolysosomes (TAPE) is an innate immune regulator modulating the TLR3 and TLR4 signaling pathways. J Biol Chem. 2011;286:7043–7051. doi: 10.1074/jbc.M110.164632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen KR, Chang CH, Huang CY, Lin CY, Lin WY, Lo YC, Yang CY, Hsing EW, Chen LF, Shih SR, et al. TBK1-associated protein in endolysosomes (TAPE)/CC2D1A is a key regulator linking RIG-I-like receptors to antiviral immunity. J Biol Chem. 2012;287:32216–32221. doi: 10.1074/jbc.C112.394346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross-Disorder Group of the Psychiatric Genomics Consortium; Genetic Risk Outcome of Psychosis (GROUP) Consortium. Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet. 2013;381:1371–1379. doi: 10.1016/S0140-6736(12)62129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Developmental Disabilities Monitoring Network Surveillance Year 2010 Principal Investigators . Prevalence of autism spectrum disorder among children aged 8 years—autism and developmental disabilities monitoring network, 11 sites, United States, 2010. MMWR Surveill Summ. 2014;63(Suppl 2):1–21. [PubMed] [Google Scholar]
- Devlin B, Scherer SW. Genetic architecture in autism spectrum disorder. Curr Opin Genet Dev. 2012;22:229–237. doi: 10.1016/j.gde.2012.03.002. [DOI] [PubMed] [Google Scholar]
- Dixit E, Boulant S, Zhang Y, Lee ASY, Odendall C, Shum B, Hacohen N, Chen ZJ, Whelan SP, Fransen M, et al. Peroxisomes are signaling platforms for antiviral innate immunity. Cell. 2010;141:668–681. doi: 10.1016/j.cell.2010.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher CM, Knoblich JA. The conserved c2 domain protein lethal (2) giant discs regulates protein trafficking in Drosophila. Dev Cell. 2006;11:641–653. doi: 10.1016/j.devcel.2006.09.014. [DOI] [PubMed] [Google Scholar]
- Gavaldà N, Gutierrez H, Davies AM. Developmental switch in NF-kappaB signalling required for neurite growth. Development. 2009;136:3405–3412. doi: 10.1242/dev.035295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez H, O’Keeffe GW, Gavaldà N, Gallagher D, Davies AM. Nuclear factor kappa B signaling either stimulates or inhibits neurite growth depending on the phosphorylation status of p65/RelA. J Neurosci. 2008;28:8246–8256. doi: 10.1523/JNEUROSCI.1941-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann A, Baltimore D. Circuitry of nuclear factor kappaB signaling. Immunol Rev. 2006;210:171–186. doi: 10.1111/j.0105-2896.2006.00375.x. [DOI] [PubMed] [Google Scholar]
- Jaekel R, Klein T. The Drosophila Notch inhibitor and tumor suppressor gene lethal (2) giant discs encodes a conserved regulator of endosomal trafficking. Dev Cell. 2006;11:655–669. doi: 10.1016/j.devcel.2006.09.019. [DOI] [PubMed] [Google Scholar]
- Lim ET, Raychaudhuri S, Sanders SJ, Stevens C, Sabo A, MacArthur DG, Neale BM, Kirby A, Ruderfer DM, Fromer M, et al. Rare complete knockouts in humans: population distribution and significant role in autism spectrum disorders. Neuron. 2013;77:235–242. doi: 10.1016/j.neuron.2012.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinelli N, Hartlieb B, Usami Y, Sabin C, Dordor A, Miguet N, Avilov SV, Ribeiro EA, Jr, Göttlinger H, Weissenhorn W. CC2D1A is a regulator of ESCRT-III CHMP4B. J Mol Biol. 2012;419:75–88. doi: 10.1016/j.jmb.2012.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattson MP, Meffert MK. Roles for NF-kappaB in nerve cell survival, plasticity, and disease. Cell Death Differ. 2006;13:852–860. doi: 10.1038/sj.cdd.4401837. [DOI] [PubMed] [Google Scholar]
- Meffert MK, Chang JM, Wiltgen BJ, Fanselow MS, Baltimore D. NF-kappa B functions in synaptic signaling and behavior. Nat Neurosci. 2003;6:1072–1078. doi: 10.1038/nn1110. [DOI] [PubMed] [Google Scholar]
- Mir A, Kaufman L, Noor A, Motazacker MM, Jamil T, Azam M, Kahrizi K, Rafiq MA, Weksberg R, Nasr T, et al. Identification of mutations in TRAPPC9, which encodes the NIK- and IKK-beta-binding protein, in non-syndromic autosomal-recessive mental retardation. Am J Hum Genet. 2009;85:909–915. doi: 10.1016/j.ajhg.2009.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochida GH, Mahajnah M, Hill AD, Basel-Vanagaite L, Gleason D, Hill RS, Bodell A, Crosier M, Straussberg R, Walsh CA. A truncating mutation of TRAPPC9 is associated with autosomal-recessive intellectual disability and postnatal microcephaly. Am J Hum Genet. 2009;85:897–902. doi: 10.1016/j.ajhg.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura A, Naito M, Tsuruo T, Fujita N. Freud-1/Aki1, a novel PDK1-interacting protein, functions as a scaffold to activate the PDK1/Akt pathway in epidermal growth factor signaling. Mol Cell Biol. 2008;28:5996–6009. doi: 10.1128/MCB.00114-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura A, Arai H, Fujita N. Centrosomal Aki1 and cohesin function in separase-regulated centriole disengagement. J Cell Biol. 2009;187:607–614. doi: 10.1083/jcb.200906019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson EC, Kim S, Walsh CA. Impaired neuronal positioning and dendritogenesis in the neocortex after cell-autonomous Dab1 suppression. J Neurosci. 2006;26:1767–1775. doi: 10.1523/JNEUROSCI.3000-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Mahony A, Raber J, Montano M, Foehr E, Han V, Lu S-M, Kwon H, LeFevour A, Chakraborty-Sett S, Greene WC. NF-kappaB/Rel regulates inhibitory and excitatory neuronal function and synaptic plasticity. Mol Cell Biol. 2006;26:7283–7298. doi: 10.1128/MCB.00510-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Riordan KJ, Huang I-C, Pizzi M, Spano P, Boroni F, Egli R, Desai P, Fitch O, Malone L, Ahn HJ, et al. Regulation of nuclear factor kappaB in the hippocampus by group I metabotropic glutamate receptors. J Neurosci. 2006;26:4870–4879. doi: 10.1523/JNEUROSCI.4527-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou XM, Lemonde S, Jafar-Nejad H, Bown CD, Goto A, Rogaeva A, Albert PR. Freud-1: a neuronal calcium-regulated repressor of the 5-HT1A receptor gene. J Neurosci. 2003;23:7415–7425. doi: 10.1523/JNEUROSCI.23-19-07415.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe O, Rio M, Carioux A, Plaza JM, Guigue P, Molinari F, Boddaert N, Bole-Feysot C, Nitschke P, Smahi A, et al. Combination of linkage mapping and microarray-expression analysis identifies NF-kappaB signaling defect as a cause of autosomal-recessive mental retardation. Am J Hum Genet. 2009;85:903–908. doi: 10.1016/j.ajhg.2009.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salama-Cohen P, Arévalo MA, Meier J, Grantyn R, Rodríguez-Tébar A. NGF controls dendrite development in hippocampal neurons by binding to p75NTR and modulating the cellular targets of Notch. Mol Biol Cell. 2005;16:339–347. doi: 10.1091/mbc.E04-05-0438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin HM, Kim MH, Kim BH, Jung SH, Kim YS, Park HJ, Hong JT, Min KR, Kim Y. Inhibitory action of novel aromatic diamine compound on lipopolysaccharide-induced nuclear translocation of NF-kappaB without affecting IkappaB degradation. FEBS Lett. 2004;571:50–54. doi: 10.1016/j.febslet.2004.06.056. [DOI] [PubMed] [Google Scholar]
- Troost T, Jaeckel S, Ohlenhard N, Klein T. The tumour suppressor Lethal (2) giant discs is required for the function of the ESCRT-III component Shrub/CHMP4. J Cell Sci. 2012;125:763–776. doi: 10.1242/jcs.097261. [DOI] [PubMed] [Google Scholar]
- Usami Y, Popov S, Weiss ER, Vriesema-Magnuson C, Calistri A, Göttlinger HG. Regulation of CHMP4/ESCRT-III function in human immunodeficiency virus type 1 budding by CC2D1A. J Virol. 2012;86:3746–3756. doi: 10.1128/JVI.06539-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu TW, Chahrour MH, Coulter ME, Jiralerspong S, Okamura-Ikeda K, Ataman B, Schmitz-Abe K, Harmin DA, Adli M, Malik AN, et al. Using whole-exome sequencing to identify inherited causes of autism. Neuron. 2013;77:259–273. doi: 10.1016/j.neuron.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M, Li XD, Chen Z. CC2D1A, a DM14 and C2 domain protein, activates NF-kappaB through the canonical pathway. J Biol Chem. 2010;285:24372–24380. doi: 10.1074/jbc.M109.100057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M, Raingo J, Chen ZJ, Kavalali ET. Cc2d1a, a C2 domain containing protein linked to nonsyndromic mental retardation, controls functional maturation of central synapses. J Neurophysiol. 2011;105:1506–1515. doi: 10.1152/jn.00950.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.