Abstract
Recent efforts at stratifying medulloblastomas based on their molecular features have revolutionized our understanding of this morbidity. Collective efforts by multiple independent groups have subdivided medulloblastoma from a single disease into four distinct molecular subgroups characterized by disparate transcriptional signatures, mutational spectra, copy number profiles and, most importantly, clinical features. We present a summary of recent studies that have contributed to our understanding of the core medulloblastoma subgroups, focusing largely on clinically relevant discoveries that have already, and will continue to, shape research.
Keywords: chromatin, genomics, medulloblastoma, molecular classification, pediatric brain tumors, subgroups
Medulloblastoma subgroup identification: a brief history of the confusion
During the past 5 or 6 years, multiple independent efforts at profiling the medulloblastoma transcriptome have led to the discovery and description of distinct molecular subgroups of this disease. However, bioinformatic analyses of gene-expression datasets generated from moderate (n = 46) to large (n = 194) cohorts of primary cases have revealed inconsistencies in the number of molecular subgroups identified. To make things even less clear, each research group has assigned their own nomenclatures to these subgroups, causing an ever-growing degree of confusion among the pediatric brain tumor community.
Four principal expression array-based analyses have arguably had the greatest influence on our definition of the molecular subgroups of medulloblastoma during this relatively brief history [1–4]. In 2006, Thompson et al. were the first to introduce the modern concept of transcriptionally discrete subgroups of medulloblastoma, describing five distinct subgroups in a cohort of 46 cases [1]. These subgroups were designated A–E, and most notable were the ‘B’ and ‘D’ subgroups with activation of the WNT and Sonic hedgehog (SHH) pathways, respectively. Two years later, Kool et al. essentially recapitulated Thompson’s finding of five medulloblastoma subgroups, this time in a series of 62 cases, once again assigned the letter designations A–E, but with no continuity with the nomenclature utilized in the Thompson study [2]. The WNT and SHH subgroups, respectively referred to as ‘A’ and ‘B’ by Kool et al., were readily identified as standalone sample clusters, whereas the remaining ‘C’, ‘D’, and ‘E’ subgroups were shown to be more closely related. In 2010, independent studies led by our group in Toronto, Canada, and by Scott Pomeroy (Cho et al.) in Boston (MA, USA) published seemingly back-to-back medulloblastoma subgroup manuscripts [3,4]. Our expression analysis of 103 primary tumors demonstrated the greatest statistical support for four distinct variants of the disease (labeled as ‘WNT’, ‘SHH’, ‘Group C’, and ‘Group D’), whereas the Boston study of 194 cases discriminated six molecular subtypes. In the latter report, subgroups were designated as ‘C1–C6’, with ‘C6’ representing the WNT cluster and ‘C3’ the SHH cluster. The remaining subgroups were more extensively characterized than in the previous publications, with ‘C1’/’C5’ and ‘C2’/’C4’ clusters representative of two unique subgroups with apparent substructure.
While it is evident from these findings that there are inconsistencies in the true nature/structure of the molecular subgroups of medulloblastoma and equal, if not greater, debate on the nomenclature of these subgroups, it has been firmly established that medulloblastoma comprises more than a single disease. There has been a general agreement in the community regarding the existence of WNT- and SHH-activated subgroups, fueled by their identification in all of the aforementioned studies and through extensive use of representative animal models that faithfully recapitulate the disease [5,6]. However, the true number of non-WNT/non-SHH subgroups has remained a divisive issue. Non-WNT/non-SHH medulloblastomas may represent one large subgroup [7,8] or, rather, four distinct subgroups as suggested in Cho et al.’s study [4]. Finally, in late 2010, a consensus was reached by leaders in the community at a closed meeting held in Boston, MA, USA. Based on the data presented in the recent Toronto and Boston series, as well as validation in subsequently published [9] and unpublished reports by independent groups, it was agreed that current evidence is most supportive of four core medulloblastoma subgroups, to be named WNT, SHH, Group 3 and Group 4 [10]. The somewhat generic names of Group 3 and Group 4 were thought to be most appropriate until the biology of these subgroups are better understood (Figure 1) [10].
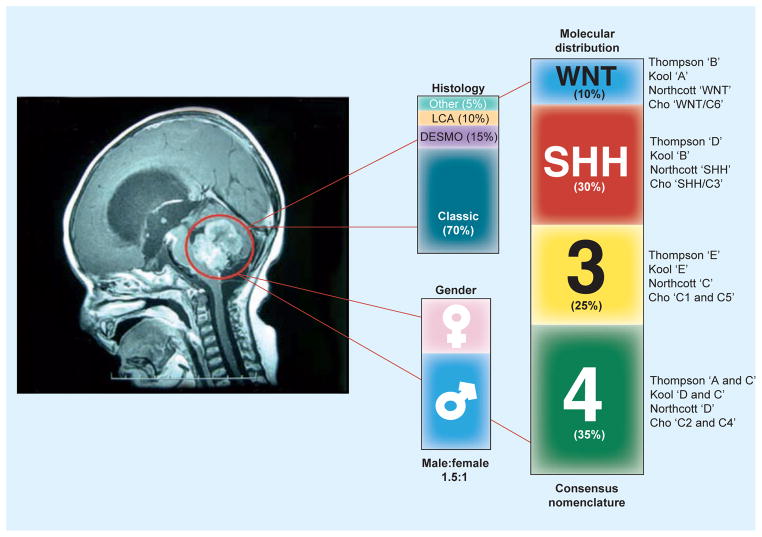
Figure 1.
Histological and gender distribution of medulloblastomas, as a single disease, as well as the prevalence of each of the consensus molecular subgroups: WNT (~10%), SHH (~30%), Group 3 (~25%) and Group 4 (~35%), across the four major studies that contributed to the discovery and definition of the molecular heterogeneity of medulloblastoma.
DESMO: Desmoplastic; LCA: Large-cell anaplastic; SHH: Sonic hedgehog.
Clinical significance
The clinical relevance associated with stratifying medulloblastoma based on their molecular and/or histological characteristics predates the publication of studies by Thompson, Kool, Northcott, and Cho that have collectively formulated much of our current understanding. In 2005, Ellison et al. provided early evidence that suggested that medulloblastomas with activated β-catenin belonged to a unique subgroup of the disease with a superior prognosis [11]. In this important study, 109 childhood medulloblastomas entered into the International Society of Pediatric Oncology/United Kingdom Children’s Cancer Study Group Primitive Neuroectodermal Tumor 3 trial were examined for β-catenin nucleopositivity and in some cases mutation status. Patients exhibiting tumors with nucleopositive β-catenin had 5-year overall survival (OS) rates of 92%, compared with only 65% for nucleonegative cases. This exceptionally good prognosis among cases with activated β-catenin reported in the European study was subsequently recapitulated in the North American St Jude Medulloblastoma-96 trial reported by Gajjar et al. in 2006 [12]. In this study, 100% (10/10) of patients falling into this ‘early’ WNT subgroup were disease-free at 5 years, compared with only 68% (40/59) of non-WNT tumors. The association of an excellent prognosis with WNT medulloblastoma has since been validated in nearly all patient cohorts examined [3,4,7,9,13–16], thus making this important subgroup a candidate for possible de-escalated therapy in future clinical trials [17].
Molecular and pathological links to clinical outcomes for what is now accepted as the SHH subgroup were also evident prior to conventional subgrouping. In 2002, Pomeroy et al.’s seminal work published in Nature demonstrated that desmoplastic medulloblastomas are transcriptionally distinct from the more prevalent classic histology and exhibit activation of SHH target genes [18]. During the past decade, numerous clinical studies have investigated the role of histology in predicting medulloblastoma outcome, with desmoplastic/nodular histology confirmed to be a positive prognostic indicator in early childhood cases [19–22], although results have been conflicting. This connection between SHH activation, desmoplastic/nodular histology and an improved patient outcome has now been verified in the context of the four medulloblastoma subgroups. Specifically, nearly all reported desmoplastic medulloblastomas belong to the SHH subgroup. Furthermore, in the large cohorts examined on tissue microarrays (TMAs) and published by Northcott et al. [3] and Remke et al. [9,13], the 5-year OS probability for SHH cases was in excess of 80%. Among the 112 Children’s Oncology Group cases (>3 years old) profiled in the Cho series for which outcome data were available, the SHH tumors exhibited a slightly diminished 6-year OS probability of approximately 65% [4]. This lower survival rate may be explained by the fact that children <3 years of age were excluded from this survival analysis, an age group in which the majority of patients belong to the SHH subgroup. The use of SHH tumors as a prognostic factor is largely based on pediatric cohorts. It is of particular importance to highlight that many prognostic markers are age-dependent in the SHH subgroup, including M-stage (adult only) and desmoplasia (pediatric only) [23]. The use of SHH tumors as a prognostic factor must be established using distinct cohorts of purely pediatric and purely adult cases to effectively establish the clinical significance of these distinct SHH variants.
Perhaps the most striking association between medulloblastoma subgroups and patient outcome came to light with the identification of the Group 3 subgroup [3,4]. Group 3 medulloblastomas are the most aggressive and malignant subgroup described to date, exhibiting 5-year OS probabilities ranging from approximately 20 to 30% [3,4,9]. This subgroup encompasses many molecular and pathological features that have been ascribed to medulloblastomas with a poor prognosis, including MYC amplification [15,16,24–27], large-cell anaplastic (LCA) histology [12,15,16,22,25,28] and metastatic disease [7,15,16,22,25,29]. In fact, the association between poor outcome and metastasis may be driven by the high prevalence of M+ disease in Group 3 tumors – we recently demonstrated that the power of M-stage as a predictor of patient outcome is lost when patients are stratified according to their molecular subgroup, with both M+ and M− Group 3 patients exhibiting a similarly dismal outcome [3].
A relevant clinical observation made by the Cho et al. study pertained to the separation of Group 3 medulloblastoma into two distinct subclasses, ‘C1’ and ‘C5’ [4]. Exposing this substructure revealed a pronounced difference in the 6-year OS, with rates of 21 versus 74% for ‘C1’ and ‘C5’, respectively. These trends suggest that the morbidity associated with Group 3 medulloblastomas is conferred by the ‘C1’ subclass, which harbors frequent MYC amplifications and is distinguished by the presence of metastasis.
Much like Group 3 medulloblastoma, our knowledge of the clinical behavior of Group 4 tumors is still rudimentary. Group 4 is the most common of all medulloblastoma subgroups, contributing to approximately 35% of all cases and thus accounting for a large percentage of what has often been referred to as non-WNT/non-SHH medulloblastoma in recent years. Group 4 tumors tend to have an ‘intermediate’ prognosis compared with the other subgroups, with reported OS rates of approximately 75–90% [3,4,9,13]. In a very recent study by Remke et al., clinical heterogeneity was observed within the Group 4 subgroup, identifying FSTL5 as a biomarker of high-risk Group 4 patients [9]. Future identification of similar biomarkers or, more likely, combinations of biomarkers will potentially lead to further dissection of high-risk patients within the core subgroups of medulloblastoma (Figure 2) [30].
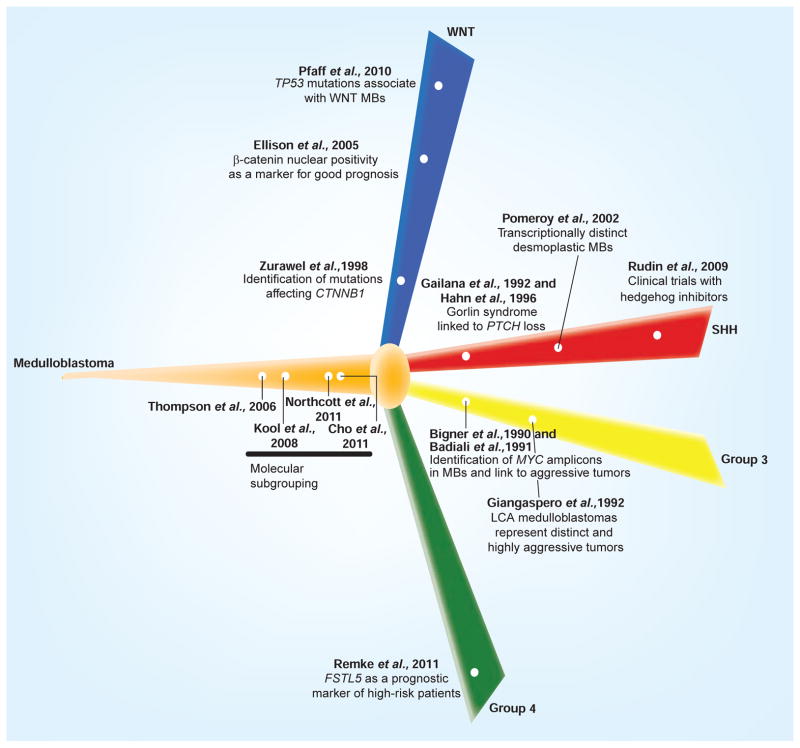
Figure 2.
Chronological reorganization of selected landmark papers that have significantly impacted our molecular or clinical understanding of the medulloblastoma subgroups consensus.
LCA: Large-cell anaplastic; MB: Medulloblastoma; SHH: Sonic hedgehog.
Demographics
Age
Although medulloblastoma is the most common malignant pediatric brain tumor, the disease does occur in adults, albeit quite rarely compared with the more prevalent glioblastomas that represent the prototypical brain tumor of adulthood. Several original reports and epidemiological studies have shown that pediatric and adult medulloblastomas are genetically distinct and can behave quite differently from a clinical standpoint, suggesting that the two may represent distinct forms of the disease with potentially disparate origins [31–33]. Observations made in recent molecular subgrouping studies may account for some of these notable clinical differences that exist between younger versus older patients afflicted with medulloblastoma.
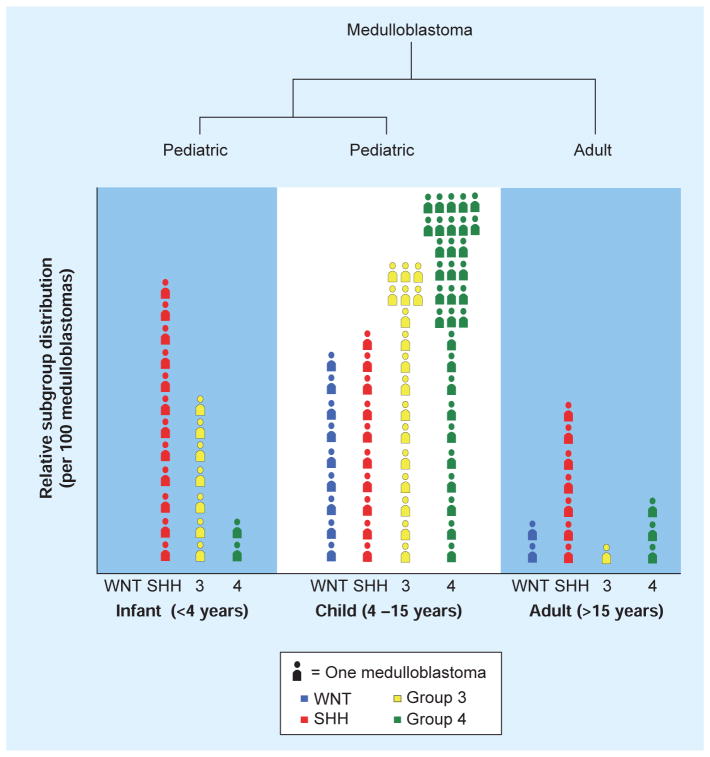
Medulloblastoma subgroups demonstrate a rather disparate distribution across distinct patient age groups. WNT tumors are found mainly in children and sometimes in adults, and are rarely seen in infants. SHH medulloblastomas are dominant in both infants and adults, accounting for more than approximately 60% of patients from either age group. By contrast, SHH tumors are underrepresented in childhood, constituting only approximately 10–15% of these cases. Group 3 medulloblastomas are found in pediatric patients but are exceptionally rare in older teenagers and almost never occur in adults. Group 4 cases are found in all age groups but appear to represent the bulk of childhood and adolescent medulloblastomas (Figure 3).
Figure 3.
Relative frequency of the four molecular subgroups of medulloblastoma across: infants (≤4 years of age), children (4–15 years of age) and adult patients (≥15 years of age) per 100 tumors, demonstrating both the prevalence (SHH: ~60% of infants) and rarity (Group 3: ~1–2% of adults) of each molecular subgroup and their association with age.
SHH: Sonic hedgehog.
The uneven distribution of medulloblastoma subgroups across infants, children and adult patients has significant biological and clinical implications. Despite excellent survival rates repeatedly reported for WNT medulloblastomas, Remke et al. recently demonstrated that the improved survival of WNT patients is only applicable to pediatric cases, as adult WNT tumors exhibited a diminished OS rate of approximately 80% in the cohort examined [13]. While SHH medulloblastomas of different age groups do not display any significant clinical differences in their OS [13,23], our group has demonstrated that differences exist in the incidence of metastasis (higher in pediatric SHH), tumor localization (more frequently located in the vermis in pediatric SHH) and a trend towards a higher incidence in female patients in pediatric SHH versus adult SHH cases [23]. Furthermore, many prognostic markers demonstrate an age-dependent relationship, highlighting the differences between pediatric and adult SHH tumors [23].
Among all published cohorts with available subgroup associations, literally only a handful of patients over the age of 18 years have been assigned to Group 3, rendering this subgroup largely a pediatric disease. Indeed, in their paper characterizing the subgroups of adult medulloblastoma, Remke et al. concluded that adult medulloblastoma consists of just three distinct molecular variants: WNT, SHH and Group 4 [13]. Furthermore, in a very recent meta-analysis study carried out by Kool et al., which included molecular and clinical data on 550 medulloblastoma cases, only four out of 177 (2.2%) adults were classified as Group 3, reaffirming the rarity of this subgroup in adults [34]. Although we are only beginning to uncover the molecular and cellular basis of this highly aggressive subgroup, it is of particular interest that Group 3 is confined to younger patients, further substantiating the hypothesis that pediatric and adult medulloblastomas are distinct diseases. Finally, while Group 4 medulloblastomas are found across all age groups, survival analyses performed in the adult study by Remke and colleagues indicate that not all Group 4 share the same prognosis, with adult Group 4 patients exhibiting a pronounced reduction in OS (~20%) compared with their pediatric counterparts [13].
Gender
It has long been reported that medulloblastoma is more commonly diagnosed in males, with an accepted male:female ratio of approximately 1.5:1 (Figure 1). Although this ratio has been repeatedly observed across patient cohorts, little insight has been gained in terms of possible mechanism(s) responsible for this gender bias. Recent integration of molecular subgrouping with gender information has shown that males and females exhibit distinct frequencies with respect to their molecular subgroup makeup. Indeed, differences in subgroup distribution may explain the disparate responses to treatment that have been observed between males and females in the clinic [35].
Based on the combined cohorts of the recent studies published by Cho et al. [4] and our group [3], multiple relevant and significant observations related to subgroup distribution according to gender come to light. The male:female ratio for the combined cohort (n = 291) is 1.53:1, which is consistent with the accepted gender ratio for this malignancy. However, these trends change rather dramatically when examined in a subgroup-specific context, with male:female ratios of 0.5:1 and 2.8:1 for WNT and Group 3, respectively. These differences translate into a threefold increase in the prevalence of WNT tumors among females compared with males, occurring in 12 and 4% of females and males, respectively. Likewise, SHH tumors constitute a higher percentage of female medulloblastomas, accounting for 39% of cases versus only 23% of males. Moreover, Group 3 and Group 4 tumors account for 73% of male medulloblastomas versus only 48% of female cases. Collectively, data from these two cohorts show an uneven distribution of medulloblastoma subgroups according to patient gender. Future studies examining larger cohorts of patients will determine the validity of these reported distributions.
Histology
According to the 2007 WHO classification of tumors of the CNS [36], medulloblastoma consists of five histological variants, including (listed in order of prevalence) classic, desmoplastic/nodular, ana-plastic, large-cell and medulloblastoma with extensive nodularity (MBEN). The anaplastic and large-cell variants are typically amalgamated as a single LCA phenotype and regarded as a more clinically aggressive form of the disease compared with the other variants [37]. By contrast, the MBEN phenotype is enriched in very young infants and has recently been correlated with a very good prognosis [38–40]. The histological subtypes are not evenly distributed among the subgroups, the most notable of which are the desmoplastic variants that appear to be almost entirely restricted to SHH tumors [2–4,7]. Indeed, Ellison et al. reported that all desmoplastic tumors correlated with a SHH immunophenotype in a large study of 235 medulloblastomas investigated on a medulloblastoma TMA [7]. Likewise, MBENs have recently been shown to be strongly linked to Gorlin syndrome, and thus the SHH subgroup due to underlying PTCH1 germline mutations [39]. Moreover, germline SUFU mutations have been recurrently catalogued in this rare subtype, further substantiating their association with SHH medulloblastoma [41]. WNT tumors are almost always of classic histology, although exceptionally rare LCA cases have also been reported. LCA tumors have been identified in all subgroups of medulloblastoma but have a significantly higher incidence in Group 3 tumors, in agreement with the poor outcome associated with this subgroup (Figure 1) [3,4]. Interestingly, the LCA histological subtype has been shown to correlate with MYC expression [42] and, reciprocally, over-expression (or amplification) of MYC leads to the development of an LCA phenotype, further linking these two markers of poor prognosis with the causal aggressiveness of Group 3 tumors [43].
Genomics
Transcriptomics
It could be argued that the ‘molecular signature’ of medulloblastoma subgroups is reflected in their underlying transcriptional profiles. Indeed, their discovery, much like the discovery of the major subtypes of GBM, can be credited to large-scale gene-expression profiling efforts that divulged the transcriptional heterogeneity of these diseases [44–46]. Despite considerable variation in the size of patient cohorts, gene-expression array platforms employed and unique combinations of bioinformatic tools, the consensus among the genes and pathways that are enriched among the subgroups has remained consistent.
WNT medulloblastomas express exorbitantly high levels of genes in the WNT pathway, including WIF1, DKK1, DKK2, DKK4, LEF1, AXIN2, WNT11, WNT16, FZD6 and FZD10 [1–4,47]. Several of these targets are known inhibitors of WNT signaling and are upregulated as part of a negative-feedback loop secondary to aberrant WNT activation. MYC is also expressed in WNT tumors and is a known target of the WNT cascade. Other transcriptional themes enriched in the WNT subgroup include TGF-β signaling, axonal guidance signaling and gene sets involved in the epithelial–mesenchymal transition.
SHH medulloblastomas, as implied by their nomenclature, are characterized by aberrant expression of a plethora of known SHH pathway genes, such as HHIP, GLI1, GLI2, GLI3, PTCH2, BOC and ATOH1 [1–4]. WNT pathway genes SFRP1, SFRP4 and SFRP5 are also highly expressed in SHH tumors, as are members of the SOX gene family including SOX2, SOX9 and SOX13. Comparing the transcriptomes of pediatric SHH tumors to their adult counterparts, we [23] and others [48] recently noted very distinct expression patterns between these age groups, with adult SHH tumors expressing elevated levels of HOX family genes (i.e., HOXA5, HOXA9 and HOXA2) and genes involved in tissue development, whereas pediatric SHH tumors showed an enrichment of gene sets related to extracellular matrix function. The disparate transcriptional profiles observed between these two age groups of SHH medulloblastoma suggest they have distinct underlying biologies, and as a result may respond differently to current targeted therapies designed to abrogate SHH pathway activation [49,50].
Group 3 and Group 4 medulloblastomas remain quite poorly characterized compared with WNT and SHH tumors. Group 3 medulloblastomas tend to exhibit a retinal or phototransduction gene-expression signature, characterized by genes such as GABRA5, NRL and CRX [2–4], whereas Group 4 medulloblastomas are characterized by expression of neuronal factors including GRM1 and GRM8, as well as potassium channel genes KCNA1 and KCNA5 [2–4]. In Cho et al.’s classification study, the Group 3 (‘C1’/’C5’) tumors were said to be GABAergic and Group 4 (‘C2’/’C4’) tumors to be glutamatergic with respect to their transcriptional profiles, in addition to the aforementioned phototransduction/neuronal signature distinction [4]. Perhaps the most striking difference between Group 3 and Group 4 at a gene expression level relates to MYC, which is expressed at very high levels in Group 3 and at only a modest baseline in Group 4 [2,3]. The aberrant expression of MYC reported in the poor-prognosis Group 3 subgroup is consistent with several previous reports that have linked elevated MYC activity with an inferior clinical outcome. Notably, in the Cho et al.’s study, aberrant MYC levels were most prevalent in the poor-outcome ‘C1’ subclass when compared with ‘C5’ [4]. This important distinction further supports the concept of both biological and clinical heterogeneity within the core subgroups, providing rationale for dissecting them beyond the core hierarchy.
Cytogenetics
The molecular subgroups of medulloblastoma are easily distinguished as a result of their divergent transcriptional profiles. However, what is responsible for these unique transcriptional programs? The current body of evidence suggests that the medulloblastoma subgroups may originate from distinct cellular lineages [51–53]. However, the discriminatory karyotypes that characterize these subgroups also have strong influences on the observed gene-expression patterns. The most notable cytogenetic events that are enriched in the different subgroups are discussed below.
Monosomy 6 was the first cytogenetic aberration linked to a medulloblastoma subgroup [1,14], now confirmed in nearly all WNT tumors karyotyped to date. Interestingly, Thompson et al. associated the loss of chromosome 6 with WNT tumors somewhat indirectly, by virtue of the majority of genes on this chromosome exhibiting reduced expression in WNT tumors [1]. Since this discovery, monosomy 6 has become a defining feature of the WNT subgroup and is rarely (if ever) found in non-WNT tumors. As a result of the WNT-specific nature of this aberration, some have utilized chromosome 6 genomic status as a surrogate marker to identify or confirm WNT subgroup affiliation. Of equal importance, mutations in CTNNB1 almost always coincide with monosomy 6, although rare exceptions of exclusivity for these events have been reported. Since WNT tumors have an exceptionally good prognosis, intensive efforts are currently being employed to determine an ideal ‘test’ for identifying these patients, including both CTNNB1 mutational screening and chromosome 6q FISH. Outside of these genetic events, the WNT genome is extremely ‘quiet’ and essentially balanced, with very few gains and losses reported elsewhere.
Deletion of chromosome 9q, long suspected to be secondary to PTCH1 mutation on 9q22, is found in approximately a third of SHH medulloblastomas. Loss of 9q appears to be highly restricted to SHH – found only in this subgroup in both the recent Cho et al. and Northcott et al. studies [3,4]. Additional over-represented copy number changes that have been reported in SHH tumors include gain of chromosomes 2 and 3, and losses of chromosomes 10q and 20p. Importantly, 9q and 10q deletions are generally mutually exclusive events, suggesting genetic heterogeneity within the SHH subgroup, some of which may be explained by underlying molecular differences between pediatric and adult SHH tumors [23,48], including a reduced frequency of 10q loss in adult cases [23]. These results, in conjunction with age-dependent prognostic markers, demonstrate that pediatric and adult cases of SHH medulloblastoma are biologically and clinically distinct [23].
From a karyotypic perspective, Group 3 and 4 share a number of common genomic features, including gain of chromosomes 7, 17q and 18, and loss of chromosomes 8, 11p and the X chromosome in females. Isochromosome 17q (i[17q]), the most frequently reported structural aberration in medulloblastoma [54,55], is prominent in both Group 3 and Group 4; however, it represents a defining feature of Group 4 tumors, as it is present in up to 80% of this subgroup, whereas Group 3 medulloblastomas display this aberration less frequently. Despite their genomic similarities, discriminatory cytogenetic events do exist between these subgroups, including gain of chromosome 1q (Group 3), and chromosome 4 (Group 4), and losses of chromosomes 5q, 10q and 16q that are more prevalent in Group 3 tumors.
Oncogenes, tumor suppressors & medulloblastoma subgroups
CTNNB1
First published in a study by Zurawel et al. in 1998 [56], mutations affecting GSK3β phosphorylation sites in exon 3 of CTNNB1 have since been reported in approximately 5–10% of medulloblastomas investigated [53–55]. By virtue of representing the apex of the WNT signaling pathway, it should have come as no surprise that CTNNB1 mutations are confined to what is now known as the WNT subgroup. Indeed, the prevalence of CTNNB1 mutations in subgroup ‘B’ reported by Thompson et al. [1] and later subgroup ‘A’ reported by Kool et al. [2] undoubtedly led to the definition of what is now accepted as the WNT subgroup.
TP53
Mutations in the TP53 tumor-suppressor gene have historically been reported in approximately 10% of sporadic medulloblastomas [53–55], a frequency that is considerably lower than that observed in glioma, where mutations in the TP53 signaling pathway are believed to contribute to upwards of 85% of cases [46]. Li-Fraumeni patients who intrinsically harbor TP53 germline mutations develop a wide range of cancers of different histologies, including medulloblastoma [53,57]. Although there have been conflicting reports regarding the prognostic significance of TP53 mutation in medulloblastoma [58–61], recent studies have suggested an association between TP53 mutation and the WNT subgroup. Pfaff et al. confirmed the mutational status of TP53 in a series of 310 medulloblastomas, established a mutation frequency of 6.8% (21/310), and showed that TP53 mutation frequently co-occurred with monosomy 6 and CTNNB1 mutation (7/21 TP53-mutated tumors), both of which are defining characteristics of WNT medulloblastomas [59]. Using immunohistochemistry-based classification, the authors also showed that TP53 mutations were found in WNT, SHH, and Group 4 tumors, but not in the prognostically inferior Group 3 subgroup. In a subsequent correspondence from Steve Clifford’s group in the UK, a similar connection between TP53 mutation and WNT subgroup affiliation was implicated, with three out of five TP53-mutated cases assigned to the WNT subgroup in a series of 50 tumors investigated [60].
PTCH1
Historically, PTCH1 has been regarded as the ‘prototypical’ tumor-suppressor gene in medulloblastoma, reported to be mutated in approximately 8–10% of sporadic cases and known to be the underlying genetic basis of Gorlin syndrome, a hereditary cancer syndrome in which individuals harboring PTCH1 germline mutation are predisposed to develop medulloblastoma [53,57]. Much like CTNNB1 mutations typifying the WNT subgroup, mutations of PTCH1 encoding the PATCHED transmembrane receptor (resulting in constitutive activation of the SHH signaling cascade) are intimately linked to the SHH subgroup. PTCH1 sequence analysis has ultimately influenced the definition of the SHH subgroup – collectively, PTCH1 mutations were observed in nine out of 27 SHH tumors reported in the Thompson and Kool studies and not found in any of the non-SHH cases [1,2]. Other components of the SHH pathway, including SUFU and SMO, are known to be mutated in medulloblastoma and contribute to medulloblastoma pathogenesis [62,63]. Although mutations affecting these genes are presumably restricted to the SHH subgroup, this correlation has yet to be comprehensively evaluated.
MYC family
Proto-oncogenes belonging to the MYC family, including MYCN, MYC and MYCL1, comprise the most frequent targets of high-level amplification reported in medulloblastoma [15,64]. Importantly, patients harboring amplification of either MYC or MYCN have been repeatedly shown to exhibit significantly compromised progression-free survival and OS probabilities across multiple patient and clinical trial cohorts [15,16,65,66]. Although MYC and MYCN were among the first oncogenes reported as being amplified in medulloblastoma, the association of these oncogenes to a given subgroup has only recently come to light. To date, virtually all reported MYC amplifications have been linked to Group 3 tumors [3,4]. In contrast to the more restricted MYC-amplified cases, MYCN-amplified cases are found in either SHH or Group 4 medulloblastomas [3,4,65]. Amplifications affecting MYCL1 are exceedingly rare in medulloblastoma and have yet to be connected to a particular subgroup(s). While initial reports demonstrated universal poor prognosis for both MYC and MYCN amplifications [15], recent studies have demonstrated that tumors with MYCN amplifications have a much greater genetic and clinical heterogeneity than originally believed [67].
MLL2 & histone-modifying genes
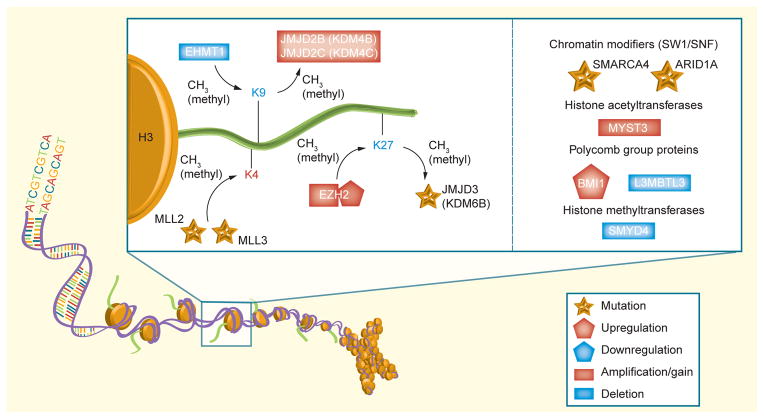
Numerous recent genomic profiling efforts have uncovered both recurrent copy number aberrations and DNA sequence variants that target genes encoding histone-modifying proteins (Figure 4). In 2009, we first reported recurrent homozygous deletions of EHMT1, a histone 3, lysine 9 methyltransferase on chromosome 9q34 [64]. In the same study, we reported recurrent amplification of JMJD2C, a jumonji family histone lysine demethylase on chromosome 9p24. Although patient subgroup information was not available at the time of this study, the observed somatic deletions of EHMT1 occurred in the context of monosomy 9q, a genetic event confined to the SHH subgroup.
Figure 4.
Schematic representation of chromatin-modifying proteins identified as mutated, amplified/deleted, or over-/under-expressed in medulloblastoma
This important pathway affects a number of lysine residues (K4, K9 and K27), altering gene expression and inhibiting differentiation in a tumor-specific context.
More recently, a landmark study by Parsons et al., which involved whole-exome sequencing of 22 medulloblastomas, identified inactivating mutations of MLL2 and MLL3, both histone lysine methyltransferases, in 16% of medulloblastomas [68]. Additional chromatin-modifying genes, particularly SMARCA4, ARID1A and KDM6B, were also found to be somatically mutated in this study, although at a much lower frequency. Although this study had limited information regarding molecular subgroup status, mutations in MLL2 and MLL3 appeared to be distributed across subgroups.
Other medulloblastoma oncogenes & tumor suppressors
OTX2 is among the most frequent targets of focal copy-number gain reported in medulloblastoma, accounting for approximately 10% of all cases [64]. Recently, Adamson et al. demonstrated that these aberrations are restricted to the Group 3 and Group 4 subgroups [69], thus potentially making OTX2 an attractive candidate for molecularly targeted therapy in non-WNT/non-SHH tumors.
The TSC1 tumor-suppressor gene on chromosome 9q34 is both mutated and recurrently focally deleted in medulloblastoma [64,68,70]. In collaboration with Bhatia et al., we recently showed that these aberrations are highly over-represented in SHH medulloblastoma [64,70], implicating cooperation of aberrant SHH signaling and deregulation of the mTOR pathway in SHH-driven tumors. Since both of these pathways are actionable, therapeutic strategies combining inhibitors of both cascades may prove to be effective in future targeted therapies of this subgroup.
Technical aspects of medulloblastoma subgroup classification
Given what has recently been uncovered with respect to the clinical ramifications of medulloblastoma subgroups, an immense amount of interest is currently focused on the development and implementation of techniques for reliably assigning clinical samples to their appropriate subgroup. All of the key gene-expression array studies described throughout this review essentially used the same approach for defining the subgroups – profiling moderate-to-large cohorts on conventional oligonucleotide arrays using high-quality RNA extracted from fresh-frozen tissue. Unsupervised class discovery approaches such as hierarchical clustering and non-negative matrix factorization performed on the most variant genes among the cohorts has proven to be a robust method for discriminating medulloblastoma subgroups [1–4,9]. Furthermore, application of various class prediction algorithms and novel bioinformatic tools such as SubClass Mapping [71] and Metagene Projection [72] have allowed groups to define published series of array data in terms of their own concept of medulloblastoma subgroups [3,4,23]. Although these array-based methods have been informative, they are not practical for conditions typically encountered in a clinical setting where access to high-quality tumor RNA and expertise capable of performing the complex bioinformatics involved in microarray data processing are unlikely to be available.
To circumvent these limitations related to sample quality and available technologies, multiple groups have extracted information gained from the previous gene-expression array studies and chosen to focus on subsets of signature genes as molecular markers of the medulloblastoma subgroups [8,73]. Since virtually all clinical medulloblastoma samples are at some stage prepared as formalin-fixed paraffin-embedded (FFPE) material, IHC for top subgroup-specific signature genes has become both a convenient and effective method for medulloblastoma classification. To date, a variety of different markers have been tested for their ability to demarcate a particular subgroup using IHC, with some of these now effectively utilized by multiple independent laboratories. For instance, nuclear CTNNB1 staining is now widely used as more or less a definitive marker of WNT medulloblastoma [3,7,11–14,47,48,74], although antibodies against DKK1 [1,3,9] and DKK4 [1] have also demonstrated efficacy in identifying WNT tumors. Similarly, SFRP1 [1,3,9,12,13,48], GLI1 [1,3,48] and, more recently, GAB1 [7] have all shown promise as molecular markers of SHH medulloblastoma. In collaboration with the team in Heidelberg, Germany, the authors have now published multiple reports whereby just four commercially available antibodies were used to establish subgroup status of several hundred FFPE medulloblastomas present on TMAs [3,9,23,65]. These markers include DKK1 (WNT), SFRP1 (SHH), NPR3 (Group 3) and KCNA1 (Group 4) and have proven to be extremely reliable markers of the four medulloblastoma subgroups. Despite these supportive published data, the transferability of our four antibody assay across the globe may be limited due to factors related to differences in sample preparation and fixation techniques used in different pathology laboratories, as well as potential lot-to-lot variability of the current antibody sources, most of which are rabbit polyclonal in origin. As such, novel molecular-based approaches for subgrouping medulloblastoma that are rapid, cost effective and compatible with FFPE-derived material routinely available in the clinical setting are warranted. Recently, we developed an assay based on NanoString Technology® that incorporated a 22-gene signature consisting of the most highly expressed subgroup-specific signature genes in order to predict medulloblastoma subgroup affiliation. This approach led to the accurate classification of medulloblastoma subgroups in approximately 98% (127/130) of fresh-frozen cases with known subgroup affiliation and, importantly, was highly compatible with FFPE-derived RNA from recent cases [73].
Preclinical models of medulloblastoma subgroups
Given the considerable progress that has been made in dissecting the molecular and clinical heterogeneity of medulloblastoma subgroups, the shift now moves towards the generation of rational and faithful preclinical models of the four subgroups. These models will be an essential tool for gaining further insight into the cellular origins of the subgroups and testing molecularly targeted therapies designed to inhibit the oncogenes and signaling pathways deregulated within them. Although multiple excellent reviews have recently summarized preclinical medulloblastoma models, particularly mouse models, we briefly discuss the current status of this topic here, highlighting the degree to which these published models match the subgroups described in the human disease. For more extensive reviews on the subject of medulloblastoma mouse models, the reader is encouraged to refer to other recent and more comprehensive publications [5,6,75].
Until only recently, most, if not all, mouse models of medulloblastoma relied on genetic perturbation of the SHH signaling pathway. The Ptch1+/− mouse is by far the most well characterized and has served as a workhorse for modeling the disease for >10 years [76–78]. Heterozygous loss of Ptch1 results in the generation of medulloblastomas within 3–6 months with a penetrance of approximately 15%. Breeding these mice with those on a Tp53-null background to generate Ptch1+/−; Tp53−/− mice increases the incidence to nearly 100%. Sporadic mouse medulloblastomas have also been successfully produced through manipulation of other SHH signaling components, including transgenic over-expression of Smo (i.e., ND2-SmoA1) [79,80], deletion of Sufu (i.e., Sufu+/−; Tp53−/−) [81], and through targeted deletion of Ptch1 in different cell types [82]. All of the above models, in addition to several others not mentioned here, faithfully represent the human SHH subgroup, which is not surprising given their underlying genotypes. A multitude of studies strongly suggests that these tumors are derived from cerebellar granule neurons, are typically of classic histology and are characterized by elevated SHH signaling [53,83]. Furthermore, multiple recent publications have described a histological phenotype consistent with what is observed in the human MBEN subtype (which appear to be over-represented in the SHH subgroup) [39,41] when the SHH pathway is activated in combination with PI3K pathway perturbation in transgenic mice [84,85]. Given the relative paucity of MBEN among human medulloblastomas, the availability of a potentially reliable preclinical model of this subtype may provide alternative avenues for gaining an improved understanding of this rare subtype.
In 2010, Gibson et al. successfully introduced what was arguably the first bona fide model of non-SHH mouse medulloblastoma [52]. Targeted expression of activated β-catenin (Blbp-Cre; Ctnnb1+/ lox[ex3]) and concurrent loss of Tp53 to cells of the lower rhombic lip/dorsal brainstem resulted in approximately 15% of mice developing medulloblastomas that were anatomically and molecularly distinct from the previous SHH models. Comparison of gene-expression data from these CTNNB1-driven mouse tumors with data derived from human cases demonstrated a significant correlation with the WNT subgroup, establishing this model as the first to accurately model a subgroup, other than SHH.
Another elegant model, also published in 2010, used an inducible system to bidirectionally overexpress both Mycn and luciferase (known as GTML) in the developing cerebellum [86]. Tumors derived from this genotype occurred in up to 75% of mice by 200 days, produced tumors of either classical or LCA pathologies, originated independently of cerebellar granule neuron progenitor cells, and only rarely (~5%) expressed SHH signature genes – strongly suggesting that GTML tumors are more often representative of non-SHH human medulloblastoma.
Very recently, two independent laboratories published back-to-back papers in Cancer Cell showing that aberrant expression of Myc combined with abrogation of Tp53 activity could faithfully recapitulate tumors resembling human Group 3 medulloblastoma [87–89]. These highly complimentary studies used an ex vivo approach to introduce Myc into either neural stem cells [88] or cerebellar progenitors [87] and transplanted the Myc-expressing cells intracranially into immunocompromised mice. When combined with loss of wild-type Tp53, either through the use of dominant negative Tp53 [88] or Tp53 deletion [87], both of these strategies yielded highly penetrant, aggressive medulloblastomas with a short latency. Importantly, tumors derived from these animals were reminiscent of human LCA medulloblastoma, with a histology common to Group 3, and transcriptionally matched expression data derived from human Group 3 tumors. The requirement for Tp53 loss-of-function in both of these models as a mechanism to evade Myc-driven apoptosis is a topic of some debate, if these models are to be considered accurate representations of human Group 3 moving forward. Although MYC amplification and overexpression are highly specific to Group 3, direct inactivation of TP53 via mutation or focal deletion is rarely seen in this subgroup, occurring much more frequently in WNT and SHH tumors, as described earlier [59,60]. Despite the general lack of TP53 mutations in Group 3, chromosome 17p deletion (chromosome 17p being the genomic location of TP53) is quite prevalent, found in upwards of 50% of cases from this subgroup [3,4,34]. Moreover, the TP53 pathway may be affected by mutations in genes other than TP53 in Group 3, although this has yet to be thoroughly investigated. Nonetheless, these new Myc-driven models of medulloblastoma will undoubtedly serve as a valuable resource for future dissection of Group 3 biology and as a platform for preclinical testing in this poor-prognosis subgroup.
Using the sleeping beauty (SB) transposon system, Wu et al. recently performed ‘saturation’ mutagenesis of Math1-expressing CGNPs in an attempt to uncover novel medulloblastoma oncogenes and tumor suppressors [90]. Using the SB system combined with next-generation sequencing, Wu et al. were able to identify genes affected by common insertion of the SB transposon (also known as common insertion sites) and thus implicated in the maintenance, progression and metastasis of medulloblastoma. Indeed, using either Ptch1+/− or Tp53−/− sensitized backgrounds, the SB model system employed in this study induced medulloblastomas with nearly 100% incidence and a very high rate of leptomeningeal spread. The high incidence of metastasis allowed for a direct comparison of common insertion sites found in the primary tumors versus the metastases and resulted in the observation of an alarmingly low degree of overlap between the two compartments. The translational impact of these findings is of paramount interest to those treating the human disease, given that the majority of basic research to date has been focused on mutational events found in primary tumors, while metastases account for the bulk of the morbidity seen in medulloblastoma patients. In other words, if the development of future targeted therapies is based solely on information learned from studying the primary tumors, the likelihood of these treatments effectively curing the patients is speculative, given the apparent genetic differences between the primaries and the metastases.
With WNT, SHH and Group 3 medulloblastoma now successfully modeled in the mouse, the community is rapidly striving to generate an accurate model of Group 4 [89]. Elucidation of the cellular origin(s) of this subgroup and proper functional evaluation of candidate Group 4 oncogenes and tumor suppressors await the development of this highly desired reagent. These details are underscored by the fact that 35–40% of medulloblastomas belong to this subgroup and it continues to prevail as the subgroup we currently know the least about from both a genetic and cellular perspective.
Expert commentary
A genomic approach to large cohort studies has revolutionized our contemporary understanding of medulloblastoma. From original reports of small blue cells of the cerebella, medulloblastomas can and must now be recognized as four disparate molecular subgroups with specific biological and clinical properties. Throughout this review, we have highlighted some of the major cytogenetic, genomic and demographic characteristics associated with each molecular subgroup. WNT, a clinically less aggressive form of the disease, is largely characterized by monosomy 6, CTNNB1 mutations, nuclear β-catenin and excellent survival outcomes (>90%). SHH medulloblastomas represent the most-characterized subgroup, with constitutive activation of SHH pathway and an approximately 65% OS, although recent evidence suggests that these trends are highly dependent on patient age. SHH tumors can present with 9q loss and PTCH mutation, and this subgroup is enriched for the desmoplastic histological subtype. Group 4 medulloblastomas remain a largely uncharacterized subgroup with similar clinical characteristics to SHH tumors, often presenting with isochromosome 17q. Group 3 tumors, which largely occur in younger patients, are the most aggressive and most metastatic form of the disease, often presenting with histological (LCA) and genomic (MYC amplification) features associated with dismal outcome. With this new-found knowledge in hand, it will be critical to perform integrative clinical and genomic means of patient stratification, rapidly adapting our treatment to accommodate the ever-changing genomic landscape of medulloblastoma.
Five-year view
Given the present accepted subdivision of medulloblastoma – from a single entity into four distinct molecular subgroups, with disparate genetic, cytogenetic and transcriptional changes – and the advent of multiple, independent technologies to permit the rapid identification of subgroup association, the next generation of clinical trials will undoubtedly be focused on treating medulloblastoma according to its molecular subgroup. Initially, the disparate OS rates that have been retrospectively associated with each molecular variant will need to be evaluated in the context of prospective clinical trial settings. Important and ‘hot’ topics that are currently under debate among pediatric neuro-oncologists are centered on the de-escalation of therapy for WNT medulloblastomas and the intensification of treatment or targeted therapy for patients with Group 3 tumors [17,91]. The effectiveness of targeted therapies has already been demonstrated with the availability of orally bioactive SHH pathway inhibitors, such as GDC-0449 and NVP-LDE225, which repress SHH signaling by targeting the SMO receptor [92,93]. These inhibitors have demonstrated considerable effect on tumor growth, causing promising, albeit transient, remission of SHH medulloblastomas [92]. While targeted inhibition of SMO on its own has resulted in resistance and patient relapse, preliminary results suggest that combinatorial targeted therapies could provide an improved response, especially if the mechanisms of resistance are fully elucidated [50,93].
Over the next 5 years, the authors’ understanding of medulloblastoma biology will continue to advance at an unprecedented rate. Improvements in sequencing technologies, and early efforts such as those reported by Parsons et al. [68], have led to a wave of whole-genome and whole-exome DNA sequencing and RNA sequencing studies. These efforts will undoubtedly describe the full mutational spectra associated with each molecular variant of medulloblastoma, probably cataloguing the true frequency of subgroup-specific mutations and identifying novel mechanisms of pathogenesis. Mutational data will be complimented by advances in our appreciation of the extent to which copy-number aberrations occur in medulloblastoma and between the medulloblastoma subgroups. We expect a major shift from discovery to validation as the first sequencing projects approach completion. This will require a concerted transformation in the field towards a more functional approach aimed at understanding the importance and relevance of the mutations and copy-number events identified during the current discovery era. Moreover, a major emphasis will be placed on identifying the cell of origin for Group 3 and Group 4 tumors and mimicking subtypes of these molecular subgroups in animal models. Furthermore, the overwhelming tumor-specific convergence on chromatin modifier genes advocates for the continued investigation, and possible use, of histone deacetylase inhibitors for treating medulloblastoma in the future [94–96].
Multiple, independent efforts led to the discovery of four core medulloblastoma subgroups. Now, with this molecular knowledge in hand, we have entered a new age in our understanding of this disease. The next 5 years of medulloblastoma research will continue on this trajectory, with refined tools and renewed interest, driving forward clinical advances in the management of this disease.
Key issues.
M edulloblastoma is no longer considered as a single disease and is now generally accepted to comprise four distinct subgroups: WNT, Sonic hedgehog (SHH), Group 3 and Group 4.
Group 3 medulloblastomas exhibit the worst prognosis of the four subgroups, while SHH and Group 4 represent intermediate-prognosis subgroups and WNT medulloblastomas currently have the best outlook in terms of patient survival.
Demographic and clinical variables, including age, gender, and histology, are not equal among medulloblastoma subgroups, possibly accounting for clinical differences traditionally associated with these variables.
Subgroups of medulloblastoma exhibit vastly different transcriptional and genomic profiles that imply distinct underlying biology and will be critical to consider in the future development of subgroup-specific targeted therapies.
There is currently a great need for the further development of highly effective and accurate methods for discerning medulloblastoma subgroup affiliation from clinical samples, particularly those preserved as formalin-fixed paraffin-embedded material.
-
The next 5 years of medulloblastoma research will involve two major waves:
A discovery phase, during which thousands of samples will be subjected to next-generation sequencing to comprehensively catalog all mutations found in medulloblastoma and especially those that are subgroup-restricted.
A validation phase, which will involve the generation of faithful preclinical models for each of the four subgroups, mimicking the genetic events observed in the human disease and subsequent utilization of these models for molecular cellular, and biochemical assays, especially those evaluating novel targeted therapies.
Footnotes
For reprint orders, please contact reprints@expert-reviews.com
Financial & competing interests disclosure
MD Taylor is supported by the NIH (grant CA159859 to MD Taylor, B Weiss and R Wechsler-Reya). The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as:
• of interest
•• of considerable interest
- 1.Thompson MC, Fuller C, Hogg TL, et al. Genomics identifies medulloblastoma subgroups that are enriched for specific genetic alterations. J Clin Oncol. 2006;24(12):1924–1931. doi: 10.1200/JCO.2005.04.4974. [DOI] [PubMed] [Google Scholar]
- 2.Kool M, Koster J, Bunt J, et al. Integrated genomics identifies five medulloblastoma subtypes with distinct genetic profiles, pathway signatures and clinicopathological features. PLoS ONE. 2008;3(8):e3088. doi: 10.1371/journal.pone.0003088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3••.Northcott PA, Korshunov A, Witt H, et al. Medulloblastoma comprises four distinct molecular variants. J Clin Oncol. 2011;29(11):1408–1414. doi: 10.1200/JCO.2009.27.4324. Principal medulloblastoma subgrouping paper that helped lead to the current consensus four-subgroup classification scheme of the disease. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4••.Cho YJ, Tsherniak A, Tamayo P, et al. Integrative genomic analysis of medulloblastoma identifies a molecular subgroup that drives poor clinical outcome. J Clin Oncol. 2011;29(11):1424–1430. doi: 10.1200/JCO.2010.28.5148. Significant medulloblastoma subgrouping paper that was first to describe the identity of a poor-prognosis subtype of Group 3 largely driven by Myc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Markant SL, Wechsler-Reya RJ. Personalized mice: modelling the molecular heterogeneity of medulloblastoma. Neuropathol Appl Neurobiol. 2012;38(3):228–240. doi: 10.1111/j.1365-2990.2011.01235.x. [DOI] [PubMed] [Google Scholar]
- 6.Wu X, Northcott PA, Croul S, Taylor MD. Mouse models of medulloblastoma. Chin J Cancer. 2011;30(7):442–449. doi: 10.5732/cjc.011.10040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ellison DW, Dalton J, Kocak M, et al. Medulloblastoma: clinicopathological correlates of SHH, WNT, and non-SHH/ WNT molecular subgroups. Acta Neuropathol. 2011;121(3):381–396. doi: 10.1007/s00401-011-0800-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schwalbe EC, Lindsey JC, Straughton D, et al. Rapid diagnosis of medulloblastoma molecular subgroups. Clin Cancer Res. 2011;17(7):1883–1894. doi: 10.1158/1078-0432.CCR-10-2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Remke M, Hielscher T, Korshunov A, et al. FSTL5 is a marker of poor prognosis in non-WNT/non-SHH medulloblastoma. J Clin Oncol. 2011;29(29):3852–3861. doi: 10.1200/JCO.2011.36.2798. [DOI] [PubMed] [Google Scholar]
- 10.Taylor MD, Northcott PA, Korshunov A, et al. Molecular subgroups of medulloblastoma: the current consensus. Acta Neuropathol. 2012;123(4):465–472. doi: 10.1007/s00401-011-0922-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ellison DW, Onilude OE, Lindsey JC, et al. United Kingdom Children’s Cancer Study Group Brain Tumour Committee. β-Catenin status predicts a favorable outcome in childhood medulloblastoma: the United Kingdom Children’s Cancer Study Group Brain Tumour Committee. J Clin Oncol. 2005;23(31):7951–7957. doi: 10.1200/JCO.2005.01.5479. [DOI] [PubMed] [Google Scholar]
- 12.Gajjar A, Chintagumpala M, Ashley D, et al. Risk-adapted craniospinal radiotherapy followed by high-dose chemotherapy and stem-cell rescue in children with newly diagnosed medulloblastoma (St Jude Medulloblastoma-96): long-term results from a prospective, multicentre trial. Lancet Oncol. 2006;7(10):813–820. doi: 10.1016/S1470-2045(06)70867-1. [DOI] [PubMed] [Google Scholar]
- 13.Remke M, Hielscher T, Northcott PA, et al. Adult medulloblastoma comprises three major molecular variants. J Clin Oncol. 2011;29(19):2717–2723. doi: 10.1200/JCO.2011.34.9373. [DOI] [PubMed] [Google Scholar]
- 14.Clifford SC, Lusher ME, Lindsey JC, et al. Wnt/Wingless pathway activation and chromosome 6 loss characterize a distinct molecular sub-group of medulloblastomas associated with a favorable prognosis. Cell Cycle. 2006;5(22):2666–2670. doi: 10.4161/cc.5.22.3446. [DOI] [PubMed] [Google Scholar]
- 15.Pfister S, Remke M, Benner A, et al. Outcome prediction in pediatric medulloblastoma based on DNA copy-number aberrations of chromosomes 6q and 17q and the MYC and MYCN loci. J Clin Oncol. 2009;27(10):1627–1636. doi: 10.1200/JCO.2008.17.9432. [DOI] [PubMed] [Google Scholar]
- 16.Ellison DW, Kocak M, Dalton J, et al. Definition of disease-risk stratification groups in childhood medulloblastoma using combined clinical, pathologic, and molecular variables. J Clin Oncol. 2011;29(11):1400–1407. doi: 10.1200/JCO.2010.30.2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramaswamy V, Northcott PA, Taylor MD. FISH and chips: the recipe for improved prognostication and outcomes for children with medulloblastoma. Cancer Genet. 2011;204(11):577–588. doi: 10.1016/j.cancergen.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 18••.Pomeroy SL, Tamayo P, Gaasenbeek M, et al. Prediction of central nervous system embryonal tumour outcome based on gene expression. Nature. 2002;415(6870):436–442. doi: 10.1038/415436a. Seminal article discriminating different histologies of pediatric brain tumors based on underlying gene-expression profiles. [DOI] [PubMed] [Google Scholar]
- 19.Rutkowski S, Bode U, Deinlein F, et al. Treatment of early childhood medulloblastoma by postoperative chemotherapy alone. N Engl J Med. 2005;352(10):978–986. doi: 10.1056/NEJMoa042176. [DOI] [PubMed] [Google Scholar]
- 20.Leary SE, Zhou T, Holmes E, Geyer JR, Miller DC. Histology predicts a favorable outcome in young children with desmoplastic medulloblastoma: a report from the children’s oncology group. Cancer. 2011;117(14):3262–3267. doi: 10.1002/cncr.25856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.von Bueren AO, von Hoff K, Pietsch T, et al. Treatment of young children with localized medulloblastoma by chemotherapy alone: results of the prospective, multicenter trial HIT 2000 confirming the prognostic impact of histology. Neuro-oncology. 2011;13(6):669–679. doi: 10.1093/neuonc/nor025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rutkowski S, von Hoff K, Emser A, et al. Survival and prognostic factors of early childhood medulloblastoma: an international meta-analysis. J Clin Oncol. 2010;28(33):4961–4968. doi: 10.1200/JCO.2010.30.2299. [DOI] [PubMed] [Google Scholar]
- 23.Northcott PA, Hielscher T, Dubuc A, et al. Pediatric and adult sonic hedgehog medulloblastomas are clinically and molecularly distinct. Acta Neuropathol. 2011;122(2):231–240. doi: 10.1007/s00401-011-0846-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.von Hoff K, Hartmann W, von Bueren AO, et al. Large cell/anaplastic medulloblastoma: outcome according to myc status, histopathological, and clinical risk factors. Pediatr Blood Cancer. 2010;54(3):369–376. doi: 10.1002/pbc.22339. [DOI] [PubMed] [Google Scholar]
- 25.Lamont JM, McManamy CS, Pearson AD, Clifford SC, Ellison DW. Combined histopathological and molecular cytogenetic stratification of medulloblastoma patients. Clin Cancer Res. 2004;10(16):5482–5493. doi: 10.1158/1078-0432.CCR-03-0721. [DOI] [PubMed] [Google Scholar]
- 26.Bigner SH, Friedman HS, Vogelstein B, Oakes WJ, Bigner DD. Amplification of the c-myc gene in human medulloblastoma cell lines and xenografts. Cancer Res. 1990;50(8):2347–2350. [PubMed] [Google Scholar]
- 27.Badiali M, Pession A, Basso G, et al. N-myc and c-myc oncogenes amplification in medulloblastomas. Evidence of particularly aggressive behavior of a tumor with c-myc amplification. Tumori. 1991;77(2):118–121. doi: 10.1177/030089169107700205. [DOI] [PubMed] [Google Scholar]
- 28.Eberhart CG, Kepner JL, Goldthwaite PT, et al. Histopathologic grading of medulloblastomas: a Pediatric Oncology Group study. Cancer. 2002;94(2):552–560. doi: 10.1002/cncr.10189. [DOI] [PubMed] [Google Scholar]
- 29.von Hoff K, Hinkes B, Gerber NU, et al. Long-term outcome and clinical prognostic factors in children with medulloblastoma treated in the prospective randomised multicentre trial HIT’91. Eur J Cancer. 2009;45(7):1209–1217. doi: 10.1016/j.ejca.2009.01.015. [DOI] [PubMed] [Google Scholar]
- 30.Gilbertson RJ. Finding the perfect partner for medulloblastoma prognostication. J Clin Oncol. 2011;29(29):3841–3842. doi: 10.1200/JCO.2011.37.5709. [DOI] [PubMed] [Google Scholar]
- 31.Korshunov A, Remke M, Werft W, et al. Adult and pediatric medulloblastomas are genetically distinct and require different algorithms for molecular risk stratification. J Clin Oncol. 2010;28(18):3054–3060. doi: 10.1200/JCO.2009.25.7121. [DOI] [PubMed] [Google Scholar]
- 32.Brandes AA, Franceschi E, Tosoni A, et al. Adult neuroectodermal tumors of posterior fossa (medulloblastoma) and of supratentorial sites (stPNET) Crit Rev Oncol Hematol. 2009;71(2):165–179. doi: 10.1016/j.critrevonc.2009.02.002. [DOI] [PubMed] [Google Scholar]
- 33.Spreafico F, Massimino M, Gandola L, et al. Survival of adults treated for medulloblastoma using paediatric protocols. Eur J Cancer. 2005;41(9):1304–1310. doi: 10.1016/j.ejca.2005.02.022. [DOI] [PubMed] [Google Scholar]
- 34•.Kool M, Korshunov A, Remke M, et al. Molecular subgroups of medulloblastoma: an international meta-analysis of transcriptome, genetic aberrations, and clinical data of WNT, SHH, Group 3, and Group 4 medulloblastomas. Acta Neuropathol. 2012;123(4):473–484. doi: 10.1007/s00401-012-0958-8. Comprehensive characterization of the transcriptional, genetic, and clinical features of 550 medulloblastomas classified according to molecular subgroup. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Curran EK, Sainani KL, Le GM, Propp JM, Fisher PG. Gender affects survival for medulloblastoma only in older children and adults: a study from the Surveillance Epidemiology and End Results Registry. Pediatr Blood Cancer. 2009;52(1):60–64. doi: 10.1002/pbc.21832. [DOI] [PubMed] [Google Scholar]
- 36.Louis DN, Ohgaki H, Wiestler OD, et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114(2):97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Giangaspero F, Rigobello L, Badiali M, et al. Large-cell medulloblastomas. A distinct variant with highly aggressive behavior. Am J Surg Pathol. 1992;16(7):687–693. [PubMed] [Google Scholar]
- 38.Suresh TN, Santosh V, Yasha TC, et al. Medulloblastoma with extensive nodularity: a variant occurring in the very young-clinicopathological and immunohistochemical study of four cases. Childs Nerv Syst. 2004;20(1):55–60. doi: 10.1007/s00381-003-0855-5. [DOI] [PubMed] [Google Scholar]
- 39.Garrè ML, Cama A, Bagnasco F, et al. Medulloblastoma variants: age-dependent occurrence and relation to Gorlin syndrome – a new clinical perspective. Clin Cancer Res. 2009;15(7):2463–2471. doi: 10.1158/1078-0432.CCR-08-2023. [DOI] [PubMed] [Google Scholar]
- 40.Giangaspero F, Perilongo G, Fondelli MP, et al. Medulloblastoma with extensive nodularity: a variant with favorable prognosis. J Neurosurg. 1999;91(6):971–977. doi: 10.3171/jns.1999.91.6.0971. [DOI] [PubMed] [Google Scholar]
- 41.Brugières L, Remenieras A, Pierron G, et al. High frequency of germline SUFU mutations in children with desmoplastic/nodular medulloblastoma younger than 3 years of age. J Clin Oncol. 2012;30(17):2087–2093. doi: 10.1200/JCO.2011.38.7258. [DOI] [PubMed] [Google Scholar]
- 42.Eberhart CG, Kratz J, Wang Y, et al. Histopathological and molecular prognostic markers in medulloblastoma: c-myc, N-myc, TrkC, and anaplasia. J Neuropathol Exp Neurol. 2004;63(5):441–449. doi: 10.1093/jnen/63.5.441. [DOI] [PubMed] [Google Scholar]
- 43.Stearns D, Chaudhry A, Abel TW, Burger PC, Dang CV, Eberhart CG. c-myc overexpression causes anaplasia in medulloblastoma. Cancer Res. 2006;66(2):673–681. doi: 10.1158/0008-5472.CAN-05-1580. [DOI] [PubMed] [Google Scholar]
- 44.Phillips HS, Kharbanda S, Chen R, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9(3):157–173. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 45.Verhaak RG, Hoadley KA, Purdom E, et al. Cancer Genome Atlas Research Network. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McLendon R, Friedman A, Bigner D, et al. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fattet S, Haberler C, Legoix P, et al. Beta-catenin status in paediatric medulloblastomas: correlation of immunohistochemical expression with mutational status, genetic profiles, and clinical characteristics. J Pathol. 2009;218(1):86–94. doi: 10.1002/path.2514. [DOI] [PubMed] [Google Scholar]
- 48.Al-Halabi H, Nantel A, Klekner A, et al. Preponderance of sonic hedgehog pathway activation characterizes adult medulloblastoma. Acta Neuropathol. 2011;121(2):229–239. doi: 10.1007/s00401-010-0780-0. [DOI] [PubMed] [Google Scholar]
- 49.Low JA, de Sauvage FJ. Clinical experience with Hedgehog pathway inhibitors. J Clin Oncol. 2010;28(36):5321–5326. doi: 10.1200/JCO.2010.27.9943. [DOI] [PubMed] [Google Scholar]
- 50.Metcalfe C, de Sauvage FJ. Hedgehog fights back: mechanisms of acquired resistance against Smoothened antagonists. Cancer Res. 2011;71(15):5057–5061. doi: 10.1158/0008-5472.CAN-11-0923. [DOI] [PubMed] [Google Scholar]
- 51.Gilbertson RJ. Mapping cancer origins. Cell. 2011;145(1):25–29. doi: 10.1016/j.cell.2011.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52•.Gibson P, Tong Y, Robinson G, et al. Subtypes of medulloblastoma have distinct developmental origins. Nature. 2010;468(7327):1095–1099. doi: 10.1038/nature09587. Identification of the putative cell of origin for WNT subgroup medulloblastomas. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gilbertson RJ, Ellison DW. The origins of medulloblastoma subtypes. Annu Rev Pathol. 2008;3:341–365. doi: 10.1146/annurev.pathmechdis.3.121806.151518. [DOI] [PubMed] [Google Scholar]
- 54.Northcott PA, Rutka JT, Taylor MD. Genomics of medulloblastoma: from Giemsa-banding to next-generation sequencing in 20 years. Neurosurg Focus. 2010;28(1):E6. doi: 10.3171/2009.10.FOCUS09218. [DOI] [PubMed] [Google Scholar]
- 55.Dubuc AM, Northcott PA, Mack S, Witt H, Pfister S, Taylor MD. The genetics of pediatric brain tumors. Curr Neurol Neurosci Rep. 2010;10(3):215–223. doi: 10.1007/s11910-010-0103-9. [DOI] [PubMed] [Google Scholar]
- 56.Zurawel RH, Chiappa SA, Allen C, Raffel C. Sporadic medulloblastomas contain oncogenic beta-catenin mutations. Cancer Res. 1998;58(5):896–899. [PubMed] [Google Scholar]
- 57.Taylor MD, Mainprize TG, Rutka JT. Molecular insight into medulloblastoma and central nervous system primitive neuroectodermal tumor biology from hereditary syndromes: a review. Neurosurgery. 2000;47(4):888–901. doi: 10.1097/00006123-200010000-00020. [DOI] [PubMed] [Google Scholar]
- 58.Tabori U, Baskin B, Shago M, et al. Universal poor survival in children with medulloblastoma harboring somatic TP53 mutations. J Clin Oncol. 2010;28(8):1345–1350. doi: 10.1200/JCO.2009.23.5952. [DOI] [PubMed] [Google Scholar]
- 59.Pfaff E, Remke M, Sturm D, et al. TP53 mutation is frequently associated with CTNNB1 mutation or MYCN amplification and is compatible with long-term survival in medulloblastoma. J Clin Oncol. 2010;28(35):5188–5196. doi: 10.1200/JCO.2010.31.1670. [DOI] [PubMed] [Google Scholar]
- 60.Lindsey JC, Hill RM, Megahed H, et al. TP53 mutations in favorable-risk Wnt/ Wingless-subtype medulloblastomas. J Clin Oncol. 2011;29(12):e344–e346. doi: 10.1200/JCO.2010.33.8590. author reply e347. [DOI] [PubMed] [Google Scholar]
- 61.Gessi M, von Bueren AO, Rutkowski S, Pietsch T. p53 expression predicts dismal outcome for medulloblastoma patients with metastatic disease. J Neurooncol. 2012;106(1):135–141. doi: 10.1007/s11060-011-0648-8. [DOI] [PubMed] [Google Scholar]
- 62.Taylor MD, Liu L, Raffel C, et al. Mutations in SUFU predispose to medulloblastoma. Nat Genet. 2002;31(3):306–310. doi: 10.1038/ng916. [DOI] [PubMed] [Google Scholar]
- 63.Yauch RL, Dijkgraaf GJ, Alicke B, et al. Smoothened mutation confers resistance to a Hedgehog pathway inhibitor in medulloblastoma. Science. 2009;326(5952):572–574. doi: 10.1126/science.1179386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64•.Northcott PA, Nakahara Y, Wu X, et al. Multiple recurrent genetic events converge on control of histone lysine methylation in medulloblastoma. Nat Genet. 2009;41(4):465–472. doi: 10.1038/ng.336. Original article reporting epigenetic deregulation resulting from copy-number alterations in medulloblastoma. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Korshunov A, Remke M, Kool M, et al. Biological and clinical heterogeneity of MYCN-amplified medulloblastoma. Acta Neuropathol. 2012;123(4):515–527. doi: 10.1007/s00401-011-0918-8. [DOI] [PubMed] [Google Scholar]
- 66.Ryan SL, Schwalbe EC, Cole M, et al. MYC family amplification and clinical risk-factors interact to predict an extremely poor prognosis in childhood medulloblastoma. Acta Neuropathol. 2012;123(4):501–513. doi: 10.1007/s00401-011-0923-y. [DOI] [PubMed] [Google Scholar]
- 67.Korshunov A, Remke M, Kool M, et al. Biological and clinical heterogeneity of MYCN-amplified medulloblastoma. Acta Neuropathol. 2011;123(4):515–527. doi: 10.1007/s00401-011-0918-8. [DOI] [PubMed] [Google Scholar]
- 68••.Parsons DW, Li M, Zhang X, et al. The genetic landscape of the childhood cancer medulloblastoma. Science. 2011;331(6016):435–439. doi: 10.1126/science.1198056. The first genome-wide sequencing paper of medulloblastoma based on Sanger sequencing technology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Adamson DC, Shi Q, Wortham M, et al. OTX2 is critical for the maintenance and progression of Shh-independent medulloblastomas. Cancer Res. 2010;70(1):181–191. doi: 10.1158/0008-5472.CAN-09-2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bhatia B, Northcott PA, Hambardzumyan D, et al. Tuberous sclerosis complex suppression in cerebellar development and medulloblastoma: separate regulation of mammalian target of rapamycin activity and p27 Kip1 localization. Cancer Res. 2009;69(18):7224–7234. doi: 10.1158/0008-5472.CAN-09-1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hoshida Y, Brunet JP, Tamayo P, Golub TR, Mesirov JP. Subclass mapping: identifying common subtypes in independent disease data sets. PLoS ONE. 2007;2(11):e1195. doi: 10.1371/journal.pone.0001195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tamayo P, Scanfeld D, Ebert BL, Gillette MA, Roberts CW, Mesirov JP. Metagene projection for cross-platform, cross-species characterization of global transcriptional states. Proc Natl Acad Sci USA. 2007;104(14):5959–5964. doi: 10.1073/pnas.0701068104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Northcott PA, Shih DJ, Remke M, et al. Rapid, reliable, and reproducible molecular sub-grouping of clinical medulloblastoma samples. Acta Neuropathol. 2012;123(4):615–626. doi: 10.1007/s00401-011-0899-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rogers HA, Miller S, Lowe J, Brundler MA, Coyle B, Grundy RG. An investigation of WNT pathway activation and association with survival in central nervous system primitive neuroectodermal tumours (CNS PNET) Br J Cancer. 2009;100(8):1292–1302. doi: 10.1038/sj.bjc.6604979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lau J, Schmidt C, Markant SL, Taylor MD, Wechsler-Reya RJ, Weiss WA. Matching mice to malignancy: molecular subgroups and models of medulloblastoma. Childs Nerv Syst. 2012;28(4):521–532. doi: 10.1007/s00381-012-1704-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zurawel RH, Allen C, Wechsler-Reya R, Scott MP, Raffel C. Evidence that haploinsufficiency of Ptch leads to medulloblastoma in mice. Genes Chromosomes Cancer. 2000;28(1):77–81. [PubMed] [Google Scholar]
- 77.Goodrich LV, Milenkovic L, Higgins KM, Scott MP. Altered neural cell fates and medulloblastoma in mouse patched mutants. Science. 1997;277(5329):1109–1113. doi: 10.1126/science.277.5329.1109. [DOI] [PubMed] [Google Scholar]
- 78.Wetmore C, Eberhart DE, Curran T. The normal patched allele is expressed in medulloblastomas from mice with heterozygous germ-line mutation of patched. Cancer Res. 2000;60(8):2239–2246. [PubMed] [Google Scholar]
- 79.Hatton BA, Villavicencio EH, Tsuchiya KD, et al. The Smo/Smo model: hedgehog-induced medulloblastoma with 90% incidence and leptomeningeal spread. Cancer Res. 2008;68(6):1768–1776. doi: 10.1158/0008-5472.CAN-07-5092. [DOI] [PubMed] [Google Scholar]
- 80.Hallahan AR, Pritchard JI, Hansen S, et al. The SmoA1 mouse model reveals that notch signaling is critical for the growth and survival of sonic hedgehog-induced medulloblastomas. Cancer Res. 2004;64(21):7794–7800. doi: 10.1158/0008-5472.CAN-04-1813. [DOI] [PubMed] [Google Scholar]
- 81.Lee Y, Kawagoe R, Sasai K, et al. Loss of suppressor-of-fused function promotes tumorigenesis. Oncogene. 2007;26(44):6442–6447. doi: 10.1038/sj.onc.1210467. [DOI] [PubMed] [Google Scholar]
- 82.Yang ZJ, Ellis T, Markant SL, et al. Medulloblastoma can be initiated by deletion of Patched in lineage-restricted progenitors or stem cells. Cancer Cell. 2008;14(2):135–145. doi: 10.1016/j.ccr.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Behesti H, Marino S. Cerebellar granule cells: insights into proliferation, differentiation, and role in medulloblastoma pathogenesis. Int J Biochem Cell Biol. 2009;41(3):435–445. doi: 10.1016/j.biocel.2008.06.017. [DOI] [PubMed] [Google Scholar]
- 84.Castellino RC, Barwick BG, Schniederjan M, et al. Heterozygosity for Pten promotes tumorigenesis in a mouse model of medulloblastoma. PLoS ONE. 2010;5(5):e10849. doi: 10.1371/journal.pone.0010849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hambardzumyan D, Becher OJ, Rosenblum MK, Pandolfi PP, Manova-Todorova K, Holland EC. PI3K pathway regulates survival of cancer stem cells residing in the perivascular niche following radiation in medulloblastoma in vivo. Genes Dev. 2008;22(4):436–448. doi: 10.1101/gad.1627008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Swartling FJ, Grimmer MR, Hackett CS, et al. Pleiotropic role for MYCN in medulloblastoma. Genes Dev. 2010;24(10):1059–1072. doi: 10.1101/gad.1907510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87•.Kawauchi D, Robinson G, Uziel T, et al. A mouse model of the most aggressive subgroup of human medulloblastoma. Cancer Cell. 2012;21(2):168–180. doi: 10.1016/j.ccr.2011.12.023. Development of one of the first Group 3 mouse models of medulloblastoma. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88•.Pei Y, Moore CE, Wang J, et al. An animal model of MYC-driven medulloblastoma. Cancer Cell. 2012;21(2):155–167. doi: 10.1016/j.ccr.2011.12.021. Development of one of the first Group 3 mouse models of medulloblastoma. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Eberhart CG. Three down and one to go: modeling medulloblastoma subgroups. Cancer Cell. 2012;21(2):137–138. doi: 10.1016/j.ccr.2012.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wu X, Northcott PA, Dubuc A, et al. Clonal selection drives genetic divergence of metastatic medulloblastoma. Nature. 2012;482(7386):529–533. doi: 10.1038/nature10825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Leary SE, Olson JM. The molecular classification of medulloblastoma: driving the next generation clinical trials. Curr Opin Pediatr. 2012;24(1):33–39. doi: 10.1097/MOP.0b013e32834ec106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rudin CM, Hann CL, Laterra J, et al. Treatment of medulloblastoma with hedgehog pathway inhibitor GDC-0449. N Engl J Med. 2009;361(12):1173–1178. doi: 10.1056/NEJMoa0902903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Buonamici S, Williams J, Morrissey M, et al. Interfering with resistance to smoothened antagonists by inhibition of the PI3K pathway in medulloblastoma. Sci Transl Med. 2010;2(51):51ra70. doi: 10.1126/scitranslmed.3001599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lee SJ, Krauthauser C, Maduskuie V, Fawcett PT, Olson JM, Rajasekaran SA. Curcumin-induced HDAC inhibition and attenuation of medulloblastoma growth in vitro and in vivo. BMC Cancer. 2011;11:144. doi: 10.1186/1471-2407-11-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Spiller SE, Ditzler SH, Pullar BJ, Olson JM. Response of preclinical medulloblastoma models to combination therapy with 13-cis retinoic acid and suberoylanilide hydroxamic acid (SAHA) J Neurooncol. 2008;87(2):133–141. doi: 10.1007/s11060-007-9505-1. [DOI] [PubMed] [Google Scholar]
- 96.Häcker S, Karl S, Mader I, et al. Histone deacetylase inhibitors prime medulloblastoma cells for chemotherapy-induced apoptosis by enhancing p53-dependent Bax activation. Oncogene. 2011;30(19):2275–2281. doi: 10.1038/onc.2010.599. [DOI] [PubMed] [Google Scholar]