Abstract
CPEB is a translational regulatory sequence-specific RNA binding protein that controls germ cell development. Here we show that CPEB heterozygous female mice are fertile but contain disorganized mammary epithelial cells in which ZO-1 and claudin-3, apical tight junction proteins, are mis-localized. CPEB depletion from mammary epithelial cells disrupts ZO-1 apical localization and tight junction distribution; conversely, ectopic expression of CPEB enhances ZO-1 localization. CPEB and ZO-1 mRNA are co-localized apically and ZO-1 3’UTR binding sites for CPEB are necessary for RNA localization. In a 3-dimensional culture system that models lumen-containing mammary ducts, depletion of CPEB or ZO-1 impairs central cavity formation, indicating a loss of cell polarity. Cavity formation in ZO-1 depleted cells is rescued when they are transduced with ZO-1 mRNA containing, but not lacking, CPEB binding sites. Our data demonstrates that CPEB-mediated ZO-1 mRNA localization is essential for tight junction assembly and mammary epithelial cell polarity.
Introduction
The asymmetric distribution of molecules in polarized cells is a hallmark of metazoan development1–3. For example, one characteristic of the anterior-posterior axis of Drosophila oocytes is the concentration of bicoid RNA at the anterior pole and oskar RNA at the posterior pole4. In Xenopus, oocytes are polarized along an animal-vegetal axis where RNAs5,6 and organelles7 are asymmetrically distributed. Polarization in neurons is evident not only by axonal and dendritic extensions, but also by the mRNAs that they contain8,9. Localization of molecules and subcellular structures allows cells to respond quickly and locally to environmental cues and provides a means of differentiation when cellular components are unequally distributed to cells as they divide.
In mice, epithelial cells lining the lumen of several tissues are highly polarized. The mammary gland, for example, develops as a branching network of interconnecting tubular ducts that culminate in alveoli or terminal end buds (TEB). The lumen of the ducts and TEBs become hollow when the interior-most cells undergo apoptosis10 in response to reproductive hormones11,12. The remaining epithelial cells that line the ducts become polarized with apical (luminal) and baso-lateral surfaces. To ensure exclusivity in the types of solutes that can passage between the lumen and the baso-lateral bloodstream, tight junctions are formed between cells near the apical surface13. Among the many factors that comprise tight junctions are the claudins, a family of 24 integral membrane proteins whose extracellular loop domains interact with one another between cells to form a selective molecular seal. The intracellular tails of the claudins contain PDZ domains that interact with the PDZ domains of the zonal occludens (ZO) proteins 1–3, members of the MAGUK (membrane-associated guanylate kinase-like homologs) family of proteins. ZO-1 and ZO-2 are essential genes14 that determine where intercellular claudin-claudin polymerization occurs and as a consequence, where tight junctions are formed. Epithelia lacking ZO-1 and ZO-2 form no tight junctions and thus the discriminating barrier preventing molecular mixing between luminal and baso-lateral regions is destroyed14.
CPEB is a sequence-specific RNA binding protein that regulates polyadenylation-induced translation in a variety of cell types including germ cells15,16, neurons17,18, and primary diploid fibroblasts19,20. CPEB binds the cytoplasmic polyadenylation element (CPE), a 3’ UTR sequence, as well as several factors including Gld2; a poly(A) polymerase; PARN, a deadenylating enzyme, symplekin, a scaffold protein upon which the RNP complex assembles, and several other factors21–23. In the nucleus, CPEB binds CPE-containing pre-mRNAs24, which like most pre-mRNAs contain long poly(A) tails, and escorts them to the cytoplasm where they associate with other members of the cytoplasmic polyadenylation complex. CPEB-bound PARN and Gld2 are constitutively active, but because PARN activity is particularly robust, the poly(A) tails are shortened. An external signal elicits CPEB phosphorylation25, which in turn induces the expulsion of PARN from the RNP complex resulting Gld2-catalyzed polyadenylation22.
Here we use both in vivo and in vitro models to show that CPEB controls tight junction assembly and cell polarity by recruiting ZO-1 mRNA to the apical region of mammary epithelial cells. The invovlement of CPEB in these processes establishes RNA localization by this protein as a new essential pathway for mammary cell development.
Results
Reduced terminal end bud cavitation in CPEB deficient mice
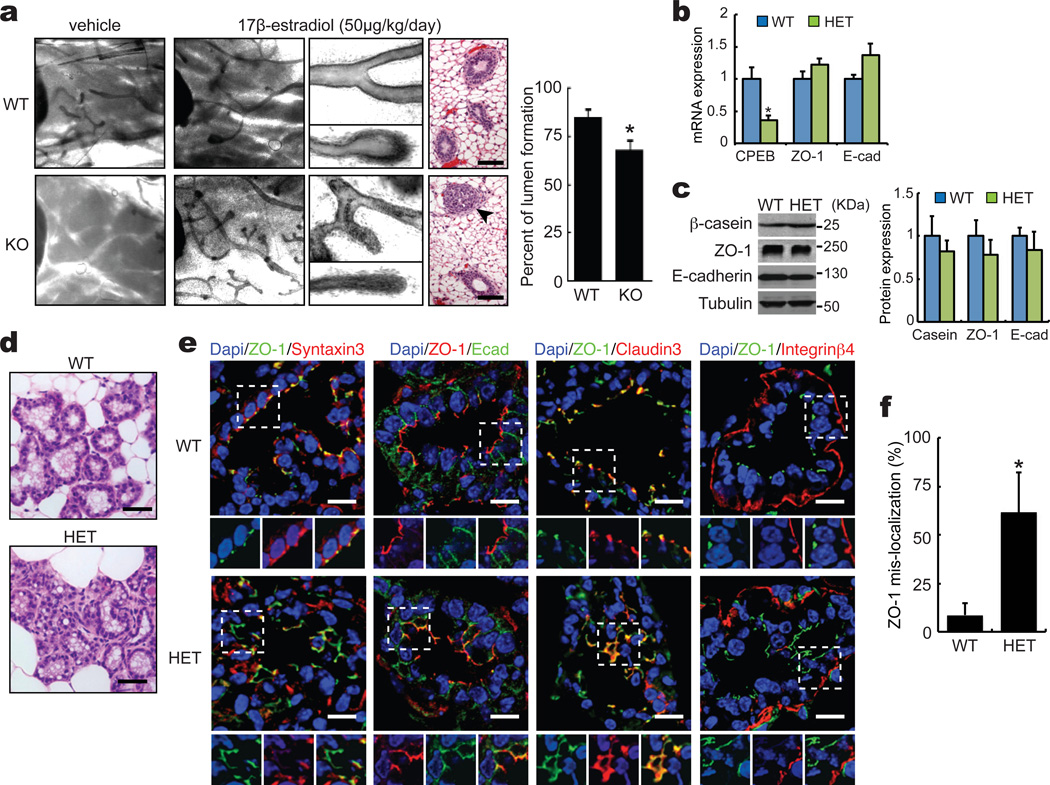
The oocytes of CPEB knockout mice fail to progress beyond the pachytene stage of meiosis due to inefficient translation of synaptonemal complex protein mRNAs16,26,27. The ovaries from these sterile animals are rudimentary and do not secrete normal levels of reproductive hormones, primarily estrogen; consequently, the mammary ducts do not proliferate and elongate after birth. Although injection of 17β-estradiol for 5 days resulted in development of the mammary epithelia (Fig. 1a) as well as the uterus (Supplementary Figure S1a), the lumen of the mammary ducts improperly contained cells (Fig. 1a, WT 15.2% vs KO 32.2%), suggesting that CPEB might be involved in establishing or maintaining mammary epithelial cell polariy (Supplementary Figure S1b and c). There were no obvious defects in cell morphology in the kidney, intestine, or liver.
Figure 1. Disruption of breast epithelial cell polarity in CPEB knockout and heterozygous mice.
(a) WT and CPEB KO mice were injected with vehicle or 17β-estradiol for 5 days followed by isolation of the mammary gland, staining with Carmine’s solution and examination in whole-mount, or fixation, paraffin embedding, and staining with hematoxylin and eosin Y. Scale bar is 50 µm. The asterisk indicates statistical significance (p<0.05, Student’s t test, 3 replicates, error bars are standard error of the mean). (b and c) Quantitative RT-PCR and western blots of RNAs and proteins, respectively, in the mammary gland from WT and CPEB heterozygous (Het) mice. The asterisk indicates statistical significance (p<0.05, Student’s t test, 3 replicates). (d) Sections of mammary gland from WT and CPEB heterozygous mice stained with hematoxylin and eosin Y. Scale bar is 50 µm. (e) Immunostaining of WT and CPEB heterozygous mammary gland for ZO-1, syntaxin-3, claudin-3, E-cadherin, and integrin-β4 (in this and all subsequent immunostaining, DAPI co-staining indicates the nucleus) . The boxes in the large merged images denote areas that are magnified and where the two fluorescent channels are separate as well as merged. Scale bar is 10 µm. (f) The percent of mis-localized ZO-1 mRNA in WT and CPEB KO mammary gland is also indicated. The asterisk indicates statistical significance (p<0.01, Student’s t test, 3 replicates).
To investigate the possible loss of polarity further, mammary epithelia from WT and CPEB heterozygous females, which are fertile, were probed for ZO-1 and E-cadherin, two proteins involved in apical-basal epithelial cell polarity; the expression level of neither factor was altered in the heterozygote (Fig. 1b, c). However, the mammary epithelium from the heterozygote was often disorganized (Fig. 1d).
Syntaxin-3 is an apical (lumenal) protein, ZO-1 and claudin-3 are essential tight junction proteins located laterally toward the apical region, E-cadherin is an adherence junction protein located toward the baso-lateral region, and integrin β4 is a basal protein. In WT mammary epithelia, these proteins segregated to their respective compartments, as expected (Fig. 1e). However, in the heterozygoous epithelia, ZO-1 had a reduced co-localization with syntaxin-3 while it had an increased co-localization with E-cadherin; ZO-1 and claudin-3 remained co-localized. In WT epthelia, ZO-1 and E-cadherin immunostaining overlapped 8% of the time; in heterozygous epithelia, 61% of the time (Fig. 1f). Integrin-β4 localization was unaffected by the partial loss of CPEB (Fig. 1e). These data indicate that apical tight junction assembly might be mis-localized to baso-lateral regions. Thus, in vivo, CPEB modulates tight junction assembly and possibly cell polarity.
CPEB mediates ZO-1 localization to the apical region
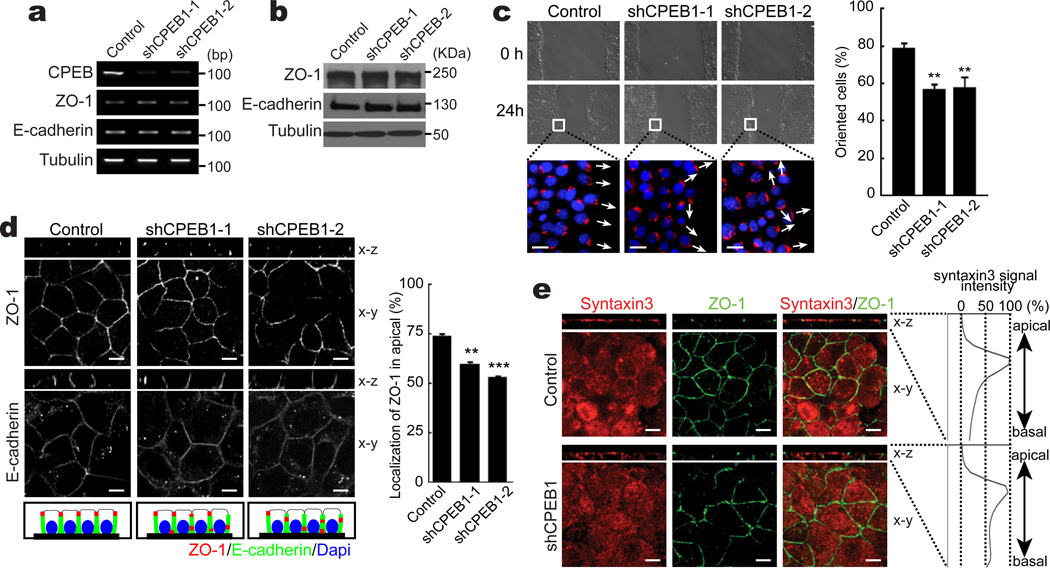
To examine tight junction assembly and cell polarity in a more defined system, we used EpH4 mouse mammary cells, which were infected with lentiviruses expressing GFP only, or GFP plus two different shRNAs targeting CPEB. Both shRNAs effectively depleted CPEB from the cells without affecting ZO-1 or E-cadherin RNA or protein levels (Fig. 2a, b and Supplementary Figure S2a). One measure of cell polarity as well as cell migration is the scratch assay, in which a wound inflicted on confluent cells is examined for repopulation. Although the rate of cell migration was unaffected by CPEB depletion, the polarity of the cells was significantly altered. As the peripheral cells migrated toward the wound, the Golgi apparatus became positioned on the leading side of the nucleus. Fig. 2c demonstrates that in CPEB-depleted cells, the orientation of the Golgi toward the wound, as detected by GM130 immunostaining, was significantly reduced.
Figure 2. CPEB regulates tight junctions and cell polarity of mammary epithelial cells.
(a and b) EpH4 cells were infected with lentiviruses expressing GFP only or GFP plus two different shRNAs against CPEB. RT-PCR indicates the level of CPEB RNA depletion and the western blots show the levels of ZO-1, E-cadherin, or tubulin after CPEB depletion. (c) Scratch assay following CPEB knockdown. EpH4 cells infected with control or two different CPEB knockdown viruses were plated and a scratch assay performed to measure cell migration. DIC imagines of cells 0 and 24 hours after the scratch and immunostaining for GM130, a Golgi marker indicating cell polarity, are shown. The arrows indicate Golgi orientation. The histogram shows the quantification of cells that were orented to the scratch. The two asterisks indicate statistical significance from control (p<0.01, Student’s t test, 3 replicates). (d) Confluent EpH4 cells infected with control or CPEB knockdown viruses were immunostained for ZO-1 and E-cadherin. The edge-on perspective of the x-z axis shows the location of ZO-1 in apical, lateral, and basal regions. Scale bar is 10 µm. The diagram illustrates the effect of CPEB depletion on ZO-1 localization and the histogram shows the quantification of ZO-1 in the apical portion of cells in the x-z axis. The asterisks indicate statistical significance (** refers to p<0.01; *** refers to p<0.001, Student’s t test, 3 replicates). (e) Confluent EpH4 cells infected with control or CPEB knockdown viruses were immunostained for ZO-1 and syntaxin-3. The graphs representing flourescence intensity of synatxin-3 in the apical to basal direction. Scale bar is 10 µm. Error bars are standard error of the mean.
When grown in monoloayer, EpH4 cells form epithelial sheets with clearly defined tight junctions as imaged by ZO-1 immunostaining. When the images were examined along the x-z axis, ZO-1 staining was detected exclusively on the apical layer. Conversely, E-cadherin staining was mostly baso-lateral (Fig. 2d). Depletion of CPEB resulted in mis-localization of ZO-1 and claudin-3 to mid-lateral and baso-lateral regions; E-cadherin localization was unaffected (Fig. 2d and Supplementary Figure S3, seen most clearly from the edge-on perspective of the x-z axis). A diagram of the micrographic images and quantification of the results of ZO-1 localization in the apical region are shown in Fig.2d. This ZO-1 mis-localization was not due to retardation of tight junction assembly (Supplementary Figure S2b).
Another indicator of tight junction assembly is cell permeability. In one assay, FITC-dextran applied apically to cells is measured in the medium bathing the basal surface. Relative to control, CPEB depletion resulted in >2-fold increase in cell permeability, indicating disruption of tight junctions (Supplementary Figure S2c). Taken together, these results indicate that CPEB is involved in tight junction assembly.
To examine the involvement of CPEB in cell polarity, EpH4 cells were immuno-stained for syntaxin-3. Fig. 2e demonstrates that in control cells, syntaxin-3 was confined to the apical surface (when viewed along the x-z axis). In CPEB-depleted cells, however, syntaxin-3 was detected in apical and basal regions. Thus, CPEB mediates not only tight junction assembly, but cell polarity as well.
Ectopic CPEB rescues tight junctions
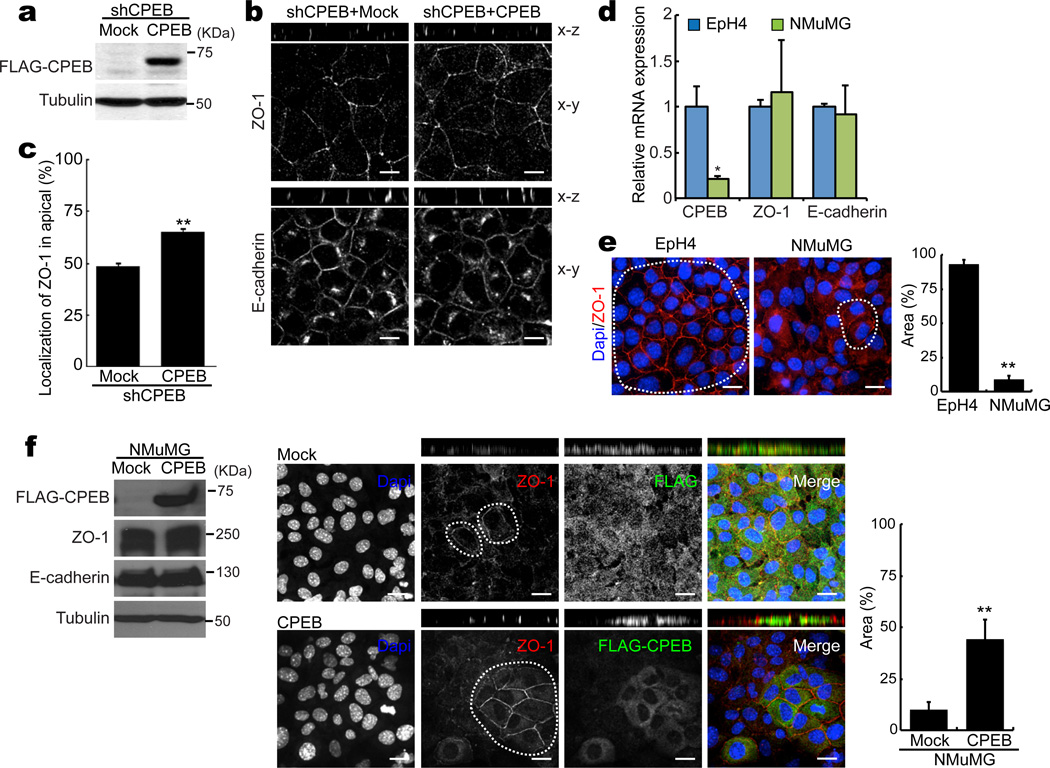
To determine whether CPEB might rescue tight junction formation, EpH4 cells that were depleted of CPEB by lentivirus-mediated knockdown were subsequently infected with a retrovirus expressing FLAG-tagged CPEB. Fig. 3a shows the expression of FLAG-CPEB by a western analysis. As presented initially in Fig. 2d, Fig. 3b shows that depletion of CPEB resulted in mis-localization of ZO-1 to lateral regions (note the distribution of ZO-1 along the x-z axis). However, expression of exogenous CPEB in the depleted cells restored the apical position of ZO-1, thus demonstrating rescue of tight junctions by ectopic CPEB (Fig. 3b, c).
Figure 3. CPEB induces ZO-1 localization to the apical surface.
(a) CPEB-depleted EpH4 cells were infected with retrovirus-expressing FLAG-tagged CPEB and examined for CPEB expression by western blotting for the FLAG epitope. (b) Immunocytochemistry of ZO-1 and E-cadherin in EpH4 cells in the x-y and x-z orientations following CPEB depletion and in some cases, following ectopic CPEB expression as well. Scale bar is 10 µm. (c) Quantification of apical ZO-1 expression. The two asterisks refer to statistical significance (p<0.01, Student’s t test, 3 replicates) (d) Comparison of CPEB, ZO-1, and E-cadherin expression in EpH4 and NMuMG cells by quantitative RT-PCR. The asterisk refers to statistical significance (p<0.05, Student’s t test, 3 replicates). (e) ZO-1 localization in EpH4 and NMuMG mammary breast epithelial cells. The circles in both cell types indicate regions that are “EpH4-like” with ZO-1 strongly localized to apical regions. The histogram refers to the percentage of the areas that show ZO-1 apical staining. The two asterisks refer to statistical significance (p<0.01, Student’s t test, 3 replicates). Scale bar is 10 µm. (f) Expression levels of FLAG-CPEB, ZO-1, E-cadherin, and tubulin in mock and FLAG-CPEB infected cells. Also shown are NMuMG cells containing ectopically-expressed FLAG-CPEB where ZO-1 protein and FLAG-CPEB were detected by immunostaining. ZO-1 localization was also examined in mock (FLAG only) transduced cells. The circles refer to areas with strong ZO-1 localization to apical regions, and thus resemble EpH4 cells. The percent of the area of the cells that resembles the EpH4-like ZO-1 localization was quantified, which is displayed in the histogram. The two asterisks refer to statistical significance (p<0.01, Student’s t test, 3 replicates). Scale bar is 10 µm. Error bars are standard error of the mean
We noticed that a second murine mammary epithelial cell line, NMuMG, contained about half the amount of CPEB as EpH4 cells (Fig. 3d). Moreover, ZO-1 was not as tightly localized to apical tight junctions in NMuMG cells compared to EpH4 cells (Fig. 3e). This observation suggested that ectopic expression of CPEB in NMuMG cells might induce strong ZO-1 apical localization. Fig. 3f shows the expression of FLAG-CPEB in these cells, which had little effect on the overall expression of ZO-1. However, ZO-1 in the CPEB-transduced cells was more strongly localized to the apical regions compared to neighboring cells that did express ectopic CPEB. Moreover, in mock (FLAG only) transduced cells, ZO-1 localization was not enhanced (Fig. 3f). The ZO-1 localization in the NMuMG cells that expressed ectopic CPEB resembled that of the relatively CPEB-rich EpH4 cells. Thus, ZO-1 localization, but not overall expression, is dependent upon CPEB.
ZO-1 localization and cell polarity require CPEB
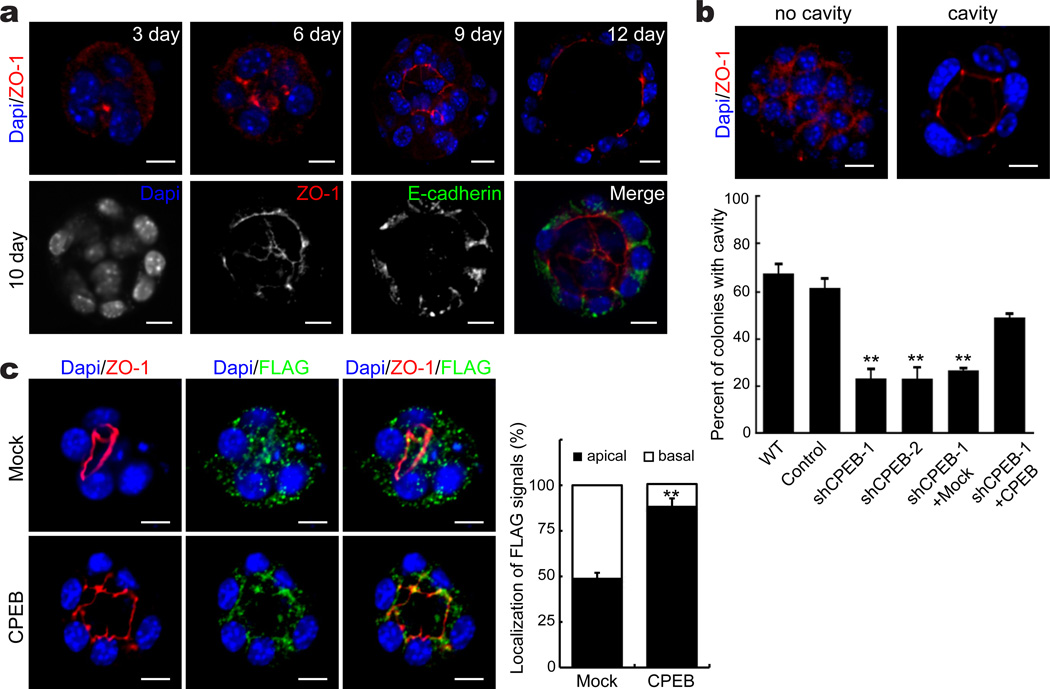
EpH4 cells grown in an anchorage-independent manner are polarized and contain central cavities; consequently, they have been used as in vitro models to examine mammary duct formation28. As expected, ZO-1 was detected in the apical region (Fig 4a, b). Cavity formation was strongly reduced when CPEB was depleted, but was rescued when CPEB was ectopically expressed (Fig. 4b). In CPEB-depleted colonies that did not form cavities, ZO-1 was randomly distributed even though steady state levels were uanffected.
Figure 4. ZO-1 localization to apical tight junctions and cavity formation require CPEB.
(a) EpH4 cells were grown in suspension in matrigel for 3–12 days. Cavity formation was monitored by immunostaining for ZO-1 and E-cadherin and examined by confocal microscopy. Scale bar = 10µm. (b) Analysis of cavity formation in EpH4 cells cultured in matrigel for 12 days. Scale bar is 10 µm. The histogram shows the percent of colonies with cavities following the indicated treatments. The two asterisks refer to statistical significance (p<0.01, Student’s t test, 3 replicates). (c) FLAG-CPEB as well as ZO-1 expression in cells that form cavities. Mock refers to cells expressing FLAG only (a nonsense protein that terminated at a stop codon in vector sequences). Scale bar is 10 µm. The histogram shows quantification of FLAG in the apical portion of cells. The asterisks refers to statistical significance (p<0.01, Student’s t test, 3 replicates). Error bars are standard error of the mean.
We next infected cells with FLAG-CPEB, followed by cavity formation and immunostaining for the FLAG epitope. Compared to the FLAG epitope alone (this construct generated a nonsense protein that terminated at a stop codon in vector sequences), CPEB was at least partly co-localized with ZO-1 at the apical region of cells, significantly more that then FLAG epitope only (Fig. 4c). Thus, CPEB as well as ZO-1 is asymmetrically distributed to the apical region of cavity forming cells.
We also investigated whether CPEB was important for tight junction assembly of kidney or intestinal cells grown in vitro. Madin Darby kidney cells (MDCK) and Caco-2 human colorectal epithelial cells, which contain very little CPEB RNA (only 17% and <1%, respectively, of the amount in mammary cells), were infected with lentiviruses expressing shRNA for CPEB. Supplementary Figure S4a–d shows that these cells had normal cavity formation and ZO-1 RNA localization, indicating that CPEB involvement in these process may be restricted to mammary cells (see Discussion).
CPEB and ZO-1 and E-cadherin mRNAs are localized apically
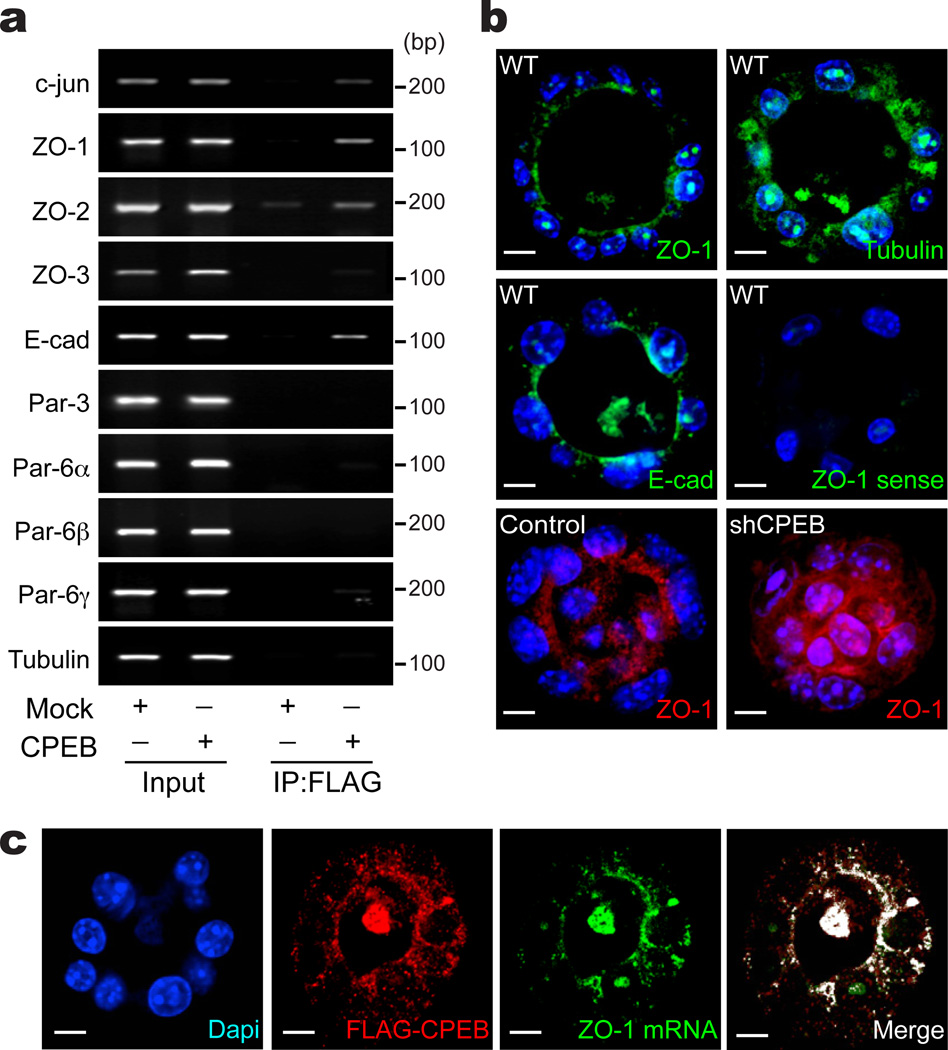
Two observations suggest that CPEB might be involved in ZO-1 mRNA localization: CPEB is often apically co-localized with ZO-1 and ZO-1 mRNA contains 5 conserved CPEs in its 3’ UTR (Supplementary Figure S5). To examine this possibility, FLAG-CPEB transduced EpH4 cells were subjected to FLAG co-immunoprecipitation and RNA analysis by RT-PCR (Fig. 5a). As expected, c-jun mRNA, but not tubulin mRNA, was co-immunoprecipitated with CPEB as shown previously29. ZO-1, ZO-2, and E-cadherin mRNAs, all of which also contain CPEs, were co-immunoprecipitated with CPEB. Other mRNAs involved in establishing cell polarity (Par-3, Par-6α, Par-6β, Par-6γ) were not co-precipitated, nor was ZO-3 mRNA. Next, cells grown without anchorage that formed a central cavity were subjected to RNA fluorescence in situ hybridization (FISH). Fig. 5b demonstrate that ZO-1 and E-cadherin mRNAs, but not tubulin mRNA, were localized apically. A ZO-1 sense probe yielded no signal. Thus, not only is ZO-1 mRNA localized apically as is ZO-1 protein, but surprisingly so too is E-cadherin mRNA even though E-cadherin protein is located baso-laterally. Depletion of CPEB abolished the apical localization of ZO-1mRNA (Fig. 5b). Finally, a double detection of transduced FLAG-CPEB (by immunocytochemistry) and ZO-1 mRNA (by FISH) shows that these two molecules were apically localized in cavity-forming cells (Fig. 5c). Thus, CPEB is necessary for ZO-1 mRNA localization during as cavity formation.
Figure 5. CPEB and ZO-1 RNA co-localize at the apical region.
(a) RNAs that co-immunoprecipitate with FLAG-CPEB. Input represents 10% of total RNA. Mock refers to FLAG immunoprecipitation from cells that were infected with FLAG only. (b) FISH for ZO-1, tubulin and E-cadherin mRNAs in cavity forming cells. ZO-1 RNA was also probed with a sense strand. FISH for ZO-1 mRNA in control and CPEB knockdown cells is also shown. Scale bar is 10 µm. (c) Double staining for ZO-1 mRNA by FISH and FLAG-CPEB by immunofluorescence. Scale bar is 10 µm.
The CPE is necessary for apical RNA localization
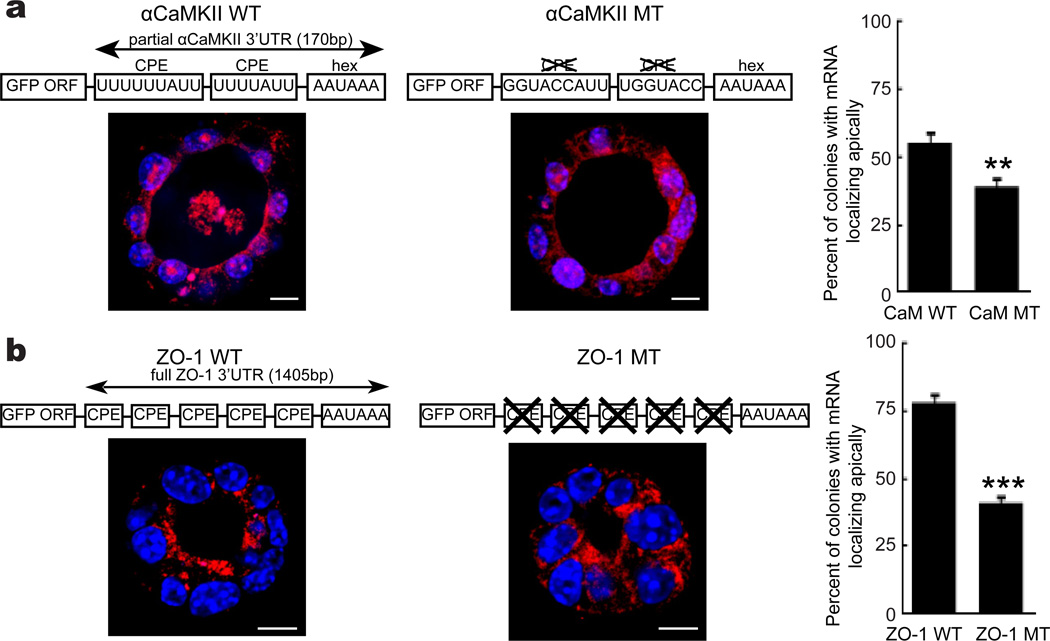
To investigate whether the CPE is involved in localizing RNA apically, GFP was fused to a portion of αCaMKII mRNA 3’ UTR, which contains two CPEs17 and which has been shown be localized to dendrites in a CPEB-dependent manner30. RNA FISH performed on cavity forming cells transduced with this construct demonstrates that the CPE mediated apical RNA localization (compare αCamKII WT to αCaMKII mutant where the CPEs were replaced by an irrelevant sequence) (Fig. 6a). In a similar experiment, GFP fused to the complete ZO-1 3’UTR was also apically localized; in contrast, mutation of 5 CPEs in the ZO-1 3’ UTR significantly reduced the number of colonies with apical GFP RNA localization (Fig. 6b). These results demonstrate that the CPE is necessary to localize RNA to the apical portion of cells arrayed in 3 dimensions.
Figure 6. The CPE is necessary and sufficient for RNA localization to the apical surface.
(a) The GFP open reading frame was fused to a portion of the αCaMKII 3’ UTR that contains two CPEs and a polyadenylation hexanucleotide AAUAAA. For comparison, GFP was fused to an identical 3’UTR had the CPEs replaced with an irrelevant sequence. These constructs were transduced into cells followed by culture in matrigel and FISH for GFP. Scale bar is 10 µm. The histogram shows the percent of colonies where the ectopic mRNA was localized apically. The two asterisks refer to statistical significance (p<0.01, Student’s t test, 3 replicates). (b) Cells transduced with the GFP coding region fused to the entire ZO-1 3’ UTR, or the ZO-1 3’ UTR mutating 5 CPEs, were grown on matrigel and processed for GFP FISH. Scale bar is 10 µm. The percent of colonies with GFP RNA localized to the apical surface is shown in the histogram. The three asterisks refer to statistical significance (p<0.001, Student’s t test, 3 replicates). Error bars are standard error of the mean.
CPE-dependent ZO-1 RNA localization and cavity formation
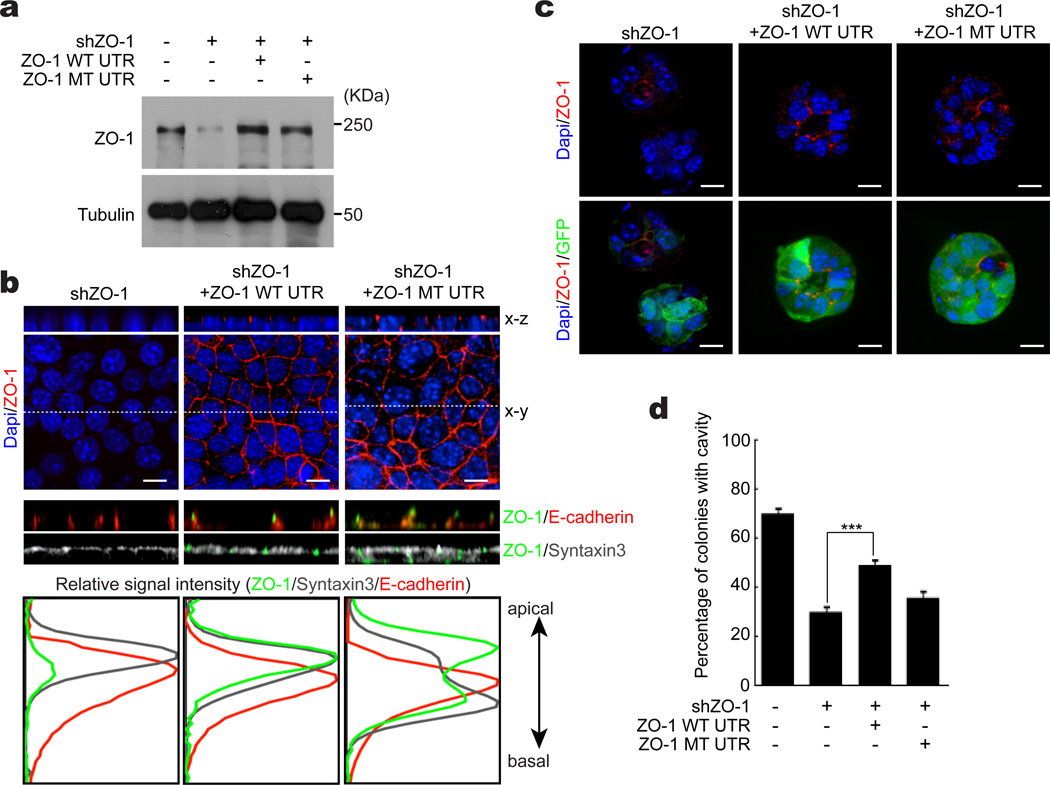
We determined whether ZO-1 mRNA apical localization, which is dependent upon the 3’ UTR CPEs as well as CPEB, is necessary for cavity formation. Cells infected with a lentiviruse containing an shRNA for ZO-1 were subsequently infected with a retrovirus expressing the entire ZO-1 coding sequence (1745 amino acids) with its normal 3’ UTR (1405 bases), or a UTR where the 5 CPEs were mutated. Figure 7a shows ~88% shRNA-mediated reduction of ZO-1, which was restored to normal levels by transduction of ZO-1 open reading frame containing its normal 3’ UTR or a UTR where the 5 CPEs were mutated. When grown as a monolayer and imaged along the x-z axis, ectopic ZO-1 derived from the mRNA with its WT 3’ UTR was localized to the apical surface and was indistinguishable from endogenous ZO-1 (Fig. 7b). In contrast, ecotopic ZO-1 derived from mRNA with the mutant 3’UTR was detected apically and baso-laterally, demonstrating that the 3’ UTR CPEs are necessary for apical localization of ZO-1. In these same cells, additional immunostining demonstrates that ZO-1 RNA with a mutant 3’UTR disrupted syntaxin-3 apical localization, indicating that mislocalization of ZO-1, but not depletion of ZO-1, reduces cell polarity (Fig. 7b).
Figure 7. CPE control of ZO-1 RNA localization is necessary for cell polarity.
(a) EpH4 cells were infected with lentivirus expressing shRNA for ZO-1 were also transduced with retrovirus expressing the entire ZO-1 coding region plus its normal WT 3’ UTR, or an identical 3’ UTR where 5 CPEs were mutated. The Western blot shows the levels of ZO-1, with tubulin serving as a loading control. (b) Cells expressing the constructs noted in panel a were grown on a monolayer and then immunostained for ZO-1, E-cadherin, and syntaxin-3. Confocal images are shown along the x-y and x-z axes, where the relative fluorescence intensities of the proteins were quantified. Scale bar is 10 µm. (c) Cells expressing the same constructs as in panel b were grown in an anchorage-independent manner. The cells were immunostainnd for ZO-1 and GFP. In the confocal images of the colonies, green indicates cells infected with shZO-1 and GFP expressing virus, red indicates immunostaining for ZO-1, and blue indicates DAPI. Scale bar is 10 µm. (d) The histogram shows the percent of colonies with cavities following the indicated treatments. The three asterisks refer to statistical significance (p<0.001, Student’s t test, 3 replicates). Error bars are standard error of the mean.
When grown in an anchorage-independent manner, 70% of colonies contained cavities, which was reduced to 30% when ZO-1 was depleted. In Fig. 7c, the GFP signal was a marker of the degree of ZO-1 depletion, indeed, high GFP expressing cells had reduced cavity formation. Ectopic expression of CPE-containing ZO-1 mRNA increased the colonies with cavities to 49% (p<0.001 Student’s t test, 3 replicates) , compared to shZO-1 alone, while expression of ZO-1 mRNA lacking the CPEs only modestly increased cavity formation (35%), which was statistically insignificant (Fig. 7d). Finally, depletion of ZO-1 had no discernible effect on E-cadherin levels, although it was not localized due to a lack of cavity formation. However, ectopic expression of ZO-1 with its wild type 3’ UTR (but not its CPE-lacking 3’ UTR) in the ZO-1 shRNA expressing cells not only restored cavity formation, it largely restored E-cadherin localization to the basal-lateral region (Supplementary Figure S6). Thus, CPEB control of ZO-1 mRNA localization is necessary for mammary epithelial cell polarity.
Because CPEB was first identified as a polyadenylation/translation factor, we used control and CPEB-depleted EpH4 cells to analyze the instantaneous synthesis of ZO-1 and E-cadherin by 35S-methionine/35S-cysteine pulse labeling followed by immunoprecipitation. In these same cells, we also examined the polyadenylation of ZO-1 mRNA and, as controls, tubulin and β-casein mRNAs. Neither the synthesis of ZO-1 or E-cadherin nor the polyadenylation of ZO-1 mRNA was affected by CEB depletion (Supplementary Figure S7) As expected, depletion of CPEB elicted reduced polyadenylation of casein mRNA31. Going further, we ectopically expressed CPEB with an alanine substitution for threonine 171, which renders the protein incapable of promoting polyadenylation21,25,26. In NMuMG cells, CPEB and CPEB T171A both promoted tight junction assembly as evidenced by enhanced ZO-1 immunostaining (Supplementary Figure S8; Fig. 3). Finally, we depleted the CPEB-associated Gld2 poly(A) polymerase21 from EpH4 cells, which also had no observable effect on tight junction assembly or cavity formation (Supplementary Figure S8). Thus, CPEB control of mRNA localization, but not polyadenylation or translation, mediates tight junction assembly and cell polarity.
Discussion
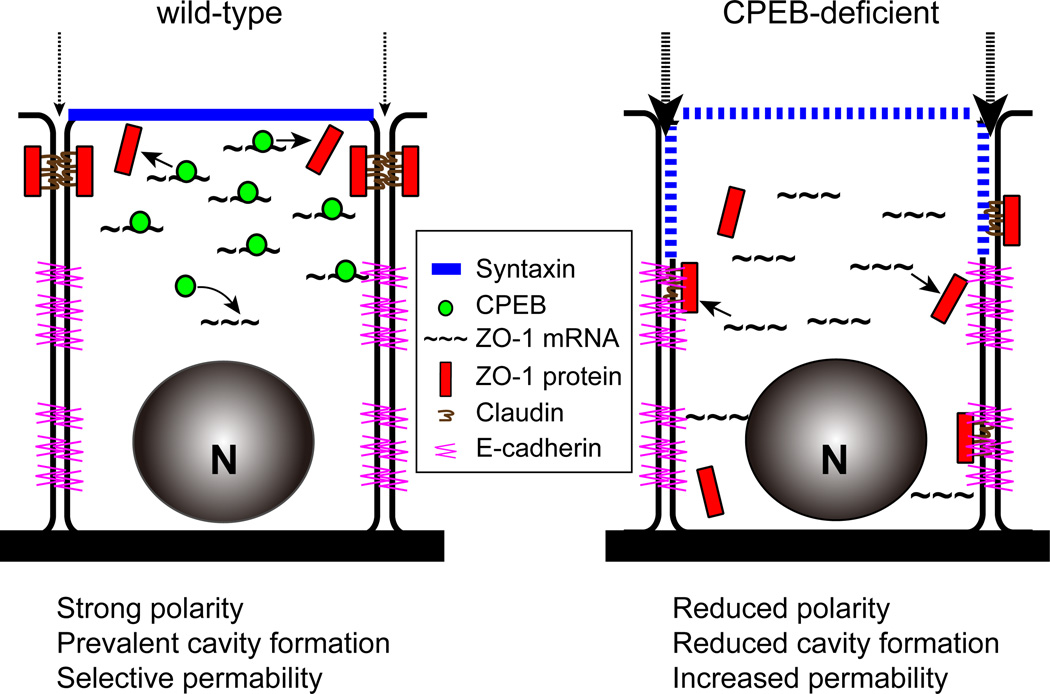
CPEB knockout mice are sterile and thus do not secrete the hormones necessary for mammary duct differentiation. CPEB heterozygous mice are fertile yet their mammary ducts contain disorganized and incompletely formed terminal end buds (TEB). Although injection of estradiol into CPEB KO mice induces mammary formation, the lumen of the ducts and their TEBs are sometimes not fully developed. Because lumen and TEB differentiation require the establishment of tight junctions on the luminal (apical)-lateral regions of adjoining cells, we surmised that CPEB might be involved in mammary epithelial cell polarity. Indeed, CPEB is essential for polarity of mammary cells grown in monolayer (anchorage-dependent) and most profoundly when grown in an anchorage-independent manner. CPEB control of polarity is mediated through ZO-1 mRNA localization; depletion of CPEB reduces apical tight junction assembly and ZO-1 localization, although it has no effect on overall ZO-1 levels or ZO-1 RNA poly(A) tail length. The ZO-1 3’ UTR contains 5 CPEs that direct reporter RNAs to the apical surface. Most importantly, depletion of ZO-1 inhibits cell polarization in a 3D culture system, which is rescued when ZO-1 mRNA containing but not lacking the CPEs is transduced into cells. These results define CPEB as a critical regulator of mammary cell polarity (Fig. 8).
Figure 8. Model depicting CPEB control of apical ZO-1 mRNA localization.
The thin and thick dashed arrows refer to strong and weak tight junction barriers to the passage of solutes in wild type and CPEB-deficient cells, repectively.
Apical concentration of proteins in mammalian epithelial cells generally occurs by protein sorting rather than by mRNA localization, usually in association with the trans-Golgi network or endosomes32–34. Indeed, there is but a single example, in this case in Drosophila epithelial cells of the follicle, where an alternately spliced isoform of stardust A mRNA is localized to the apical domain through an association with dynein35. At least in some cases, mRNA localization is thought to be determined in the nucleus, in association with exon junction complex (EJC) or other proteins36–38. Upon protein remodeling (association or dissociation of factors) in the cytoplasm, the RNP, most likely in a translationally dormant form, coalesces with a motor protein for transport on microtubules to the ultimate destination39,40. In neurons, Xenopus oocytes, and mammalian cell lines, CPEB shuttles between nucleus and cytoplasm24,41, and likely does so in epithelial cells where it probably binds mRNA before or during export. Moreover, in neuronal dendrites, CPEB associates with kinesin and dynein is transported in both retrograde and anterograde directions30. We would thus anticipate that in mammary epithelial cells, CPEB also interacts with a molecular motor for apical localization.
In addition to ZO-1 mRNA, CPEB binds ZO-2 mRNA, which encodes a protein that has at least a partially redundant function14. This observation, plus the fact that the CPE-containing PTEN is critically important for mammary epithelial cell polarity42 suggests that CPEB control of polarity might involve these mRNAs in addition to ZO-1 (indeed, perhaps CPEB modifies the polyadenylation/translation of other such mRNAs). Moreover, it is paradoxical that CPEB binds to and co-localizes with E-cadherin mRNA in an apical domain even though E-cadherin protein resides baso-laterally. We surmise that E-cadherin is synthesized apically but selectively transported to baso-lateral regions. In a larger sense, the presence of a CPE in an mRNA’s 3’ UTR and the site of residence of the encoded protein do not necessarily go hand-in-hand. For example, the mRNA for a major apical polarity protein Par-3 has no CPE while mRNAs for all three isoforms of apical Par-6 have CPEs yet none are co-immunoprecipitated by CPEB (Figure 5a) and thus if they are localized, they are unlikley to be directly controlled by CPEB. Many but not all of the claudin mRNAs contain CPEs, yet probably all of these proteins are apical. Virtually all CPE-containing mRNAs contain recognition sites for different binding proteins and miRNAs, which may displace CPEB or act singly or in combination to impart localization patterns different from ZO-1 mRNA, or indeed no localization at all.
In CPEB knockout mice, we detected no disruption of polarity in kidney or intestinal cells. Analysis of expression databases revealed that the level of CPEB is considerably lower in these tissues compared to mammary gland. Indeed, CPEB in MDCK (canine kidney) and Caco2 (human intestinal) epithelial cells was also low compared with EpH4 and NmuMG mammary epithelial cells. Moreover, CPEB depletion from MDCK and Caco2 cells had no discernable effect on ZO-1 localization or cell polarity. Why only mammary cell tight junctions require CPEB is not clear, but it may be related to the fact that mammary cells, but not kidney or intestinal cells, require hormonal sitmulation to induce lumen formation. Perhaps CPEB responds to hormonal signaling to promote apical ZO-1 RNA localization. Indeed, cavity formation in vitro with EpH4 and NMuMG mammary cells requires hormones (insulin, corticosterone, and prolactin) in the culture medium, which is not the case with MDCK or Caco2 cells (Supplementary Figure S4).
Our results generally agree with those of Umeda et al (2004) 43, who showed that depletion of ZO-1 did little to disrupt polarity in monolayer cells even though solute permeability was slightly increased. However, while those investigators did not examine the effect of ZO-1 deficiency in cells grown in 3 dimensions, Sourisseau et al (2006) 44 did find that ZO-1 deficiency destroyed polarity in cells grown in matrigel. It thus seems that 3 dimensional cell polarity is particularly sensitive to the levels and location of ZO-1 and perhaps other proteins as well.
The requirement of tight junctions for establishing and maintaining cell polarity is controversial; indeed some epithelial cells polarize even in the absence of tight junctions. On the other hand, consider the case of Par-3, a major cell polarity protein important for tight junction assembly45,46. Ectopic expression of Par-6 disrupts the localization of Par-3 at cell–cell adhesion sites, which in turn alters tight junction assembly as determined by mis-localization of ZO-145. Similar experiments also show that ZO-1 mis-localization is linked to disruption of the cell polarity protein complex47. These data are consistent with our observations that mis-localized ZO-1 disrupts cell polarity eventhough deletion of ZO-1 has no effect on this process in a 2D monolayer culture system. Perhaps cells lacking tight junctions compensate for cell polarity by an alternate mechanism, at least when cultured in monolayer. However, once tight junctions are mis-localized, cells lose polarity. Thus, CPEB supplies ZO-1 mRNA to apical sites to promote tight junction assembly, and may coordinate with the Par3-aPKC pathway to establish cell polarity in mammary epithelial cells.
Methods
Mouse strains and antbodies
CPEB knockout and heterozygous mice were generated as previously described16. All experiments with mice were performed according to the guidelines of the Institutional Animal Care and Use Committee of University of Massachusetts Medical School. The following antibodies were used: anti-ZO-1 (Invitrogen, for western blot 1:1000, for immunofluorescence, IF 1:100), anti-integrinβ4 (for IF 1:200), anti-E-cadherin (for western blot 1:2000, for IF 1:200), and anti-GM130 (BD Transduction, for IF 1:200)), anti-claudin3 (Abcam, for IF 1:100)), anti-syntaxin3 (Synaptic system, for IF 1:100), anti-β-casein (SantaCruz, for western blot 1:500), anti-HA (Covance, for IF 1:200), and anti-FLAG (for western blot 1:1000, for IF 1:100) and anti-tubulin (Sigma, for western blot 1:5000).
Whole mount mammary gland analysis
Inguinal mammary glands (L4 and R4) were collected from WT and CPEB KO female mice (8 weeks) injected with 50 µg/kg 17β-estradiol or vehicle (0.15M NaCl) for 5 days and from WT and CPEB heterozygous mice on day 1 of lactation. For whole-mount staining, the gland was fixed in Carnoy’s solution (10% glacial acetic acid, 30% chloroform and 60% absolute ethanol), and subsequently stained in 0.2% carmine alum. For histological analysis, the gland was fixed in 4% paraformaldehyde, embedded in paraffin, sectioned, and stained with Hematoxylin-Eosin.
Mammary gland tissues were frozen using liquid nitrogen. Frozen sections were fixed in 4% paraformaldehyde, and culture cells were fixed in 3% formaldehyde for 30 min. They were permeabilized with 0.5% Triton X-100/PBS for 10 min, blocked with 2% BSA/PBS for 1 h, treated with primary antibodies for 1 hr, and incubated with secondary antibodies for 1 hr. They were then mounted with ProLong Gold plus DAPI (Invitrogen). Images were captured using an inverted confocal microscope.
For FISH, 3D cultured cells were fixed in 3% formaldehyde for 30 min, dehydrated by successive washes in 25%, 50%, 75% and 100% methanol/PBST, and then rehydrated using the reverse order. Cells were acetylated in 0.25% acetic anhydride in 0.1 M triethanolamine for 10 min, treated with 10 µg/ml proteinase K for 10 min, stopped digestion in 2 mg/ml glycine/PBS for 10 min, and postfixed in 0.2% glutaraldehyde/3% formaldehyde for 20 min. Cells were prehybridized in NorthernMax hybridization buffer (Ambion), and incubated with 100 ng/ml fluorescein-labeled riboprobe overnight at 60°C. After washes with 50% formamide, 1% SDS in 5xSSC pH 4.5 and 50% formamide, 1% Tween 20 in 2xSSC pH4.5 at 60°C, cells were blocked with 2% BSA/PBS for 1 hr, and incubated with Alexa-594 conjugated anti-fluorescein antibody for 1 hr. Images were captured using an inverted confocal microscope.
Plasmid construction
Lentiviral shRNAs targeting mouse CPEB and ZO-1 were cloned into pLentiLox 3.7 vector; shCPEB-1: GTC GTG TGA CTT TCA ATA A, shCPEB-2: GTC CCA GAG ACC CTC TAA A, shZO1-1: GAG CGG GCT ACC TTA CTG A, shZO1-2: GTG CAT CTG AGA TGT TTG A. Human CPEB tagged with HA was cloned into pOZ-C retrovirus vector, and mouse CPEB and human ZO-1 tagged with 3xFLAG were cloned into pBABE retrovirus vector. Mouse CamKII 3’UTR with CPE (WT) or mutated CPE (MT) were cloned downstream of GFP in pBABE Mouse ZO-1 3’UTR with CPE (WT) or mutated CPE (MT) were cloned downstream of GFP or human ZO-1 tagged with 3xFLAG in pBABE, respectively.
Cell culture and virus infection
Mouse mammary gland epithelial cells (EpH4 and NmuMG), dog kidney epithelial cells (MDCK), and human colorectal epithelial cells (Caco2) were maintained in DMEM containing 5 µg/ml insulin, 10% FBS and antibiotic-antimycotic (Invitrogen). The lentiviral and retroviral constructs were co-transfected with each packaging plasmids into 293T cells by the calcium phosphate transfection method. Virus-containing supernatants were collected 48 hours after transfection, filtered to eliminate cells and target cells were infected in the presence of 6 µg/ml polybrene. Infected cells were selected with puromycin or sorted on a fluorescence activated cell sorter by GFP signal intensity.
For matrigel 3D cultures, cells were trypsinized, resuspended in assay media, and plated as single cells on 100% Matrigel-coated glass coverslips in 24-well tissue culture plates. Assay media was DMEM supplemented with 2% Matrigel, 5 µg/ml insulin, 1 µg/ml hydrocortisone and 3 µg/ml prolactin, and changed every 3 days. To visualize changes in morphology of cavity structures, immunofluorescence for ZO-1 was conducted.
For calcium switching, cells were grown on glass coverslips until a monolayer was formed, and cultured in a low calcium medium, S-MEM supplemented with 5 µM CaCl2 and 10% FBS that had been pretreated with Chelex resin (Bio-Rad) overnight. To begin the calcium switching, the medium was replaced with normal DMEM with 10% FBS.
For the wound scratch assays, EpH4 cells were grown in monolayers on glass coverslips and wounded with a pipette tip. After 24 hr, cell migration and orientation was assessed.
For the paracellular flux assay, cells were plated on Transwell filters 12 mm in diameter (Corning) for 6 days, and 10 µg/ml FITC-dextran (Sigma) was added to the medium in the apical compartment. After 1 hr incubation, 100 µl aliquot of the medium was collected from the apical and basal compartment and the paracellular tracer flux was measured as the amount of FITC-dextran in the medium with a fluorometer.
Immunoprecipitation of ribonucleoprotein complexes
Cells were lysed in polysome lysis buffer, 10mM HEPES-KOH pH7.0, 100mM KCl, 5mM MgCl2, 0.5% NP-40, 1mM DTT, protease inhibitor cocktail tablets (Roche), and 100 U/ml RNaseOut (Invitrogen). The lysate was incubated with anti-FLAG antibody or normal IgG at 4°C for 6 h, and further incubated with protein-G magnetic beads at 4°C for 14 hr. The beads were washed five times with polysome buffer and treated with 0.3 mg/ml protease K at 50 °C for 30 min. RNA was isolated using TRIZOL reagent (Invitrogen).
Poly(A) and translation analysis
For the poly(A) tail (PAT) assay, cDNA was synthesized with oligo(dT) anchor primer (5’-GCG AGC TCC GCG GCC GCG-T12-3’), and subsequent PCR was conducted with anchor primer (5’-GCG AGC TCC GCG GCC GCG-3’) and specific primer for ZO-1 (5’-TAT GGC ACA TCA GCA CGA TT-3’), tubulin (5’-CTG CTT CCA CAG GGA TGT TT-3’) and β-casein (5’-TTG AAT TCG ACC CAT GGA CT-3’) RNA in the presence of 32P-ATP.
For translational analysis, control and shCPEB1 infected EpH4 cells were cultured in methionine and cysteine-free media for 45 min and then cultured in media containing 100 µCi 35S-methionine and 35S-cysteine for 20 min. The cells were lysed in polysome lysis buffer containing protease inhibitor. The lysate was incubated with anti-ZO-1, anti-E-cadherin antibody or normal IgG at 4°C for 6 hr, and further incubated with anti-mouse IgG magnetic beads at 4°C for 14 h. The beads were resuspended with SDS gel loading buffer for analysis by electrophoresis and phosphorimaging.
Statistical analysis
Data are presented as mean ± s.e.m., and Student’s t-test (two-tailed) was used to compare between groups (p<0.05 was considered significant).
Supplementary Material
Acknowledgements
We thank Maria Ivshina for reagents and valuable advice, Paul Furcinitti of the Digital Light Microscopy Core and Richard Konz, Jr. of the Flow Cytometry Core for assistance and use of their facilities, and Lori Lorenz for reading the manuscript. K.N. was supported by JSPS Postdoctoral Fellowships for Research Abroad. T.U. was supported by a postdoctoral fellowship from FRAXA Research Foundation. The work was supported by grants from the NIH (AG30323 and HD37267). Additional core support from the Diabetes Endocrinology Research Center (DK32520) is gratefully acknowledged.
Footnotes
Contributions
K.N. performed all the experiments and T.U. generated the lentiviruses. K.N. and J.D.R. designed the experiments and J.D.R. wrote the manuscript.
Competing financial interests
The authors declare no competing financial interests
References
- 1.Holt CE, Bullock SL. Subcellular mRNA localization in animal cells and why it matters. Science. 2009;326:1212–1216. doi: 10.1126/science.1176488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.St Johnston D, Ahringer J. Cell polarity in eggs and epithelia: parallels and diversity. Cell. 2010;141:757–774. doi: 10.1016/j.cell.2010.05.011. [DOI] [PubMed] [Google Scholar]
- 3.Meignin C, Davis I. Transmitting the message: intracellular mRNA localization. Curr Opin Cell Biol. 2010;22:112–119. doi: 10.1016/j.ceb.2009.11.011. [DOI] [PubMed] [Google Scholar]
- 4.Becalska AN, Gavis ER. Lighting up mRNA localization in Drosophila oogenesis. Development. 2009;136:2493–2503. doi: 10.1242/dev.032391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Czaplinski K, Singer RH. Pathways for mRNA localization in the cytoplasm. Trends Biochem Sci. 2006;31:687–693. doi: 10.1016/j.tibs.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 6.Lewis RA, Mowry KL. Ribonucleoprotein remodeling during RNA localization. Differentiation. 2007;75:507–518. doi: 10.1111/j.1432-0436.2007.00159.x. [DOI] [PubMed] [Google Scholar]
- 7.Claussen M, Horvay K, Pieler T. Evidence for overlapping, but not identical, protein machineries operating in vegetal RNA localization along early and late pathways in Xenopus oocytes. Development. 2004;131:4263–4273. doi: 10.1242/dev.01283. [DOI] [PubMed] [Google Scholar]
- 8.Lin AC, Holt CE. Function and regulation of local axonal translation. Curr Opin Neurobiol. 2008;18:60–68. doi: 10.1016/j.conb.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang DO, Martin KC, Zukin RS. Spatially restricting gene expression by local translation at synapses. Trends Neurosci. 2010;33:173–182. doi: 10.1016/j.tins.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mailleux AA, Overholtzer M, Schmelzle T, Bouillet P, Strasser A, Brugge JS. BIM regulates apoptosis during mammary ductal morphogenesis, and its absence reveals alternative cell death mechanisms. Dev Cell. 2007;12:221–234. doi: 10.1016/j.devcel.2006.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reginato MJ, Muthuswamy SK. Illuminating the center: mechanisms regulating lumen formation and maintenance in mammary morphogenesis. J Mammary Gland Biol Neoplasia. 2006;11:205–211. doi: 10.1007/s10911-006-9030-4. [DOI] [PubMed] [Google Scholar]
- 12.Lelièvre SA. Tissue polarity-dependent control of mammary epithelial homeostasis and cancer development: an epigenetic perspective. J Mammary Gland Biol Neoplasia. 2010;15:49–63. doi: 10.1007/s10911-010-9168-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Furuse M. Molecular basis of the core structure of tight junctions. Cold Spring Harb Perspect Biol. 2010;2:a002907. doi: 10.1101/cshperspect.a002907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Umeda K, Ikenouchi J, Katahira-Tayama S, Furuse K, Sasaki H, Nakayama M, Matsui T, Tsukita S, Furuse M, Tsukita S. ZO-1 and ZO-2 independently determine where claudins are polymerized in tight-junction strand formation. Cell. 2006;126:741–754. doi: 10.1016/j.cell.2006.06.043. [DOI] [PubMed] [Google Scholar]
- 15.Hake LE, Richter JD. CPEB is a specificity factor that mediates cytoplasmic polyadenylation during Xenopus oocyte maturation. Cell. 1994;79:617–627. doi: 10.1016/0092-8674(94)90547-9. [DOI] [PubMed] [Google Scholar]
- 16.Tay J, Richter JD. Germ cell differentiation and synaptonemal complex formation are disrupted in CPEB knockout mice. Dev Cell. 2001;1:201–213. doi: 10.1016/s1534-5807(01)00025-9. [DOI] [PubMed] [Google Scholar]
- 17.Wu L, Wells D, Tay J, Mendis D, Abbott MA, Barnitt A, Quinlan E, Heynen A, Fallon JR, Richter JD. CPEB-mediated cytoplasmic polyadenylation and the regulation of experience-dependent translation of alpha-CaMKII mRNA at synapses. Neuron. 1998;21:1129–1139. doi: 10.1016/s0896-6273(00)80630-3. [DOI] [PubMed] [Google Scholar]
- 18.Huang YS, Jung MY, Sarkissian M, Richter JD. N-methyl-D-aspartate receptor signaling results in Aurora kinase-catalyzed CPEB phosphorylation and alpha CaMKII mRNA polyadenylation at synapses. EMBO J. 2002;21:2139–2148. doi: 10.1093/emboj/21.9.2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Groisman I, Ivshina M, Marin V, Kennedy NJ, Davis RJ, Richter JD. Control of cellular senescence by CPEB. Genes Dev. 2006;20:2701–2712. doi: 10.1101/gad.1438906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burns DM, Richter JD. CPEB regulation of human cellular senescence, energy metabolism, and p53 mRNA translation. Genes Dev. 2008;22:3449–3460. doi: 10.1101/gad.1697808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barnard DC, Ryan K, Manley JL, Richter JD. Symplekin and xGLD-2 are required for CPEB-mediated cytoplasmic polyadenylation. Cell. 2004;119:641–651. doi: 10.1016/j.cell.2004.10.029. [DOI] [PubMed] [Google Scholar]
- 22.Kim JH, Richter JD. Opposing polymerase-deadenylase activities regulate cytoplasmic polyadenylation. Mol Cell. 2006;24:173–183. doi: 10.1016/j.molcel.2006.08.016. [DOI] [PubMed] [Google Scholar]
- 23.Kim JH, Richter JD. RINGO/cdk1 and CPEB mediate poly(A) tail stabilization and translational regulation by ePAB. Genes Dev. 2007;21:2571–2579. doi: 10.1101/gad.1593007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin CL, Evans V, Shen S, Xing Y, Richter JD. The nuclear experience of CPEB: implications for RNA processing and translational control. RNA. 2010;16:338–348. doi: 10.1261/rna.1779810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mendez R, Hake LE, Andresson T, Littlepage LE, Ruderman JV, Richter JD. Phosphorylation of CPE binding factor by Eg2 regulates translation of c-mos mRNA. Nature. 2000;404:302–307. doi: 10.1038/35005126. [DOI] [PubMed] [Google Scholar]
- 26.Tay J, Hodgman R, Sarkissian M, Richter JD. Regulated CPEB phosphorylation during meiotic progression suggests a mechanism for temporal control of maternal mRNA translation. Genes Dev. 2003;17:1457–1462. doi: 10.1101/gad.1071403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Racki WJ, Richter JD. CPEB controls oocyte growth and follicle development in the mouse. Development. 2006;133:4527–4537. doi: 10.1242/dev.02651. [DOI] [PubMed] [Google Scholar]
- 28.Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30:256–268. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
- 29.Zearfoss NR, Alarcon JM, Trifilieff P, Kandel E, Richter JD. A molecular circuit composed of CPEB-1 and c-Jun controls growth hormone-mediated synaptic plasticity in the mouse hippocampus. J Neurosci. 2008;28:8502–8509. doi: 10.1523/JNEUROSCI.1756-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang YS, Carson JH, Barbarese E, Richter JD. Facilitation of dendritic mRNA transport by CPEB. Genes Dev. 2003;17:638–653. doi: 10.1101/gad.1053003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Choi KM, Barash I, Rhoads RE. Insulin and prolactin synergistically stimulate beta-casein messenger ribonucleic acid translation by cytoplasmic polyadenylation. Mol Endocrinol. 2004;18:1670–1686. doi: 10.1210/me.2003-0483. [DOI] [PubMed] [Google Scholar]
- 32.Mostov K, Su T, ter Beest M. Polarized epithelial membrane traffic: conservation and plasticity. Nat Cell Biol. 2003;5:287–293. doi: 10.1038/ncb0403-287. [DOI] [PubMed] [Google Scholar]
- 33.Rodriguez-Boulan E, Kreitzer G, Müsch A. Organization of vesicular trafficking in epithelia. Nat Rev Mol Cell Biol. 2005;6:233–247. doi: 10.1038/nrm1593. [DOI] [PubMed] [Google Scholar]
- 34.Sharma N, Low SH, Misra S, Pallavi B, Weimbs T. Apical targeting of syntaxin 3 is essential for epithelial cell polarity. J Cell Biol. 2006;173:937–948. doi: 10.1083/jcb.200603132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Horne-Badovinac S, Bilder D. Dynein regulates epithelial polarity and the apical lolcalization of stardust A mRNA. Plos Genetics. 2008;4:40–51. doi: 10.1371/journal.pgen.0040008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Farina KL, Singer RH. The nuclear connection in RNA transport and localization. Trends Cell Biol. 2002;12:466–472. doi: 10.1016/s0962-8924(02)02357-7. [DOI] [PubMed] [Google Scholar]
- 37.Gu W, Pan F, Zhang H, Bassell GJ, Singer RH. A predominantly nuclear protein affecting cytoplasmic localization of beta-actin mRNA in fibroblasts and neurons. J Cell Biol. 2002;156:41–51. doi: 10.1083/jcb.200105133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shen Z, St-Denis A, Chartrand P. Cotranscriptional recruitment of She2p by RNA pol II elongation factor Spt4-Spt5/DSIF promotes mRNA localization to the yeast bud. Genes Dev. 2010;24:1914–1926. doi: 10.1101/gad.1937510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lewis RA, Mowry KL. Ribonucleoprotein remodeling during RNA localization. Differentiation. 2007;75:507–518. doi: 10.1111/j.1432-0436.2007.00159.x. [DOI] [PubMed] [Google Scholar]
- 40.Messitt TJ, Gagnon JA, Kreiling JA, Pratt CA, Yoon YJ, Mowry KL. Multiple kinesin motors coordinate cytoplasmic RNA transport on a subpopulation of microtubules in Xenopus oocytes. Dev Cell. 2008;15:426–436. doi: 10.1016/j.devcel.2008.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kan MC, Oruganty-Das A, Cooper-Morgan A, Jin G, Swanger S, Bassell G, Florman H, van Leyen K, Richter JD. CPEB4 is a cell survival protein retained in the nucleus upon ischemia or endoplasmic reticulum calcium depletion. Mol Cell Biol. 2010;30:5658–5671. doi: 10.1128/MCB.00716-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martin-Belmonte F, Gassama A, Datta A, Yu W, Rescher U, Gerke V, Mostov K. PTEN-mediated apical segregation of phosphoinositides controls epithelial morphogenesis through Cdc42. Cell. 2007;128:383–397. doi: 10.1016/j.cell.2006.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Umeda K, Matsui T, Nakayama M, Furuse K, Sasaki H, Furuse M, Tsukita S. Establishment and characterization of cultured epithelial cells lacking expression of ZO-1. J Biol Chem. 2004;279:44785–44794. doi: 10.1074/jbc.M406563200. [DOI] [PubMed] [Google Scholar]
- 44.Sourisseau T, Georgiadis A, Tsapara A, Ali RR, Pestell R, Matter K, Balda MS. Regulation of PCNA and cyclin D1 expression and epithelial morphogenesis by the ZO-1-regulated transcription factor ZONAB/DbpA. Mol Cell Biol. 2006;26:2387–2398. doi: 10.1128/MCB.26.6.2387-2398.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Joberty G, Petersen C, Gao L, Macara IG. The cell-polarity protein Par6 links Par3 and atypical protein kinase C to Cdc42. Nature Cell Biol. 2000;2:531–539. doi: 10.1038/35019573. [DOI] [PubMed] [Google Scholar]
- 46.Chen X, Macara IG. Par-3 controls tight junction assembly through the Rac exchange factor Tiam1. Nat Cell Biol. 2005;7:262–269. doi: 10.1038/ncb1226. [DOI] [PubMed] [Google Scholar]
- 47.Hurd TW, Gao L, Roh MH, Macara IG, Margolis B. Direct interaction of two polarity complexes implicated in epithelial tight junction assembly. Nat Cell Biol. 2003;5:137–142. doi: 10.1038/ncb923. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.