Abstract
Specific wavelengths of light can exert various physiological changes in plants, including effects on responses to disease incidence. To determine whether specific light wavelength had effects on rotting disease caused by Pseudomonas putida 229, soybean sprouts were germinated under a narrow range of wavelengths from light emitting diodes (LEDs), including red (650–660), far red (720–730) and blue (440–450 nm) or broad range of wavelength from daylight fluorescence bulbs. The controls were composed of soybean sprouts germinated in darkness. After germination under different conditions for 5 days, the soybean sprouts were inoculated with P. putida 229 and the disease incidence was observed for 5 days. The sprouts exposed to red light showed increased resistance against P. putida 229 relative to those grown under other conditions. Soybean sprouts germinated under red light accumulated high levels of salicylic acid (SA) accompanied with up-regulation of the biosynthetic gene ICS and the pathogenesis- related (PR) gene PR-1, indicating that the resistance was induced by the action of SA via de novo synthesis of SA in the soybean sprouts by red light irradiation. Taken together, these data suggest that only the narrow range of red light can induce disease resistance in soybean sprouts, regulated by the SA-dependent pathway via the de novo synthesis of SA and up-regulation of PR genes.
Introduction
Soybean (Glycine max L. Merr.) is one of the most important crops in terms of providing oil and protein [1]; accordingly, soybean has been used as a model system for the seed developmental process [2]. The consumption of soy food exerts high benefits on human health including reduced incidence of coronary heart disease, reduced risk of breast and prostate cancers, improved bone health and relief of menopausal symptoms [3]. Soybeans are utilized in a variety of foods including soymilk, soy yoghurt, tofu, miso, soy sauce, soy flour, soy cheese, green and dried soybeans, soybean sprouts, and other fermented food products [4]. Additionally, soybean sprouts are commonly consumed in Northeast Asian countries including Korea, China and Japan as vegetables in soups, salads and side dishes [5].
Any type of reduction in soybean yield mainly occurs during the sprout stage of development [6]. Different diseases cause loss of soybean production, and causal organisms responsible for such losses include fungi and bacteria such as Rhizoctonia sp., Pseudomonas sp., Phytopthora sp. and Bradyrhizobium japonicum [1,7–9]. Furthermore, sprouts have been shown to be the means of transmission of a number of food borne outbreaks of infection [6,10,11]. These occurrences of infection integrated salmonella poisoning and Escherichia coli 0157 infection, and implicated all kinds of seed sprouts, including alfalfa, clover, cress, mung bean, radish and soybean [6,11]. Thus, microbial infection of soybean sprouts has large negative impacts on soybean production and the soybean sprout industry.
Light plays an essential role in plant growth and development together with host defensive mechanisms. Subjecting plants to specific wavelengths of light such as ultraviolet (UV) and red light can induce plants to develop higher levels of disease resistance against pathogens. UV-C irradiation leads to accumulation of phytoalexin hydroxyphaseollin, which helps soybean plants develop resistance against P. megasperma var. sojae [12]. UV irradiation also facilitates accumulation of the phytoalexins sakuranetin and oryzalexin F in rice leaves, which might help increase resistance against microbial pathogens [13,14]. Red light treatment of pepper, pumpkin, and tomato seedlings led to development of resistance against P. capsici [15]. Furthermore, systemic disease resistance against root-knot nematode Meloidogyne javanica and a bacterial disease, P. syringae pv. Tomato DC 3000, was induced by pretreatment of Arabidopsis with red light [16].
Salicylic acid (SA) is a small phenolic compound produced by both prokaryotes and eukaryotes [17]. Biotic and abiotic stimuli increase the endogenous levels of SA in plants, which helps induce the defense mechanism [17]. SA-mediates plant immune response to systemic acquired resistance (SAR), which limits the growth of biotrophic and necrotrophic virulent pathogens and favors long-term protection against a broad spectrum of microorganisms [18–21]. P. fluorescens WCS417r triggered SAR by accumulating endogenous SA in radish but not in Arabidopsis which induced systemic resistance through SA-independent signaling pathway [22,23]. The endogenously increased SA describes the state of SAR by inducing the expression levels of pathogenesis-related (PR) genes, such as PR-1, PR-2 and PR-5, which are considered to be the effector genes for SAR [24,25]. SA does not always induce resistance to pathogens, shown in the suppressed resistance by SA addition in the induced resistance in the broad beans by red light against Botrytis cinerea [26]. The SA-dependent defense system is working specifically based on the relation between a plant host species and a pathogen species. In tomato plants, SA-dependent defense pathway induced resistances to Botrytis cinerea but not to Oidium neolycopersici, however, in the tobacco it induced resistance to O. neolycopersici but not to B.cinerea [27]. Therefore, plant defense responses activated by SA-dependent pathway depend on a specific host-pathogen system, not on a commonly shared system even differed in a closely related host-pathogen relation.
Jasmonic acid (JA) and SA are essential plant hormones in plant-pathogen defense signaling pathway regulating induction of PR genes [28]. These two plant hormones signaling pathways are mutually antagonistic or show no modulation in one another induction [29,30]. Silver leaf whitefly infested Arabidopsis leaves induced the SA-regulated PR-1 gene transcripts, but no or very low expression of PDF1.2, a JA-regulated gene marker [30]. SA treatment blocked the biosynthesis of JA in tomato leaves and production of JA was inhibited by SA application in wounded tobacco plants [31,32]. Exogenous application of SA induced the expression of acidic PR genes, but the expression of the genes was inhibited by JA application. In contrast, JA application induced the expression of basic PR genes, which expression was hampered by SA treatment [28]. All evidence indicates that SA and JA exert antagonistic effects on plant-pathogen defense signaling.
Recently, the availability of light emitting diodes (LEDs) has made it possible to study the effects of narrow ranges of light wavelength on plant growth and development. Previously, the roles of a certain wavelength on the physiology of plants were studied by employing light filters. However, LED technology has enabled replacement of filters with LEDs that emit single wavelengths of light. Use of LEDs as light sources can easily make the emitting wavelength and light intensity selectable; therefore, LED application to plant experiments has resulted in a great increase in accumulation of knowledge regarding the effects of individual light wavelengths on plant growth and development [33].
In this study, we irradiated soybean sprouts with a narrow range of light wavelength to determine (i) whether soybean sprouts germinated under continuous irradiation with red light gained resistance against Pseudomonas putida 229, a rotting bacterium of soybean sprouts and (ii) the underlying mechanism for the induced resistance, especially for SA-mediated resistance.
Materials and Methods
Germination of soybean sprouts under irradiation with different wavelengths of light
Seeds of soybean [Glycine max (L.) Merr. cv. Pungsan] were provided by Dr. Euiho Park, who had preserved soybean stocks in a seed storage room maintained with low moisture and low temperature. The soybeans used in this experiment were harvested from the field of Yeungnam University in the Fall of 2012. Seeds were washed in double distilled water (ddH2O), sterilized by dipping and shaking in 70% ethanol for 30 s, and then rinsed with ddH2O at least five times. Next, the sterilized seeds were soaked in ddH2O for 4 h at 25°C to initiate germination, after which the surface water was removed gently by hand shaking and the seeds were placed on wet tissue papers of uniform thickness in a plastic wicker tray. The trays with soybean seeds were placed in separate sectors of a chamber at 25°C and 50% humidity. Each sector was then irradiated with different light sources, including LEDs emitting light at 440, 660, or 730 nm (blue, red, and far-red, respectively) and daylight fluorescence (DLF) bulbs (three band lamp, 11W, Alim Industry, Republic of Korea). The red, far-red, blue and fluorescent light sources had light intensities of 46.10, 2.19, 35.01 and 10.85 μM photons /m2s, respectively. The control was composed of soybean seeds germinated under the same conditions without any light treatment. The soybean seeds were grown for 5 days while providing the proper amount of ddH2O daily to prevent the seeds from drying out.
Bacterial inoculation
A single colony of P. putida 229, a pathogenic bacteria that causes soybean sprout rotting, was grown in nutrient broth (NB) media (Difco Nutrient Broth, Becton, Dickinson and Company, Sparks, USA) at 25°C for 2 days with shaking at 100 rpm. After removing the root tips (3cm from the end) from soybean sprouts grown for 5 days with a sterile scalpel, the remaining parts were inoculated by immersion in P. putida 229 culture adjusted to an OD600 of 1.0 for 8 h. The soybean sprouts were recovered from the culture solution, after which the culture drops on the surface were removed by gentle hand shaking, and samples were incubated on wet-tissue paper at 25°C in darkness for 5 days, during which time they were observed for disease incidence. The disease incidence was calculated by dividing the number of the infected soybean hypocotyls by the total number of hypocotyls inoculated in each treatment. Fifteen hypocotyls were inoculated with P. putida 229 in each treatment, and those experiments were repeated four times.
To prepare the samples for SA and JA analysis and gene expression, 5 day old soybean sprouts inoculated with P. putida 229 were subjected to the following treatments: control (0 h inoculation + 0 h incubation), 6 h treatment (3 h inoculation + 3 h incubation), 16 h treatment (8 h inoculation + 8 h incubation) and 32 h treatment (8 h inoculation + 24 h incubation). Only hypocotyls were collected and used for the extraction of SA, JA and total RNA.
Analysis of the contents of SA and JA
The extraction of SA from the soybean hypocotyls was conducted according to the method described by Marek et al. [34], with slight modification. Briefly, 0.5 g of hypocotyls were ground with a mortar and pestle, mixed with 3 mL of 90% methanol, and then centrifuged at 14,000g at 4°C for 10 min, after which the supernatant was collected. The pellets were re-extracted with 1.5 mL of 100% methanol, after which the supernatant was combined with the previously collected supernatant. The combined supernatant samples were then concentrated to a final volume of around 250 μL using a speed vacuum (miVac DUO concentrator, New York, USA), then resuspended in 1 mL of hydrolysis buffer (0.1 M sodium acetate buffer, pH 5.5).
Next, the mixture was split into two equal volumes and analyzed for free SA and glucose-conjugated SA (salicylic acid 2-O-β-D-glucoside, SAG) in two separate tubes. To determine the amount of SAG, 10 units of β- glucosidase (Sigma-Aldrich, St. Louis, MO USA) were added to the tube. Following incubation at 37°C for 1.5 h, 625 μL of 10% TCA was added to both tubes, after which the samples were centrifuged at 14,000 g and 4°C for 10 min. The supernatant was then transferred to a fresh tube and mixed with 1 mL extraction solvent composed of ethylacetate: cyclohexane (1:1 ratio). Only the top organic phase was transferred into a fresh tube, after which it was concentrated to a final volume of ∼125 μL using a speed vacuum. Next, the residue was resuspended in 0.5 mL of 0.2 M sodium acetate buffer (pH 5.5), after which it was centrifuged at 14,000 g at 4°C for 10 min. The supernatant was subsequently removed and filtered using a 0.45 μm nylon filter (Chemco Scientific, Japan).
Extraction of JA was performed according to the method described by Muller and Bosch [35] with minor modifications. In brief, 100 mg of frozen soybean hypocotyls were ground with a motor and pestle using liquid nitrogen, and mixed with 2 mL of extraction solvents composed of methanol: isopropanol (20:80 ratio) with 1% glacial acetic acid. The solvent-mixed samples were then sonicated at 4–10°C for 30 min and centrifuged at 10,000 g at 4°C for 10 min. The supernatant was collected in a glass vial, and the residue was re-extracted with 1 mL of extraction solvent by following the same procedure. The newly collected supernatant was added into the original supernatant in the same glass vial, and dried under nitrogen gas stream. The dried samples were mixed with 500 μL of methanol and filtered using the 0.45 μm nylon filter (Chemco Scientific, Japan).
After the extraction of SA and JA, both the compound amount were determined using an HPLC with a Denali C18 120A 5μ, 150mm × 4.6 mm column (Grace Davison Discovery Sciences, Illinois, USA) and an UV detector (YL9100, Young-Lin, Republic of Korea). The mobile phase used for SA and JA analysis was 0.2 M sodium acetate buffer (pH 5.5) in 10% methanol at a flow rate of 0.80 mL mˉ1 and acetonitrile: water (25:75) with 0.1% trifluoroacetic acid (TFA) at a flow rate of 1 mL mˉ1, respectively. The amount of SA and JA were determined by comparing the area for the corresponding peaks with the standard curve drawn using different concentrations of free SA (Duchefa Biochemie, Netherlands) and JA (Sigma-Aldrich, St. Louis, USA).
RNA extraction and semi-quantitative RT-PCR
Total RNA was isolated from the hypocotyls under the same conditions as the SA and JA extraction using the Tri-Reagent solution (Molecular Research Center, Inc., Ohio, USA) according to the manufacturer’s protocol. Next, 50 ng of total RNA were used for semi-quantitative RT-PCR (TITANIUM one step RT-PCR kit, Takara Bio Inc., Japan) on a PCR machine (XP Thermal Cycler, BIOER, Japan). The primers for the candidate genes were designed using the Primer3 (v.0.4.0) program (http://bioinfo.ut.ee/primer3-0.4.0/) [36,37]. The primers for the genes in the biosynthesis of SA and JA and for the PR genes are summarized in Table 1. The genes selected for the SA synthesis were phenylalanine ammonium lyase (PAL) and isochorismate synthase (ICS). The genes for the JA synthesis included acyl-CoA oxidase (ACX) and 3-ketoacyl-CoA thiolase (KAT). The PR-1 and PR-4 genes were selected for the SA and JA- dependent PR genes, respectively. Actin gene was used as a reference to verify equal amounts of total RNA. Semi-quantitative RT-PCR was conducted under the following reaction conditions: initial denaturation at 95°C for 5 min, followed by 30 cycles of 95°C for 20 s, 50–54°C for 30 s and 72°C for 40 s, and then final extension at 72°C for 5 min. The PCR products were visualized by electrophoresis on 1.2% agarose gel.
Table 1. Primers used for PCR and their amplicon sizes.
| Genes (NCBI accession no.) | Primer pairs | Amplicon size (bp) |
|---|---|---|
| Isochorismate synthase (ICS) (AW596452.1) | Forward; 5’–CAACAGAAGAGGCACAACTT– 3’ | 215 |
| Reverse; 5’–GAGTTCTAGCTCATCCCACTC– 3’ | ||
| Phenylalanine ammonium lyase (PAL) (X52953.1) | Forward; 5’ –GGAGTCTCTATGGACAACACAC –3’ | 194 |
| Reverse; 5’ –TGGAGTTCAGAGCAGTAAGAAG– 3’ | ||
| Acyl-CoA Oxidase (ACX) (NM_001250062.1) | Forward; 5’ –CTGGTCTTTCTATCACTGGAAG– 3’ | 216 |
| Reverse; 5’–CATCACTTCCATAGTCAGGTTC– 3’ | ||
| 3-ketoacyl-CoA thiolase (KAT) (AY383736.1) | Forward; 5’– CAGATAGAGGAATTGAGTGCAG –3’ | 198 |
| Reverse; 5’– GCCTATTCACACGATCTACTGT –3’ | ||
| Pathogenesis- related1 (PR-1) (AF136636.1) | Forward; 5’ –TGATGTTGCCTACGCTCAAG –3’ | 137 |
| Reverse; 5’ –AAGCAGCAACCGTATCATCC– 3’ | ||
| Pathogenesis- related4 (PR-4) (Z11977.1) | Forward; 5’ –GCTTGCGGGTGACAAATAC– 3’ | 96 |
| Reverse; 5’ –ACACTCCCACGTCCAAATC– 3’ | ||
| Actin (U60500.1) | Forward; 5’ –GAGAGAGGATACTCCTTCAGC –3’ | 204 |
| Reverse; 5’ –GAACAGTACTTCTGGGCAAC –3’ |
Statistical analysis
All numeric data represent the means of three samples ± the standard deviation (SD). The variance of the sample data was identified by Duncan’s test using the Statistical analysis software (SAS) version 9.1 (SAS Inc., Cary, NC, USA).
Results
Growth of soybean sprouts under different germinating conditions
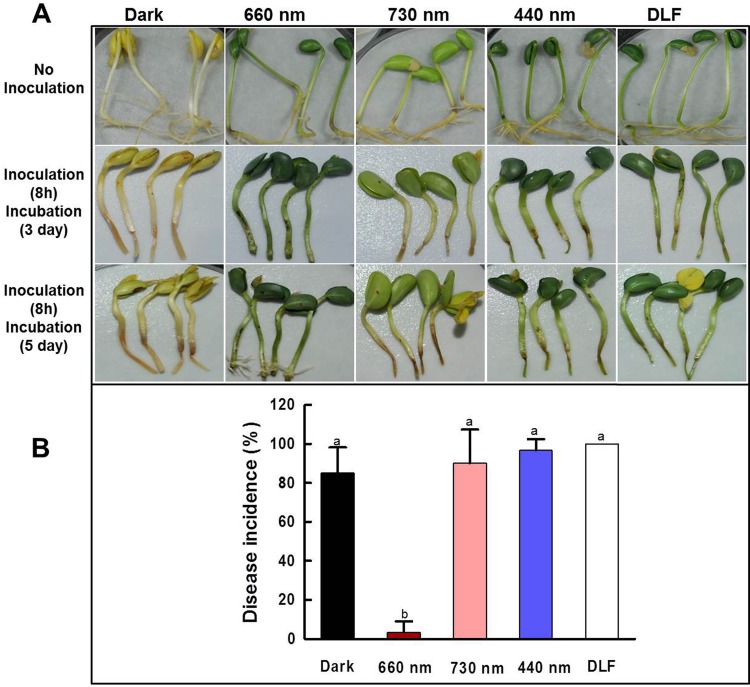
All soybean seeds germinated under the aforementioned conditions; however, they developed different colors and hypocotyl growth (Fig. 1A). The sprouts germinated under red, blue and white light were dark green, while those germinated under far-red light were light green (Fig. 1A). The sprouts germinated in darkness had yellow seeds and white hypocotyls due to the lack of chlorophylls in the absence of photosynthesis. Additionally, the sprouts germinated under far-red light had shorter hypocotyls than those grown under other conditions (Fig. 1A).
Fig 1. Effects of different wavelengths on the growth of soybean sprouts and disease susceptibility to Pseudomonas putida 229.
Soybean sprouts were germinated for 5 days under different conditions. (A) The soybean sprouts germinated in darkness, red light, far-red light, blue light and fluorescence daylight. The sprouts tips were cut, inoculated in bacterial culture for 8 h, and then incubated for 5 days in darkness. The disease symptoms were observed after 3 and 5 days of incubation in darkness. (B) Comparison of disease incidence in soybean sprouts germinated in darkness and under different light irradiation for 5 days. The experiments were repeated four times. The different letter indicates a significant difference (P<0.01).
Red light-induced resistance of soybean sprouts to P. putida 229
Soybean sprouts germinated under different conditions were incubated in darkness for a total of 5 days after inoculation (DAI) of P. putida 229 to measure the disease incidence. Except for sprouts germinated under red light, other sprouts germinated in darkness and under other wavelengths of light rotted at 5 DAI with P. putida 229 (Fig. 1A). The soybean sprouts germinated under red light had not developed any symptoms of disease at 3 DAI (Fig. 1A), and had fully recovered from the disease and grew adventitious root at 5 DAI (Fig. 1A). The soybean sprouts germinated under red light had only 3.3% disease incidence, which was significantly very lower than those germinated in darkness or under other wavelengths (Fig. 1B). The soybean sprouts germinated in darkness and under blue light, far red light and daylight fluorescence had 85%, 96.7%, 90% and 100% disease incidence, respectively. (Fig. 1B).
Expression analysis of genes in the biosynthetic pathway of SA and JA
After observation of disease resistance in soybean sprouts, we examined the expression levels of genes in the pathway of SA and JA biosynthesis to determine which plant hormone was responsible for the increased resistance in soybean sprouts. Furthermore, the expression levels of the PR-1 and PR-4 genes induced by SA and JA, respectively, were analyzed to confirm that the defense molecule was induced by red light irradiation.
The biosynthesis of SA takes place from chorismic acid via the enzymatic reaction of ICS or PAL Wildermuth et al. [38]. From chorismic acid, the ICS or PAL is the committed step for further SA biosynthesis [30], therefore, the genes of ICS and PAL were selected to determine the expression levels for the de novo synthesis of SA by the infection of P. putida 229. When the expression levels of the ICS gene were measured, the ICS gene appeared to be highly expressed in soybean sprouts grown under red light (0 h + 0h) (Fig. 2A); therefore, the gene was responsible for the higher levels of SA in sprouts grown under red light. The expression levels in the soybean sprouts grown in darkness or under red light decreased after being incubated with P. putida 229 (3 h + 3 h and 8 h + 8 h), but returned to the original expression levels in soybean sprouts grown under red light after 24 h (Fig. 2A). Expression of the PAL gene did not differ significantly among soybean sprouts grown in darkness or under red light, or in response to infection with P. putida 229 (Fig. 2A). To confirm that the increased levels of SA affected expression of the PR gene, we measured the expression level of the PR-1 gene, which is a downstream marker of SA accumulation. After incubation for 24 h (8 h + 24 h), the gene was up-regulated to a greater degree in soybean sprouts grown under red light than in those grown in darkness, but gene expression was not detected in un-infected sprouts (0 h + 0 h) (Fig. 2A).
Fig 2. Expression profile of genes in the biosynthetic pathways of (A) Salicylic acid and (B) jasmonic acid, and their pathogenesis-related (PR) genes.
Abbreviation: ICS, isochorismate synthase; PAL, phenylalanine ammonium lyase; ACX, acyl-CoA oxidase; KAT, 3-ketoacyl-CoA thiolase. The actin gene was used as the reference gene for equal usage of total RNA.
In the JA biosynthesis, 3-oxo-2-[2’(Z)-pentenyl]-cyclopentane-1-octanoic acid (OPC8:0) is converted to 2-trans-enoyl-CoA by acyl-CoA oxidase (ACX), then synthesized to JA by 3-ketoacyl-CoA thiolase (KAT) [39], therefore, the two enzymes were selected to determine the gene expression. Both ACX and KAT genes were expressed in soybeans grown in darkness and under red light, but their expression levels did not change in response to P. putida 229 infection (Fig. 2B). Concordant with the data describing genes involved in the biosynthesis of JA, the expression level of the PR-4 gene induced by high JA content did not change, but was expressed at high levels with or without P. putida 229 infections (Fig. 2B).
Free- and conjugated- SA accumulation in soybean sprouts irradiated with red light
To clarify the mechanism of resistance against the P. putida 229 in soybean sprouts germinated under red light, we measured the endogenous content of both free SA and SAG in soybean sprouts. Soybean sprouts germinated for 5 days in darkness or under red light were used as uninoculated controls or inoculated with P. putida 229 for 8 h and then subjected to incubation for 24 h in darkness (Fig. 3). The SA content of soybean sprouts germinated in darkness and under red light irradiation did not differ significantly (Fig. 3A). In contrast, the sprouts germinated under red light contained SA content more than 2.0 times greater than those germinated in darkness after P. putida 229 inoculation, representing a significant difference (Fig. 3A). The level of SAG in the soybean sprouts grown under red light was slightly higher than in soybeans grown in darkness; however, the level did not differ significantly before and after inoculation (Fig. 3B).
Fig 3. Content of free and conjugated salicylic acids.
(A) Free salicylic acid (SA) and (B) glucose-conjugated salicylic acid (SA glucoside, SAG) in soybean sprouts grown in darkness or under red light irradiation. The SA and SAG contents of germinated soybean sprouts were measured without inoculation of Pseudomonas putida 229 (control), or after inoculation for 8 h followed by incubation in darkness for 24 h (inoculation). The experiments were repeated two times. The different letter indicates a significant difference (P<0.05).
JA contents in soybean sprouts irradiated with red light
The content of JA in soybean sprouts was determined to elucidate whether the disease resistance in the soybean sprouts germinated under red light was induced by JA-dependent pathway, though the expression patterns of ACX and KAT gene in the JA biosynthesis pathway implied that the resistance might not have been induced by high accumulation of JA. JA content was measured for the soybean sprouts germinated for 5 days in darkness or under red light as uninoculated samples, or for them inoculated with P. putida 229 for 8 h and then subjected to incubation for 24 h in darkness as inoculated samples (Fig. 4). JA content of uninoculated hypocotyls germinated in darkness or under red light was not significantly different. Both sprouts significantly increased the amount of JA after inoculation, however, each soybean sprout germinated in darkness or under red light did not differ the content of JA in the hypocotyls inoculated with P. putida 229 (Fig. 4).
Fig 4. Content of jasmonic acids (JA).

Content of JA in soybean sprouts germinated in darkness or under red light was analyzed. JA contents in the soybean hypototyls were measured without inoculation of Pseudomonas putida 229 (control), or with inoculation for 8 h followed by incubation in darkness for 24 h (inoculation). These experiments were repeated two times. Different letter indicates a significant difference (P<0.05).
Discussion
Light is essential for the lives of plant species, providing energy for their survival and working as a signal to develop differentially and exert physiological changes. Great effort has been made to understand the roles of different light wavelengths on plant disease resistance [15,40,41]. Induced disease resistance against Phytophthora capsici has been observed in many plants, including pumpkin, pepper and tomato seedlings, in response to the application of red light treatment [15]. In broad bean, red light treatment suppressed the leaf spot disease caused by Alternaria tenuissima [40]. Irradiation of Nicotiana benthamiana with different wavelengths of light induced resistance against wildfire disease, especially by blue and red wavelengths [41].
Soybean yield is mainly decreased by the attack of several fungal, bacterial and viral diseases [42]. Several chemicals including 2,6-dichloroisonicotinic acid (INA), benzothiadiazole (BTH) and humic acids have been used to control these diseases [42,43]. Although these chemicals are effective at controlling such diseases, they can be expensive and exert adverse effects on the environment. Therefore, in this study, we used an environmentally friendly method that could be useful for identification of genes involved in resistance against disease infection. Furthermore, most studies of soybean have been conducted using seeds; therefore, investigations of the protection of soybean sprouts against disease infection are warranted. As shown in Fig. 1, red light irradiation increased resistance against P. putida 229 in soybean sprouts, which is in agreement with the results of earlier studies conducted using rice, Arabidopsis and broad bean [44–46]. The disease resistance induced by light was solely dependent on a specific range of wavelength, and intrusion of other wavelength might diminish the effect, as shown in the disease susceptibility in the soybean sprouts grown under the daylight fluorescence bulbs, which emitted mixed wavelength of red, green and blue area.
Plants develop resistance against pathogens by the induction of plant hormones such as SA, JA and ethylene [47]. The two major pathogen defense signaling pathways can be classified into (i) SA dependent pathway and (ii) SA-independent pathway that involves JA [29]. The signaling pathways triggered by SA and JA molecules are mutually antagonistic for the resistance against pathogens infection [29]. SA is well known for its important roles in pathogen resistance and SAR [48,49]. Mutants defective in SA synthesis showed increased susceptibility to powdery mildew [27]. In our study, the soybean sprouts germinated under red light contained 1.7 times higher SA contents than those germinated in darkness. When soybean sprouts were inoculated with P. putida, those germinated under red light accumulated significantly higher levels of SA (more than 2.0 fold) than those germinated in darkness (Fig. 3). These results suggest that SA-mediated defense responses are involved in the enhanced resistance to P. putida 229 in soybean sprouts germinated under red light. Similarly, cucumber plants treated with red light accumulated significantly higher SA levels only after Sphaerotheca fuliginea inoculation, however, no significant difference was observed when compared to red light irradiated and control plants without inoculation [50]. Previously, non-infected soybean leaves were shown to induce SAR activity via the SA-dependent pathway against P. syringae pv. glucinea through prior infection with Phytophthora sojae [51]. The non-significant expression levels of the ACX, KAT and PR-4 gene but significant expression levels of the ICS and PR-1 gene confirm that increased resistance is induced by the SA-dependent pathway. The involvement of solely the SA-dependent pathway for the induced resistance was confirmed by no difference between the content of JA in soybean sprouts germinated for 5 days in darkness or under red light (Fig. 4).
The SA-mediated defense was characterized by the endogenous synthesis of SA and activation of several PR genes encoding PR proteins. It has been proposed that SA production in plants occurs via the ICS-mediated pathway rather than the PAL-mediated pathway (Wildermuth et al. [38]. The increased levels of SA in soybean sprouts germinated under red light in the present study can be explained by up-regulation of the expression level of the ICS gene. The ICS gene in the soybean sprouts germinated under red light decreased the expression levels after 8 h of incubation, but these levels were recovered after 24 h of incubation in darkness. Moreover, the ICS gene expression level in soybean sprouts germinated under red light were significantly higher than those germinated in darkness after 24 h of incubation (Fig. 2A). The expression levels of the ICS gene in soybean sprouts germinated in darkness gradually decreased during the 24 h incubation period after the infection, which can be explained as a compatible interaction between the host plant and the pathogen that leads to a successful infection [52]. Indeed, the compatible relation as the successful infection was observed as rotting in 5 DAI for the soybean sprouts germinated in darkness (Fig. 1A). The recovered expression of the ICS gene in sprouts germinated under red light may imply the development of incompatibility in the sprouts in response to red light irradiation. In an incompatible interaction, plants gain resistance after pathogen infection by inducing different responses such as oxidative burst and SA accumulation [52,53], and our data clearly indicated that more SA accumulated in soybean sprouts germinated under red light irradiation (Fig. 3A). The expression levels of the PR-1 and PR-4 genes were also measured to determine the mechanism for the increased levels of disease resistance in soybean sprouts germinated under red light. PR-1 gene expression levels were highly up-regulated by P. putida 229 inoculation in soybean sprouts germinated under red light, whereas PR-4 gene expression levels were not changed (Fig. 2). PR-1 is a molecular marker of the SA-dependent SAR pathway [48]; therefore, the high accumulation of the PR-1 gene by P. putida infection suggests that the newly developed resistance in soybean sprouts germinated under red light is due to SAR by higher levels of SA.
Taken together, the expression levels of the ICS and PR-1 gene and high levels of endogenous SA in soybean sprouts germinated under red light indicate that the resistance against the bacterial pathogen occurs via SAR. Further studies using molecular approaches and metabolome analysis could lead to identification of genes and associated biochemical pathways of key regulatory factors involved in the induction of resistance by red light irradiation.
Data Availability
All relevant data are within the paper.
Funding Statement
This research was conducted under a research fund provided by Yeungnam University. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Libault M, Farmer A, Joshi T, Takahashi K, Langley RJ, et al. (2010) An integrated transcriptome atlas of the crop model Glycine max, and its use in comparative analyses in plants. Plant J 63: 86–99. 10.1111/j.1365-313X.2010.04222.x [DOI] [PubMed] [Google Scholar]
- 2. Cannon SB, May GD, Jackson SA (2009) Three sequenced legume genomes and many crop species: rich opportunities for translational genomics. Plant physiol 151: 970–977. 10.1104/pp.109.144659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Xiao CW (2008) Health effects of soy protein and isoflavones in humans. J Nutr 138: 1244S–1249S. [DOI] [PubMed] [Google Scholar]
- 4. Holt S (1997) Soya; The health food of the next millennium. J Kor Soybean Dig 14: 77–90. [Google Scholar]
- 5. Liu K (1997) Nonfermented oriental soyfoods. Soybeans: In: Liu K (ed) Soybeans: chemistry, technology, and utilization. International Thomson Publishing, New York, pp 137–217. [Google Scholar]
- 6. Robertson LJ, Johannessen GS, Gjerde BK, Loncarevic S (2002) Microbiological analysis of seed sprouts in Norway. Int J Food Microbiol 75: 119–126. [DOI] [PubMed] [Google Scholar]
- 7. Doupnik B Jr (1993) Soybean production and disease loss estimates for north central United States from 1989 to 1991. Plant Dis 77:1170–1172. [Google Scholar]
- 8. Kang SG, Park E, Do KS (2009) Identification of a pathogen-Induced glycine max transcription factor GmWRKY1. Plant Pathol J 25: 381–388. [Google Scholar]
- 9. Tu JC (1978) Protection of soybean from severe Phytophthora root rot by Rhizobium. Physiol Plant Pathol 12: 233–240. [Google Scholar]
- 10.Harmon S, Kautter D, Solomon H (1987) Bacillus cereus contamination of seeds and vegetable sprouts grown in a home sprouting kit. J Food Protect. [DOI] [PubMed]
- 11. Taormina PJ, Beuchat LR, Slutsker L (1999) Infections associated with eating seed sprouts: an international concern. Emerging Infect Dis 5: 626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bridge MA, Klarman W (1973) Soybean phytoalexin, hydroxyphaseolin, induced by ultraviolet irradiation. Phytopathol 63: 606–608. [Google Scholar]
- 13. Kodama O, Miyakawa J, Akatsuka T, Kiyosawa S (1992) Sakuranetin, a flavanone phytoalexin from ultraviolet-irradiated rice leaves. Phytochemistry 31: 3807–3809. [Google Scholar]
- 14. Kato H, Kodama O, Akatsuka T (1994) Oryzalexin F, a diterpene phytoalexin from UV-irradiated rice leaves. Phytochemistry 36: 299–301. [Google Scholar]
- 15. Islam S, Babadoost M, Honda Y (2002) Effect of red light treatment of seedlings of pepper, pumpkin, and tomato on the occurrence of Phytophthora damping-off. HortScience 37: 678–681. 12399237 [Google Scholar]
- 16. Islam SZ, Babadoost M, Bekal S, Lambert K (2008) Red Light‐induced Systemic Disease Resistance against Root‐knot Nematode Meloidogyne javanica and Pseudomonas syringae pv. tomato DC 3000. J Phytopathol 156: 708–714. [Google Scholar]
- 17. An C, Mou Z (2011) Salicylic Acid and its Function in Plant Immunity F. J Integ Plant Biol 53: 412–428. 10.1111/j.1744-7909.2011.01043.x [DOI] [PubMed] [Google Scholar]
- 18. Malamy J, Carr JP, Klessig DF, Raskin I (1990) Salicylic acid: a likely endogenous signal in the resistance response of tobacco to viral infection. Science 250: 1002–1004. [DOI] [PubMed] [Google Scholar]
- 19. Métraux J, Signer H, Ryals J, Ward E, Wyss-Benz M, et al. (1990) Increase in salicylic acid at the onset of systemic acquired resistance in cucumber. Science 250: 1004–1006. [DOI] [PubMed] [Google Scholar]
- 20. Jia C, Zhang L, Liu L, Wang J, Li C, et al. (2012) Multiple phytohormone signalling pathways modulate susceptibility of tomato plants to Alternaria alternata f. sp. lycopersici. J Exp Bot 64: 637–650. 10.1093/jxb/ers360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sanchez L, Courteaux B, Hubert J, Kauffmann S, Renault J-H, et al. (2012) Rhamnolipids elicit defense responses and induce disease resistance against biotrophic, hemibiotrophic, and necrotrophic pathogens that require different signaling pathways in Arabidopsis and highlight a central role for salicylic acid. Plant Physiol 160: 1630–1641. 10.1104/pp.112.201913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Leeman M, Van Pelt J, Den Ouden F, Heinsbroek M, Bakker P, et al. (1995) Induction of systemic resistance byPseudomonas fluorescens in radish cultivars differing in susceptibility to fusarium wilt, using a novel bioassay. Eur J Plant Pathol 101: 655–664. [Google Scholar]
- 23. Pieterse C, Van Wees S, Hoffland E, Van Pelt JA, Van Loon LC (1996) Systemic resistance in Arabidopsis induced by biocontrol bacteria is independent of salicylic acid accumulation and pathogenesis-related gene expression. Plant Cell 8: 1225–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ward ER, Uknes SJ, Williams SC, Dincher SS, Wiederhold DL, et al. (1991) Coordinate gene activity in response to agents that induce systemic acquired resistance. Plant Cell 3: 1085–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Durrant W, Dong X (2004) Systemic acquired resistance. Annl Rev Plant Biol 42: 185–209. [DOI] [PubMed] [Google Scholar]
- 26. Khanam NN, Ueno M, Kihara J, Honda Y, Arase S (2005) Suppression of red light-induced resistance in broad beans to Botrytis cinereaby salicylic acid. Physiol Mol Plant Pathol 66: 20–29. [Google Scholar]
- 27. Achuo E, Audenaert K, Meziane H, Höfte M (2004) The salicylic acid‐dependent defence pathway is effective against different pathogens in tomato and tobacco. Plant Pathol 53: 65–72. [PubMed] [Google Scholar]
- 28. Niki T, Mitsuhara I, Seo S, Ohtsubo N, Ohashi Y (1998) Antagonistic effect of salicylic acid and jasmonic acid on the expression of pathogenesis-related (PR) protein genes in wounded mature tobacco leaves. Plant Cell Physiol 39: 500–507. [Google Scholar]
- 29. Kunkel BN, Brooks DM (2002) Cross talk between signaling pathways in pathogen defense. Curr Opin Plant Biol 5: 325–331. [DOI] [PubMed] [Google Scholar]
- 30. Zarate SI, Kempema LA, Walling LL (2007) Silverleaf whitefly induces salicylic acid defenses and suppresses effectual jasmonic acid defenses. Plant Physiol 143: 866–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pena-Cortés H, Albrecht T, Prat S, Weiler EW, Willmitzer L (1993) Aspirin prevents wound-induced gene expression in tomato leaves by blocking jasmonic acid biosynthesis. Planta 191: 123–128. [Google Scholar]
- 32. Sano H, Seo S, Koizumi N, Niki T, Iwamura H, et al. (1996) Regulation by cytokinins of endogenous levels of jasmonic and salicylic acids in mechanically wounded tobacco plants. Plant Cell Physiol 37: 762–769. [Google Scholar]
- 33. Noda S, Fujita M (2009) Light-emitting diodes: Photonic crystal efficiency boost. Nature Phot 3: 129–130. [Google Scholar]
- 34. Marek G, Carver R, Ding Y, Sathyanarayan D, Zhang X, et al. (2010) A high-throughput method for isolation of salicylic acid metabolic mutants. Plant methods 6: 21 10.1186/1746-4811-6-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Müller M, Munné-Bosch S (2011) Rapid and sensitive hormonal profiling of complex plant samples by liquid chromatography coupled to electrospray ionization tandem mass spectrometry. Plant Methods 7: 37 10.1186/1746-4811-7-37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, et al. (2012) Primer3—new capabilities and interfaces. Nucleic Acids Res 40: e115–e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Koressaar T, Remm M (2007) Enhancements and modifications of primer design program Primer3. Bioinformatics 23: 1289–1291. [DOI] [PubMed] [Google Scholar]
- 38. Wildermuth MC, Dewdney J, Wu G, Ausubel FM (2001) Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414: 562–565. [DOI] [PubMed] [Google Scholar]
- 39. Schaller A, Stintzi A (2009) Enzymes in jasmonate biosynthesis–structure, function, regulation. Phytochemistry 70: 1532–1538. 10.1016/j.phytochem.2009.07.032 [DOI] [PubMed] [Google Scholar]
- 40. Rahman M, Honda Y, Arase S (2003) Red‐Light‐Induced Resistance in Broad Bean (Vicia faba L.) to Leaf Spot Disease Caused by Alternaria tenuissima. J Phytopathol 151: 86–91. [Google Scholar]
- 41. Ahn S-Y, Kim S, Baek K-H, Yun H (2013) Inhibiting wildfire and inducing defense-related gene expression by LED treatment on Nicotiana benthamiana . J Plant Pathol 95: 477–483. [Google Scholar]
- 42. Abdel-Monaim MF, Ismail ME, Morsy KM (2011) Induction of systemic resistance of Benzothiadiazole and Humic acid in soybean plants against Fusarium wilt disease. Mycobiology 39: 290–298. 10.5941/MYCO.2011.39.4.290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Vallad GE, Goodman RM (2004) Systemic acquired resistance and induced systemic resistance in conventional agriculture. Crop Sci 44: 1920–1934. [Google Scholar]
- 44. Guo A, Reimers P, Leach J (1993) Effect of light on incompatible interactions between Xanthomonas oryzae pv oryzae and rice. Physiol Mol Plant Pathol 42: 413–425. [Google Scholar]
- 45. Islam S, Honda Y, Arase S (1998) Light‐induced resistance of broad bean against Botrytis cinerea. J Phytopathol 146: 479–485. [Google Scholar]
- 46. Zeier J, Pink B, Mueller MJ, Berger S (2004) Light conditions influence specific defence responses in incompatible plant–pathogen interactions: uncoupling systemic resistance from salicylic acid and PR-1 accumulation. Planta 219: 673–683. [DOI] [PubMed] [Google Scholar]
- 47. Dong X (1998) SA, JA, ethylene, and disease resistance in plants. Curr Opin Plant Biol 1: 316–323. [DOI] [PubMed] [Google Scholar]
- 48. Borsani O, Valpuesta V, Botella MA (2001) Evidence for a role of salicylic acid in the oxidative damage generated by NaCl and osmotic stress in Arabidopsis seedlings. Plant Physiol 126: 1024–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Shirasu K, Nakajima H, Rajasekhar VK, Dixon RA, Lamb C (1997) Salicylic acid potentiates an agonist-dependent gain control that amplifies pathogen signals in the activation of defense mechanisms. Plant Cell 9: 261–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wang H, Jiang YP, Yu HJ, Xia XJ, Shi K, et al. (2010) Light quality affects incidence of powdery mildew, expression of defence-related genes and associated metabolism in cucumber plants. Eur J Plant Pathol 127: 125–135. [Google Scholar]
- 51. Sandhu D, Tasma IM, Frasch R, Bhattacharyya MK (2009) Systemic acquired resistance in soybean is regulated by two proteins, orthologous to Arabidopsis NPR1. BMC Plant Biol 9: 105 10.1186/1471-2229-9-105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Heil M, Bostock RM (2002) Induced systemic resistance (ISR) against pathogens in the context of induced plant defences. Ann Bot 89: 503–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Carviel JL, Wilson DC, Isaacs M, Carella P, Catana V, et al. (2014) Investigation of Intercellular Salicylic Acid Accumulation during Compatible and Incompatible Arabidopsis-Pseudomonas syringae Interactions Using a Fast Neutron-Generated Mutant Allele of EDS5 Identified by Genetic Mapping and Whole-Genome Sequencing. PloS one 9: e88608 10.1371/journal.pone.0088608 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.