Abstract
Fluorogen-activating-proteins (FAPs) are a novel platform of fluorescence biosensors utilized for protein discovery. The technology currently demands molecular manipulation methods that limit its application and adaptability. Here, we highlight an alternative approach based on universal affinity reagents for protein detection. The affinity reagents were engineered as bi-partite fusion proteins, where the specificity moiety is derived from IgG-binding proteins –Protein-A or Protein-G – and the signaling element is a FAP. In this manner, primary antibodies provide the antigenic selectivity against a desired protein in biological samples, while FAP affinity reagents target the constant region (Fc) of antibodies and provide the biosensor component of detection. Fluorescence results using various techniques indicate minimal background and high target specificity for exogenous and endogenous proteins in mammalian cells. Additionally, FAP-based affinity reagents provide enhanced properties of detection previously absent using conventional affinity systems. Distinct features explored in this report include: (1) unfixed signal wavelengths (excitation and emission) determined by the particular fluorogen chosen, (2) real-time user controlled fluorescence on-set and off-set, (3) signal wavelength substitution while performing live analysis, and (4) enhanced resistance to photobleaching.
Keywords: FAP, Fluorogen, Affinity Reagents, Biosensors
Introduction
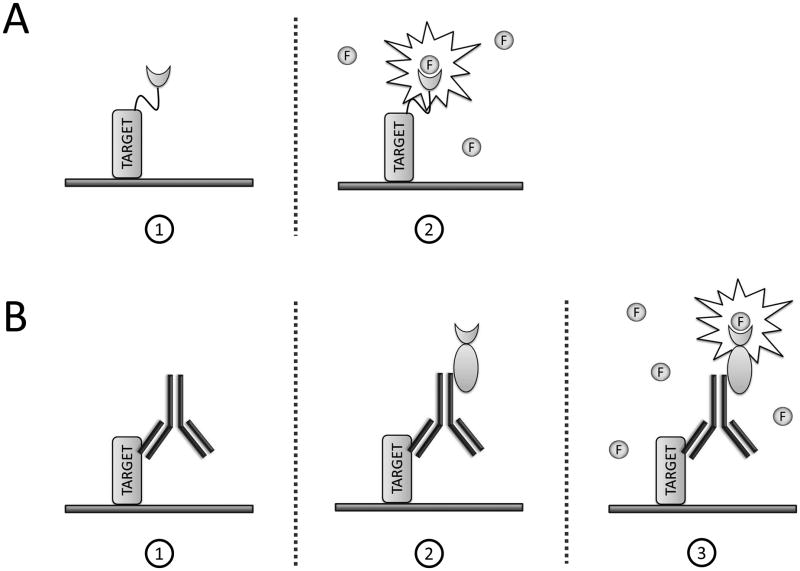
Fluorogen-activating proteins (FAPs) are polypeptides that bind small organic molecules (fluorogens) that are non-fluorescent in solution, but highly fluorescent when bound by the FAP (Szent-Gyorgyi et al., 2008). Single chain antibodies (scFv's) with FAP activity were recently described and successfully employed in drug discovery (Holleran et al., 2012; Wu et al., 2012), as well as, in studies of cellular phenomena, including receptor dynamics (Fisher et al., 2010; Holleran et al., 2010; Saunders et al., 2012; Wu et al., 2013), pH gradient-flux for vesicular traffic monitoring (Grover et al., 2012), and synapse formation (Shruti et al., 2012). In all cases, the FAP was expressed from a recombinant gene that encoded a protein fusion between the FAP and the protein of interest (Fig. 1A). This approach results in two significant setbacks: 1) time and labor regarding quality control and generation of each recombinant protein, and 2) artificial protein expression from a non-native promoter, typically altering protein regulation and abundance in the cell.
Figure 1.
Methods for protein discovery utilizing FAP-technology. A: Current recombinant protein approach: (1) Target protein is genetically fused to FAP, and (2) the fluorogen presented in the medium binds its cognate FAP, resulting in fluorescence signal. B: Protein labeling using universal affinity FAP reagents: (1) Antibody binds target with high specificity, then (2) the FAP affinity reagent binds the constant region (Fc) of the antibody, and (3) the fluorogen presented in the medium binds its cognate FAP, resulting in fluorescence signal.
To address these limitations we developed FAP-based affinity reagents, which offer capabilities of immediate protein tagging and fluorescence labeling, as well as, long-term storage and usage. Instead of fusing FAPs with full-length antibodies (multimeric proteins), Fabs, scFvs, or affibodies, where each target protein would require a unique FAP reagent, we derived a universal method: a single FAP-reagent able to target a multiplicity of different proteins. The mechanism utilizes the diverse pool of readily available commercial antibodies to provide antigenic specificity against the target protein – recombinant or native. Next, a secondary reagent, consisting of a FAP fused to an immunoglobulin-binding domain (derived from Staphylococcus aureus ProteinA or Streptococcus ProteinG), binds the Fc-region of antibodies. The complete set of components – analyte, primary antibody, secondary reagent, and fluorogen – produce the detection complex shown in Figure 1B.
In this manuscript we present a novel FAP labeling system where fluorogen-activating-proteins are fused to immunoglobulin-binding domains for immunodetection. As a result, when tested against cell-surface or intra-cellular antigens the affinity reagents demonstrate high target specificity and minimal signal background. In addition, FAP-based reagents deliver fluorescence manipulation features previously absent with conventional affinity systems.
Materials and Methods
Plasmid Construction
Protein expression plasmid pKM260 was modified at NheI and EcoRV sites via insertion of annealed overlapping oligos that resulted in a two-module expression system. After the hexa-histidine tag, the first module is spanned by two unique SfiI restriction sites and is linked by ten amino acids to the second module, which contains two unique DraIII restriction sites. All FAPs used in this study were previously described (Ozhalici-Unal et al. 2008; Szent-Gyorgyi et al. 2008; Szent-Gyorgyi et al. 2010). The ZZ-peptide was a gift from Dr. Georgiou, University of Texas, and the C1-C3 domains of Protein G (PrG) were commercially synthesized (Genscript). The nucleotide overhangs of each module insert were generated as previous (Holleran et al., 2010). The pDisplaySacLac2 plasmid was used for all transient transfections (Holleran et al., 2010) with FAPs ligated at SfiI restriction sites.
Optical Spectroscopy
Analyses were performed using a Safire2 plate reader (TECAN) in transparent, flat-bottom, 96-well microtiter plates. The excitation/emission wavelengths were 514/555nm for TO1-2p fluorogen, 610/655nm for DIR fluorogen, and 635/665nm for MG-2p fluorogen. For in-vitro assays, measurements were performed with 500nM protein and 1uM fluorogen in phosphate-buffer-saline (PBS). Live cell assays were performed with 106 cells per well in PBS in presence of fluorogen. All samples were measured in triplicates.
Cell labeling using FAP-reagents
For live suspension cells, incubation consisted of primary antibody in PBS plus 0.5% calf serum for 30 minutes on ice. After wash, cells were incubated in PBS plus 0.5% calf serum and 500 nM FAP-reagent for 30 minutes on ice. For live adherent cells, the same protocol was performed as above, with conditions at room temperature instead of ice. In addition, a 1-step labeling protocol was developed with similar efficacy as the 2-step labeling protocol mentioned here (Fig. S5).
For intracellular labeling, cells were fixed with 4% paraformaldehyde (Sigma-Aldrich) in PBS for 15 minutes (in dark, at room temperature), then washed and simultaneously detergent permeabilized and blocked using PBS plus 0.25% saponin (Sigma-Aldrich) and 3% calf serum for 30 minutes at room temperature. Cells were labeled with primary antibody in PBS plus 0.5% calf serum, 0.25% saponin for 1 hour on ice. After wash, the cells were labeled with FAP-reagent in PBS plus 0.5% calf serum, 0.25% saponin, for 1 hour on ice. Concentrations of primary antibody and FAP-reagent were standardized for each intracellular staining.
The following commercial primary antibodies were used in this report: anti-CXCR4 (A00995, Genscript), anti-HA (ChIP-grade, ab9110, Abcam), anti-EEA1 (A01514, GenScript), anti-lamin(A/C) (A01455, GenScript), and anti-tubulin-β (ab15568, Abcam)
Confocal Microscopy
Cells were plated on 35mm glass-bottom dishes (MatTek) and imaged in PBS plus fluorogen. Micrographs were obtained using a Carl Zeiss LSM 510 Meta/UV DuoScan inverted spectral confocal microscope using 40× or 60× objectives. Settings for TO1-2p fluorogen include a 488 nm excitation laser and a 505-550 nm emission filter, for DIR fluorogen a 561nm excitation laser and 575 nm LP emission filter, and for MG-2p fluorogen a 633 nm excitation laser and 650 nm LP emission filter. The illumination intensities and gain settings were held constant across samples; image analysis was performed with minimal processing using ImageJ software (NIH).
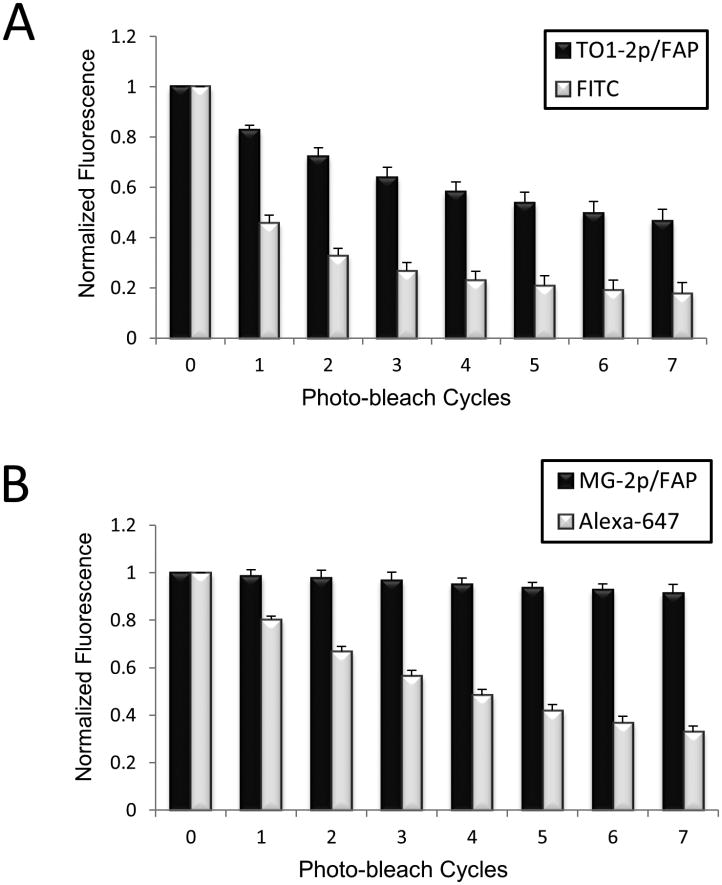
Cell surface photobleaching
Each photo-bleach cycle equals 20 iterations of 100% laser power on six Z-planes per image. A total of 7 sequential cycles were performed, interrupted by 30 seconds of image acquisition. Settings were maintained constant across all comparison samples. A 488nm laser was used for TO-FAP and FITC (ab6798, Abcam), and a 633nm laser for MG-FAP and Alexa647 (p21462, Invitrogen). Maximum pixel intensities were determined by collapsing the Z-planes onto one image after each photobleach cycle. Mean values of all images per cycle were normalized to initial (unbleached) maximum pixel intensity values.
Flow-cytometry
Data were collected using FACS Vantage SE Flow Cytometer with FACS Diva option (Becton Dickinson). Settings for TO1-2p fluorogen include an argon 488-nm excitation laser with a 530/30 nm emission filter, and for MG-2p fluorogen a HeNe 623-nm excitation laser with a 685/35 nm emission filter. Measurements were performed in PBS plus fluorogen, with total acquired events greater than 104 per sample. Gating strategies for all experiments are described in Supplemental Figure 2. Data analysis was carried out using FACSDiva Software v5.0.2 (Becton Dickinson).
Statistics
A student's t-test was used to quantify statistical significance. In figures the error bars denote standard deviation of the mean, and single stars represent significant differences (p < 0.05).
Supplemental Material
Additional methods and supplemental figures are provided online.
Results
Purified FAP-ZZ affinity reagents are determined functional in-vitro
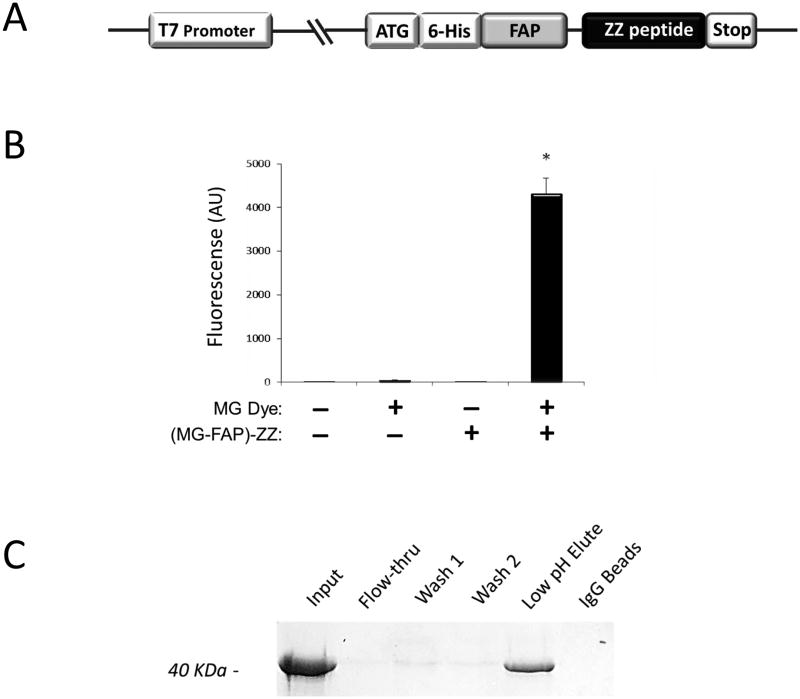
To obtain FAP affinity reagents that bind to immunoglobulins, we generated a set of bi-partite fusion proteins: At the N-termini several different FAPs (Table S1) were fused via a small linker to the ZZ-peptide – a synthetic analog derived from Staphylococcus aureus Protein-A – at the C-termini (Fig. 2A). The advantages of choosing the ZZ-peptide as a fusion protein include its small size, simple folding structure, high aqueous solubility, and the strong affinity for the Fc-region of IgGs – a characteristic of Protein-A itself (Boström et al., 2005; Braisted and Wells, 1996; Jendeberg et al., 1995). Consequently, the FAP-ZZ reagents were expressed in E.coli and purified via affinity chromatography utilizing the N-terminal 6-histidine tag (Fig 2A).
Figure 2.
A purified FAP-ZZ affinity reagent is determined functional in-vitro. A: Schematic of fusion protein in expression plasmid: N-terminal hexa-histidine (6-His) tag is followed by a FAP domain connected via a small linker to the ZZ peptide. B: Fluorimetry data using 500 nM (MG-FAP)-ZZ protein and 1 uMMG-2p fluorogen in PBS. C: SDS–PAGE gel image of IgG-chromatography with purified (MG-FAP)-ZZ reagent
In order to determine FAP activity, the FAP-ZZ fusion proteins were assayed by optical spectroscopy in the presence or absence of cognate fluorogen. Detection of strong red-fluorescence occurred only when (MG-FAP)-ZZ and cognate fluorogen were together present; while the no-fluorogen or no-protein controls lacked signal (Fig. 2B). Equivalent results were obtained for the (DIR-FAP)-ZZ and (TO-FAP)-ZZ fusions (Fig. S3).
The FAP-ZZ fusion proteins were also assayed by IgG affinity chromatography to determine their ZZ-peptide activity. Figure 2C shows results for the (MG-FAP)-ZZ protein; the protein binds to IgG at neutral pH and is eluted at low pH, the expected behavior for the ZZ-peptide. Similar results were obtained for the (DIR-FAP)-ZZ and (TO-FAP)-ZZ fusions (Fig. S4). Thus, the fusion proteins retained activity of their constituent domains (FAP or ZZ-peptide) in-vitro.
FAP-ZZ affinity reagents target surface proteins on live cells with specificity and low background
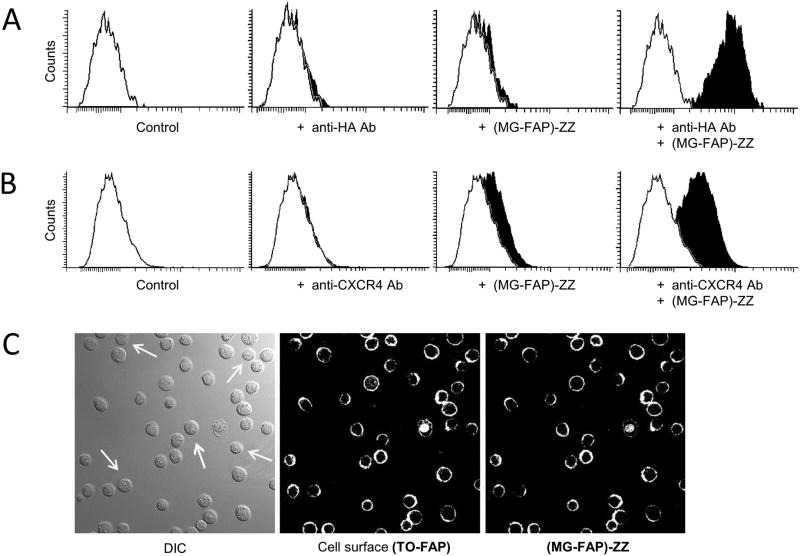
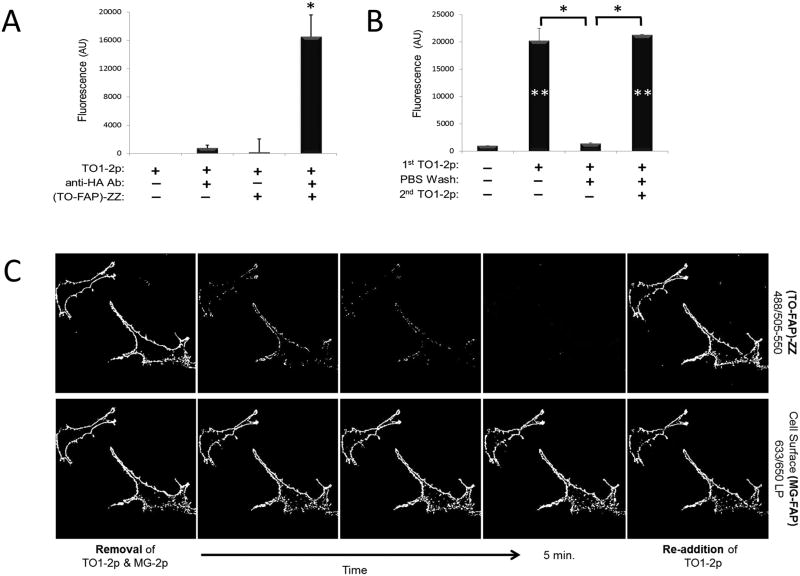
Next, we asked whether FAP affinity reagents would target cell-surface proteins on live cells. Here the antigen was the hemagglutinin (HA) epitope present at the N-terminus of a cell surface TO-FAP (HL1.0.1-TO1) – anchored to the plasma membrane by a membrane-spanning domain (Fisher et al., 2010). The cells were sequentially labeled using anti-HA antibody and (MG-FAP)-ZZ reagent. Only cells incubated with antibody and (MG-FAP)-ZZ affinity reagent (in presence of MG-fluorogen) exhibited strong fluorescence, whereas control cells that lacked either or both reagents showed absence of signal (Fig. 3A). Confocal microscopy observations of the same cells (Fig. 3C) stayed consistent with the flow-cytometry results. In this case, the cells also received TO-fluorogen to visualize the endogenous TO-FAP (center image), and untagged cells in the population (white arrows) served as negative controls. The micrographs demonstrate strong co-labeling, while untagged cells lacked fluorescence detection for both channels.
Figure 3.
FAP-ZZ affinity reagent targets surface proteins on living cells. A: Flow-cytometry histograms from live HA-tagged U937 cells labeled with anti-HA antibody and (MGFAP)-ZZ affinity reagent at the cell surface. B: Flow-cytometry histograms from live HEK293 cells labeled with anti-CXCR4 antibody and (MGFAP)-ZZ affinity reagent at the cell surface. Each histogram represents a minimum of 10K live cell events in the presence of 100nM MG-2p fluorogen. C: Micrographs of live U937 cells HA-tagged to TO-FAP (HL1.0.1-TO1) at the surface, and then labeled with anti-HA antibody and (MG-FAP)-ZZ affinity reagent. Images were acquired using a 40_ objective in the presence of both, 100nM MG-2p and TO1-2p, fluorogens. White arrows indicate cells without HA-tagged TO-FAP (HL1.0.1-TO1).
Similar results were obtained when we addressed a native cell surface protein. Here the target protein was the chemokine receptor CXCR4, which is known to be expressed on the surface of HEK 293 cells (Tanner et al., 2007). Cells were incubated with anti-CXCR4 antibody against the extracellular N-terminus of CXCR4, followed by incubation with (MG-FAP)-ZZ affinity reagent. Flow cytometry results indicated fluorescence only for cells incubated with antibody and (MG-FAP)-ZZ affinity reagent (in presence of MG-fluorogen), while control cells that lacked either or both reagents showed absence signal (Fig. 3B). In this case, the detection of signal per cell for native CXCR4 was lower than the HA-epitope tagged cells previously described; however, this is expected because recombinant tagged proteins generally utilize active promoters. Taken together, the experiments demonstrate FAP-ZZ affinity reagents as suitable for detection of proteins at the surface of living cells.
FAP-ZZ affinity reagents target surface and intracellular proteins on fixed cells with specificity and low background
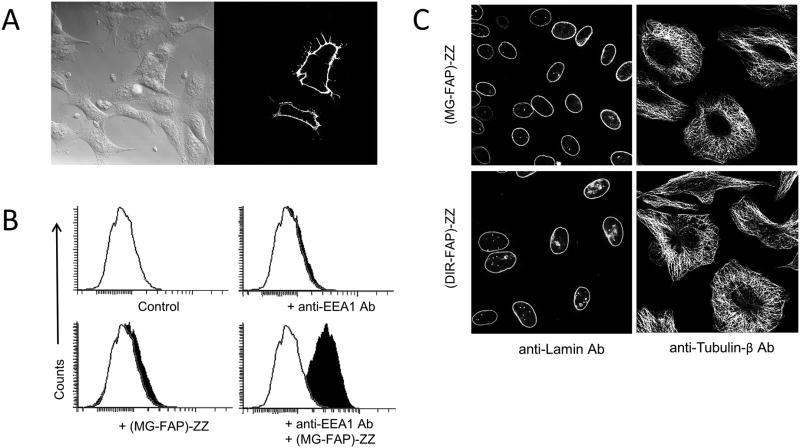
Next, we asked whether FAP-ZZ affinity reagents would detect proteins from paraformaldehyde-fixed cells. HEK 293 cells were transiently transfected with an expression vector encoding an HA-tagged surface protein, and then labeled with anti-HA antibody and (MG-FAP)-ZZ reagent. Confocal microscopy results detected fluorescence only for the transfected cells (Fig. 4A), while non-transfected cells in the population remained dark, indicating reagent specificity at the surface of fixed cells.
Figure 4.
FAP-ZZ affinity reagents target surface and intracellular proteins on fixed cells. A: Micrograph of fixed (non-permeabilized) HEK293 cells transiently expressing HAepitope at the surface. Cells were labeled with anti-HA antibody and (MG-FAP)-ZZ affinity reagent; then imaged with a 40× objective in presence of 50 nM MG-2p fluorogen. B: Flowcytometry histograms of fixed and permeabilized U937 cells labeled using anti-EEA1 (early-endosome-antigen-1) antibody and (MG-FAP)-ZZ affinity reagent. Each histogram represents a minimum of 10K live cell events in the presence of 25 nM MG-ester fluorogen. C:Micrographs of fixed and permeabilized HEK293 cells labeled with anti-lamin or anti-1-tubulin primary antibodies, and then by (MG-FAP)-ZZ or (DIR-FAP) ZZ secondary reagents. Images were acquired with a 60_ objective in the presence of 25 nM MG-ester or DIR fluorogens.
We then proceeded to test intracellular protein labeling. Figure 4B shows flow-cytometry histograms for fixed and permeabilized cells incubated with anti-EEA1 (an endosomal protein) antibody, followed by incubation with (MG-FAP)-ZZ reagent. As previously observed using live cells, signal was only detected for the complete combination of reagents.
Commercial antibodies in combination with FAP-ZZ reagents were used to visualize intracellular proteins by confocal microscopy using fixed and permeabilized cells. Here the proteins laminA/C and β-tubulin were addressed, each with two different FAP-ZZ reagents, (MG-FAP)-ZZ and (DIR-FAP)-ZZ (Fig. 4C). In each case, visual inspection of target structures (nuclear envelope and microtubule cytoskeleton) demonstrated target specificity and absence of background. This suggests that the assayed FAP affinity reagents and cognate fluorogens are suitable for intracellular studies. We also note that the two fluorogens, MG-ester and DIR, have rather different excitation and emission spectra (Fig. S1), and may facilitate multi-labeling studies. In summary, the experimental results demonstrate FAP-ZZ reagents as suitable biosensors for intracellular studies due to their low background and high specificity.
FAP-ZZ affinity reagents display enhanced resistance to photobleaching when compared to conventional fluorescent secondary reagents
In one set of experiments, live cells were labeled with a primary antibody, and then labeled with either (TO-FAP)-ZZ reagent or a commercial FITC-labeled secondary (both secondary reagents have comparable excitation/emission). After several cycles of photobleaching, the results indicated a two-fold photostability advantage for the (TO-FAP)-ZZ reagent when compared to the FITC-reagent (Fig. 5A). We also compared red-channel reagents where live cells were labeled with a primary antibody, and then labeled with either (MG-FAP)-ZZ reagent or a commercial Alexa-647 secondary (both secondary reagents have comparable excitation/emission). Here the FAP reagent demonstrated superior resistance to photobleaching. In fact, the (MG-FAP)-ZZ reagent displayed negligible loss in fluorescence – over seven photobleach cycles – during which the Alexa fluorophore lost about three-fourths of its signal (Fig. 5B). The results indicate that FAP affinity reagents exhibit improved photostability when compared to conventional fluorophore-based systems.
Figure 5.
FAP-ZZ affinity reagents exhibit enhanced photostability. A: Live HAtagged HEK293cells were labeled with anti-HA primary antibody, and then with either (TO-FAP)-ZZ reagent or FITC-conjugated secondary at the cell surface. n=13 samples for TO-FAP labeled cells in the presence of 500nM TO1-2p, and n=10 samples for FITC labeled cells. B: Live HA-tagged HEK293 cells were labeled with anti-HA primary antibody, and then with either (MG-FAP)-ZZ affinity reagent or Alexa647-conjugated secondary at the cell surface. n=10 samples for MG-FAP labeled cells in the presence of 100nM MG-2p, and n=15 samples for Alexa647 labeled cells.
FAP-ZZ affinity reagents permit manipulation of fluorescence signal
As mentioned, FAP-fluorogen complexes are noncovalent. For some FAP-fluorogen pairs, including (MG-FAP)-ZZ reagent, exchange of bound fluorogen with fluorogen in the medium occurs slowly, whereas for other pairs, including (TO-FAP)-ZZ reagent, exchange is rapid. In the latter case, one would expect that washing labeled cells into fluorogen-free medium would lead to rapid loss of signal. Live cells fluorimetry data revealed fluorescence signal loss upon replacement with fluorogen-free medium, and gain of signal when cells were returned to fluorogen-containing medium (Fig. 6B).
Figure 6.
Fluorescence signal manipulation from a FAP-ZZ affinity reagent. A: Fluorescence measurement graph of live HA-tagged U937 cells labeled with anti-HA antibody and (TO-FAP)-ZZ reagent in presence of 500 nM TO1-2p fluorogen. B: Graph of fluorescence measurement after addition, removal and re-addition of fluorogen in solution. Cells were labeled as in (A); then placed in fluorogen-free medium, and lastly, placed in medium containing 500 nM TO1-2p fluorogen (__corrected for background using unlabeled cells with fluorogen). C: Time-lapse micrographs of live HEK 293 cells HA-tagged to MG-FAP at the surface, and then labeled with anti-HA antibody and (TO-FAP)-ZZ affinity reagent. Cells were initially placed in medium containing both, 100 nM MG-2p and 500nM TO1-2p fluorogens; then placed in fluorogen-free medium, and lastly, placed in medium containing only 500 nM TO1-2p fluorogen.
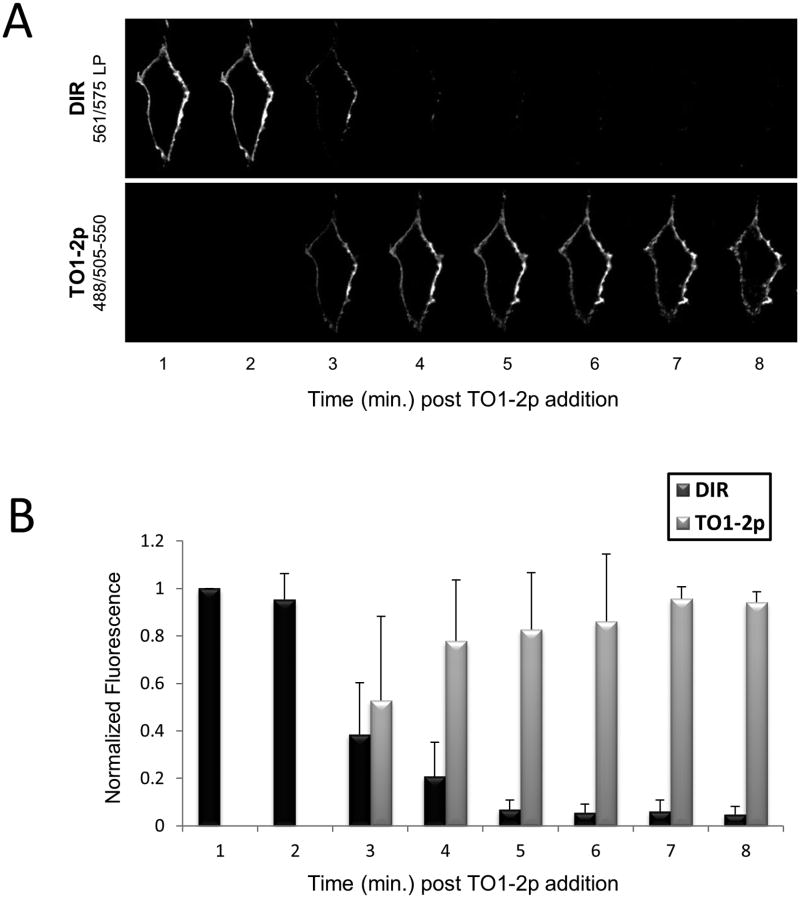
Similarly, live HEK 293 cells were labeled at the cell surface using (TO-FAP)-ZZ reagent against an HA-epitope fused to a cell endogenous membrane-anchored MG-FAP. In this case, the cells were imaged in the presence of two fluorogens; MG-fluorogen was included in the medium to interact with the cell surface MG-FAP, while TO-fluorogen was added to interact with (TO-FAP)-ZZ reagent targeting the HA-epitope. Cells were imaged; then washed into fluorogen-free medium (PBS) and further imaged at one minute intervals. After five minutes, fresh TO-fluorogen was re-added, and the cells were imaged yet again. During the five minutes in which both fluorogens were absent from the medium, signal from the TO-FAP diminished to undetectable levels, and when TO-fluorogen was re-introduced, signal rapidly returned (Fig. 6C). On the other hand, MG-fluorescence persisted throughout the experiment, and remained unperturbed, even at two-hours later time point (data not shown). The result stayed consistent with the very slow dissociation kinetics for MG-fluorogen with MG-FAP (Table S1). Taken together, our observations reveal a method for user-controlled reversible fluorescence.
A single FAP-ZZ reagent provides multi-spectral detection and dynamic color switching
Certain FAPs are described as “promiscuous” because they possess the ability to form fluorescent complexes with fluorogens of more than one color (Ozhalici-Unal et al., 2008). This is true for the FAP in (TO-FAP)-ZZ reagent, which can activate a green fluorogen (TO1-2p) or a red fluorogen (DIR). Since this FAP also displays rapid fluorogen exchange with the medium, as illustrated in Figure 6, we used the (TO-FAP)-ZZ reagent to demonstrate color switching via replacement of one fluorogen with another on the same cell population.
Live cells were labeled with primary antibody and (TO-FAP)-ZZ reagent, and then placed in the presence of DIR fluorogen. A molar excess of TO-fluorogen was then added to the medium and cells were immediately imaged using DIR and TO channels at every one minute interval. As shown in figure 7A, the red DIR signal (upper panels) rapidly disappeared and was replaced by the green TO signal (lower panels). Data quantification reveals a fast equilibrium (approx. 7 minutes), where DIR signal lowers to levels below detection while TO signal rapidly increases and then stabilizes (Fig. 7B). Taken together, a single FAP-ZZ reagent offers multi-spectral fluorescence with the ability for real-time color switching.
Figure 7.
Multi-spectral detection and dynamic color switching from a FAP-ZZ reagent. A: Micrographs of live HA-tagged HEK 293 cells labeled with primary antibody and (TO-FAP)-ZZ reagent at the surface. Cells were initially placed in 200 nM DIR fluorogen medium; after addition of 1 uM TO1-2p fluorogen, time series image acquisition followed. B: Graph of normalized mean fluorescence intensities from captured images at every 1 min interval (n=5 images).
A FAP fusion reagent based on Protein-G labels cell surface epitopes
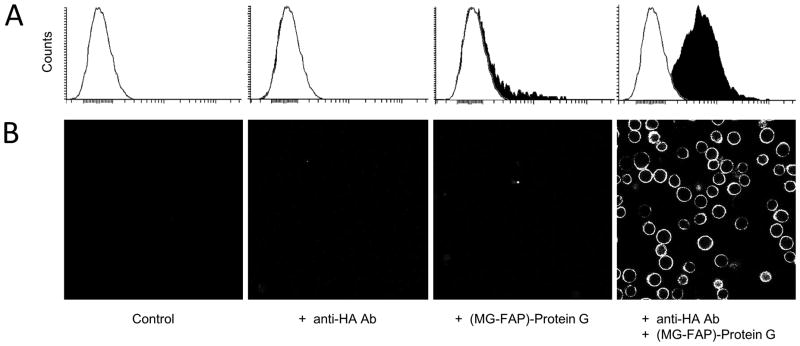
In addition to FAP-ZZ fusion proteins, we asked whether other classes of IgG-binding elements, besides the ZZ-peptide, would generate functional affinity reagents when genetically fused to FAPs. We engineered and purified a FAP-fusion protein using the C1-C3 domains of Streptococcus Protein-G (Guss et al., 1986; Sjobringso et al., 1991). Similar to FAP-ZZ reagents, the FAP/Protein-G fusion protein – (MG-FAP)-PrG – retained both, the IgG-binding and fluorogen-activating properties of its constituent parts in-vitro (Fig. S3 and S4). Live cell analysis against a surface epitope using flow-cytometry and confocal imaging informed of high target specificity and minimal background (Fig. 8). However, further inspection of the FAP-PrG reagent presented evidence of nonspecific (i.e., antibody-independent) binding to live cells when used at excess concentrations, a property absent using FAP-ZZ reagents (Fig. S7). This observation informs of the requirement for FAP-PrG reagent optimization (via titration methods) prior to protein labeling studies. In summary, FAP-reagents are not limited to the ZZ-peptide, but may form functional protein fusions with diverse classes of IgG-binding elements.
Figure 8.
A FAP/Protein G fusion reagent targets a surface marker on live cells. A: Flow-cytometry histograms of live U937 cells HA-tagged at the surface, and labeled with anti-HA antibody and (MG-FAP)-PrG affinity reagent. Each of the panels represents a minimum of 10K live cell events. B: Micrographs of same cells as (A) acquired with a 40_ objective. All data were measured in presence of 100 nM MG-2p fluorogen.
Discussion
Fluorogen activating proteins (FAPs) provide a number of special features as reporters of protein location and abundance in living cells (Szent-Gyorgyi et al., 2008). To date, however, use of FAP technology demands expression of the FAP as a fusion partner with the protein of interest, which entails construction of an appropriate recombinant gene and introduction of the gene into cells prior to analysis – a process that is time and labor intensive.
The work reported here was motivated by a desire to make FAP detection possible for almost any protein. Accordingly, a set of soluble reagents were engineered: FAP fusions to the ZZ-peptide, derived from S. aureus ProteinA, or C1-C3 domains of Streptococcus ProteinG, both of which provide specific binding to the constant region (Fc) of numerous IgG-subclasses (Boström et al., 2005; Tashiro and Montelione, 1995). Since IgG's are available against tens of thousands of different proteins, the reagents, in principle, expand the reach of FAP biosensor technology to any of their targets.
When compared to conventional affinity systems, FAP affinity reagents offer several advantages regarding fluorescence detection. One benefit is enhanced photostability (Fig. 5). The FAP-fluorogen complex forms via non-covalent interactions, and permits equilibrium exchange of the fluorogen between the FAP and its surrounding medium. In this manner, oxidative cleavage of the fluorogen (as a FAP complex) forces a thermodynamically favorable exchange for an unbleached fluorogen species, and prevents loss of fluorescence signal (Shank et al. 2009; Silva et al. 2007). Additional protection against photo-damage is provided by the FAP-fluorogen complex itself. Protein engulfment of the fluorogen forms a protective shell against oxidative damage when compared to free forms of fluorogen (data unpublished). Additionally, improved photostability may be attained via molecular alterations of the fluorogens. As previously shown, fluorogen chemical substitutions using electron withdrawing groups, such as cyano and fluoro, demonstrate improved stability against photo-induced damage, and also prolonged fluorescence detection as FAP complexes (Shank et al., 2009). Taken together, FAP technology as affinity reagents provides enhanced photostability, which is a favorable attribute for multiplicity of assays requiring longevity of signal.
A secondary advantage of FAP affinity reagents involves modular signal detection. In this report we illustrate a user controlled on-set and off-set of fluorescence via manipulation of fluorogen concentration in the cellular medium (Fig. 6). In this manner, fluorescence is reversible in either direction. On the other hand, such property is currently unmanageable with conventional fluorophore-based methods, since fluorescence is permanent upon labeling. Even harsh approaches that remove fluorescence at the cellular surface, such as, protease-assisted protein cleavage, lead only to a one directional removal of signal – and may jeopardy cell viability. Hence, modular fluorescence using FAP-reagents may benefit applications requiring protein turnover at the cellular membrane, protein trafficking studies, and post cell-labeling assays that demand removal of signal.
A third advantage of the FAP-based affinity reagents is multi-spectral fluorescence from a single reagent. The property of FAP promiscuity, affinity for more than one fluorogen partner, offers alternate excitation and emission wavelengths with broad spectral ranges (Ozhalici-Unal et al., 2008). This report demonstrates a dynamic fluorogen exchange assay using a promiscuous FAP affinity reagent and two different fluorogens, where rapid fluorescence switching occurs from red to green channels of detection (Fig. 7). On the other hand, conventional systems provide fixed and immutable fluorescence – each labeling reagent is defined by a single detection wavelength. Therefore, instead of “one reagent, one color”, a single FAP affinity reporter provides an expanded and versatile palette of colors. Such property may prove advantageous for two-color pulse-chase studies, dynamic in-vivo assays, as well as, flexibility for cellular co-labeling.
It should be noted that FAP technology was previously determined as bright as fluorescent proteins (Szent-Gyorgyi et al., 2008); as a result, fluorophore based systems prove comparably brighter (Fig. S8). In addition, FAP crystallography analyses indicate only one fluorogen binds the protein scaffold (Senutovitch et al., 2013; Szent-Gyorgyi et al., 2013), while fluorophore labeled antibodies contain several fluorophores per reagent – further accentuating their fluorescence brightness. In this regard, FAP affinity reagents may prove disadvantageous for detection of low abundance antigens when compared to conventional fluorophore systems. A current approach to improve FAP signal intensity utilizes fluorogen modifications. Examples, such as dendron-based fluorogens, employ intra-molecular energy transfer from donors to acceptors and significantly increase fluorescence emission to six-fold levels than the fluorogens presented in this report (Szent-Gyorgyi et al. 2010). A second approach for improved fluorescence may involve directed evolution of the FAP protein scaffold. As previously demonstrated, methods of random mutagenesis coupled with fluorescence selection methods generated FAP isolates with improved brightness in the cytoplasm of cells, as well as, stronger FAP-fluorogen binding at the surface of yeast cells (Szent-Gyorgyi et al. 2008; Yates et al. 2012). Consequently, directed evolution may prove advantageous for FAP reagents with reduced signal. In summary, FAP technology offers a malleable bi-partite approach for improving or altering the desired properties of detection.
Conclusion
This report unveils a novel class of universal affinity reagents based on FAP technology. When used for protein discovery, the affinity reagents exhibit high selectivity, and minimal background. Additionally, FAP based affinity reagents demonstrate improved properties of fluorescence detection. Accordingly, we unveil an innovative biosensor platform for protein discovery that advances the current field of affinity reporters.
Supplementary Material
Acknowledgments
We thank Yehuda Creeger and Haibing Teng for instrument training and assistance (flow-cytometry and confocal microscopy respectively). We also thank MBIC chemists for synthesizing the fluorogens used in this report. Funding for this work came from NIH National Center for Research Resources Grant U54RR022241.
Abbreviations
- FAP
fluorogen activating protein
- MG
malachite green
- TO
thiazole orange
- DIR
dimethyl indole red
Footnotes
The authors have declared no conflict of interest.
Works Cited
- Boström T, Nilvebrant J, Hober S. Purification Systems Based on Bacterial Surface Proteins. Proteins 2005 [Google Scholar]
- Braisted AC, Wells JA. Minimizing binding protein. 1996;93:5688–5692. doi: 10.1073/pnas.93.12.5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher GW, Adler Sa, Fuhrman MH, Waggoner AS, Bruchez MP, Jarvik JW. Detection and quantification of beta2AR internalization in living cells using FAP-based biosensor technology. Journal of biomolecular screening. 2010;15:703–9. doi: 10.1177/1087057110370892. http://www.ncbi.nlm.nih.gov/pubmed/20488980. [DOI] [PubMed] [Google Scholar]
- Grover A, Schmidt BF, Salter RD, Watkins SC, Waggoner AS, Bruchez MP. Genetically encoded pH sensor for tracking surface proteins through endocytosis. Angewandte Chemie (International ed in English) 2012;51:4838–42. doi: 10.1002/anie.201108107. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3538816&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guss B, Eliasson M, Olsson a, Uhlén M, Frej aK, Jörnvall H, Flock JI, Lindberg M. Structure of the IgG-binding regions of streptococcal protein G. The EMBO journal. 1986;5:1567–75. doi: 10.1002/j.1460-2075.1986.tb04398.x. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1166981&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holleran J, Brown D, Fuhrman MH, Adler Sa, Fisher GW, Jarvik JW. Fluorogen-activating proteins as biosensors of cell-surface proteins in living cells. Cytometry Part A : the journal of the International Society for Analytical Cytology. 2010;77:776–82. doi: 10.1002/cyto.a.20925. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2945705&tool=&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holleran JP, Glover ML, Peters KW, Bertrand Ca, Watkins SC, Jarvik JW, Frizzell Ra. Molecular medicine. Vol. 18. Cambridge, Mass.: 2012. Pharmacological rescue of the mutant cystic fibrosis transmembrane conductance regulator (CFTR) detected by use of a novel fluorescence platform; pp. 685–96. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3388127&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jendeberg L, Persson B, Andersson R, Karlsson R, Uhlén M, Nilsson B. Kinetic analysis of the interaction between protein A domain variants and human Fc using plasmon resonance detection. Journal of molecular recognition : JMR. 1995;8:270–8. doi: 10.1002/jmr.300080405. http://www.ncbi.nlm.nih.gov/pubmed/8588944. [DOI] [PubMed] [Google Scholar]
- Ozhalici-Unal H, Pow CL, Marks Sa, Jesper LD, Silva GL, Shank NI, Jones EW, Burnette JM, Berget PB, Armitage Ba. A rainbow of fluoromodules: a promiscuous scFv protein binds to and activates a diverse set of fluorogenic cyanine dyes. Journal of the American Chemical Society. 2008;130:12620–1. doi: 10.1021/ja805042p. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2633110&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders MJ, Szent-Gyorgyi C, Fisher GW, Jarvik JW, Bruchez MP, Waggoner AS. Methods. Vol. 57. San Diego, Calif.: 2012. Fluorogen activating proteins in flow cytometry for the study of surface molecules and receptors; pp. 308–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senutovitch N, Stanfield RL, Bhattacharyya S, Rule GS, Wilson A, Armitage BA, Waggoner AS, Berget PB. NIH Public Access. 2013;51 doi: 10.1021/bi201422g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shank NI, Zanotti KJ, Lanni F, Berget PB, Armitage Ba. Enhanced photostability of genetically encodable fluoromodules based on fluorogenic cyanine dyes and a promiscuous protein partner. Journal of the American Chemical Society. 2009;131:12960–9. doi: 10.1021/ja9016864. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2744899&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shruti S, Urban-Ciecko J, Fitzpatrick Ja, Brenner R, Bruchez MP, Barth AL. The brain-specific Beta4 subunit downregulates BK channel cell surface expression. PloS one. 2012;7:e33429. doi: 10.1371/journal.pone.0033429. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3306404&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva GL, Ediz V, Yaron D, Armitage Ba. Experimental and computational investigation of unsymmetrical cyanine dyes: understanding torsionally responsive fluorogenic dyes. Journal of the American Chemical Society. 2007;129:5710–8. doi: 10.1021/ja070025z. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2535610&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjobringso U, Bjorcksli L, Kasternll W. Streptococcal Protein G. Biochemistry. 1991;266:399–405. [Google Scholar]
- Szent-Gyorgyi C, Stanfield RL, Andreko S, Dempsey A, Ahmed M, Capek S, Waggoner A, Wilson Ia, Bruchez MP. Malachite Green Mediates Homodimerization of Antibody VL Domains to Form a Fluorescent Ternary Complex with Singular Symmetric Interfaces. Journal of molecular biology. 2013 doi: 10.1016/j.jmb.2013.08.014. http://www.ncbi.nlm.nih.gov/pubmed/23978698. [DOI] [PMC free article] [PubMed]
- Szent-Gyorgyi C, Schmidt BaF, Creeger Y, Fisher GW, Zakel KL, Adler S, Fitzpatrick JaJ, Woolford Ca, Yan Q, Vasilev KV, Berget PB, Bruchez MP, Jarvik JW, Waggoner A. Fluorogen-activating single-chain antibodies for imaging cell surface proteins. Nature biotechnology. 2008;26:235–40. doi: 10.1038/nbt1368. http://www.ncbi.nlm.nih.gov/pubmed/18157118. [DOI] [PubMed] [Google Scholar]
- Szent-Gyorgyi C, Schmidt BF, Fitzpatrick JaJ, Bruchez MP, Scheme S. Fluorogenic dendrons with multiple donor chromophores as bright genetically targeted and activated probes. Journal of the American Chemical Society. 2010;132:11103–9. doi: 10.1021/ja9099328. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2920033&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner S, Shen Z, Ng J, Florea L, Guigó R, Briggs SP, Bafna V. Improving gene annotation using peptide mass spectrometry. Genome research. 2007;17:231–9. doi: 10.1101/gr.5646507. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1781355&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tashiro M, Montelione GT. Structures of bacterial immunoglobulin-binding domains and their complexes with immunoglobulins. Current opinion in structural biology. 1995;5:471–81. doi: 10.1016/0959-440x(95)80031-x. http://www.ncbi.nlm.nih.gov/pubmed/8528763. [DOI] [PubMed] [Google Scholar]
- Wu Y, Tapia PH, Fisher GW, Simons PC, Strouse JJ, Foutz T, Waggoner AS, Jarvik J, Sklar La. Discovery of regulators of receptor internalization with high-throughput flow cytometry. Molecular pharmacology. 2012;82:645–57. doi: 10.1124/mol.112.079897. http://www.ncbi.nlm.nih.gov/pubmed/22767611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Tapia PH, Fisher GW, Waggoner AS, Jarvik J, Sklar LA. High-throughput flow cytometry compatible biosensor based on fluorogen activating protein technology. Cytometry Part A : the journal of the International Society for Analytical Cytology. 2013 doi: 10.1002/cyto.a.22242. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.