Abstract
Continuous (clustered) proportion data often arise in various domains of medicine and public health where the response variable of interest is a proportion (or percentage) quantifying disease status for the cluster units, ranging between zero and one. However, because of the presence of relatively disease-free as well as heavily diseased subjects in any study, the proportion values can lie in the interval [0, 1]. While beta regression can be adapted to assess covariate effects in these situations, its versatility is often challenged because of the presence/excess of zeros and ones because the beta support lies in the interval (0, 1). To circumvent this, we augment the probabilities of zero and one with the beta density, controlling for the clustering effect. Our approach is Bayesian with the ability to borrow information across various stages of the complex model hierarchy and produces a computationally convenient framework amenable to available freeware. The marginal likelihood is tractable and can be used to develop Bayesian case-deletion influence diagnostics based on q-divergence measures. Both simulation studies and application to a real dataset from a clinical periodontology study quantify the gain in model fit and parameter estimation over other ad hoc alternatives and provide quantitative insight into assessing the true covariate effects on the proportion responses.
Keywords: augmented beta, Bayesian, outliers, periodontal disease, q-divergence
1. Introduction
Clinical studies often generate proportion data where the response of interest is continuous and confined in the interval (0, 1), such as percentages, proportions, fractions, and rates [1]. Examples include proportion of nucleotides that differ for a given sequence or gene in foot-and-mouth disease [2], the percent decrease in glomerular filtration rate at various follow-up times since baseline [3], and so on. With fidelity to the usual Gaussian assumptions for model errors, one might here be tempted to fit a linear regression model to assess the response–covariate relationship [4]. However, this leads to misleading conclusions by ignoring the range constraints in the responses. The logistic-normal model in [5], which assumes normal distribution for logit-transformed proportion responses, can provide a computationally convenient framework, but it suffers from an interpretation problem given that the expected value of response is not a simple logit function of the covariates. In this context, the beta regression (BR) proposed in [6] can accomplish direct modeling of covariates under a generalized linear model specification, leading to easy interpretation. The beta density [7] is extremely flexible and can take on a variety of shapes to account for non-normality and skewness in proportion data. The BR model considers a specific re-parameterization of the associated beta density parameters and connects the covariates with the mean and precision of the density through appropriate link functions. Despite its versatility, its potential is limited for proportion responses with support in (0, 1).
The motivating data example for this paper comes from a clinical study [8], where the clinical attachment level (or, CAL), a clinical marker of periodontal disease (PD), is measured at each of the six sites of a subject’s tooth. The underlying statistical question here is to estimate the functions that model the dependence of the ‘proportion of diseased sites corresponding to a specific tooth type (represented by incisors, canines, premolars, and molars)’ with the covariables. Figure 1 (left panel) plots the raw (unadjusted) density histogram of the proportion responses aggregated over subjects and tooth types. The responses lie in the closed interval [0, 1] where 0 and 1 represent ‘completely disease-free’ and ‘highly diseased’ cases, respectively. Although BR might be applicable here post (ad hoc) re-scaling [9] of the data from [0, 1] to the interval (0, 1), various limitations are observed working on a transformed scale [10]. These re-scalings might provide a nice working solution for small proportions of zeros and ones, but sensitivity toward parameter estimation can be considerable with higher proportions. This inefficiency is only aggravated because of the presence of additional clustering (tooth within mouth/subject) in the data, as in our case. Hence, from a practical perspective, there is a need to seek an appropriate theoretical model that avoids data transformations yet is capable of handling the challenges the data present. To circumvent this, we propose an efficient generalized linear mixed model (GLMM) framework by augmenting the probabilities of occurrence of zeros and ones to the BR model via a zero-and-one-augmented beta (ZOAB) random effects (ZOAB-RE) model, which can accommodate the subject-level clustering.
Figure 1.
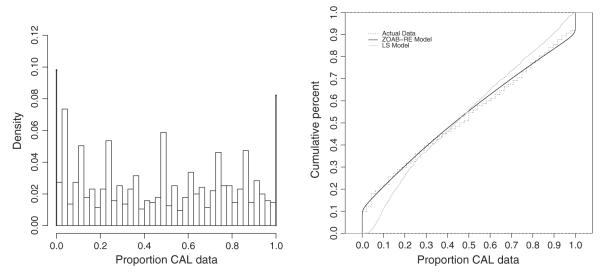
The left panel plots the (raw) density histogram, aggregated over subjects and tooth types for the periodontal disease data. The ‘pins’ at the extremes represent the proportion of zeros (9.8%) and ones (8.1%). The right panel presents the empirical cumulative distribution function of the real data, and that obtained after fitting the zero-and-one-augmented beta random effects (ZOAB-RE) model (model 1) and the lemon-squeezer (LS) model (model 3).
There have been various specifications of the BR model. The BR model in [6] re-parameterizes the beta density parameters and connects the data covariates to the response mean via a logit link, assuming that the data precision is constant (nuisance) across all observations. This was subsequently modified by linking the covariates to the dispersion parameter via the variable dispersion BR model in [9]. Very recently, Verkuilen and Smithson [11] used Gauss–Hermite quadrature to calculate maximum likelihood (ML) estimates and a Gibbs sampler for Bayesian estimation in the context of BR models for correlated proportion data. Also, Figueroa-Zuiga et al. [12] presented a Bayesian approach to the correlated BR model through Gibbs samplers and used the deviance information criterion (DIC) [13], expected AIC (EAIC), and expected BIC (EBIC) for model selection. However, to the best of our knowledge, there are no studies that utilize a Bayesian paradigm to model clustered (correlated) proportion data where the proportions lie in the interval [0,1]. Our proposition ‘augments’ point masses at zero and one to a continuous (beta) density that does not include zero and one in its support, similar in spirit to [14]. In addition, following the pioneering work of Cook [15], we develop case-deletion and local influence diagnostics to assess the effect of outliers on the parameter estimates. Our approach is Bayesian, with the ability to borrow information across various stages of the complex model hierarchy, and produces a computationally convenient framework amenable to available freeware like OpenBUGS ([16]).
The rest of the article proceeds as follows. After a brief introduction to the BR model, Section 2 introduces the ZOAB-RE model and develops the Bayesian estimation scheme. Section 3 applies the proposed ZOAB-RE model to the motivating data and uses Bayesian model selection to select the best model. It also summarizes and discusses the estimation of the fixed effects, other model parameters, and outlier detections. Section 4 presents simulation studies to assess finite sample performance of our model with another competing transformation-based model under model misspecification and also to study the efficiency of the influence diagnostic measures to detect outliers. Conclusions and future developments appear in Section 5.
2. Statistical model and Bayesian inference
2.1. Beta regression model
The beta distribution is often the model of choice for fitting continuous data restricted in the interval (0, 1) because of the flexibility it provides in terms of the variety of shapes it can accommodate. The probability density function of a beta distributed random variable Y parameterized in terms of its mean μ and a precision parameter ϕ is given by
| (1) |
where Γ(.) denotes the gamma function, E(Y) = μ, and . Therefore, for a fixed value of the mean μ, higher values of ϕ lead to a reduction of Var(Y), and vice versa. If Y has PDF as in (1), we write Y ~ beta(μϕ; (1 – μ)ϕ). Next, to connect the covariate vector xi to the random sample Y1, . . . , Yn of Y, we use a suitable link function g1 that maps the mean interval (0, 1) onto the real line. This is given as , where β is the vector of regression parameters, and the first element of xi is 1 to accommodate the intercept. The precision parameter ϕi is either assumed constant [6] or regressed onto the covariates [9] via another link function h1, such that , where zi is a covariate vector (not necessarily similar to xi) and α is the corresponding vector of regression parameters. Similar to xi, zi also accommodates an intercept. Both g1 and h1 are strictly monotonic and twice differentiable. Choices of g1 include the logit specification g1(μi) = log{μi/(1 – μi)}, the probit function g1(μi = Φ−1 (μi) where Φ(.) is the standard normal density, and the complementary log–log function g1(μi) = log{−log(1 – μi)}, among others, and those of h1 include the log function h1(ϕi) = log(ϕ), the square-root function , and the identity function h1(ϕi) = ϕi (with special attention to the positivity of the estimates) [17]. Estimation follows via either the (classical) ML route [6, 9] through Gauss–Hermite quadratures available in the betareg library in R [18], or Bayesian [2] through Gibbs sampling.
2.2. Zero-and-one augmented beta random effects model
The BR model described earlier only applies to observations that are independent, and moreover, it is suitable only for responses lying in (0, 1). However, for our PD dataset, the responses pertaining to a particular subject are clustered in nature and lie bounded in [0, 1]. We now develop a ZOAB model to address both the bounded support problem and the data clustering. Our proposition comprises a three-part mixture distribution, with degenerate point masses at 0 and 1, and a beta density to have the support of Yi ∈ [0, 1]. Thus, Y ~ ZOAB(p0i , p1i, μi, ϕ), if the density of Yi, i = 1, . . . , n, follows:
| (2) |
where p0i ≥ 0 denotes the probability Yi = 0, p1i ≥ 0 denotes the probability Yi = 1, 0 ≤ p0i + p1i ≤ 1, and f(yi∣μi, ϕ) is given in (1). The mean and variance of Yi is given by
For clustered data, the ZOAB-RE model is defined as follows. Let Y1, . . . , Yn be n independent continuous random vectors, where is the vector of length ni for the sample unit i, with the components yij ∈ [0, 1]. Next, the covariates can be regressed onto a suitably transformed μij, p0ij, and p1ij, such that
| (3) |
| (4) |
| (5) |
where , , ; Xi, W0i, and W0i are design matrices of dimension p × ni, r × ni, and s × ni, corresponding to the vectors of fixed effects β = (β1, . . . , βp)˕, ψ = (ψ1, . . . , ψr)˕ and ρ = (ρ1, . . . , ρs)˕ respectively; and Zi is the design matrix of dimension q × ni corresponding to REs vector bi = (bi1, . . . , biq)˕. Choice of link functions for g1, g2, and g3 here remain the same as for g1 in Section 2.1. For the sake of interpretation, we prefer to use the logit link. Note that in our model development, the dispersion parameter ϕ is chosen as constant and the regressions onto p0i and p1i are free of REs to avoid over-parameterization. However, it is certainly possible to regress ϕ onto covariates through an appropriate link function (say, log). Also, p0ij and p1ij can be treated as constants across all sample units. To this end, we define our ZOAB-RE model as Yij · ZOAB-RE(p0ij, p1ij, μij, ϕ) i = 1, . . . . n, j = 1, . . . , ni.
2.3. Data likelihood
Let Ω = (β, ψ, ρ, ϕ) denote the parameter vector in this ZOAB-RE model. The primary goal here is to estimate Ω and to derive inference on β adjusting for the effects of clustering. Our observed sample for n subjects is .y1, X1, Z1, W01, W11), . . . , (yn, Xn, Zn, W0n, W1n), with yi as the response vector for subject i. The joint data likelihood (without integrating out the random-effects bi) is given as
| (6) |
where
☉Ai indicates the product of the elements of Ai, with , with , Dki is a diagonal matrix of dimension ni×ni whose j-th element of the diagonal is the indicator function I{yij=k}, k = 0, 1, j = 1, . . . , ni, Ini is the identity matrix with dimension ni × ni and Bi is a diagonal matrix of dimension ni × ni whose j-th element of the diagonal is and , Xij and Zij correspond to the j-th column of the matrices Xi and Zi, respectively.
Although one can certainly pursue a classical estimation route using ML methods following [19], a Bayesian treatment of our model has not been considered earlier in the literature. Recent developments in Markov chain Monte Carlo (MCMC) methods facilitate easy and straightforward implementation of the Bayesian paradigm through conventional software such as OpenBUGS. Hence, we consider a Bayesian estimation framework that can accommodate full parameter uncertainty through appropriate prior choices supported by proper sensitivity investigations. This framework can provide a direct probability statement about a parameter through credible intervals (CIs) [20]. Next, we investigate the choice of priors for our model parameters to conduct Bayesian inference.
2.4. Priors, hyperpriors, and posterior distributions
We specify practical weakly informative prior opinion on the fixed-effects regression parameters β, ψ, ρ, ϕ (dispersion parameter) and the random effects bi. Specifically, we assign independent and identically distributed (i.i.d) Normal(0, precision = 0.01) priors on the elements of β, ψ, and ρ, which centers the ‘odds-ratio’ type inference at 1 with a sufficiently wide 95% interval. Priors for ϕ μ Gamma(0.1, 0.01) and bi are Normal with zero mean and precision = ), where σb ~ Unif(0, 100) [21]. Although multivariate specifications (multivariate zero mean vector with inverted-Wishart covariance) are certainly possible, we stick to simple (and independent) choices. For cases where p0 and p1 are considered constants across all subjects, we allocate the Dirichlet prior with hyperparameter α = (α1, α2, α3) for the probability vector (p0, p1, 1 – p0 – p1), where αs ~ Gamma(1, 0.001), s = 1, 2, 3.
The posterior conclusions are based on the joint posterior distribution of all the model parameters (conditional on the data) and obtained by combining the likelihood given in (6) and the joint prior densities using the Bayes’ theorem:
| (7) |
where , μj(.), j = 0, . . . , 5 denote the prior/hyperprior distributions on the model parameters as described earlier. The relevant MCMC steps (combination of Gibbs and Metropolis-within-Gibbs sampling) were implemented using the BRugs package [22], which connects the R with the OpenBUGS software. After discarding 50,000 burn-in samples, we used 50,000 more samples (with spacing of 10) from two independent chains with widely dispersed starting values for posterior summaries. Convergence was monitored via MCMC chain histories, autocorrelation and cross-correlation, density plots, and the Brooks–Gelman–Rubin potential scale reduction factor RO , all available in the R coda library [23]. Associated BRugs code is available on request from the corresponding author.
2.5. Bayesian model selection and influence diagnostics
We use the conditional predictive ordinate (CPO) statistic [24] for our model selection derived from the posterior predictive distribution (PPD). A summary statistic obtained from the CPO is the log pseudomarginal likelihood (LPML) [24]. Larger values of LPML indicate better fit. Because the harmonic-mean identity used in the CPO computation can be unstable [25], we consider a more pragmatic route and compute the CPO (and associated LPML) statistics using 500 non-overlapping blocks of the Markov chain, each of size 2000 post-convergence (i.e., after discarding the initial burn-in samples), and report the expected LPML computed over the 500 blocks. Some other measures, like the DIC, EAIC, and EBIC [24], can also be used. Because of the mixture framework in our ZOAB-RE model, we use the DIC3 [26] measure as an alternative to the DIC [13]. Model selection follows the ‘lower is better’ law; that is, the model with the lowest value for these criteria gets selected.
To determine model adequacy after selecting the best model, we apply the Bayesian p-value [27] that utilizes some discrepancy measures based on PPD. Samples from the PPD (denoted by ypr) are replicates of the observed model generated data y; hence, there is some signal of model inadequacy if the observed value is extreme relative to the reference PPD. Because of the clustered nature of our data, we consider the sum statistic T(y, θ) = sum(y) as our discrepancy measure. Then, the Bayesian p-value pB is calculated as the number of times T(ypr, θ) exceeds T(y, θ) out of L simulated draws, that is, pB = Pr(T(ypr, θ) ≥ T(y, θ)∣y). A very large p-value (> 0.95) or a very small one (< 0.05) signals model misspecification.
In addition, some influence diagnostic measures are developed to study the impact of outliers on fixed-effects parameter estimates caused by data perturbation schemes based on case-deletion statistics [28], and the q-divergence measures [29–31] between posterior distributions. We use three choices of these divergences, namely the Kullback–Leibler (KL) divergence, the J distance (symmetric version of the KL divergence), and the L1 distance. We use the calibration method [32] to obtain the cut-off values as 0.90, 0.83, and 1.32 for the L1, KL, and J distances, respectively.
3. Data analysis and findings
In this section, we apply our proposed ZOAB-RE model to the PD data. We start with a short description of the dataset. A study [8] assessing the status and progression of PD among Gullah-speaking African-Americans with type-2 diabetes was conducted at the Medical University of South Carolina (MUSC) via a detailed questionnaire focusing on demographics as well as social, medical, and dental history. CAL was recorded at each of the six tooth sites per tooth for 28 teeth (considered full dentition, excluding the four third molars). With 290 subjects, we focus on quantifying the extent and severity of PD for the tooth types (four canines and eight each of incisors, premolars, and molars). Our response variable is as follows: ‘proportion of diseased tooth sites (with CAL value > 3 mm) for each of the four tooth types’. This gives rise to a clustered data framework where each subject records four observations corresponding to the four tooth types. Missing teeth were considered ‘missing due to PD’, where all sites for that tooth contributed to the diseased category. Subject-level covariables in this dataset include gender (0 = male, 1 = female), age of subject at examination (in years, ranging from 26 to 87 years), glycosylated hemoglobin (HbA1c) status indicator (0 = controlled, < 7%; 1 = uncontrolled, ≥ 7%), and smoking status (0 = non-smoker, 1 = smoker). The smoker category is composed of both the current and past smokers. We also considered a tooth-level variable representing each of the four tooth types, with ‘canine’ as the baseline. As observed in the density histogram in Figure 1 (left panel), the data are continuous in the range [0, 1]. Because of the presence of a substantial number of zeros (114, 9.8%) and ones (94, 8.1%), BR might be inappropriate here. Hence, we resort to the ZOAB-RE model, controlling for subject-level clustering.
From (3), we now have with g1 as the logit link, β˕ = (β0, . . . , β7) with β0 as the intercept and β1, . . . , β7 as the regression parameters, and = (1, Genderi, Agei, HbA1ci, Smokeri, Incisori, Premolari, Molari), and bi is the subject-level random effect term. To improve convergence, we standardized ‘age’ by subtracting its mean and dividing by its standard deviation. Note that, here, the model covariates are regressed onto μij, p0ij, and p1ij, but it is also possible to consider p0 and p1 constants across all subjects. This leads to our choice of two competing models:
We also fit a non-augmented BR model by transforming the data points y to y′ via the lemon-squeezer (LS) transformation given by y′ = [y(N–1)+1/2]/N [9], where N is the total number of observations, and fit the previous regressions to μi with the logit link. This is our model 3, or the LS model. Although other link functions (such as probit, cloglog, etc) are available, we currently restrict ourselves to the symmetric logit link whose adequacy is assessed later. Note that models 1 and 2, which fit the same dataset, can be compared using the model choice criteria described in Subsection 2.5, but not model 3 because it considers a transformed dataset. Hence, model 3 is assessed using plots of empirical cumulative distribution functions (ECDFs) of the fitted values to determine how closely the fits resemble the true data.
In the absence of historical data/experiment, our prior choices follow the specifications described in Section 2.4. Table I presents the DIC3, LPML, EAIC, and EBIC values calculated for models 1 and 2. Notice that model 1 (our ZOAB-RE model with regression onto μij, p0ij, and p1ij) outperforms model 2 for all criteria. From Figure 1 (right panel), it is also clear that the ECDFs from the fitted values using model 1 represent the true data more closely than those using model 3. Considering these, we select model 1 as our best model. With respect to goodness-of-fit assessment, pB = 0.798, which indicates no overall lack of fit. Figure 2 plots the posterior parameter means and the 95% CIs for the regression onto μ for models 1–3. The gray intervals in Figure 2 contain zero (the non-significant covariates), while the black intervals do not include zero (the significant ones at 5% level). The covariates gender, age, and the tooth types (incisor, premolar, and molar) significantly explain the proportion responses. Conditional on the set of other covariates and REs, parameter interpretation can be expressed in terms of the corresponding covariate effect directly on μij, specifically the ratio . Here, μij is the ‘expected proportion of diseased sites’, and 1 – μij is the complement, that is, the ‘expected remaining proportion to being completely diseased’, both conditional on μij not being zero or one. Hence, the results in Table II can be expressed as the number of times the ratio is higher/lower with every unit increase (for a continuous covariate, such as age) or a change in category say from 0 to 1 (for a discrete covariate, say gender). For example, this ratio for age (a strong predictor of PD) is (1.4, 95% CI = [1.2, 1.6]). For gender, we conclude that this ratio is 40% lower for men as compared with women. Although study recruitment design was gender blind, women participated at a higher rate than the men, not unusual for studies on this population [33, 34], and further patient navigator techniques are being developed to achieve better gender balance. The other significant covariates can be interpreted similarly. For example, this ratio is 8.5 times higher for the posteriorly located molars as compared with anteriorly placed canines (the baseline).
Table I.
Model comparison using DIC3, LPML, EAIC, and EBIC criteria.
| Model |
||
|---|---|---|
| Criterion | 1 | 2 |
| DIC3 | 993 | 1243.5 |
| LPML | −500:5 | −623:7 |
| EAIC | 992:7 | 1231 |
| EBIC | 1124:2 | 1286.6 |
Figure 2.
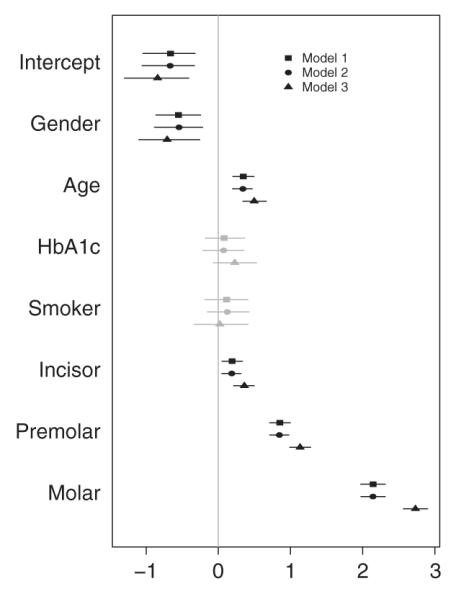
Posterior mean and 95% credible intervals (CIs) of parameter estimates from models 1 to 3. CIs that include zero are gray, and those that do not include zero are black.
Table II.
The values are the number of times higher/lower the ratio of the conditional ‘expected proportion of diseased sites’ (denoted by μij) is, to the ‘expected remaining proportion to complete disease’ (denoted by 1 − μij), conditional on this proportion not being zero or one, with one unit increase in the covariates.
| Parameter | Model 1 | Model 2 | Model 3 |
|---|---|---|---|
| Intercept | 0.5 | 0.5 | 0.4 |
| Gender | 0.6 | 0.6 | 0.5 |
| Age | 1.4 | 1.4 | 1.6 |
| HbA1c | 1.1 | 1.1 | 1.3 |
| Smoker | 1.1 | 1.1 | 1 |
| Incisor | 1.2 | 1.2 | 1.4 |
| Premolar | 2.3 | 2.3 | 3.1 |
| Molar | 8.5 | 8.5 | 15.3 |
The mean estimates (standard deviations) of ϕ for the models 1, 2, and 3 are 7.6 (0.42), 7.6 (0.43), and 4.6 (0.26), respectively, and those of for the models 1, 2, and 3 are 1.2 (0.13), 1.2 (0.13), and 1.8 (0.18), respectively. Based on these and from Table II, we conclude there is little difference between models 1 and 2 with respect to the estimates of β, ϕ, and . The main advantage of model 1 is that it identifies significant covariates related to free PD and completely diseased tooth types, which is not available in model 2. However, the estimates of premolar, molar, ϕ, and 2b obtained from model 3 are greater than those obtained from models 1 and 2, with the highest difference being for molar. Interestingly, the estimates of from model 3 are smaller (greater) than those from models 1 and 2, implying that augmenting leads to a lower (estimated) variance of Y than the transformation-based model 3.
Figure 3 plots the posterior parameter means and the 95% CIs of the parameters used to model p0 (left panel) and p1 (right panel) for model 1. Gender, age, and the type of tooth significantly explain free of PD, while gender, age, and molar significantly explain the completely diseased category. Table III presents the number of times higher/lower of the odds for free of PD (second column) and completely diseased (third column). For example, the odds of a tooth type free of PD are 2.9 times greater for men than for women, while the odds of a completely diseased molar are about 13 times than that of a (baseline) canine. Interestingly, the odds of a completely diseased tooth type are 2.5 times higher for a unit increase in age. Interpretation for the other parameters is similar.
Figure 3.
Posterior mean and 95% credible intervals (CIs) of parameter estimates for p0ij (left panel) and p1ij (right panel) from model 1. CIs that include zero are gray, and those that do not include zero are black.
Table III.
The values corresponding to p0ij represent odds of having a ‘disease-free’ versus ‘diseased’ tooth type, while those for p1ij denote odds of ‘completely diseased’ versus ‘diseased and disease-free’ tooth types.
| Parameter | p 0ij | p 1ij |
|---|---|---|
| Intercept | 0.2 | 0.03 |
| Gender | 2.9 | 0.5 |
| Age | 0.6 | 2.5 |
| HbA1c | 0.7 | 1.4 |
| Smoker | 0.7 | 0.7 |
| Incisor | 0.5 | 1.4 |
| Premolar | 0.08 | 1.3 |
| Molar | 0.005 | 13.3 |
To investigate the adequacy of the logit link for our regression, we consider an empirical approach via plots of the linear predictor versus the predicted probability [14], as depicted in Figure 4. We consider ηij from model 1 and divide it into 10 intervals containing roughly an equal number of observations. We plot the distribution of the inverse-logit transformed linear predictors (denoted by the black box plots) representing the fitted mean μij of the non-zero-one responses. Next, we overlay the empirical distributions of the observed non-zero-one responses represented by the gray box plots. From Figure 4, we observe no evidence of link misspecification; that is, the shapes of the fitted and observed trends are similar. As mentioned earlier, one can definitely fit other link functions, but the convenient interpretations in terms of μij are no longer valid for these fits.
Figure 4.
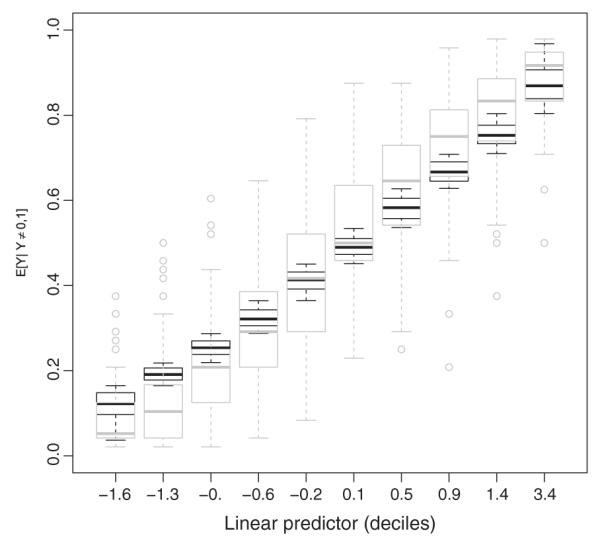
Observed and fitted relationship between the linear predictor ηij and the (conditional) non-zero-one mean μij. Modeled logit relationships are represented by black box plots, while the empirical proportions by gray box plots.
We also conducted a sensitivity analysis on the prior assumptions for the random-effects precision and the fixed-effects precision parameter. In particular, we allowed σb ~ Uniform(0, k), where k ∈ {10, 50}, and also the typical inverse-gamma choice for the precision ~ Gamma(k, k), where k ∈ {0.001, 0.1}. We also chose the normal precision on the fixed-effects to be 0.1, 0.25 (which reflects an odds ratio in between e−4 and e4), and 0.001. We checked the sensitivity in the posterior estimates of β by changing one parameter at a time and refitting model 1. Although slight changes were observed in parameter estimates and model comparison values, the results appeared to be robust and did not change our conclusions regarding the best model, inference (and sign) of the fixed effects, and the influential observations.
Finally, to determine the effect of possible influential observations, we computed the q-divergence measures for model 1. In particular, the subjects with ID numbers 135, 159, 174, and 285 were considered influential because the values of the L1, KL, and J distances exceeded the specified thresholds. The subjects 135, 159, and 285 have higher proportion responses for all tooth types (with Yij > 0.75) than for the corresponding mean proportions across all subjects. On the contrary, subject 174 is free of PD (Yij = 0) across all tooth types. To quantify the impact of these observations on the covariate effects, we refit the model by first removing these subjects successively and then as a whole. Compared with other covariates, the estimate of molar for the regression onto p0ij was impacted substantially. A minor impact on smoker for regression onto p0ij was also observed when all influential observations were removed. Overall, parameter significance and signs of the coefficients remained the same. Henceforth, we assert to use the estimates obtained from fitting model 1 to the full data without removing these subjects.
4. Simulation studies
In this section, we conduct two finite sample simulation studies. For the first, we plan to investigate the consequences on the (regression) parameter estimation under model misspecification via mean squared error (MSE), relative bias (RB), and coverage probability (CP) for (a) the ZOAB-RE model (model 1) and (b) the LS model (model 3) for varying sample sizes. In the second, we evaluate the efficiency of the q-divergence measures to detect atypical observations in the ZOAB-RE model.
4.1. Simulation 1
We generate ξij ~ Normal(μij, 1), where i = 1, . . . , n (the number of subjects), j = 1, . . . , 5 (indicating cluster of size 5 for each subject), with location parameter μij modeled as μij = β0 + β1xij + bi, and bi ~. Then, . We choose various sample sizes n = 50, 100, 150, and 200. The explanatory variables xij are generated as independent draws from a Uniform(0, 1), and regression parameters and variance components are fixed at β0 = −0.5, β1 = 0.5, and σ2 = 2. This generates data from a logit-normal model with yij ∈ (0, 1). Next, we can have two sets of p0 and p1, namely case a: p0 D 0.01, p1 = 0.01, and case b: p0 = 0.1, p1 = 0.08 (representative of the real data). The final step is to allocate the zeros, ones, and the yij ∈ (0, 1) with probabilities p0, p1, and (1 – p0 – p1), which is achieved via multinomial sampling. To keep the simulation design simple, we do not consider the regressions onto p0 and p1.
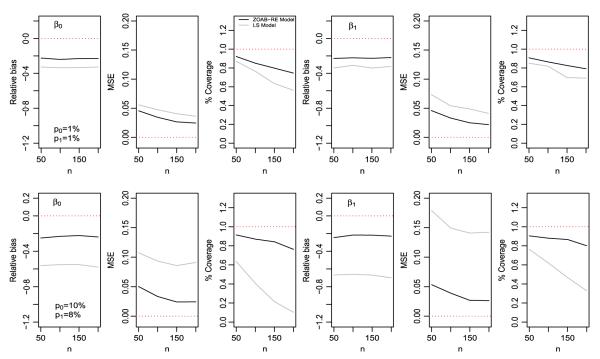
In the first simulation study, we simulated 500 such datasets and fitted the ZOAB-RE and the SL models with similar prior choices as in the data analysis. With our parameter space , and θs an element of θ, we calculate the MSE as MSE , the RB as , and the 95% CP as Cp where I is the indicator function such that θs lies in the interval , with and as the estimated lower and upper 95% CIs, respectively. Figure 5 presents a visual comparison of the parameters β0 and β1 for varying sample sizes and proportions p0 and p1, where the black and gray lines represent the ZOAB-RE model and the LS model, respectively.
Figure 5.
Relative bias, mean squared error (MSE) and coverage probability of β0 and β1 after fitting the zero- and-one-augmented beta random effects (ZOAB-RE) (black line) and lemon-squeezer (LS) (gray line) models, with p0 = p1 = 1% (upper panel) and p0 = 10%, p1 = 8% (lower panel).
As expected, both panels of Figure 5 reveal that the absolute values of RB for both β0 and β1 are much larger for the SL model than the ZOAB-RE model, with the RB increasing with increasing p0 and p1 (case b). We observe similar behavior for MSE and CP; that is, both the parameters from the ZOAB-RE model are estimated with lower MSE and higher CP as compared with the corresponding ones from the LS model, with the performance of the LS model getting worse with increasing proportions of extreme values. Clearly, when data are generated from a misspecified (augmented logit-normal) model, the LS model seems to produce a considerable impact on the regression parameter estimates as compared with the more robust ZOAB-RE model. For the sake of brevity, the MSE, RB, and CP for the other parameters (p0, p1, ) are not presented here, but we discuss the results. The proportions p0 and p1 are estimated with positive RB. Interestingly, for , the RB remains negative for all cases, with the absolute value of the RB increasing with increasing sample size mainly for the LS model. This might occur because the LS transformation induces lower variability in the data leading to an underestimated and RB. With this increase in RB, the 95% CI does not include the true value of , and hence, the CP is mostly 0 for higher n (150 and 200) for both models in case a, and also for all sample sizes for the LS model in case b. We conclude that under model misspecification, applying the LS transformation may not be adequate even for a moderate number of zeros and ones, with the performance deteriorating further as the proportion of extremes increases.
4.2. Simulation 2
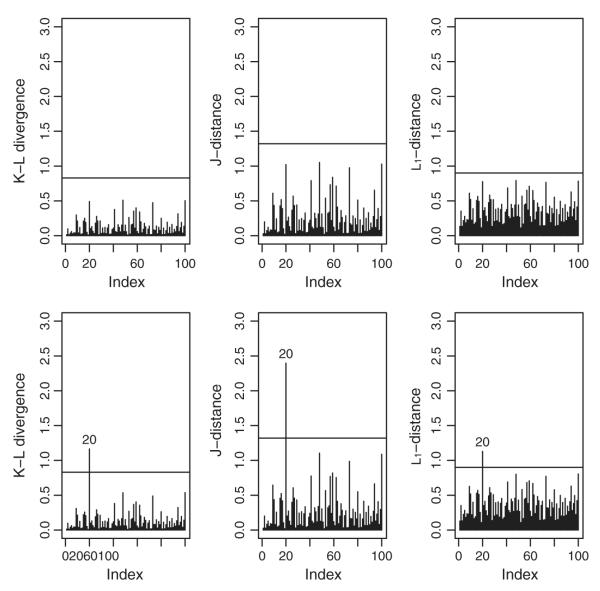
Here, we simulated one dataset with 100 subjects using the same data generation scheme as in Simulation 1. We perturb the response vector for ID #20 via y20 = y20 + 2SD(y20), where SD stands for standard deviation. If an element of the perturbed vector was greater than 1, we assigned 1 there. Figure 6 presents the q-divergence measures, both without perturbation (upper panel) and with perturbation (lower panel). We conclude from here that the divergence measures can correctly detect the influential (perturbed) observations.
Figure 6.
The q-divergence measures (KL, J, and L1 distance) without perturbation (upper panel), and after perturbing subject ID #20 (lower panel) for the simulated data.
5. Conclusions
Motivated by the classical development in [19], we developed a model for clustered responses in [0, 1] and applied it to an interesting PD dataset. Our model allows the parameters p0ij, p1ij, and μij to depend on covariates, leading to identifying covariates that are significant to explain disease-free, progressing with disease, and completely diseased tooth types. We also developed tools for outlier detection using q-divergence measures and quantified their effect on the posterior estimates of the model parameters. Both simulation studies and real data application justify seeking an appropriate theoretical model over utilizing ad hoc data transformations for proportion data. Note that the proposition in [19] (without any random effects) is termed ‘inflated beta distributions’. Typically, for cases of value-inflation, such as the zero-inflated counts in [35] or the zero-inflated (longitudinal) continuous data as in [36], inflation occurs when the probability mass of a value exceeds what is allowed by the proposed (underlying) distribution. This is certainly not the case here, and following [14], we prefer to call it an ‘augmented’ model over an ‘inflated’ model. Our model can be fitted using standard available software packages, such as R and OpenBUGS, with easy access to practitioners in the field.
It is of interest to investigate the presence of thick/heavy tails in the underlying ZOAB-RE proposition and to model the random-effect term bi using robust alternatives (say, the t-density) over the normal density as in [12]. For our dataset, the results were very similar using a t-density, and hence, we did not consider it any further.
Our current analysis considers clustered cross-sectional periodontal proportion data. Often, these study subjects can be randomized to dental treatments and subsequent longitudinal follow-ups, leading to a clustered-longitudinal framework, where one might be interested in estimating the profiles (both overall and subject-level) in the proportion of diseased surfaces for the four tooth types with time. Our ZOAB-RE can certainly be extended to such situations with proper consideration to the GLMM REs specification. Other propositions available in the literature on modeling clustered (or longitudinal) proportion responses include simplex mixed-effects models [4], robust transformation models [3, 37], and so on. How these models compare with ours and ways to adapt these to proportion responses in [0, 1] are components of future research and will be considered elsewhere.
Acknowledgements
We thank the editor, associate editor, and two referees whose constructive comments led to an improved presentation. We also thank the Center for Oral Health Research at MUSC for providing the motivating data and Prof. Elizabeth Slate for interesting insights on clinical interpretations. Galvis acknowledges the support from CAPES/CNPq – IEL Nacional – Brasil. Bandyopadhyay acknowledges the support from the US National Institutes of Health grants UL1TR000114 (CTSA award), P30 CA77598 (University of Minnesota Masonic Cancer Center grant), R03DE021762, and R03DE023372. Lachos was supported by grants 305054/2011-2 from CNPq–Brazil and 2011/17400-6 from FAPESP–Brazil.
References
- 1.Kieschnick R, McCullough BD. Regression analysis of variates observed on (0, 1): percentages, proportions and fractions. Statistical Modelling. 2003;3(3):193–213. [Google Scholar]
- 2.Branscum AJ, Johnson WO, Thurmond MC. Bayesian beta regression: applications to household expenditure data and genetic distance between foot-and-mouth disease viruses. Australian & New Zealand Journal of Statistics. 2007;49(3):287–301. [Google Scholar]
- 3.Song PXK, Tan M. Marginal models for longitudinal continuous proportional data. Biometrics. 2000;56(2):496–502. doi: 10.1111/j.0006-341x.2000.00496.x. [DOI] [PubMed] [Google Scholar]
- 4.Qiu Z, Song PXK, Tan M. Simplex mixed-effects models for longitudinal proportional data. Scandinavian Journal of Statistics. 2008;35(4):577–596. [Google Scholar]
- 5.Aitchison J. The Statistical Analysis of Compositional Data. Journal of the Royal Statistical Society. Series B (Methodological) 1982;44(2):139–177. [Google Scholar]
- 6.Ferrari S, Cribari-Neto F. Beta regression for modelling rates and proportions. Journal of Applied Statistics. 2004;31(7):799–815. [Google Scholar]
- 7.Johnson N, Kotz S, Balakrishnan N. Continuous Univariate Distributions. Vol. 2. John Wiley & Sons; New York: 1994. [Google Scholar]
- 8.Fernandes J, Salinas C, London S, Wiegand R, Hill E, Slate E, Grewal J, Werner P, Sanders J, Lopes-Virella M. Prevalence of periodontal disease in Gullah African American diabetics. Journal of Dental Research. 2006;85:997. [Google Scholar]
- 9.Smithson M, Verkuilen J. A better lemon squeezer? Maximum-likelihood regression with beta-distributed dependent variables. Psychological Methods. 2006;11(1):54. doi: 10.1037/1082-989X.11.1.54. [DOI] [PubMed] [Google Scholar]
- 10.Lachos VH, Bandyopadhyay D, Dey DK. Linear and nonlinear mixed-effects models for censored HIV viral loads using normal/independent distributions. Biometrics. 2011;67(4):1594–1604. doi: 10.1111/j.1541-0420.2011.01586.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Verkuilen J, Smithson M. Mixed and mixture regression models for continuous bounded responses using the beta distribution. Journal of Educational and Behavioral Statistics. 2012;37(1):82–113. [Google Scholar]
- 12.Figueroa-Zuiga J, Arellano-Valle RB, Ferrari SL. Mixed beta regression: a bayesian perspective. Computational Statistics & Data Analysis. 2013;61:137–147. DOI: 10.1016/j.csda.2012.12.002. [Google Scholar]
- 13.Spiegelhalter DJ, Best NG, Carlin BP, van der Linde A. Bayesian measures of model complexity and fit. Journal of the Royal Statistical Society-Series B. 2002;64(4):583–639. [Google Scholar]
- 14.Hatfield LA, Boye ME, Hackshaw MD, Carlin BP. Multilevel Bayesian models for survival times and longitudinal patient-reported outcomes with many zeros. Journal of the American Statistical Association. 2012;107:875–885. [Google Scholar]
- 15.Cook RD. Assessment of local influence. (Series B).Journal of the Royal Statistical Society. 1986;48:133–169. [Google Scholar]
- 16.Thomas A, OHara B, Ligges U, Sturtz S. Making BUGS open. R News. 2006;6(1):12–17. [Google Scholar]
- 17.Simas A, Barreto-Souza W, Rocha A. Improved estimators for a general class of beta regression models. Computational Statistics and Data Analysis. 2010;54(2):348–366. [Google Scholar]
- 18.Zeileis A, Cribari-Neto F, Grn B. Beta regression in R. Journal of Statistical Software. 2010;34(2):1–24. [Google Scholar]
- 19.Ospina R, Ferrari S. Inflated beta distributions. Statistical Papers. 2010;51(1):111–126. [Google Scholar]
- 20.Dunson D. Commentary: practical advantages of Bayesian analysis of epidemiologic data. American Journal of Epidemiology. 2001;153(12):1222. doi: 10.1093/aje/153.12.1222. [DOI] [PubMed] [Google Scholar]
- 21.Gelman A. Prior distributions for variance parameters in hierarchical models. Bayesian Analysis. 2006;1(3):515–534. [Google Scholar]
- 22.Thomas A, O’Hara B, Ligges U, Sturtz S. Making BUGS Open. R News. 2006;6(1):12–17. [Google Scholar]
- 23.Cowles MK, Carlin BP. Markov chain Monte Carlo convergence diagnostics: a comparative review. Journal of the American Statistical Association. 1996;91(434):883–904. [Google Scholar]
- 24.Carlin B, Louis T. Bayesian Methods for Data Analysis (Texts in Statistical Science) Chapman and Hall/CRC; New York: 2008. [Google Scholar]
- 25.Raftery A, Newton M, Satagopan J, Krivitsky P. Estimating the integrated likelihood via posterior simulation using the harmonic mean identity (with discussion) In: Berger J, Dawid A, Heckerman D, Smith A, West M, editors. Bayesian Statistics 8. Vol. 8. Oxford University Press; London, UK: 2007. pp. 1–45. [Google Scholar]
- 26.Celeux G, Forbes F, Robert CP, Titterington DM. Deviance information criteria for missing data models. Bayesian Analysis. 2006;1(4):651–673. [Google Scholar]
- 27.Gelman A, Carlin JB, Stern HS, Rubin D. Bayesian Data Analysis. Chapman & Hall/CRC; Boca Raton, FL: 2004. [Google Scholar]
- 28.Cook RD, Weisberg S. Residuals and Influence in Regression. Chapman & Hall/CRC; Boca Raton, FL: 1982. [Google Scholar]
- 29.Csisz I. Information-type measures of difference of probability distributions and indirect observations. Studia Scientiarum Mathematicarum Hungarica. 1967;2:299–318. [Google Scholar]
- 30.Weiss R. An approach to Bayesian sensitivity analysis. (Series B (Methodological)).Journal of the Royal Statistical Society. 1996;58(4):739–750. [Google Scholar]
- 31.Lachos VH, Castro LM, Dey DK. Bayesian inference in nonlinear mixed–effects models using normal independent distributions. Computational Statistics & Data Analysis. 2013;64:237–252. [Google Scholar]
- 32.Peng F, Dey DK. Bayesian analysis of outlier problems using divergence measures. The Canadian Journal of Statistics. 1995;23:199–213. [Google Scholar]
- 33.Johnson-Spruill I, Hammond P, Davis B, McGee Z, Louden D. Health of Gullah families in South Carolina with type 2 diabetes: diabetes self-management analysis from project sugar. The Diabetes Educator. 2009;35(1):117–123. doi: 10.1177/0145721708327535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bandyopadhyay D, Reich BJ, Slate EH. Bayesian modeling of multivariate spatial binary data with applications to dental caries. Statistics in Medicine. 2009;28(28):3492–3508. doi: 10.1002/sim.3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lachenbruch PA. Analysis of data with excess zeros. Statistical Methods in Medical Research. 2002;11(4):297–302. doi: 10.1191/0962280202sm289ra. [DOI] [PubMed] [Google Scholar]
- 36.Ghosh P, Albert PS. A Bayesian analysis for longitudinal semicontinuous data with an application to an acupuncture clinical trial. Computational Statistics & Data Analysis. 2009;53(3):699–706. doi: 10.1016/j.csda.2008.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang P, Qiu Z, Fu Y, Song PXK. Robust transformation mixed-effects models for longitudinal continuous proportional data. Canadian Journal of Statistics. 2009;37(2):266–281. [Google Scholar]