Abstract
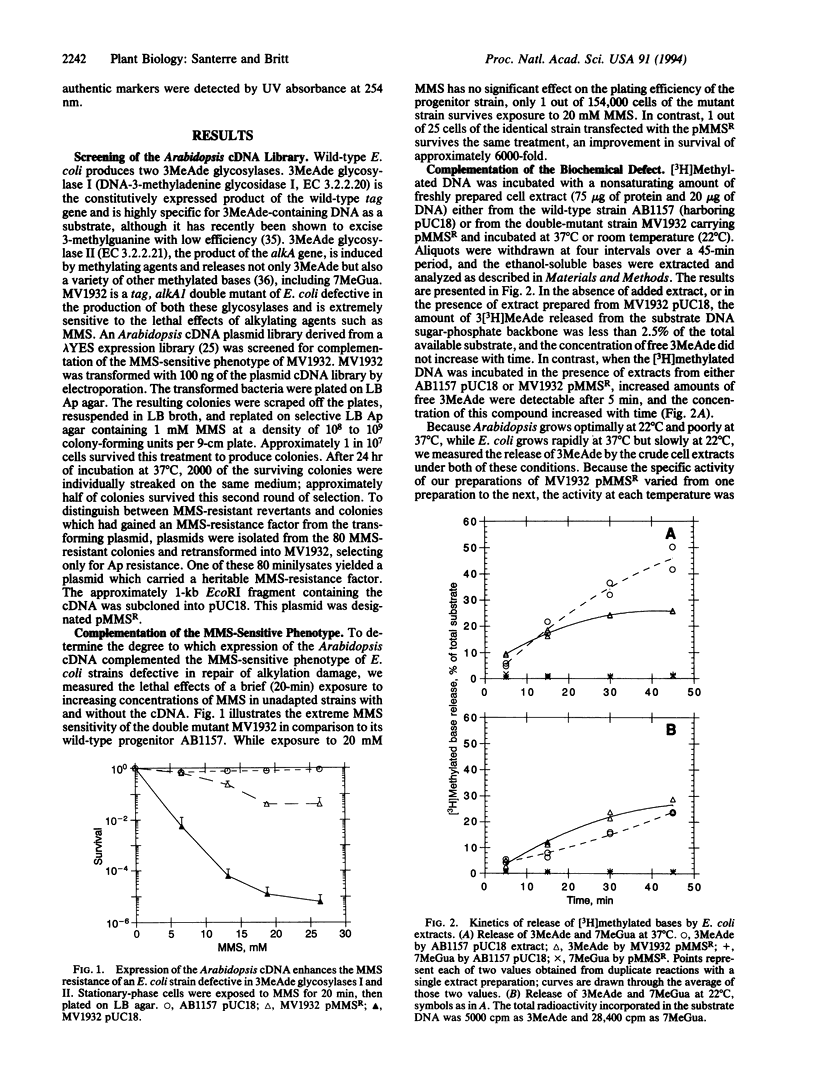
We have isolated an Arabidopsis thaliana cDNA that complements the methyl methanesulfonate-sensitive phenotype of an Escherichia coli double mutant deficient in 3-methyladenine glycosylases (DNA-3-methyladenine glycosidases I and II, EC 3.2.2.20 and 3.2.2.21, respectively, encoded by tag and alkA). Expression of the Arabidopsis cDNA enhances the methyl methanesulfonate resistance of the E. coli double mutant by nearly four orders of magnitude. The cDNA corresponds to a single-copy, nuclearly encoded sequence which specifies a predicted 28.1-kDa protein with a charge of +8 at pH 7.0. Enzymatic analysis of extracts prepared from the transformed mutants indicates that the cDNA encodes a 3-methyladenine glycosylase. The predicted amino acid sequence of the Arabidopsis glycosylase has significant homology to other eukaryotic 3-methyladenine glycosylases.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bagg A., Kenyon C. J., Walker G. C. Inducibility of a gene product required for UV and chemical mutagenesis in Escherichia coli. Proc Natl Acad Sci U S A. 1981 Sep;78(9):5749–5753. doi: 10.1073/pnas.78.9.5749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berdal K. G., Bjørås M., Bjelland S., Seeberg E. Cloning and expression in Escherichia coli of a gene for an alkylbase DNA glycosylase from Saccharomyces cerevisiae; a homologue to the bacterial alkA gene. EMBO J. 1990 Dec;9(13):4563–4568. doi: 10.1002/j.1460-2075.1990.tb07909.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjelland S., Bjørås M., Seeberg E. Excision of 3-methylguanine from alkylated DNA by 3-methyladenine DNA glycosylase I of Escherichia coli. Nucleic Acids Res. 1993 May 11;21(9):2045–2049. doi: 10.1093/nar/21.9.2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Cerutti H., Osman M., Grandoni P., Jagendorf A. T. A homolog of Escherichia coli RecA protein in plastids of higher plants. Proc Natl Acad Sci U S A. 1992 Sep 1;89(17):8068–8072. doi: 10.1073/pnas.89.17.8068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarti D., Ibeanu G. C., Tano K., Mitra S. Cloning and expression in Escherichia coli of a human cDNA encoding the DNA repair protein N-methylpurine-DNA glycosylase. J Biol Chem. 1991 Aug 25;266(24):15710–15715. [PubMed] [Google Scholar]
- Chen J., Derfler B., Maskati A., Samson L. Cloning a eukaryotic DNA glycosylase repair gene by the suppression of a DNA repair defect in Escherichia coli. Proc Natl Acad Sci U S A. 1989 Oct;86(20):7961–7965. doi: 10.1073/pnas.86.20.7961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke N. D., Kvaal M., Seeberg E. Cloning of Escherichia coli genes encoding 3-methyladenine DNA glycosylases I and II. Mol Gen Genet. 1984;197(3):368–372. doi: 10.1007/BF00329931. [DOI] [PubMed] [Google Scholar]
- Elledge S. J., Mulligan J. T., Ramer S. W., Spottswood M., Davis R. W. Lambda YES: a multifunctional cDNA expression vector for the isolation of genes by complementation of yeast and Escherichia coli mutations. Proc Natl Acad Sci U S A. 1991 Mar 1;88(5):1731–1735. doi: 10.1073/pnas.88.5.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelward B. P., Boosalis M. S., Chen B. J., Deng Z., Siciliano M. J., Samson L. D. Cloning and characterization of a mouse 3-methyladenine/7-methyl-guanine/3-methylguanine DNA glycosylase cDNA whose gene maps to chromosome 11. Carcinogenesis. 1993 Feb;14(2):175–181. doi: 10.1093/carcin/14.2.175. [DOI] [PubMed] [Google Scholar]
- Evensen G., Seeberg E. Adaptation to alkylation resistance involves the induction of a DNA glycosylase. Nature. 1982 Apr 22;296(5859):773–775. doi: 10.1038/296773a0. [DOI] [PubMed] [Google Scholar]
- Foster P. L., Eisenstadt E. Induction of transversion mutations in Escherichia coli by N-methyl-N'-nitro-N-nitrosoguanidine is SOS dependent. J Bacteriol. 1985 Jul;163(1):213–220. doi: 10.1128/jb.163.1.213-220.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher P. E., Brent T. P. Partial purification and characterization of 3-methyladenine-DNA glycosylase from human placenta. Biochemistry. 1982 Dec 7;21(25):6404–6409. doi: 10.1021/bi00268a013. [DOI] [PubMed] [Google Scholar]
- Gavel Y., von Heijne G. A conserved cleavage-site motif in chloroplast transit peptides. FEBS Lett. 1990 Feb 26;261(2):455–458. doi: 10.1016/0014-5793(90)80614-o. [DOI] [PubMed] [Google Scholar]
- Karran P., Lindahl T., Ofsteng I., Evensen G. B., Seeberg E. Escherichia coli mutants deficient in 3-methyladenine-DNA glycosylase. J Mol Biol. 1980 Jun 15;140(1):101–127. doi: 10.1016/0022-2836(80)90358-7. [DOI] [PubMed] [Google Scholar]
- Kieber J. J., Tissier A. F., Signer E. R. Cloning and Characterization of an Arabidopsis thaliana Topoisomerase I Gene. Plant Physiol. 1992 Aug;99(4):1493–1501. doi: 10.1104/pp.99.4.1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson K., Sahm J., Shenkar R., Strauss B. Methylation-induced blocks to in vitro DNA replication. Mutat Res. 1985 Jun-Jul;150(1-2):77–84. doi: 10.1016/0027-5107(85)90103-4. [DOI] [PubMed] [Google Scholar]
- Loechler E. L., Green C. L., Essigmann J. M. In vivo mutagenesis by O6-methylguanine built into a unique site in a viral genome. Proc Natl Acad Sci U S A. 1984 Oct;81(20):6271–6275. doi: 10.1073/pnas.81.20.6271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lütcke H. A., Chow K. C., Mickel F. S., Moss K. A., Kern H. F., Scheele G. A. Selection of AUG initiation codons differs in plants and animals. EMBO J. 1987 Jan;6(1):43–48. doi: 10.1002/j.1460-2075.1987.tb04716.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Male R., Helland D. E., Kleppe K. Purification and characterization of 3-methyladenine-DNA glycosylase from calf thymus. J Biol Chem. 1985 Feb 10;260(3):1623–1629. [PubMed] [Google Scholar]
- Male R., Nes I. F., Kleppe K. Purification and properties of 3-methyladenine-DNA glycosylase from L-cells. Eur J Biochem. 1981 Dec;121(1):243–248. doi: 10.1111/j.1432-1033.1981.tb06455.x. [DOI] [PubMed] [Google Scholar]
- O'Connor T. R., Laval F. Isolation and structure of a cDNA expressing a mammalian 3-methyladenine-DNA glycosylase. EMBO J. 1990 Oct;9(10):3337–3342. doi: 10.1002/j.1460-2075.1990.tb07534.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor T. R., Laval J. Human cDNA expressing a functional DNA glycosylase excising 3-methyladenine and 7-methylguanine. Biochem Biophys Res Commun. 1991 May 15;176(3):1170–1177. doi: 10.1016/0006-291x(91)90408-y. [DOI] [PubMed] [Google Scholar]
- Pang Q., Hays J. B., Rajagopal I. A plant cDNA that partially complements Escherichia coli recA mutations predicts a polypeptide not strongly homologous to RecA proteins. Proc Natl Acad Sci U S A. 1992 Sep 1;89(17):8073–8077. doi: 10.1073/pnas.89.17.8073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang Q., Hays J. B., Rajagopal I., Schaefer T. S. Selection of Arabidopsis cDNAs that partially correct phenotypes of Escherichia coli DNA-damage-sensitive mutants and analysis of two plant cDNAs that appear to express UV-specific dark repair activities. Plant Mol Biol. 1993 Jun;22(3):411–426. doi: 10.1007/BF00015972. [DOI] [PubMed] [Google Scholar]
- Pruitt R. E., Meyerowitz E. M. Characterization of the genome of Arabidopsis thaliana. J Mol Biol. 1986 Jan 20;187(2):169–183. doi: 10.1016/0022-2836(86)90226-3. [DOI] [PubMed] [Google Scholar]
- Rebeck G. W., Samson L. Increased spontaneous mutation and alkylation sensitivity of Escherichia coli strains lacking the ogt O6-methylguanine DNA repair methyltransferase. J Bacteriol. 1991 Mar;173(6):2068–2076. doi: 10.1128/jb.173.6.2068-2076.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riazuddin S., Lindahl T. Properties of 3-methyladenine-DNA glycosylase from Escherichia coli. Biochemistry. 1978 May 30;17(11):2110–2118. doi: 10.1021/bi00604a014. [DOI] [PubMed] [Google Scholar]
- Sakumi K., Sekiguchi M. Structures and functions of DNA glycosylases. Mutat Res. 1990 Sep-Nov;236(2-3):161–172. doi: 10.1016/0921-8777(90)90003-n. [DOI] [PubMed] [Google Scholar]
- Samson L., Derfler B., Boosalis M., Call K. Cloning and characterization of a 3-methyladenine DNA glycosylase cDNA from human cells whose gene maps to chromosome 16. Proc Natl Acad Sci U S A. 1991 Oct 15;88(20):9127–9131. doi: 10.1073/pnas.88.20.9127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sargentini N. J., Smith K. C. Much of spontaneous mutagenesis in Escherichia coli is due to error-prone DNA repair: implications for spontaneous carcinogenesis. Carcinogenesis. 1981;2(9):863–872. doi: 10.1093/carcin/2.9.863. [DOI] [PubMed] [Google Scholar]
- Shure M., Wessler S., Fedoroff N. Molecular identification and isolation of the Waxy locus in maize. Cell. 1983 Nov;35(1):225–233. doi: 10.1016/0092-8674(83)90225-8. [DOI] [PubMed] [Google Scholar]
- Singhal R. K., Hinkle D. C., Lawrence C. W. The REV3 gene of Saccharomyces cerevisiae is transcriptionally regulated more like a repair gene than one encoding a DNA polymerase. Mol Gen Genet. 1992 Dec;236(1):17–24. doi: 10.1007/BF00279638. [DOI] [PubMed] [Google Scholar]
- Smith K. C. Spontaneous mutagenesis: experimental, genetic and other factors. Mutat Res. 1992 Aug;277(2):139–162. doi: 10.1016/0165-1110(92)90002-q. [DOI] [PubMed] [Google Scholar]
- Vaughan P., Sedgwick B., Hall J., Gannon J., Lindahl T. Environmental mutagens that induce the adaptive response to alkylating agents in Escherichia coli. Carcinogenesis. 1991 Feb;12(2):263–268. doi: 10.1093/carcin/12.2.263. [DOI] [PubMed] [Google Scholar]
- Waterman M. S., Eggert M. A new algorithm for best subsequence alignments with application to tRNA-rRNA comparisons. J Mol Biol. 1987 Oct 20;197(4):723–728. doi: 10.1016/0022-2836(87)90478-5. [DOI] [PubMed] [Google Scholar]
- Xiao W., Samson L. In vivo evidence for endogenous DNA alkylation damage as a source of spontaneous mutation in eukaryotic cells. Proc Natl Acad Sci U S A. 1993 Mar 15;90(6):2117–2121. doi: 10.1073/pnas.90.6.2117. [DOI] [PMC free article] [PubMed] [Google Scholar]




