Summary
Background
Incorporation of secretory proteins into ER-derived vesicles involves recognition of cytosolic signals by the COPII coat protein, Sec24. Additional cargo diversity is achieved through cargo receptors, which include the Erv14/Cornichon family that mediate export of transmembrane proteins despite the potential for such clients to directly interact with Sec24. The molecular function of Erv14 thus remains unclear, with possible roles in COPII-binding, membrane domain chaperoning and lipid organization.
Results
Using a targeted mutagenesis approach to define the mechanism of Erv14 function, we identify conserved residues in the second transmembrane domain of Erv14 that mediate interaction with a subset of Erv14 clients. We further show that interaction of Erv14 with a novel cargo-binding surface on Sec24 is necessary for efficient trafficking of all of its clients. However, we also determine that some Erv14 clients also engage directly an adjacent cargo-binding domain of Sec24, suggesting a novel mode of dual interaction between cargo and coat.
Conclusions
We conclude that Erv14 functions as a canonical cargo receptor that couples membrane proteins to the COPII coat, but that maximal export requires a bivalent signal that derives from motifs on both the cargo protein and Erv14. Sec24 can thus be considered a coincidence detector that binds simultaneously to multiple signals to drive packaging of polytopic membrane proteins. This mode of dual signal binding to a single coat protein might serve as a general mechanism to trigger efficient capture, or may be specifically employed in ER export to control deployment of nascent proteins.
Introduction
Biogenesis of integral membrane proteins and secreted soluble proteins initiates in the endoplasmic reticulum (ER). Once appropriately folded, these proteins are packaged into COPII vesicles, named for the coat machinery that generates them [1]. Although some proteins exit the ER stochastically via bulk flow [2, 3], more efficient ER egress relies on sorting signals that interact with the COPII coat subunit, Sec24 [4–10]. Yeast and mammalian Sec24 paralogs contain multiple cargo-binding sites, each of which recognizes distinct motifs [4, 10–13]. Despite this detailed characterization of Sec24 interaction with a subset of cargo proteins, our broader understanding of how the full repertoire of diverse secretory proteins is trafficked remains incomplete.
Expanded diversity in cargo selection is achieved at least in part by receptors that link multiple cargo proteins to the coat [14, 15]. In yeast, Erv29 recruits a subset of soluble secretory and vacuolar proteins that cannot directly contact Sec24 [16]. Mammalian ERGIC-53 and related family members similarly package multiple secreted glycoproteins [17], including clotting factors [18]. Lumenally-oriented GPI-anchored proteins, which are also subject to topological constraints, employ the p24 complex [19]. The conserved Erv14/cornichon family (Erv14 in yeast, cornichon in Drosophila, CNIH in mammals) is required for efficient ER export of numerous endomembrane proteins [20–26], most of which are polytopic and reside in the late secretory pathway [22]. Why membrane proteins that have the potential to interact directly with the COPII coat require a cargo receptor has long been a puzzle. One model is that Erv14-dependent cargo proteins evolved by genetic recombination that repositioned their sorting signals into the ER lumen [15]. Indeed, appending ER export signals to cytoplasmic domains of Erv14/cornichon clients bypasses the requirement for the receptor [22, 26]. Furthermore, the COPII-binding signal on Erv14 is required for ER export of at least one cargo, Axl2 [21]. However, some yeast Erv14 clients possess their own sorting signals (eg. Gap1 [5]; Yor1 [27]), raising the question of how important the COPII coupling function of Erv14 is.
In addition to coupling cargoes to the COPII coat, Erv14 has also been proposed to chaperone long transmembrane domains (TMDs), which are characteristic of plasma membrane proteins [28]. Since the bilayer of the ER is relatively thin, the potential for hydrophobic mismatch while plasma membrane proteins transit through this organelle might drive a requirement for such a chaperone. Indeed, TMD length seems to play an important role in determining the rate of ER export and Erv14-dependency for at least one client, Mid2 [22]. Whether this TMD-length dependence solely reflects a cargo selection mechanism or has broader importance for protein stability remains to be determined, especially since only a subset of plasma membrane proteins appears to require Erv14 for ER export [22].
We previously identified a requirement for Erv14 in trafficking of the ABC transporter, Yor1 [29]. Intrigued by why this membrane protein, which contains its own ER export motif [27], would need a cargo receptor, we sought to further dissect the mechanisms by which Erv14 contributes to cargo packaging. We find that Erv14-COPII interaction is universally required for export of all Erv14 clients. Conversely, residues within the second TMD of Erv14 appear to define a discrete binding site that mediates interaction with only a subset of Erv14 clients. In this respect, Erv14 seems to act as a classical cargo receptor, binding simultaneously to cargo proteins and the COPII coat to drive ER export. Yet multiple cargoes also showed a separate specific requirement for Sec24, suggesting that dual ER export signals are a common feature for Erv14-dependent polytopic membrane proteins. Finally, we identify the site on Sec24 that likely recognizes Erv14, and propose that Sec24 can be considered a coincidence detector that binds simultaneously to two signals: one on the cargo protein itself and another on Erv14. By using multiple weak sorting signals, the coat may efficiently capture polytopic membrane proteins at the ER membrane but still ensure coat release from the vesicle as prior to fusion with the Golgi. Furthermore, this mode of Sec24-client interaction might serve to regulate ER export in the context of protein quality control such that only correctly assembled proteins present a dual signal and thus fully engage the coat.
Results
Erv14 and Erv15 act redundantly in ER export of Yor1 to promote Sec24-mediated transport
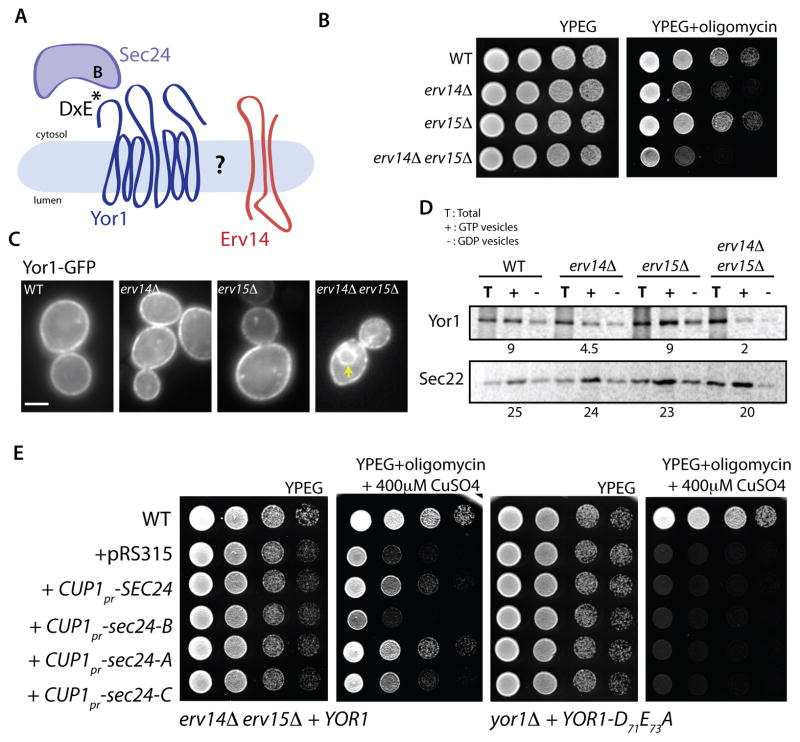
We recently identified a role for Erv14 in ER export of the yeast ABC transporter, Yor1 [29]. This was surprising since Yor1 possesses a canonical di-acidic ER export signal that mediates interaction with the Sec 24 B-site [27]. Furthermore, Yor1 was not identified in a systematic screen for Erv14 clients [22], raising the question of the precise nature of the relationship between Yor1 and Erv14 (Figure 1A). We first considered that Yor1 may have eluded detection as an Erv14 client because of functional redundancy with Erv15, a closely related paralog [30]. Indeed, an erv14Δ erv15Δ double mutant strain was highly sensitive to the Yor1 substrate, oligomycin (Figure 1B), indicative of impaired delivery of Yor1 to the plasma membrane and thus reduced clearance of the toxin from the cytosol. Furthermore, Yor1-GFP was readily detected in the ER in the erv14Δ erv15Δ double mutant whereas ER retention was not apparent in the single mutant strains, consistent with its absence from the PAIRS analysis of Erv14 clients [22] (Figure 1C).
Figure 1. Erv14 and Erv15 are necessary for ER export of Yor1 but can be bypassed by over-expression of SEC24.
(A) Schematic representation of the Yor1-Sec24-Erv14 associations; Yor1 has a DxE motif that interacts with the Sec24 B-site. (B) Serial dilutions of the yeast strains indicated were tested for sensitivity to oligomycin (0.4 μg/ml). (C) Localization of Yor1-GFP in the strains indicated was examined by epifluoresence microscopy (Scale bar = 2um). Arrowheads indicate perinuclear ER (D) Packaging of Yor1-HA into COPII vesicles was examined in vitro using membranes isolated from the strain backgrounds indicated. Radiolabeled cells were permeabilized and incubated with COPII proteins in the presence of either GTP (+) or GDP (−). HA-tagged Yor1 and Erv14 were immunoprecipitated from total (T) and vesicle fractions, then analyzed by SDS-PAGE. The percentage of cargo in the vesicle fraction was quantified relative to that in the donor membranes. An independent cargo protein, Sec22, was also examined to confirm efficient generation of vesicles. Numbers indicate budding efficiency (% total). (E) Serial dilutions of erv14Δ erv15Δ strains transformed with the indicated variants of SEC24 under the control of the copper-inducible CUP1 promoter were spotted on media containing oligomycin (0.4 μg/ml) and CuSO4 (400 μM). Partial rescue of the oligomycin sensitivity of the erv14Δ erv15Δ strain could be observed upon induction of wild-type SEC24, sec24-A and sec24-C. In contrast, over-expression of sec24-B failed to restore drug resistance (left panels). Yor1 engagement with Sec24 is essential for rescue since no oligomycin resistance was observed in a yor1Δ strain expressing the sorting signal mutant of Yor1 (YOR1-D71A E73A) and transformed with the same SEC24 constructs (right panels). See also Figure S1.
We quantified defects in Yor1 traffic using in vitro budding assays that reconstitute vesicle formation from permeabilized cells to measure capture of newly synthesized cargo into COPII vesicles [27]. Radiolabeled wild-type, erv14Δ, erv15Δ, and erv14Δ erv15Δ membranes expressing HA-tagged Yor1 were incubated with purified COPII proteins and either GTP or GDP. Vesicles were separated from donor membranes by differential centrifugation and cargo proteins were immunoprecipitated from total membrane and vesicle fractions. Relative to wild-type, erv15Δ cells showed no defect in packaging of Yor1 into vesicles whereas in erv14Δ cells, Yor1 capture was reduced by ~50%. This defect was further exacerbated in the erv14Δ erv15Δ cells (Figure 1D). These data imply that both Erv14 and Erv15 have the capacity to act as cargo adaptors for Yor1, but that Erv15 alone is not sufficient to complement the defects associated with loss of Erv14. Indeed, when we placed the ERV15 ORF under the control of the ERV14 promoter in erv14Δ and erv14Δ erv15Δ strains, Erv15 was able to fully complement the oligomycin sensitivity of both strains (Figure S1A) and restored packaging of Yor1-HA into COPII vesicles in vitro (Figure S1B). This rescue phenotype is consistent with the known expression levels of Erv14 and Erv15, with Erv15 undetectable at steady state [31].
We next asked what function Erv14/Erv15 might perform in the context of Yor1 biogenesis. We tested whether Erv14 is a folding chaperone for Yor1 by monitoring susceptibility to limited proteolysis, which reflects the folding status of the assembled protein [32, 33](Figure S1C). We treated membranes isolated from wild-type and erv14Δ erv15Δ strains expressing Yor1-HA with increasing concentrations of trypsin and examined the profile of HA-tagged protein fragments that resulted. No difference was apparent in Yor1 fragmentation, either at steady state or from membranes that had been radiolabeled to monitor newly synthesized Yor1-HA (Figure S1C), suggesting that Erv14 does not influence the global folding of Yor1.
We next explored the relationship between Yor1, Erv14 and Sec24 genetically by testing whether overexpression of Sec24 could compensate for lack of Erv14. We reasoned that if Erv14 functions as a cargo receptor to recruit the COPII coat, then excess Sec24 might drive ER export of Yor1 (via its diacidic motif) by mass action. Indeed, Sec24 driven by a copper-inducible promoter partially rescued the oligomycin sensitivity of an erv14Δ erv15Δ mutant (Figure 1E, left panels). Overexpression of Sec24 could simply increase the number of vesicles made, which might stochastically increase capture of Yor1. However, over-expression of the Sec24 B-site mutant failed to rescue the oligomycin sensitivity of the erv14Δ erv15Δ strain (Figure 1E, left panels), suggesting that specific cargo-coat interaction is important, and arguing against bulk flow. Similarly, Sec24 overexpression was unable to rescue the oligomycin-sensitive phenotype of a yor1Δ strain expressing a Yor1 mutant that lacks the DxE motif (Figure 1E, right panels) or various misfolded alleles of Yor1. We note that the Sec24 A-site mutant gave slightly better rescue than wild-type Sec24. Perhaps by depleting A-site specific cargo, additional space is available to accommodate more Yor1 under these partial rescue conditions. Our interpretation of the Sec24 overexpression experiments is that ER export of Yor1 can be driven by specific interaction with Sec24, but that Erv14 provides additional affinity between client and coat and thereby enhances export. In these conditions, we expect that excess Sec24 engages with Sec23, which is more abundant than Sec24, perhaps at the expense of Sec23-Iss1 and Sec23-Lst1 complexes.
COPII binding by Erv14 is universally required for client trafficking
If Erv14 promotes Yor1-Sec24 interaction, then Yor1 traffic should be impaired when the COPII-binding signal of Erv14 is mutated, as has been previously reported for the Erv14 client, Axl2 [21]. A cytosolic motif (IFRTL) in Erv14 is required for COPII binding and ER export of Erv14 [21] (Figure 2A, lower panel) and is at least partially conserved in Erv15 and other orthologs (Figure S2A). We examined the effect of mutating the Erv14-IFRTL motif in the context of either Erv14-GFP or Erv14-HA. The tags alone had no impact on Erv14 function as assessed by oligomycin resistance (Figure S2B). Mutation of the Erv14-IFRTL signal conferred oligomycin sensitivity, suggesting impaired Yor1 traffic (Figure S2C). Furthermore, in vitro packaging of Yor1 into COPII vesicles was compromised in the context of the Erv14-IFRTL mutant (Figure 2A, upper panel; Figure S2D).
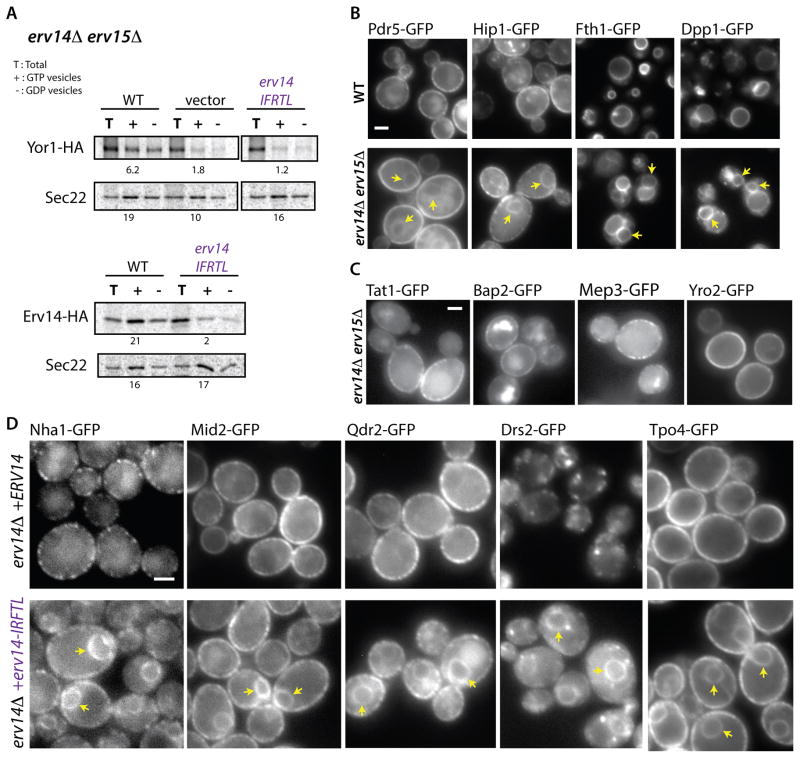
Figure 2. ER export of Erv14 clients relies on Erv14-Sec24 interaction.
(A) Packaging of Yor1-HA or Erv14-HA into COPII vesicles was examined in vitro from membranes isolated from the strain backgrounds indicated and as described in Figure 1D. Numbers indicate budding efficiency (% total). (B and C) Summary of the PAIRS analysis: wide-field micrographs of the indicated cargo proteins tagged with GFP, expressed in wild-type or erv14Δ erv15Δ cells (Scale bar = 2um). (B) Examples of novel Erv14 clients. Note that Fth1 and Dpp1 are vacuolar membrane proteins. Arrowheads indicate perinuclear ER (C) A selection of plasma membrane proteins that do not require Erv14 for ER export. (D) GFP-tagged versions of the indicated Erv14 clients were examined in an erv14Δ erv15Δ strain expressing either wild-type ERV14 (top panels) or erv14-IFTRL (lower panels). All proteins showed perinuclear localization (arrowheads) consistent with ER-retention in the presence of erv14-IFTRL (Scale bar=2μm). See also Figures S2 and S3.
We next tested the universal importance of COPII binding with respect to Erv14 function. We first sought to more fully define the client repertoire of Erv14, bearing in mind the functional redundancy between Erv14 and Erv15. We turned to the powerful PAIRS approach [22] to monitor the localization of 180 GFP-tagged membrane proteins (Supplementary Data File 1) in an erv14Δ erv15Δ background. We identified 16 additional proteins whose transport out of the ER was dependent on Erv14/Erv15 (Figure S3A). Mirroring previous findings [22, 23], the vast majority of new clients are polytopic transmembrane proteins that reside in the late secretory pathway, with a strong preference for the plasma membrane (Figure 2B, two left columns; Supplementary Data File 1), although other steady-state localizations such as vacuolar membrane are also represented (Figure 2B, two right columns). Importantly, approximately half of the plasma membrane proteome remained unaffected by the absence of Erv14/Erv15 (Figure S3B; Supplementary Data File 1). Erv14 independence was reconfirmed for several of these non-clients by PCR-mediated integration of the GFP tag at the C-terminus of the cargo proteins in a fresh erv14Δ erv15Δ strain, ruling out potential artifacts associated with the mating-based approach (Figure 2C). We conclude that there is a degree of specificity in Erv14 function, whereby only some polytopic proteins require assistance in leaving the ER. Importantly, expression of Erv14-IFRTL caused ER retention of all the clients we tested (Figure 2D), demonstrating that the ability of Erv14 to engage the COPII coat is universally essential for its function.
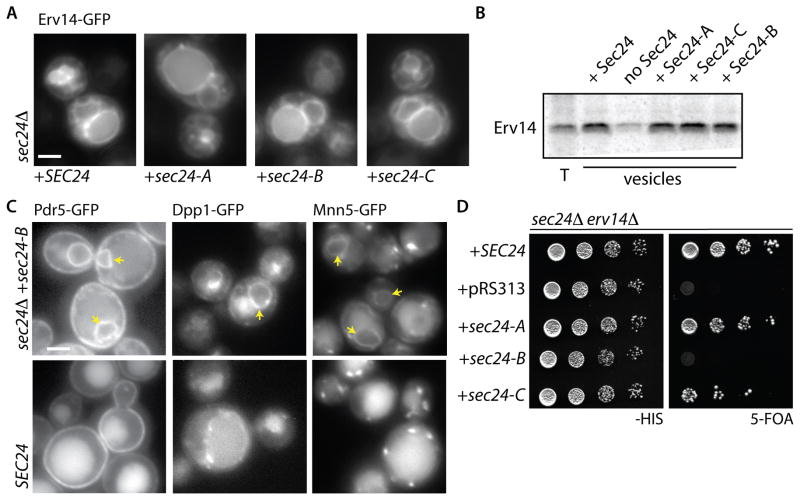
We next probed the relationship between Erv14 and Sec24, testing whether the 3 known binding surfaces on yeast Sec24 (the A-, B-, and C-sites) are responsible for traffic of Erv14. We expressed Erv14-GFP in strains expressing wild-type or mutant forms of Sec24 as the sole copy of this essential protein. In wild-type cells, Erv14-GFP was found in both the ER and the vacuole limiting membrane (Figure 3A, left panel). The proportion of Erv14-GFP in both the ER and vacuole was unchanged in any of the Sec24 mutant strains, suggesting that none of these sites contribute to ER export of Erv14 (Figure 3A). This conclusion was confirmed using the in vitro vesicle formation assay, where incorporation of Erv14-HA into COPII vesicles was comparable regardless of the form of Sec24 used (Figure 3B), suggesting that the IFRTL export motif of Erv14 engages Sec24 via an area distinct from the three known binding sites.
Figure 3. Erv14 engagement with the COPII coat is independent of known sites on Sec24 but a subset of Erv14 clients rely on the B-site for ER export.
(A) Erv14-GFP was expressed in a sec24Δ strain that co-expressed wild-type or cargo-binding mutants of SEC24 as indicated. In all strains Erv14-GFP was detected in both ER and vacuolar membranes with no obvious increase in the proportion of ER localized signal (Scale bar =2μm). (B) Capture of Erv14-HA into COPII vesicles was monitored in vitro as described for Figure 1D using wild-type or mutant versions of Sec24 as indicated. Erv14 was equally detected in the vesicle fractions regardless of the Sec24 variant used. (C) The localization of GFP-tagged clients was examined in presence of wild-type (lower panels) or cargo-binding mutants of SEC24 (top panels): Pdr5-GFP, Dpp1-GFP and Mnn5-GFP were ER-retained in cells expressing the B-site mutant of Sec24. Arrowheads indicate perinuclear ER (D) Yeast strains bearing deletions in SEC24 and ERV14 and expressing wild-type SEC24 from a URA3-marked plasmid were transformed with the indicated variants of SEC24 and serial dilutions were spotted onto medium containing 5-FOA, which counterselects for the URA3-marked wild-type version. The B-site mutant of Sec24 was unable to sustain growth as sole copy when ERV14 was deleted. See also Figure S4.
Given that some Erv14 clients possess sorting signals, we tested how widespread this phenomenon might be by examining localization of a variety of Erv14 clients in Sec24 mutant backgrounds. The majority of clients did not use any of the defined sites on Sec24 (Figure S4A). However, a small number of Erv14 clients were ER-retained in the Sec24 B-site mutant, similar to Yor1 [27]. These included functionally diverse clients, Pdr5, Dpp1 and Mnn5 (Figure 3C). Thus, for a subset of Erv14 clients, efficient capture into COPII vesicles is driven by both a direct interaction with the Sec24 B-site and indirect contact with the coat via the Erv14-IFRTL motif, which engages a different region of Sec24. For those Erv14 clients that do not engage the known sites on Sec24, it remains to be determined whether ER export is mediated solely by Erv14 or if they also engage unidentified Sec24 sites (see Figure 7).
Figure 7. Sec24 as a coincidence detector that binds two signals to drive ER export.
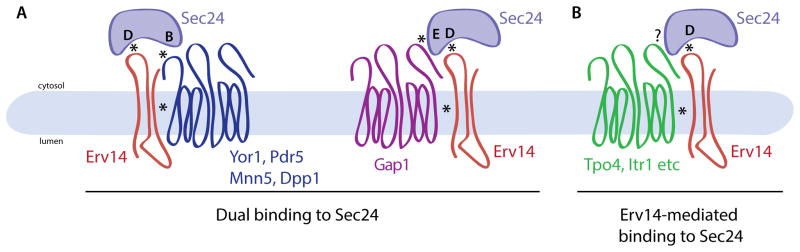
(A) Yor1 and other clients interact directly with the Sec24 B-site and indirectly with the Sec24 D-site via the IFRTL motif of Erv14 (left). Gap1 also employs Erv14 but binds to Sec24 via an unknown additional E-site (right). (B) Some Erv14 clients (e.g. Tpo4, Itr1, etc.) do not use known sites on Sec24, but may employ unknown sites on Sec24 or may rely exclusively on Erv14-Sec24 interactions to drive export.
If a subset of Erv14 clients relies on dual interaction with both Sec24 and Erv14, then combined defects in both Sec24 cargo-binding and Erv14 function should have a phenotypic cost. We tested this by deleting ERV14 in strains expressing cargo-binding mutant forms of SEC24. Each of these mutants is permissive for growth when present as the sole copy of Sec24 (Figure S4B, left panel), but the Sec24 B-site mutant could not support viability in the absence of Erv14 (Figure 3D). Conversely, there was no phenotypic cost when the receptor for soluble cargo proteins, Erv29, was deleted (Figure S4B, right panel). Our interpretation of this specific genetic interaction is that the combined abrogation of both Erv14 and the Sec24 B-site causes a significant deficit in ER export of membrane proteins such that viability is not supported.
Erv14 binds a novel site on the membrane-proximal surface of Sec24
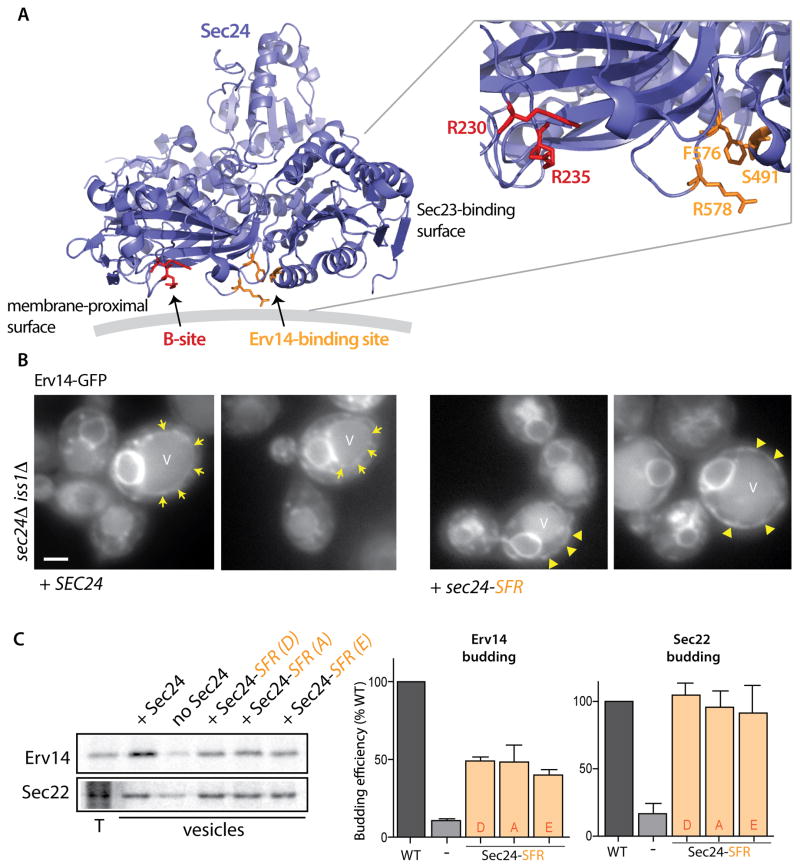
To identify residues on Sec24 that might contribute to Erv14 recognition, we mutated surface-exposed residues on Sec24 and screened for phenotypes that might indicate Yor1 trafficking defects. We identified three residues (S491, F576 and R578) that form a patch on the membrane-proximal face of Sec24 (Figure 4A), adjacent to the B-site [10]. Mutation of all three residues, which delineate what we now call the D-site, to alanine rendered cells sensitive to oligomycin, consistent with a Yor1 trafficking defect (Figure S5A). Of these residues, F576 and R578 seem to be most important since the double mutation alone was significantly oligomycin sensitive (Figure S5A). We examined the localization of Erv14-GFP in a strain that expressed the Sec24-SFR triple mutant in the absence of both Sec24 and its close paralog, Iss1/Sfb2 since these residues are conserved between the two isoforms, and indeed are also conserved in human Sec24a/b isoforms. In the presence of wild-type Sec24, Erv14-GFP localized both to the ER and the vacuole membrane (Figure 4B, arrows denote vacuole limiting membrane; Figure S5B), whereas in the presence of sec24-SFR, Erv14-GFP was depleted from the vacuole membrane and was more abundant in cortical ER (Figure 4B, arrowheads denote cortical ER; Figure S5C). To confirm Erv14 trafficking defects, we purified the mutant forms of Sec24 and used them in in vitro budding reactions. In the presence of the mutant forms of Sec24, Erv14 was less abundant in the vesicle fraction, consistent with a role for this site in recognizing Erv14 (Figure 4C). The residual packaging of Erv14 indicates the possible contribution of additional residues on the surface of Sec24, which remain to be identified via structural analysis. Importantly, a control protein, Sec22, was unaffected by any of the mutations that affect the D-site of Sec24, (Figure 4C).
Figure 4. Identification of a novel binding site on Sec24 required for ER exit of Erv14.
(A) Ribbon diagram of Sec24 showing the location of the B-site (Arg230, Arg235: red) and a new site (Ser491, Phe576 and Arg578: orange) involved in Erv14 packaging. This figure was generated using PyMOL (http://www.pymol.org). The gray line represents the lipid bilayer. (B) Erv14-GFP localization was monitored by epifluorescence microscopy in sec24Δ iss1Δ strains expressing wild-type SEC24 (left panels, arrows indicate vacuolar membrane) or the sec24-SFR mutant (right panel, arrowheads indicate cortical ER) as the sole copy (Scale bar =2μm). (C) Incorporation of Erv14 into COPII vesicles was monitored as described in Figure 1D using wild-type or mutant versions of Sec24 (S491D F576A R578A, S491A F576A R578A or S491E F576A R578A) as indicated. Quantification (right panel; n=3; errors bars represent SD) showed that the abundance of Erv14-HA in the vesicle fraction was reduced in the context of mutant versions of SEC24, where Sec22 was unaffected. See also Figure S5.
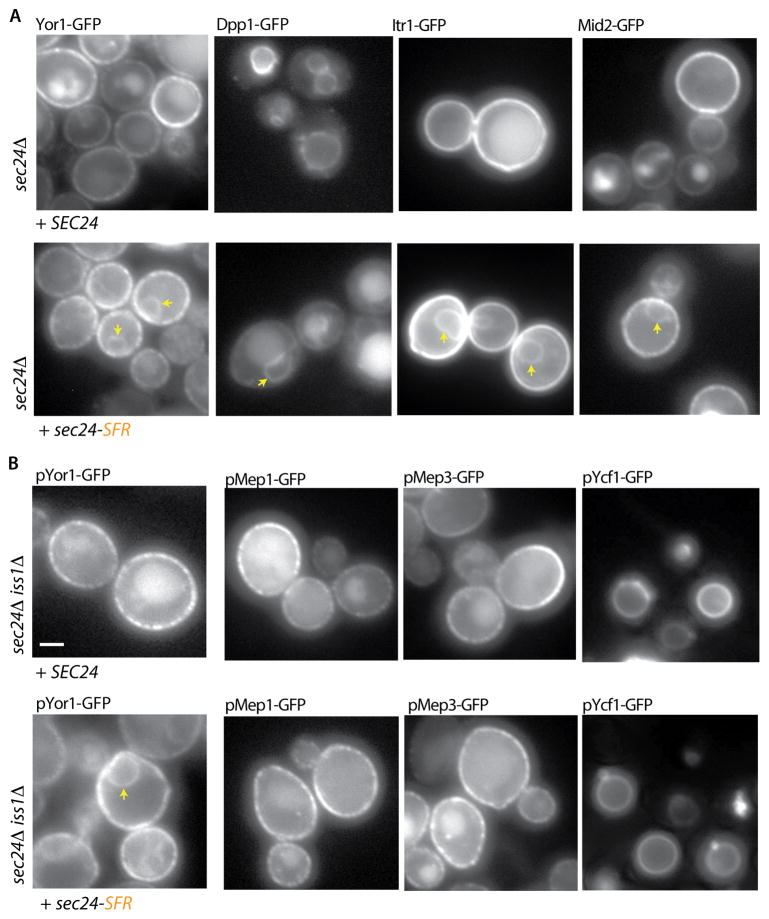
Consistent with our model that the Sec24 S491, F576 and R578 residues correspond to the Erv14 binding site, all Erv14 clients that we tested were ER-retained when this mutant was the sole copy of Sec24 (Figure 5A). This included both B-site Sec24 clients like Yor1-GFP and Dpp1-GFP and B-site independent cargoes like Itr1-GFP and Mid2-GFP, suggesting that the Erv14-binding site is not simply indirectly affecting the B-site. Importantly, non-Erv14 clients were unaffected by this mutation (Figure 5B). We tested localization of plasmid-borne copies of a subset of plasma membrane proteins that do not require Erv14 for traffic in a sec24Δ iss1Δ double mutant, finding that pMep1-GFP, pMep3-GFP and pYcf1-GFP were normally localized when Sec24-SFR was expressed (Figure 5B). If this new site on Sec24 corresponds to the Erv14-binding site, then we should observe synthetic lethality when this site is combined with B-site mutations, similar to the phenotype described above (Figure 3D). Indeed, when the F576A and R578A mutations were combined with R230A/R235A mutations, cells grew poorly at all growth temperatures (Figure S6A). Together, these data support a model whereby Erv14 binds to a novel cargo-interaction site on the membrane-proximal surface of Sec24 to mediate efficient ER export of its full repertoire of clients. For a subset of clients, interaction with the Sec24 B-site is also required for export, positioning Sec24 as a coincidence detector that binds simultaneously to signals that derive from both Erv14 and its client. In support of coincident cargo binding at the B- and D-sites, we observed cross-complementation of the Sec24-B and Sec24-D site mutants: cells were viable when individual B- and D-site mutants were expressed on separate plasmids, suggesting that trafficking is only impaired when these lesions occur on the same protein (Figure S6B).
Figure 5. Mutation of the Erv14 binding site on Sec24 selectively impairs ER export of Erv14 clients.
(A) GFP-tagged clients were introduced into a sec24Δ strain expressing expressing wild-type SEC24 (top panels) or the sec24-SFR mutant. Perinuclear ER retention (arrowheads) was detected for all cargo proteins (Scale bar =2μm). (B) Plasmids bearing GFP-tagged versions of Yor1, Mep1, Mep3 or Ycf1 were introduced into sec24Δ iss1Δ strains expressing wild-type SEC24 (top panels) or the sec24-SFR mutant (lower panel): Yor1-GFP was detected in the perinuclear ER (arrowhead) in the presence of the sec24-SFR mutant whereas the localization of Mep1-GFP, Mep3-GFP and Ycf1-GFP was comparable to that observed with wild-type SEC24. See also Figure S6.
The second transmembrane domain of Erv14 contains a specific cargo-binding site
We next sought to dissect the mechanism by which Erv14 binds its clients. We reasoned that residues important for Erv14 function should be conserved between the two yeast paralogs and among their homologs in different kingdoms (Figure S2A). We generated a series of point mutants (Table S2) in constructs expressing Erv14-HA or Erv14-GFP, identifying sites that impacted oligomycin sensitivity (Figure S2C). None of these mutations destabilized Erv14 at steady state, suggesting that they don’t globally induce misfolding (Figure S2F). The lumenal Erv14 mutant, erv14-DYPE, showed intermediate incorporation into ER-derived vesicles, with a ~50% reduction compared to wild-type (Figure S2E), which parallels the weaker oligomycin phenotype and Yor1 export defect observed for this particular allele (Figures S2C and S2D, respectively). Conversely, mutation of three conserved residues in the second TMD, F62, L63 and N74 (referred to as FLN), dramatically impaired Yor1-HA packaging in vitro leaving Erv14-HA packaging unaffected (Figures S2D and S2E), suggesting that this site modulates Yor1 export independent of Erv14 ER egress.
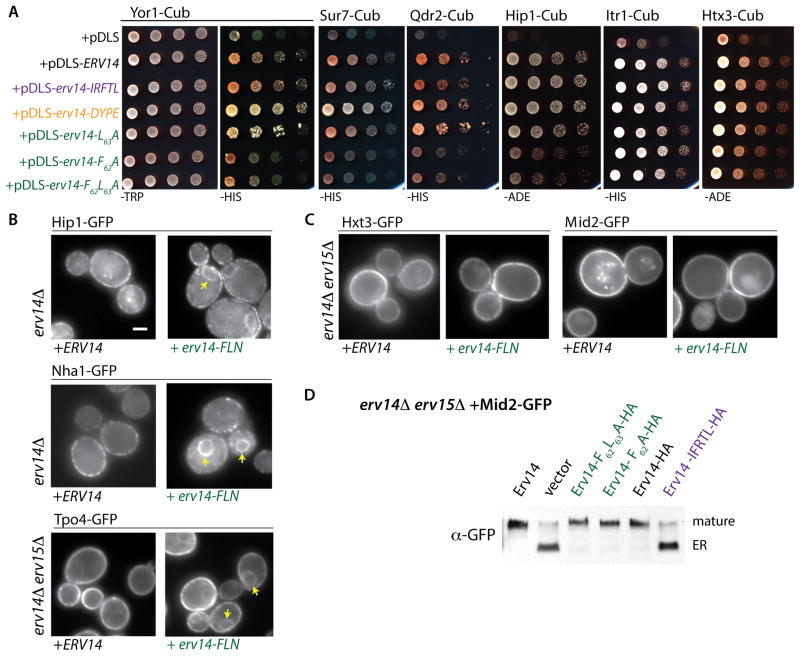
One obvious potential role for the FLN residues of Erv14 is as a cargo-binding site. We sought to test this by monitoring Erv14-client interaction using the Membrane Yeast Two-Hybrid (MYTH) approach [34]. The chromosomal copies of various clients were tagged with a C-terminal fragment of ubiquitin (Cub) followed by a transcription factor, and plasmids bearing fusions of the N-terminal ubiquitin fragment with various forms of Erv14 (Nub-Erv14) were introduced into these strains [35]. Physical proximity of the bait and prey proteins permits assembly of ubiquitin that is proteolytically cleaved, liberating the transcription factor and driving expression of selectable markers (histidine or adenine prototrophy in this case). Relative to a vector control, Erv14 and Yor1 showed robust interaction that was not perturbed by mutations in either the COPII-binding IFRTL motif or the lumenal DYPE site (Figure 6A). Conversely, mutation of the TMD2 residues, F62 and L63, dramatically reduced the interaction between Yor1-Cub and Nub-Erv14 (Figure 6A). Similar effects were observed for additional Erv14 clients, Sur7-Cub, Qdr2-Cub and Hip1-Cub, with the TMD2 mutants specifically abolishing interaction (Figure 6A). In contrast, these TMD2 mutations did not cause any detectable growth defect when co-expressed with Itr1-Cub and Hxt3-Cub (Figure 6A), which suggests that Erv14 TMD2 does not drive interaction with these clients.
Figure 6. Mutations in TMD2 of Erv14 impair binding with a subset of clients.
(A). Interactions between Erv14 and its clients were analyzed using the membrane yeast two-hybrid split ubiquitin system. The indicated clients were tagged with the CYT (Cub-(YFP)LexA-VP16) cassette in the yeast strain THY AP4 (lexA::HIS3 lexA::ADE2). Each client-Cub strain was subsequently transformed with plasmids expressing the indicated versions of ERV14 tagged with Nub. Serial dilutions were spotted onto SD-TRP plates (control) and SD-HIS or SD-ADE to test for interaction. Nub-ERV14, Nub-erv14-IFTRL and Nub-erv14-DYPE conferred equivalent interaction on selective media whereas the TMD2 mutations, F62A and F62AL63A, both decreased interaction with Yor1-CYT, Sur7-CYT, Qdr2-CYT and Hip-CYT, but not with Itr1-CYT or Hxt3-CYT. (B and C) Wide-field fluorescence micrographs of Tpo4-GFP, Nha1-GFP and Mid2-GFP expressed in erv14Δ or erv14Δ erv15Δ strains expressing the indicated variants of ERV14. Arrowheads delineate perinuclear ER (Scale bar =2μm). (D) Anti-GFP immunoblots of whole cell lysates from erv14Δ erv15Δ strains co-expressing Mid2-GFP and the indicated Erv14 variants. Mid2 undergoes post-ER glycosylation resulting in a slowly migrating species (mature). Expression of erv14-IFRTL caused accumulation of the ER form of Mid2 (ER). In contrast, no maturation defects were observed for the erv14-F62A and erv14-F62AL63A mutants. See also Figure S2, Table S2 and Table S3.
We confirmed the differential TMD2-dependent interaction between Erv14 and its clients by immunoprecipitating HA-tagged Erv14 from digitonin-solubilized microsomal membranes expressing Erv14-HA or Erv14-F62AL63A-HA. To improve client recovery with each construct, the COPII binding signal was also mutated to enrich for ER-retained Erv14 [21]. Mass spectrometry of the precipitated material verified that the TMD2 mutation impairs binding with a subset of Erv14 clients (Table S3). Furthermore, we observed differential effects of the TMD2 mutations on the localization of various clients, with impaired ER export of Hip1-GFP, Tpo4-GFP and Nha1-GFP (Figure 6B) but normal localization of Hxt3-GFP and Mid2-GFP (Figure 6C). Mid2-GFP traffic was also assessed by immunoblot, which permits the ER-retained protein to be distinguished from the larger Golgi-modified form [22]. In an erv14Δ erv15Δ strain, Mid2-GFP is predominantly in the ER form; the TMD2 mutant form of Erv14 rescues Golgi modification to the same extent as wild-type Erv14, whereas the COPII binding mutant form of Erv14 was unable to restore traffic (Figure 6D). Taken together, these data indicate that TMD2 of Erv14 is responsible for the mobilization of multiple cargo clients, and likely corresponds to a direct site of interaction with many, but not all, clients.
Discussion
Having recently identified Erv14 as regulator of ER export of Yor1 [29], we sought to understand why a membrane protein with its own ER export signal also requires a cargo receptor. Our data position Erv14 as classical cargo receptor, binding to both cargo proteins and the COPII coat to maximize efficient ER export (Figure 7). The COPII-binding motif of Erv14 [21] is necessary for efficient trafficking of all of its clients, and is recognized by a site on Sec24 that specifically impacts Erv14 clients when mutated. The positioning of the Erv14-binding site on the membrane-proximal surface of Sec24 adjacent to the B-site supports a model of dual Sec24 interaction with both Erv14- and cargo-borne signals. We therefore propose a novel mechanism for Sec24 action, whereby coincident detection of multiple export signals is required for efficient capture into COPII vesicles.
Coat adaptors in other vesicle trafficking events also function as coincidence detectors, although there are some differences between these examples and the Sec24 system we describe here. In the case of the AP-2 adaptor complex that drives clathrin-mediated endocytosis, AP-2 subunits bind simultaneously to inositol phospholipids and endocytic sorting signals [36]. This dual interaction occurs following large conformational changes in the AP-2 holocomplex, and different polypeptide subunits of the AP-2 complex contribute distinct binding specificities [37]. Coincident detection of lipid-based signals and protein-based sorting motifs has also been identified for sorting nexins that mediate endosomal traffic [38]. The dual signal recognition by Sec24 that we describe here is distinct in that it is mediated by two protein-based signals on cargo proteins themselves. Furthermore, the two sorting signals interact with a single coat adaptor that is thought to function as a static cargo-binding platform rather than an allosteric coat component [10, 13].
A dual requirement for interaction with both Erv14 and Sec24 is not unique to Yor1 and the other Erv14/Sec24 clients identified here (Mnn5, Pdr5 and Dpp1). Vesicular stomatitis virus glycoprotein (VSV-G) employs the Erv14 ortholog, CNIH4, for normal ER exit [39] in addition to a di-acidic motif on its cytoplasmic tail [40]. Furthermore, a requirement for dual signals might extend to additional Erv14 clients since either disruption of Erv14/Erv15 or abrogation of export signals confers an incomplete block in ER export [40–42], The positioning of the Erv14-binding site on a central region of Sec24 seems compatible with simultaneous binding of cargo to additional surfaces that remain to be characterized. Indeed, yeast Gap1 depends on both Erv14 [23] and its own sorting motif [5], which interacts with an unknown site on Sec24 [4]. In our analysis, Gap1 would thus be mistakenly characterized as relying exclusively on Erv14 for ER export. Membrane proteins that are not Erv14 clients might possess two independent ER export signals, or may use higher affinity interactions with Sec24 that preclude the need for the additional action of Erv14. The advantage of presenting the coat with multiple signals may stem from the relatively weak affinities between Sec24 and its cargo [10]. Similar principles may apply to membrane proteins that oligomerize prior to ER export; assembly of monomers would serve to concentrate and orient a simple sorting signal in order to maximize COPII binding. ERGIC-53 conforms to this model, where hexamerization is driven by lumenal and TMD determinants that drive orientation of a cytosolic ER export signal [43].
A requirement for multiple COPII signals might also contribute to additional aspects of polytopic membrane protein biogenesis. For example, Erv14 could ensure protein quality control by specifically recognizing properly assembled TMDs, which would then trigger engagement of the COPII coat and ER export. Such context-dependent use of ER export signals occurs in yeast, where traffic of the Erv41/Erv46 complex depends on the correct positioning of both COPII binding signals [44]. An important aspect of polytopic membrane protein biogenesis is the physical environment of the ER membrane: membrane proteins that reside in the late secretory pathway, as most Erv14 clients do, possess longer TMDs than ER resident proteins [28]. The physical requirements associated with driving proteins with long hydrophobic spans into a curved nascent vesicle membrane might impose an extra energetic burden on the coat system and thereby require additional avidity for the coat. Interestingly, Erv14 appears to employ multiple modes of cargo binding: we identified a discrete site of client interaction contained within TMD2 of Erv14, whereas other clients seem to bind via lumenal domains [26] or by the physical length of the client TMD [22]. These different binding modes might reflect the different needs of the various clients, making Erv14 a truly multifunctional “escort” that not only couples cargo to the COPII coat but also promotes selection of distinct client proteins [45].
Our dissection of Sec24-Erv14-client interactions sheds new light on the complexity of ER export signals and the different strategies that have evolved to accommodate efficient sorting of the wide variety of secretory cargoes. Further mechanistic investigation of the interactions between Sec24, Erv14 and its clients will be central to dissecting how ER export adapts to the diversity of cargo that must navigate the early secretory pathway.
Materials and Methods
Yeast growth, strain and plasmid construction
Yeast cultures were grown at 30°C in standard rich media (YPD: 1% yeast extract, 2% peptone, and 2% glucose) or synthetic complete media (SC: 0.67% yeast nitrogen base and 2% glucose, supplemented with amino acids appropriate for auxotrophic growth). When needed G418 (Calbiochem) or Nourseothricin (WERNER BioAgents) were added to 200 μg/ml. In cases where G418 was required in a SD-based medium, yeast nitrogen base without ammonium sulfate (Conda Pronadisa) and mono-sodium glutamate (Sigma-Aldrich) were used as nitrogen source. For testing sensitivity to oligomycin, tenfold serial dilutions of saturated cultures were applied to YPEG plates (1% yeast extract, 2% peptone, 3% ethanol, and 3% glycerol) supplemented with oligomycin (final concentrations from 0.2 μg/ml to 0.4 μg/ml; Sigma-Aldrich). Genetic modifications at the CAN1 locus were selected on SD-ARG medium supplemented with canavanine (50 μg/ml; Sigma-Aldrich). For strain and plasmid construction, please see Supplemental Experimental Procedures.
Live Cell Imaging
Cells were grown in liquid media to mid-log phase. Images were taken on a Zeiss AxioImager Z1 upright fluorescent microscope with a 100x 1.4 NA oil immersion objective (Carl Zeiss Ltd.). Images were collected using AxioVision Rel software (Carl Zeiss Ltd.) and processed with Adobe Photoshop.
Secretome analysis and Robotic Manipulations
All genetic manipulations were performed using SGA techniques as previously described [22]. Briefly, the query strains SPY81 or SPY82, in some cases bearing plasmids containing ERV14 wild-type or mutants, were mated on rich media plates with strains selected from the yeast GFP library [46]. After diploid selection, sporulation was induced by transferring cells to nitrogen starvation plates. Haploid cells containing all desired mutations were selected on plates containing all markers alongside the toxic amino acid derivatives canavanine (Sigma-Aldrich) to select against remaining diploids. The SGA procedure was conducted using a RoToR bench-top colony arrayer (Singer Instruments).
Limited Proteolysis
Yor1 conformation was analyzed as described previously [27]. Briefly wild type or erv14Δ erv15Δ mutant cells expressing wild-type Yor1-HA were harvested during mid-log phase, resuspended in buffer B88 (20 mM HEPES, pH 6.8, 250 mM sorbitol, 150 mM KOAc, 5 mM MgOAc) and divided into four 25-μl reactions (2.5 OD600/reaction). Each reaction was treated with a final concentration of 0, 25, 50, or 100 ng/μl trypsin (Sigma-Aldrich) for 10 min on ice. Digestion was terminated by addition of 0.2 μg/ml (final concentration) soybean trypsin inhibitor (Sigma-Aldrich) and incubated on ice for 15 min. Proteins were separated by SDS-PAGE and the pattern of Yor1 fragments analyzed by immmunoblot by using an anti-HA antibody. For analysis during early stages of Yor1 biogenesis, radiolabeled semi-intact cells were used as the source of membranes [47]; spheroplasts were treated with trypsin as described above, and Yor1 fragments were immunoprecipitated and analyzed by SDS-PAGE and PhosphorImage.
In Vitro Vesicle Budding
Sar1, Sec23/24, and Sec13/31 were prepared as described previously [1]. Radiolabeled semi-intact cells were prepared as described previously [47]. Briefly, cells were grown to mid-log phase prior to Met/Cys starvation (10 min) and addition of Express protein labeling mix (~70μCi/OD600 of cells; MP Biomedicals). Cells were labeled for 15 min at 30°C, then killed and converted to spheroplasts, which were washed once with low acetate B88 (50 mM KOAc,) and twice with B88 before incubation with COPII proteins (10 μg/ml Sar1, 10 μg/ml Sec23/24, and 20 μg/ml Sec13/31) either in the presence of 0.1 mM GTP with a 10× ATP regeneration system or 0.1 mM GDP. Vesicles were separated from donor membranes by centrifugation at 16,000 rpm (5 min), solubilized with 1% SDS (final concentration), and cargo proteins were immunoprecipitated and analyzed by SDS-PAGE and PhosphorImage.
Erv14 Co-immunoprecipitation and mass spectrometry
Microsomal membranes were purified as described [1] from strains expressing HA-tagged forms of Erv14-IFRTL and Erv14-IFRTL-FL. An untagged strain served as a negative control. Membranes (500 μg total membranes) were resuspended in 70mM Tris, pH 7.5, 210mM NaCl and solubilized with digitonin (1.5% final concentration) for 30 minutes on ice. Insoluble material was removed by centrifugation and the lysate incubated with anti-HA resin (Sigma-Aldrich) for 60 minutes at 4°C. IPs were washed with Tris/NaCl/digitonin and proteins analyzed by SDS-PAGE on Tris-Tricine gels followed by Colloidal Coomassie staining. Gel lanes were excised in 1 mm slices and subjected to tryptic digest and mass spectrometry. Mass spectrometry data were analyzed using Scaffold software.
Supplementary Material
Figure S1 (related to Figure 1). Both Erv14 and Erv15 are necessary for Yor1 trafficking but do not contribute to Yor1 folding. (A) The phenotype of erv14Δ or erv14Δ erv15Δ mutants expressing ERV14 or ERV15 under the control of the ERV14 promoter was tested by serial dilutions onto YPEG + oligomycin (0.4 μg/ml). (B) Incorporation of Yor1 into COPII vesicles was measured from membranes isolated from erv14Δ erv15Δ cells that express either ERV14 or ERV15 under the control of the ERV14 promoter as indicated. (C) Microsomal membranes (left panel) or radiolabelled semi intact cells (center panel) isolated from wild-type or erv14Δ erv15Δ cells expressing wild-type Yor1-HA were treated with increasing amounts of trypsin followed by SDS-PAGE and detection of HA-tagged proteolytic fragments. Misfolding of Yor1 results in the appearance of multiple lower molecular weight bands, as observed for a steady-state experiment using a misfolded allele of Yor1-HA (Yor1*-HA; right panel).
Figure S2 (related to Figure 2 and Figure 6). Structure-function analysis of Erv14 (A) Multiple sequence alignment between Erv14 and Erv15 from Saccharomyces cerevisiae (Sc) and their Cornichon orthologs in human (Hs), mouse (Mm) and Drosophilia (Dm), generated by Multalin (http://npsapbil.ibcp.fr/cgibin/npsa_automat.pl?page=/NPSA/npsa_multalin.html). Conserved and similar residues are indicated in red and green, respectively. Boxes delimit the positions of three TMDs previously defined by Powers et al. (2002). The purple stars specify the residues analyzed in this study. (B) Ten-fold serial dilutions of erv14Δ erv15Δ cells expressing untagged, HA- or GFP-tagged versions of ERV14 from a CEN plasmid or untagged ERV14 from a 2μ plasmid (o/e) were tested for sensitivity to the indicated concentrations of oligomycin. Addition of HA and GFP epitopes at the C-terminus of Erv14 did not influence complementation of the oligomycin sensitivity displayed by the erv14Δ erv15Δ strain. Over-expression of ERV14 accentuated drug sensitivity. (C) Cultures of erv14Δ erv15Δ cells bearing the plasmids indicated were spotted onto media containing increasing concentrations of oligomycin (0, 0.2 μg/ml and 0.3 μg/ml). Neither the cytosolic mutant (erv14-IFTRL) nor the TMD2 mutant (erv14-FLN) could complement oligomycin sensitivity. The lumenal mutant (erv14-YPE) showed an intermediate phenotype. (D) Incorporation of Yor1-HA into COPII vesicles was assayed as described in Fig. 1 from membranes isolated from the strains indicated. Quantification (right panel; n=3; error bars represent SD) showed that the capture of both Yor1-HA (D) and Erv14-HA (E) into the vesicle fraction was inhibited by mutation of the Erv14 IFTRL signal. Conversely, the abundance of Yor1-HA in COPII vesicles was perturbed in the context of erv14-FLN whereas packaging of erv14-FLN-HA itself was unaffected compared to wild-type ERV14-HA. Membranes expressing erv14-DYPE were partially affected both for Yor1 capture and Erv14 capture. (E) Anti-HA immunoblots of whole cell lysates expressing the indicated Erv14 variants.
Figure S3 (related to Figure 2). Erv14 traffics a subset of plasma membrane proteins. (A and B) Wide-field fluorescent micrographs of the indicated cargo proteins tagged with GFP, expressed in wild-type or erv14Δ erv15Δ cells (Scale bar =2μm). (A) Novel Erv14 clients identified in this study. (B) Plasma membrane proteins whose trafficking is independent of Erv14.
Figure S4 (related to Figure 3). Most Erv14 clients do not use known Sec24 binding sites. (A) Localization of GFP-tagged versions of the indicated Erv14 clients were examined by epifluorescence microscopy in sec24Δ strains expressing the cargo-binding site sec24 mutants indicated. None of the Sec24 mutants impaired cell surface localization of Sur7-GFP, Tpo4-GFP, Itr1-GFP, Mid2-GFP and Hxt6-GFP (Scale bar =2μm). (B) The indicated variants of SEC24 were introduced into sec24Δ or sec24Δ erv29Δ cells expressing wild-type SEC24 from a URA3-marked plasmid and serial dilutions of the transformants were spotted onto medium containing 5-FOA, which counterselects for the URA3-marked plasmid. All SEC24 mutants were viable in both sec24Δ and sec24Δ erv29Δ strains.
Figure S5 (related to Figure 4). Characterization of the Erv14 binding site (Ser491, Phe576, Arg578) on Sec24. (A) Strains expressing the indicated variants of SEC24 as the sole copy were spotted on media containing oligomycin (0.4 μg/ml) to test for Yor1 function at the plasma membrane. (B and C) Erv14-GFP localization was monitored by epifluorescence microscopy in sec24Δ iss1Δ strains expressing (B) wild-type SEC24 or (C) the sec24-SFR mutant (Scale bar =2μm).
Figure S6 (related to Figure 5). Phenotypic characterization of the Erv14 binding site (Ser491, Phe576, Arg578) on Sec24. (A) A sec24Δ strain expressing wild-type SEC24 from a URA3-marked plasmid was transformed with the indicated variants of SEC24 and serial dilutions were spotted onto medium containing 5-FOA, which counterselects for the URA3-marked wild-type version. A variant of Sec24 bearing mutations that affect both the B-site (Arg230 and Arg235) and the Erv14-binding site (Phe578 and Arg578) was unable to sustain growth at 30°C. (B) A sec24Δ strain co-expressing wild-type SEC24 on a URA3-marked plasmid and the B-site mutant sec24-R230A R235A on a HIS3-marked plasmid was transformed with the indicated variants of SEC24 on a LEU2-marked plasmid and viability was tested after counterselection of the URA3-marked wild-type in presence of 5-FOA. No growth defect could be observed upon co-expression of Sec24 mutants independently affected for the B-site or for the Erv14-binding site.
Table S2 (related to Figure 6) – Oligomycin phenotypes of ERV14 mutants
Table S3 (related to Figure 6)– Mass spectrometry of Erv14-HA co-IPs
Highlights.
ER export is driven by sorting signals that specify capture into COPII vesicles.
Erv14 is a cargo receptor that couples many membrane proteins to the COPII coat.
Erv14 binds to a novel site on the COPII coat adaptor, Sec24.
Sec24 simultaneously binds signals that derive from both Erv14 and its clients.
Acknowledgments
We thank Sean Munro, Manu Hegde and members of the Hegde lab for support and advice in relation to the mass spectrometry experiments, and Mark Skehel for performing mass spectrometry analysis. Research reported in this publication was supported by NIGMS of the National Institutes of Health under award numbers R01GM078089 and R01GM085089 (E.A.M).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Barlowe C, Orci L, Yeung T, Hosobuchi M, Hamamoto S, Salama N, Rexach MF, Ravazzola M, Amherdt M, Schekman R. COPII: A membrane coat formed by Sec proteins that drive vesicle budding from the endoplasmic reticulum. Cell. 1994;77:895–907. doi: 10.1016/0092-8674(94)90138-4. [DOI] [PubMed] [Google Scholar]
- 2.Thor F, Gautschi M, Geiger R, Helenius A. Bulk flow revisited: transport of a soluble protein in the secretory pathway. Traffic. 2009;10:1819–1830. doi: 10.1111/j.1600-0854.2009.00989.x. [DOI] [PubMed] [Google Scholar]
- 3.Martinez-Menarguez JA, Geuze HJ, Slot JW, Klumperman J. Vesicular tubular clusters between the ER and Golgi mediate concentration of soluble secretory proteins by exclusion from COPI-coated vesicles. Cell. 1999;98:81–90. doi: 10.1016/S0092-8674(00)80608-X. [DOI] [PubMed] [Google Scholar]
- 4.Miller EA, Beilharz TH, Malkus PN, Lee MC, Hamamoto S, Orci L, Schekman R. Multiple cargo binding sites on the COPII subunit Sec24p ensure capture of diverse membrane proteins into transport vesicles. Cell. 2003;114:497–509. doi: 10.1016/s0092-8674(03)00609-3. [DOI] [PubMed] [Google Scholar]
- 5.Malkus P, Jiang F, Schekman R. Concentrative sorting of secretory cargo proteins into COPII-coated vesicles. J Cell Biol. 2002;159:915–921. doi: 10.1083/jcb.200208074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Otsu W, Kurooka T, Otsuka Y, Sato K, Inaba M. A new class of endoplasmic reticulum export signal PhiXPhiXPhi for transmembrane proteins and its selective interaction with Sec24C. J Biol Chem. 2013;288:18521–18532. doi: 10.1074/jbc.M112.443325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sieben C, Mikosch M, Brandizzi F, Homann U. Interaction of the K(+)-channel KAT1 with the coat protein complex II coat component Sec24 depends on a di-acidic endoplasmic reticulum export motif. The Plant journal: for cell and molecular biology. 2008;56:997–1006. doi: 10.1111/j.1365-313X.2008.03658.x. [DOI] [PubMed] [Google Scholar]
- 8.Farhan H, Reiterer V, Korkhov VM, Schmid JA, Freissmuth M, Sitte HH. Concentrative export from the endoplasmic reticulum of the gamma-aminobutyric acid transporter 1 requires binding to SEC24D. J Biol Chem. 2007;282:7679–7689. doi: 10.1074/jbc.M609720200. [DOI] [PubMed] [Google Scholar]
- 9.Sucic S, El-Kasaby A, Kudlacek O, Sarker S, Sitte HH, Marin P, Freissmuth M. The serotonin transporter is an exclusive client of the coat protein complex II (COPII) component SEC24C. J Biol Chem. 2011;286:16482–16490. doi: 10.1074/jbc.M111.230037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mossessova E, Bickford LC, Goldberg J. SNARE selectivity of the COPII coat. Cell. 2003;114:483–495. doi: 10.1016/s0092-8674(03)00608-1. [DOI] [PubMed] [Google Scholar]
- 11.Miller EA, Liu Y, Barlowe C, Schekman R. ER-Golgi Transport Defects Are Associated with Mutations in the Sed5p-binding Domain of the COPII Coat Subunit, Sec24p. Mol Biol Cell. 2005 doi: 10.1091/mbc.E05-03-0262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mancias JD, Goldberg J. The transport signal on Sec22 for packaging into COPII-coated vesicles is a conformational epitope. Mol Cell. 2007;26:403–414. doi: 10.1016/j.molcel.2007.03.017. [DOI] [PubMed] [Google Scholar]
- 13.Mancias JD, Goldberg J. Structural basis of cargo membrane protein discrimination by the human COPII coat machinery. EMBO J. 2008;27:2918–2928. doi: 10.1038/emboj.2008.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geva Y, Schuldiner M. The back and forth of cargo exit from the endoplasmic reticulum. Curr Biol. 2014;24:R130–136. doi: 10.1016/j.cub.2013.12.008. [DOI] [PubMed] [Google Scholar]
- 15.Dancourt J, Barlowe C. Protein sorting receptors in the early secretory pathway. Annu Rev Biochem. 2010;79:777–802. doi: 10.1146/annurev-biochem-061608-091319. [DOI] [PubMed] [Google Scholar]
- 16.Belden WJ, Barlowe C. Role of Erv29p in collecting soluble secretory proteins into ER-derived transport vesicles. Science. 2001;294:1528–1531. doi: 10.1126/science.1065224. [DOI] [PubMed] [Google Scholar]
- 17.Appenzeller C, Andersson H, Kappeler F, Hauri HP. The lectin ERGIC-53 is a cargo transport receptor for glycoproteins. Nat Cell Biol. 1999;1:330–334. doi: 10.1038/14020. [DOI] [PubMed] [Google Scholar]
- 18.Nichols WC, Seligsohn U, Zivelin A, Terry VH, Hertel CE, Wheatley MA, Moussalli MJ, Hauri HP, Ciavarella N, Kaufman RJ, et al. Mutations in the ER-Golgi intermediate compartment protein ERGIC-53 cause combined deficiency of coagulation factors V and VIII. Cell. 1998;93:61–70. doi: 10.1016/s0092-8674(00)81146-0. [DOI] [PubMed] [Google Scholar]
- 19.Muniz M, Nuoffer C, Hauri HP, Riezman H. The Emp24 complex recruits a specific cargo molecule into endoplasmic reticulum-derived vesicles. J Cell Biol. 2000;148:925–930. doi: 10.1083/jcb.148.5.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Powers J, Barlowe C. Transport of axl2p depends on erv14p, an ER-vesicle protein related to the Drosophila cornichon gene product. J Cell Biol. 1998;142:1209–1222. doi: 10.1083/jcb.142.5.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Powers J, Barlowe C. Erv14p directs a transmembrane secretory protein into COPII-coated transport vesicles. Mol Biol Cell. 2002;13:880–891. doi: 10.1091/mbc.01-10-0499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herzig Y, Sharpe HJ, Elbaz Y, Munro S, Schuldiner M. A systematic approach to pair secretory cargo receptors with their cargo suggests a mechanism for cargo selection by Erv14. PLoS Biol. 2012;10:e1001329. doi: 10.1371/journal.pbio.1001329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Castillon GA, Watanabe R, Taylor M, Schwabe TM, Riezman H. Concentration of GPI-anchored proteins upon ER exit in yeast. Traffic. 2009;10:186–200. doi: 10.1111/j.1600-0854.2008.00857.x. [DOI] [PubMed] [Google Scholar]
- 24.Sauvageau E, Rochdi MD, Oueslati M, Hamdan FF, Percherancier Y, Simpson JC, Pepperkok R, Bouvier M. CNIH4 interacts with newly synthesized GPCR and controls their export from the endoplasmic reticulum. Traffic. 2014;15:383–400. doi: 10.1111/tra.12148. [DOI] [PubMed] [Google Scholar]
- 25.Castro CP, Piscopo D, Nakagawa T, Derynck R. Cornichon regulates transport and secretion of TGFalpha-related proteins in metazoan cells. J Cell Sci. 2007;120:2454–2466. doi: 10.1242/jcs.004200. [DOI] [PubMed] [Google Scholar]
- 26.Bokel C, Dass S, Wilsch-Brauninger M, Roth S. Drosophila Cornichon acts as cargo receptor for ER export of the TGFalpha-like growth factor Gurken. Development. 2006;133:459–470. doi: 10.1242/dev.02219. [DOI] [PubMed] [Google Scholar]
- 27.Pagant S, Kung L, Dorrington M, Lee MC, Miller EA. Inhibiting endoplasmic reticulum (ER)-associated degradation of misfolded Yor1p does not permit ER export despite the presence of a diacidic sorting signal. Mol Biol Cell. 2007;18:3398–3413. doi: 10.1091/mbc.E07-01-0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sharpe HJ, Stevens TJ, Munro S. A comprehensive comparison of transmembrane domains reveals organelle-specific properties. Cell. 2010;142:158–169. doi: 10.1016/j.cell.2010.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Louie RJ, Guo J, Rodgers JW, White R, Shah NA, Pagant S, Kim P, Livstone M, Dolinski K, McKinney BA, et al. A yeast phenomic model for the gene interaction network modulating CFTR-DeltaF508 protein biogenesis. Genome medicine. 2012;4:103. doi: 10.1186/gm404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakanishi H, Suda Y, Neiman AM. Erv14 family cargo receptors are necessary for ER exit during sporulation in Saccharomyces cerevisiae. J Cell Sci. 2007;120:908–916. doi: 10.1242/jcs.03405. [DOI] [PubMed] [Google Scholar]
- 31.Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O’Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 32.Pagant S, Brovman EY, Halliday JJ, Miller EA. Mapping of interdomain interfaces required for the functional architecture of Yor1p, a eukaryotic ATP-binding cassette (ABC) transporter. J Biol Chem. 2008;283:26444–26451. doi: 10.1074/jbc.M803912200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pagant S, Halliday JJ, Kougentakis C, Miller EA. Intragenic suppressing mutations correct the folding and intracellular traffic of misfolded mutants of Yor1p, a eukaryotic drug transporter. J Biol Chem. 2010;285:36304–36314. doi: 10.1074/jbc.M110.142760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stagljar I, Korostensky C, Johnsson N, te Heesen S. A genetic system based on split-ubiquitin for the analysis of interactions between membrane proteins in vivo. Proc Natl Acad Sci U S A. 1998;95:5187–5192. doi: 10.1073/pnas.95.9.5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Snider J, Kittanakom S, Damjanovic D, Curak J, Wong V, Stagljar I. Detecting interactions with membrane proteins using a membrane two-hybrid assay in yeast. Nature protocols. 2010;5:1281–1293. doi: 10.1038/nprot.2010.83. [DOI] [PubMed] [Google Scholar]
- 36.Collins BM, McCoy AJ, Kent HM, Evans PR, Owen DJ. Molecular architecture and functional model of the endocytic AP2 complex. Cell. 2002;109:523–535. doi: 10.1016/s0092-8674(02)00735-3. [DOI] [PubMed] [Google Scholar]
- 37.Jackson LP, Kelly BT, McCoy AJ, Gaffry T, James LC, Collins BM, Honing S, Evans PR, Owen DJ. A large-scale conformational change couples membrane recruitment to cargo binding in the AP2 clathrin adaptor complex. Cell. 2010;141:1220–1229. doi: 10.1016/j.cell.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ghai R, Bugarcic A, Liu H, Norwood SJ, Skeldal S, Coulson EJ, Li SS, Teasdale RD, Collins BM. Structural basis for endosomal trafficking of diverse transmembrane cargos by PX-FERM proteins. Proc Natl Acad Sci U S A. 2013;110:E643–652. doi: 10.1073/pnas.1216229110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Simpson JC, Cetin C, Erfle H, Joggerst B, Liebel U, Ellenberg J, Pepperkok R. An RNAi screening platform to identify secretion machinery in mammalian cells. Journal of biotechnology. 2007;129:352–365. doi: 10.1016/j.jbiotec.2006.12.027. [DOI] [PubMed] [Google Scholar]
- 40.Nishimura N, Balch WE. A di-acidic signal required for selective export from the endoplasmic reticulum. Science. 1997;277:556–558. doi: 10.1126/science.277.5325.556. [DOI] [PubMed] [Google Scholar]
- 41.Fiedler K, Veit M, Stamnes MA, Rothman JE. Bimodal interaction of coatomer with the p24 family of putative cargo receptors. Science. 1996;273:1396–1399. doi: 10.1126/science.273.5280.1396. [DOI] [PubMed] [Google Scholar]
- 42.Iodice L, Sarnataro S, Bonatti S. The carboxyl-terminal valine is required for transport of glycoprotein CD8 alpha from the endoplasmic reticulum to the intermediate compartment. J Biol Chem. 2001;276:28920–28926. doi: 10.1074/jbc.M103558200. [DOI] [PubMed] [Google Scholar]
- 43.Nufer O, Kappeler F, Guldbrandsen S, Hauri HP. ER export of ERGIC-53 is controlled by cooperation of targeting determinants in all three of its domains. J Cell Sci. 2003;116:4429–4440. doi: 10.1242/jcs.00759. [DOI] [PubMed] [Google Scholar]
- 44.Otte S, Barlowe C. The Erv41p-Erv46p complex: multiple export signals are required in trans for COPII-dependent transport from the ER. EMBO J. 2002;21:6095–6104. doi: 10.1093/emboj/cdf598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Herrmann JM, Malkus P, Schekman R. Out of the ER--outfitters, escorts and guides. Trends Cell Biol. 1999;9:5–7. doi: 10.1016/s0962-8924(98)01414-7. [DOI] [PubMed] [Google Scholar]
- 46.Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O’Shea EK. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 47.Kuehn MJ, Schekman R, Ljungdahl PO. Amino acid permeases require COPII components and the ER resident membrane protein Shr3p for packaging into transport vesicles in vitro. J Cell Biol. 1996;135:585–595. doi: 10.1083/jcb.135.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 (related to Figure 1). Both Erv14 and Erv15 are necessary for Yor1 trafficking but do not contribute to Yor1 folding. (A) The phenotype of erv14Δ or erv14Δ erv15Δ mutants expressing ERV14 or ERV15 under the control of the ERV14 promoter was tested by serial dilutions onto YPEG + oligomycin (0.4 μg/ml). (B) Incorporation of Yor1 into COPII vesicles was measured from membranes isolated from erv14Δ erv15Δ cells that express either ERV14 or ERV15 under the control of the ERV14 promoter as indicated. (C) Microsomal membranes (left panel) or radiolabelled semi intact cells (center panel) isolated from wild-type or erv14Δ erv15Δ cells expressing wild-type Yor1-HA were treated with increasing amounts of trypsin followed by SDS-PAGE and detection of HA-tagged proteolytic fragments. Misfolding of Yor1 results in the appearance of multiple lower molecular weight bands, as observed for a steady-state experiment using a misfolded allele of Yor1-HA (Yor1*-HA; right panel).
Figure S2 (related to Figure 2 and Figure 6). Structure-function analysis of Erv14 (A) Multiple sequence alignment between Erv14 and Erv15 from Saccharomyces cerevisiae (Sc) and their Cornichon orthologs in human (Hs), mouse (Mm) and Drosophilia (Dm), generated by Multalin (http://npsapbil.ibcp.fr/cgibin/npsa_automat.pl?page=/NPSA/npsa_multalin.html). Conserved and similar residues are indicated in red and green, respectively. Boxes delimit the positions of three TMDs previously defined by Powers et al. (2002). The purple stars specify the residues analyzed in this study. (B) Ten-fold serial dilutions of erv14Δ erv15Δ cells expressing untagged, HA- or GFP-tagged versions of ERV14 from a CEN plasmid or untagged ERV14 from a 2μ plasmid (o/e) were tested for sensitivity to the indicated concentrations of oligomycin. Addition of HA and GFP epitopes at the C-terminus of Erv14 did not influence complementation of the oligomycin sensitivity displayed by the erv14Δ erv15Δ strain. Over-expression of ERV14 accentuated drug sensitivity. (C) Cultures of erv14Δ erv15Δ cells bearing the plasmids indicated were spotted onto media containing increasing concentrations of oligomycin (0, 0.2 μg/ml and 0.3 μg/ml). Neither the cytosolic mutant (erv14-IFTRL) nor the TMD2 mutant (erv14-FLN) could complement oligomycin sensitivity. The lumenal mutant (erv14-YPE) showed an intermediate phenotype. (D) Incorporation of Yor1-HA into COPII vesicles was assayed as described in Fig. 1 from membranes isolated from the strains indicated. Quantification (right panel; n=3; error bars represent SD) showed that the capture of both Yor1-HA (D) and Erv14-HA (E) into the vesicle fraction was inhibited by mutation of the Erv14 IFTRL signal. Conversely, the abundance of Yor1-HA in COPII vesicles was perturbed in the context of erv14-FLN whereas packaging of erv14-FLN-HA itself was unaffected compared to wild-type ERV14-HA. Membranes expressing erv14-DYPE were partially affected both for Yor1 capture and Erv14 capture. (E) Anti-HA immunoblots of whole cell lysates expressing the indicated Erv14 variants.
Figure S3 (related to Figure 2). Erv14 traffics a subset of plasma membrane proteins. (A and B) Wide-field fluorescent micrographs of the indicated cargo proteins tagged with GFP, expressed in wild-type or erv14Δ erv15Δ cells (Scale bar =2μm). (A) Novel Erv14 clients identified in this study. (B) Plasma membrane proteins whose trafficking is independent of Erv14.
Figure S4 (related to Figure 3). Most Erv14 clients do not use known Sec24 binding sites. (A) Localization of GFP-tagged versions of the indicated Erv14 clients were examined by epifluorescence microscopy in sec24Δ strains expressing the cargo-binding site sec24 mutants indicated. None of the Sec24 mutants impaired cell surface localization of Sur7-GFP, Tpo4-GFP, Itr1-GFP, Mid2-GFP and Hxt6-GFP (Scale bar =2μm). (B) The indicated variants of SEC24 were introduced into sec24Δ or sec24Δ erv29Δ cells expressing wild-type SEC24 from a URA3-marked plasmid and serial dilutions of the transformants were spotted onto medium containing 5-FOA, which counterselects for the URA3-marked plasmid. All SEC24 mutants were viable in both sec24Δ and sec24Δ erv29Δ strains.
Figure S5 (related to Figure 4). Characterization of the Erv14 binding site (Ser491, Phe576, Arg578) on Sec24. (A) Strains expressing the indicated variants of SEC24 as the sole copy were spotted on media containing oligomycin (0.4 μg/ml) to test for Yor1 function at the plasma membrane. (B and C) Erv14-GFP localization was monitored by epifluorescence microscopy in sec24Δ iss1Δ strains expressing (B) wild-type SEC24 or (C) the sec24-SFR mutant (Scale bar =2μm).
Figure S6 (related to Figure 5). Phenotypic characterization of the Erv14 binding site (Ser491, Phe576, Arg578) on Sec24. (A) A sec24Δ strain expressing wild-type SEC24 from a URA3-marked plasmid was transformed with the indicated variants of SEC24 and serial dilutions were spotted onto medium containing 5-FOA, which counterselects for the URA3-marked wild-type version. A variant of Sec24 bearing mutations that affect both the B-site (Arg230 and Arg235) and the Erv14-binding site (Phe578 and Arg578) was unable to sustain growth at 30°C. (B) A sec24Δ strain co-expressing wild-type SEC24 on a URA3-marked plasmid and the B-site mutant sec24-R230A R235A on a HIS3-marked plasmid was transformed with the indicated variants of SEC24 on a LEU2-marked plasmid and viability was tested after counterselection of the URA3-marked wild-type in presence of 5-FOA. No growth defect could be observed upon co-expression of Sec24 mutants independently affected for the B-site or for the Erv14-binding site.
Table S2 (related to Figure 6) – Oligomycin phenotypes of ERV14 mutants
Table S3 (related to Figure 6)– Mass spectrometry of Erv14-HA co-IPs