Abstract
Purpose
Studies linking cholesterol levels to the development of colorectal neoplasia are inconsistent, and Mendelian randomization has been suggested as a way to help avoid problems with confounding and reverse causation.
Methods
We genotyped individuals who received a colonoscopy at Group Health (1998–2007) for 96 of 102 single-nucleotide polymorphisms (SNPs) identified by the Global Lipids Genetics Consortium. Participants included 139 advanced adenoma cases, 518 non-advanced adenoma cases, 380 non-adenomatous polyp cases, and 754 polyp-free controls. All had at least one available pre-colonoscopy lipid measurement from electronic records maintained by Group Health.
Results
Advanced adenoma cases were more likely than controls to have higher pre-colonoscopy zenith low-density lipoprotein (LDL), triglycerides (TG), and total cholesterol (TC) (odds ratio, OR, per 20 mg/dL LDL increase: 1.16, 95% confidence interval, CI, 1.03–1.30; per 40 mg/dL TG increase: 1.09, 1.03–1.16; and per 20 mg/dL TC increase: 1.09, 1.02–1.18). For these traits, genotype-polyp ORs using weighted allele scores were not statistically significant (OR per increase in score scaled to a 20 mg/dL LDL increase: 1.17, 0.78–1.75; a 40 mg/dL TG increase: 1.12, 0.91–1.38; a 20 mg/dL TC increase: 0.99, 0.71–1.38).
Conclusions
Cholesterol levels may be associated with advanced adenomas, but larger studies are warranted to determine whether this association can be attributed to genetics.
Keywords: cholesterol, colonoscopy, colorectal adenoma, colorectal hyperplastic polyp, Mendelian randomization
Introduction
It is unclear whether dyslipidemia is a risk factor for colorectal neoplasia [1–4]. It has been challenging for observational studies to determine if the co-occurrence of dyslipidemia and colorectal neoplasia is causal, given that both conditions share risk factors including high-fat diet, obesity, insulin resistance, smoking, and sedentary lifestyle [5, 6]. Mendelian randomization has been suggested as a potential solution [7–9]. Under assumptions employed in instrumental variables analysis [10], Mendelian randomization studies use the distribution of alleles in the population to simulate randomized assignment to lower or higher cholesterol over the life course.
In the case of polygenic dyslipidemia, which is common and highly hereditable [11], the Global Lipids Genetics Consortium (GLGC) genome-wide association study (GWAS) identified 102 germline single-nucleotide polymorphisms (SNPs) across 95 genes reaching genome-wide statistical significance (P<5×10−8) for associations with blood concentrations of low-density lipoprotein cholesterol (LDL), high-density lipoprotein cholesterol (HDL), triglycerides (TG), or total cholesterol (TC) [12]. Using these SNPs, we compared Mendelian randomization estimates to associations from clinical cholesterol measurements in a sample of men and women who underwent colonoscopy at Group Health, a large healthcare system in Washington State.
Materials and Methods
Study population
Participants, ages 25–79, were enrollees of Group Health for at least 3 years who received a colonoscopy for any indication from 1998–2007. Eligibility criteria and participation information have been previously described [13]. All provided informed consent to access to medical records, completed a health history interview, and were asked to provide a DNA sample [14]. Study protocols were approved by the Institutional Review Boards of Group Health Research Institute and Fred Hutchinson Cancer Research Center.
Outcome ascertainment
Biopsies collected during colonoscopy received a standardized pathology review. Adenomas were distinguished from non-adenomatous polyps, which included hyperplastic polyps, traditional serrated adenomas, and sessile serrated adenomas. An advanced adenoma was defined as any tubular, tubulovillous, or villous adenoma ≥10 mm in diameter, with ≥20% villous components, or high-grade dysplasia [15]. Participants were classified into 4 groups based on the lesion with the highest malignant potential: 1) advanced adenomas cases; 2) non-advanced adenoma cases; 3) non-adenomatous polyp cases; and 4) those without polyps (controls).
Phenotype measurement
All LDL, HDL, TG, and TC measurements from at most 20 years prior to the study colonoscopy were extracted from Group Health’s database of lab results. We determined each participant’s highest LDL, highest TG, highest TC measurement (zenith), and lowest HDL measurement (nadir). LDL or TG measurements were unavailable for about 30% of participants, as these were not routinely used to assess cardiovascular disease risk at Group Health until the later period of data collection. Information on lipid-controlling drug prescriptions dispensed at eligible pharmacies was extracted from electronic pharmacy records [16]. Medication types included both statins and non-statins (fibric acids, cholesterol absorption inhibitors, and nicotinic acid).
Genotype measurement
Genotyping was performed using a custom GoldenGate assay from Illumina (San Diego, CA, USA). Prior to genotyping, 11 of the 102 SNPs identified by the GLGC were projected to have low likelihood of success based on Illumina’s Assay Design Tool. For 5 of these SNPs (rs7515577, EVI5; rs1042034, APOB; rs9488822, FRK; rs12967135, MC4R; and rs7255436; ANGPTL4), we identified proxies with r2>0.3 from HapMap Caucasians. Six SNPs for which no proxy could be identified were excluded (rs1367117, APOB; rs13238203, TYW1B; rs4759375, SBNO1; rs2652834, LACTB; rs7241918, LIPG; rs2277862, ERGIC3), leaving 96 of the 102 SNPs available for analyses. All SNPs were tested for departures from Hardy-Weinberg equilibrium.
Statistical analyses
For each of LDL, HDL, TG, and TC, we estimated 3 primary associations: 1) lipid-polyp odds ratios (OR) with 95% confidence intervals (CI) comparing each case group to controls using polytomous logistic regression; 2) genotype-lipid associations using ordinary linear regression; and 3) genotype-polyp Mendelian randomization ORs using 2-stage linear-logistic regression [17]. We used trait-specific allele scores created by counting alleles associated with an increased mean in the GLGC GWAS, weighted by effect size from their analysis (for HDL, the score was based on alleles associated with decreased mean HDL) [18]. For missing genotypes, we imputed the mean score calculated among participants not missing genotypes. Minimally-adjusted models included age at colonoscopy, sex, race, and year of colonoscopy. Fully-adjusted models additionally included all variables listed in the footnote to Table 2. Analyses restricted to only Caucasian study participants were also considered.
Table 2.
Lipid-polyp associations for pre-colonoscopy cholesterol extremes, Group Health, 1998–2007
| Cholesterol measurement | Unit change for OR | Advanced adenoma cases (N=139) vs. controls (N=754)
|
Non-advanced adenoma cases(N=518) vs. controls (N=754)
|
Non-adenomatous polyp cases (N=380) vs. controls (N=754)
|
|||
|---|---|---|---|---|---|---|---|
| ORa (95% CI) | P | ORa (95% CI) | P | ORa (95% CI) | P | ||
| Zenith LDL | +20 mg/dL | 1.16 (1.03, 1.30) | 0.01 | 1.07 (0.99, 1.15) | 0.08 | 1.10 (1.01, 1.20) | 0.03 |
| Nadir HDL | −10 mg/dL | 0.93 (0.79, 1.09) | 0.37 | 0.94 (0.86, 1.05) | 0.28 | 0.98 (0.88, 1.10) | 0.72 |
| Zenith TG | +40 mg/dL | 1.09 (1.03, 1.16) | 0.006 | 1.03 (0.98, 1.08) | 0.21 | 1.02 (0.96, 1.07) | 0.53 |
| Zenith TC | +20 mg/dL | 1.09 (1.02, 1.18) | 0.02 | 1.03 (0.98, 1.09) | 0.29 | 1.05 (0.99, 1.11) | 0.12 |
OR is adjusted for age at colonoscopy, sex, race, data-collection period, education, BMI, NSAID use, family history of CRC, estrogen-only use (in women), estrogen-plus-progestin use (in women), cigarette smoking, alcohol consumption, diabetes mellitus, fruit servings per day, vegetable servings per day, recreational and exercise physical activity, prior endoscopy (approximately 2 years before study colonoscopy), and data-collection period.
Abbreviations: BMI, body mass index; CI, confidence interval; HDL, high-density lipoprotein cholesterol; LDL, low-density lipoprotein cholesterol; NSAID, nonsteroidal anti-inflammatory drug; OR, odds ratio; TC, total cholesterol; TG, triglycerides.
In secondary analyses, without using an allele score, we regressed genotype-lipid ORs on the association from the GLGC GWAS and tested for the statistical significance of slopes using inverse-variance-weighted linear regression [19]. To account for the possibility that only post-treatment cholesterol values may be available for some lipid-controlling drug users, we increased the zenith by 30 mg/dL for users with zenith LDL<130 mg/dL (N=90). The GLGC considered a similar approach [12]. The value 130 mg/dL delineates borderline high LDL as defined by the Adult Treatment Panel III [5], and 30 mg/dL increase represents a conservative estimate of the average treatment effect of statins from randomized controlled trials [20]. We also considered excluding these N=90 study participants, and, separately, excluding all lipid-controlling drug users (N=503).
Because we evaluated a single summary measure for each of 4 traits, we considered Bonferroni-corrected 2-sided P-values≤0.05/4=0.0125 to denote statistical significance. Analyses were performed with SAS 9.2 (Cary, NC, USA) or R 3.0.0 (Vienna, Austria).
Results
Participant characteristics
A total of 2,506 participants completed the interview. DNA was unavailable from 640 participants, and 19 participants with DNA had an insufficient amount for genotyping. A further 56 had no cholesterol measurements available from the Group Health database of lab measurements. No differences were noted in the distribution of excluded participants by case-control status. Most were Caucasian and most identified to have polyps were men (Table 1).
Table 1.
Characteristics of colorectal polyp cases and controls, Group Health, 1998–2007
| Characteristics at colonoscopy | Controls (N=754) | Advanced adenomasa (N=139) | Non-advanced adenomasb (N=518) | Non-adenomatous polypsc (N=380) |
|---|---|---|---|---|
| Age (years), N (%) | ||||
| <50 | 65 (9) | 5 (4) | 28 (5) | 19 (5) |
| 50–59 | 291 (39) | 53 (38) | 192 (37) | 177 (47) |
| 60–69 | 272 (36) | 51 (37) | 194 (37) | 133 (35) |
| 70–79 | 126 (17) | 30 (22) | 104 (20) | 51 (13) |
| Sex, N (%) | ||||
| Male | 306 (41) | 78 (56) | 277 (53) | 170 (45) |
| Female | 448 (59) | 61 (44) | 241 (47) | 210 (55) |
| Race, N (%) | ||||
| Caucasian | 656 (87) | 121 (87) | 439 (85) | 337 (89) |
| Black/African American | 20 (3) | 4 (3) | 11 (2) | 3 (1) |
| Asian/Pacific Islander | 33 (4) | 8 (6) | 27 (5) | 15 (4) |
| Other | 45 (6) | 6 (4) | 41 (8) | 25 (7) |
| Body mass index (kg/m2), N (%) | ||||
| <25 | 298 (40) | 45 (33) | 157 (31) | 160 (42) |
| 25–30 | 309 (41) | 56 (41) | 221 (43) | 142 (37) |
| ≥30 | 144 (19) | 37 (27) | 136 (26) | 78 (21) |
| Unknown | 3 | 1 | 4 | 0 |
| Previous endoscopy, N (%) d | ||||
| No | 340 (45) | 90 (65) | 281 (55) | 176 (47) |
| Yes | 408 (55) | 48 (35) | 232 (45) | 201 (53) |
| Unknown | 6 | 1 | 5 | 3 |
Adenoma cases have at least one tubular, tubulovillous, or villous adenoma ≥10 mm in diameter, with ≥20% villous components, or high-grade dysplasia.
Non-advanced adenoma cases have at least one tubular or tubulovillous adenoma, all <10 mm in diameter, with <20% villous components, and no high-grade dysplasia.
Non-adenomatous polyp cases had at least one hyperplastic polyp, traditional serrated adenoma, or sessile serrated adenoma, and no adenomas.
Colonoscopy or sigmoidoscopy performed ≥2 years before study colonoscopy.
Lipid-polyp associations
Increases in zenith LDL, zenith TG, and zenith TC were each associated with increased odds of advanced adenomas (Table 2). Associations for zenith LDL were similar in analyses involving the substituted treatment effect, excluding participants for whom we substituted a treatment effect, and also excluding all of those without record of lipid-controlling drug prescriptions (not shown).
Genotyping results
All but 3 genotyped SNPs were missing for <1% of participants. Exceptions were rs2068888 (CYP26A1) missing for 74% of participants, and rs7134375 (PDE3A) and rs4420638 (APOE/APOC1), both missing for 26% of participants due to clustering failures. For all but 1 SNP, rs4129767 (PGS1), the minor allele observed among our controls matched the minor allele reported from the GLGC GWAS. To be consistent with the GLGC GWAS, we report associations for the G allele of rs4129767 (frequency of 54% in controls, but was 49% in the GLGC GWAS).
Genotype-lipid associations
Genotype-lipid associations using GLGC-weighted allele scores were in the expected direction and highly statistically significant among our 754 controls, with P-values ranging from 1×10−6 for the regression of LDL allele score on zenith LDL to 1×10−17 for the regression of HDL allele score on nadir HDL (Table 3). A variant of CETP (rs3764261) met genome-wide statistical significance for the association with nadir HDL among controls. Allele scores were not associated with any of the covariates included as adjustment variables (not shown).
Table 3.
Genotype-lipid associations among controls, Group Health, 1998–2007
| Cholesterol measurement | Unit change in score for β | Controls (N=754)
|
||
|---|---|---|---|---|
| βa (95% CI) | F | P | ||
| Zenith LDL | +1 | 2.3 (1.4, 3.2) | 25 | 1×10−6 |
| Nadir HDL | +1 | −0.9 (−1.1, −0.7) | 77 | 1×10−17 |
| Zenith TG | +1 | 9.8 (6.6, 12.9) | 37 | 2×10−9 |
| Zenith TC | +1 | 2.5 (1.8, 3.3) | 45 | 5×10−11 |
OR is adjusted for age at colonoscopy, sex, race, and data-collection period.
Abbreviations: CI, confidence interval; HDL, high-density lipoprotein cholesterol; LDL, low-density lipoprotein cholesterol; OR, odds ratio; TC, total cholesterol; TG, triglycerides.
Genotype-polyp associations
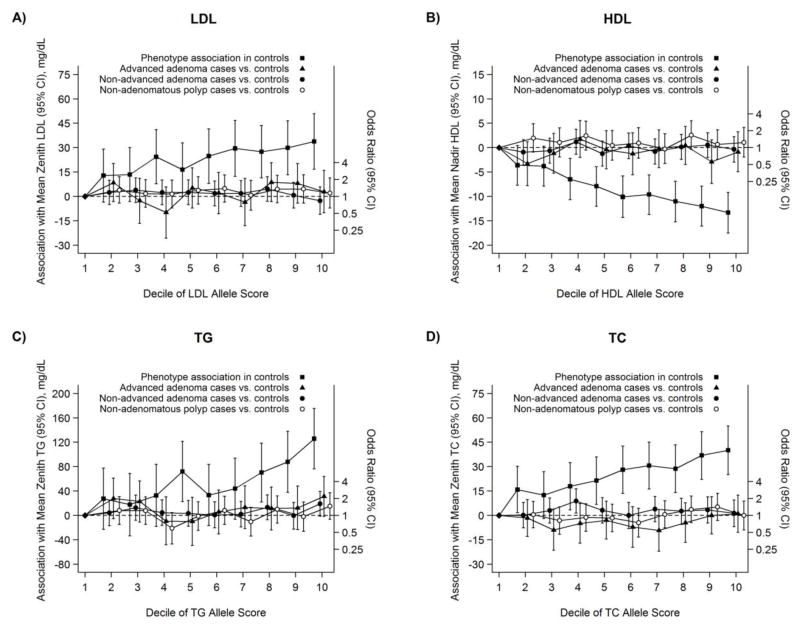
None of the Mendelian randomization estimates for genotype-polyp associations based on allele scores was statistically significant (Table 4). The genotype-lipid association appeared linear based on deciles in controls (Figure 1). Multi-SNP analyses without using allele scores also revealed no statistically significant associations. In general, polymorphisms with the largest magnitude per-allele associations with lipid phenotypes, either in the GLCG GWAS or in our controls, were not associated with colorectal polyps (Supplemental Figures 1 and 2). Analyses restricted to only Caucasian study participants were similar (not shown).
Table 4.
Genotype-polyp associations from trait-specific allele scores, Group Health, 1998–2007
| Cholesterol measurement | Unit change for OR (scaled)a | Advanced adenoma cases (N=139) vs. controls (N=754)
|
Non-advanced adenoma cases(N=518) vs. controls (N=754)
|
Non-adenomatous polyp cases (N=380) vs. controls (N=754)
|
|||
|---|---|---|---|---|---|---|---|
| ORb (95% CI) | P | ORb (95% CI) | P | ORb (95% CI) | P | ||
| LDL allele score | +20 mg/dL | 1.17 (0.78, 1.75) | 0.46 | 0.99 (0.77, 1.27) | 0.95 | 1.13 (0.86, 1.49) | 0.40 |
| HDL allele score | −10 mg/dL | 0.95 (0.60, 1.48) | 0.81 | 1.05 (0.79, 1.39) | 0.76 | 1.03 (0.75, 1.41) | 0.87 |
| TG allele score | +40 mg/dL | 1.12 (0.91, 1.38) | 0.30 | 1.09 (0.95, 1.24) | 0.21 | 1.07 (0.92, 1.23) | 0.39 |
| TC allele score | +20 mg/dL | 0.99 (0.71, 1.38) | 0.94 | 1.02 (0.83, 1.26) | 0.84 | 1.08 (0.86, 1.37) | 0.51 |
+8.7 units for LDL allele score, +11.1 units for HDL allele score, +4.1 units for TG allele score, +8.0 units for TC allele score.
OR is adjusted for age at colonoscopy, sex, race, and data-collection period.
Abbreviations: CI, confidence interval; HDL, high-density lipoprotein cholesterol; LDL, low-density lipoprotein cholesterol; OR, odds ratio; TC, total cholesterol; TG, triglycerides.
Fig. 1.
Estimated difference in mean value of the blood lipid phenotypes (zenith LDL in A, nadir HDL in B, zenith TG in C, and zenith TC in D) comparing deciles of the allele score in controls to the first decile (black squares; plotted with respect to left Y-axis), Group Health, 1998–2007. Estimated odds ratios of advanced adenomas vs. controls (black triangles), non-advanced adenomas vs. controls (black circles), and non-adenomatous polyps vs. controls (white circles) are adjusted for age, sex, race, and data-collection period; 95% CIs shown.
Discussion
We found that higher extremes in LDL, TG, and TC occurring, on average, about 4–6 years before colonoscopy depending on the trait, were associated with the prevalence of advanced adenomas, those lesions most likely to progress to invasive cancer [15]. In contrast, evidence from GWAS-identified allele scores was not strong, particularly in light of the apparent inconsistency between which SNPs were associated with lipid phenotypes and which were associated with polyps. Until larger studies can be conducted, this analysis provides a preliminary indication that genetically-influenced cholesterol levels may be unrelated to the development of colorectal neoplasms.
Mendelian randomization analyses require strong assumptions that are not readily verifiable. Alleles must function to alter blood lipid levels without unmeasured common causes of both the polymorphism and polyp occurrence, and without the alleles being involved in mechanisms that influence polyp formation separate from the mechanisms by which they alter cholesterol levels (i.e., no genetic pleiotropy) [10]. Mendelian randomization analyses of traits with complex biology can be difficult to interpret. Some SNPs we evaluated may be inappropriate for use as instrumental variables due to pleiotropy or weak-instrument bias [21]. It has been suggested that, given the strong assumptions involved, null Mendelian randomization results may be more plausible than positive results [22].
We acknowledge the modest sample size is a primary limitation [23, 24]. It is estimated that the 102 SNPs from the GLGC GWAS collectively explain approximately 12% of total variation, or about 30% of the expected genetic variation, in each lipid trait [12]. For comparison, in this same sample, an allele score comprised of 13 SNPs identified from GWAS of colorectal cancer was associated with increased prevalence of advanced adenomas with P=2×10−3 [14], despite evidence that these SNP explain far less of the heritability of colorectal cancer than the GLGC GWAS SNPs explain of the heritability of lipids [25]. Larger studies will benefit from enhanced statistical power, but the ability of such studies to harmonize pathology information, past lipid trajectories, and pharmacy data will likely be limited. Larger studies should also attempt to include additional polymorphisms from more powerful genetic association studies of lipid traits that explain more of the expected genetic variation, as cholesterol loci of potential relevance to colorectal neoplasia may not be fully represented among these 102 SNPs.
The ability to compare estimates from Mendelian randomization to those from clinical lipids measurements was a key motivation for collecting data on both genotype and phenotype. This also permitted an internal assessment of the strength of the instruments. Genes in cholesterol metabolism pathways have not been among the loci reaching genome-wide statistical significance from GWAS of colorectal polyps [26, 27] or invasive colorectal cancer [28, 29]. Thus, we used trait-specific allele scores in an attempt to operationalize sufficiently strong instruments. Use of such scores has limitations [18], however, and we chose to supplement multi-SNP analyses with single-SNP analyses.
It is not typically necessary to adjust for disease risk factors in genetic association studies, and the minimally-adjusted estimates we report align closely with the classical notions of Mendelian randomization. In order to enhance instrument purity, we could have eliminated among the set of lipid SNPs of interest, those representative of loci found to be also associated with other risk factors for colorectal polyps such as high-fat diet, obesity, insulin resistance, smoking, and sedentary lifestyle in other GWAS. Having found no clear overall signal of association with all SNPs in a score and no clear pattern of association in single-SNP analyses, retrospective pruning of the SNP list based on external information on genotypic associations with other phenotypes did not prove to be useful in identifying associations with polyps. Lastly, we acknowledge that race-adjustment alone may be insufficient to adequately control for population stratification [30], but ancestry-informative markers were not available in this sample, and the results changed little when those not of Caucasian race were excluded.
In summary, despite observing a statistically significant positive association between higher pre-colonoscopy extremes in LDL, TG, and TC and the prevalence of advanced adenomas, there was insufficient evidence to conclude that genetic variation controlling cholesterol levels may be involved in polyp formation. On one hand, although statistical power was limited, the magnitude of the point estimates for the Mendelian randomization odds ratios, particularly for the association between advanced adenomas and SNPs for LDL and TG were of comparable magnitude to those for measured zenith LDL and zenith TG. On the other hand, Mendelian randomization odds ratios did not achieve statistical significance, and SNPs known to have the largest magnitude association with blood lipids were not associated with the prevalence of polyps.
If confirmed in larger studies, a null association from Mendelian randomization analysis may suggest that clinical dyslipidemia is only a bystander to environmental or genetic causes of neoplastic growth in the colon and rectum. Dyslipidemia may be a marker of, for instance, the type of visceral adiposity, insulin resistance, or dietary exposures that, independent of biological pathways that involve cholesterol metabolism, could lead to polyp formation. Alternatively, larger studies may have the power to identify genetic associations that implicate extremes in blood cholesterol, more directly, in neoplastic pathways, consistent with the magnitude of some of our point estimates, should they be replicated with increased precision.
Supplementary Material
Acknowledgments
We thank Joseph Webster (Group Health Research Institute), Jeanne DaGloria (Fred Hutchinson Cancer Research Center), Dr. Margaret Mandelson (Group Health Research Institute), and Dr. Santica Marcovina (University of Washington) for their contributions at various stages of this research. This work was supported by the National Cancer Institute at the National Institutes of Health (grant numbers R03CA171014, T32CA009168, K05CA152715, P01CA074184, R01CA097325). Pilot funding was provided by the Division of Public Health Sciences at Fred Hutchinson Cancer Research Center.
Footnotes
Conflicts of interest
The authors declare no conflicts of interest.
References
- 1.Bayerdorffer E, Mannes GA, Richter WO, et al. Decreased high-density lipoprotein cholesterol and increased low-density cholesterol levels in patients with colorectal adenomas. Ann Intern Med. 1993;118:481–487. doi: 10.7326/0003-4819-118-7-199304010-00001. [DOI] [PubMed] [Google Scholar]
- 2.Bird CL, Ingles SA, Frankl HD, Lee ER, Longnecker MP, Haile RW. Serum lipids and adenomas of the left colon and rectum. Cancer Epidemiol Biomarkers Prev. 1996;5:607–612. [PubMed] [Google Scholar]
- 3.Kang HW, Kim D, Kim HJ, et al. Visceral obesity and insulin resistance as risk factors for colorectal adenoma: a cross-sectional, case-control study. Am J Gastroenterol. 2010;105:178–187. doi: 10.1038/ajg.2009.541. [DOI] [PubMed] [Google Scholar]
- 4.Yang MH, Rampal S, Sung J, et al. The association of serum lipids with colorectal adenomas. Am J Gastroenterol. 2013;108:833–841. doi: 10.1038/ajg.2013.64. [DOI] [PubMed] [Google Scholar]
- 5.Expert Panel on Detection, Evaluation and Treatment of High Blood Cholesterol in Adults . Executive summary of the third report of The National Cholesterol Education Program (NCEP) Expert panel on detection, evaluation, and treatment of high blood cholesterol in adults (Adult Treatment Panel III) JAMA. 2001;285:2486–2497. doi: 10.1001/jama.285.19.2486. [DOI] [PubMed] [Google Scholar]
- 6.Potter JD. Colorectal cancer: molecules and populations. J Natl Cancer Inst. 1999;91:916–932. doi: 10.1093/jnci/91.11.916. [DOI] [PubMed] [Google Scholar]
- 7.Katan MB. Apolipoprotein E isoforms, serum cholesterol, and cancer. Lancet. 1986;1:507–508. doi: 10.1016/s0140-6736(86)92972-7. [DOI] [PubMed] [Google Scholar]
- 8.Davey Smith G, Ebrahim S. ‘Mendelian randomization’: can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol. 2003;32:1–22. doi: 10.1093/ije/dyg070. [DOI] [PubMed] [Google Scholar]
- 9.Katan MB. Commentary: Mendelian Randomization, 18 years on. Int J Epidemiol. 2004;33:10–11. doi: 10.1093/ije/dyh023. [DOI] [PubMed] [Google Scholar]
- 10.Glymour MM, Tchetgen EJ, Robins JM. Credible Mendelian randomization studies: approaches for evaluating the instrumental variable assumptions. Am J Epidemiol. 2012;175:332–339. doi: 10.1093/aje/kwr323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heller DA, de Faire U, Pedersen NL, Dahlen G, McClearn GE. Genetic and environmental influences on serum lipid levels in twins. N Engl J Med. 1993;328:1150–1156. doi: 10.1056/NEJM199304223281603. [DOI] [PubMed] [Google Scholar]
- 12.Teslovich TM, Musunuru K, Smith AV, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burnett-Hartman AN, Passarelli MN, Adams SV, et al. Differences in epidemiologic risk factors for colorectal adenomas and serrated polyps by lesion severity and anatomical site. Am J Epidemiol. 2013;177:625–637. doi: 10.1093/aje/kws282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burnett-Hartman AN, Newcomb PA, Hutter CM, et al. Variation in the association between colorectal cancer susceptibility loci and colorectal polyps by polyp type. Am J Epidemiol. 2014;180:223–232. doi: 10.1093/aje/kwu114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Winawer SJ, Zauber AG. The advanced adenoma as the primary target of screening. Gastrointest Endosc Clin N Am. 2002;12:1–9. v. doi: 10.1016/s1052-5157(03)00053-9. [DOI] [PubMed] [Google Scholar]
- 16.Saunders KW, Davis RL, Stergachis A. Group Health Cooperative of Puget Sound. In: Strom BL, editor. Pharmacoepidemiology. 3. Chichester, United Kingdom: John Wiley & Sons, Ltd; 2000. pp. 247–262. [Google Scholar]
- 17.Burgess S. Identifying the odds ratio estimated by a two-stage instrumental variable analysis with a logistic regression model. Stat Med. 2013;32:4726–4747. doi: 10.1002/sim.5871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burgess S, Thompson SG. Use of allele scores as instrumental variables for Mendelian randomization. Int J Epidemiol. 2013;42:1134–1144. doi: 10.1093/ije/dyt093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thompson SG, Higgins J. How should meta-regression analyses be undertaken and interpreted? Stat Med. 2002;21:1559–1573. doi: 10.1002/sim.1187. [DOI] [PubMed] [Google Scholar]
- 20.Cholesterol Treatment Trialists’ Collaboration . Efficacy and safety of more intensive lowering of LDL cholesterol: a meta-analysis of data from 170 000 participants in 26 randomised trials. Lancet. 2010;376:1670–1681. doi: 10.1016/S0140-6736(10)61350-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cai B, Small DS, Have TR. Two-stage instrumental variable methods for estimating the causal odds ratio: analysis of bias. Stat Med. 2011;30:1809–1824. doi: 10.1002/sim.4241. [DOI] [PubMed] [Google Scholar]
- 22.VanderWeele TJ, Tchetgen Tchetgen EJ, Cornelis M, Kraft P. Methodological challenges in mendelian randomization. Epidemiology. 2014;25:427–435. doi: 10.1097/EDE.0000000000000081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pierce BL, Ahsan H, Vanderweele TJ. Power and instrument strength requirements for Mendelian randomization studies using multiple genetic variants. Int J Epidemiol. 2011;40:740–752. doi: 10.1093/ije/dyq151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brion MJ, Shakhbazov K, Visscher PM. Calculating statistical power in Mendelian randomization studies. Int J Epidemiol. 2013;42:1497–1501. doi: 10.1093/ije/dyt179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiao S, Peters U, Berndt S, et al. Estimating the heritability of colorectal cancer. Hum Mol Genet. 2014;23:3898–3905. doi: 10.1093/hmg/ddu087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang J, Carvajal-Carmona LG, Chu JH, et al. Germline variants and advanced colorectal adenomas: adenoma prevention with celecoxib trial genome-wide association study. Clin Cancer Res. 2013;19:6430–6437. doi: 10.1158/1078-0432.CCR-13-0550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Edwards TL, Shrubsole MJ, Cai Q, et al. Genome-wide association study identifies possible genetic risk factors for colorectal adenomas. Cancer Epidemiol Biomarkers Prev. 2013;22:1219–1226. doi: 10.1158/1055-9965.EPI-12-1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tenesa A, Dunlop MG. New insights into the aetiology of colorectal cancer from genome-wide association studies. Nat Rev Genet. 2009;10:353–358. doi: 10.1038/nrg2574. [DOI] [PubMed] [Google Scholar]
- 29.Peters U, Jiao S, Schumacher FR, et al. Identification of genetic susceptibility loci for colorectal tumors in a genome-wide meta-analysis. Gastroenterology. 2013;144:799–807. doi: 10.1053/j.gastro.2012.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barnholtz-Sloan JS, Chakraborty R, Sellers TA, Schwartz AG. Examining population stratification via individual ancestry estimates versus self-reported race. Cancer Epidemiol Biomarkers Prev. 2005;14:1545–1551. doi: 10.1158/1055-9965.EPI-04-0832. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.