Abstract
Background
Aldosterone, synthesized in the adrenal cortex by the enzyme CYP11B2, induces positive sodium balance and predisposes to hypertension. Various investigators, using genomic DNA analyses, have linked −344T polymorphism in the hCYP11B2 gene to human hypertension. Human CYP11B2 gene promoter has three SNPs in linkage disequilibrium: T/A at −663, T/C at −470 and C/T at −344. Variants ACT occur together and form the haplotype-I while variants TTC constitute haplotype-II. We hypothesize that these SNPs, when present together, will lead to haplotype-dependent differences in the transcriptional regulation of the hCYP11B2 gene and affect blood pressure regulation.
Methods and Results
We evaluated differences in tissue expression, in vivo, and consequential effects on blood pressure stemming from the two haplotypes. Novel transgenic (TG) mice with the hCYP11B2 gene, targeted to the mouse HPRT locus, with either haplotype-II or I variant are used in the study. Our results show increased adrenal and renal expression of hCYP11B2 in TG mice with haplotype-I, as compared to mice with haplotype-II. Importantly, we observed increased baseline blood pressure in haplotype-I TG mice, an effect accentuated by a high-salt diet. Pathophysiological impact of elevated aldosterone was corroborated by our results showing up-regulation of proinflammatory markers in renal tissues from the TG mice with haplotype-I.
Conclusions
These findings characterize haplotype-dependent regulation of the hCYP11B2 gene where −344T serves as a reporter polymorphism and show that haplotype-I leads to increased expression of hCYP11B2, with permissive effects on blood pressure and inflammatory milieu.
Keywords: gene expression/regulation, aldosterone, genetic hypertension, genetics, transgenic models, renin angiotensin system
Introduction
Renin-angiotensin-aldosterone (RAAS) axis acts in cohort to exert long-term regulation of the mean arterial pressure (MAP). Aldosterone acts on the principal cells to regulate total body Na and the ECF volume, effects that govern the set point for the MAP. Additionally, via activation of the classical mineralocorticoid receptor (MR), and a GPCR, aldosterone brings about oxidative stress and tissue remodeling. In this regard, RAAS over activity, including hyperaldosteronism, contributes to essential hypertension and end organ damage. The CYP11B2 gene encodes aldosterone synthase, the rate-limiting enzyme in the biosynthesis of aldosterone. This gene is expressed primarily in the adrenal cortex and to a limited extent in the kidneys, the brain, and the adipose tissues.
The human CYP11B2 (hCYP11B2) gene has T/C polymorphism at the −344 site in its promoter region1. The −344T allele is accompanied by higher CYP11B2 gene expression in human studies, as compared to the −344C allele2. Caucasian subjects (n= 437) 65 years and older demonstrated higher baseline blood pressure in TT vs. CC homozygotes3. Other investigators provide complementary evidence for the role of this polymorphism in cardiovascular disease: −344T allele is associated with increased blood pressure with aging in Italian subjects4; T-allele is associated with higher excretion rate of tetrahydroaldosterone in Caucasian subjects5; −344T is shown to be associated with higher blood pressure in newly diagnosed hypertensive subjects with African ancestry6; genomic DNA analysis from 406 hypertensive and 424 normotensive South-Indian subjects have found −344T-allele to be associated with hypertension (p=0.007)7; finally, homozygosity for −344C confers 17% lower risk for hypertension, with respect to homozygous TT subjects, in meta analyses8. Importantly, Iwai et al have analyzed the role of 13 polymorphisms at CYP11B2 locus in 1443 Japanese subjects and concluded that only −344T allele was associated with increased aldosterone levels9. Thus, −344C/T polymorphism is associated with increased risk of cardiovascular disease that could be a consequence of differential transcriptional regulation caused by the SNP.
The nucleotide sequence of the CYP11B2 promoter [AAGGC(C/T)C] has sequence homology with the transcription factor SF1 binding site [AAGGTCA]. Recombinant SF1 binds five times stronger to an oligonucleotide containing −344C as compared to −344T10. However, reporter constructs of hCYP11B2, with −344T or −344C, show similar baseline expression that is equally repressed when co-transfected with SF-1 expression plasmid, in human adrenocortical cells (H295R)10. This suggested to us that the −344C/T polymorphism is functionally less important and may only be a marker for other SNPs in the hCYP11B2 promoter. In this regard, we have identified two additional SNPs in the promoter of the hCYP11B2 gene that are in complete linkage disequilibrium with the −344 site. These SNPs are rs1799998 (T/C at −344), rs10087214 (C/T at −470), and rs28659182 (C/A at −663). Variant −344T almost always occurs with variants −470C and −663A (named haplotype-I), and variant −344C almost always occurs with variants −470T and −663T (named haplotype-II). Thus, for the first time, we have identified SNP blocks in the hCYP11B2 promoter that form two distinct haplotypes. These haplotypes were discovered by in silico analysis of the of the promoter variants of the hCYP11B2 gene using the Hap Map data. We have further confirmed the presence of these haplotypes in human populations by analysis of the 1000 genome database.
This study was designed to test the transcriptional regulation and physiological differences between the two haplotypes of the hCYP11B2 gene, in vivo. To this end, we have generated transgenic (TG) mice with “knocked in” hCYP11B2 gene, containing either haplotype-I or haplotype-II, at the mouse HPRT locus. These TG mice contain 11.2 Kb of DNA encompassing 2.5 Kb of the 5’-flanking region (containing either haplotype-I or haplotype-II), 2.9 Kb of the 3’-flanking region, and 5.86 Kb of the coding region containing all the nine exons and eight introns of hCYP11B2 gene. We show here, in our novel TG mice with hCYP11B2 transgene, increased CYP11B2 gene expression in renal and adrenal tissues of mice with haplotype-I (−344T), as compared to mice with haplotype-II (−344C). Importantly, the haplotype-I TG male mice have significantly increased baseline blood pressure, an effect accentuated by high salt diet, as opposed to mice with haplotype-I. Thus, this is the first study characterizing haplotype-dependent regulation of the human CYP11B2 gene, in an in vivo setting, with examination of the consequential cardiovascular effects.
Methods
Patient Selection
We have analyzed the genomic DNA from 277 normotensive (151 males and 126 females) and 295 hypertensive subjects (179 males and 116 females). All of these subjects were recruited from the outpatient department of Westchester Medical Center (Valhalla, NY). All case and control subjects gave informed consent before participating in the research. The Institutional Review Board at the New York Medical College approved the research protocol for animal use. Individuals were excluded if they had a previous history of coronary artery disease, peripheral vascular disease, cerebrovascular disease, secondary hypertension, diabetes mellitus, or renal diseases. For a period of 30 min before blood pressure (BP) measurement, no exercise, alcohol, caffeine, or smoking were allowed. BP was measured by conventional mercury sphygmomanometer. Measurements by two different observers were taken at the left arm with individuals in the seated position after 15 min of resting. The criteria for hypertension was defined as a systolic blood pressure (SBP) > 140 mm Hg, a diastolic blood pressure (DBP) > 90 mm Hg, or under antihypertensive therapy. The normotensives (with SBP/DBP < 140/90 mm Hg) without a history of hypertension and without diabetes mellitus were recruited from the same population and matched for the sex and age. Clinical data of the patients were self-reported during a detailed pre-study examination and confirmed by medical charts provided by their physicians.
Analysis of Genomic DNA in Patients
The genomic DNA was amplified using 5'-TGG AGG GTG TAC CTG TGT CA-3' as a forward primer (hCyp11B2_463F) and 5’-GTC CTG CTG GTC TGA GGA TG-3’ as a reverse primer (hCyp11B2_192R) to amplify the 270-bp 5′-flanking region of the hCYP11B2 gene containing the T/C polymorphic site at the −344 position of the promoter. The amplified fragment was analyzed by 3.5% agarose gel electrophoresis and was confirmed by direct sequencing. For a small cohort of samples, the genomic DNA was amplified using hCyp11B2_1350F 5’-ACA GCA ATG ATG CAA GGG AT-3’ as a forward primer and hCyp11B2_192R 5’-GTC CTG CTG GTC TGA GGA TG-3’ as a reverse primer to amplify the 5′-flanking region of the hCYP11B2 gene containing the polymorphic site at the −344, −470 and −663 position of the promoter. The amplified products were resolved on 2% agarose gel and subsequently sequenced and conformed by DNA sequencing.
Plasmid Construction and Cell Culture and Transient Transfection
The hCYP11B2 reporter construct containing 2.5kb promoter having −344 T (Haplotype-I) and −344C (Haplotype-II) was PCR amplified using 5’ CCG CTC GAG ATG TCA ATG GAA ACT GGA AGC TGA AAG GC 3’ as a forward primer and 5’ CCC AAG CTT TCC AAT GCT CCC TCC ACC CTG -3’ as reverse primer. The Haplotype-I was obtained from the BAC clone RP11-304E16 which contain T at −344 position and Haplotype-II was obtained from the human genomic sample containing −344C, −470T, −663T in the promoter. The restriction enzymes XhoI and HindIII sites were used in the forward and reverse primers respectively to directionally clone the amplified templates in the pGL4-basic vector lacking eukaryotic promoter and enhancer sequences (Promega). Expression vector RSV-β-gal was obtained from Promega. Plasmid DNAs for transient transfection were prepared by Qiagen midi plasmid kit using conditions described by the manufacturer. The human adrenocortical carcinoma cells (H295R) were routinely cultured as monolayer and maintained in 100-mm tissue culture dishes with the ‘complete media’ containing the basal 1:1 mixture of Dulbecco’s Modified Eagle’s and Ham’s F12 (DMEM/F12, Gibco-Invitrogen) media, supplemented with 2.5% Nu-Serum IV (BD Biosciences), 1% ITS+ Premix (insulin, 6.25 µg/mL; transferrin, 6.25 µg/mL; selenium, 6.25 ng/mL; bovine serum albumin, 1.25 mg/mL; linoleic acid, 5.35 µg/mL, BD Biosciences) and 0.5% antibiotics (100U/ml penicillin and 100 µg/ml streptomycin mix, Gibco-Invitrogen) in a humidified atmosphere of 95% air and 5% CO2 at 37°C in an incubator. The growth media was changed every other day and upon 90% confluence, the cells were plated at about 70% density and grown in the complete media in 6-well tissue culture plates under same growth conditions. For transient transfections, reporter DNA (1µg) and β-galactosidase DNA (10 ng) were mixed with pBluescript DNA to a final weight of 1.2 µg of DNA. Transient transfections were performed with Attractene transfection reagent (Qiagen) following the manufacturer's protocol. The cells were harvested 48 h post-transfection, and whole cell extracts were prepared by resuspension in 200 µl of lysis buffer (Promega). An aliquot of the cell extract was used to measure luciferase activity in a Turners Design 20/20 luminometer using a luciferase assay system (Promega) as described by the manufacturer. Luciferase activity was normalized to β-galactosidase activity that was determined using the β-glo assay system (Promega).
Preparation and Modification of hCYP11B2 plasmid for generation of TG mice
11.2 Kb DNA encompassing the hCYP11B2 gene was obtained from a BAC clone (RP11-304E16) by PCR amplification using 5’ GCGGCCGCATGTCAATGGAAACTGGAAGCTGAAAGGC 3’ and 5’ GCGGCCGCGGCTTAGGCAAGGATTTCATGACCGAG 3’ as forward and reverse primers respectively These primers have Not1 restriction enzyme site for further sub-cloning purpose (Not1 site is shown in bold letters). The amplified DNA contains 2.5 Kb of the 5’-flanking region, 2.9 Kb of the 3’-flanking region, and 5.86 Kb coding region (containing all the exons and introns) of hCYP11B2 gene. The amplified PCR product was initially cloned in to the TOPO XL Cloning vector (Invitrogen) as per the instructions of the manufacturer and sequenced completely that authenticated the integrity of the hCYP11B2 gene. This clone contains −344T allele and therefore corresponds to haplotype-1 of hCYP11B2 gene and was designated as hCYP11b2-Hap-I. The promoter sequence of this clone has a 750 bp region encompassing polymorphic sites at −344, −470,and −663 that is recognized by unique restriction enzymes Ahd1 and Psil. Thus, hCYP11b2-344-Hap-I plasmid DNA was treated with restriction enzymes Ahd1 and Psil to remove this 750 bp fragment. Then, an 1100 bp fragment was amplified from the genomic DNA of a human subject containing haplotype-II (−344C, −470T, −663T) of the hCYP11B2 gene. The amplified fragment was treated with restriction enzymes Ahd1 and Psil and 750 bp DNA fragment was gel purified. This fragment was then ligated in the original linearized plasmid hCYP11b2-Hap-I. The resulting DNA was sequenced to confirm that it contains −344C allele (haplotype-II) and was designated as hCYP11b2-Hap-II. The TopoXL vectors containing hCYP11b2-Hap-I and hCYP11b2-Hap-II were treated with Not1 restriction enzyme and released fragments were subcloned in Not-1 restriction site in pMP8SKB vector (obtained from Dr. Sarah Bronson’s laboratory) to produce pMP8phCYP11B2-Hap-I and pMP8phCYP11B2-Hap-II. This DNA contains a unique Pvu-1 site close to the Not-1 cloning site so that the cloned DNA can be linearized after Pvu-1 digestion for electroporation in BK4 stem cells. This Pvu-1 site can also be used as a diagnostic tool to analyze the cloned DNA in this vector since it will show an additional band after Pvu1-Not1 digestion. Finally the plasmids pMP8phCYP11B2-Hap-I and pMP8phCYP11B2-Hap-II containing full length CYP11B2 with either −344 T or −344C alleles were linearized with Pvu1 restriction enzyme, columns from Qiagen purified linearized fragments.
Generation of TG Mice
The linearized plasmids were used for electroporation in BK4 ES cells in Dr. Fiering’s laboratory to generate TG mice11. After electroporation, ES cells were grown in selective HAT medium and HAT resistant colonies were isolated and expanded. DNA from different ES cells was amplified using hCYP11B2 specific primers. The PCR product was analyzed by 0.8% agarose gel electrophoresis and ES cells containing hCYP11B2 gene were identified. ES cells (containing either haplotype-I or –II of the hCYP11B2 gene) were used to generate TG mice, on the C57/BL6 background, at the Dartmouth Medical Center. We have confirmed the presence of hCYP11B2 gene by PCR amplification of the tail DNAs of these TG animals using 2 sets of hCYP11B2 gene specific primers. Finally, we confirmed TG animals containing haplotype-I and haplotype-II of hCYP11B2 gene by sequence determination of the promoter region. We initially developed 3 TG lines from each construct but after analysis kept one line each for future studies. These TG mice have single copy of the hCYP11B2 gene, as determined by Q-PCR. Male TG animals were backcrossed with female C57 mice and all experiments are performed using the heterozygous male mice with C57/BL6 background. TG animals were routinely analyzed by 3 sets of human CYP11B2 gene specific primers: first from the promoter region (forward primer 5’-ACA GCA ATG ATG CAA GGG AT-3’ and reverse primer 5’-GTC CTG CTG GTC TGA GGA TG-3 amplified −1350 and −192 region of the promoter to give 1158 bp fragment), second from the coding region (forward primer 5’-TCC AGA AAA TCT ACC AGG AAC TGG C-3’ and reverse primer 5’-ATG TTC ACT GAT GCT GGC TGC-3’ amplified 3435 and 4497 region of the coding sequence to give 1062 bp fragment), and third from the 3’-flanking region (forward primer 5’-GCT GGT CAG AAG TGG GAT AGG TT-3’ and reverse primer 5’-CCC ATA AAC AAG GAA GCC ATC TCT G-3’ amplified 5606 and 7208 region of the 3’-flanking region to give 1600 bp fragment).
Blood Pressure Measurement
All experiments were performed following an IACUC approved protocol in accordance with the NIH Guide for the Care and Use of Laboratory Animals. All mice were fed with standard mice chow and had access to water ad libitum. Blood pressure (BP) was measured in the conscious state by telemetry. A radiotelemetric system from Data Science International (St. Paul, MN) was used for this procedure. Briefly, Mice were anaesthetized with ketamine and xylazine (90 and 10 mg/kg, respectively), a 2–3 cm incision was performed exposing the neck and upper thoracic region. The left carotid artery was isolated and the tip of the telemetric catheter (model TA11PA-C10) was inserted into the carotid artery and advanced into the aortic arch, with the telemetric device main body positioned into a subcutaneous pocket into the right side of the abdomen. The skin was sutured with self-dissolving sutures. The surgery was performed under aseptic conditions. After one week of recovery from the surgical procedure, BP readings were recorded every ten minutes using Data-Science instrument as described previously12. Mean BP values were calculated for every hour from the values taken over six days. Before implantation of the BP device, the zero offset of the instrument was measured and the unit was soaked in 0.9% NaCl. The data was sampled every 10 min by averaging over 10s and stored on a hard disk. Systolic (SBP), diastolic (DBP) and mean arterial pressure (MAP), and heart rate (HR) were recorded using DATAQUEST software. For statistical analyses, six days of baseline values were used and data is analyzed using one-way ANOVA for each time point. To study the impact of Na-load on blood pressure: TG mice were treated first with low Na diet (0.01% Na) for a period of two weeks, followed by a period of normalization (2 wks.), and then treated with high-salt diet (8% Na). Blood pressure was measured using telemetric probes during the last 6 days of each treatment.
Quantitative-Real-time PCR
Adrenal and kidney from 8-week-old male TG mice containing either cyp11B2 haplotype-I or haplotype-II were harvested following CO2 asphyxiation and stored in All-protect tissue reagent (Qiagen). The Total RNA was isolated using RNeasy Plus minikit (Qiagen). Quantitative real-time RT-PCR (Q-RT-PCR) was next performed using power SYBR Green master mix (Cat# 4367659, Life technologies) and an ABI 7500 Fast Real-Time PCR System thermocycler (Applied Biosystems). Human (PPH01239F) and mouse (PPM57638A) CYP11B2 specific primers, and mouse GAPDH (PPM02946E) primers were purchased from SuperArray Bioscience Corporation (MD). Following a 95°C incubation for 10 minutes, 40 cycles of PCR (95°C/15s; 60°C/1m), were then performed on an ABI 7500 Fast Real-Time PCR System with 3 µl of cDNA, 50 nM PCR primers and 12.5ul SYBR Green PCR Master Mix in 25 µl reactions. Threshold cycles (CT) for three replicate reactions were determined and relative transcript abundance calculated following normalization with mouse GAPDH.
In Vivo ChIP assay
The ChIP assay was performed using the EZ ChIP chromatin immunoprecipitation kit from Millipore. The mice were perfused with normal saline. Adrenal and Kidney tissue were excised and washed in PBS; minced into smaller pieces; fixed with 1% formaldehyde for 20 min at room temperature; and washed with chilled PBS followed by their lysis. The DNA was fragmented by sonication, and 10 µl of the chromatin solution was saved as input. A 5-µg amount of RNA pol II antibody or rabbit immunoglobulin G was added to the tubes containing 900 µl of sonicated chromatin solution, and the mixture was incubated overnight at 4 °C. The antibody complexes were captured with protein A-agarose beads and subjected to serial washes (as described in manufacturer's protocol). The chromatin fraction was extracted with SDS buffer and reverse cross-linked at 65 °C for 4–6 h. The DNA was then purified using Qiagen PCR purification columns (Cat#28106). The immunoprecipitated DNA (1 µl) and the input DNA (1 µl) were subjected to 35 cycles of PCR amplification using −213F 5’- CAT CCT CAG ACC AGC AGG ACT TG-3’ as a forward and +29R 5’- CAC ACC TCT GCC TTT GCC CTG AGT G-3’ as a reverse primer when RNA pol II antibody was used for immunoprecipitation. This amplified 242-bp fragment containing RNA pol II-binding region of the human CYP11B2 gene promoter. The PCR-amplified products were analyzed on 2% agarose gel. Further using the same primer set the samples are subjected to Quantitative-Real-time PCR using SYBR Green PCR Master Mix. Threshold cycles (CT) for three replicate reactions were determined and relative enriched DNA abundance calculated following normalization with Input DNA.
Immunoblotting
Adrenal and renal tissues from male TG mice containing either haplotype-I, haplotype-II animals were homogenized in 250ul of homogenizing buffer (10mM potassium phosphate buffer, 250mM sucrose, 1mM EDTA, NP40 and protease inhibitor cocktail, Sigma Chemical) followed by high speed centrifugation (13000 rpm) at 4°C for 20 min. Supernatant was then collected and used for Western blot analysis. Proteins (25µg) were fractionated by SDS-PAGE (10% polyacrylamide) and transferred to Immobilon-P transfer membranes (Millipore, USA). Membranes were blocked in odyssey blocking buffer (Cat# 927–40000, LI-COR Biosciences - U.S.) and immunoblotted with commercially available monoclonal antibodies for hCYP11B2 (Cat#EPR-10494, Abcam, MA, USA) and β-actin (Cat# A2228-200Ul, Sigma-Aldrich, St. Louis, MO, USA). The immune complexes were detected by using secondary antibody conjugated with IRDye800 or IRDye700 (Cat# 926–32222 and Cat#926–32213) and images were captured using an Odyssey Imaging System (LI-COR). Densitometric analysis of protein bands was performed by Gel-Pro Analyzer software from Media Cybernetics (Media Cybernetics, Inc., PA, USA). The results were averaged and normalized with beta actin.
Immunohistochemistry
Adrenals from 3 male mice per group (Hap-I, Hap-II and c57 control) were fixed in formalin immediately after dissection. Formalin-fixed tissues were embedded in paraffin blocks and sectioned to 5µm thickness. Sections for IHC (immunohistochemistry) were baked at 60°C for 1 hour prior to hydration in alcohol and treatment with Antigen Retrieval solution. Sections were then quenched with H2O2 for 30 min. Using the VECTASTAIN Elite ABC PEROXIDASE kit PK- 6101 (Vector Laboratories Inc., Burlingame, CA), sections were blocked for 1 hour with the blocking serum provided in the kit. Next all the sections were incubated with the primary anti-CYP11B2 (1:100, rabbit monoclonal, Epitomics-an abcam company, CA, USA) antibody overnight at 4°C. On day 2 sections after washing with TBS, were incubated with secondary antibody provided in the kit and finally slides were incubated with ABC reagent (also provided in the kit). Staining was done with VECTOR DAB SUBSTRATE KIT FOR PEROXIDASE (SK-4100) and after counterstaining with hematoxylin slides were mounted with cytoseal(XYL) from Thermo Scientific and cover slips from Fisher Scientific were used to cover the sections.
Aldosterone ELISA
ALPCO aldosterone ELISA kit was used to assay serum aldosterone levels in TG and C57 mice, as per manufacturer’s protocol.
Statistical Analysis
Continuous data are expressed as means ± S.E.M. The normality of the data were checked by the D’Agostino-Pearson omnibus test. The nonparametric tests were performed using Mann-Whitney U test for two groups and Kruskal Wallis test for three groups. Mixed effects regression model was used to test for differences in mean arterial blood pressure between the groups. Haplotype associations were carried out by Chi-square test using SNP and Variation suite (Golden Helix). Allele frequencies and 95% confidence interval were calculated for each polymorphism and were tested using Chi-square test. Statistical analyses were carried out using SAS, R, GraphPad Prism, SNP and variation suite. p < 0.05 was considered Statistically significant.
Results
Haplotype-I of hCYP11B2 gene is associated with Hypertension in Caucasians
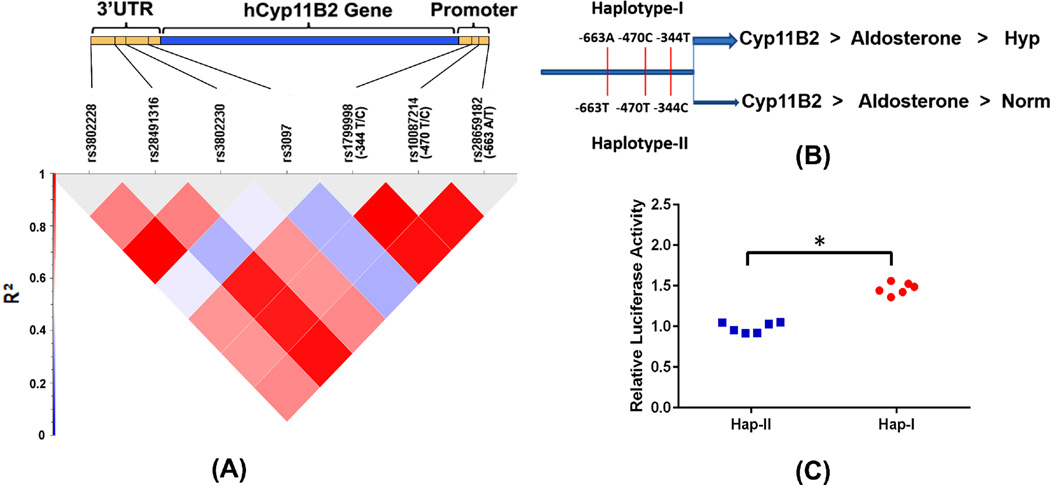
The CEU population data from 1000 genomes was used to establish the linkage disequilibrium of SNPs in the promoter of the hCYP11B2. Analysis of the 1000 genome database reveals complete linkage disequilibrium (r2=1) between the three polymorphic sites of the hCYP11B2 gene promoter (Fig. 1A). These findings were in consistent with the remaining populations from 1000 genomes and the SNPs are in complete linkage disequilibrium. We then collected patient samples and confirmed the haplotype association with human hypertension. All the samples in the study were from Caucasian population (by self-report). The genotype distributions were consistent with Hardy-Weinberg equilibrium (p=0.93). 277 normotensive (151 males and 126 females) and 295 hypertensive subjects (179 males and 116 females) were recruited from the outpatient department of the Westchester Medical Center, Valhalla, NY (mean age: 59 plus/minus 10 years). All cases were diagnosed as having primary hypertension and patients with secondary hypertension, diabetes mellitus, or ischemic heart disease were excluded. In all patient samples, the haplotype characterization is based on genotyping for the −344 polymorphic site. Complete linkage disequilibrium of SNPs, forming haplotypes II and I, is confirmed by genotyping 20 randomly selected samples for −470 and −663 polymorphic sites in addition to genotyping for −344. The frequency of the −344T allele was 0.56 (95%CI 0.52–0.60) in hypertensive subjects and 0.495 (95%CI 0.45–0.53) in normotensive subjects. The frequency of the −344C allele was 0.44 (95%CI 0.40–0.48) in hypertensive subjects and 0.505 (95%CI 0.47–0.55) in normotensive subjects. We have tested for differences in allele frequencies using Chi-square test and the test results indicated significant difference in the allele frequencies (p=0.0176). Homozygotes for −344T alleles were 100/295 in hypertensive subjects and 68/277 in normotensive subjects. On the other hand homozygotes for −344C allele were 66/295 in hypertensive subjects and 70/277 in normotensive subjects. We have used a haplotype association test using a chi square test comparing the TCA haplotype (Hap-I) with CTT haplotype (Hap-II) and found that TCA haplotype is significantly associated with hypertension (OR =1.49, 95%CI 1.21–1.82). Results of this experiment suggested that −344T allele which is part of haplotype-I of the hCYP11B2 gene is associated with increased blood pressure in our cohort.
Figure 1.
Linkage disequilibrium between the SNPs in 1Kb promoter and 3’UTR of hCyp11B2 gene (A). The CEU population data from 1000genomes was used to plot linkage disequilibrium map. Pictorial representation of SNPs constituting haplotype I and II of the hCYP11B2 gene (B). Relative luciferase activity after transient transfections in H295R cells (C). n=6. *p<0.05 vs. cells transfected with haplotype II containing pGL4 basic vector.
Transient transfection shows increased transcription of haplotype-I vs. haplotype-II
We tested haplotype-dependent transcriptional regulation of the CYP11B2 gene, in vitro, using the H295R cells. RSV-β-gal vector was used to transfect the cells with a 2.5Kb promoter of the hCYP11B2 gene, contaning either haplotype-I or haplotpe II sequence. As shown in Fig. 1B, hCYP11B2 is expressed in transfected A295R cells and haplotype-I transfected cells show significantly greater (p<0.05) expression when compared to cells transfected with haplotype-II (1C).
CYP11B2 human transgene, single copy, is expressed in TG mice with haplotype-II & I
We selectively targeted the human CYP11B2 gene to the mouse HPRT locus by employing ES cells harboring a deletion in the endogenous HPRT gene. We used a special targeting vector capable of restoring its full functionality upon homologous recombination. The use of gene targeting is essential in developing a model for studying effects of allelic gene variants in vivo because it nullifies the copy number and positional effects associated with transgene expression in TG mouse models generated by pronuclear injection. It allows reproducible insertion of the single copy of a transgene in a predetermined locus, permitting direct comparison between individually generated TG lines. As shown in Fig. S1, our TG animals contain single copy of the humanCYP11B2 gene at the HPRT locus, for both haplotype-II mice and I.
Haplotype-I of the hCYP11B2 gene increase transcriptional activity, in vivo
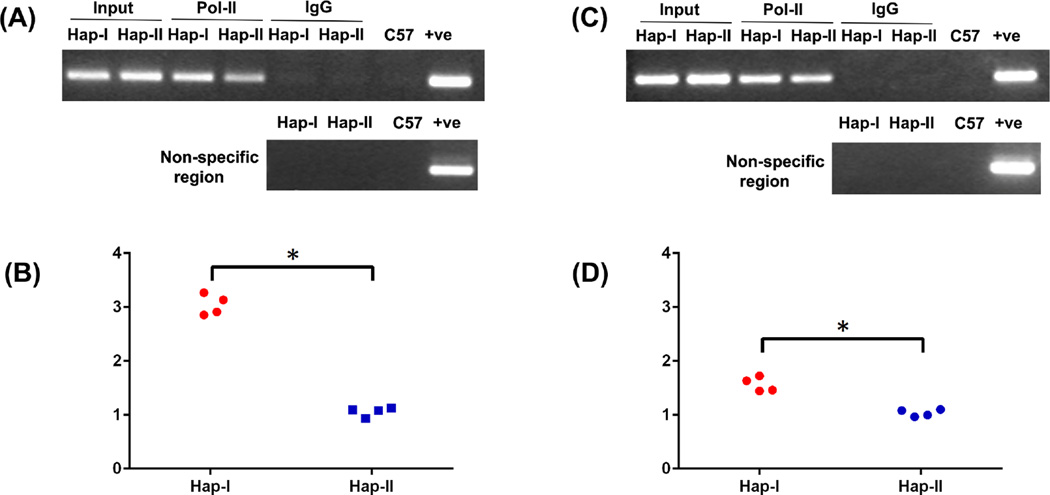
We examined differential transcriptional activity of the hCYP11B2 gene, in vivo, in our two TG lines. Chromatin immunoprecipitation (ChIP) assay was performed using Pol II-specific antibody on chromatin extracts from adrenal and renal tissues of the TG animals. As shown in Fig. 2, Pol II enrichment to the chromatin isolated from these tissues are significantly greater in mice with Haplotype-I of the hCYP11B2 gene, as compared to mice with Haplotype-II. These results show increased Pol II binding to the hCYP11B2 gene in TG mice with the haplotype-I of the hCYP11B2 gene as compared to the gene in mice with haplotype-II. The two transgenes are identical except for the specified SNPs. Thus, the ChIP experiment demonstrates haplotype-dependent increased transcription of human transgene in the haplotype-I mice.
Figure 2.
Chromatin immunoprecipitation assay shows stronger binding of Pol II to the promoter of hCYP11B2 gene from the adrenal and kidney chromatin of transgenic mice containing haplotype I as compared with haplotype II. A and C show the PCR products obtained from immunoprecipitated DNA resolved on agarose gel whereas B and D show the quantitation of immunoprecipitated DNA by real time PCR from the two haplotypes in the presence of Pol II from adrenal and kidney respectively. The Ct values obtained from the immunoprecipitated DNA were normalized to the Ct values from the input DNA. n = 4. *, p < 0.05 versus haplotype II.
CYP11B2 mRNA is increased in the Adrenals and renal tissues of TG Mice containing Haplotype-I of the hCYP11B2 gene, as compared to the Haplotype-II
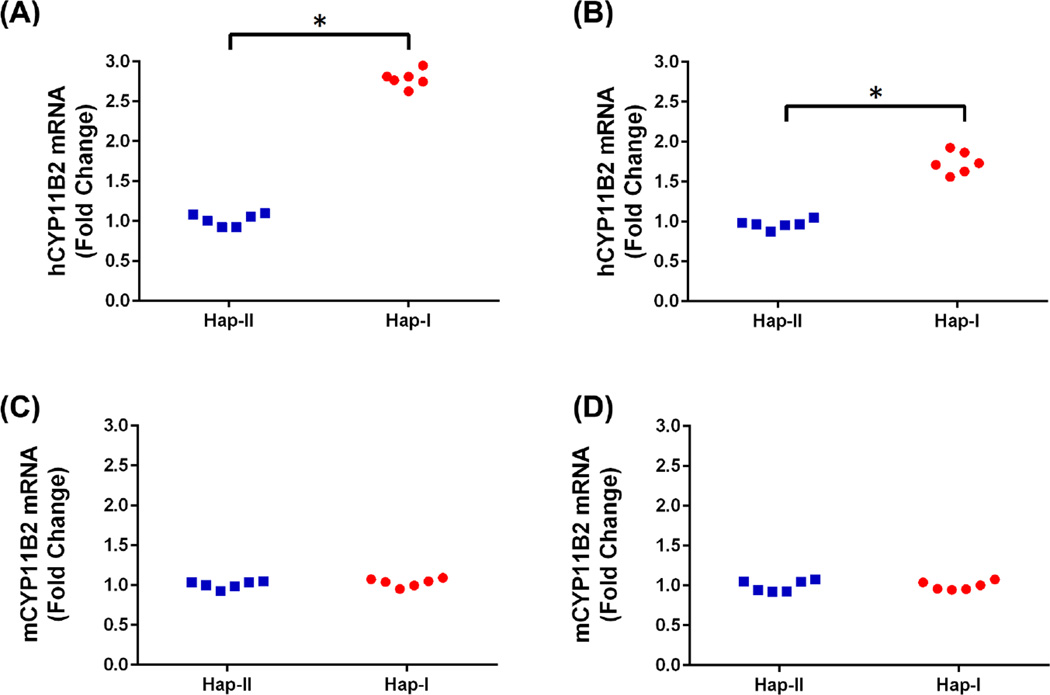
Relative gene expression of the hCYP11B2 gene was quantified in adrenal and renal tissues of the TG mice using PCR and gel electrophoresis. Significantly increased (p<0.05) hCYP11B2 gene expression is observed in adrenal (Fig. S2: 2.21 fold change) and renal (Fig. S2: 1.54 fold change) tissues of the haplotype-I TG mice as compared to the tissues from haplotype-II TG mice. No expression of the human transgene was observed in the regular C57 control mice. These experiments establish the presence of the transgene in the TG mice and its absence in the C57 controls.
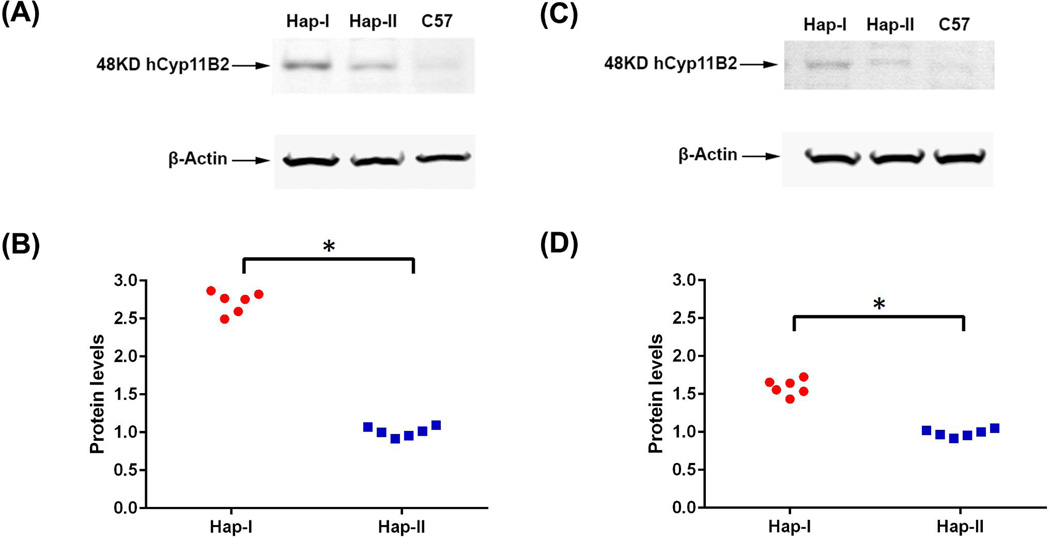
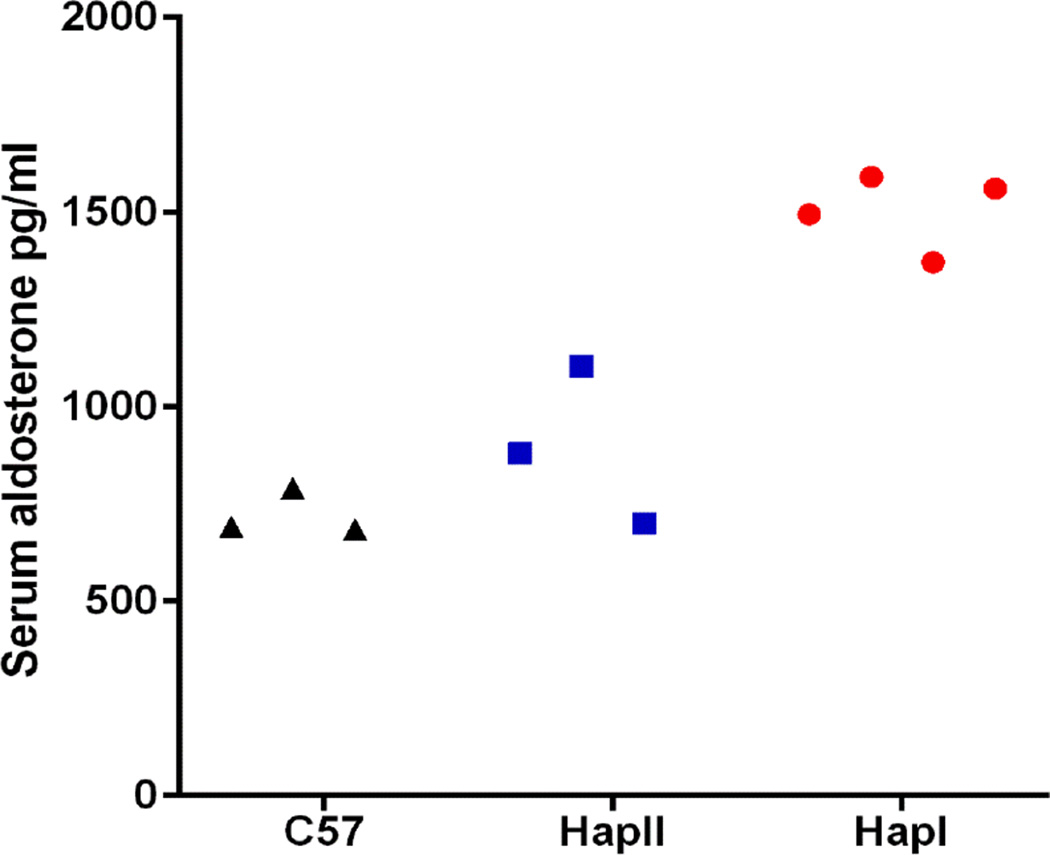
Complementary experiments were conducted to quantitate human and mouse CYP11B2 expression in adrenal and renal tissues of the TG animals. As shown in Fig. 3, renal and adrenal tissues from the TG mice with haplotype-I have significantly (p<0.05) elevated levels of the human CYP11B2 transgene as compared to mice with haplotype-II. This differential expression was not observed in the mouse CYP11B2 gene. Presence of the transgene did not affect endogenous mCYP11B2 expression as no significant difference in gene expression was observed in the two haplotypes, when compared to C57 mice (normalized CT values for mCYP11B2 in the adrenals: haplotype-I- 1.76±0.26, haplotype-II- 1.54±0.24, C57– 1.63±0.1). These results indicate that the polymorphisms in the human CYP11B2 gene predisposed it to varied transcriptional regulation in the two haplotypes. This was confirmed with immunoblot analyses of the adrenal and renal tissues (Fig. 4). Haplotype-I TG mice demonstrated significantly (p<0.05) elevated levels of the human transgene when probed with a human-specific anti-CYP11B2 antibody, as compared to tissues from the haplotype-II TG mice. Plasma aldosterone levels were also significantly (p=0.002) elevated in the haplotype-I TG mice when compared to both, plasma aldosterone levels in haplotype-II TG and C57 mice (Fig. 5). It is important to note that the transgene was not detected in naïve C57 mice and the mere presence of the transgene does not increase baseline aldosterone levels in TG mice with haplotype-II.
Figure 3.
Quantitative-RT-PCR analysis showing hCYP11B2 expression in the two haplotypes of the TG-mice. Panel A shows results in the adrenals while panel B shows results from the kidneys. n=6. * p<0.05 vs. haplotype II. Panel C (adrenal) and D (renal) show Q-RT-PCR analyses for the mCYP11B2 in these transgenic mice. n=6.
Figure 4.
hCYP11B2 Expression in adrenal gland (A) and Kidney tissue (C) of transgenic mice containing either haplotype-I or haplotype-II by Western Blot analysis. Panel A shows representative Western blot and Panel B shows quantitation of bands from the adrenal glands. Panel C shows representative Western blot and Panel D shows quantitation of bands from the kidneys. n=6. * p<0.05 vs. haplotype II.
Figure 5.
Aldosterone levels measurement by ELISA from the serum of the transgenic mice containing haplotype-I and haplotype-II of hCYP11B2 gene and control C57 mice. n = 3–4 (p=0.002).
hCYP11B2 is localized to the zona glomerulosa in the adrenal glands
Immunohistochemistry shows localization of the hCYP11B2 in the zona glomerulosa of the adrenal cortex i.e. in its native environment (Fig. S3). The color intensity is appreciably higher in cross-sections from mice with haplotype-I, as compared to sections from mice with haplotype-II. Absence of reactivity in tissue-sections from C57 mice points to the specificity of the technique in differentiating the hCYP11B2 protein.
Blood Pressure is increased in TG Mice containing Haplotype-I of hCYP11B2 gene as compared to Haplotype-II
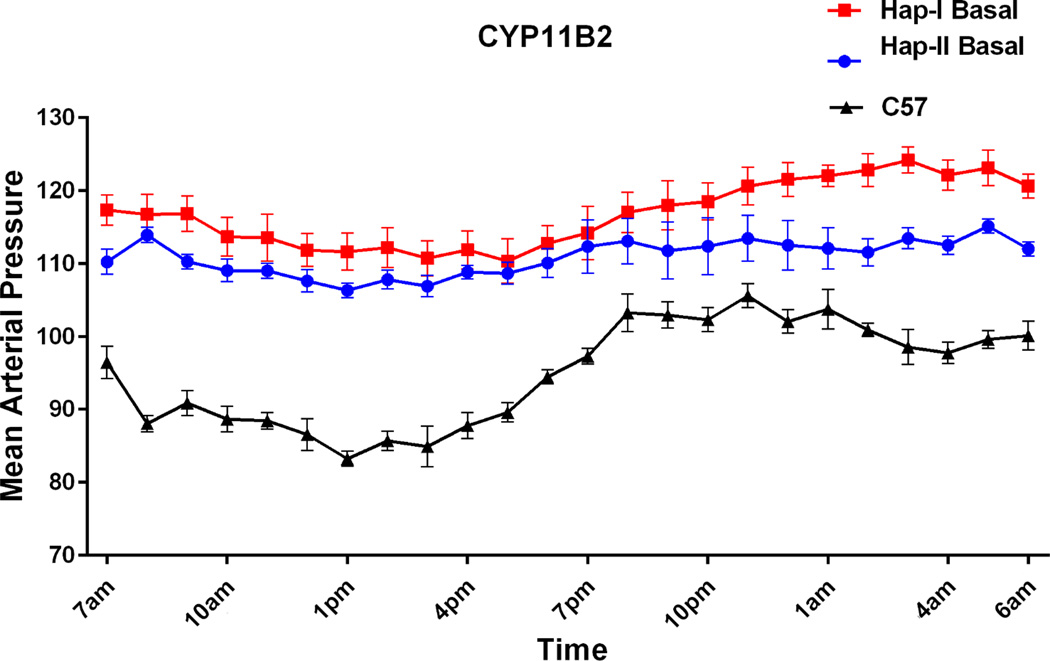
We next determined the effect of haplotype-I and haplotype-II of hCYP11B2 gene on blood pressure of TG mice. Butz and Davisson12 have provided details of blood pressure measurement using telemetry. They have shown that this is a reliable procedure for providing high-fidelity mean arterial pressure (MAP) and heart rate (HR) recordings for 50–60 days in mice weighing 22 g on average but as small as 17 g. No morbidity or mortality was observed in different strains of mice using this procedure. Importantly, different strains of mice fully recovered from anesthesia and surgery, as indicated by a return of normal circadian rhythms, after 5–7 days post-surgery. We have previously used this procedure11, 13 and also used it in current studies. Mean arterial pressure of male TG animals containing haplotype-I and haplotype-II of hCYP11B2 gene is shown in Fig. 6. Blood pressure was measured in 8–12 weeks old conscious mice for 24 hr over a period of six days (n=6). Results of this experiment showed that TG mice with haplotype-I (−344T allele)(red line) had an average increase of 7mm Hg in blood pressure over TG mice containing haplotype-II (−344C allele)(blue line). It is important to mention that blood pressure was increased by 9mmHg during the nighttime when animals were active.
Figure 6.
Mean arterial pressure tracing over 24-hrs using telemetry probes. Tracing for TG mice containing haplotype I and haplotype II of hCYP11B2 gene are shown in red and blue, and of control C57 mice is shown in black respectively. n=6. p<0.0005 between haplotype I vs. haplotype II.
Pathophysiological effects of haplotypes on salt-dependence of blood pressure and on inflammatory mediators
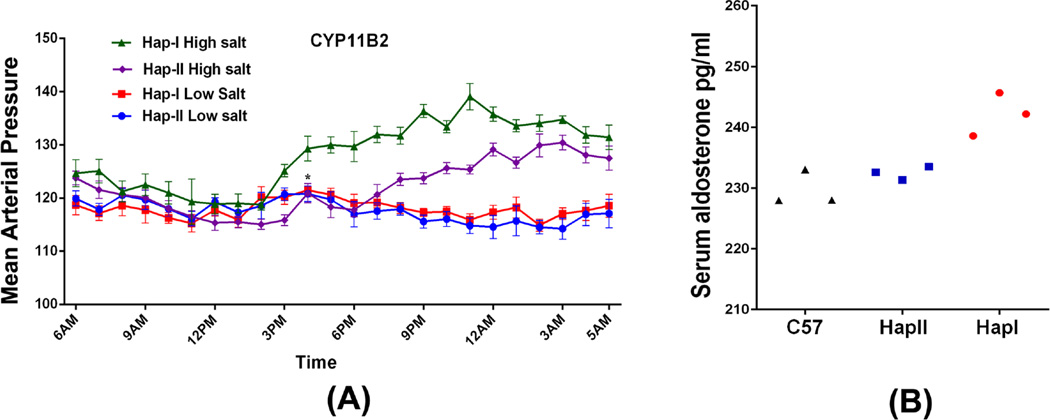
RAAS overcativity is associated with salt-dependent hypertension. We examined the effects of increased renal levels of aldosterone on blood pressure in the two TG lines, in the presence of low (0.01% NaCl) or high salt (8% NaCl) diet (two weeks of salt treatment). There was no difference in BP of TG mice containing either Hap-I or Hap-II of hCYP11B2 gene on a low salt diet (Fig. 7A). High-salt treatment increased the MAP both haplotypes however, this increase was significantly higher in TG mice haplotype-I as compared to mice with haplotype-II (Fig. 7A). On the other hand, salt treatment had no significant effect on the blood pressure in non-transgenic C57 mice (97.8±2.8 mmHg Vs. 95.5±3.5 mmHg at baseline). Na load suppresses the RAS including plasma aldosterone levels. As expected, mice treated with high-salt diet showed significant reduction in plasma aldosterone levels. It is important to note, however, that the aldosterone suppression was attenuated in TG mice with haplotype-I of the hCYP11B2 gene (Fig. 7B). These results strongly suggest that inappropriately high aldosterone levels upon Na loading could mediate increased blood pressure in haplotype-I mice.
Figure 7.
Mean arterial pressure tracing over 24-hrs using telemetry probes from the transgenic mice with haplotype II and haplotype I of the hCYP11B2 gene fed low salt and high salt diet (A). n=6. P<0.0005 between each group Hap-I High salt vs Hap-I Low salt, Hap-II High salt vs Hap-II Low salt and Hap-I high salt vs Hap-II high salt. The serum aldosterone levels after high salt diet from haplotype I, haplotype II and C57 are shown in (B). n=3. p=0.002
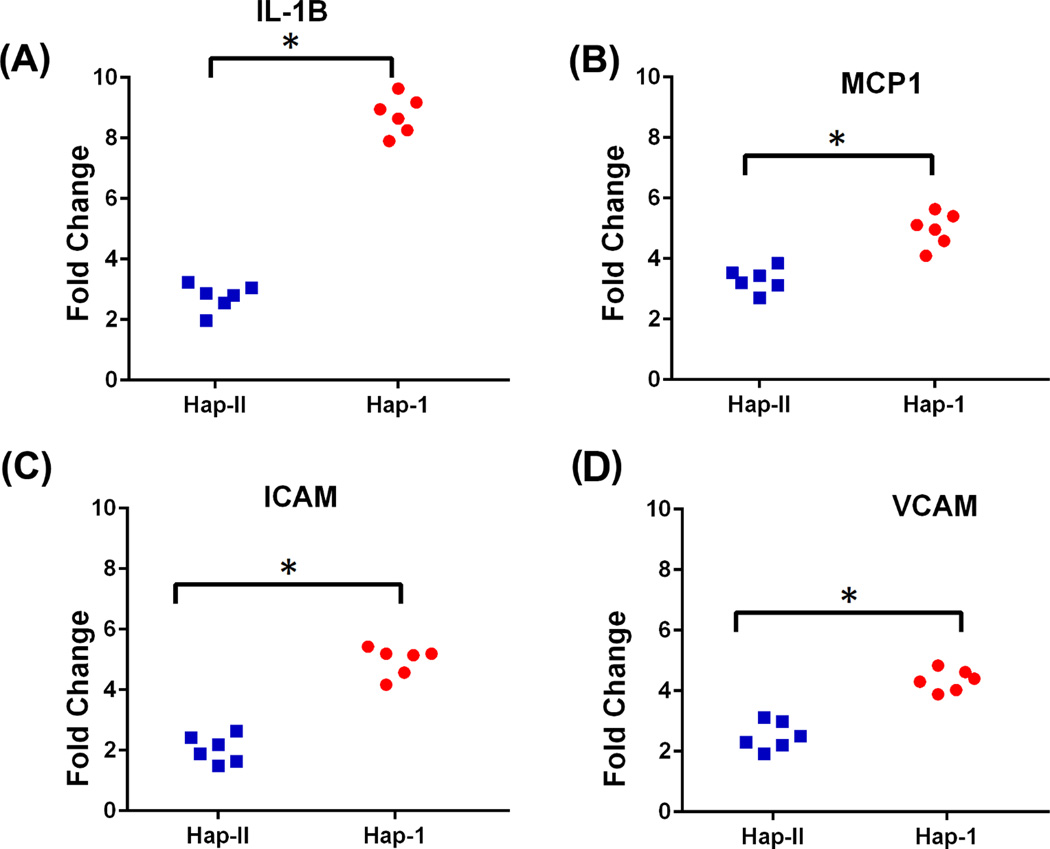
We confirmed pathophysiological impact of elevated renal aldosterone levels via assessment of inflammatory markers, previously shown to be up regulated by aldosterone. As shown in Fig. 8, renal expression of IL1β, MCP1, iCAM, and VCAM is significantly increased (p<0.05) in mice with haplotype-I of the hCYP11B2 gene when compared to mice with haplotype-II.
Figure 8.
mRNA levels of pro-inflammatory markers in the kidneys of transgenic mice containing haplotype II and I of hCYP11B2 gene. n=6. * p<0.05 vs. haplotype II.
Discussion
Hypertension is a complex disease encompassing multiple patho-physiological processes on a backdrop of genetic predisposition. Inter-individual variation of blood pressure, up to 45%, can be accounted for by genetic differences in genes regulating the physiological processes governing blood pressure. Inappropriate levels of aldosterone are observed in 15% of patients with essential hypertension14–16 that increases to 22% in patients with resistant hypertension17. In this regard, association studies have linked CYP11B2 polymorphism to human hypertension and cardiovascular diseases. The first key finding of the study identifies two polymorphic sites (−470 & −663) on the hCYP11B2 promoter that almost always occur in linkage disequilibrium with the −344 site. Together, −663A, −470C and −344T form the haplotype-I; −663T, −470T, and −344C are categorized as the haplotype-II. This is the first report identifying these linked polymorphisms in the hCYP11B2 gene. Importantly, Haplotype-I of the hCYP11B2 gene is associated with human hypertension and shows increased transcriptional activity, in vitro.
Physiological impact of enhanced transcriptional activity of the gene was tested in novel TG animals generated with either haplotype-I transgene or II. TG mice are at present the most rigorous system available for identifying and characterizing cis-acting DNA elements, and to understand the role of these elements in transcriptional regulation of a gene in different cell types. However, a transgene can integrate at different sites in the chromosome and depending on the site of integration promoter activity may vary from experiment to experiment. Additionally, different number of transgene copies may integrate in the genome of different mice and promoter activity may vary depending on the number of transgenes in a particular line. In order to overcome these limitations, Bronson et al have performed gene targeting at the HPRT locus to selectively target a single copy of the gene in a controlled fashion in the genome18. We11 and others19, 20 have used this strategy to generate TG mice containing a single copy of the human angiotensinogen (AGT) gene containing all the exons, introns, and 3' and 5' flanking regions of the gene. Results of these experiments showed that insertion of the single copy transgene upstream of HPRT locus does not affect the overall tissue and cell-specific expression or hormonal regulation of human AGT gene.
The second key finding of the study is the generation of hCYP11B2 TG mice where the transgene is targeted to the mouse HPRT locus. We show here that both haplotypes have a single copy of the transgene with observation of increased CYP11B2 expression in TG mice with haplotype-I, as compared to mice with haplotype-II. It is noteworthy that increased CYP11B2 expression was observed in both the adrenals and the renal tissues. Where classical view of the mineralocorticoids places aldosterone synthase primarily in the adrenal cortex, recently emerging evidence highlights CYP11B2 expression in extra adrenal tissues, especially kidneys. The increased tissue expression of aldosterone synthase, in haplotype-I, could depend on the increased binding affinity for the transcription factors regulating CYP11B2 gene expression. In vivo ChIP assay supports this notion by showing increased Pol II binding to the transgene in haplotype-I mice. This RNA polymerase is essential for eukaryotic transcription and increased chromatin binding of this enzyme strongly suggests enhanced transcriptional activity of the gene. Pol II binding and activity is aided by a plethora of transcription factors, both enhancers and suppressers, binding to the chromatin regions with sequence homology to the transcription factor. SNPs in the promoter and/or enhancer regions of the gene alter this binding and thus, have the potential to modulate gene transcription. In this regard, in silico analysis suggests that transcription factor GATA-1 may bind to oligonucleotides containing either −663A (in LD with −344T) or −663T (in LD with −344C). However, transcription factor AP4 binds more strongly to the oligonucleotide containing −663T. This suggests that AP4 may be interfering in transcriptional activation by GATA-1 resulting in reduced transcriptional activation of CYP11B2 gene containing −344C haplotype when compared to −344T haplotype. Similarly, transcription factors ETS, MYC, and USF have stronger homology with the promoter sequence containing −470C (in LD with −344T) as compared to −470T (in LD with −344C) of the hCYP11B2 gene. We can speculate that binding of these transcription factors will increase the expression of CYP11B2 gene containing −344T haplotype.
Aldosterone is the key regulator of Na balance via activation of the MR in the principal cells of the cortical collecting tubule. Additionally, redox and structural effects of aldosterone are now well established. Chronically elevated aldosterone induces cardiac hypertrophy and fibrosis; causes vascular remodeling, including perivascular fibrosis and reduced arterial distensibility; and increases the activity of cellular oxidases and precipitates redox imbalance21, 22. Complementary clinical studies have demonstrated favorable outcomes in patients with cardiovascular diseases being treated with aldosterone antagonists (SAVE, CONSENSUS). Also, patients with aldosterone excess have substantially higher rates of atrial fibrillation, stroke, and myocardial infarction compared with patients with essential hypertension who were matched according to their level of blood pressure elevation23. In line with this historical evidence, the third key finding of the study shows that increased CYP11B2 expression in TG mice with haplotype-I was accompanied by increased circulating levels of aldosterone with higher systolic, diastolic and mean arterial pressure. Importantly, achievement of the physiological steady state during a Na+ replete diet is critically reliant upon the withdrawal of RAS thereby, increasing the steepness of the pressure-natriuresis response and decreased renal aldosterone levels. Combined, these effects promote Na+ excretion, restoration of effective circulating volume and maintenance of the cardiovascular steady state. Haplotype-dependent overexpression of CYP11B2, apart from increasing baseline blood pressure, will also prevent excretion of Na+ during dietary excesses. This notion is supported by our findings of greater blood pressure increase in −344T haplotype, when placed on a high salt diet. Inappropriately elevated aldosterone levels have been suggested to underlie salt-sensitivity in both humans and animal models. Our results are line with these observations and provide translational bridge to the studies reporting hypertension in patients with the −344T polymorphism. African American and South Asian ancestry is a risk factor for salt-sensitive hypertension and these are the cohorts where −344T has been observed to be associated with high blood pressure. It appears plausible that the −344T haplotype-Is associated with incomplete suppression of the CYP11B2 gene during a high salt diet. This in turn, will lead to increased blood pressure in mice with this haplotype during Na+ excess, as was observed here. It is important to mention that allele frequency of −344T is much higher in people with African, African-American and South Asian ancestry as compared to Caucasians6,7.
Aldosterone also induces the synthesis of proinflammatory molecules such as MCP-1, adhesion molecules, and cytokines such as IL-6. Additionally, aldosterone increases production of reactive oxygen species, possibly through increased expression of key intracellular regulators such as NFκB. Many of the long-term structural and functional effects of aldosterone are attributed to the activation of these mediators24. Contextually, higher CYP11B2 expression in TG mice with haplotype-I is accompanied by up-regulation of these inflammatory, adhesion ad redox markers. These findings further support our hypothesis that haplotype-dependent differential regulation of the CYP11B2 gene predisposes to pathophysiological effects observed with inappropriate kevels of aldosterone.
In conclusion, we have identified novel polymorphisms in the hCYP11B2 gene that are in linkage disequilibrium and divide the population in two haplotypes, termed I and II. These polymorphisms in the gene-promoter favor Pol II binding and increase gene transcription, most likely via altered chromatin-transcription factor association. Consequential increase in tissue and plasma aldosterone bioavailability contributes to salt-sensitive hypertension and a pathophysiological setting of prooxidant and proinflammatory milieu in tissues in mice with the haplotype-I transgene. Haplotype-dependent differential regulation and expression of CYP11B2 has important clinical implications. Patients with the haplotype-I of this gene are at an increased risk of inappropriately high aldosterone levels, predisposing these patients to aldosterone-dependent long-term morbidity; especially in cohort with fluctuations in dietary Na.
Acknowledgments
Funding Sources: This work was supported by NIH grants HL81752, HL105113, and HL092558 to AK
Footnotes
Conflict of Interest Disclosures: None
References
- 1.White PC, Slutsker L. Haplotype analysis of CYP11B2. Endocr Res. 1995;21:437–442. doi: 10.3109/07435809509030459. [DOI] [PubMed] [Google Scholar]
- 2.Tanahashi H, Mune T, Takahashi Y, Isaji M, Suwa T, Morita H, et al. Association of Lys173 Arg polymorphism with CYP11B2 expression in normal adrenal glands and aldosterone-producing adenomas. J Clin Endocrinol Metab. 2005;90:6226–6231. doi: 10.1210/jc.2005-0299. [DOI] [PubMed] [Google Scholar]
- 3.Casiglia E, Tikhonoff V, Mazza A, Rynkiewicz A, Limon J, Caffi S, et al. C-344Tpolymorphism of the aldosterone synthase gene and blood pressure in the elderly: a population-based study. J Hypertens. 2005;23:1991–1996. doi: 10.1097/01.hjh.0000183119.92455.a7. [DOI] [PubMed] [Google Scholar]
- 4.Russo P, Siani A, Venezia A, Iacone R, Russo O, Barba G, et al. Interaction between the C(−344)T polymorphism of CYP11B2 and age in the regulation of blood pressure and plasma aldosterone levels: cross-sectional and longitudinal findings of the Olivetti Prospective Heart Study. J Hypertens. 2002;20:1785–1792. doi: 10.1097/00004872-200209000-00023. [DOI] [PubMed] [Google Scholar]
- 5.Davies E, Holloway CD, Ingram MC, Inglis GC, Friel EC, Morrison C, et al. Aldosterone excretion rate and blood pressure in essential hypertension are related to polymorphic differences in the aldosterone synthase gene CYP11B2. Hypertension. 1999;33:703–707. doi: 10.1161/01.hyp.33.2.703. [DOI] [PubMed] [Google Scholar]
- 6.Tiago AD, Badenhorst D, Nkeh B, Candy GP, Brooksbank R, Sareli P, et al. Impact of renin-angiotensin- aldosterone system gene variants on the severity of hypertension in patients with newly diagnosed hypertension. Am J Hypertens. 2003;16:1006–1010. doi: 10.1016/j.amjhyper.2003.07.010. [DOI] [PubMed] [Google Scholar]
- 7.Rajan S, Ramu P, Umamaheswaran G, Adithan C. Association of aldosterone synthase (CYP11B2 C- 344T) gene polymorphism & susceptibility to essential hypertension in a south Indian Tamil population. Indian J Med Res. 2010;132:379–385. [PubMed] [Google Scholar]
- 8.Sookoian S, Gianotti TF, Gonzalez CD, Pirola CJ. Association of the C-344T aldosterone synthase gene variant with essential hypertension: a meta-analysis. J Hypertens. 2007;25:5–13. doi: 10.1097/01.hjh.0000254372.88488.a9. [DOI] [PubMed] [Google Scholar]
- 9.Iwai N, Kajimoto K, Tomoike H, Takashima N. Polymorphism of CYP11B2 determines salt sensitivity in Japanese. Hypertension. 2007;49:825–831. doi: 10.1161/01.HYP.0000258796.52134.26. [DOI] [PubMed] [Google Scholar]
- 10.Bassett MH, Zhang Y, Clyne C, White PC, Rainey WE. Differential regulation of aldosterone synthase and 11beta-hydroxylase transcription by steroidogenic factor-1. J Mol Endocrinol. 2002;28:125–135. doi: 10.1677/jme.0.0280125. [DOI] [PubMed] [Google Scholar]
- 11.Jain S, Vinukonda G, Fiering SN, Kumar A. A haplotype of human angiotensinogen gene containing - 217A increases blood pressure in transgenic mice compared with-217G. Am J Physiol Regul Integr Comp Physiol. 2008;295:R1849–R1857. doi: 10.1152/ajpregu.90637.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Butz GM, Davisson RL. Long-term telemetric measurement of cardiovascular parameters in awake mice: a physiological genomics tool. Physiol Genomics. 2001;5:89–97. doi: 10.1152/physiolgenomics.2001.5.2.89. [DOI] [PubMed] [Google Scholar]
- 13.Jain S, Tillinger A, Mopidevi B, Pandey VG, Chauhan CK, Fiering SN, et al. Transgenic mice with-6A haplotype of the human angiotensinogen gene have increased blood pressure compared with-6G haplotype. J Biol Chem. 2010;285:41172–41186. doi: 10.1074/jbc.M110.167585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Young WF. Primary aldosteronism: renaissance of a syndrome. Clin Endocrinol (Oxf) 2007;66:607–618. doi: 10.1111/j.1365-2265.2007.02775.x. [DOI] [PubMed] [Google Scholar]
- 15.Rossi GP, Bernini G, Caliumi C, Desideri G, Fabris B, Ferri C, et al. A prospective study of the prevalence of primary aldosteronism in 1,125 hypertensive patients. J Am Coll Cardiol. 2006;48:2293–2300. doi: 10.1016/j.jacc.2006.07.059. [DOI] [PubMed] [Google Scholar]
- 16.Mulatero P, Stowasser M, Loh KC, Fardella CE, Gordon RD, Mosso L, et al. Increased diagnosis of primary aldosteronism, including surgically correctable forms, in centers from five continents. J Clin Endocrinol Metab. 2004;89:1045–1050. doi: 10.1210/jc.2003-031337. [DOI] [PubMed] [Google Scholar]
- 17.Calhoun DA, Nishizaka MK, Zaman MA, Thakkar RB, Weissmann P. Hyperaldosteronism among black and white subjects with resistant hypertension. Hypertension. 2002;40:892–896. doi: 10.1161/01.hyp.0000040261.30455.b6. [DOI] [PubMed] [Google Scholar]
- 18.Bronson SK, Plaehn EG, Kluckman KD, Hagaman JR, Maeda N, Smithies O. Single-copy transgenic mice with chosen-site integration. Proc Natl Acad Sci U S A. 1996;93:9067–9072. doi: 10.1073/pnas.93.17.9067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cvetkovic B, Yang B, Williamson RA, Sigmund CD. Appropriate tissue- and cell-specific expression of a single copy human angiotensinogen transgene specifically targeted upstream of the HPRT locus by homologous recombination. J Biol Chem. 2000;275:1073–1078. doi: 10.1074/jbc.275.2.1073. [DOI] [PubMed] [Google Scholar]
- 20.Cvetkovic B, Keen HL, Zhang X, Davis D, Yang B, Sigmund CD. Physiological significance of two common haplotypes of human angiotensinogen using gene targeting in the mouse. Physiol Genomics. 2002;11:253–262. doi: 10.1152/physiolgenomics.00076.2002. [DOI] [PubMed] [Google Scholar]
- 21.Fiebeler A, Luft FC. The mineralocorticoid receptor and oxidative stress. Heart Fail Rev. 2005;10:47–52. doi: 10.1007/s10741-005-2348-y. [DOI] [PubMed] [Google Scholar]
- 22.Habibi J, DeMarco VG, Ma L, Pulakat L, Rainey WE, Whaley-Connell AT, et al. Mineralocorticoid receptor blockade improves diastolic function independent of blood pressure reduction in a transgenic model of RAAS overexpression. Am J Physiol Heart Circ Physiol. 2011;300:H1484–H1491. doi: 10.1152/ajpheart.01000.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Milliez P, Girerd X, Plouin PF, Blacher J, Safar ME, Mourad JJ. Evidence for an increased rate of cardiovascular events in patients with primary aldosteronism. J Am Coll Cardiol. 2005;45:1243–1248. doi: 10.1016/j.jacc.2005.01.015. [DOI] [PubMed] [Google Scholar]
- 24.Connell JM, Mackenzie SM, Freel EM, Fraser R, Davies E. A lifetime of aldosterone excess: long-term consequences of altered regulation of aldosterone production for cardiovascular function. Endocr Rev. 2008;29:133–154. doi: 10.1210/er.2007-0030. [DOI] [PubMed] [Google Scholar]