Abstract
Immunotherapy has demonstrated impressive outcomes for some patients with cancer. However, selecting patients who are most likely to respond to immunotherapy remains a clinical challenge. Here, we discuss immune escape mechanisms exploited by cancer and present strategies for applying this knowledge to improving the efficacy of cancer immunotherapy.
Introduction
The immune system is a critical regulator of tumor biology with the capacity to support or inhibit tumor development, growth, invasion and metastasis. Strategies designed to harness the immune system are the focus of several recent promising therapeutic approaches for cancer patients. For example, adoptive T cell therapy has produced impressive remissions in patients with advanced malignancies (1). In addition, therapeutic monoclonal antibodies designed to disrupt inhibitory signals received by T cells through the cytotoxic T-lymphocyte-associated antigen 4 (CTLA-4; also known as CD152) and programmed cell death-1 (PD-1; also known as CD279) molecules are demonstrating long-term survival benefits for some patients with metastatic melanoma (2, 3). However, not all tumors appear to respond to these immunomodulatory maneuvers. This observation emphasizes the heterogeneity of cancer and suggests the existence of additional immunoregulatory mechanisms in many patients. A major challenge for cancer immunotherapy, therefore, lies in understanding these resistance mechanisms for selecting patients who are most likely to benefit.
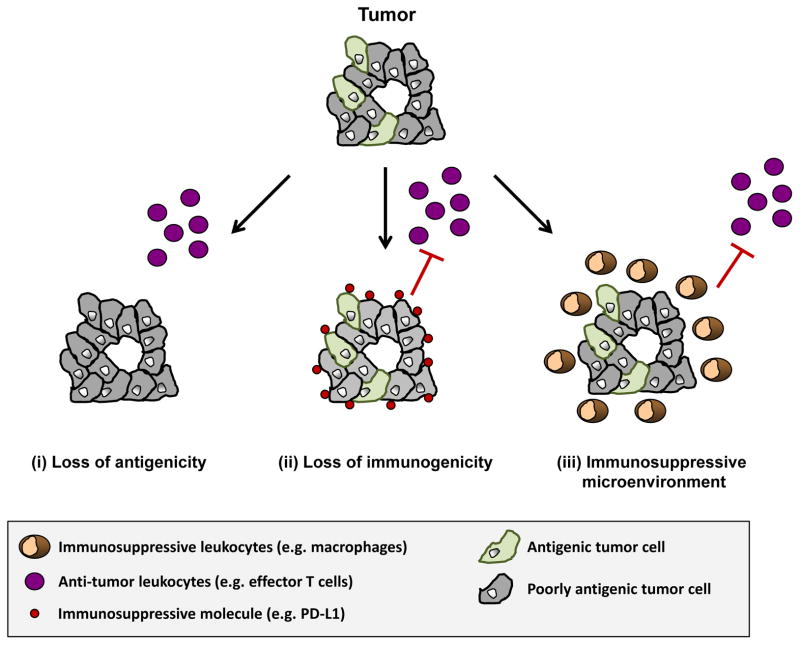
A central tenet of cancer immunotherapy is that the immune system actively surveys for malignant transformation and can be induced to recognize and eliminate malignant cells. This premise was initially proposed by Thomas and Burnet in 1957 in the immunosurveillance hypothesis which postulated a role for the immune system in controlling the development and outgrowth of nascent transformed cells (4., 5). This hypothesis has since been refined based on knowledge that the immune system cannot only protect against tumor development but can also select for tumors with decreased antigenicity and/or immunogenicity and therefore, promote tumor outgrowth. In this process termed “cancer immunoediting”, cancer clones evolve to avoid immune-mediated elimination by leukocytes that have anti-tumor properties (6). However, some tumors may also escape elimination by recruiting immunosuppressive leukocytes which orchestrate a microenvironment that spoils the productivity of an anti-tumor immune response (7). Thus, although the immune system can be harnessed, in some cases, for its anti-tumor potential, clinically relevant tumors appear to be marked by an immune system that actively selects for poorly immunogenic tumor clones and/or establishes a microenvironment that suppresses productive anti-tumor immunity (Figure 1).
Figure 1. Immune escape mechanisms in cancer.
Clinically apparent tumors must evolve mechanisms to evade immune elimination. During this process, nascent transformed cells may be selected for based on (i) a loss of antigenicity and/or (ii) a loss of immunogenicity. Loss of antigenicity may be achieved through the acquisition of defects in antigen processing and presentation or through the loss of immunogenic tumor antigens leading to a lack of immunogenic peptides presented in the context of a peptide/MHC complex. Although a loss of antigenicity is also associated with a loss of immunogenicity, malignant cells can gain additional immunosuppressive properties, such as expression of PD-L1 or secretion of suppressive cytokines (e.g. IL-10, TGF-β), which further reduces their immunogenicity. (iii) Tumors may also escape immune elimination by orchestrating an immunosuppressive microenvironment. Malignant transformation induced by alterations in oncogenes and tumor suppressor genes can lead to the recruitment of an immune response that suppresses anti-tumor immunity.
Immune signatures for cancer immunotherapy are actively being explored across many clinical studies (8). However, unlike small molecule inhibitors, for which the presence or absence of a target mutation may predict response, the efficacy of immunotherapy relies on the functional competence of multiple immunological elements. For example, strategies designed to harness the anti-tumor potential of endogenous T cells rely on the proper execution of a series of steps which have been described in the cancer immunity cycle (9). In this cycle, tumor cells must release immunogenic tumor antigens for the priming and activation of tumor-specific T cells. Tumor-reactive T cells must then infiltrate tumor tissue and recognize cancer cells in the context of a peptide-MHC complex to induce cancer cell death. To evade immune mediated elimination, tumors must then develop strategies that disrupt this cycle. Therefore, predicting the clinical benefit of T cell immunotherapy is likely to require an understanding of each of these steps as it relates to a patient’s individual tumor. Here, we discuss tumor antigenicity, tumor immunogenicity and the tumor microenvironment as key elements of this cycle which may be used to predict clinical benefit with T cell immunotherapy and guide the development of rational combinations.
Antigenicity
The ability of the immune system to distinguish between normal and malignant cells is fundamental to cancer immunotherapy and relies, in part, on malignant cells retaining sufficient antigenicity. Tumors can express a variety of non-mutated and mutated antigens which have the potential to elicit tumor-specific immune responses (10). However, to avoid immune-mediated elimination, cancer cells may lose their antigenicity. Loss of antigenicity can arise due to the immune selection of cancer cells which lack or mutate immunogenic tumor antigens as well as through the acquisition of defects or deficiencies in antigen presentation (e.g. loss of major histocompatibility (MHC) expression or dysregulation of antigen processing machinery) (6). As a result, knowledge of the antigenicity of malignant cells may inform the potential susceptibility of a cancer to immune elimination by endogenous T cells.
Tumor antigens can be derived from viral proteins, proteins encoded by cancer-germline genes, differentiation antigens and proteins arising from somatic mutations or gene rearrangements (10). However, because malignant cells across patients and even within the same patient can be quite different, it remains unclear how to effectively quantify the antigenicity of a cancer. One approach might be to determine the frequency of somatic mutations within a cancer as a predictor of neoantigens. Until recently though, it was unclear whether mutated proteins that produce “neoantigens” could even serve as effective targets for cancer immunotherapy. Recent studies have now demonstrated that T cells recognizing neoantigens infiltrate tumor tissue and can mediate strong tumor regressions after ex vivo expansion and adoptive transfer (11-13). These findings raise the possibility that tumor mutational frequency might correlate with responsiveness to immunotherapy (14).
Mutational analysis of solid tumor malignancies has recently revealed significant variability with some immunogenic cancers, such as melanoma, displaying a high mutation rate and other non-immunogenic cancers, such as PDAC, showing a low mutation rate (15, 16). While this finding has sometimes been used to explain the poor immunogenicity of cancers like PDAC, it is also important to note that some cancers, such as renal and bladder, despite being characterized by few somatic mutations have demonstrated significant responsiveness to immunotherapy (3). Thus, at this time it remains unclear whether mutational status alone can be used to effectively identify patients with highly antigenic tumors. Certainly, not all neoantigens produced by mutations will necessarily be strongly immunogenic and non-mutated antigens may also represent key targets for cancer immunotherapy (10).
Even if a tumor expresses sufficient immunogenic antigens, immune detection is dependent on the capacity to present antigen in the context of a peptide-MHC complex. Thus, a more effective approach to addressing antigenicity may be to assess the capacity of malignant cells to present antigen. For example, tumors which lose MHC expression or acquire defects in antigen presentation may escape immune-mediated elimination by tumor-specific T cells. To this end, downregulation of MHC class I molecules has been found in approximately 20-60% of common solid malignancies including melanoma, lung, breast, renal, prostate, and bladder cancers (17). Moreover, changes in MHC expression are observed to correlate with the clinical course in some malignancies (17, 18). Beyond MHC alterations, components of antigen presentation machinery have also been found to be dysregulated in human tumors at the epigenetic, transcriptional and post-transcriptional levels (17).
For more than a decade, it has been recognized that intact antigen presentation machinery, including MHC expression, in malignant cells is critical for T cell dependent anti-tumor immunity (19, 20). More recently, this knowledge has been underscored by findings showing that MHC class I and II molecules can be used as independent prognostic factors for colorectal cancer (21) and for predicting the efficacy of immunotherapy in bladder cancer (22) and chemotherapy in ovarian cancer (23). Together, we believe that this data strongly argues a role for MHC class I/II testing of tumor tissue as a measure of antigenicity in order to identify patients who are most likely to benefit from T cell immunotherapy. We are mindful, though, that there are likely to be challenges with this approach including the capacity of MHC expression to be enhanced in response to cytokines, such as interferons, which may be induced by T cell immunotherapy. In addition, tumor biopsies to assess for MHC expression are unlikely to account for the significant level of cancer cell heterogeneity that can been seen within tumors and even between tumor lesions within the same patient. Despite these potential drawbacks though, we propose that knowledge of the antigenicity of a patient’s cancer informed through MHC testing may provide key insights into how to effectively personalize immunotherapy.
Immunogenicity
Tumors which retain sufficient antigenicity for immune recognition can escape elimination by decreasing their immunogenicity. For example, IFN-γ produced by tumor infiltrating lymphocytes can induce the upregulation of the immunoinhibitory molecule PD-L1 on malignant cells (24). Across a variety of tumor types, membranous PD-L1 expression by tumor cells has been shown to strongly correlate with lymphocyte-rich regions of a tumor and with objective responses to anti-PD-1 antibody therapy (25). These findings suggest that PD-L1 expression may be used to define a tumor that is responsive to immunotherapy. However, not all PD-L1+ tumors are associated with immune infiltrates and some PD-L1+ tumors do not respond to anti-PD-1 immunotherapy (24). Thus, additional markers of tumor immunogenicity will be needed which may involve other immune checkpoint molecules (e.g. galectin 9) expressed on tumor cells and surrounding stromal cells and/or the expression of negative regulatory markers present on tumor-infiltrating lymphocytes (e.g. PD-1, LAG-3, TIM-3, VISTA, CD244, CD160, and BTLA) (26, 27). Similar to PD-1/PD-L1, these molecular pathways can act to fine-tune the cellular fate of tumor-infiltrating T cells. In fact, it is likely that some redundancy exists between these immunoinhibitory pathways and PD-1/PD-L1 such that individual or combinations of therapies that disrupt signaling through these alternative immunoregulatory pathways may also alter the balance between activation and inhibitory signals received by T cells and lead to productive anti-tumor T cell immunity (28). The challenge, though, will be in understanding how to select patients for a particular immunomodulatory therapy as combination therapies may unveil increased toxicity (29). As such, it may be necessary to administer therapies sequentially particularly since modulation of the immune microenvironment due to improved T cell function may induce alternative mechanisms of immune evasion (30). This concept implies that the immunogenicity of cancer as suggested by the immunoediting theory is a dynamic process that is shaped by the phenotype of the surrounding microenvironment.
Tumor Microenvironment
Leukocyte infiltration into tumor tissue and recognition of malignant cells is necessary for successful immune mediated elimination. However, significant variability in the leukocyte infiltrate can be seen across tumors of different tissue types (31). For example, effector T cells are observed to infiltrate tumor tissue in some solid malignancies such as melanoma and breast carcinoma, but are rarely observed to infiltrate other malignancies such as pancreatic ductal adenocarcinoma (PDAC). While the immune response seen in each of these tumor types may initially act to inhibit tumor development, increasing evidence suggests that in some tumors, such as breast carcinoma and PDAC, tumor-infiltrating leukocytes may coordinate an active site of “immune privilege” which spoils the productivity of anti-tumor immunity (32). In support of this hypothesis, strategies that reverse immune suppression through elimination of immunosuppressive cell populations have been found to restore T cell infiltration into tumor tissue and the capacity of T cells to mediate anti-tumor activity (33-35). These findings suggest that some tumors may retain sufficient antigenicity and immunogenicity for recognition by tumor-specific T cells but evade immune elimination by orchestrating a suppressive microenvironment.
The ability of tumors to orchestrate an immunosuppressive microenvironment is dependent on reciprocal interactions between the tumor and host. This microenvironment has clear implications in shaping the cellular fate of tumor-infiltrating lymphocytes (8). As a result, strategies to redirect the phenotype of the tumor microenvironment from immunosuppressive to immunostimulatory may hold promise for enhancing the efficacy of T cell immunotherapy. Achieving this therapeutic goal, though, will require knowledge of the mechanisms of immune suppression exploited by tumors. However, immunosuppressive mechanisms that are active in tumors are likely to be highly influenced by the complexity of the cellular response that surrounds malignant cells. For example, immune suppressive mechanisms active in tumors infiltrated by T cells may be absent from tumors wherein T cells are scarce and myeloid cells dominate. This plasticity of the tumor microenvironment may explain, in part, the challenge faced by single agent immunotherapies and further may complicate the use of immune signatures obtained prior to therapy for fully guiding treatment decision making as additional mechanisms of immunoresistance may emerge during treatment. This phenomenon of emerging resistance is reminiscent of human immunodeficiency virus (HIV) (36) and suggests that combination therapy will be necessary in many cases to prevent the emergence of immune escape variants.
One example of treatment-induced immunoresistance seen in cancer involves the production of indoleamine 2,3-dioxygenase (IDO) within the tumor microenvironment in response to IFN-γ produced by anti-tumor T cells (30). IDO is an enzyme that catalyzes the degradation of the essential amino acid tryptophan to kynurenine and in doing so, drives T cell suppression. The role of IDO as an immune resistance mechanism to immune checkpoint therapy (including anti-CTLA-4 and anti-PD-1 blocking antibodies) is supported by preclinical data (37) and is the basis for an ongoing clinical study investigating combination therapy with CTLA-4 blocking antibody ipilimumab and INCB024360, a selective inhibitor of IDO (NCT01604889).
We hypothesize that the conditional state of the tumor microenvironment is pliable and that an optimal state exists for enhanced responsiveness to T cell immunotherapy. The capacity of single agent immune checkpoint blockade with anti-CTLA-4 and anti-PD-1 blocking antibodies to induce sustained remissions in some patients argues that some tumors may already exist in this optimal state. The ideal goal then is to induce a shift in the tumor microenvironment toward this optimal state for tumors that would otherwise be minimally or unresponsive to T cell immunotherapy. One approach to describing this state would be to perform molecular profiling of the tumor microenvironment. For example, melanomas characterized by a preexisting T cell-inflamed tumor microenvironment appear to display a distinct molecular profile characterized by high expression of three immunosuppressive mechanisms including IDO, PD-L1 and Foxp3+ regulatory T cells (30). Applying a similar molecular approach to tumors that are sensitive versus resistant to immunotherapy across a diverse range of cancers may provide insight into how to shift the conditional state of a tumor from immunoresistant to immunosensitive.
In addition to molecular profiling, immune profiling of cancer may also be an effective approach to define the conditional state of the tumor microenvironment for guiding treatment decisions. Immune profiling of cancer has been evaluated in many solid malignancies as a prognostic factor. In breast carcinoma, an immune signature consisting of CD68high/CD4high/CD8low cells correlates with reduced survival (33). This inverse relationship between CD68 and CD8 cells that infiltrate breast carcinomas supports a role for innate immunity in regulating T cell immunosurveillance in cancer. In colorectal carcinoma, high densities of CD8+ and CD45RO+ cells predicts decreased tumor recurrence and improved survival in patients with early-stage disease (38). Moreover, examination of the type, density and location of leukocytes infiltrating tumor tissues has been shown to correlate as a better predictor of survival than histopathological methods in patients with stage I to stage IV colorectal carcinoma (39). This observation forms the rationale for the Immunoscore which classifies tumors based on their densities of CD8+ and CD45RO+ T cells and their location within the tumor microenvironment (i.e. invasive margin versus tumor center) (40). Overall, these findings are encouraging and support the application of immune profiling as a prognostic factor. Moreover, it will also be important to understand the role of immune profiling for selecting patients who are likely to respond to immunotherapy.
Tailoring cancer immunotherapy
Strategies to enhance the efficacy of immunotherapy will need to consider immune escape mechanisms exploited by cancer. As discussed above, malignant cells can evade immune elimination through loss of antigenicity and/or loss of immunogenicity and by coordinating an immunosuppressive microenvironment. The degree to which a tumor exploits these mechanisms of immune evasion may vary by tumor type and even by tumor lesion. Variability in immune escape mechanisms exploited by cancer can be inferred from the treatment response heterogeneity seen to date with currently investigated immunotherapies. Overall, it is likely that cancer immunotherapy will need to consider tumor heterogeneity when selecting patients most likely to benefit.
The tailoring of cancer immunotherapy to a particular patient is an active area of investigation largely being explored as adoptive cell therapy using tumor-infiltrating lymphocytes or engineering T cells with chimeric antigen receptors (1). However, the application of immunomodulatory strategies such as immune checkpoint blockade and vaccine therapies will also require personalization. For instance, only a fraction of patients with melanoma respond to single agent immune checkpoint blockade (i.e. anti-CTLA-4 and anti-PD-1/PD-L1 immunotherapy). A lack of response, though, does not necessarily imply that immune checkpoint blockade has no role. For example, some patients who do not respond to anti-CTLA-4 blocking antibodies have demonstrated responses to anti-PD-1/PD-L1 immunotherapy (41). This finding supports investigation of combination therapies for which impressive response rates have been observed when anti-PD-1 antibodies are combined with anti-CTLA-4 antibodies in patients with melanoma (29). Similarly, promising results have been seen in PDAC when vaccination strategies designed to induce tumor-specific T cells are combined with immunomodulatory approaches (i.e. anti-CTLA-4 blockade) (42). Thus, selection of the appropriate immunotherapeutic or combination of immunotherapeutics may require a personalized approach.
For tumors which retain sufficient antigenicity and immunogenicity, immunotherapy will need to focus on strategies that boost tumor-specific immunity (e.g. vaccines and/or adoptive therapy with TILs) and enhance T cell killing of tumor cells (e.g. immune checkpoint blockade using anti-CTLA-4 and anti-PD-L1/PD-1 antibodies). However, it is likely that these approaches will have limited efficacy for tumors which harbor defects in antigen processing and presentation and therefore, cannot be recognized by T cells. For these tumors, strategies that re-direct innate immunity (e.g. NK cells or macrophages) with anti-tumor properties or alternatively, incorporate the adoptive cell therapy of T cells engineered to express chimeric antigen receptors (CAR) that recognize tumor specific proteins may be more effective due to their lack of dependency on MHC-restricted antigen presentation.
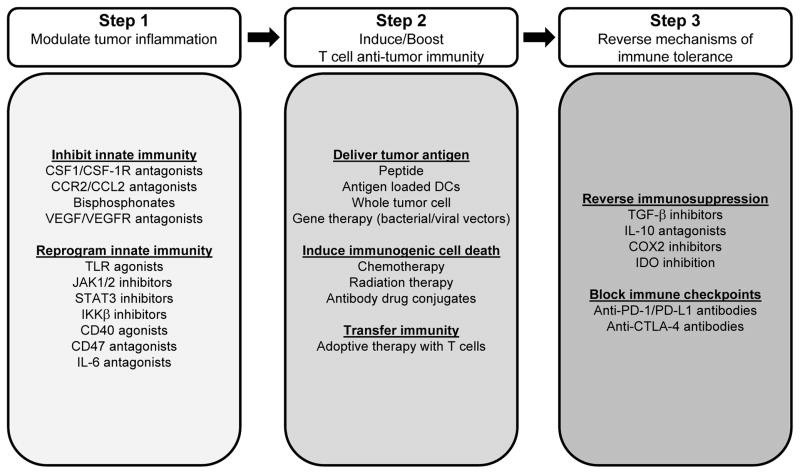
For tumors dominated by immune suppressive cell populations with a weak lymphocyte infiltrate, so-called “immune privileged” tumors, strategies that dismantle immune suppression to permit the induction of tumor-specific T cells and their trafficking to tumors will be critical. This may require a multi-step interventional process in which treatments designed to modulate the tumor microenvironment and reverse immunoregulatory mechanisms established by innate immunity are combined with approaches that invoke tumor-specific T cell immunity. In support of this approach, Highfill et al. recently demonstrated a role for inhibiting the trafficking of myeloid suppressor cells into tumors by disruption of CXCR2 signaling as a strategy for unveiling the anti-tumor activity of anti-PD-1 therapy (34). However, restoring T cell immunosurveillance in “immune privileged” tumors may eventually lead to immunoselection of malignant cells with a loss of antigenicity and/or immunogenicity. As a result, combinatorial approaches may be the key to producing sustained clinical benefit (Figure 2).
Figure 2. A multi-targeted approach to cancer immunotherapy.
A three-step process is proposed for restoring productive immunosurveillance in cancer. In Step 1, the tumor microenvironment is targeted by inhibiting pro-tumor signaling pathways engaged by tumor-associated leukocytes or by polarizing tumor-associated leukocytes with anti-tumor properties. The goal of this step is to establish an environment that is conducive to T cell priming, trafficking, and activation. In Step 2, T cell anti-tumor immunity is established by delivering tumor antigen in the form of a vaccine, by inducing immunogenic cell death or by transferring immunity with ex vivo activated tumor reactive lymphocytes. In Step 3, immunosuppressive mechanisms that restrain T cell effector function are reversed including blocking immune checkpoints that are involved in maintaining peripheral T cell tolerance. The selection of strategies from each ‘Step’ may vary for individual tumors and may be influenced by tumor antigenicity, tumor immunogenicity and the immune profile of the tumor microenvironment.
Concluding remarks
The immune system has a pivotal role in shaping tumor biology. Strategies that restore the capacity of the immune system to recognize and eliminate malignant cells have produced clinical benefit, but only for some patients. This treatment response heterogeneity may reflect the ability of tumors to adapt to immune pressure through the loss of antigenicity and immunogenicity as well as through their ability to establish an immunosuppressive microenvironment. Therefore, distinct therapeutic strategies, depending on the mechanism of immune evasion exploited by cancer, may be required for restoring productive cancer immunosurveillance. Classification of tumors based on their antigenicity and immunogenicity and on molecular and immune profiling of the tumor microenvironment may be necessary to effectively personalize cancer immunotherapy and improve clinical outcomes for more patients.
Translational Relevance.
Immunotherapy is a promising treatment modality for cancer. However, predicting which patients will benefit remains a challenge. Fundamental to the outgrowth of malignant cells is their ability to evade immune elimination and therefore, understanding the mechanisms exploited by a cancer may help guide the application of immunotherapy. Here, we propose that the tailoring of immunotherapy to patients may require knowledge of tumor antigenicity and immunogenicity as well as the composition of the tumor microenvironment. In our discussion, we describe challenges and future directions in applying this knowledge to enhancing the efficacy of immunotherapy by improving patient selection and by informing the development of rational combination therapies.
Acknowledgments
Financial Support: This work was supported by grants from the National Institutes of Health (K08 CA138907), Grant 2013107 from the Doris Duke Charitable Foundation, and the Damon Runyon Cancer Research Foundation for which Gregory L. Beatty is the Nadia’s Gift Foundation Innovator of the Damon Runyon-Rachleff Innovation Award (DRR-15-12).
Footnotes
Disclosure of potential conflicts of interest: G.L.B declares receipt of research funding from Novartis. The authors have no additional financial interests.
References
- 1.Kalos M, June CH. Adoptive T cell transfer for cancer immunotherapy in the era of synthetic biology. Immunity. 2013;39:49–60. doi: 10.1016/j.immuni.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hamid O, Robert C, Daud A, Hodi FS, Hwu WJ, Kefford R, et al. Safety and tumor responses with lambrolizumab (anti-PD-1) in melanoma. The New England journal of medicine. 2013;369:134–44. doi: 10.1056/NEJMoa1305133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. The New England journal of medicine. 2012;366:2443–54. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burnet FM. The concept of immunological surveillance. Prog Exp Tumor Res. 1970;13:1–27. doi: 10.1159/000386035. [DOI] [PubMed] [Google Scholar]
- 5.Thomas L. On immunosurveillance in human cancer. Yale J Biol Med. 1982;55:329–33. [PMC free article] [PubMed] [Google Scholar]
- 6.Schreiber RD, Old LJ, Smyth MJ. Cancer immunoediting: integrating immunitys roles in cancer suppression and promotion. Science. 2011;331:1565–70. doi: 10.1126/science.1203486. [DOI] [PubMed] [Google Scholar]
- 7.Ferrone C, Dranoff G. Dual roles for immunity in gastrointestinal cancers. J Clin Oncol. 2010;28:4045–51. doi: 10.1200/JCO.2010.27.9992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gajewski TF, Schreiber H, Fu YX. Innate and adaptive immune cells in the tumor microenvironment. Nat Immunol. 2013;14:1014–22. doi: 10.1038/ni.2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen DS, Mellman I. Oncology meets immunology: the cancer-immunity cycle. Immunity. 2013;39:1–10. doi: 10.1016/j.immuni.2013.07.012. [DOI] [PubMed] [Google Scholar]
- 10.Coulie PG, Van den Eynde BJ, van der Bruggen P, Boon T. Tumour antigens recognized by T lymphocytes: at the core of cancer immunotherapy. Nat Rev Cancer. 2014;14:135–46. doi: 10.1038/nrc3670. [DOI] [PubMed] [Google Scholar]
- 11.Tran E, Turcotte S, Gros A, Robbins PF, Lu YC, Dudley ME, et al. Cancer immunotherapy based on mutation-specific CD4+ T cells in a patient with epithelial cancer. Science. 2014;344:641–5. doi: 10.1126/science.1251102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Robbins PF, Lu YC, El-Gamil M, Li YF, Gross C, Gartner J, et al. Mining exomic sequencing data to identify mutated antigens recognized by adoptively transferred tumor-reactive T cells. Nat Med. 2013;19:747–52. doi: 10.1038/nm.3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Rooij N, van Buuren MM, Philips D, Velds A, Toebes M, Heemskerk B, et al. Tumor exome analysis reveals neoantigen-specific T-cell reactivity in an ipilimumab-responsive melanoma. J Clin Oncol. 2013;31:e439–42. doi: 10.1200/JCO.2012.47.7521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Snyder A, Makarov V, Merghoub T, Yuan J, Zaretsky JM, Desrichard A, et al. Genetic Basis for Clinical Response to CTLA-4 Blockade in Melanoma. The New England journal of medicine. 2014 doi: 10.1056/NEJMoa1406498. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matsushita H, Vesely MD, Koboldt DC, Rickert CG, Uppaluri R, Magrini VJ, et al. Cancer exome analysis reveals a T-cell-dependent mechanism of cancer immunoediting. Nature. 2012;482:400–4. doi: 10.1038/nature10755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr, Kinzler KW. Cancer genome landscapes. Science. 2013;339:1546–58. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Campoli M, Ferrone S. HLA antigen changes in malignant cells: epigenetic mechanisms and biologic significance. Oncogene. 2008;27:5869–85. doi: 10.1038/onc.2008.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang CC, Campoli M, Ferrone S. Classical and nonclassical HLA class I antigen and NK Cell-activating ligand changes in malignant cells: current challenges and future directions. Advances in cancer research. 2005;93:189–234. doi: 10.1016/S0065-230X(05)93006-6. [DOI] [PubMed] [Google Scholar]
- 19.Garrido F, Cabrera T, Concha A, Glew S, Ruiz-Cabello F, Stern PL. Natural history of HLA expression during tumour development. Immunology today. 1993;14:491–9. doi: 10.1016/0167-5699(93)90264-L. [DOI] [PubMed] [Google Scholar]
- 20.Johnsen AK, Templeton DJ, Sy M, Harding CV. Deficiency of transporter for antigen presentation (TAP) in tumor cells allows evasion of immune surveillance and increases tumorigenesis. J Immunol. 1999;163:4224–31. [PubMed] [Google Scholar]
- 21.Sconocchia G, Eppenberger-Castori S, Zlobec I, Karamitopoulou E, Arriga R, Coppola A, et al. HLA class II antigen expression in colorectal carcinoma tumors as a favorable prognostic marker. Neoplasia. 2014;16:31–42. doi: 10.1593/neo.131568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kitamura H, Torigoe T, Honma I, Sato E, Asanuma H, Hirohashi Y, et al. Effect of human leukocyte antigen class I expression of tumor cells on outcome of intravesical instillation of bacillus calmette-guerin immunotherapy for bladder cancer. Clin Cancer Res. 2006;12:4641–4. doi: 10.1158/1078-0432.CCR-06-0595. [DOI] [PubMed] [Google Scholar]
- 23.Shehata M, Mukherjee A, Deen S, Al-Attar A, Durrant LG, Chan S. Human leukocyte antigen class I expression is an independent prognostic factor in advanced ovarian cancer resistant to first-line platinum chemotherapy. Br J Cancer. 2009;101:1321–8. doi: 10.1038/sj.bjc.6605315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taube JM, Anders RA, Young GD, Xu H, Sharma R, McMiller TL, et al. Colocalization of inflammatory response with B7-h1 expression in human melanocytic lesions supports an adaptive resistance mechanism of immune escape. Science translational medicine. 2012;4:127ra37. doi: 10.1126/scitranslmed.3003689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taube JM, Klein AP, Brahmer JR, Xu H, Pan X, Kim JH, et al. Association of PD-1, PD-1 ligands, and other features of the tumor immune microenvironment with response to anti-PD-1 therapy. Clin Cancer Res. 2014 doi: 10.1158/1078-0432.CCR-13-3271. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Inozume T, Hanada K, Wang QJ, Ahmadzadeh M, Wunderlich JR, Rosenberg SA, et al. Selection of CD8+PD-1+ lymphocytes in fresh human melanomas enriches for tumor-reactive T cells. J Immunother. 2010;33:956–64. doi: 10.1097/CJI.0b013e3181fad2b0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. 2012;12:252–64. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Le Mercier I, Chen W, Lines JL, Day M, Li J, Sergent P, et al. VISTA Regulates the Development of Protective Antitumor Immunity. Cancer research. 2014;74:1933–44. doi: 10.1158/0008-5472.CAN-13-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wolchok JD, Kluger H, Callahan MK, Postow MA, Rizvi NA, Lesokhin AM, et al. Nivolumab plus ipilimumab in advanced melanoma. The New England journal of medicine. 2013;369:122–33. doi: 10.1056/NEJMoa1302369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spranger S, Spaapen RM, Zha Y, Williams J, Meng Y, Ha TT, et al. Up-regulation of PD-L1, IDO, and T(regs) in the melanoma tumor microenvironment is driven by CD8(+) T cells. Science translational medicine. 2013;5:200ra116. doi: 10.1126/scitranslmed.3006504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Coussens LM, Zitvogel L, Palucka AK. Neutralizing tumor-promoting chronic inflammation: a magic bullet? Science. 2013;339:286–91. doi: 10.1126/science.1232227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mellor AL, Munn DH. Creating immune privilege: active local suppression that benefits friends, but protects foes. Nat Rev Immunol. 2008;8:74–80. doi: 10.1038/nri2233. [DOI] [PubMed] [Google Scholar]
- 33.DeNardo DG, Brennan DJ, Rexhepaj E, Ruffell B, Shiao SL, Madden SF, et al. Leukocyte complexity predicts breast cancer survival and functionally regulates response to chemotherapy. Cancer Discov. 2011;1:54–67. doi: 10.1158/2159-8274.CD-10-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Highfill SL, Cui Y, Giles AJ, Smith JP, Zhang H, Morse E, et al. Disruption of CXCR2-mediated MDSC tumor trafficking enhances anti-PD1 efficacy. Science translational medicine. 2014;6:237ra67. doi: 10.1126/scitranslmed.3007974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lesokhin AM, Hohl TM, Kitano S, Cortez C, Hirschhorn-Cymerman D, Avogadri F, et al. Monocytic CCR2(+) myeloid-derived suppressor cells promote immune escape by limiting activated CD8 T-cell infiltration into the tumor microenvironment. Cancer research. 2012;72:876–86. doi: 10.1158/0008-5472.CAN-11-1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Laskey SB, Siliciano RF. A mechanistic theory to explain the efficacy of antiretroviral therapy. Nature reviews. 2014;12:772–80. doi: 10.1038/nrmicro3351. [DOI] [PubMed] [Google Scholar]
- 37.Holmgaard RB, Zamarin D, Munn DH, Wolchok JD, Allison JP. Indoleamine 2,3-dioxygenase is a critical resistance mechanism in antitumor T cell immunotherapy targeting CTLA-4. J Exp Med. 2013;210:1389–402. doi: 10.1084/jem.20130066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pages F, Berger A, Camus M, Sanchez-Cabo F, Costes A, Molidor R, et al. Effector memory T cells, early metastasis, and survival in colorectal cancer. The New England journal of medicine. 2005;353:2654–66. doi: 10.1056/NEJMoa051424. [DOI] [PubMed] [Google Scholar]
- 39.Galon J, Costes A, Sanchez-Cabo F, Kirilovsky A, Mlecnik B, Lagorce-Pages C, et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science. 2006;313:1960–4. doi: 10.1126/science.1129139. [DOI] [PubMed] [Google Scholar]
- 40.Galon J, Angell HK, Bedognetti D, Marincola FM. The continuum of cancer immunosurveillance: prognostic, predictive, and mechanistic signatures. Immunity. 2013;39:11–26. doi: 10.1016/j.immuni.2013.07.008. [DOI] [PubMed] [Google Scholar]
- 41.Weber JS, Kudchadkar RR, Yu B, Gallenstein D, Horak CE, Inzunza HD, et al. Safety, efficacy, and biomarkers of nivolumab with vaccine in ipilimumab-refractory or -naive melanoma. J Clin Oncol. 2013;31:4311–8. doi: 10.1200/JCO.2013.51.4802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Le DT, Lutz E, Uram JN, Sugar EA, Onners B, Solt S, et al. Evaluation of ipilimumab in combination with allogeneic pancreatic tumor cells transfected with a GM-CSF gene in previously treated pancreatic cancer. J Immunother. 2013;36:382–9. doi: 10.1097/CJI.0b013e31829fb7a2. [DOI] [PMC free article] [PubMed] [Google Scholar]