Key Points
inv(3)/t(3;3) disease exhibits high rates of activated RAS/RTK signaling, epigenetic modifier, splice, and transcription factor mutations.
AML and MDS with inv(3)/t(3;3) display similar mutational and gene expression profiles and should be considered a single molecular entity.
Abstract
Myeloid malignancies bearing chromosomal inv(3)/t(3;3) abnormalities are among the most therapy-resistant leukemias. Deregulated expression of EVI1 is the molecular hallmark of this disease; however, the genome-wide spectrum of cooperating mutations in this disease subset has not been systematically elucidated. Here, we show that 98% of inv(3)/t(3;3) myeloid malignancies harbor mutations in genes activating RAS/receptor tyrosine kinase (RTK) signaling pathways. In addition, hemizygous mutations in GATA2, as well as heterozygous alterations in RUNX1, SF3B1, and genes encoding epigenetic modifiers, frequently co-occur with the inv(3)/t(3;3) aberration. Notably, neither mutational patterns nor gene expression profiles differ across inv(3)/t(3;3) acute myeloid leukemia, chronic myeloid leukemia, and myelodysplastic syndrome cases, suggesting recognition of inv(3)/t(3;3) myeloid malignancies as a single disease entity irrespective of blast count. The high incidence of activating RAS/RTK signaling mutations may provide a target for a rational treatment strategy in this high-risk patient group.
Introduction
Acute myeloid leukemia (AML) with inv(3)(q21q26.2) or t(3;3)(q21;q26.2) [inv(3)/t(3;3)] is a distinct disease entity in the current World Health Organization classification.1 High therapy resistance is the common feature of myeloid malignancies, particularly AML with 3q21/3q26 aberrations, manifesting as low rates of complete remission and subsequent failure of current treatment strategies.2-4 Appearance of the characteristic 3q aberrations also indicates disease progression and portends adverse outcome in myelodysplastic syndrome (MDS) and chronic myeloid leukemia (CML).5-7 Therapy resistance in this subtype of malignancies is linked to the inappropriate activation of the proto-oncogene ecotropic viral integration-1 (EVI1) as a consequence of the chromosome 3 rearrangements. EVI1 is a hematopoietic stemness factor and transcription factor with chromatin-remodeling activity.8-10 EVI1 is also overexpressed in approximately 11% of all AML cases in the absence of 3q aberrations and represents an independent adverse prognostic factor in these patients.11 We and others have shown that, as a consequence of inv(3)/t(3;3) rearrangements, EVI1 becomes activated via structural repositioning of a distal GATA2 enhancer from 3q21 to the EVI1 locus at 3q26.12,13 Relocation of the enhancer additionally confers reduced and monoallelic GATA2 expression in this AML subtype. Notably, GATA2 deficiency has been shown to impair hematopoietic stem cell frequency and fitness,14-16 and Evi1 activation in murine inv(3)/t(3;3) models is followed by leukemia onset after a long latency of 6 months.13 Hence, we hypothesize that additional cooperating genetic events, other than EVI1 and GATA2 deregulation, are required for full leukemic transformation, resulting in a myeloid disease with dismal outcome. Full understanding of the complete spectrum of molecular defects associated with this highly refractory AML subtype may provide additional rationale for treatment and to overcome therapeutic nihilism in this incurable disease category. Therefore, within this study, we sought to extend the molecular characterization of myeloid disorders with inv(3)/t(3;3) aberrations by next-generation sequencing (NGS).
Methods
Patient samples
From the combined study groups of the Dutch-Belgian Cooperative Trial Group for Hematology-Oncology and the German-Austrian AML Study Group we selected 32 AML (including 2 cell lines MUTZ-3 and UCSD-AML1), 4 CML-BC (including 2 cell lines HNT-34 and MOLM-1), and 5 MDS cases for NGS analysis. Included patients harbored an inv(3)/t(3;3) aberration on chromosome banding analysis (supplemental Table 1 available on the Blood Web site) subsequently confirmed by NGS analysis. Cultured CD3+ T cells from diagnostic bone marrow served as whole-exome sequencing (WES; n = 10) germline control. Written informed consent was obtained from all individuals in accordance with the Declaration of Helsinki. All trials were approved by the institutional review boards of Erasmus University Medical Center and the University of Ulm. All samples were sequenced on the Illumina HiSeq 2500 system and processed as described previously.12
3q-capture sequencing
From the collected patient material, the genomic DNA was sheared with the Covaris S2 device (Covaris) with default settings. Subsequently, the sample libraries were prepared using the TruSeq DNA Sample Preparation Guide (Illumina). The target chromosomal regions 3q21.1-3q26.2 (∼40 Mb) were captured by using custom in-solution oligonucleotide baits (Nimblegen SeqCap EZ Choice XL). The final sample libraries were subjected to paired-end sequencing (2 × 100 bp) and were aligned against the human genome 19 (hg19) using the Burrows Wheeler Aligner (BWA) with default settings.17 Exact breakpoint positions were determined with Breakdancer v1.1.18 Exact breakpoint sequences were resolved by extracting proximal reads supporting or spanning the breakpoint using the identified breakpoint positions and an algorithm able to extract the relevant reads from BAM files by using the Samtools API.19 Relevant reads were identified by their discordant distance to the paired mate read as a result of the inv(3)/t(3;3) aberration (supporting reads) or being a member of a cluster of truncated reads with the same clipping position (spanning reads). The extracted reads were subsequently used as input for the de novo assembler Velvet v1.0.1720 with default settings, and the assembled region was validated with UCSC Blat.21 If resolved, the breakpoint sequences of 3q21 and 3q26 were used for the estimation of the variant allele frequency (VAF) to infer the cellular prevalence of the inv(3)/t(3;3) aberration. All 3q-capture sequencing (3q-Seq) reads were aligned against the resolved breakpoint sequences of 3q21 and 3q26 and their respective native wild-type sequences. The VAF was estimated by comparing the total number of reads aligning on the breakpoint sequence to the total number of reads aligning to the respective native wild-type sequence.
RNA-Seq and whole-exome sequencing
From the collected patient material, total RNA was extracted with phenol-chloroform and subsequently transcribed by using Superscript II RT (Invitrogen). Shearing of the cDNA was performed with the Covaris S2 device (Covaris) with the default settings and was further constructed according to the TruSeq RNA Sample Preparation v2 Guide (Illumina). The sample libraries were subjected to paired-end sequencing (2 × 75 bp) and aligned against hg19 using TopHat v2.22 Genomic DNA from patients and in vitro cultured control CD3+ T cells were processed similar to 3q-Seq protocols and captured by exome bead capture (SeqCap EZ Human Exome Library v3.0). The sample libraries were paired-end sequenced (2 × 100 bp) and subsequently aligned against hg19 using BWA with default settings.17
Overall, we performed whole-transcriptome sequencing (RNA-Seq) on 41 and WES on 10 out of these 41 inv(3)/t(3;3) myeloid malignancies. Read and alignment statistics for RNA-Seq and WES data are found in supplemental Figure 1A-C and supplemental Table 4. On average, we observed a medium to high coverage for the targeted exome in WES data (∼62×), sufficient to detect mutations with a VAF of 10% or more. Reads generated for RNA-Seq analyses predominately fell within transcribed regions (∼52%) (ie, ribosomal genes, coding sequence, and UTRs), according to the RefSeq Transcriptome database, and, on average, 91% of the reads could be aligned to hg19. Gene expression profiles (GEP) for 24 inv(3)/t(3;3) patients were constructed for differential expression, cluster, and principle component analyses with the DESeq2 package.23 Copy number variation (CNV) profiles from the WES data were calculated by CNVsvd (M.A.S., R.H, and P.J.M.V., manuscript in preparation; supplemental Figure 2). Briefly, per patient the total number of fragments was determined for each exon or determined from consecutive 500 nucleotide-wide windows for large exons. The estimation of CNVs is hampered by systematic variance introduced by sequence technology bias or repetitive and homologous sequences, which can be observed in all sequenced cases. By using a control reference data set under the assumption that these cases have a normal karyotype (ie, the in vitro cultured CD3+ T cells), the local variance composition can be captured. These estimated local variance components can be used to attenuate the systematic variance in all sequenced cases. Finally, the normalized count statistics were used for the estimation of the CNV WES profile.
Variant detection
RNA-Seq data were preprocessed for variant detection by splitting the exon boundary spanning reads using the Genome Analysis Toolkit.24 Subsequently, the variants were determined with the Samtools API and MuTect for RNA-Seq and WES data.19,25 The detected variants were annotated with AnnoVar26 and further characterized by multiple read statistics determined by an in-house developed algorithm. In brief, the algorithm determines for each variant the VAF, local read statistics based on the alignment and base qualities, mutation likelihood given the local sequence context, recurrence given the catalog of somatic mutations in cancer, recurrence determined from population-based sequencing efforts (1000 genomes project), and, when available, the likelihood of the mutation given the same set of read statistics in a control sample. The validity of our approach combining WES data with RNA-Seq data to infer variants is substantiated by the observation that nonsense-mediate decay was negligible for mutant allele detection, as was demonstrated by similar VAFs of mutant disease alleles observed within cases characterized by both WES and RNA-Seq (supplemental Figure 3). Frameshift and premature stop codon–introducing mutations were selected and dichotomized on their location in the gene body. Mutations located in the terminal exon or approximately 50 bp from the exon boundary of the penultimate exon should theoretically be unaffected by nonsense-mediate decay, whereas stop codon–introducing mutations situated in other locations of the gene body should be affected. Finally, variants were examined when they were recurrently detected in >2 patients or if they were previously linked to leukemogenesis or cancer pathogenesis.27,28 All listed variants were validated by Sanger sequencing, except for FLT3-ITD, which was determined by reverse-transcription polymerase chain reaction.
Allelic imbalance of GATA2
In total, 30 inv(3)/t(3;3) cases accommodated informative heterozygous single-nucleotide variants (SNVs) in the GATA2 locus according to the 3q-Seq data. We previously showed that the inv(3)/t(3;3) causes monoallelic expression of GATA2 from the nonrearranged allele.12 Subsequently, we determined the allelic contribution of the genotypes of the heterozygous SNV in the matched RNA-Seq case. The average of the allelic contribution was taken when multiple heterozygous SNVs were accommodated in the GATA2 locus. The polar histogram was constructed with the R package “phenotypicForest.”29
Clonality analysis
The VAFs of the acquired mutations were estimated from the 10 paired inv(3)/t(3;3) myeloid malignancies characterized with WES. The VAF of the inv(3)/t(3;3) aberration was estimated from the 3q-Seq data unless the breakpoints could not be resolved or no 3q-Seq data were available. In these cases, the cytogenetically determined inv(3)/t(3;3) positive metaphases were used. The VAFs were corrected by the local CNV, determined by CNVsvd, and possible loss-of-heterozygosity ascertained by determining the loss of proximal heterozygous SNVs with respect to the control WES data. The clonal architecture was illustrated in violin plots. In brief, the density of mutations with a similar VAF was determined by a kernel-density approach and is represented by the width of the graph. These plots were generated by the R package “easyGgplot2.”30
Results
Mutant disease allele categorization
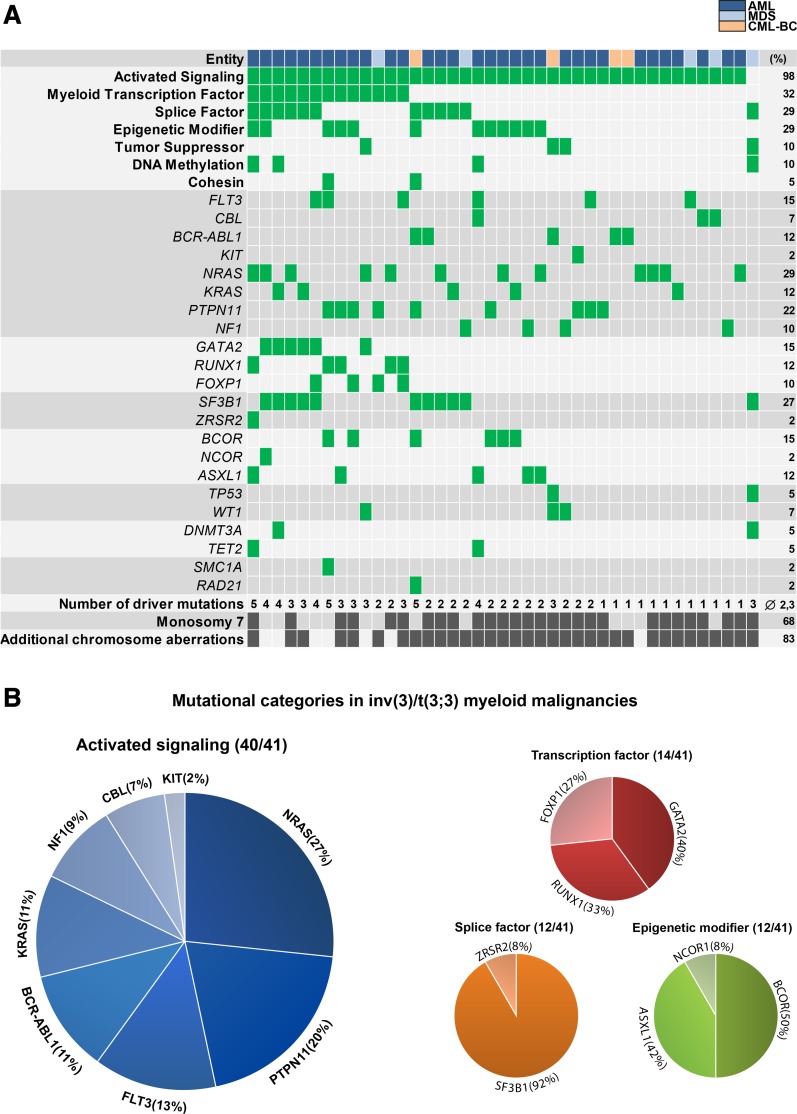
We first assigned mutations to mutational categories to discern patterns of mutations within inv(3)/t(3;3) myeloid disease (Figure 1A).28 All identified mutations were confirmed to be somatic in samples with available paired T-cell control (10/41 cases). In addition to the “hardwired” deregulated expression of EVI1 and GATA2, all 41 samples contained at least one additional mutation in one of the categories relevant for leukemia pathogenesis (average 2.3 category mutations per sample [Figure 1A and supplemental Tables 2 and 3]). Notably, all AML and CML-BC, as well as 4 out of 5 MDS samples contained mutations in genes activating RAS/RTK signaling, amounting to an incidence of 98% of all malignancies with an inv(3)/t(3;3). Furthermore, mutations were frequently found in myeloid transcription factor genes (32%), splice factor–encoding genes (29%), epigenetic modifier genes (29%), tumor-suppressor genes (10%), DNA-methylation genes (10%), and cohesin-complex genes (5%) (Figure 1A).
Figure 1.
Mutational spectrum observed in inv(3)/t(3;3) myeloid malignancies. (A) Distribution of acquired mutations in 41 inv(3)/t(3;3) myeloid malignancies in conjunction with the aggregation of the mutations into mutational categories: mutant (green), positive for cytogenetic abnormality (dark gray), wild-type (gray). (B) Distribution of mutations within mutational categories present in >10% of the inv(3)/t(3;3) myeloid malignancies.
Complementing previous reports on the high incidence of NRAS mutations in inv(3)/t(3;3) AML,3,6 we found on aggregate 47% of all samples containing mutations directly affecting RAS, that is, NRAS (27%), KRAS (11%), and NF1 (9%) (Figure 1B). These mutations were mutually exclusive and also largely nonoverlapping with any other mutation affecting signaling pathways involving RAS (ie, PTPN11 [20%], FLT3 [13%], CBL [7%], KIT [2%], and BCR-ABL1 [12%]) (Figure 1B). GATA2 was the most commonly mutated transcription factor in inv(3)/t(3;3) myeloid malignancies (15%; 5 AML and 1 MDS patient) and occurred in all cases in 1 of the 2 GATA2 zinc-finger domains. RUNX1 mutations were present in 12% and did not coincide with GATA2 mutations; however, mutations in the splice factor–encoding gene SF3B1 (27%) were enriched in GATA2-mutated samples. Mutations in GATA2, SF3B1, and RUNX1 were established to be somatic in all cases with control material available. Interestingly, we detected novel truncating mutations and CNVs, resulting in the loss of 1 copy of the transcription factor FOXP1 on 3p14.1 (10%), which is recurrently involved in chromosomal aberrations within lymphoma,31 but its association with AML pathogenesis is unknown. The predominant monosomal karyotype within inv(3)/t(3;3) myeloid malignancies, mainly conferred by monosomy 7 (68%), is contrasted by the low incidence of TP53 mutations (5%) (Figure 1A), which had been suggested to be involved in the etiology of complex and of monosomal karyotype AML.32,33
No mutational pattern alluded to the high coincidence of the loss of chromosome 7 in inv(3)/t(3;3) myeloid disease (Figure 1 and supplemental Tables 2 and 3). However, previous reports have indicated that haploinsufficiency for CUX1 (located on 7q22.1), a gene strongly downregulated in our cohort of inv(3)/t(3;3)/-7 patients (supplemental Table 5), activates phosphoinositide 3-kinase (PI3K) signaling by transcriptional downregulation of the PI3K inhibitor PI3KIP1,34 and could therefore be an important cooperating lesion in inv(3)/t(3;3)/monosomy 7 myeloid syndromes.35
To date, no independent prognostic factor within the inv(3)/t(3;3) AML subset has been identified as a result of its low incidence and the extremely short median survival of inv(3)/t(3;3) AML patients (10 months).3 Baseline patient characteristics and clinical outcome data were available in 21 individuals with inv(3)/t(3;3) AML. The high frequency of RAS/RTK pathway mutations allowed us to perform an exploratory analysis within this small patient cohort. There were no statistically significant differences in patient characteristics, nor overall survival (OS) and event-free survival in cases with RAS mutations (NRASmut, KRASmut, NF1mut) compared with cases with other mutations activating signaling pathways (supplemental Figure 4). The median OS of RASmut patients was 9.8 months vs 8.9 months of other RTKmut patients (median OS 9.8 months).
Clonality analysis
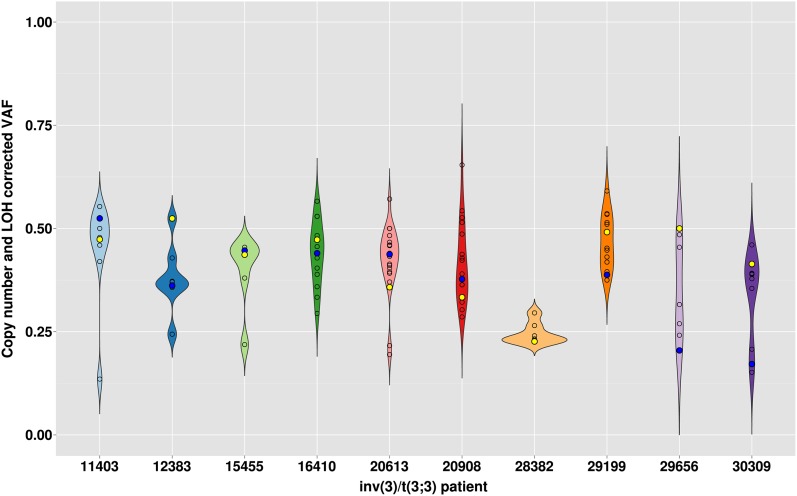
To address the question of whether the highly overrepresented RAS/RTK pathway mutations and other recurrent somatic alterations in inv(3)/t(3;3) AML co-occurred in the same dominant clone, we assessed the allelic ratios of the EVI1-rearranged and mutant-candidate disease alleles (Figure 2). WES analysis in conjunction with germline T-cell control was available from 10 AML patients. Cytogenetic evaluation of blast percentage and NGS read count estimation of the percentage of the 3q21q26.2 fusion (allele frequency) were concordant. In 2 cases (AML 20908 and 29656) without available 3q-Seq data, cytogenetics served to estimate the percentage of the inv(3) allele. The inv(3)/t(3;3) aberrations were detectable in the majority of cases (7/10 cases) in as much as 100% of the cells (ie, resulting in an allelic ratio of the heterozygous 3q21q26.2 fusion allele of approximately 0.5), reflecting high blast percentage in these cases. The RAS and RTK mutations were mainly found in the dominant EVI1–rearranged clone, and a similar pattern is found for all other identified alterations (eg, in transcription factor, splice factor, epigenetic modifier genes), which mostly co-occur at a similar frequency as the RAS/RTK mutations. However, in AML 12383 (PTPN11 mutation) and AMLs 29656 and 30309 (both NF1-mutated), the 3q-rearrangement was found in the major clone, whereas the RAS pathway mutations were present in only about half of these cells. In 2 cases (AML 20613 and 20908), the inv(3)/t(3;3) aberrations were less frequent than other concomitant mutations. In the inv(3) MDS case 28382 without any detected activating signaling mutation, the allelic ratio of the inv(3) was about 0.25, suggesting that both dysplastic-appearing cells as well as myeloblasts (blast percentage as per cytologic evaluation <20%) carried both the inv(3) aberration and coincident gene mutations (SF3B1, TP53, DNMT3A; see Figure 1A). Together, these data suggest that the inv(3) or t(3;3) aberration is the primary genetic hit in this subset of malignancies, with high proportion of clones harboring concurrent activating signaling mutations. Owing to the very short survival of these patients and general failure to achieve complete remission, no time-course monitoring could be performed to reveal clonal evolution.
Figure 2.
Clonal architecture inferred from somatic mutations observed in 10 inv(3)/t(3;3) myeloid malignancies. Distribution of estimated VAF determined from whole-exome sequencing. The width of the graph represents the density of mutations with similar VAFs. The yellow dot denotes the VAF of the 3q-aberration [(inv(3)/t(3;3)], the blue dot denotes the VAF of the RAS/RTK-associated mutation, and open circles denote the VAF of all other mutations.
Expression of mutant GATA2
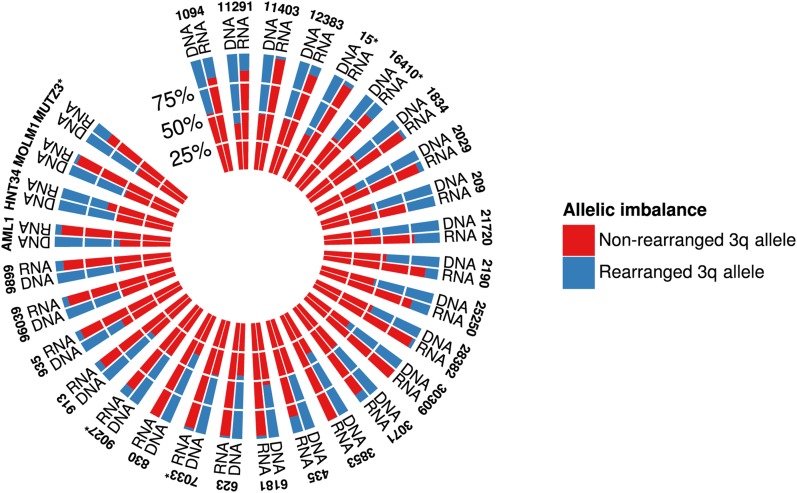
The inv(3)/t(3;3) chromosomal rearrangements separate an upstream GATA2 enhancer from 3q21 and fuse it to the 3q26.2/EVI1 locus, thereby acquiring features of a monoallelic super-enhancer on the rearranged 3q allele.12,13,36,37 Integrative analysis of RNA-seq with 3q-capture DNA-seq data using informative, heterozygous SNVs (single-nucleotide polymorphisms plus somatic mutations) revealed almost exclusive monoallelic expression of the mutant GATA2 alleles (Figure 3), as shown in the polar plot by the contribution of the rearranged 3q and nonrearranged 3q allele read counts for GATA2 in 30 inv(3)/t(3;3) cases, including cell lines available for analysis. This observation indicates that the remaining active, nonrearranged GATA2 allele acquired the mutation, whereas the nonmutated GATA2 allele was silenced as a result of the chromosomal rearrangement. Thus, in our inv(3)/t(3;3) AML cohort, heterozygous GATA2 mutations were “functionally” hemizygous as a result of monoallelic GATA2 silencing.
Figure 3.
Polar histogram plot of allelic imbalance of GATA2 expression observed in RNA-Seq. For each patient, the average VAF is estimated from informative heterozygous SNVs from 3q-Seq data. Average RNA-Seq GATA2 VAF is estimated from the same SNV positions. Asterisks denote the presence of a somatic GATA2 mutation in the indicated sample.
Gene expression and mutation patterns in AML and MDS
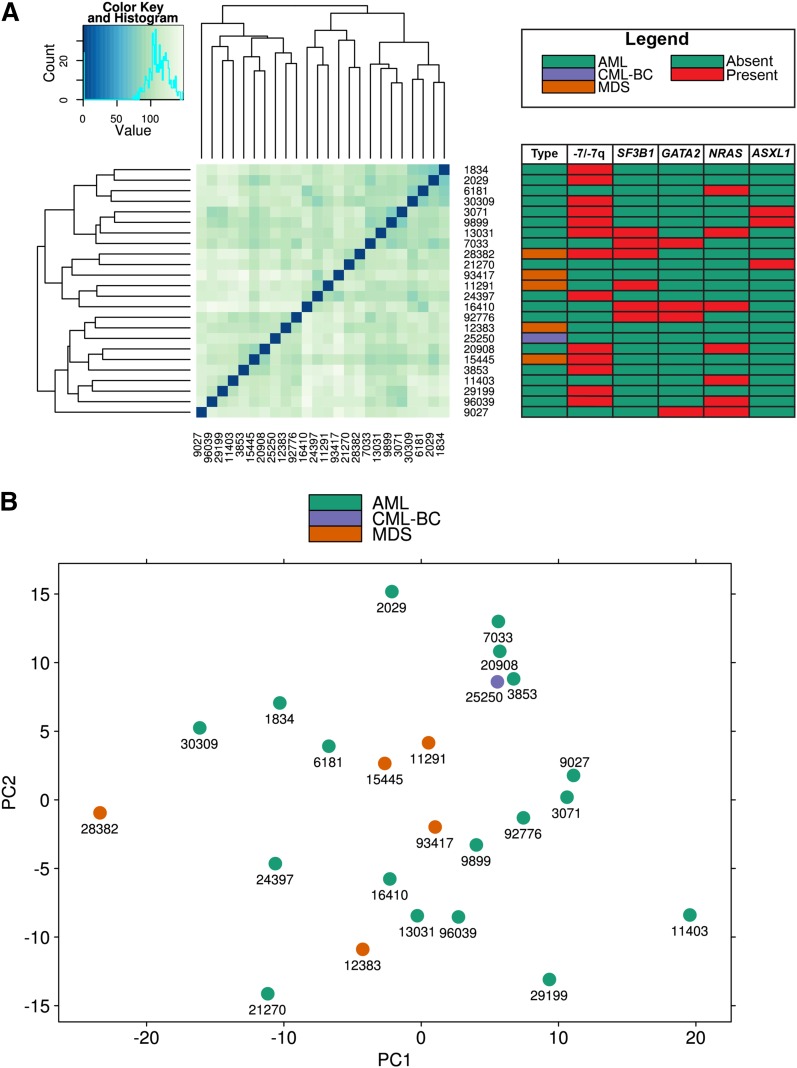
It is a matter of debate whether MDS with the distinct inv(3)(q21q26.2) or t(3;3)(q21;q26.2) should be regarded as AML, irrespective of blast percentage in the bone marrow, similar to the current World Health Organization (WHO) guidelines applied in the diagnosis of core-binding factor AML with inv(16)/t(16;16) or t(8;21) and of acute promyelocytic leukemia with t(15;17).1,5,6,38 In an effort to discriminate MDS and AML with inv(3)/t(3;3) based on gene expression programs and the spectrum of coincident gene mutations, we performed cluster and principle component analyses (Figure 4A-B). No cluster formation emerged, neither based on the MDS/AML dichotomy nor any other unsubstantiated group within our data set. Furthermore, we performed a differential expression analysis to infer genes that could differentiate between MDS and AML. In summary, after Benjamini-Hochberg correction for multiple testing, we could only detect 2 differentially expressed genes (C11orf45: P = .0009, CILP: P = .04) without a documented role in leukemogenesis. In addition, we observed that MDS patients with inv(3)/t(3;3) are as equally therapy resistant as their AML counterparts in a small set of cases analyzed (data not shown). In conclusion, we were unable to detect cluster formation, indicating the strong homogeneity of inv(3)/t(3;3) myeloid malignancies based on GEPs and the pattern of cooperating genetic lesions.
Figure 4.
GEP analysis. (A) Clustering of GEPs using the Euclidean distance metric and hierarchical clustering. The adjacent table details the acquired mutation of the clustered malignancies. (B) Principle component analysis performed on the GEPs of 24 inv(3)/t(3;3) myeloid malignancies displays no discrimination between AML and MDS.
Discussion
Collectively, we present data that suggest a common genetic background of myeloid malignancies harboring inv(3) or t(3;3) and show that RAS alterations and activating RTK mutations are more frequent in this disease subset than has been previously reported.3,6,39,40 The spectrum of secondary genetic lesions is generally found in the same EVI1-rearranged dominant clone. No unique cluster within inv(3)/t(3;3) myeloid malignancies could be identified, neither by gene expression or mutation profiling nor by analysis of patient characteristics or clinical outcome. Thus, our data further support the notion that inv(3)/t(3;3) myeloid disorders could be categorized as AML, irrespective of blast count, similar to WHO AML categories t(8;21), inv(16)/t(16;16), or t(15;17), which is also suggested by the molecular pathobiology common to all inv(3)/t(3;3) myeloid malignancies.12,13
Reclassification of the currently annotated WHO AML subtype inv(3)/t(3;3); RPN1-EVI1–rearranged as inv(3)/t(3;3); GATA2-EVI1–rearranged AML is supported by the observation that GATA2 allelic imbalances and monoallelic expression of heterozygous GATA2 mutations occur because of the distinct chromosomal rearrangements. Whether this and other myeloid transcription factor alterations contribute to disease biology and the highly adverse clinical phenotype of inv(3)/t(3;3) patients remains to be shown, although GATA2 and other transcription factor disturbances have been described to be preleukemic lesions.28,41-45 Of note, myeloid malignancies with inv(3) or t(3;3) define yet another subset of AML with high enrichment of GATA2 mutations next to CEBPA-mutated AML.46,47
We included CML cases in blast crisis with an inv(3)/t(3;3) under the assumption that CML-BC closely resembles AML biology.48 The BCR-ABL1 fusion is an RTK mutant that in itself activates RAS pathways and is the first event in transformation of myeloid precursors, as opposed to MDS and AML cells first acquiring inv(3)/t(3;3).49,50 Despite the difference of the biology and the etiology of CML, the mutational spectrum of inv(3)/t(3;3) CML-BC cells appeared to be same, as was further suggested by transcriptome analysis, which showed that GEP of the one CML-BC case did not differ from that of AML and MDS cases. However, the small number of inv(3)/t(3;3) MDS and CML cases in our study preclude conclusive assessment of the role of inv(3)/t(3;3) with regard to clinical phenotype.
In summary, inv(3)/t(3;3) myeloid malignancies harbor a common set of molecular alterations (ie, EVI1 and GATA2 deregulation coupled with mutations activating key signaling pathways). The dependence on constitutive RAS/RTK signaling activity of inv(3)/t(3;3)-transformed AML cells might be the molecular correlate of the observed high white blood cell counts in this disease subset. Also, in view of the negative impact of GATA2 deficiencies on proliferation and regeneration of myeloid progenitors,15,41,51,52 these activated signaling mutations may be indispensable for survival and propagation of inv(3)/t(3;3)-transformed myeloid progenitors. The high mutational burden of inv(3)/t(3;3) cells compared with other AML subtypes27 (supplemental Table 2) could also provide clues about why inv(3)/t(3;3) malignancies invariably associate with an extremely poor prognosis. Because these rare inv(3)/t(3;3) myeloid malignancies form a highly unmet medical need, novel therapeutic approaches could be derived from the observation of constitutive activation of the MAPK pathway in almost 100% of these tumors. Exploiting signaling pathways therapeutically by using FLT3- or PI3K-inhibitors53 or hypothetically by interfering with RAS-signaling, possibly in combination with BET-inhibitors,12 may serve as valuable adjuncts to the scarce armamentarium of chemotherapeutic drugs effective in this subset of malignancies.
Acknowledgments
This work was supported by grants from the Deutsche Forschungsgemeinschaft (GR3955/1-1) (S.G.), the Lady Tata Memorial Trust (S.G.), the Center for Translational Molecular Medicine (GR03O-102) (M.A.S.), an EHA Research Fellowship (S.G.), and the Worldwide Cancer Research (formerly AICR) (12-1309) (E.B.).
Footnotes
The online version of this article contains a data supplement.
There is an Inside Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: S.G., M.A.S., R.D., and P.J.M.V. designed research, performed experiments, analyzed and interpreted data, and wrote the manuscript; S.G., A.Z., M.H., R.H., E.M.J.B., and C.E. generated NGS libraries and performed Sanger and Illumina sequencing; H.B.B., B.L., K.D., H.D., and P.J.M.V. collected specimens and clinical data; H.B.B., K.D., H.D., and P.J.M.V. performed cytogenetic and molecular analyses of leukemia samples.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Peter J. M. Valk, Department of Hematology Erasmus University Medical Center, Wytemaweg 80, Room Nc806, 3015 CN Rotterdam, The Netherlands; e-mail: p.valk@erasmusmc.nl.
References
- 1.Vardiman JW, Thiele J, Arber DA, et al. The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: rationale and important changes. Blood. 2009;114(5):937–951. doi: 10.1182/blood-2009-03-209262. [DOI] [PubMed] [Google Scholar]
- 2.Grigg AP, Gascoyne RD, Phillips GL, Horsman DE. Clinical, haematological and cytogenetic features in 24 patients with structural rearrangements of the Q arm of chromosome 3. Br J Haematol. 1993;83(1):158–165. doi: 10.1111/j.1365-2141.1993.tb04647.x. [DOI] [PubMed] [Google Scholar]
- 3.Lugthart S, Gröschel S, Beverloo HB, et al. Clinical, molecular, and prognostic significance of WHO type inv(3)(q21q26.2)/t(3;3)(q21;q26.2) and various other 3q abnormalities in acute myeloid leukemia. J Clin Oncol. 2010;28(24):3890–3898. doi: 10.1200/JCO.2010.29.2771. [DOI] [PubMed] [Google Scholar]
- 4.Testoni N, Borsaru G, Martinelli G, et al. 3q21 and 3q26 cytogenetic abnormalities in acute myeloblastic leukemia: biological and clinical features. Haematologica. 1999;84(8):690–694. [PubMed] [Google Scholar]
- 5.Cui W, Sun J, Cotta CV, Medeiros LJ, Lin P. Myelodysplastic syndrome with inv(3)(q21q26.2) or t(3;3)(q21;q26.2) has a high risk for progression to acute myeloid leukemia. Am J Clin Pathol. 2011;136(2):282–288. doi: 10.1309/AJCP48AJDCKTHUXC. [DOI] [PubMed] [Google Scholar]
- 6.Haferlach C, Bacher U, Haferlach T, et al. The inv(3)(q21q26)/t(3;3)(q21;q26) is frequently accompanied by alterations of the RUNX1, KRAS and NRAS and NF1 genes and mediates adverse prognosis both in MDS and in AML: a study in 39 cases of MDS or AML. Leukemia. 2011;25(5):874–877. doi: 10.1038/leu.2011.5. [DOI] [PubMed] [Google Scholar]
- 7.Johansson B, Fioretos T, Mitelman F. Cytogenetic and molecular genetic evolution of chronic myeloid leukemia. Acta Haematol. 2002;107(2):76–94. doi: 10.1159/000046636. [DOI] [PubMed] [Google Scholar]
- 8.Cattaneo F, Nucifora G. EVI1 recruits the histone methyltransferase SUV39H1 for transcription repression. J Cell Biochem. 2008;105(2):344–352. doi: 10.1002/jcb.21869. [DOI] [PubMed] [Google Scholar]
- 9.Goyama S, Yamamoto G, Shimabe M, et al. Evi-1 is a critical regulator for hematopoietic stem cells and transformed leukemic cells. Cell Stem Cell. 2008;3(2):207–220. doi: 10.1016/j.stem.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 10.Perkins AS, Fishel R, Jenkins NA, Copeland NG. Evi-1, a murine zinc finger proto-oncogene, encodes a sequence-specific DNA-binding protein. Mol Cell Biol. 1991;11(5):2665–2674. doi: 10.1128/mcb.11.5.2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gröschel S, Lugthart S, Schlenk RF, et al. High EVI1 expression predicts outcome in younger adult patients with acute myeloid leukemia and is associated with distinct cytogenetic abnormalities. J Clin Oncol. 2010;28(12):2101–2107. doi: 10.1200/JCO.2009.26.0646. [DOI] [PubMed] [Google Scholar]
- 12.Gröschel S, Sanders MA, Hoogenboezem R, et al. A single oncogenic enhancer rearrangement causes concomitant EVI1 and GATA2 deregulation in leukemia. Cell. 2014;157(2):369–381. doi: 10.1016/j.cell.2014.02.019. [DOI] [PubMed] [Google Scholar]
- 13.Yamazaki H, Suzuki M, Otsuki A, et al. A remote GATA2 hematopoietic enhancer drives leukemogenesis in inv(3)(q21;q26) by activating EVI1 expression. Cancer Cell. 2014;25(4):415–427. doi: 10.1016/j.ccr.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsu AP, Johnson KD, Falcone EL, et al. GATA2 haploinsufficiency caused by mutations in a conserved intronic element leads to MonoMAC syndrome. Blood. 2013;121(19):3830–3837. doi: 10.1182/blood-2012-08-452763. S3831-S3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lim KC, Hosoya T, Brandt W, et al. Conditional Gata2 inactivation results in HSC loss and lymphatic mispatterning. J Clin Invest. 2012;122(10):3705–3717. doi: 10.1172/JCI61619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ling KW, Ottersbach K, van Hamburg JP, et al. GATA-2 plays two functionally distinct roles during the ontogeny of hematopoietic stem cells. J Exp Med. 2004;200(7):871–882. doi: 10.1084/jem.20031556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen K, Wallis JW, McLellan MD, et al. BreakDancer: an algorithm for high-resolution mapping of genomic structural variation. Nat Methods. 2009;6(9):677–681. doi: 10.1038/nmeth.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li H, Handsaker B, Wysoker A, et al. 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18(5):821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kent WJ. BLAT—the BLAST-like alignment tool. Genome Res. 2002;12(4):656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14(4):R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-Seq data with DESeq2. bioRxiv. 2014. [DOI] [PMC free article] [PubMed]
- 24.McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cibulskis K, Lawrence MS, Carter SL, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31(3):213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kandoth C, McLellan MD, Vandin F, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502(7471):333–339. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cancer Genome Atlas Research Network. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368(22):2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ladroue C. phenotypicForest 0.2. http://chrisladroue.com/phorest/. Accessed October, 13, 2014.
- 30.Kassambara A. Statistical tools for high-throughput data analysis. http://www.sthda.com/english/articles/3-easyggplot2/. Accessed October, 13, 2014.
- 31.Streubel B, Vinatzer U, Lamprecht A, Raderer M, Chott A. T(3;14)(p14.1;q32) involving IGH and FOXP1 is a novel recurrent chromosomal aberration in MALT lymphoma. Leukemia. 2005;19(4):652–658. doi: 10.1038/sj.leu.2403644. [DOI] [PubMed] [Google Scholar]
- 32.Breems DA, Van Putten WL, De Greef GE, et al. Monosomal karyotype in acute myeloid leukemia: a better indicator of poor prognosis than a complex karyotype. J Clin Oncol. 2008;26(29):4791–4797. doi: 10.1200/JCO.2008.16.0259. [DOI] [PubMed] [Google Scholar]
- 33.Rücker FG, Schlenk RF, Bullinger L, et al. TP53 alterations in acute myeloid leukemia with complex karyotype correlate with specific copy number alterations, monosomal karyotype, and dismal outcome. Blood. 2012;119(9):2114–2121. doi: 10.1182/blood-2011-08-375758. [DOI] [PubMed] [Google Scholar]
- 34.Wong CC, Martincorena I, Rust AG, et al. Chronic Myeloid Disorders Working Group of the International Cancer Genome Consortium. Inactivating CUX1 mutations promote tumorigenesis. Nat Genet. 2014;46(1):33–38. doi: 10.1038/ng.2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McNerney ME, Brown CD, Wang X, et al. CUX1 is a haploinsufficient tumor suppressor gene on chromosome 7 frequently inactivated in acute myeloid leukemia. Blood. 2013;121(6):975–983. doi: 10.1182/blood-2012-04-426965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grass JA, Jing H, Kim SI, et al. Distinct functions of dispersed GATA factor complexes at an endogenous gene locus. Mol Cell Biol. 2006;26(19):7056–7067. doi: 10.1128/MCB.01033-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koche RP, Armstrong SA. Genomic dark matter sheds light on EVI1-driven leukemia. Cancer Cell. 2014;25(4):407–408. doi: 10.1016/j.ccr.2014.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Döhner H, Estey EH, Amadori S, et al. European LeukemiaNet. Diagnosis and management of acute myeloid leukemia in adults: recommendations from an international expert panel, on behalf of the European LeukemiaNet. Blood. 2010;115(3):453–474. doi: 10.1182/blood-2009-07-235358. [DOI] [PubMed] [Google Scholar]
- 39.Renneville A, Roumier C, Biggio V, et al. Cooperating gene mutations in acute myeloid leukemia: a review of the literature. Leukemia. 2008;22(5):915–931. doi: 10.1038/leu.2008.19. [DOI] [PubMed] [Google Scholar]
- 40.Tsurumi S, Nakamura Y, Maki K, et al. N-ras and p53 gene mutations in Japanese patients with myeloproliferative disorders. Am J Hematol. 2002;71(2):131–133. doi: 10.1002/ajh.10188. [DOI] [PubMed] [Google Scholar]
- 41.de Pater E, Kaimakis P, Vink CS, et al. Gata2 is required for HSC generation and survival. J Exp Med. 2013;210(13):2843–2850. doi: 10.1084/jem.20130751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hahn CN, Chong CE, Carmichael CL, et al. Heritable GATA2 mutations associated with familial myelodysplastic syndrome and acute myeloid leukemia. Nat Genet. 2011;43(10):1012–1017. doi: 10.1038/ng.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nishimoto N, Arai S, Ichikawa M, et al. Loss of AML1/Runx1 accelerates the development of MLL-ENL leukemia through down-regulation of p19ARF. Blood. 2011;118(9):2541–2550. doi: 10.1182/blood-2010-10-315440. [DOI] [PubMed] [Google Scholar]
- 44.Mardis ER, Ding L, Dooling DJ, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009;361(11):1058–1066. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Watanabe-Okochi N, Kitaura J, Ono R, et al. AML1 mutations induced MDS and MDS/AML in a mouse BMT model. Blood. 2008;111(8):4297–4308. doi: 10.1182/blood-2007-01-068346. [DOI] [PubMed] [Google Scholar]
- 46.Green CL, Tawana K, Hills RK, et al. GATA2 mutations in sporadic and familial acute myeloid leukaemia patients with CEBPA mutations. Br J Haematol. 2013;161(5):701–705. doi: 10.1111/bjh.12317. [DOI] [PubMed] [Google Scholar]
- 47.Greif PA, Dufour A, Konstandin NP, et al. GATA2 zinc finger 1 mutations associated with biallelic CEBPA mutations define a unique genetic entity of acute myeloid leukemia. Blood. 2012;120(2):395–403. doi: 10.1182/blood-2012-01-403220. [DOI] [PubMed] [Google Scholar]
- 48.Radich JP. The Biology of CML blast crisis. Hematology (Am Soc Hematol Educ Program) 2007:384–391. doi: 10.1182/asheducation-2007.1.384. [DOI] [PubMed] [Google Scholar]
- 49.Calabretta B, Perrotti D. The biology of CML blast crisis. Blood. 2004;103(11):4010–4022. doi: 10.1182/blood-2003-12-4111. [DOI] [PubMed] [Google Scholar]
- 50.Sawyers CL, McLaughlin J, Witte ON. Genetic requirement for Ras in the transformation of fibroblasts and hematopoietic cells by the Bcr-Abl oncogene. J Exp Med. 1995;181(1):307–313. doi: 10.1084/jem.181.1.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hsu AP, Sampaio EP, Khan J, et al. Mutations in GATA2 are associated with the autosomal dominant and sporadic monocytopenia and mycobacterial infection (MonoMAC) syndrome. Blood. 2011;118(10):2653–2655. doi: 10.1182/blood-2011-05-356352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rodrigues NP, Janzen V, Forkert R, et al. Haploinsufficiency of GATA-2 perturbs adult hematopoietic stem-cell homeostasis. Blood. 2005;106(2):477–484. doi: 10.1182/blood-2004-08-2989. [DOI] [PubMed] [Google Scholar]
- 53.Park S, Chapuis N, Tamburini J, et al. Role of the PI3K/AKT and mTOR signaling pathways in acute myeloid leukemia. Haematologica. 2010;95(5):819–828. doi: 10.3324/haematol.2009.013797. [DOI] [PMC free article] [PubMed] [Google Scholar]