The era of precision medicine has changed the treatment paradigm for some cancers, but for others, the term precision is a stretch. New targets and therapies have broadened choices for patients with lung cancer or melanoma; however, in colorectal cancer (CRC), for example, the evolution of biomarkers has been primarily to inform whom not to treat rather than to identify the patients who will benefit from treatment. The refinements in CRC have been limiting rather than expansive; by broadening the definition of RAS mutations, fewer patients have multiple targeted options.
The epidermal growth factor receptor (EGFR) antibody cetuximab was first approved a decade ago for use with FOLFIRI (folinic acid, fluorouracil, and irinotecan) chemotherapy in metastatic CRC (mCRC), based on the findings of the CRYSTAL (Cetuximab Combined With Irinotecan in First-Line Therapy for Metastatic Colorectal Cancer) trial.1 Shortly thereafter, after the results of the PRIME (Panitumumab Randomized Trial in Combination With Chemotherapy for Metastatic Colorectal Cancer to Determine Efficacy) trial, a second EGFR antibody, panitumumab, was approved for use with FOLFOX (folinic acid, fluorouracil, and oxaliplatin) in a similar patient population (2006).2 It would be another 3 years before the US Food and Drug Administration (FDA) acknowledged emerging data suggesting that the most common activating mutations in KRAS, at codons 12 and 13 in exon 2, were negative predictors for treatment benefit from EGFR antibodies. In 2009, the FDA restricted the use of cetuximab and panitumumab to the roughly 60% of patients without KRAS exon 2 mutations. Enrichment for KRAS codon 12 and 13 wild-type tumors increased response rates to approximately 60%1,2 and spurred a quest to further characterize the downstream effectors of EGFR.
After no definitive predictive roles were identified for activating BRAF and PIK3CA mutations or for loss of PTEN expression, the focus shifted back to RAS.3 First, perspective narrowed as questions arose concerning whether all KRAS codon 12 and 13 mutations were equivalent; retrospective analyses suggested that patients with KRAS G13D mutations may derive a modest benefit from EGFR antibodies, intermediate between that of KRAS wild-type and codon 12–mutated tumors.4–6 However, the effect was not robust enough to merit practice change.7
Meanwhile, interest was growing in KRAS exon 3 and 4 mutations, found to be oncogenic and associated with resistance to EGFR-targeted agents in preclinical models.8,9 These findings have yielded a bounty of secondary analyses of phase II to III clinical trials—studies large enough to investigate the effect of so-called new, extended, or expanded RAS mutations.10–19 In the August 1, 2014, issue of Lancet Oncology, Heinemann et al20 reported on expanded RAS mutations in the FIRE-3 trial, corroborating the current trend.
The definition of expanded RAS has evolved. For the moment, the field has settled on KRAS and NRAS codons 12 and 13 (exon 2), 59 and 61 (exon 3), and 117 and 146 (exon 4). Mutations in KRAS and NRAS are typically mutually exclusive. Among five trials of first-line therapy for mCRC, including FIRE-3, the prevalence of expanded RAS mutations in tumor specimens, previously determined to be wild type at exon 2 of KRAS, ranged from 15% to 27%.12,15,16,19,20 Studies at both extremes of mutation prevalence used BEAMing (beads, emulsions, amplification, and magnetics) technology, with a 5% mutated/wild-type allele sensitivity cutoff. A meta-analysis of nine randomized controlled trials evaluating EGFR antibodies in all lines of mCRC therapy found that approximately 20% of KRAS exon 2 wild-type tumors harbored an expanded RAS mutation.21 By exon, the estimated prevalence of mutations was as follows: 4.3% in exon 3 and 6.7% in exon 4 of KRAS, and 3.8% in exon 2, 4.8% in exon 3, and 0.5% in exon 4 of NRAS.21
Meta-analysis revealed that EGFR antibody efficacy was superior for all–RAS wild-type tumors compared with the expanded RAS-mutant subgroup with respect to both progression-free survival and overall survival. Conversely, efficacy was not significantly different between the expanded RAS-mutant and KRAS exon 2–mutant subgroups.21 In FIRE-3 specifically, median progression-free survival was inferior among patients with expanded RAS mutations who received cetuximab rather than bevacizumab plus FOLFIRI (hazard ratio, 2.22; 95% CI, 1.28 to 3.86; P = .004).20 Although lack of benefit for EGFR inhibitor treatment in the setting of expanded RAS mutations was a consistent finding, statistically significant detrimental effects of EGFR antibodies were not observed reproducibly across studies. In CALGB (Cancer and Leukemia Group B)/SWOG (Southwest Oncology Group) 80405, as distinct from FIRE-3, the all–RAS wild-type subgroup associated with improved outcome, independent of treatment containing cetuximab or bevacizumab, suggesting that expanded RAS mutations may be prognostic (A.P. Venook, personal communication, October 2014). Common to all trials, CIs around hazard ratios for the expanded RAS subset were large, reflecting small numbers of patients.
Do these trials provide sufficient evidence to mandate routine testing for expanded RAS mutations in all patients with mCRC? The European Commission has updated prescribing indications for panitumumab and cetuximab, restricting use to patients with RAS wild-type mCRC. In the United States, package inserts state that EGFR antibodies are not indicated for treatment of KRAS mutation–positive mCRC, without reference to NRAS or specific KRAS codons.22,23 The recently updated National Comprehensive Cancer Network colon cancer guidelines state that “whenever possible,” non–exon 2 KRAS and NRAS mutation status should be determined, and patients with “any known” KRAS or NRAS mutation should not receive cetuximab or panitumumab.24 Questions remain about how to integrate the new guidelines into clinical practice.
Which RAS Mutations Matter?
Because of the rarity of specific RAS mutations evaluated, and because analyses have pooled the effects of expanded RAS mutations from as many as 10 individual KRAS and NRAS codons, it is difficult to determine to what degree each specific mutation confers lack of sensitivity to EGFR antibodies. No single expanded RAS mutation occurs at a frequency of > 6%, and some RAS mutations evaluated occur at frequencies of ≤ 1% (ie, mutations at codons 59 and 117 in KRAS and NRAS and at codon 146 in NRAS). Without the creation of a big-data warehouse from tested and treated patients, it is unlikely that the contribution of any one rare RAS mutation will ever be definitively determined. Indeed, there is still debate over whether KRAS G13D mutations (approximately 8% of CRCs) confer the same degree of resistance as codon 12 mutations. Furthermore, as whole-exome or targeted-exome sequencing replaces hot-spot mutational analysis, the identification of rare RAS mutations of unknown significance outside of these codons may create additional confusion.
Because not all individual RAS mutations are necessarily created equal, we may not be able to safely assume that any RAS mutation will predict lack of response to EGFR antibodies. Our confidence in the utility of any one specific RAS mutation as a negative predictive biomarker may have to rely in part on indirect clinical and preclinical data. For example, we can be more assured that an individual RAS mutation will predict lack of response to EGFR antibodies if that same mutation has been identified in patients with CRCs originally classified as RAS wild type who developed acquired resistance to EGFR antibodies. Mutations in codons 12, 13, 61, and 146 and NRAS codons 12, 13, and 61 have been observed in the setting of acquired resistance.25–28 Preclinical data may also be helpful. For example, KRAS codon 117 mutations have been shown to be activating and able to confer resistance to EGFR inhibitors in preclinical models.8,9 Conversely, there are few preclinical data to support a functional role for the rare codon 59 RAS mutation. Therefore, we should exercise caution when considering whether to withhold an EGFR antibody from a patient with a codon 59 mutation or another rare RAS mutation about which little is known.
What Are Best Practices to Test for RAS Mutations?
The current landscape of molecular testing has incorporated a variety of testing platforms, each with varying technical performance, cost, and feasibility.29 Currently, the only FDA-approved test for use with cetuximab and panitumumab is a KRAS companion diagnostic that tests for mutations on KRAS codons 12 and 13 (using Scorpion Amplified Refractory Mutation System [ARMS] polymerase chain reaction [PCR] methodology, with reported sensitivity of approximately 1% to 5%). Alternate technologies, including BEAMing assays and digital PCR, have improved sensitivity to the range of 0.1% for tissue samples, representing the ability to detect one heterozygous mutant cancer cell in a background of 500 wild-type cells. Many next-generation sequencing platforms in clinical use can reliably detect allele frequencies in the range of 5% to 10%, although lower allele frequencies are commonly seen and variably reported in clinical results. Data using these high-sensitivity assays have employed various allele frequencies for calling mutations, with many of the recent retrospective studies applying a 5% threshold. When KRAS mutations are present in CRC, they are typically present in the vast majority if not all of the cancer cells (mutations in KRAS are approximately 95% concordant in paired CRC metastases and primary tumors).30 Nonetheless, retrospective studies using higher-sensitivity research assays such as digital PCR have suggested that tumors with rare KRAS-mutant clones may be present in up to 20% to 30% of patient cases determined to be KRAS wild type by prior clinical testing.31–33 At least one study has suggested that these rare clones may provide a reservoir for acquired resistance to EGFR monoclonal antibodies.26 An analysis of various thresholds for allele frequency from the CRYSTAL study demonstrated trends toward greater discrimination of benefit down to a threshold of 0.1% mutant RAS, although even this large study was underpowered to robustly address this question.31 Because it remains possible that patients whose tumors harbor rare RAS-mutant cells might still derive meaningful initial benefit from EGFR antibodies, exceedingly sensitive detection thresholds may not be appropriate in restricting use of EGFR antibodies.
An additional barrier to the interpretation of the allele frequency results from the variable handling of tumor tissue before DNA extraction. Microdissection of the tumor tissue from the glass slide increases the purity of the cancer cell DNA by approximately two- to 10-fold, but this is labor intensive. In the current FDA-approved assay, microdissection is recommended only for patient cases where < 20% of the cells on the slide are cancerous, but the estimation of percentage of tumor cells has substantial interobserver variation.34 In most retrospective studies, no microdissection was performed, adding to uncertainty about the true mutant allele frequency in the tumors. Additional retrospective studies will need to incorporate microdissection with quantification to appropriately address the impact of allele frequency, including correlation with clinical outcomes.
Should BRAF Be Included in Testing Panel?
Activating BRAF codon 600 (exon 15) mutations occur in 5% to 10% of CRCs and are mutually exclusive from RAS mutations, with rare exceptions.13 Retrospective analyses of several large randomized trials have not defined a clear role for BRAF V600 as a negative predictive biomarker for response to EGFR antibodies.12,13,16,35 Although limited by sample size, it is notable that no study has shown a statistically significant benefit from the addition of an EGFR antibody to chemotherapy in patients with BRAF V600–mutant CRC. Furthermore, these analyses have confirmed prior observations that patients with BRAF V600–mutant CRC have a poor prognosis and do not respond well to standard therapies. Therefore, consideration of clinical trials as an early option for patients with BRAF V600–mutant CRC may be of greater clinical importance than whether such patients should receive EGFR antibodies. Indeed, several recently reported clinical trials evaluating RAF inhibitor–based combinations, including with EGFR antibodies, in patients with BRAF V600–mutant CRC have shown promising preliminary results, with some studies achieving response rates close to 40%.36–38
In summary, we recommend expanded RAS mutation testing as part of the initial workup for mCRC, because this approach will identify an additional approximately 11% of patients with CRC who are unlikely to benefit from EGFR antibodies. We advise using a platform sensitive enough to detect mutations at an allele frequency threshold of ≤ 5% (Fig 1). Although no definitive role for BRAF as a negative predictive biomarker has been established, we recommend routine testing for BRAF V600 mutations because ongoing BRAF-directed clinical trials offer a promising alternative. Finally, in our pursuit of expanded RAS testing, we must be cautious not to expand so rapidly that we outpace the existing data by basing clinical decisions on rare RAS mutations, such as codon 59 or other infrequent RAS mutations, about which little is presently understood. Continued efforts to fine-tune our approach to the molecular classification of CRC will hone our ability to deliver precise and effective care to our patients.
Fig 1.
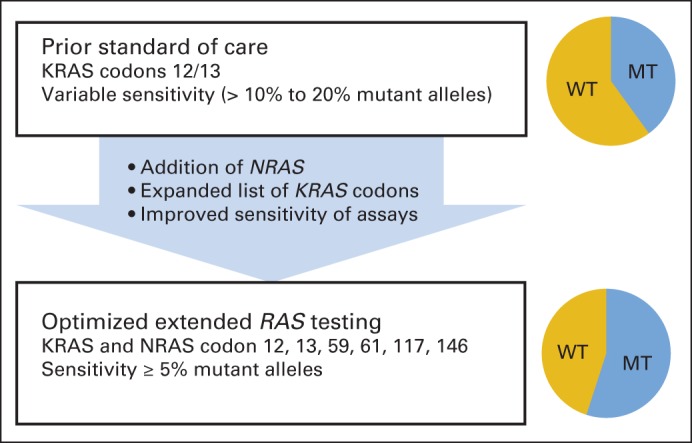
Advances in RAS testing. Optimal clinical implementation of RAS testing involves expansion of number of tested codons to include less common mutations and use of assays with sufficient sensitivity for RAS-mutant alleles. To date, preponderance of clinical data reported on expanded RAS mutations has used 5% threshold for detection of mutated (MT)/wild-type (WT) alleles; therefore, this represents a reasonable threshold while additional analyses are conducted.
Acknowledgment
Supported by the Masin Foundation and National Cancer Institute (NCI) and National Institutes of Health Award No. K08CA175143 (C.E.A.) and by NCI Award Nos. R01CA172670 and R01CA184843 (S.K.). We thank Stan Hamilton, MD, and Alan Venook, MD, for their critical review of the manuscript.
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Disclosures provided by the authors are available with this article at www.jco.org.
AUTHOR CONTRIBUTIONS
Manuscript writing: All authors
Final approval of manuscript: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Expanded RAS: Refining the Patient Population
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or jco.ascopubs.org/site/ifc.
Chloe E. Atreya
Speakers' Bureau: Bayer Diagnostics
Research Funding: GlaxoSmithKline
Ryan B. Corcoran
Honoraria: GlaxoSmithKline
Consulting or Advisory Role: Avidity Nanomedicines
Scott Kopetz
Consulting or Advisory Role: Amgen, Roche, GlaxoSmithKline, Janssen, Bristol-Myers Squibb, Agendia, Merrimack, Sysmex, Bayer
Research Funding: Roche, Pfizer, Bristol-Myers Squibb, Amgen, Merrimack, GlaxoSmithKline, sanofi-aventis, Eli Lilly
REFERENCES
- 1.Van Cutsem E, Köhne CH, Hitre E, et al. Cetuximab and chemotherapy as initial treatment for metastatic colorectal cancer. N Engl J Med. 2009;360:1408–1417. doi: 10.1056/NEJMoa0805019. [DOI] [PubMed] [Google Scholar]
- 2.Douillard JY, Siena S, Cassidy J, et al. Randomized, phase III trial of panitumumab with infusional fluorouracil, leucovorin, and oxaliplatin (FOLFOX4) versus FOLFOX4 alone as first-line treatment in patients with previously untreated metastatic colorectal cancer: The PRIME study. J Clin Oncol. 2010;28:4697–4705. doi: 10.1200/JCO.2009.27.4860. [DOI] [PubMed] [Google Scholar]
- 3.Karapetis CS, Jonker D, Daneshmand M, et al. PIK3CA, BRAF, and PTEN status and benefit from cetuximab in the treatment of advanced colorectal cancer: Results from NCIC CTG/AGITG CO.17. Clin Cancer Res. 2014;20:744–753. doi: 10.1158/1078-0432.CCR-13-0606. [DOI] [PubMed] [Google Scholar]
- 4.De Roock W, Jonker DJ, Di Nicolantonio F, et al. Association of KRAS p.G13D mutation with outcome in patients with chemotherapy-refractory metastatic colorectal cancer treated with cetuximab. JAMA. 2010;304:1812–1820. doi: 10.1001/jama.2010.1535. [DOI] [PubMed] [Google Scholar]
- 5.Tejpar S, Celik I, Schlichting M, et al. Association of KRAS G13D tumor mutations with outcomes in patients with metastatic colorectal cancer treated with first-line chemotherapy with or without cetuximab. J Clin Oncol. 2012;30:3570–3577. doi: 10.1200/JCO.2012.42.2592. [DOI] [PubMed] [Google Scholar]
- 6.Morelli MP, Kopetz S. Hurdles and complexities of codon 13 KRAS mutations. J Clin Oncol. 2012;30:3565–3567. doi: 10.1200/JCO.2012.43.6535. [DOI] [PubMed] [Google Scholar]
- 7.Schirripa M, Lonardi S, Cremolini C, et al. Phase II study of single-agent cetuximab in KRAS G13D mutant metastatic colorectal cancer (mCRC) J Clin Oncol. 2014;32(suppl 15s):219s. doi: 10.1093/annonc/mdv385. abstr 3524. [DOI] [PubMed] [Google Scholar]
- 8.Janakiraman M, Vakiani E, Zeng Z, et al. Genomic and biological characterization of exon 4 KRAS mutations in human cancer. Cancer Res. 2010;70:5901–5911. doi: 10.1158/0008-5472.CAN-10-0192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith G, Bounds R, Wolf H, et al. Activating K-Ras mutations outwith “hotspot” codons in sporadic colorectal tumours: Implications for personalised cancer medicine. Br J Cancer. 2010;102:693–703. doi: 10.1038/sj.bjc.6605534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maughan TS, Adams RA, Smith CG, et al. Addition of cetuximab to oxaliplatin-based first-line combination chemotherapy for treatment of advanced colorectal cancer: Results of the randomised phase 3 MRC COIN trial. Lancet. 2011;377:2103–2114. doi: 10.1016/S0140-6736(11)60613-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seymour MT, Brown SR, Middleton G, et al. Panitumumab and irinotecan versus irinotecan alone for patients with KRAS wild-type, fluorouracil-resistant advanced colorectal cancer (PICCOLO): A prospectively stratified randomised trial. Lancet Oncol. 2013;14:749–759. doi: 10.1016/S1470-2045(13)70163-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Douillard JY, Oliner KS, Siena S, et al. Panitumumab-FOLFOX4 treatment and RAS mutations in colorectal cancer. N Engl J Med. 2013;369:1023–1034. doi: 10.1056/NEJMoa1305275. [DOI] [PubMed] [Google Scholar]
- 13.Peeters M, Oliner K, Parker A, et al. Massively parallel tumor multigene sequencing to evaluate response to panitumumab in a randomized phase 3 study of metastatic colorectal cancer. Clin Cancer Res. 2013;19:1902–1912. doi: 10.1158/1078-0432.CCR-12-1913. [DOI] [PubMed] [Google Scholar]
- 14.Patterson SD, Peeters M, Siena S, et al. Comprehensive analysis of KRAS and NRAS mutations as predictive biomarkers for single agent panitumumab (pmab) response in a randomized, phase III metastatic colorectal cancer (mCRC) study (20020408) J Clin Oncol. 2013;31(suppl 15s):242s. abstr 3617. [Google Scholar]
- 15.Ciardiello F, Lenz HJ, Kohne CH, et al. Treatment outcome according to tumor RAS mutation status in CRYSTAL study patients with metastatic colorectal cancer (mCRC) randomized to FOLFIRI with/without cetuximab. J Clin Oncol. 2014;32(suppl 15s):214s. abstr 3506. [Google Scholar]
- 16.Bokemeyer C, Kohne CH, Ciardiello F, et al. Treatment outcome according to tumor RAS mutation status in OPUS study patients with metastatic colorectal cancer (mCRC) randomized to FOLFOX4 with/without cetuximab. J Clin Oncol. 2014;32(suppl 15s):214s. abstr 3505. [Google Scholar]
- 17.Brodowicz T, Vrbanec D, Kaczirek K, et al. FOLFOX4 plus cetuximab administered weekly or every two weeks in first-line treatment of patients with KRAS and NRAS wild-type (wt) metastatic colorectal cancer (mCRC) J Clin Oncol. 2014;(suppl):32. doi: 10.1093/annonc/mdt116. abstr LBA391. [DOI] [PubMed] [Google Scholar]
- 18.Peeters M, Oliner KS, Price TJ, et al. Analysis of KRAS/NRAS mutations in phase 3 study 20050181 of panitumumab (pmab) plus FOLFIRI versus FOLFIRI for second-line treatment (tx) of metastatic colorectal cancer (mCRC) J Clin Oncol. 2014;(suppl):32. abstr LBA387. [Google Scholar]
- 19.Schwartzberg LS, Rivera F, Karthaus M, et al. PEAK: A randomized, multicenter phase II study of panitumumab plus modified fluorouracil, leucovorin, and oxaliplatin (mFOLFOX6) or bevacizumab plus mFOLFOX6 in patients with previously untreated, unresectable, wild-type KRAS exon 2 metastatic colorectal cancer. J Clin Oncol. 2014;32:2240–2247. doi: 10.1200/JCO.2013.53.2473. [DOI] [PubMed] [Google Scholar]
- 20.Heinemann V, von Weikersthal LF, Decker T, et al. FOLFIRI plus cetuximab versus FOLFIRI plus bevacizumab as first-line treatment for patients with metastatic colorectal cancer (FIRE-3): A randomised, open-label, phase 3 trial. Lancet Oncol. 2014;15:1065–1075. doi: 10.1016/S1470-2045(14)70330-4. [DOI] [PubMed] [Google Scholar]
- 21.Sorich MJ, Wiese MD, Rowland A, et al. Extended RAS mutations and anti-EGFR monoclonal antibody survival benefit in metastatic colorectal cancer: A meta-analysis of randomized controlled trials. Ann Oncol. doi: 10.1093/annonc/mdu378. [epub ahead of print on August 12, 2014] [DOI] [PubMed] [Google Scholar]
- 22.Erbitux package insert. Branchburg, NJ: ImClone Systems; 2013. [Google Scholar]
- 23.Vectibix package insert. Thousand Oaks, CA: Amgen; 2013. [Google Scholar]
- 24.Benson AB, 3rd, Venook AP, Bekaii-Saab T, et al. Colon cancer, version 3.2014. J Natl Compr Canc Netw. 2014;12:1028–1059. doi: 10.6004/jnccn.2014.0099. [DOI] [PubMed] [Google Scholar]
- 25.Misale S, Yaeger R, Hobor S, et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature. 2012;486:532–536. doi: 10.1038/nature11156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Diaz LA, Jr, Williams RT, Wu J, et al. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature. 2012;486:537–540. doi: 10.1038/nature11219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morelli M, Overman M, Dasari A, et al. Heterogeneity of acquired KRAS and EGFR mutations in colorectal cancer patients treated with anti-EGFR monoclonal antibodies. J Clin Oncol. 2013;31(suppl 15s):216s. abstr 3512. [Google Scholar]
- 28.Morelli M, Overman M, Sanchez E, et al. Frequency of concurrent gene mutations and copy number alterations in circulating cell-free DNA (cfDNA) from refractory metastatic CRC patients. J Clin Oncol. 2014;32(suppl 15s):724s. abstr 11117. [Google Scholar]
- 29.Franklin WA, Haney J, Sugita M, et al. KRAS mutation: Comparison of testing methods and tissue sampling techniques in colon cancer. J Mol Diagn. 2010;12:43–50. doi: 10.2353/jmoldx.2010.080131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vakiani E, Janakiraman M, Shen R, et al. Comparative genomic analysis of primary versus metastatic colorectal carcinomas. J Clin Oncol. 2012;30:2956–2962. doi: 10.1200/JCO.2011.38.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van Cutsem E, Lenz HJ, Kohne CH, et al. Outcome according to tumor RAS mutation status in CRYSTAL study patients with metastatic colorectal cancer randomized to FOLFIRI with or without cetuximab as first-line treatment. Presented at 16th World Congress Gastrointestinal Conference; June 25-28, 2014; Barcelona, Spain. [Google Scholar]
- 32.Dono M, Massucco C, Chiara S, et al. Low percentage of KRAS mutations revealed by locked nucleic acid polymerase chain reaction: Implications for treatment of metastatic colorectal cancer. Mol Med. 2012;18:1519–1526. doi: 10.2119/molmed.2012.00175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Molinari F, Felicioni L, Buscarino M, et al. Increased detection sensitivity for KRAS mutations enhances the prediction of anti-EGFR monoclonal antibody resistance in metastatic colorectal cancer. Clin Cancer Res. 2011;17:4901–4914. doi: 10.1158/1078-0432.CCR-10-3137. [DOI] [PubMed] [Google Scholar]
- 34.Song S, Nones K, Miller D, et al. Qpure: A tool to estimate tumor cellularity from genome-wide single-nucleotide polymorphism profiles. PLoS One. 2012;7:e45835. doi: 10.1371/journal.pone.0045835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bokemeyer C, Van Cutsem E, Rougier P, et al. Addition of cetuximab to chemotherapy as first-line treatment for KRAS wild-type metastatic colorectal cancer: Pooled analysis of the CRYSTAL and OPUS randomised clinical trials. Eur J Cancer. 2012;48:1466–1475. doi: 10.1016/j.ejca.2012.02.057. [DOI] [PubMed] [Google Scholar]
- 36.Bendell J, Atreya C, Andre T, et al. Efficacy and tolerability in an open-label phase I/II study of MEK inhibitor trametinib (T), BRAF inhibitor dabrafenib (D), and anti-EGFR antibody panitumumab (P) in combination in patients (pts) with BRAF V600E mutated colorectal cancer (CRC) J Clin Oncol. 2014;32(suppl 15s):216s. abstr 3515. [Google Scholar]
- 37.Van Geel R, Elez E, Bendell J, et al. Phase I study of the selective BRAFV600 inhibitor encorafenib (LGX818) combined with cetuximab and with or without the -specific PI3K inhibitor BYL719 in patients with advanced BRAFmutant colorectal cancer. J Clin Oncol. 2014;32(suppl 15s):216s. abstr 3514. [Google Scholar]
- 38.Hong D, Morris VK, Fu S, et al. Phase 1B study of vemurafenib in combination with irinotecan and cetuximab in patients with BRAF-mutated advanced cancers and metastatic colorectal cancer. J Clin Oncol. 2014;32(suppl 15s):216s. abstr 3516. [Google Scholar]


