Abstract
Malaria causes nearly 1 million deaths annually. Recent emergence of multidrug resistance highlights the need to develop novel therapeutic interventions against human malaria. Given the involvement of sugar binding plasmodial proteins in host invasion, we set out to identify such proteins as targets of small glycans. Combining multidisciplinary approaches, we report the discovery of a small molecule inhibitor, NIC, capable of inhibiting host invasion through interacting with a major invasion-related protein, merozoite surface protein-1 (MSP-1). This interaction was validated through computational, biochemical, and biophysical tools. Importantly, treatment with NIC prevented host invasion by Plasmodium falciparum and Plasmodium vivax—major causative organisms of human malaria. MSP-1, an indispensable antigen critical for invasion and suitably localized in abundance on the merozoite surface represents an ideal target for antimalarial development. The ability to target merozoite invasion proteins with specific small inhibitors opens up a new avenue to target this important pathogen.
Keywords: malaria, Plasmodium falciparum, Plasmodium vivax, glycan mimetic small molecules, chemical biology, mass spectrometry, red blood cell, host invasion, merozoite surface proteins
Malaria remains a major health concern for the developing world. Although recent trends suggest a progressive decline, nearly 220 million malaria infections (causing approximately 700 000 deaths) occurred in 2010 alone (http://www.who.int/malaria/world_malaria_report_2011/en/). These data largely focus on Plasmodium falciparum infections in Africa and gravely underestimate the contribution of Plasmodium vivax, which may account for another 200 million infections in Asia and South America. Thus, malaria against which vaccine development efforts are still ongoing [1] exerts an enormous burden on global human health [2]. Efforts to treat malaria largely rely on chemotherapeutic approaches to block parasitic replication besides preventive measures targeting the mosquito vector. However, recent emergence of resistance against chloroquine [3] and reduced efficacy of artemisinin [4] point to the critical need for exploring new set of antimalarial targets.
Merozoite entry into erythrocytes is driven by complex recognition events between parasitic proteins and sugar moieties on the host red blood cells (RBCs) [5, 6]. Merozoites bind to sialic acid residues on RBC receptors for invasion [7], however, sialic acid-independent invasion pathways have also been elucidated [8, 9]. Although several plasmodial proteins play critical roles during invasion, merozoite surface proteins (MSPs) and apical membrane antigens (AMAs) have been the most studied. Among MSPs, MSP-1 is considered a prime candidate for vaccine development [10, 11]. It is required for the attachment of merozoites to host receptors such as Band 3 [12] and for normal parasitic development [13]. MSP-1 undergoes 2 proteolytic processing steps during merozoite maturation [14]. Following cleavage, MSP-1 complex remains noncovalently attached to the merozoite surface until invasion, except for a small fragment of 19-kDa from the C-terminus (MSP-119) that remains attached to the merozoite even after invasion. Antibodies targeting MSP-119 epitopes [15, 16] block merozoite invasion, illustrating a definite invasion-associated role of this domain. In addition, MSP-119 is involved in normal parasitic development [16] and food vacuole formation [17].
Glycan derivatives interfere with replication and invasion of P. falciparum [18–20]. Especially, sulfated glycosaminoglycans are known to block merozoite [21] and sporozoite invasion [22]. It has been shown that heparin blocks merozoite invasion by binding to a specific domain of MSP-1 termed MSP-133 thereby preventing the secondary proteolytic processing [23]. Although MSP-119 does not bind to heparin-like glycosaminoglycan oligosaccharides [23], its ability to bind to small molecules has not been investigated. In this study, we report the identification of a small molecule, 2-butyl-5-chloro-3-(4-nitro-benzyl)-3H-imidazole-4-carbaldehyde (NIC) capable of binding to MSP-119 of human malaria parasites: P. falciparum and P. vivax. Using different approaches, we demonstrate that NIC interacts with EGF-like domains present on MSP-119. These observations collectively point to the possibility of targeting plasmodial invasion proteins for antimalarial drug development.
MATERIALS AND METHODS
Parasites and Reagents
Collection of blood for Plasmodium culture was verified and approved by the Institutional Review Board (IRB) of National University of Singapore (NUS). P. falciparum strain 3D7 was used for all experiments unless otherwise stated. Transgenic parasites - D10-PfM3′ and D10-PcMEGF [24] were gifted by Paul Gilson and Danny Wilson (Burnet Institute, Australia). Various recombinant MSP-119 (MRA 47, MRA 48, MRA 55, and MRA 60), antibodies (MRA 3, MRA 16), and parasites (Dd2, K1, T9-94, W2-mef, and FCR3) were obtained from Malaria Research and Reference Reagent Resource Center (MR4). Parasites were synchronized by sorbitol selection or MACS separation (Miltenyi Biotec). Parasitemia was scored by flow cytometric counting (LSRII flow cytometer- BD Biosciences) after staining with Hoechst. Dose-response curve generation and half maximal inhibitory concentration (IC50) determination were performed with GraphPad Prism 5 using a four-parameter logistic curve (variable slope).
Virtual Screening and Design of Novel MSP-1 Ligands
Insight II (Accelrys) was used for molecular modeling [25]. Automatic docking, clustering and energy minimizations [26] were performed using LigandFit module. A cavity detection algorithm and Monte Carlo conformational search algorithm were used for generating ligand poses consistent with active site shapes. Ligand poses at these sites were selected based on shape matching, using a cutoff distance of 3 Å. Structure based high-throughput virtual screening method of LigandFit (DS version 2.5) was used to identify potential ligands that interact with MSP-1 (PDB ID: 1CEJ) using NCBI database [27]. Glycan-mimetic small molecules such as KI-105, DMBO, BIC, NIC, CIC, and DIC were synthesized as reported [28]. These compounds were purified and characterized using Agilent 1200 series LC system (Agilent ZORBAX, Eclipse-XDB-C18, 5 µm, 4.6 mm × 150 mm).
Docking of Small Molecule Ligands to MSP-119
All ligands were subjected to LigandFit docking to determine reasonable conformations for each low energy conformer at the binding site. Initially, crystal structures of MSP-119 from P. falciparum [29] (PDB ID: 1CEJ), P. vivax [30] (PDB ID: 2NPR) and P. knowlesi [31] (PDB ID: 1N1I) were used for docking. Further, homology modeling of the MSP-119 domains for P. chabaudi, P. malariae, and P. ovale were performed using ‘build homology modeling’ protocol by DS version 2.5.
SPR Measurements
SPR measurements were conducted using a Biacore T-200 instrument (Biacore, GE Healthcare). MSP-119 peptide was directly immobilized onto a flow cell on a CM5-S sensor chip (Biacore, GE Healthcare) through standard amine coupling [32]. An empty flow cell was used as control. Surface was activated for 7 minutes with 1:1 mixture of 0.2 M N-ethyl-N'-[3-(diethylamino)propyl]carbodiimide (EDC) and 0.05 M N-hydroxysuccinimde (NHS). Different interactants (in 10 mM sodium acetate, pH 4.0) was injected across the activated surface at 10 µL/min until desired immobilization level of approximately 3000 RU was achieved. The surface was blocked with 7-minute injection of 1 M ethanolamine-HCl, pH 8.5. Small molecules were screened against the immobilized protein at a flow rate of 30 µL/min, with a 60-second association phase and a 5-minute dissociation phase, in running buffer (20 mM Na2HPO4-NaH2PO4, pH 7.4, 150 mM NaCl, 5% DMSO). Measurements for affinity determination were performed under similar conditions using 2-fold dilutions of 10 µM. Sensorgrams were double referenced [33] and evaluated with Scrubber 2 software. Equilibrium responses against concentration were fitted to a simple 1:1 binding isotherm using a global Rmax. Each experiment was carried out at least 3 times. To validate the selectivity of binding and to approximately estimate the affinity constants, simulation exercise were carried out as reported [34].
Localization and Affinity-enrichment of the Target Using NIC-Biotin
NIC- biotin was synthesized by amine coupling of the aldehyde group. Late-stage schizonts (approximately 45 hpi) were treated with NIC-biotin for 4 hours, and aliquots were fixed (0.1% paraformaldehyde/phosphate-buffered saline [PBS]) and permeabilized (0.2% Triton X100/PBS) in presence of Casein. After washing, samples were incubated with phycoerythrin-conjugated streptavidin (Invitrogen) and Hoechst for 30 minutes. Fluorescence microscopy was performed using an LSM 710 confocal microscope (Carl Zeiss). For affinity purifying NIC-reactive proteins, schizonts (approximately 48 hpi) were fractionated into extraparasitic and parasitic fractions by extraction with 0.02% saponin and 1% Triton X-100 sequentially. Samples were then diluted 1:5 with 20 mM sodium acetate buffer (pH 5.5), treated with 50 µM NIC-biotin for 2 hours, and incubated with streptavidin-agarose beads (Thermo Scientific). After removing unbound proteins by centrifugation, pellet was washed with PBS and proteins extracted for SDS-PAGE. Coomassie staining was carried out to visualize polypeptides. Protein bands were excised and subjected to MALDI/TOF-TOF analyses (4800 Proteomics Analyser- Applied Biosystems). MS data were searched using MASCOT v 2.1 (Matrix Science Ltd, London, UK) against NCBI Database. For examining target-specificity, affinity enrichment experiments were performed with (1) nonbiotinylated NIC, (2) in competition with excess of NIC (100 µM), and (3) denatured (SDS/heat) sample. Proteins bound to streptavidin beads were collected, resolved on SDS-PAGE for Western blotting using anti-MSP-1 antibody.
Plasmodium vivax Ex Vivo Invasion Assays
During February 2012, samples from P. vivax infected patients at Shoklo Malaria Research Unit (SMRU), Mae Sot region (Thailand) were collected after written informed consent. Cord blood was taken from 3 post healthy delivery placentas. The samples were collected as per ethical guidelines in the approved protocols: OXTREC 027-025 (University of Oxford, Centre for Clinical Vaccinology and Tropical Medicine, UK) and MUTM 2008-215 from the Ethics Committee of the Faculty of Tropical Medicine, Mahidol University, Bangkok. Leukocytes and platelets were removed using a CF11 column ([35]) before conducting P. vivax invasion assays as described elsewhere [36]. An inert chemical intermediate of the synthetic scheme, 2-butyl-5-chloro-4-carbaldehyde (BCI) at a concentration of 200 µM and 25 µg/mL of anti-Duffy receptor DARC antibody (gift by Drs Colin and Bertrand; Institut National de la Transfusion Sanguine, 75015 Paris, France) served as negative and positive controls respectively. To determine invasion rates, a 3 color cytometery method [37] using a field flow cytometer (BD Accuri C6) was used. Flow cytometery data was verified with Giemsa-stained smears using microscopy.
RESULTS
Identification of NIC as a Potential Ligand Against EGF-like Domains of Plasmodial MSP-1
Antibodies targeting EGF-like domains on the C-terminal motif (MSP-119) prevent host invasion. We therefore set out to identify small molecules that can bind to MSP-119 domain as invasion blockers. First, using the solution structure [29] a virtual screening was performed to prioritize small molecules that could interact with MSP-119 of P. falciparum. Based on this screening, we chose KI-105 [38] and DMBO [39] that are known to interact with EGF-like domain containing proteins in other systems as promising ligands to interact with MSP-119. However, neither KI-105 nor DMBO exhibited any antimalarial activity against P. falciparum. Thus, we synthesized and evaluated other related scaffolds such as BIC, NIC, CIC, and DIC (Supplementary Figure 1A) as binders of MSP-119. Among these, it was earlier demonstrated that NIC binds to vascular endothelial growth factor (VEGF) resulting in reduced cancer cell proliferation [40].
Based on these computational analyses, we posited that NIC could bind with a higher affinity to EGF-like domains present on MSP-119 compared to other structurally related compounds (Table 1). The energy-minimized structure of NIC is highlighted in Figure 1A. The ribbon diagram showing the structure of EGF-like domain of PfMSP-119 in complex with NIC is shown as a ball-and-stick model (Figure 1B). Based on the structural complex of NIC with PfMSP-119, the key residues positioned to make contact with NIC included Lys29, Cys30, Leu31, Leu32, Asn33, Tyr34, Lys35, and Gln36. NIC was likely to form additional bonds with Glu51, Asn52, Asn53, Gly54 and Gly55 (Figure 1C). Also, it appears that NIC has the unique ability to bind to MSP-119 through its ionic nitro-group on the benzene ring making it the most suitable ligand. Sequence alignment of residues within the EGF-like domains of MSP-119 from multiple plasmodia revealed remarkable conservation of key residues that are positioned to make contact with NIC (Figure 1D). Many of these conserved residues were within epitopes recognized by invasion-blocking antibodies against MSP-1.
Table 1.
Summary of Docking and Scoring Analyses Among the Selected Small Molecule Scaffolds Against PfMSP-119
| Ligand | MW | PLP1 | PLP2 | LigScore1 | LigScore2 | Jain | PMF | LIE | Dock Score |
|---|---|---|---|---|---|---|---|---|---|
| NIC | 321.7 | 82.9 | 65.0 | 2.6 | 1.2 | −0.3 | 36.0 | 1.4 | 45.0 |
| CIC | 339.2 | 46.4 | 46.7 | 1.9 | 3.2 | 0.6 | 27.1 | −3.4 | 33.5 |
| BIC | 415.7 | 58.0 | 43.1 | 1.3 | 4.1 | −1.9 | 55.9 | −1.3 | 37.6 |
| DIC | 283.7 | 66.5 | 71.6 | 2.4 | 3.2 | 3.3 | −18.0 | − | 27.6 |
| KI-105 | 379.3 | 55.6 | 42.3 | 1.1 | 3.9 | 2.4 | 44.6 | −2.1 | 38.6 |
| DMBO | 373.4 | … | … | … | … | … | … | … | … |
Abbreviations: DS, dock score; Jain, empirical scoring function through an evaluation of the structures and binding affinities of a series of protein-ligand complexes; LIE, ligand internal energy; PLP1, piecewise linear potential 1; PLP2, piecewise linear potential 2; PMF, potential of mean force.
Figure 1.
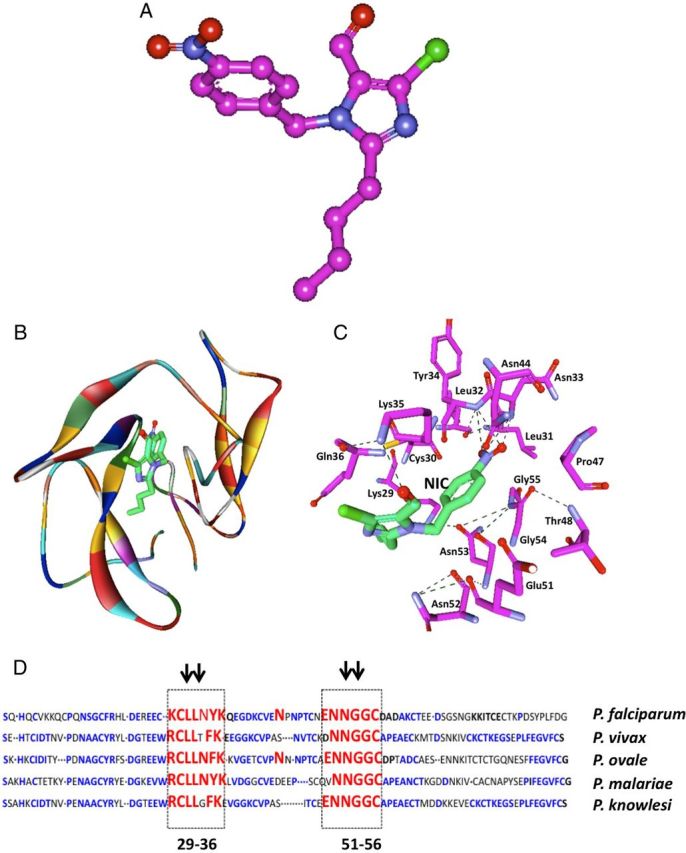
Prediction of NIC as a small molecule scaffold with potential binding affinity to MSP-119 of Plasmodium falciparum. A, energy minimized structure of NIC highlighting its ionic functional groups (highlighted with arrowheads) is shown. B, Ribbon diagram of the structure of EGF-like domain of PfMSP-119 in complex with NIC is shown. Secondary structure of the protein is shown with α-helices and β-strands, whereas NIC is represented in ball-and-stick model (green for carbon, blue for oxygen, white for hydrogen, and yellow for sulphur). C, Interaction map showing the key amino acids at PfMSP-119 that are positioned to make interactions with NIC. The hydrogen bonding is shown in red. D, Sequence alignment of C-terminal residues of MSP-119 of various plasmodial species: conserved amino acid residues are in blue, residues that are likely to interact with NIC are highlighted in red. Abbreviation: NIC, 2-butyl-5-chloro-3-(4-nitro-benzyl)-3H-imidazole-4-carbaldehyde.
NIC Specifically Interacts with PfMSP-119
Computational analyses predicted that among KI-105 variants (BIC, NIC, CIC, and DIC), NIC had the highest binding affinity to MSP-119. This was validated via surface plasmon resonance (SPR) measurements using recombinant PfMSP-119, where none of the small molecules showed detectable binding to PfMSP-119 except NIC (Figure 2A). Notably, BCI, which served as the negative control as well as heparin, showed no measurable interaction with MSP-119 as expected from previous reports [23].
Figure 2.

NIC interacts specifically with MSP-119 of Plasmodium falciparum. A, SPR Interaction analysis of NIC over immobilized PfMSP-119. Immobilization was performed by amine coupling to a CM5 sensor chip to the level of 3000 RU. NIC (concentrations) was injected sequentially over the relevant and control surfaces. The sensorgrams were fit using the steady state affinity model from Scrubber software. Each experiment was carried out at least 3 times to confirm the results. B, Experimental values of Req for the corrected sensorgrams were compared to the simulated values for KD = 10 µM and 50 µM. The value for the affinity constant cannot be determined from equilibrium analysis (see results) and was estimated to be between these 2 values (37 µM as calculated from the fit). C, Fluorescent microscopy analyses using NIC-biotin reveals the target on merozoite surface, co-localizing with MSP-119-GFP. D, Affinity purification of proteins using 50 µM NIC-biotin shows specific enrichment of MSP-119. Magnet purified parasites were fractionated into extraparasitic and merozoite fractions and incubated with NIC-biotin. SDS-PAGE analysis of biotinylated proteins showed specific enrichment of a polypeptide later confirmed to be MSP-119 by mass spectrometry analysis and Western blot. E, Affinity pull-out experiments of parasitic material using 50 µM NIC-biotin (lane A), 50 µM NIC (lane B), 100 µM NIC followed by 50 µM NIC-biotin (lane C), and under denaturing conditions (lane D). The latter 3 conditions significantly reduced the extent of PfMSP-119 enrichment. Abbreviation: NIC, 2-butyl-5-chloro-3-(4-nitro-benzyl)-3H-imidazole-4-carbaldehyde.
We determined the affinity constant from a fit to a simple binding isotherm in an equilibrium analysis, with an estimated value in the tens of micromolar range (approximately 37 µM; Figure 2B). Because we could not reach saturation of the surface due to lack of solubility of NIC at high concentrations (under conditions used for SPR measurements), and thus the calculated value of Rmax could not be precise, we chose to show the range for the value of affinity constant by simulating the Req values vs the NIC concentration for affinities of 25 and 50 µM and plotting the experimental values for Req at different concentrations of NIC, as adopted elsewhere [34]. Figure 2B shows that the values of responses for the measured interaction between NIC and PfMSP-119 do fall between 25 and 50 µM.
To validate the interaction of NIC with its parasitic target, we first conducted co-localization experiments using NIC-biotin (Supplementary Figure 1B and 1C) that has comparable invasion inhibitory effects on malaria parasites as NIC (Supplementary Figure 1D). Using a parasite line that expresses MSP-119-GFP, we observed co-localization of NIC-biotin with MSP-1 in late-stage schizonts (Figure 2C). In contrast, ring or trophozoite stage parasites when very little MSP-1 is present showed no detectable labeling with NIC-biotin (Supplementary Figure 1E). However, due to the dispersed nature of GFP distribution in these parasites [41], it was not possible to conclusively document co-localization. Hence, fluorescence microscopy was repeated using antibodies against MSP-119 together with NIC-biotin (Supplementary Figure 1F); further supporting an interaction between NIC and MSP-119.
Next, we used NIC-biotin to identify parasitic targets. Schizont-stage parasites were fractionated into infected red cell/extraparasitic (iRBC) and parasitic fractions by differential extraction using saponin [42]. Further, each sample was incubated with 50 µM NIC-biotin and then with streptavidin-agarose beads to affinity purify proteins that interact with NIC. Aliquots from unbound and bound fractions from each sample were resolved on SDS-PAGE and polypeptides visualized by Coomassie staining. Enrichment of a low molecular weight protein (Figure 2D) was observed from the parasitic fraction and was identified as MSP-119 through mass spectrometry (Supplementary Figure 1G). Importantly, no other proteins, including MSP-133, the target of heparin as reported previously [23], were enriched in the NIC-bound fraction. Specificity of NIC-biotin binding to MSP-119 was confirmed by conditions such as (a) performing affinity-enrichment from parasitic extracts with NIC, (b) pretreatment of the extract with 100 µM NIC prior to incubation with NIC-biotin, and (c) by performing NIC-biotin incubation after denaturing the sample with SDS/heat (Figure 2E). As expected, pretreatment with excess NIC significantly reduced (30%–50%) the affinity enrichment of MSP-119 by NIC-biotin. Due to the multiple steps involved in this experiment where each sample was processed individually, these results are primarily qualitative.
NIC Blocks P. falciparum Replication by Impairing Merozoite Invasion
Assays were performed on several P. falciparum strains to evaluate the effect of the small molecules on erythrocytic development. Trophozoite-stage parasites (approximately 24 hpi) were allowed to grow until the next cycle in presence of inhibitors. DMSO and BCI served as negative controls; 48 hours later, samples were collected, stained with Hoechst, and parasitemia scored by flow cytometry. Among the scaffolds tested, only NIC inhibited parasite replication (Figure 3A) with a median IC50 of 21.7 µM. Potential nonspecific interaction of NIC with red cell proteins was ruled out by preincubating RBCs with NIC for 2 hours and washing them prior to mixing with late-stage parasites. RBCs that were pretreated with NIC supported normal parasite invasion and growth (Figure 3B). Microscopic evaluation of Giemsa-stained smears revealed that NIC treatment resulted in impaired merozoite invasion. Clumps of agglutinated merozoites were observed in smears prepared from postrupture time points (Figure 3C) after NIC treatment. NIC showed comparable inhibitory potential on a number of plasmodial strains tested (Table 2) demonstrating a conserved target.
Figure 3.
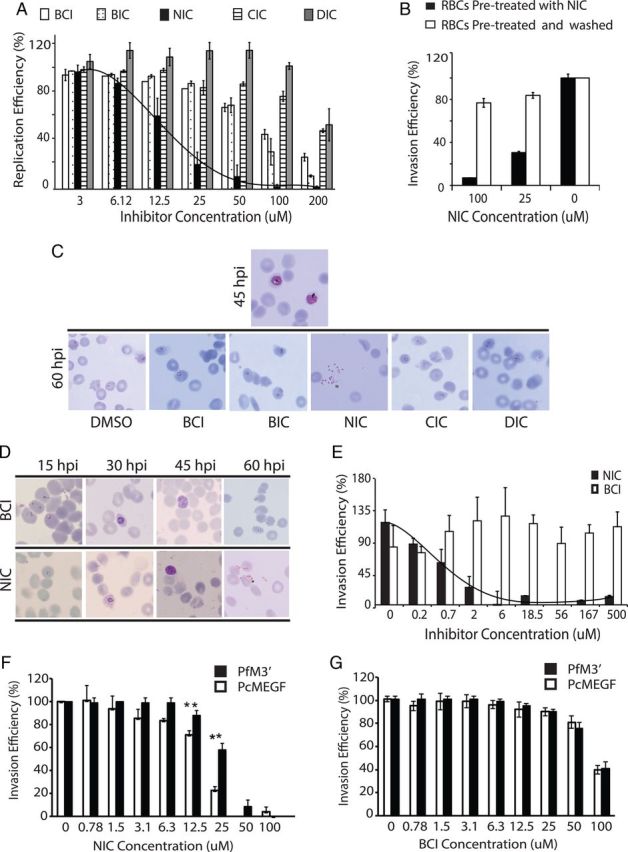
Evaluation of invasion-blocking properties of NIC in Plasmodium falciparum. A, Dose-dependent effect of 4 major small molecule scaffolds on the replication of P. falciparum; NIC shows a significant and specific inhibition of parasite replication (transition from schizonts to ring) with an average IC50 of 21.7 µM, error bars show standard deviation, the figure represents compiled data from 3 independent experiments. B,RBCs preincubated with NIC and washed prior to parasite infection, supports normal invasion and growth ruling out nonspecific binding of NIC to red cell proteins. C, Microscopic analysis of representative Giemsa-stained smears from inhibitor-treated parasites (DMSO, BCI, BIC, NIC, DIC, and CIC each at 25 µM) demonstrate that treatment with NIC resulted in an invasion defective phenotype, clumps of extracellular merozoites that were unable to invade was observed. None of the other inhibitors induced similar phenotypes at the tested concentration. D, Stage-specific effect of NIC (25 µM) on P. falciparum development demonstrating a specific inhibitory effect on invasion. E, The inhibitory effect of NIC on red cell invasion by purified merozoites, the figure shows combined data from 4 independent experiments and error bars indicate standard deviation. F, Transgenic parasites (PcMEGF) that contain MSP-119 from Plasmodium chabaudi (PcMSP-119) at schizont stages were allowed to rupture and invade in presence of NIC. These parasites showed increased sensitivity to NIC compared to wild-type (PfM3′) parasites. The figure represent combined results from 3 independent experiments, error bars show standard deviation. G, Transgenic parasites (PcMEGF) that contain MSP-119 from Plasmodium chabaudi (PcMSP-119) when allowed to rupture and invade in presence of BCI, showed no differences in their sensitivity. The figure represents combined results from 3 independent experiments; error bars show standard deviation. Abbreviation: NIC, 2-butyl-5-chloro-3-(4-nitro-benzyl)-3H-imidazole-4-carbaldehyde.
Table 2.
Effect of NIC on Various Strains of Plasmodium falciparum, Based on Replication Assays (Using Schizont-Stage Parasites)
| Parasite Strains | IC50 (µM) |
|
|---|---|---|
| BCI | NIC | |
| 3D7 | >200 | 21.7 ± 6.2 |
| Dd2 | >200 | 21.0 ± 0.1 |
| FCR3 | >200 | 20.0 ± 0.4 |
| K1 | >200 | 22.5 ± 6.5 |
| T994 | >200 | 23.6 ± 2.4 |
| W2-mef | >200 | 21.6 ± 8.3 |
| W2-mef (NM) | >200 | 22.3 ± 6.6 |
The values represent compiled results from 3 independent experiments.
Abbreviations: BCI, 2-butyl-5-chloro-4-carbaldehyde; IC50, half maximal inhibitory concentration; NIC, 2-butyl-5-chloro-3-(4-nitro-benzyl)-3H-imidazole-4-carbaldehyde.
Further, ring stage parasites (approximately 12 hpi) were treated with 20 μM NIC and phenotypes monitored at specific intervals (15, 30, 45, and 60 hpi) using Giemsa-stained smears. By 60 hpi, all the schizonts had ruptured and established rings in the control (DMSO) treatments, whereas merozoites failed to invade in presence of NIC (Figure 3D). The invasion inhibition by NIC was confirmed by live video microscopy [43]. In presence of NIC, the schizonts appeared to burst slowly and despite making initial contact with RBCs, merozoites failed to invade even after several minutes (Supplementary Videos 2A and 2B). Next, invasion inhibition assays were conducted by mixing purified merozoites [44] with fresh RBCs in presence of NIC. This experiment revealed a strong and specific invasion inhibition in presence of NIC (Figure 3E). Negative control (BCI) showed no significant inhibitory effects on invasion even at 200 μM. Although there are minor differences in the sensitivity to the inhibitors of purified merozoites compared to intact schizonts, these experiments provide strong evidence for the invasion inhibitory activity of NIC. Furthermore, schizont-stage parasites (approximately 45 hpi) were exposed for 2 hours to NIC, and NIC was removed by washing. These parasites were able to invade efficiently, revealing NIC's specific inhibitory effect solely on invasion (Supplementary Figure 1H).
Computational docking revealed that NIC was likely to have superior binding affinity to MSP-119 of P. chabaudi (Supplementary 3A) compared to PfMSP-119. Hence, we used a transgenic strain of P. falciparum (PcMEGF) that has PcMSP-119 swapped in [24] to evaluate its sensitivity to NIC. Dose-dependent effect of NIC on replication of PcMEGF was examined together with the control parasites (PfM3′) that contain the original PfMSP-119. This revealed significantly higher sensitivity of the PcMEGF parasites to NIC (Figure 3F) compared to PfM3′, in agreement with the computational predictions. BCI-treated parasites (PfM3′ and PcMEGF) served as controls and showed no apparent differences in sensitivity (Figure 3G). These data functionally validate the interaction between NIC and MSP-119, leading to impairment of invasion.
NIC Inhibits P. vivax Invasion
The residues within EGF-like domains of MSP-119, are significantly conserved among multiple plasmodia. Computational analyses predicted that MSP-119 from several human malaria parasites were likely to bind to NIC (Supplementary 3B). Hence, we inspected the interaction of NIC with PvMSP-119, EGF-like domains of which are critical for reticulocyte invasion [45] by P. vivax. SPR measurements using recombinant PvMSP-119 confirmed specific interaction of NIC with the target (Figure 4A), however with lower affinity compared to PfMSP-119 (Figure 4B). The effect of NIC on ex vivo invasion of P. vivax into human reticulocytes was tested using a recently developed assay [36]. Purified schizonts from clinical isolates were mixed with enriched reticulocytes in presence of different amounts of NIC. BCI-treated parasites served as negative control while an anti- Duffy antibody (2C3) that blocks invasion at 25 µg/mL concentration served as positive control. After 24 hours, newly invaded reticulocytes were counted by a field-based flow cytometry assay [36] (Figure 4C). A concentration-dependent inhibition of invasion by NIC on P. vivax merozoites was observed with an average IC 50 of 23.6 µM. BCI showed no inhibition even at 100 µM concentration. These results were further verified by microscopic inspection of Giemsa-stained blood smears (Figure 4D). In BCI-treated samples, robust invasion was observed (Figure 4D, upper panel), whereas very few ring-stage infections were found in samples that were incubated with 25 or 100 µM NIC (Figure 4D, middle and lower panels). Finally, 6 independent analyses were carried out using different field isolates—all of them demonstrated dose-dependent inhibition of invasion caused by NIC (Figure 4E), with a Median IC50 of 23.6 µM (IQR 16.9–27.24). These observations are consistent with other results and strongly support the idea that NIC has broad affinity to MSP119 from multiple forms of human malaria.
Figure 4.
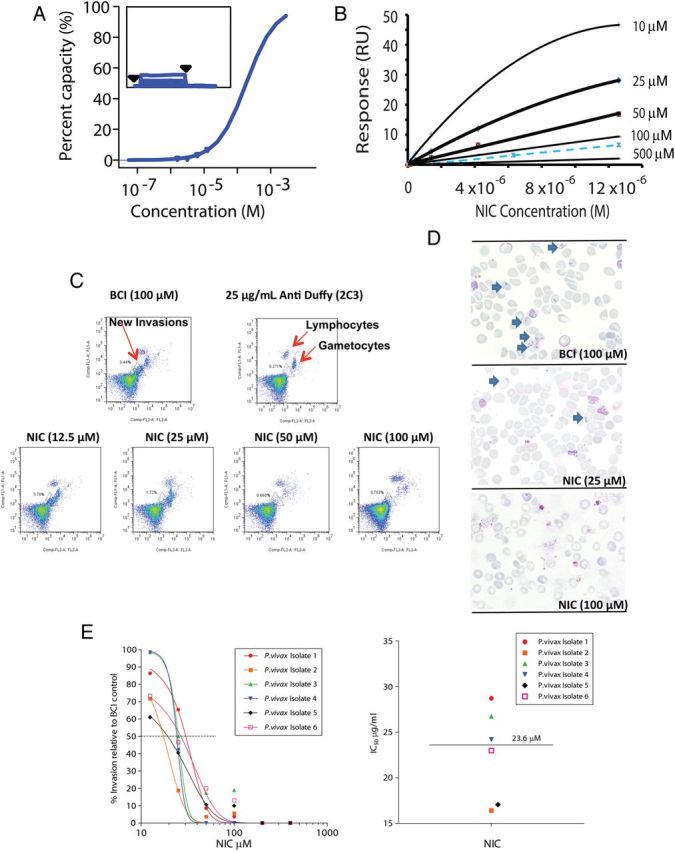
NIC binds to PvMSP-119 and shows anti-invasive activity against clinical isolates of Plasmodium vivax. A, SPR measurements using recombinant PvMSP-119 reveal specific binding of NIC with the target antigen. Immobilization, the binding assay and the determination of the affinity were performed as in Figure 2. B, Experimental values of Req for the corrected sensorgrams were compared to the simulated values for KD = 100 µM and 500 µM. The value for the affinity constant cannot be determined from equilibrium analysis (see results) and was estimated to be between these 2 values. C, An ex vivo assay on 6 Thai isolates of P. vivax showed that NIC had a dose-dependent inhibitory effect on the invasion of human cord reticulocytes by P. vivax, as determined by flow cytometry; BCI as well as an antibody against Duffy protein served as negative and positive controls, respectively. D, Microscopic analyses of the Giemsa-stained smears collected from inhibitor-treated cultures reveal blockage of invasion by P. vivax in presence of NIC. E, Compiled data derived from the treatment with NIC of 6 Thai P. vivax isolates analyzed using a nonlinear sigmiodal model provide a median IC50 of 23.6 µM. Abbreviations: IC50, half maximal inhibitory concentration; NIC, 2-butyl-5-chloro-3-(4-nitro-benzyl)-3H-imidazole-4-carbaldehyde.
DISCUSSION
Host invasion by plasmodia is facilitated by coordinated action of merozoite surface proteins [46], proteases [47, 48], and signaling molecules such as calcium [49]. Extracellular merozoites have a short life span; they become nonviable within minutes of release making strategies targeting invasion an attractive proposition. Prior research explored invasion-related proteases [47, 48] as well as antibodies for potential therapeutic development. However, the likely off-target effects of protease inhibitors and permeability limitations of antibodies hinder these approaches significantly.
Chemical biology tools allow detailed and targeted investigation of complex biological events, such as malaria parasite invasion. MSP-1 is the most abundant invasion protein and binds to glycan residues on RBC to initiate host entry. The smaller fragment derived from MSP-1 processing; MSP-119 is critical for invasion and parasite development. It shows less polymorphism (within its EGF-like domains) highlighting suitability as a pan-plasmodial target. We conducted in silico and in vitro screening to identify a small molecule, NIC with significant binding affinity to both recombinant and parasitic PfMSP-119 that inhibits merozoite invasion. Although at this stage we do not fully understand the detailed molecular mechanism leading to invasion inhibition by NIC binding to MSP-1, our results represent a major advancement in the hunt for antimalarials with pan-species efficacy. NIC was able to bind to MSP-1 and inhibit invasion not only upon merozoite release but also within late-stage schizonts. This highlights the permeability advantages of small molecules over therapeutic antibodies. Efforts to optimize molecular structures from the NIC lineage with stronger/irreversible binding properties to MSP-1 may provide a platform to further consider these molecules for in vivo experiments and detailed pharmacological profiling.
The abundance, accessible cellular localization, and indispensable nature of invasion proteins make them promising drug targets. In the Plasmodial genome, there are several proteins with putative EGF-like domains. Other members of the MSP family, MSP4 and MSP8 as well as lesser-known proteins such as PF3D7-1403200 and PF3D7-1002300 (www.plasmodb.org) contain such domains. It would be desirable to study these proteins/domains as candidate targets to design small molecule scaffolds, individually or as a “group target.” Designing small molecules to target invasion proteins offer a viable option in the development of a resistance-free drug against human malaria.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://jid.oxfordjournals.org). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgments. The authors thank MR4 for various recombinant proteins and antibodies and Brendon Crabb, Paul Gilson, and Dany Wilson for providing transgenic parasites (D10-PfM3 and D10-PcMEGF). The support from the core facility at the Department of Biological Sciences-NUS in performing mass spectrometry analysis is thankfully acknowledged.
Authors contributions. R. C. designed, performed, and coordinated research, analyzed the data, and wrote the article. B. designed and performed experiments related to chemical synthesis, performed computational analysis and assisted in manuscript writing. B. R. was involved in P. vivax study design, performed P. vivax invasion assays, analyzed the data, verified results, and assisted in article preparation. K. L. performed P. falciparum experiments and assisted in article preparation. Y. H. performed SPR analyses. L. M. assisted with P. falciparum experiments. A. C. assisted with P. falciparum experiments. K. G. performed microscopy analysis. L. R. was involved in P. vivax study design and verified results. F. N. provided reagents and other tools for research, contributed to ethical clearance. S. G. S. was involved in SPR study design, provided research tools, and verified SPR results. R. S. was involved in study design, provided tools for research, and verified results. M. D. was involved in study design, provided tools for research, and verified results. S. S. was involved in study design, provided tools for research, and verified the results. P. R. P. contributed to study design, provided tools for research, verified the results, and wrote the article.
Financial support. The authors acknowledge Singapore-MIT Alliance for Research and Technology (SMART) Centre funded by The National Research Foundation (NRF) for financial support. R. C. was partially supported by the start-up research grant (SRG LSC 2013 049) from Singapore University of Technology and Design (SUTD). M. D. and S. S. acknowledge partial support from the National Institutes of Health (NIH) grant R01HL094270.
Potential conflicts of interest. All authors: No potential conflicts of interest.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Agnandji ST, Lell B, Soulanoudjingar SS, et al. First results of phase 3 trial of RTS, S/AS01 malaria vaccine in African children. N Engl J Med. 2011;365:1863–75. doi: 10.1056/NEJMoa1102287. [DOI] [PubMed] [Google Scholar]
- 2.Murray CJ, Rosenfeld LC, Lim SS, et al. Global malaria mortality between 1980 and 2010: a systematic analysis. Lancet. 2012;379:413–31. doi: 10.1016/S0140-6736(12)60034-8. doi:10.1016/S0140-6736(12)60034-8. [DOI] [PubMed] [Google Scholar]
- 3.Mita T, Tanabe K, Kita K. Spread and evolution of Plasmodium falciparum drug resistance. Parasitol Int. 2009;58:201–9. doi: 10.1016/j.parint.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 4.O'Brien C, Henrich PP, Passi N, Fidock DA. Recent clinical and molecular insights into emerging artemisinin resistance in Plasmodium falciparum. Curr Opin Infect Dis. 2011;24:570–7. doi: 10.1097/QCO.0b013e32834cd3ed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Breuer WV, Kahane I, Baruch D, Ginsburg H, Cabantchik ZI. Role of internal domains of glycophorin in Plasmodium falciparum invasion of human erythrocytes. Infect Immun. 1983;42:133–40. doi: 10.1128/iai.42.1.133-140.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mayer DC, Cofie J, Jiang L, et al. Glycophorin B is the erythrocyte receptor of Plasmodium falciparum erythrocyte-binding ligand, EBL-1. Proc Natl Acad Sci U S A. 2009;106:5348–52. doi: 10.1073/pnas.0900878106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sim BK, Chitnis CE, Wasniowska K, Hadley TJ, Miller LH. Receptor and ligand domains for invasion of erythrocytes by Plasmodium falciparum. Science. 1994;264:1941–4. doi: 10.1126/science.8009226. [DOI] [PubMed] [Google Scholar]
- 8.Duraisingh MT, Maier AG, Triglia T, Cowman AF. Erythrocyte- binding antigen 175 mediates invasion in Plasmodium falciparum utilizing sialic acid-dependent and independent pathways. Proc Natl Acad Sci U S A. 2003;100:4796–801. doi: 10.1073/pnas.0730883100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spadafora C, Awandare GA, Kopydlowski KM, et al. Complement receptor 1 is a sialic acid-independent erythrocyte receptor of Plasmodium falciparum. PLoS Pathog. 2010;6:e1000968. doi: 10.1371/journal.ppat.1000968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Langermans JA, Hensmann M, van Gijlswiik M, et al. Preclinical evaluation of a chimeric malaria vaccine candidate in Montanide ISA 720: immunogenicity and safety in rhesus macaques. Hum Vaccin. 2006;2:222–6. doi: 10.4161/hv.2.5.3276. [DOI] [PubMed] [Google Scholar]
- 11.Ellis RD, Martin LB, Schaffer D, et al. Phase 1 trial of the Plasmodium falciparum blood stage vaccine MSP1 (42)- C1/Alhydrogel with and without CPG 7909 in malaria naïve adults. PLoS One. 2010;5:e8787. doi: 10.1371/journal.pone.0008787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goel VK, Li X, Chen H, Liu SC, Chishti AH, Oh SS. Band 3 is a host receptor binding merozoite surface protein 1 during the Plasmodium falciparum invasion of erythrocytes. Proc Natl Acad Sci U S A. 2003;100:5164–9. doi: 10.1073/pnas.0834959100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Child MA, Epp C, Bujard H, Blackman MJ. Regulated maturation of malaria merozoite surface protein-1 is essential for parasite growth. Mol Microbiol. 2010;78:187–202. doi: 10.1111/j.1365-2958.2010.07324.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holder AA, Blackman MJ, Burghaus PA, et al. A malaria merozoite surface protein (MSP1)-structure, processing and function. Mem Inst Oswaldo Cruz. 1992;87(suppl 3):37–42. doi: 10.1590/s0074-02761992000700004. [DOI] [PubMed] [Google Scholar]
- 15.Chappel JA, Holder AA. Monoclonal antibodies that inhibit Plasmodium falciparum invasion in vitro recognise the first growth factor-like domain of merozoite surface protein-1. Mol Biochem Parasitol. 1993;60:303–11. doi: 10.1016/0166-6851(93)90141-j. [DOI] [PubMed] [Google Scholar]
- 16.Moss DK, Remarque EJ, Faber BW, et al. Plasmodium falciparum 19- kilodalton merozoite surface protein 1 (MSP1)-specific antibodies that interfere with parasite growth in vitro can inhibit MSP1 processing, merozoite invasion, and intracellular parasite development. Infect Immun. 2012;80:1280–7. doi: 10.1128/IAI.05887-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dluzewski AR, Ling IT, Hopkins JM, et al. Formation of the food vacuole in Plasmodium falciparum: a potential role for the 19 kDa fragment of merozoite surface protein 1 (MSP1(19)) PLoS One. 2008;3:e3085. doi: 10.1371/journal.pone.0003085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xiao L, Yang C, Patterson PS, Udhayakumar V, Lal AA. Sulfated polyanions inhibit invasion of erythrocytes by plasmodial merozoites and cytoadherence of endothelial cells to parasitized erythrocytes. Infect Immun. 1996;64:1373–8. doi: 10.1128/iai.64.4.1373-1378.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kisilevsky R, Crandall I, Szarek WA, et al. Short-chain aliphatic polysulfonates inhibit the entry of Plasmodium into red blood cells. Antimicrob Agents Chemother. 2002;46:2619–26. doi: 10.1128/AAC.46.8.2619-2626.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adams Y, Freeman C, Schwartz-Albiez R, Ferro V, Parish CR, Andrews KT. Inhibition of Plasmodium falciparum growth in vitro and adhesion to chondroitin-4-sulfate by the heparan sulfate mimetic PI-88 and other sulfated oligosaccharides. Antimicrob Agents Chemother. 2006;50:2850–2. doi: 10.1128/AAC.00313-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dalton JP, Hudson D, Adams JH, Miller LH. Blocking of the receptor-mediated invasion of erythrocytes by Plasmodium knowlesi malaria with sulfated polysaccharides and glycosaminoglycans. Eur J Biochem. 1991;195:789–94. doi: 10.1111/j.1432-1033.1991.tb15767.x. [DOI] [PubMed] [Google Scholar]
- 22.Basseri HR, Doosti S, Akbarzadeh K, Nateghpour M, Whitten MM, Ladoni H. Competency of Anopheles stephensimysorensis strain for Plasmodium vivax and the role of inhibitory carbohydrates to block its sporogenic cycle. Malar J. 2008;7:131. doi: 10.1186/1475-2875-7-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boyle MJ, Richards JS, Gilson PR, Chai W, Beeson JG. Interactions with heparin-like molecules during erythrocyte invasion by Plasmodium falciparum merozoites. Blood. 2010;115:4559–68. doi: 10.1182/blood-2009-09-243725. [DOI] [PubMed] [Google Scholar]
- 24.O'Donnell RA, de Koning-Ward TF, Burt RA, et al. Antibodies against merozoite surface protein (MSP)-1(19) are a major component of the invasion-inhibitory response in individuals immune to malaria. J Exp Med. 2001;193:1403–12. doi: 10.1084/jem.193.12.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu G, Robertson DH, Brooks CL, III, Vieth M. Detailed analysis of grid-based molecular docking: a case study of CDOCKER- A CHARMm-based MD docking algorithm. J Comput Chem. 2003;24:1549–62. doi: 10.1002/jcc.10306. [DOI] [PubMed] [Google Scholar]
- 26.Venkatachalam CM, Jiang X, Oldfield T, Waldman M. LigandFit: a novel method for the shape-directed rapid docking of ligands to protein active sites. J Mol Graph Model. 2003;21:289–307. doi: 10.1016/s1093-3263(02)00164-x. [DOI] [PubMed] [Google Scholar]
- 27.Shoichet BK. Virtual screening of chemical libraries. Nature. 2004;432:862–5. doi: 10.1038/nature03197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gaonkar SL, Lokanatha Rai KM, Suchetha Shetty N. Microwave-assisted synthesis and evaluation of anti- inflammatory activity of new series of N- substituted 2-butyl-5- chloro- 3 H-imidazole- 4- carbaldehyde derivatives. Med Chem Res. 2009;18:221–30. [Google Scholar]
- 29.Morgan WD, Birdsall B, Frenkiel TA, et al. Solution structure of an EGF module pair from the Plasmodium falciparum merozoite surface protein 1. J Mol Biol. 1999;289:113–22. doi: 10.1006/jmbi.1999.2753. [DOI] [PubMed] [Google Scholar]
- 30.Babon JJ, Morgan WD, Kelly G, Eccleston JF, Feeney J, Holder AA. Structural studies on Plasmodium vivax merozoite surface protein-1. Mol Biochem Parasitol. 2007;153:31–40. doi: 10.1016/j.molbiopara.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 31.Garman SC, Simcoke WN, Stowers AW, Garboczi DN. Structure of the C-terminal domains of merozoite surface protein-1 from Plasmodium knowlesi reveals a novel histidine binding site. J Biol Chem. 2003;278:7264–9. doi: 10.1074/jbc.M210716200. [DOI] [PubMed] [Google Scholar]
- 32.Johnsson B, Löfås S, Lindquist G. Immobilization of proteins to a carboxymethyldextran-modified gold surface for biospecific interaction analysis in surface plasmon resonance sensors. Anal Biochem. 1991;198:268–77. doi: 10.1016/0003-2697(91)90424-r. [DOI] [PubMed] [Google Scholar]
- 33.Myszka DG. Improving biosensor analysis. J Mol Recognit. 1999;12:279–84. doi: 10.1002/(SICI)1099-1352(199909/10)12:5<279::AID-JMR473>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 34.Wei L, Jiang P, Yau YH, et al. Residual structure in islet amyloid polypeptide mediates its interactions with soluble insulin. Biochemistry. 2009;48:2368–76. doi: 10.1021/bi802097b. [DOI] [PubMed] [Google Scholar]
- 35.Sriprawat K, Kaewpongsri S, Suwanarusk R, et al. Effective and cheap removal of leukocytes and platelets from Plasmodium vivax infected blood. Malar J. 2009;8:115. doi: 10.1186/1475-2875-8-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Russell B, Suwanarusk R, Borlon C, et al. A reliable ex vivo invasion assay of human reticulocytes by Plasmodium vivax. Blood. 2011;118:e74–81. doi: 10.1182/blood-2011-04-348748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Malleret B, Claser C, Ong AS, et al. A rapid and robust tri-color flow cytometry assay for monitoring malaria parasite development. Sci Rep. 2011;1:118. doi: 10.1038/srep00118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ishida K, Wierzba MK, Teruya T, Simizu S, Osada H. Novel heparansulfate mimetic compounds as antitumor agents. Chem Biol. 2004;11:367–77. doi: 10.1016/j.chembiol.2004.02.015. [DOI] [PubMed] [Google Scholar]
- 39.Basappa, Murugan S, Kavitha CV, et al. A small oxazine compound as an anti-tumor agent: a novel pyranoside mimetic that binds to VEGF, HB- EGF, and TNF-α. Cancer Lett. 2010;297:231–43. doi: 10.1016/j.canlet.2010.05.016. [DOI] [PubMed] [Google Scholar]
- 40.Basappa, Sugahara K, Thimmaiah KN, Bid HK, Houghton PJ, Rangappa KS. Anti-tumor activity of a novel HS-mimetic vascular endothelial growth factor binding small molecule. PLoS ONE. 2012;7:e39444. doi: 10.1371/journal.pone.0039444. doi:10.1371/journal.pone.0039444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wilson DW, Crabb BS, Beeson JG. Development of fluorescent Plasmodium falciparum for in vitro growth inhibition assays. Malar J. 2010;9:152. doi: 10.1186/1475-2875-9-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chandramohanadas R, Davis PH, Beiting DP, et al. Apicomplexan parasites co-opt host calpains to facilitate their escape from infected cells. Science. 2009;324:794–7. doi: 10.1126/science.1171085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Treeck M, Zacherl S, Herrmann S, et al. Functional analysis of the leading malaria vaccine candidate AMA- 1 reveals an essential role for the cytoplasmic domain in the invasion process. PLoS Pathog. 2009;5:e1000322. doi: 10.1371/journal.ppat.1000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boyle MJ, Wilson DW, Richards JS, et al. Isolation of viable Plasmodium falciparum merozoites to define erythrocyte invasion events and advance vaccine and drug development. Proc Natl Acad Sci U S A. 2010;107:14378–83. doi: 10.1073/pnas.1009198107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Han HJ, Park SG, Kim SH, et al. Epidermal growth factor-like motifs 1 and 2 of Plasmodium vivax merozoite surface protein 1 are critical domains in erythrocyte invasion. Biochem Biophys Res Commun. 2004;320:563–70. doi: 10.1016/j.bbrc.2004.06.008. [DOI] [PubMed] [Google Scholar]
- 46.Iyer J, Grüner AC, Rénia L, Snounou G, Preiser PR. Invasion of host cells by malaria parasites: a tale of two protein families. Mol Microbiol. 2007;65:231–49. doi: 10.1111/j.1365-2958.2007.05791.x. [DOI] [PubMed] [Google Scholar]
- 47.Greenbaum DC, Baruch A, Grainger M, et al. A role for the protease falcipain 1 in host cell invasion by the human malaria parasite. Science. 2002;298:2002–6. doi: 10.1126/science.1077426. [DOI] [PubMed] [Google Scholar]
- 48.Koussis K, Withers-Martinez C, Yeoh S, et al. A multifunctional serine protease primes the malaria parasite for red blood cell invasion. EMBO J. 2009;28:725–35. doi: 10.1038/emboj.2009.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Singh S, Alam MM, Pal-Bhowmick I, Brzostowski JA, Chitnis CE. Distinct external signals trigger sequential release of apical organelles during erythrocyte invasion by malaria parasites. PLoS Pathog. 2010;6:e1000746. doi: 10.1371/journal.ppat.1000746. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


