Abstract
Background. Rifaximin, a nonabsorbable antibiotic that decreases lipopolysaccharide (LPS) in cirrhotics, may decrease the elevated levels of microbial translocation, T-cell activation and inflammation in human immunodeficiency virus (HIV)-positive immune nonresponders to antiretroviral therapy (ART).
Methods. HIV-positive adults receiving ART for ≥96 weeks with undetectable viremia for ≥48 weeks and CD4+ T-cell counts <350 cells/mm3 were randomized 2:1 to rifaximin versus no study treatment for 4 weeks. T-cell activation, LPS, and soluble CD14 were measured at baseline and at weeks 2, 4, and 8. Wilcoxon rank sum tests compared changes between arms.
Results. Compared with no study treatment (n = 22), rifaximin (n = 43) use was associated with a significant difference between study arms in the change from baseline to week 4 for CD8+T-cell activation (median change, 0.0% with rifaximin vs +0.6% with no treatment; P = .03). This difference was driven by an increase in the no-study-treatment arm because there was no significant change within the rifaximin arm. Similarly, although there were significant differences between study arms in change from baseline to week 2 for LPS and soluble CD14, there were no significant changes within the rifaximin arm.
Conclusions. In immune nonresponders to ART, rifaximin minimally affected microbial translocation and CD8+T-cell activation.
Trial registration number. NCT01466595.
Keywords: HIV, immune nonresponders to ART, microbial translocation, immune activation, inflammation, rifaximin
Immune activation plays a central role in human immunodeficiency virus (HIV) pathogenesis. The frequency of activated CD8+ T cells, as defined by the up-regulation of HLA-DR and CD38 on the cell surface, predicts disease progression in untreated HIV-positive persons [1] and a CD4+ T-cell increase with antiretroviral treatment (ART) [2, 3]. ART leads to a decline in immune activation in most HIV-infected patients, yet levels of activation do not normalize in most individuals [4, 5]. Moreover, up to 25% of individuals starting ART fail to achieve CD4+ T-cell counts >350 cells/mm3 even after 2 years of virally suppressive treatment, and these patients have higher levels (percentages) of activated (HLA-DR+CD38+) CD8+ T cells than patients with better immune restoration [6]. A 5% increase in activated CD8+ T cells is associated with 35 fewer CD4+ T cells gained during ART [2]. Incomplete immune recovery with ART is also associated with increased mortality, opportunistic infections, and cardiovascular disease and malignancies [7, 8].
Many mechanisms have been proposed to account for the elevated levels of immune activation, including the direct effects of HIV, the effects of coinfections such as cytomegalovirus, loss of normal immunoregulatory responses, and HIV-mediated destruction of mucosal barriers with chronic systemic exposure to gut microbial elements. HIV-infected individuals sustain a rapid and profound depletion of gut mucosal CD4+ T cells as early as a few days after infection [9]. These changes lead to defects in mucosal immune and epithelial barrier function that allows the translocation of gut microbial products, such as lipopolysaccharide (LPS) [10]; LPS levels are elevated in the plasma of HIV-infected individuals and associated with increased levels of immune activation [10]. ART does not fully reverse the deficits in gut mucosal CD4+ T cells [11, 12], and both the levels of gut microbial translocation and immune activation predict the extent of immune reconstitution with ART [6, 13]. Taken together, these data support a model that attributes CD4+ T-cell loss to activation driven partly by translocated gut microbial products. In addition, LPS levels are associated with risk of myocardial infarction [14], whereas levels of soluble CD14 (sCD14), another marker for microbial translocation, are associated with mortality in treated HIV-positive persons [15].
Antibiotics have been used in simian immunodeficiency virus-infected macaques to decrease plasma LPS levels, albeit transiently [10]. Rifaximin, a nonabsorbed oral antibiotic used to improve symptoms of hepatic encephalopathy in patients with cirrhosis, has also been shown to reduce plasma LPS levels. The administration of 28 days of 1200 mg/d of rifaximin led to a 50% decline in plasma LPS levels in alcoholic cirrhotics [16]. In addition, the use of rifaximin has been shown to be safe in alcoholic cirrhotics.
We conducted AIDS Clinical Trials Group (ACTG) protocol A5286, a multicenter randomized, open-label pilot study of 4 weeks of treatment with rifaximin versus no study treatment in ART-treated HIV-infected subjects with incomplete immune recovery. We hypothesized that rifaximin use would be safe and would reduce gut microbial translocation and, consequently, would lower immune activation levels in HIV-infected subjects with incomplete CD4+ T-cell recovery during ART.
METHODS
Study Subjects and Design
Institutional review board approval was obtained by each ACTG site. Subjects provided written informed consent and included HIV-infected subjects aged 18–65 years who were receiving ART for ≥96 weeks before study entry, had plasma HIV RNA levels below the limit of detection for ≥48 weeks before study entry, and had screening CD4+ T-cell counts <350 cells/mm3. They were randomized 2:1 to treatment with rifaximin (550 mg by mouth twice daily for 4 weeks) or to no study treatment. Patients with a history of inflammatory bowel disease, Clostridium difficile colitis, or chronic liver disease; recent or current use of antimicrobials (except prophylaxis of opportunistic infections), immune modulator, or probiotic treatment (including probiotic yoghurt); active diarrhea; or active substance abuse were excluded. Fasting blood samples were collected at preentry, entry, and weeks 2, 4, and 8 after entry to measure markers of cellular activation, inflammation, gut microbial translocation, and coagulation. Safety and medication adherence assessments were performed at postentry visits. Cryopreserved blood samples from 20 age-matched HIV-negative men from the Multicenter AIDS Cohort Study were assayed for biomarker levels and served as controls.
Flow Cytometry
Characterization of T-cell activation and proliferation were performed in batch by multicolor flow cytometry on cryopreserved peripheral blood mononuclear cells. Frozen peripheral blood mononuclear cells were removed from liquid nitrogen storage and thawed rapidly in a 37°C water bath, washed in phosphate-buffered saline, and stained immediately. The cells were stained for cell viability with Aqua Live/Dead cell stain kit (Invitrogen) before cell surface staining. Cell surface markers were stained with the following antibodies: CD3 v450, CD8 APC-H7, HLA-DR PE, CD38 APC (all BD Biosciences), and CD4 PE-Texas Red (Invitrogen). Samples were then permeabilized (BD Biosciences) and stained for Ki-67 AF700. After staining, all cells were fixed in 1% formaldehyde and analyzed within 24 hours on a LSR2 flow cytometer (BD) using FACS Diva software v6.1.1. Analysis of flow cytometry data was performed by a single technician using FlowJo software (Tree Star).
Soluble Markers
To evaluate markers of microbial translocation and monocyte activation, frozen serum samples were thawed and analyzed in batches. Serum concentrations of LPS and sCD14 were quantified using the Limulus Amebocyte Lysate (LAL) kit (Lonza) and the human sCD14 enzyme-linked immunosorbent assay (ELISA) kit (R&D), respectively, per manufacturers’ instructions. Serum samples for the LAL were heat inactivated and diluted 1:10 with LAL Reagent Water (Lonza). Commercially available ELISA kits were used to determine plasma levels of C-reactive protein (CRP; R&D), interleukin 6 (IL-6; R&D), sCD163 (R&D), and d-dimer (Sekisui Diagnostics) according to manufacturers' instructions. Duplicates of 20% of the samples were included in each ELISA plate. Results were analyzed using a Biotek ELX800 ELISA reader and Gen5 software (version 2.01.12). Plasma concentrations of soluble tumor necrosis factor α receptor II (sTNFR-II) were quantified using a fluorescent multiplexed immunoassay platform (Meso Scale Discovery).
Statistical Analysis
The study was powered to detect a 5% treatment effect in the primary end point: change from baseline to week 4 in the percentage of HLA-DR+CD38+ CD8+ T cells. A sample size of 63 evaluable subjects was estimated to provide 80% power to detect this treatment effect.
All statistical tests were 2-sided at the .05 nominal level of significance without adjustments for multiple testing. Exact Wilcoxon rank sum tests were used to evaluate differences between study arms, and 95% confidence intervals were estimated using the Hodges–Lehmann method. Rank-based (Spearman) correlations were used to assess correspondence between responses. Baseline measurements were compared between subjects HIV-negative controls using exact Wilcoxon rank sum tests. Tests of interaction involved comparing nonparametric estimates of treatment effect [17] between strata, using standard errors [18].
Baseline values were the average of entry and preentry values. Soluble biomarkers were log10 transformed. Primary adverse events included all serious adverse events as defined by ICH guidelines. Targeted protocol events were defined as all laboratory abnormalities of grade ≥2, signs and symptoms defined by the 2004 Division of AIDS (DAIDS) grading table, and all diagnoses identified by the ACTG criteria for clinical events, particularly a diagnosis of C. difficile colitis.
The primary analysis of efficacy was an as-treated analysis limited to subjects in both arms who have data for baseline and week 4, remain on study treatment through week 4 (allowing ≤6 missed doses), and did not change ART, use prohibited medications, or have virologic failure (2 consecutive plasma HIV-1 RNA levels ≥40 copies/mL) during this time period. Secondary analyses were also as-treated analyses, including the same subjects as in the primary analysis. For analyses of measurements after week 4, data were excluded after a subject changed ART, used prohibited medications, or had confirmed virologic failure (as defined above).
RESULTS
Study Subjects
Seventy-three subjects were enrolled between October 2011 and July 2012, including 49 in the rifaximin arm and 24 in the no-study-treatment arm. In the rifaximin arm, 2 subjects discontinued the study owing to side effects (grade ≤2), 1 was taking a prohibited medication, and 3 missed the week 4 visit. In the no-study-treatment arm, 1 subject withdrew consent and 1 had virologic failure. Forty-three subjects in the rifaximin and 22 in the no-study-treatment arm, respectively, were included in the as-treated analysis.
Baseline demographic, immunologic, and virologic characteristics were well balanced between the 2 arms (Table 1). The subjects' baseline levels of CD8+ and CD4+ T-cell activation and proliferation, and levels of soluble markers of microbial translocation and inflammation, with the exception of sCD163 and CRP, were significantly higher than in HIV-negative controls (see Supplementary Table 1 for control demographics), and the subjects' d-dimer levels were significantly lower compared with controls (Table 2).
Table 1.
Baseline Demographic, Immunologic, and Virologic Characteristics of the 65 Subjects Included in the As-Treated Analyses
| Baseline Characteristic | All Subjects (N = 65) |
Rifaximin Arm (n = 43) |
No-Treatment Arm (n = 22) |
|---|---|---|---|
| Age, median (IQR), y | 50 (44–55) | 48 (43–56) | 51 (45–55) |
| Age ≥60 y, No. (%) | 7 (11) | 4 (9) | 3 (14) |
| Race/ethnicity, No. (%) | |||
| White non-Hispanic | 31 (48) | 19 (44) | 12 (55) |
| Black non-Hispanic | 20 (31) | 17 (40) | 3 (14) |
| Hispanic | 13 (20) | 7 (16) | 6 (27) |
| Other | 1 (2) | 0 (0) | 1 (5) |
| Sex, No. (%) | |||
| Male | 59 (91) | 39 (91) | 20 (91) |
| Female | 6 (9) | 4 (9) | 2 (9) |
| Cotrimoxazole use, No. (%) | 29 (45) | 21 (49) | 8 (36) |
| Baseline CD4+ T-cell counta | |||
| Median (IQR), cells/mm3 | 236 (179–284) | 251 (179–286) | 223 (167–280) |
| ≥350 cells/mm3, No. (%) | 1 (2) | 0 (0) | 1 (5) |
| Nadir CD4+ T-cell count, median (IQR), cells/mm3 | 41 (15–88) | 50 (22–71) | 40 (10–88) |
| Entry HIV-1 RNA level below lower limit of assay, No. (%) | 65 (100) | 43 (100) | 22 (100) |
| Interval since 1st undetectable HIV-1 RNA level, median (IQR), y | 3.8 (2.4–7.3) | 3.3 (2.2–7.3) | 3.9 (2.8–8.8) |
Abbreviations: HIV-1, human immunodeficiency virus type 1; IQR, interquartile range.
a Mean of preentry and entry counts.
Table 2.
Baseline Immune Biomarker Levels in Subjects Compared With HIV-Negative Age-Matched Controls
| Biomarkers | HIV-Negative Controls (n = 20) |
Pooled Study Arms at Baseline (N = 65) |
P Value |
|---|---|---|---|
| Median (IQR) | Median (IQR ) | ||
| Cellular activation markers | |||
| HLA-DR+ CD38+ CD8+ T cells, % | 4.05 (2.68–6.04) | 8.14 (5.18–13.70) | <.001 |
| HLA-DR+ CD38+ CD4+ T cells, % | 1.76 (1.46–2.42) | 4.95 (3.64–6.06) | <.001 |
| Ki-67+ CD8+ T cells, % | 0.50 (0.31–0.74) | 0.71 (0.52–0.99) | .01 |
| Ki-67+ CD4+ T cells, % | 0.58 (0.46–1.11) | 1.65 (1.39–2.50) | <.001 |
| Gut microbial translocation markers | |||
| LPS, pg/mL | 84.0 (70.7–91.3) | 103.7 (73.7–121.2) | .04 |
| sCD14, ng/mL | 1270.9 (1100.2–1464.0) | 1808.9 (1604.8–2161.6) | <.001 |
| Soluble markers | |||
| IL-6, pg/mL | 0.9 (0.6–1.4) | 1.2 (0.9–2.2) | .02 |
| CRP, ng/mL | 1126.9 (3467–3176) | 1446.9 (731–2989) | .35 |
| d-dimer, ng/mL | 192.2 (157–240) | 129.6 (96–214) | .02 |
| sTNFR-II, pg/mL | 2995.8 (2319–3777) | 4214.0 (3186–6354) | <.001 |
| sCD163, ng/mL | 658.4 (610–772) | 611.4 (438–744) | .28 |
Abbreviations: CRP, C-reactive protein; HIV, human immunodeficiency virus; IL-6, interleukin 6; IQR, interquartile range; LPS, lipopolysaccharide; sCD14, soluble CD14; sCD163, soluble CD163; sTNFR-II, soluble tumor necrosis factor α receptor II.
The antiretroviral regimens were balanced between the 2 study arms. None of the subjects had a change or interruption in entry antiretroviral regimen during the study. Three subjects in the rifaximin arm missed ≤3 doses of study treatment and were included in the as-treated analyses.
Effects of Rifaximin on Cellular Immune Activation
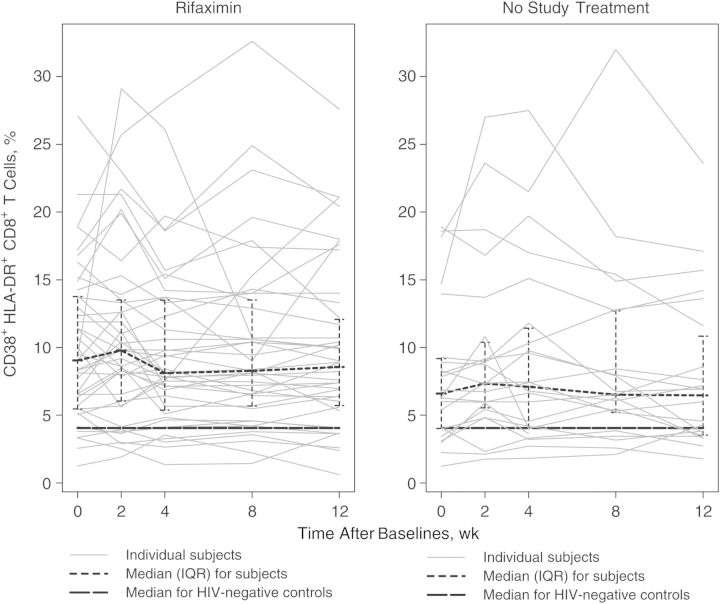
The study's primary end point was the change in CD8+ T-cell activation (percentage of HLA-DR+CD38+ T-cells) from baseline to week 4. The median (interquartile range) change for the rifaximin arm was 0.00% (−1.70% to 1.00%), and the median change for the no-study-treatment arm was 0.64% (0.11%–1.48%) (Table 3). The changes were significantly different when comparing the 2 arms (P = .03), but the within-arm change in the rifaximin arm was not significant (Table 3 and Figure 1). The difference between the arms was driven by an increase in CD8+ T-cell activation in the no-treatment arm. There were no significant differences between the 2 arms in the changes from baseline to week 2 or week 8.
Table 3.
Median Changes From Baseline in Cellular Biomarkers and Median Log10 Change From Baseline in Soluble Biomarkers
| Biomarker | Week After Baseline | Treatment Arm | Subjects, No. | Median (IQR) Change From Baseline | 95% CI for Median Change | P Value |
|---|---|---|---|---|---|---|
| Cellular activation markers | ||||||
| HLA-DR+CD38+ CD8+ T cells, % | 2 | Rifaximin | 42 | 0.01 (−1.32 to 1.88) | −.63 to 1.12 | .34 |
| No study treatment | 22 | 0.30 (−0.25 to 1.98) | −.10 to 1.99 | |||
| 4 | Rifaximin | 43 | 0.00 (−1.70 to 1.00) | −.99 to 0.33 | .03 | |
| No study treatment | 22 | 0.64 (0.11–1.48) | .11 to 1.51 | |||
| 8 | Rifaximin | 41 | 0.01 (−1.84 to 0.89) | −1.11 to .50 | .72 | |
| No study treatment | 21 | −0.19 (−0.85 to 0.86) | −1.05 to 1.32 | |||
| Ki-67+ CD8+ T cells, % | 2 | Rifaximin | 42 | 0.00 (−0.26 to 0.26) | −.13 to .13 | .11 |
| No study treatment | 22 | 0.12 (−0.09 to 0.44) | .01 to .41 | |||
| 4 | Rifaximin | 43 | −0.12 (−0.27 to 0.12) | −.21 to .00 | .01 | |
| No study treatment | 22 | 0.12 (−0.07 to 0.27) | −.02 to .27 | |||
| 8 | Rifaximin | 41 | −0.07 (−0.18 to 0.11) | −.17 to .07 | .08 | |
| No study treatment | 21 | 0.12 (−0.04 to 0.23) | −.06 to .23 | |||
| Microbial translocation and monocyte activation markers | ||||||
| LPS, log10 pg/mL | 2 | Rifaximin | 43 | −0.01 (−0.09 to 0.02) | −.05 to .01 | .01 |
| No study treatment | 22 | 0.03 (−0.03 to 0.11) | −.01 to .15 | |||
| 4 | Rifaximin | 43 | 0.00 (−0.08 to 0.05) | −.04 to .02 | .44 | |
| No study treatment | 22 | −0.01 (−0.04 to 0.05) | −.02 to .05 | |||
| 8 | Rifaximin | 42 | −0.01 (−0.12 to 0.06) | −.06 to .03 | .22 | |
| No study treatment | 21 | 0.02 (−0.02 to 0.08) | −.03 to .08 | |||
| sCD14, log10 ng/mL | 2 | Rifaximin | 43 | −0.00 (−0.06 to 0.04) | −.04 to .02 | .03 |
| No study treatment | 22 | 0.05 (−0.02 to 0.13) | −.00 to .09 | |||
| 4 | Rifaximin | 43 | −0.03 (−0.07 to 0.06) | −.05 to .01 | .97 | |
| No study treatment | 22 | −0.03 (−0.05 to 0.01) | −.07 to .02 | |||
| 8 | Rifaximin | 42 | −0.04 (−0.09 to 0.03) | −.08 to −.01 | .049 | |
| No study treatment | 21 | 0.01 (−0.05 to 0.08) | −.03 to .07 | |||
| Soluble biomarkers | ||||||
| d-dimer, log10 ng/mL | 2 | Rifaximin | 42 | −0.03 (−0.09 to 0.09) | −.05 to .04 | .30 |
| No study treatment | 22 | 0.00 (−0.03 to 0.11) | −.02 to .08 | |||
| 4 | Rifaximin | 43 | −0.00 (−0.12 to 0.06) | −.07 to .03 | .33 | |
| No study treatment | 22 | −0.03 (−0.14 to 0.04) | −.11 to .01 | |||
| 8 | Rifaximin | 42 | −0.04 (−0.17 to 0.14) | −.09 to .04 | .23 | |
| No study treatment | 21 | 0.04 (−0.07 to 0.10) | −.04 to .14 | |||
| IL-6, log10 pg/mL | 2 | Rifaximin | 43 | 0.02 (−0.11 to 0.15) | −.07 to .07 | .94 |
| No study treatment | 22 | −0.05 (−0.11 to 0.20) | −.10 to .20 | |||
| 4 | Rifaximin | 43 | −0.03 (−0.14 to 0.08) | −.08 to .03 | .33 | |
| No study treatment | 22 | 0.05 (−0.13 to 0.12) | −.06 to .13 | |||
| 8 | Rifaximin | 42 | −0.05 (−0.18 to 0.07) | −.12 to .01 | .02 | |
| No study treatment | 21 | 0.05 (−0.08 to 0.18) | −.02 to .19 | |||
| CRP, log10 ng/mL | 2 | Rifaximin | 43 | 0.00 (−0.26 to 0.21) | −.14 to .08 | .14 |
| No study treatment | 22 | 0.04 (−0.09 to 0.32) | −.04 to .27 | |||
| 4 | Rifaximin | 43 | −0.08 (−0.29 to 0.15) | −.20 to .03 | .73 | |
| No study treatment | 22 | −0.09 (−0.21 to 0.16) | −.22 to .12 | |||
| 8 | Rifaximin | 42 | −0.04 (−0.26 to 0.13) | −.18 to .03 | .046 | |
| No study treatment | 21 | 0.09 (−0.02 to 0.28) | −.06 to .31 | |||
| sTNFR-II, log10 pg/mL | 2 | Rifaximin | 42 | −0.03 (−0.11 to 0.03) | −.08 to .01 | .04 |
| No study treatment | 22 | 0.04 (−0.06 to 0.17) | −.03 to .13 | |||
| 4 | Rifaximin | 42 | −0.04 (−0.17 to 0.09) | −.09 to .02 | .08 | |
| No study treatment | 22 | 0.07 (−0.05 to 0.13) | −.06 to .11 | |||
| 8 | Rifaximin | 42 | 0.04 (−0.07 to 0.12) | −.02 to .08 | .35 | |
| No study treatment | 21 | −0.01 (−0.15 to 0.10) | −.11 to .06 | |||
| sCD163, log10 ng/mL | 2 | Rifaximin | 43 | 0.01 (−0.04 to 0.05) | −.02 to .02 | .31 |
| No study treatment | 22 | 0.02 (−0.03 to 0.07) | −.01 to .06 | |||
| 4 | Rifaximin | 43 | 0.00 (−0.03 to 0.04) | −.02 to .02 | .30 | |
| No study treatment | 22 | 0.02 (−0.04 to 0.09) | −.01 to .07 | |||
| 8 | Rifaximin | 42 | 0.01 (−0.03 to 0.04) | −.02 to .03 | .48 | |
| No study treatment | 21 | 0.04 (−0.06 to 0.09) | −.02 to .08 | |||
Abbreviations: CI, confidence interval. CRP, C-reactive protein; HIV, human immunodeficiency virus; IL-6, interleukin 6; IQR, interquartile range; LPS, lipopolysaccharide; sCD14, soluble CD14; sCD163, soluble CD163; sTNFR-II, soluble tumor necrosis factor α receptor II.
Figure 1.
Percentages of CD38+HLA-DR+ CD8+ T cells per subject over time in each arm. Abbreviation: IQR, interquartile range.
There was also a significant difference between the 2 study arms in the change in CD8+ T-cell proliferation (percentage of Ki-67+ CD8+ T cells) from baseline to week 4 (P = .01), but changes within each arm were not significant. The median (interquartile range) change for the rifaximin arm was −0.12% (−0.27% to 0.12%), and the median change for the no-study-treatment arm was 0.12% (−0.07% to 0.27%). There were no significant differences in the change from baseline to week 2 or 8. We did not observe significant differences in changes in CD4+ T-cell count or measures of CD4+ T-cell activation (percentage of HLA-DR+CD38+ CD4+ T cells) or proliferation (percentage of Ki-67+ CD4+ T cells) at any week (data not shown). These results suggest that rifaximin weakly affected CD8+ T-cell activation and proliferation after 4 weeks of therapy.
A significant proportion of subjects had activation levels that overlapped with those of controls. In post hoc analyses, we stratified subjects based on whether their preentry percentage of HLA-DR+CD38+ CD8+ T cells was higher or lower than the pooled median value, and we evaluated differences between study arms in changes in T-cell activation and microbial translocation from entry to week 4 (Table 4). A trend toward a significant difference between the arms was seen in change from baseline to week 4 in the percentage of HLA-DR+CD38+ CD8+ T cells, and a significant difference between the arms was seen in the percentage of Ki-67+ CD8+ T cells among those with higher preentry activation; treatment effects were not seen in those with lower preentry activation. When we stratified by baseline levels of either LPS or sCD14 (high vs low, with the median as the cutoff), we also saw a similar pattern with Ki-67+ CD8+ T cells among those in the high baseline sCD14 stratum but not in the low sCD14 stratum or either LPS stratum. When we applied the same approach to changes in LPS or sCD14, we did not see any significant differences (data not shown), although we had less statistical power to detect differences between the arms for these test results within subgroups, so these results should be interpreted cautiously.
Table 4.
Median Change From Baseline to Week 4 in CD8+ T-Cell Activation and Proliferation Stratified by High or Low Levels of Preentry CD8+ T-Cell Activation, LPS, or sCD14
| Stratum | Treatment Arm | Subjects, No. | Median Change (IQR) | 95% CI for Median Change | P Value (Between Arms) | P Value (Interaction) |
|---|---|---|---|---|---|---|
| Change in HLA-DR+CD38+ CD8+ T cells from entry to wk 4, % | ||||||
| Strata based on median preentry CD8+ T-cell activation | ||||||
| High | Rifaximin | 23 | −1.00 (−2.32 to 0.96) | −1.95 to .56 | .08 | .049 |
| No study treatment | 8 | 1.45 (−0.57 to 2.23) | −.76 to 7.30 | |||
| Low | Rifaximin | 18 | 0.64 (−0.17 to 1.87) | −.09 to 1.54 | .33 | |
| No study treatment | 13 | 0.27 (−0.08 to 0.65) | −.36 to .96 | |||
| Strata based on median baseline LPS | ||||||
| High | Rifaximin | 25 | −0.63 (−1.91 to 0.80) | −1.91 to .50 | .09 | .58 |
| No study treatment | 8 | 0.86 (0.15 to 1.78) | −.86 to 2.01 | |||
| Low | Rifaximin | 18 | 0.27 (−0.70 to 1.21) | −.70 to .81 | .25 | |
| No study treatment | 14 | 0.64 (−0.30 to 1.47) | −.30 to 1.97 | |||
| Strata based on median baseline sCD14 | ||||||
| High | Rifaximin | 23 | −0.08 (−1.91 to 0.80) | −1.20 to .33 | .11 | .85 |
| No study treatment | 10 | 0.56 (0.11 to 0.80) | −.06 to 1.13 | |||
| Low | Rifaximin | 20 | 0.13 (−1.66 to 1.28) | −1.83 to 1.25 | .16 | |
| No study treatment | 12 | 1.31 (−0.56 to 2.30) | −.48 to 2.92 | |||
| Change in Ki-67+ CD8+ T cells from baseline to wk 4, % | ||||||
| Strata based on median preentry CD8+ T-cell activation | ||||||
| High | Rifaximin | 23 | −0.18 (−0.50 to 0.04) | −.55 to −.07 | .009 | .10 |
| No study treatment | 8 | 0.24 (−0.08 to 0.27) | −.25 to 1.42 | |||
| Low | Rifaximin | 18 | 0.00 (−0.13 to 0.27) | −.13 to .18 | .68 | |
| No study treatment | 13 | 0.09 (−0.07 to 0.23) | −.11 to .23 | |||
| Strata based on median baseline LPS | ||||||
| High | Rifaximin | 25 | −0.13 (−0.28 to 0.06) | −.31 to −.01 | .081 | .67 |
| No study treatment | 8 | 0.25 (−0.09 to 0.27) | −.22 to .28 | |||
| Low | Rifaximin | 18 | 0.00 (−0.17 to 0.13) | −.30 to .12 | .18 | |
| No study treatment | 14 | 0.10 (−0.07 to 0.26) | −.06 to .44 | |||
| Strata based on median baseline sCD14 | ||||||
| High | Rifaximin | 23 | −0.13 (−0.27 to 0.14) | −.21 to .03 | .04 | .60 |
| No study treatment | 10 | 0.12 (0.00 to 0.27) | −.22 to .36 | |||
| Low | Rifaximin | 20 | −0.03 (−0.37 to 0.09) | −.49 to .08 | .16 | |
| No study treatment | 12 | 0.16 (−0.17 to 0.27) | −.15 to .43 | |||
Abbreviations: CRP, C-reactive protein; IQR, interquartile range; LPS, lipopolysaccharide; sCD14, soluble CD14.
Effects of Rifaximin on Gut Microbial Translocation
Significant differences were seen between study arms in the change from baseline to week 2 for LPS (median log10 change for rifaximin vs no-study-treatment arm, −0.01 vs +0.03 pg/mL; P = .01) and sCD14 (median log10 change, −0.003 vs +0.05 ng/mL; P = .03) (Table 3); however, the within-arm changes were not significant. At week 4, the differences in the change from baseline between arms were no longer significant for either LPS or sCD14. At week 8, rifaximin use was associated with a small decline in sCD14 (median log10 change for rifaximin vs no-study-treatment arm, −0.04 vs +0.01 ng/mL; P = .049), but not in LPS. These results suggest that rifaximin therapy weakly affected markers of microbial translocation and monocyte activation.
Effects of Rifaximin on Soluble Markers of Inflammation and Coagulation
We noted a significant difference between the 2 arms in the change from baseline to week 2 for sTNFR-II, but not in IL-6, CRP, sCD163, or d-dimer (Table 3). There were no significant differences in the changes from baseline to week 4 in any of the soluble biomarkers. We observed significant differences between the 2 arms in the change from baseline to week 8 for IL-6 and CRP, but not in sTNFR-II, sCD163, or d-dimer. None of the within-arm changes were significant.
Associations Between Biomarkers
We also determined the associations between the levels of biomarkers of activation, inflammation, coagulation, and microbial translocation, as well the correlations between changes in these biomarkers with rifaximin treatment. Among subjects' baseline biomarker levels, there were significant positive correlations between sCD14 and IL-6, CRP, and d-dimer, and between LPS and d-dimer (Table 5). It is noteworthy that we did not find significant correlations between the percentage of HLA-DR+CD38+ or Ki-67+ CD8+ T cells and either LPS or sCD14 levels.
Table 5.
Spearman Correlations Between Biomarker Levels at Baseline and Between Changes From Baseline to Week 4 in Biomarkers
| Biomarker | Correlation with LPS |
Correlation with sCD14 |
||||
|---|---|---|---|---|---|---|
| Subjects, No. | r | P Value | Subjects, No. | r | P Value | |
| Correlation between biomarkers at baseline | ||||||
| HLA-DR+CD38+ CD8+ T cells | 65 | −0.03 | .80 | 65 | −0.10 | .41 |
| Ki-67+ CD8+ T cells | 65 | 0.18 | .16 | 65 | 0.08 | .52 |
| IL-6 | 65 | 0.21 | .10 | 65 | 0.32 | .009 |
| LPS | 65 | 0.19 | .13 | |||
| CRP | 65 | 0.16 | .19 | 65 | 0.26 | .04 |
| sCD14 | 65 | 0.19 | .13 | |||
| d-dimer | 65 | 0.34 | .006 | 65 | 0.32 | .009 |
| sTNFrII | 65 | 0.02 | .86 | 65 | 0.10 | .44 |
| Correlation between changes in biomarkers from baseline to wk 4 | ||||||
| HLA-DR+CD38+ CD8+ T cells | 43 | 0.12 | .46 | 43 | −0.01 | .95 |
| Ki-67+ CD8+ T cells | 43 | 0.06 | .70 | 43 | 0.00 | .98 |
| IL-6 | 43 | −0.20 | .20 | 43 | 0.32 | .04 |
| LPS | 43 | −0.01 | .96 | |||
| CRP | 43 | −0.29 | .06 | 43 | 0.30 | .05 |
| sCD14 | 43 | −0.01 | .96 | |||
| d-dimer | 43 | −0.05 | .76 | 43 | 0.23 | .14 |
| sTNFrII | 42 | 0.02 | .90 | 42 | −0.11 | .47 |
Abbreviations: CRP, C-reactive protein; IL-6, interleukin 6; LPS, lipopolysaccharide; sCD14, soluble CD14; sTNFrII, soluble tumor necrosis factor α receptor II.
When considering correlations between changes in these biomarkers with treatment (Table 5), we did not find significant correlations between changes in the percentage of HLA-DR+CD38+ CD8+ T cells and either LPS or sCD14. However, we found significant positive correlations between changes in sCD14 and IL-6 from baseline to week 4 (r = 0.32; P = .04), but not between other soluble biomarkers, including between sCD14 and LPS.
Safety
There were no deaths. Thirty-six subjects (49%) reported a primary adverse event, 27 from the rifaximin arm and 9 from the no-study-treatment arm. There was no significant difference in maximum primary event grade between the 2 study arms. None of the grade 3 or 4 events were related to study treatment. One subject discontinued owing to grade 2 anorexia and another owing to intolerance. Primary adverse events that were possibly or definitely related to study treatment included nausea, constipation, flatulence, anorexia, stomach ache, “feeling sick,” and elevated lipase (Supplementary Table 2).
DISCUSSION
Rifaximin use was associated with a small but significant difference between the 2 arms in LPS levels after 2 weeks but not after 4 weeks. However, the within-arm changes were not significant. Thus, rifaximin seems minimally effective when used as a modulator of gut microbial translocation among immune nonresponders to virally suppressive ART. Systemic antibiotic use has a transient effect on LPS levels in simian immunodeficiency virus–infected macaques [10]. This transient effect may be related to the replacement of gut bacteria with antibiotic-resistant isolates. As with the use of other antibiotics, rifaximin-resistant bacteria emerge with chronic rifaximin use [19]. Moreover, a recent study showed no significant changes in microbial abundance with rifaximin use for 8 weeks [20]. HIV infection is associated with a gut microbial dysbiosis [21–24] that may be difficult to modulate with antimicrobials alone [25]. An approach using sevelamer, an endotoxin-binding agent, did not show a reduction in either LPS or sCD14 among untreated HIV-positive persons [26]. Nutritional supplements that modulate the gut microbiome have had mixed effects on systemic immune activation and inflammation when used in treatment-naive HIV-positive persons [27–29].
Our findings are unlike those observed in patients with cirrhosis, in whom rifaximin use has been associated with a significant decrease in LPS levels after 4 and 8 weeks of treatment [16, 20]. The mechanism of elevated LPS probably differs between cirrhosis and HIV infection, with factors beyond microbial translocation playing a role in the former. Furthermore, increased LPS levels among those with persistently low CD4+ T-cell counts despite ART may be more likely to be caused by impairments in immune cells [30] rather than increased gut microbial replication. Finally, LPS has historically been a difficult biomarker to measure precisely, making cross-study comparisons difficult. The effect of rifaximin on sCD14, a marker of monocyte activation by LPS, is more complicated, as we saw both early and posttreatment rifaximin effects. Whereas the former could be related to LPS-driven changes, the latter may be related to inhibition by rifaximin of monocyte activation [31].
We also found that rifaximin use had a marginal impact on CD8+ T-cell activation and proliferation after 4 weeks of study treatment. Although the week 4 effect on activation may be a delayed effect of the early transient effect on LPS and sCD14, we did not find a correlation between changes in levels of these markers and changes in measures of CD8+ T-cell activation. Moreover, baseline levels of either LPS or sCD14 did not correlate with baseline levels of immune activation in our subjects, in contrast to some previous observations [10, 13] but consistent with others [32]. It is possible that persistently elevated cellular immune activation in immune nonresponders is in part due to reasons other than gut microbial translocation.
In addition, there was substantial overlap between the levels of T-cell activation in the study population compared with HIV-negative controls. Thus, contrary to our expectations, lack of an immune response to ART as we defined it may not be reliably associated with elevated immune activation. When we stratified the changes in activation or gut microbial translocation by baseline levels of the percentages of HLA-DR+CD38+ CD8+ T cells, we found a trend toward a difference between study arms among those with higher levels of the latter, suggesting that this study may not have enrolled the optimal population to demonstrate a therapeutic reduction of activation. The lower d-dimer and sCD163 levels among our subjects compared with controls also support this idea. Studies that select HIV-positive persons with elevated activation or inflammation at entry may be needed to rigorously test the effect of immunomodulatory approaches.
Rifaximin may have direct anti-inflammatory effects that are independent of its effect on the gut microbiome and microbial translocation [20, 31, 33]. Recent studies have shown that elevated inflammation is a more important predictor of non–AIDS-defining outcomes than T-cell activation among HIV-positive persons receiving virally suppressive ART [15, 34, 35]. Decreases of 0.12 log10 and 0.07 log10 in IL-6 and sCD14, respectively, are associated with a 25% reduction in risk of a non-AIDS morbid event. In this study, we saw small but significant posttreatment effects on IL-6, CRP, and sCD14, and the changes in IL-6 and CRP correlated with changes in sCD14. This raises the possibility that longer use of rifaximin may decrease the levels of inflammatory markers that have been associated with outcomes in virally suppressed HIV-infected persons. Whether these rifaximin-related changes in inflammatory markers are due to direct effects on inflammatory pathways remains unclear and needs further study.
The treatment-related differences between study arms were small and transient, and the changes within each study arm were mostly insignificant, suggesting the possibility that the significant differences we saw were due to random fluctuations in immune markers that are unrelated to treatment. In retrospect, the paucity of study time points, the short duration of pretreatment follow-up, and the limited precision of the selection criteria, along with the absence of meaningful rifaximin-mediated effects on either LPS or sCD14, constrained our ability to answer our hypothesis.
In summary, rifaximin was safe and well-tolerated in HIV-infected persons with a suboptimal CD4+ T-cell response to virally suppressive ART. In this patient population, rifaximin had a marginal impact on gut microbial translocation and monocyte and T-cell activation. Correlations were not detected between either microbial translocation or inflammation markers and T-cell activation levels at baseline or between changes in microbial translocation and T-cell activation. Because microbial translocation and T-cell activation are both elevated in treated HIV-positive subjects with poor immune reconstitution [6, 13], our findings suggest a need to investigate the extent to which these processes are linked mechanistically. Rather, changes in sCD14 were more closely linked to changes in systemic inflammatory markers (IL-6 and CRP) associated with non-AIDS comorbid conditions and mortality in ART-treated HIV-positive persons. Whether the prolonged use of rifaximin would affect the pathways represented by these markers needs to be clarified. More importantly, to help identify interventions to improve long-term outcomes, we need better ways of defining virally suppressed ART-treated HIV-infected persons with elevated levels of inflammation and activation, who are at increased risk for morbid events.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://jid.oxfordjournals.org). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgments. We thank the A5286 participants, the ACTG clinical trial sites that enrolled and followed up the participants, and the ACTG. The other members of the A5286 team, besides the authors, include John W. Mellors, MD (virologist); Ali Keshavarzian, MD, Benigno Rodriguez, MD, Mariam Aziz, MD, Rachel Presti, MD, Steven Deeks, MD (investigators); Ruth Ebiasah, PharmD, MS, RPh (DAIDS pharmacist); Laurie Myers, MS, MT (data manager); LuAnn Borowski, MSc, Jill Plants, BS (laboratory technologists); David A. Palm, MS (CSS representative); Derek Weibel, MS Ed (laboratory data manager); and Beverly Putnam, RN, ANP (field representative). The following sites participated in A5286: Alabama Therapeutics Clinical Research Site (CRS) (Elizabeth Lindsey, RN, and Amy Player, PharmD), Beth Israel Deaconess Medical Center ACTG CRS (Mary Albrecht, MD, and Andrea Kershaw, NP), Brigham and Women's Hospital ACTG CRS (Paul Sax, MD, and Cheryl Keenan, RN), Case CRS (Patricia Walton, RN, and Jane Baum, RN), Cornell CRS (Todd Stroberg, RN, and Valery Hughes, NP), Duke University Medical Center Adult CRS, Georgetown University CRS (Laura Coster, MD, and Princy N. Kumar, MD), HIV Prevention & Treatment CRS (Michael T. Yin and Jolene Noel-Connor), Hospital of the University of Pennsylvania CRS (Pablo Tebas, MD, and Aleshia Thomas, RN, BSN), Institute of Human Virology Baltimore Treatment CRS (Charles E. Davis, Jr, MD, and Robert R. Redfield, Jr, MD), Massachusetts General Hospital ACTG CRS (Amy Sbrolla, RN, and Teri Flynn, ANP), MetroHealth CRS (Traci Davis, RN, and Kim Whitely, RN), New Jersey Medical School CRS (Baljinder Singh, MA, and Shobha Swaminathan, MD), Northwestern University CRS (Donna McGregor and Frank Palella, MD), NYU and Bellevue HIV/AIDS Clinical Trials Unit (Judith Aberg, MD, and Karen Cavanagh, RN), Puerto Rico–ACTU (Jorge L. Santana Bagur, MD, and Olga Méndez Flores, MD), Rush CRS (Janice Fritsche and Beverly Sha, MD), Stanford CRS (Debbie Slamowitz, RN, ACRN, and Sandra Valle, PA-C), The Miriam Hospital ACTG CRS (Karen Tashima, MD, and Helen Patterson, LPN), Ohio State University AIDS CRS (Heather Harber, RN, and Michael Para, MD), The Ponce de Leon Center CRS (Molly Eaton, MD, and Dale Maddox, RN, BSN), UCLA CARE Center CRS (Judith Currier, MD, and Vanessa Cajahuaringa), UCSF AIDS CRS (Annie Luetkemeyer, MD, and Jay Dwyer, RN), University of Cincinnati CRS (Carl J. Fichtenbaum, MD, and Michelle Saemann, RN), University of Colorado Hospital CRS (Graham Ray and Thomas Campbell), University of Miami HIV/ACTU (Margaret A. Fischl, MD, and Hector Bolivar, MD), University of North Carolina AIDS CRS (Jonathan Oakes and Miriam Chicurel-Bayard), University of Pittsburgh CRS (Christine Tripoli, BSN, and D. Renee Weinman, BS), University of Rochester/Trillium Health (Mary Adams, RN, and Christine Hurley, RN), University of Washington AIDS CRS (Shelia Dunaway, MD, and Sheryl Storey, PA-C), and Washington University CRS (Michael Klebert, PhD, RN, ANP-BC, and Michael Royal, BS, RPh).
Disclaimer. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Financial support. This work was supported by grants from the National Institute of Health (AI068636 to the ACTG Network Leadership; AI068634 and AI068636 to ACTG Statistical Data Analysis Center [E. S. C. and R. J. B.]; AI068636 to A. L. L.; AI069452, A1025439, AI069424, AI069447, AI069419, AI069418, AI069532, AI069470, AI069494, AI069511-08, AI069439, AI069471, AI069418, AI069556, AI069477, AI069494, AI069439, AI069501, AI069471, AI069412, AI069412, AI069412, AI069502, AI069494, AI69501, AI069423, AI069415, AI069481, AI069534, AI069503 and AI069432 to the ACTG clinical research sites; TR001082, RR024160, TR001070, TR000439 and TR001111 to the Clinical and Translational Science Centers; and Al050404, AI073961, AI50410 and AI045008 to the Centers for AIDS Research).
Potential conflicts of interest. All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Liu Z, Cumberland WG, Hultin LE, Kaplan AH, Detels R, Giorgi JV. CD8+ T-lymphocyte activation in HIV-1 disease reflects an aspect of pathogenesis distinct from viral burden and immunodeficiency. J Acquir Immune Defic Syndr Hum Retrovirol. 1998;18:332–40. doi: 10.1097/00042560-199808010-00004. [DOI] [PubMed] [Google Scholar]
- 2.Hunt PW, Martin JN, Sinclair E, et al. T cell activation is associated with lower CD4+ T cell gains in human immunodeficiency virus-infected patients with sustained viral suppression during antiretroviral therapy. J Infect Dis. 2003;187:1534–43. doi: 10.1086/374786. [DOI] [PubMed] [Google Scholar]
- 3.Anthony KB, Yoder C, Metcalf JA, et al. Incomplete CD4 T cell recovery in HIV-1 infection after 12 months of highly active antiretroviral therapy is associated with ongoing increased CD4 T cell activation and turnover. J Acquir Immune Defic Syndr. 2003;33:125–33. doi: 10.1097/00126334-200306010-00002. [DOI] [PubMed] [Google Scholar]
- 4.French MA, King MS, Tschampa JM, da Silva BA, Landay AL. Serum immune activation markers are persistently increased in patients with HIV infection after 6 years of antiretroviral therapy despite suppression of viral replication and reconstitution of CD4+ T cells. J Infect Dis. 2009;200:1212–5. doi: 10.1086/605890. [DOI] [PubMed] [Google Scholar]
- 5.Gandhi RT, Spritzler J, Chan E, et al. Effect of baseline- and treatment-related factors on immunologic recovery after initiation of antiretroviral therapy in HIV-1-positive subjects: results from ACTG 384. J Acquir Immune Defic Syndr. 2006;42:426–34. doi: 10.1097/01.qai.0000226789.51992.3f. [DOI] [PubMed] [Google Scholar]
- 6.Lederman MM, Calabrese L, Funderburg NT, et al. Immunologic failure despite suppressive antiretroviral therapy is related to activation and turnover of memory CD4 cells. J Infect Dis. 2011;204:1217–26. doi: 10.1093/infdis/jir507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chene G, Sterne JA, May M, et al. Prognostic importance of initial response in HIV-1 infected patients starting potent antiretroviral therapy: analysis of prospective studies. Lancet. 2003;362:679–86. doi: 10.1016/s0140-6736(03)14229-8. [DOI] [PubMed] [Google Scholar]
- 8.van Lelyveld SF, Gras L, Kesselring A, et al. Long-term complications in patients with poor immunological recovery despite virological successful HAART in Dutch ATHENA cohort. AIDS. 2012;26:465–74. doi: 10.1097/QAD.0b013e32834f32f8. [DOI] [PubMed] [Google Scholar]
- 9.Brenchley JM, Schacker TW, Ruff LE, et al. CD4+ T cell depletion during all stages of HIV disease occurs predominantly in the gastrointestinal tract. J Exp Med. 2004;200:749–59. doi: 10.1084/jem.20040874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brenchley JM, Price DA, Schacker TW, et al. Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat Med. 2006;12:1365–71. doi: 10.1038/nm1511. [DOI] [PubMed] [Google Scholar]
- 11.Chun TW, Nickle DC, Justement JS, et al. Persistence of HIV in gut-associated lymphoid tissue despite long-term antiretroviral therapy. J Infect Dis. 2008;197:714–20. doi: 10.1086/527324. [DOI] [PubMed] [Google Scholar]
- 12.Guadalupe M, Reay E, Sankaran S, et al. Severe CD4+ T-cell depletion in gut lymphoid tissue during primary human immunodeficiency virus type 1 infection and substantial delay in restoration following highly active antiretroviral therapy. J Virol. 2003;77:11708–17. doi: 10.1128/JVI.77.21.11708-11717.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang W, Lederman MM, Hunt P, et al. Plasma levels of bacterial DNA correlate with immune activation and the magnitude of immune restoration in persons with antiretroviral-treated HIV infection. J Infect Dis. 2009;199:1177–85. doi: 10.1086/597476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pedersen KK, Pedersen M, Troseid M, et al. Microbial translocation in HIV infection is associated with dyslipidemia, insulin resistance, and risk of myocardial infarction. J Acquir Immune Defic Syndr. 2013;64:425–33. doi: 10.1097/QAI.0b013e31829f919d. [DOI] [PubMed] [Google Scholar]
- 15.Sandler NG, Wand H, Roque A, et al. Plasma levels of soluble CD14 independently predict mortality in HIV infection. J Infect Dis. 2011;203:780–90. doi: 10.1093/infdis/jiq118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vlachogiannakos J, Saveriadis AS, Viazis N, et al. Intestinal decontamination improves liver haemodynamics in patients with alcohol-related decompensated cirrhosis. Aliment Pharmacol Ther. 2009;29:992–9. doi: 10.1111/j.1365-2036.2009.03958.x. [DOI] [PubMed] [Google Scholar]
- 17.Brumback LC, Pepe MS, Alonzo TA. Using the ROC curve for gauging treatment effect in clinical trials. Stat Med. 2006;25:575–90. doi: 10.1002/sim.2345. [DOI] [PubMed] [Google Scholar]
- 18.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44:837–45. [PubMed] [Google Scholar]
- 19.Kothary V, Scherl EJ, Bosworth B, et al. Rifaximin resistance in Escherichia coli associated with inflammatory bowel disease correlates with prior rifaximin use, mutations in rpoB, and activity of Phe-Arg-β-naphthylamide-inhibitable efflux pumps. Antimicrob Agents Chemother. 2013;57:811–7. doi: 10.1128/AAC.02163-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bajaj JS, Heuman DM, Sanyal AJ, et al. Modulation of the metabiome by rifaximin in patients with cirrhosis and minimal hepatic encephalopathy. PLoS One. 2013;8:e60042. doi: 10.1371/journal.pone.0060042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mutlu EA, Keshavarzian A, Losurdo J, et al. A compositional look at the human gastrointestinal microbiome and immune activation parameters in HIV infected subjects. PLoS Pathog. 2014;10:e1003829. doi: 10.1371/journal.ppat.1003829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dillon SM, Lee EJ, Kotter CV, et al. An altered intestinal mucosal microbiome in HIV-1 infection is associated with mucosal and systemic immune activation and endotoxemia. Mucosal Immunol. 2014;7:983–94. doi: 10.1038/mi.2013.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vujkovic-Cvijin I, Dunham RM, Iwai S, et al. Dysbiosis of the gut microbiota is associated with HIV disease progression and tryptophan catabolism. Sci Transl Med. 2013;5:193ra91. doi: 10.1126/scitranslmed.3006438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lozupone CA, Li M, Campbell TB, et al. Alterations in the gut microbiota associated with HIV-1 infection. Cell Host Microbe. 2013;14:329–39. doi: 10.1016/j.chom.2013.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gonzalez-Hernandez LA, Jave-Suarez LF, Fafutis-Morris M, et al. Synbiotic therapy decreases microbial translocation and inflammation and improves immunological status in HIV-infected patients: a double-blind randomized controlled pilot trial. Nutr J. 2012;11:90. doi: 10.1186/1475-2891-11-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sandler NG, Zhang X, Bosch RJ, et al. Sevelamer does not decrease lipopolysaccharide or soluble CD14 levels but decreases soluble tissue factor, low-density lipoprotein (LDL) cholesterol, and oxidized LDL cholesterol levels in individuals with untreated HIV infection. J Infect Dis. 2014 doi: 10.1093/infdis/jiu305. pii: jiu305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cahn P, Ruxrungtham K, Gazzard B, et al. The immunomodulatory nutritional intervention NR100157 reduced CD4+ T-cell decline and immune activation: a 1-year multicenter randomized controlled double-blind trial in HIV-infected persons not receiving antiretroviral therapy (the BITE Study) Clin Infect Dis. 2013;57:139–46. doi: 10.1093/cid/cit171. [DOI] [PubMed] [Google Scholar]
- 28.Gori A, Rizzardini G, Van't Land B, et al. Specific prebiotics modulate gut microbiota and immune activation in HAART-naive HIV-infected adults: results of the "COPA" pilot randomized trial. Mucosal Immunol. 2011;4:554–63. doi: 10.1038/mi.2011.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schunter M, Chu H, Hayes TL, et al. Randomized pilot trial of a synbiotic dietary supplement in chronic HIV-1 infection. BMC Complement Altern Med. 2012;12:84. doi: 10.1186/1472-6882-12-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lim A, Amini A, D'Orsogna LJ, et al. Antibody and B-cell responses may control circulating lipopolysaccharide in patients with HIV infection. AIDS. 2011;25:1379–83. doi: 10.1097/QAD.0b013e328348a789. [DOI] [PubMed] [Google Scholar]
- 31.Rosette C, Buendia-Laysa F, Jr, Patkar S, et al. Anti-inflammatory and immunomodulatory activities of rifamycin SV. Int J Antimicrob Agents. 2013;42:182–6. doi: 10.1016/j.ijantimicag.2013.04.020. [DOI] [PubMed] [Google Scholar]
- 32.Abad-Fernandez M, Vallejo A, Hernandez-Novoa B, et al. Correlation between different methods to measure microbial translocation and its association with immune activation in long-term suppressed HIV-1-infected individuals. J Acquir Immune Defic Syndr. 2013;64:149–53. doi: 10.1097/QAI.0b013e31829a2f12. [DOI] [PubMed] [Google Scholar]
- 33.Kalambokis GN, Mouzaki A, Rodi M, Tsianos EV. Rifaximin improves thrombocytopenia in patients with alcoholic cirrhosis in association with reduction of endotoxaemia. Liver Int. 2012;32:467–75. doi: 10.1111/j.1478-3231.2011.02650.x. [DOI] [PubMed] [Google Scholar]
- 34.Tenorio AR, Zheng Y, Bosch RJ, et al. Soluble markers of inflammation and coagulation but not T-cell activation predict non-AIDS-defining morbid events during suppressive antiretroviral treatment. J Infect Dis. 2014;210:1248–59. doi: 10.1093/infdis/jiu254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hunt PW, Sinclair E, Rodriguez B, et al. Gut epithelial barrier dysfunction and innate immune activation predict mortality in treated HIV infection. J Infect Dis. 2014;210:1228–38. doi: 10.1093/infdis/jiu238. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.