Abstract
β-1,3-glucan is a major cell wall component of Pneumocystis cysts. We have characterized endo-β-1,3-glucanase (Eng) from 3 species of Pneumocystis. The gene eng is a single-copy gene that encodes a protein containing 786 amino acids in P. carinii and P. murina, and 788 amino acids in P. jirovecii, including a signal peptide for the former 2 but not the latter. Recombinant Eng expressed in Escherichia coli was able to solubilize the major surface glycoprotein of Pneumocystis, thus potentially facilitating switching of the expressed major surface glycoprotein (Msg) variant. Confocal immunofluorescence analysis of P. murina–infected mouse lung sections localized Eng exclusively to the cyst form of Pneumocystis. No Eng was detected after mice were treated with caspofungin, a β-1,3-glucan synthase inhibitor that is known to reduce the number of cysts. Thus, Eng is a cyst-specific protein that may play a role in Msg variant expression in Pneumocystis.
Keywords: cell wall; endo-β-1,3-glucanase; Eng; glucan; Pneumocystis
Pneumocystis is an opportunistic pathogen that causes pneumonia in human immunodeficiency virus-infected and other immunocompromised patients [1, 2]. This organism is known to be genetically diverse; each host species is infected by at least 1 genetically distinct strain that represent Pneumocystis species, with P. jirovecii infecting humans, P. carinii and P. wakefieldiae infecting rats, and P. murina infecting mice [3–6]. Two major stages have been identified morphologically in the life cycle of Pneumocystis: cysts and trophic forms. The cyst has a cell wall that has an electron-lucent layer consisting primarily of β-1,3 glucan, which in fungi is important in maintaining cell-wall integrity. The trophic form has no detectable β-1,3 glucan [7].
Enzymes that synthesize and degrade glucans play a critical role in fungal life cycles, as exemplified by the efficacy of echinocandins, which are inhibitors of β-1,3 glucan synthase, in the treatment of infections caused by Candida and other fungal species. Endo-β-1,3-glucanase is an enzyme (EC 3.2.1.39) involved in the degradation of β-1,3 glucan and belongs to glycoside hydrolase family 81 [8]. Two forms of endo-β-1,3-glucanase, Eng1 and Eng2, have been characterized from yeast. Eng1 is an extracellular protein that contains a signal peptide, while Eng2 is an intracellular protein with no signal peptide [8, 9]. Endo-β-1,3-glucanases have been shown to be important for cell separation in yeast [8–11].
Cloning and expression of a P. carinii endo-β-1,3-glucanase was recently reported [12]; based on the absence of a predicted signal peptide, it was categorized as a homologue of Eng2. In prior studies, we and others have shown that the major surface glycoprotein (Msg) of Pneumocystis can be solubilized by treating organisms with endo-β-1,3-glucanase [13, 14]. Because Msg confers on Pneumocystis the potential for antigenic variation [15], we were interested in determining if this recently identified Eng homologue would be able to release Msg, which could potentially provide a mechanism to facilitate switching Msg variants in a given organism. We thus undertook to further characterize this protein from multiple Pneumocystis species, and to examine its functionality.
MATERIALS AND METHODS
Pneumocystis DNA and RNA Preparation
Pneumocystis carinii or P. murina organisms were isolated from the lungs of immunosuppressed rats or mice by Ficoll-Hypaque density gradient centrifugation as previously reported [16]. Of note, our colony appears to be exclusively P. carinii; we have never identified P. wakefieldiae genes in rat Pneumocystis isolates. For P. jirovecii, genomic DNA or RNA was isolated from autopsy lung samples. Genomic DNA was isolated using QIAamp DNA mini kit (Qiagen, Valencia, California), and total RNA was extracted using RNeasy mini kit (Qiagen). Human and animal experimentation guidelines of the National Institutes of Health were followed in the conduct of these studies.
Polymerase Chain Reaction Amplification of eng
Polymerase chain reaction (PCR) was performed using HotStar Taq (Qiagen) or AccuPrime Pfx (Invitrogen, Carlsbad, California). Partial P. carinii and P. jirovecii eng DNA sequences were obtained from published data [12, 17]. Partial genomic sequence (approximately 2 kb) of P. murina eng was obtained by the amplification of genomic DNA using 2 sets of primers (GK42pc and GK53pc; GK52pc and GK51pc) designed from the conserved region of the P. carinii eng DNA sequence.
Complete eng genomic DNA and complementary DNA (cDNA) sequences were obtained by inverse PCR or RNA ligase mediated rapid amplification of cDNA ends (RLM-RACE) [18]. RNA isolated from partially purified P. carinii or P. murina organisms and P. jirovecii–infected lung samples was subjected to RLM-RACE using the First Choice RLM-RACE kit (Ambion Inc, Austin, Texas) according to the manufacturer's protocol. Reverse transcription PCR and 3′-RACE amplifications were performed as described previously [19]. Additional methods are provided in the Supplementary Materials.
Sequencing
PCR products were purified using the PCR purification kit (Edge Biosystem, Gaithersburg, Maryland) or QIAquick Gel Extraction Kit (Qiagen), and were sequenced directly or after subcloning using ABI 3100 Genetic Analyzer (Applied Biosystems, Foster City, California) or commercially (MacrogenUSA, Rockville, Maryland).
Southern and Northern Blot Analysis
For Southern blot analysis, P. carinii genomic DNA was digested with Xba1 or EcoR1 restriction enzymes, separated on 1% agarose gel, transferred to a Nytran membrane, and hybridized with oligonucleotide probes (GK28pc, GK29pc, or GK19pc) labeled using DIG Oligonucleotide Tailing Kit (Roche Diagnostics Corporation, Indianapolis, Indiana). Before rehybridization, the blot was stripped at 37°C using a solution containing 0.2M NaOH and 0.1% sodium dodecyl sulfate (SDS).
For northern blot analysis, total RNA extracted from partially purified P. carinii organisms was subjected to agarose gel electrophoresis in the presence of formaldehyde, transferred to a Nytran membrane, and hybridized with GK29pc or GK19pc as described for Southern blot analysis. Before reprobing, the blot was stripped at 80°C using a buffer containing 0.05M Tris-HCl pH 7.5, 50% deionized formamide, and 5% SDS.
Protein Expression and Refolding
Pneumocystis carinii Eng recombinant protein was expressed in Escherichia coli. An approximately 1100 bp region from the 3′-end of P. carinii eng cDNA (spanning 1387 bp–2535 bp, GenBank #KM056671), which included the entire catalytic domain based on sequence homology to Saccharomyces cerevisiae Eng, was amplified and cloned into pET32 (EMD Biosciences, San Diego, California). Recombinant protein was induced with 1 mM isopropyl-β-D-thiogalactopyranoside for 3 hours at 37°C and refolded as described previously [20]. Vector with no insert was processed in a similar manner.
Digestion of Pneumocystis Cell Wall With Eng Protein
Ficoll-purified P. carinii organisms were incubated overnight at 50°C with the refolded Eng or control (no insert) in a buffer containing 0.05 M sodium acetate, pH 5.0. The supernatant obtained after centrifugation was subjected to Western blot analysis using monoclonal antibody RA-E7, which recognizes P. carinii Msg (a kind gift of Drs Walzer and Linke) [21].
Peptide Antibodies
A mixture of 2 synthetic peptides, KKWLLYVFPKEKSE (corresponds to 277–290 amino acids of P. murina Eng) and PYSPSAKKPSYSKEAL (corresponds to 421–436 amino acids of P. murina Eng) were used to commercially raise antibodies in rabbits (PickCell Laboratories BV, Amsterdam, Netherlands). The antibody preparations were affinity purified using recombinant Eng protein.
SDS–Polyacrylamide Gel Electrophoresis and Western Blot Analyses
Protein extracts obtained from partially purified P. carinii organisms or recombinant Eng protein preparations were subjected to SDS–polyacrylamide gel electrophoresis (PAGE) (Invitrogen) and electroblotting. Blots were probed with anti-Eng antibody or, for Msg detection, monoclonal antibody RA-E7 [21] followed by horseradish peroxidase-conjugated secondary antibodies.
Animal Treatment Studies
CD40L-KO mice (B6;129S2-Tnfsf5tm1Imx/J), which are highly susceptible to Pneumocystis infection [22], were obtained from The Jackson Laboratory (Bar Harbor, Maine) and subsequently bred at the National Institutes of Health. Ten CD40L-KO mice were randomly divided into 3 groups. Group 1 (n = 4) received caspofungin (3 mg/kg intraperitoneally) once daily for 8 days, together with trimethoprim-sulfamethoxazole (48 mg/kg trimethoprim and 240 mg/kg sulfamethoxazole orally) for the last 3 days; group 2 (n = 3) received trimethoprim-sulfamethoxazole at the same dose for 3 days; group 3 (n = 3) received no drug. After sacrifice, the lungs were removed and sections were placed in phosphate buffered saline for quantitation of Pneumocystis burden, RNAlater for gene expression studies, and Histochoice preservative for immunofluorescence studies.
Immunofluorescence and Confocal Microscopic Analysis
Immunofluorescent labeling of P. murina-infected lung tissue sections was performed by Histoserv, Inc (Germantown, Maryland). Lungs were colabeled with affinity-purified rabbit anti-Eng antibody, mouse anti-Pneumocystis antibody 4D7 [23], and mouse β-1,3-glucan monoclonal antibody (Biosupplies Australia Pty Ltd, Bundoora, VIC, Australia). Alexa Fluor 594–labeled donkey antirabbit immunoglobin G (IgG) (Invitrogen) was used for Eng detection, biotin conjugated antimouse IgG and Alexa Fluor 488-conjugated streptavidin (Invitrogen) were used for the labeling of β-1,3-glucan, and Alexa Fluor 555–labeled donkey antimouse IgG (Invitrogen) was used to detect Pneumocystis organisms. Nuclei were labeled with DAPI fluorescent stain and slides mounted in Vectashield (Vector Labs, Burlingame, California). Images were captured on a TCS SP5 White Light Laser Confocal System (Leica Microsystems, Buffalo Grove, Illinois) and deconvolved using Huygens Essential, version 4.5.1p2 (Scientific Volume Imaging, Hilversum, the Netherlands); further postprocessing, including colocalization analysis, was done in Imaris 7.6.3 (Bitplane, Zurich, Switzerland) and final figure compilation in Indesign CS6 (Adobe, San Jose, California).
Real-Time PCR
cDNA was reverse transcribed from total RNA extracted from lung samples using High Capacity RNA to cDNA kit (Applied Biosystems); eng expression was quantitated by real-time PCR using primers GK31pm and GK33pm with iTaq Universal SYBR Green Supermix (Biorad, Hercules, California) and ViiA 7 Real-Time PCR system (Applied Biosystems) according to the manufacturer's instructions. Change in eng expression was calculated by the relative quantification (ΔΔCT) method, standardized to P. murina 18S ribosomal RNA (rRNA) levels.
RESULTS
Cloning of the eng Gene From Pneumocystis
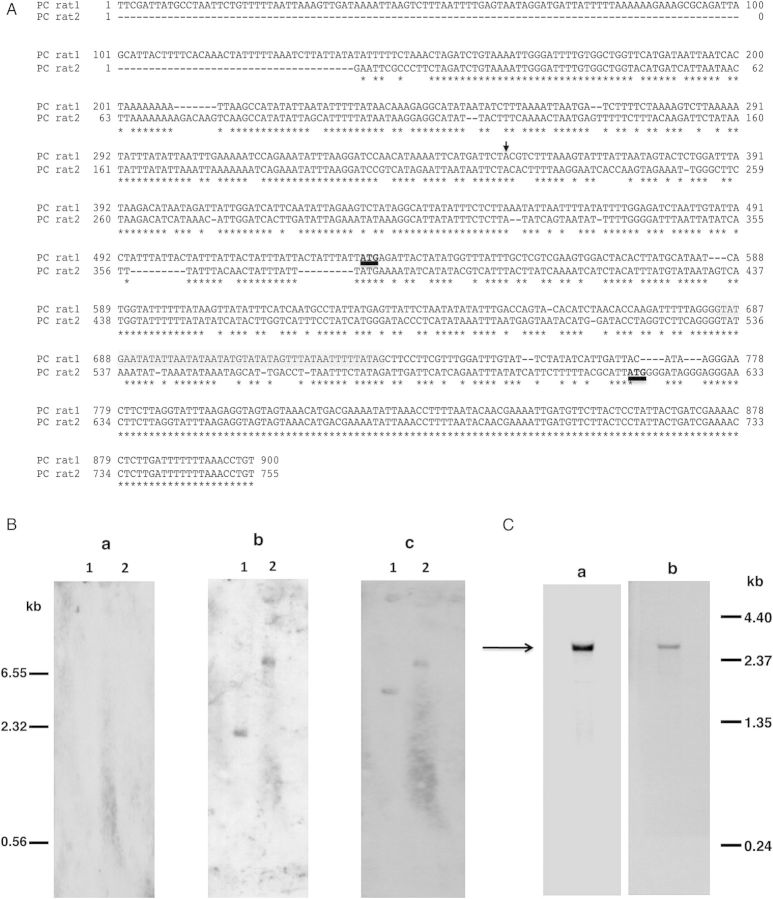
Given our long-standing interest in the Msg family of surface proteins, we were interested in determining whether the previously reported eng2 gene [12] encodes a functional enzyme that can release Msg from intact organisms. We thus undertook to express recombinant protein and examine its function. Initially, we tried to amplify the complete P. carinii eng cDNA using primers GK1pc and GK2pc designed from the published eng2 sequence (GenBank #EU814521), without success. We subsequently were able to amplify approximately 2100 bp from the 3′-end of the gene using primers GK24pc and GK10pc, but had no success in amplifying approximately 600 bp of the 5′-end of the gene. We were ultimately able to successfully amplify the 5′-end of the gene by inverse PCR, and to identify the transcription start site by 5′-RACE using cDNA. As shown in Figure 1A, the eng sequence that we obtained (PC rat1) differed substantially from the published sequence (PC rat2) for the first 627 bp, but beyond that point was a 100% match.
Figure 1.
A, Alignment of 5′-region of P. carinii eng (PC rat1) sequence with the published sequence (PC rat2) [12]. The translation start site, ATG, is shown in bold and underlined. Conserved sequences are denoted by the asterisks and the intron is highlighted in gray. The transcription start site is indicated by the arrow. B, Southern blot analysis of P. carinii genomic DNA. Genomic DNA extracted from partially purified P. carinii organisms was digested with Xba1 (lane 1) or EcoR1 (lane 2). The blot was hybridized successively with 3 different digoxigenin-labeled oligonucleotides, with stripping of the blot between hybridizations. No hybridization signal was observed with oligonucleotide GK28pc designed from PC rat2 (panel a). Oligonucleotide GK29pc designed from PC rat1 hybridized to a single band with both Xba1 and EcoR1 digests (panel b). Also, oligonucleotide GK19pc designed from the 3′-end common to ratpc1 and ratpc2, hybridized to a single band with both Xba1 and EcoR1 digests (panel c). An internal restriction site accounts for the size differences in the bands that hybridized with the 5′-end and the 3′-end oligonucleotides in the Xba1 digests. C, Northern blot analysis of total RNA from P. carinii. (Panel a) Hybridization with oligonulceotide GK29pc resulted in a band of approximately 2.6 kb (indicated by the arrow). (Panel b) The blot in panel a was hybridized, after stripping, with oligonucleotide GK19pc, giving an RNA band of the same size as in panel a.
To verify this new sequence, Southern and northern blot analyses were performed. In Southern blot analysis with P. carinii genomic DNA digested with restriction enzymes Xba1 or EcoR1 (Figure 1B), no hybridization signal was detected with oligo GK28pc designed from the 5′-end of the published eng sequence (PC rat2; Figure 1B, panel a), while a single band was seen for both Xba1 (lane 1) and EcoR1 (lane 2) with GK29pc, designed from the 5′-end of the new eng sequence (PC rat1; Figure 1B, panel b). Hybridization was also observed when the same blot was reprobed with GK19pc, which is designed from the 3′-end and is identical in both sequences (Figure 1B, panel c). Southern blot analysis also demonstrated that P. carinii eng is a single copy gene. In northern blotting analysis with RNA extracted from partially purified P. carinii organisms, both probes GK29pc and GK19pc detected a band of approximately 2.6 kb, consistent with the size expected from the cDNA sequence (Figure 1C).
We next determined the P. murina eng sequence and confirmed the P. jirovecii eng sequence. A partial eng sequence for P. murina was obtained by amplifying genomic DNA using primers designed from the regions of P. carinii eng gene that were conserved when aligned with yeast eng sequences. The complete eng sequence was assembled from sequences obtained by inverse PCR, 5′-RACE and 3′-RACE techniques. A partial eng sequence for P. jirovecii was obtained from the published P. jirovecii genome data [17]; 5′-RACE was performed to determine the 5′-untranslated region.
The accuracy of eng genomic and cDNA sequences for 3 Pneumocystis species was confirmed by sequencing the entire coding region of eng amplified from genomic DNA and cDNA from each species. The alignment of eng genomic and cDNA sequences from all 3 species is shown in the Supplementary Figure 1. Comparison of genomic and cDNA sequences identified 5 introns for P. carinii and P. murina, and 4 introns for P. jirovecii. Introns are in identical locations in all 3 species, with the exception that the first intron present in P. carinii and P. murina was absent in P. jirovecii. The GenBank accession numbers for the P. carinii, P. murina, and P. jirovecii eng genomic and cDNA sequences are KM056670-75. We could identify only 1 eng gene encoding a protein belonging to the glycoside hydrolase family 81 in each Pneumocystis species. Because it is not clear if Pneumocystis Eng is more closely related to Eng1 or Eng2 in yeast (see below), we have designated the Pneumocystis endo-β-1,3-glucanase genes described in this paper simply as eng.
Deduced Amino Acid Sequences of Pneumocystis Eng
The cDNA sequences of P. carinii and P. murina eng each encode a protein of 786 amino acids, while P. jirovecii eng cDNA encodes a protein of 788 amino acids. Figure 2 shows the alignment of the deduced amino acid sequences of Eng from all 3 Pneumocystis species together with those of 2 yeast, Saccharomyces cerevisiae and Schizosaccharomyces pombe. P. carinii Eng showed 76% identity to that of P. murina and both showed 58%–60% identity to that of P. jirovecii. The predicted catalytic sites are highly conserved in all 3 species as well as in yeast [8]. Pneumocystis Eng showed 38%–40% and 27%–29% identity to S. pombe Eng2 and Eng1, respectively, and 34% and 23% identity to S. cerevisiae Eng2 and Eng1, respectively.
Figure 2.
Alignment of deduced amino acid sequences of Pneumocystis Eng with Eng1 and Eng2 from S. cerevisiae and S. pombe. Identical amino acid residues are boxed. The putative catalytic active sites are denoted by asterisks [8]. Alignment was performed using Clustal W.
Both P. carinii and P. murina Eng contains a signal peptide predicted by SignalP 3.0, similar to yeast Eng1 [9], despite the greater sequence similarity to yeast Eng2 [11]. In contrast, no signal peptide was predicted in P. jirovecii Eng.
Characterization of Pneumocystis Eng Expression and Enzyme Activity
To examine Eng expression, we utilized a polyclonal antibody generated against a mixture of 2 synthetic peptides designed from a conserved region of the P. murina Eng protein. Western blot analysis of P. carinii extracts showed that this antibody recognized an approximately 105 kDa protein band (Figure 3A), which is slightly larger than the predicted size of approximately 90 kDa.
Figure 3.
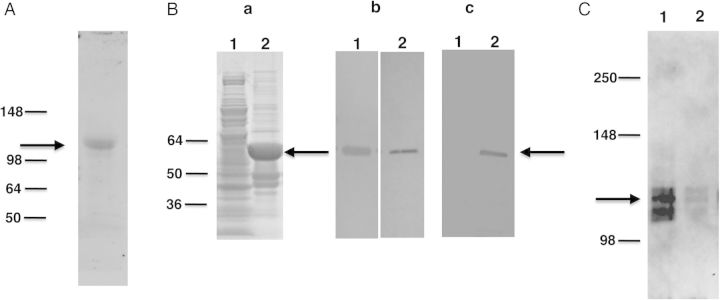
Western blot analysis of P. carinii Eng. A, Protein extracts prepared from partially purified P. carinii organisms probed with anti-Eng antibody. Immunoreactivity to an approximately 105 kDa band (indicated by arrow) was detected. B, Recombinant P. carinii Eng expressed in E. coli as a His-tag fusion protein was analyzed by SDS-PAGE and Western blotting. (Panel a) Recombinant protein was analyzed by SDS-PAGE gel stained with Coomassie Blue. Extracts from bacterial cells expressing recombinant protein showed a band of the expected size (approximately 60 kDa) indicated by the arrow (lane 2). This band was not seen with bacterial cells transformed with vector alone (lane 1). (Panel b) Western blot analysis of the bacterial cells expressing Eng protein, showing an immunoreactive band of the expected size when probed with anti-Eng antibody (lane 1) and anti-His tag antibody (lane 2). (Panel c) Western blot analysis of purified, refolded recombinant protein. An approximately 60 kDa band was detected with anti-His tag antibody (lane 2), but no immunoreactivity was observed in the vector-alone control (lane 1). C, The refolded recombinant Eng protein was tested for its ability to release Pneumocystis Msg. P. carinii organisms were incubated with the refolded protein or negative control sample. The supernatant obtained after centrifugation was analyzed for the presence of Msg by Western blot using a monoclonal antibody against P. carinii Msg. Strong immunoreactivity for Msg (approximately 115 kDa, shown by arrow) was seen in supernatant digested with recombinant Eng protein (lane 1). Only a weak signal was detected in supernatant incubated with protein preparation from the vector alone control (lane 2) or with buffer alone (data not shown). Abbreviations: Msg, major surface glycoprotein; SDS-PAGE, sodium dodecyl sulfate–polyacrylamide gel electrophoresis.
To examine Eng function, a fragment of the P. carinii eng cDNA (spanning 1387–2535 bp) that encodes amino acid residues 404–786, which includes the putative catalytic active sites, was expressed as a His tag fusion protein in E. coli. SDS-PAGE analysis of the bacterial extract (Figure 3B, panel a) expressing recombinant protein showed a prominent band of approximately 60 kDa protein, consistent with the expected size; this band was not seen in the control cell lysate with vector alone. By Western blot analysis (Figure 3B, panel b), the expressed protein showed immunoreactivity to the polyclonal anti-Eng antibody, as well as a His-tag antibody.
Most of the recombinant protein was insoluble. To obtain functional protein, recombinant protein was solubilized with 8M urea and then refolded, as previously described [20]. Control vector with no insert was processed in a similar manner. Western blot analysis of refolded protein (Figure 3B, panel c) showed an immunoreactive band of approximately 60 kDa when probed with anti-His tag antibody, while no band was detected for the no-insert control.
The refolded Eng protein was analyzed for its ability to digest Pneumocystis cell wall and release Msg as has been shown in previous studies with commercial β-1,3-glucanase [13, 14]. Pneumocystis carinii organisms were incubated with the refolded Eng protein or negative control, and release of Msg was analyzed by Western blot using a monoclonal antibody [21]. Strong immunoreactivity with Msg (approximately 115 kDa) was detected in the supernatant obtained after digestion with refolded recombinant Eng protein (Figure 3C), indicating that, like other endo-β-1,3-glucanases, the refolded Eng protein has the ability to release Msg. Minimal activity was detected with the protein preparation from bacteria transformed with the control vector (Figure 3C).
Immunohistochemical Analysis of Eng
To localize the expression of Eng in P. murina organisms, infected mouse lung sections were labeled with affinity-purified anti-Eng antibody together with an anti-Pneumocystis monoclonal antibody (4D7), which reacts with all stages of the organism, as well as an anti-β-1,3 glucan antibody, which reacts only with the cyst form, and then examined by confocal microscopy. Based on colocalization of the Eng labeling exclusively with β-1,3 glucan, Eng expression is limited to Pneumocystis cysts (Figure 4A, top panel, and Supplementary Figure 2). Preincubation of the anti-Eng antibody with the peptides used for immunization resulted in decrease or loss of immunoreactivity (data not shown).
Figure 4.
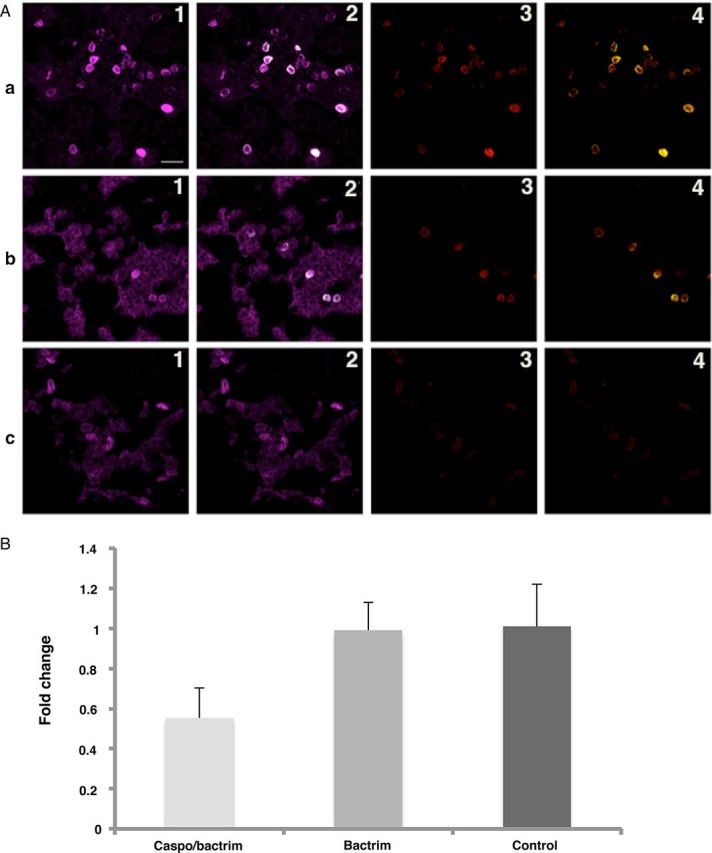
Immunohistochemical and real-time PCR analysis of P. murina Eng. A, Confocal immunofluorescence microscopic analysis of triple immunofluorescence labeling of P. murina–infected mouse lung tissue sections using anti-Eng, antiglucan and anti-Pneumocystis (4D7) antibodies. (Panel a) Immunofluorescence labeling of P. murina–infected lung tissue sections from a mouse with no drug treatment. (Panel b) Immunofluorescence labeling of P. murina–infected lung tissue sections from a mouse treated with trimethoprim-sulfamethoxazole only. (Panel c) Immunofluorescence labeling of P. murina–infected lung tissue sections from a mouse treated with both caspofungin and trimethoprim-sulfamethoxazole. (1.) Labeling of Pneumocystis using 4D7 antibody; magenta color indicates immunoreactivity. (2.) Anti-Eng fluorescence colocalizes with anti-Pneumocystis (4D7) labeling; colocalization is seen in white (mathematically generated colocalization output); note that not all organisms are colabeled with the anti-Eng antibody. (3.) Labeling of cysts with antiglucan antibody; red color indicates immunoreactivity. (4.) Anti-Eng fluorescence colocalizes with antiglucan labeling; colocalization is seen in yellow (mathematically generated colocalization output); note that all anti-Eng labeling colocalizes with antiglucan labeling. In panel c, the absence of cysts due to caspofungin treatment is associated with an absence of anti-Eng reactivity. Bar in 1a is 10 microns. A more detailed figure showing the individual labeling and other merged images is provided in Supplementary Figure 2. B, Real-time PCR analysis of eng mRNA expression in P. murina. RNA extracted from lung tissues collected from P. murina–infected mouse lungs was reverse transcribed and eng expression was analyzed by real-time PCR using P. murina 18S rRNA as the endogenous control. X-axis indicates groups based on drug treatment. Y-axis shows the fold change in P. murina eng mRNA expression compared to control. The animals treated with a combination of caspofungin and trimethoprim-sulfamethoxazole decreased the eng mRNA expression to approximately 50%. Treatment with trimethoprim-sulfamethoxazole alone did not result in any change of eng mRNA expression compared to the control. Similar results were seen when using caspofungin alone in a separate experiment (data not shown). Abbreviations: mRNA, messenger RNA, PCR, polymerase chain reaction.
Because caspofungin treatment dramatically reduces the number of cysts in P. murina–infected mice [24] and the expression of Eng appears to be localized to cysts, we determined whether the expression of Eng decreases with caspofungin treatment. We utilized lung samples from a study in which P. murina–infected animals were untreated or treated with caspofungin for 8 days plus trimethoprim-sulfamethoxazole for the last 3 days or trimethoprim-sulfamethoxazole alone for 3 days. Compared to the no-treatment control (Figure 4A, top panel), trimethoprim-sulfamethoxazole treatment alone for 3 days had no impact on the number of cysts or on Eng expression (Figure 4A, middle panel). In contrast, caspofungin plus trimethoprim-sulfamethoxazole treatment markedly reduced the number of cysts, and no Eng labeling was observed (Figure 4A, bottom panel). In a separate experiment, treatment with caspofungin alone also resulted in loss of Eng labeling (data not shown). These observations further support that Eng expression is limited to the cyst form of Pneumocystis.
We also examined whether eng messenger RNA (mRNA) expression in P. murina is decreased during caspofungin treatment of infected mice. Real-time PCR analysis demonstrated an approximately 50% decrease in eng mRNA expression in animals treated with a combination of caspofungin and trimethoprim-sulfamethoxazole, while treatment with trimethoprim-sulfamethoxazole alone did not result in any change of eng mRNA expression compared to the no-treatment control (Figure 4B). Treatment with caspofungin alone for 21 days resulted in an approximately 50% decrease in eng mRNA expression (data not shown).
DISCUSSION
We have characterized the endo-β-1,3-glucanase from 3 Pneumocystis species (P. carinii, P. murina, and P. jirovecii) and have shown that by immunofluorescence it is localized to cysts. The gene is transcribed as an approximately 2.6 kb mRNA that encodes a protein containing 786 amino acids in P. carinii and P. murina, and 788 amino acids in P. jirovecii. This protein shows homology to yeast Eng1 and Eng2 proteins and belongs to glycoside hydrolase family 81.
The cDNA sequence of P. carinii eng that we have identified differs at the 5′-end from the previously reported eng2 gene [12], which we were unable to clone or detect by Southern blot, while the P. carinii eng sequence reported here was supported by Southern and northern blot analyses. The reason for the differences between these 2 sequences is not clear, but potentially may be related to differences in the rat Pneumocystis strain or species (P. carinii vs P. wakefieldiae) used in our study and the previous study.
Eng1 and Eng2 are the 2 forms of endo-β-1,3-glucanase belonging to glycoside hydrolase family 81 (GH 81) that have been characterized from yeast [8, 9, 11]. Alignment of deduced amino acid sequences of Eng from Pneumocystis with Eng1 and Eng2 from yeast showed that a region of approximately 650 amino acids containing the catalytic sites characteristic of the GH 81 family is conserved in Pneumocystis [8]. In yeast, Eng1 is an extracellular protein with a signal peptide, while Eng2 is an intracellular protein with no signal peptide [9]. Of note, while the Eng proteins from all 3 Pneumocystis species showed greater identity with yeast Eng2 (34%–40%) than yeast Eng1 (23%–29%), P. carinii and P. murina Eng sequences contain signal peptides, similar to yeast Eng1. Interestingly, P. jirovecii Eng, like the previously reported P. carinii Eng [12], does not have a predicted signal peptide. Given that each Pneumocystis species has a single eng gene per genome, and that its relationship to the yeast eng genes is ambiguous, we have designated the endo-β-1,3-glucanase encoded by this gene as Eng, rather than Eng1 or Eng2.
Msg is the predominant cell surface protein expressed in Pneumocystis [15, 25, 26], and is encoded by a multicopy gene family that confers on Pneumocystis the potential for antigenic variation. We were interested in characterizing Pneumocystis Eng because Msg has been reported to be released from Pneumocystis by enzyme preparations containing β-1,3-glucanase from various sources [13, 14]. Consistent with these reports, the recombinant Eng prepared in the present study was also able to release Msg from Pneumocystis as demonstrated by Western blotting. Thus, one possible role for Eng in Pneumocystis is to facilitate release of Msg, which may allow a rapid change in the Msg isoform expressed by the organism.
Endo-β-1,3-glucanase is involved in cell separation in yeast [9, 11]. In S. cerevisiae, Eng1 and Eng2 deletion mutants showed defects in cell separation, resulting in the clumping of cells. It has been shown that the cell separation defect in S. cerevisiae Eng2 mutants can be rectified by complementing with Pneumocystis endo-β-1,3-glucanase [12].
Immunofluorescence analysis of P. murina–infected mouse lung tissue sections showed that Eng is exclusively expressed in the cyst form of Pneumocystis. This finding differs from the observation of Villegas et al [12] who demonstrated reactivity of P. carinii Eng with the trophic form by electron microscopy; this again may be related to differences in the Pneumocystis species studied or the methodologies utilized. Moreover, caspofungin treatment of P. murina–infected mice eliminated the reactivity of anti-Eng antibody, while greatly reducing the number of cyst forms, further supporting the expression of Eng exclusively in cysts. The mechanisms leading to this loss of reactivity are unclear, but may simply be related to cyst death, or alternatively may result from regulatory mechanisms associated with decreased synthesis of glucans in the presence of caspofungin.
Characterization of Eng from different Pneumocystis species further highlights the diversity in this family of organisms. Further studies of glucan metabolism as well as other metabolic pathways should provide insights into the unique biology of different members of this atypical fungal opportunist.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://jid.oxfordjournals.org). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgments. The authors thank the National Institute of Allergy and Infectious Diseases (NIAID) Research Technology Branch Bioimaging Section for their technical assistance, and Rene Costello and Howard Mostowski for providing animal care.
Financial support. This work was supported by the Intramural Research Programs of the National Institutes of Health (NIH) Clinical Center and the National Institute of Allergy and Infectious Diseases, NIH.
Potential conflicts of interest. All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Kovacs JA, Masur H. Evolving health effects of Pneumocystis: one hundred years of progress in diagnosis and treatment. JAMA. 2009;301:2578–85. doi: 10.1001/jama.2009.880. [DOI] [PubMed] [Google Scholar]
- 2.Thomas CF, Jr, Limper AH. Current insights into the biology and pathogenesis of Pneumocystis pneumonia. Nat Rev Microbiol. 2007;5:298–308. doi: 10.1038/nrmicro1621. [DOI] [PubMed] [Google Scholar]
- 3.Stringer JR, Stringer SL, Zhang J, Baughman R, Smulian AG, Cushion MT. Molecular genetic distinction of Pneumocystis carinii from rats and humans. J Eukaryot Microbiol. 1993;40:733–41. doi: 10.1111/j.1550-7408.1993.tb04468.x. [DOI] [PubMed] [Google Scholar]
- 4.Sinclair K, Wakefield AE, Banerji S, Hopkin JM. Pneumocystis carinii organisms derived from rat and human hosts are genetically distinct. Mol Biochem Parasitol. 1991;45:183–4. doi: 10.1016/0166-6851(91)90042-5. [DOI] [PubMed] [Google Scholar]
- 5.Mazars E, Guyot K, Durand I, et al. Isoenzyme diversity in Pneumocystis carinii from rats, mice, and rabbits. J Infect Dis. 1997;175:655–60. doi: 10.1093/infdis/175.3.655. [DOI] [PubMed] [Google Scholar]
- 6.Ma L, Imamichi H, Sukura A, Kovacs JA. Genetic divergence of the dihydrofolate reductase and dihydropteroate synthase genes in Pneumocystis carinii from 7 different host species. J Infect Dis. 2001;184:1358–62. doi: 10.1086/324208. [DOI] [PubMed] [Google Scholar]
- 7.Nollstadt KH, Powles MA, Fujioka H, Aikawa M, Schmatz DM. Use of beta-1,3-glucan-specific antibody to study the cyst wall of Pneumocystis carinii and effects of pneumocandin B0 analog L-733,560. Antimicrob Agents Chemother. 1994;38:2258–65. doi: 10.1128/aac.38.10.2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martin-Cuadrado AB, Fontaine T, Esteban PF, et al. Characterization of the endo-beta-1,3-glucanase activity of S. cerevisiae Eng2 and other members of the GH81 family. Fungal Genet Biol. 2008;45:542–53. doi: 10.1016/j.fgb.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 9.Baladron V, Ufano S, Duenas E, Martin-Cuadrado AB, del Rey F, Vazquez de Aldana CR. Eng1p, an endo-1,3-beta-glucanase localized at the daughter side of the septum, is involved in cell separation in Saccharomyces cerevisiae. Eukaryot Cell. 2002;1:774–86. doi: 10.1128/EC.1.5.774-786.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Encinar del Dedo J, Duenas E, Arnaiz Y, del Rey F, Vazquez de Aldana CR. {beta}-glucanase Eng2 is required for ascus wall endolysis after sporulation in the fission yeast Schizosaccharomyces pombe. Eukaryot Cell. 2009;8:1278–86. doi: 10.1128/EC.00148-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martin-Cuadrado AB, Duenas E, Sipiczki M, Vazquez de Aldana CR, del Rey F. The endo-beta-1,3-glucanase eng1p is required for dissolution of the primary septum during cell separation in Schizosaccharomyces pombe. J Cell Sci. 2003;116:1689–98. doi: 10.1242/jcs.00377. [DOI] [PubMed] [Google Scholar]
- 12.Villegas LR, Kottom TJ, Limper AH. Characterization of PCEng2, a {beta}-1,3-endoglucanase homolog in Pneumocystis carinii with activity in cell wall regulation. Am J Respir Cell Mol Biol. 2010;43:192–200. doi: 10.1165/rcmb.2009-0131OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Linke MJ, Cushion MT, Walzer PD. Properties of the major antigens of rat and human Pneumocystis carinii. Infect Immun. 1989;57:1547–55. doi: 10.1128/iai.57.5.1547-1555.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lundgren B, Lipschik GY, Kovacs JA. Purification and characterization of a major human Pneumocystis carinii surface antigen. J Clin Invest. 1991;87:163–70. doi: 10.1172/JCI114966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kovacs JA, Powell F, Edman JC, et al. Multiple genes encode the major surface glycoprotein of Pneumocystis carinii. J Biol Chem. 1993;268:6034–40. [PubMed] [Google Scholar]
- 16.Kovacs JA, Halpern JL, Swan JC, Moss J, Parrillo JE, Masur H. Identification of antigens and antibodies specific for Pneumocystis carinii. J Immunol. 1988;140:2023–31. [PubMed] [Google Scholar]
- 17.Cisse OH, Pagni M, Hauser PM. De novo assembly of the Pneumocystis jirovecii genome from a single bronchoalveolar lavage fluid specimen from a patient. mBio. 2012;4:e00428–12. doi: 10.1128/mBio.00428-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kutty G, Ma L, Kovacs JA. Characterization of the expression site of the major surface glycoprotein of human-derived Pneumocystis carinii. Mol Microbiol. 2001;42:183–93. doi: 10.1046/j.1365-2958.2001.02620.x. [DOI] [PubMed] [Google Scholar]
- 19.Kutty G, Achaz G, Maldarelli F, et al. Characterization of the meiosis-specific recombinase Dmc1 of Pneumocystis. J Infect Dis. 2010;202:1920–9. doi: 10.1086/657414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kutty G, Hernandez-Novoa B, Czapiga M, Kovacs JA. Pneumocystis encodes a functional S-adenosylmethionine synthetase gene. Eukaryot Cell. 2008;7:258–67. doi: 10.1128/EC.00345-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Linke MJ, Sunkin SM, Andrews RP, Stringer JR, Walzer PD. Expression, structure, and location of epitopes of the major surface glycoprotein of Pneumocystis carinii f. sp. carinii. Clin Diagn Lab Immunol. 1998;5:50–7. doi: 10.1128/cdli.5.1.50-57.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hernandez-Novoa B, Bishop L, Logun C, et al. Immune responses to Pneumocystis murina are robust in healthy mice but largely absent in CD40 ligand-deficient mice. J Leukoc Biol. 2008;84:420–30. doi: 10.1189/jlb.1207816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mei Q, Wang Q, Chen Y, Liu Y, Fan W, Li P. Experimental and clinical study on pneumocystosis. IV. Use of a 4D7 monoclonal antibody to detect Pneumocystis carinii by immunoperoxidase staining. Zhongguo Ji Sheng Chong Xue Yu Ji Sheng Chong Bing Za Zhi. 1995;13:130–3. [PubMed] [Google Scholar]
- 24.Cushion MT, Linke MJ, Ashbaugh A, et al. Echinocandin treatment of pneumocystis pneumonia in rodent models depletes cysts leaving trophic burdens that cannot transmit the infection. PLOS One. 2010;5:e8524. doi: 10.1371/journal.pone.0008524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haidaris PJ, Wright TW, Gigliotti F, Haidaris CG. Expression and characterization of a cDNA clone encoding an immunodominant surface glycoprotein of Pneumocystis carinii. J Infect Dis. 1992;166:1113–23. doi: 10.1093/infdis/166.5.1113. [DOI] [PubMed] [Google Scholar]
- 26.Garbe TR, Stringer JR. Molecular characterization of clustered variants of genes encoding major surface antigens of human Pneumocystis carinii. Infect Immun. 1994;62:3092–101. doi: 10.1128/iai.62.8.3092-3101.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.