Abstract
Multiple sclerosis (MS), a chronic disorder of the central nervous system and common cause of neurological disability in young adults, is characterized by moderate but complex risk heritability. Here we report the results of a genome-wide association study performed in a 1000 prospective case series of well-characterized individuals with MS and group-matched controls using the Sentrix® HumanHap550 BeadChip platform from Illumina. After stringent quality control data filtering, we compared allele frequencies for 551 642 SNPs in 978 cases and 883 controls and assessed genotypic influences on susceptibility, age of onset, disease severity, as well as brain lesion load and normalized brain volume from magnetic resonance imaging exams. A multi-analytical strategy identified 242 susceptibility SNPs exceeding established thresholds of significance, including 65 within the MHC locus in chromosome 6p21.3. Independent replication confirms a role for GPC5, a heparan sulfate proteoglycan, in disease risk. Gene ontology-based analysis shows a functional dichotomy between genes involved in the susceptibility pathway and those affecting the clinical phenotype.
INTRODUCTION
Multiple sclerosis (MS) is a chronic and often disabling neurological disease associated with inflammation, loss of myelin and ensuing axonal damage in the central nervous system (CNS) (1). It is a relatively common disease, affecting approximately 1–2 per 1000 individuals; ancestry, latitude, sex, and age influence risk and incidence (2,3). Etiologically, MS is characterized by a polygenic heritable component that involves multifaceted interactions with environmental factors (4). Two genes, HLA-DRB1 and IL7R (CD127), have been unambiguously associated with disease susceptibility and/or protection following their identification as candidates by function (5–9). Additional disease genes were discovered through genome-wide association analysis (10,11), most notably the independently replicated IL2RA (CD25), CD58 (LFA3), CLEC16A (KIAA0350) and EVI5 (12–15), demonstrating the power of this approach to identify even the modest genetic effects that contribute to this complex neurological trait.
Clinical manifestations of MS are diverse and vary from benign to a rapidly evolving and ultimately incapacitating disease. Concordance in families for some early and late clinical features suggests that in addition to susceptibility, genomic variants influence disease severity or other aspects of the phenotype (16–19). It has been difficult to discern, however, whether phenotypic diversity reflects true etiological heterogeneity (20), modifying roles of specific genes (19), or some combination of the two. Unfortunately, published reports proposing specific genetic influences on the natural history of MS relied almost exclusively on small series of cases, retrospective clinical assessment and, in some cases, non-validated phenotypic endpoints. Furthermore, the confounding effects of drug treatment and population stratification have not been generally considered. It is also important to recognize that the aggregate contribution of individual germ line variants to the disease course may be quite modest. This is highlighted by observations that the clinical expression of MS can be different between monozygotic twin siblings. Nevertheless, the demonstration of even a modest genetic effect of a known gene on the course of MS could help elucidate fundamental mechanisms of disease expression and yield a major therapeutic opportunity.
To identify additional common variants associated with MS risk and phenotype, we conducted a genome-wide association study (GWAS) using the Sentrix® HumanHap550 BeadChip platform from Illumina in a 1000 prospective case series of well-characterized individuals with MS and matched controls. After stringent quality control, we compared allele frequencies for 551 642 SNPs in 978 cases and 883 controls and assessed genotypic influences on susceptibility, age of onset, disease severity, brain lesion load and normalized brain volume.
RESULTS
A total of 2068 individuals were enrolled within a 12 month period and genotyped using the Illumina Sentrix® HumanHap550 BeadChip platform. Following quality control filtering of SNPs and samples, the final dataset included 978 MS cases and 883 controls of European ancestry. Approximately, half of these were enrolled by the US site, with one-quarter from each of the two European sites. To assess bias introduced by population stratification, the entire dataset and each case–control group were scanned for the presence on non-random distribution of alleles using STRUCTURE (v.2.) (21) and principal component analysis (data not shown). This analysis resulted in the exclusion of 36 individuals (15 cases and 21 controls) enrolled at the US site and three cases from the EU sites. We then assessed the median distribution of test statistics with the genomic control parameter λGC (22): San Francisco λGC =1.03, Basel λ GC =1.02, Amsterdam λ GC =1.01; all sites combined, λ GC =1.04. The results show that the variance inflation factors due to subpopulation structure within each cohort are less than 3%, and the pooled variance inflation factor is still less than 4%. In any case, origin of the sample was entered into the logistic regression model as a covariate to minimize the effects of the pooled variance inflation.
Table 1 lists demographic and clinical features of the study subjects. Overall, there was a high degree of demographic similarity among samples from the three sites, including frequency of HLA-DRB1*1501 carriers, but significant differences among sites were noted for some of the clinical variables used in the analysis of genotype–phenotype correlations. For all analyses, samples were pooled into a single dataset to increase statistical power (23).
Table 1.
Summary of demographic and clinical characteristics of study participants at the three sites at time of analysis
| Variable | Total |
San Francisco |
Amsterdam |
Basel |
||||
|---|---|---|---|---|---|---|---|---|
| Patients | Controls | Patients | Controls | Patients | Controls | Patients | Controls | |
| Female, n (%) | 653 (67) | 584 (66) | 334 (69) | 287 (66) | 137 (60) | 148 (62) | 182 (69) | 149 (71) |
| Male, n (%) | 325 (33) | 299 (34) | 153 (31) | 147 (34) | 92 (40) | 90 (38) | 80 (31) | 62 (29) |
| Female:Male ratio | 2.0:1 | 2.0:1 | 2.2:1 | 2.0:1 | 1.5:1 | 1.6:1 | 2.3:1 | 2.4:1 |
| Age at onset of disease, mean (range) | 34 (11–61) | 34 (11–61) | 35 (16–60) | 32 (11–59) | ||||
| Age at time of analysis, mean (range) | 47 (21–69) | 46 (21–74) | 46 (21–69) | 46 (23–69) | 48 (25–67) | 45 (24–68) | 47 (21–69) | 47 (21–74) |
| DRB1*1501+ individuals, n (%) | 474 (48) | 190 (22) | 224 (46) | 86 (20) | 118 (52) | 59 (25) | 132 (50) | 45 (21) |
| Disease duration (years) mean | 11.5 | 10.7 | 11.4 | 12.9 | ||||
| Median (range) | 9.0 (0–47) | 8.0 (0–46) | 10.0 (1–34) | 11.0 (0–47) | ||||
| Disease type, n (%) | ||||||||
| Relapsing remitting | 659 (67.9) | 343 (70.4) | 141 (61.8) | 175 (68.4) | ||||
| Secondary progressive | 137 (14.1) | 46 (9.5) | 46 (20.2) | 45 (17.5) | ||||
| Primary progressive | 72 (7.4) | 18 (3.7) | 29 (12.7) | 25 (9.8) | ||||
| Clinical isolated syndrome | 100 (10.3) | 79 (16.2) | 10 (4.4) | 11 (4.3) | ||||
| Unknown | 3 (0.3) | 1 (0.2) | 2 (0.9) | 0 (0) | ||||
| DMT, n (%)a | 432 (54) | 245 (63) | 60 (32) | 127 (58) | ||||
| EDSS, n (%)b | ||||||||
| <3 | 440 (55.4) | 279 (71.9) | 58 (31.0) | 103 (47.0) | ||||
| 3 to <6 | 271 (34.1) | 86 (22.2) | 95 (50.8) | 90 (41.1) | ||||
| 6–6.5 | 68 (8.6) | 22 (5.7) | 24 (12.8) | 22 (10.1) | ||||
| ≥7 | 15 (1.9) | 1 (0.3) | 10 (5.4) | 4 (1.8) | ||||
| Mean (SD) | 2.74 (1.81) | 2.06 (1.64) | 3.74 (1.72) | 3.07 (1.67) | ||||
| MSSS, nc | 794 | 388 | 187 | 219 | ||||
| Median | 2.97 | 2.23 | 4.82 | 3.17 | ||||
| Mean (SD) | 3.45 (2.37) | 2.70 (2.21) | 4.81 (2.23) | 3.61 (2.21) | ||||
| T2 lesion load/mm3, nd | 791 | 387 | 185 | 219 | ||||
| Median | 2580 | 2164 | 3001 | 3276 | ||||
| Range | 0–81317.2 | 0–64258.3 | 0–81317.2 | 0–34984.5 | ||||
| nBPV/cm3, ne | 753 | 371 | 176 | 206 | ||||
| Mean (SD) | 1530.8 (86.11) | 1511.6 (81.91) | 1554.5 (83.17) | 1544.9 (88.55) | ||||
| Range | 1225–1767 | 1225–1704 | 1360–1750 | 1309–1767 | ||||
aDisease modifying treatment (DMT). Test of association with site: χ2 = 50.64; P < 0.0001.
bExpanded disability status scale (EDSS). Test of association with site: χ2 = 102.17; P < 0.0001.
cMultiple sclerosis severity scale (MSSS). Test of variation among sites (Kruskal–Wallis test): χ2 = 107.82; P < 0.0001.
dT2 lesion load/mm3. Test of variation among sites (Kruskal–Wallis test): χ2 = 18.84; P < 0.0001.
eNormalized brain parenchymal volume (nBPV). ANOVA of variation among sites: F = 19.77; P < 0.0001.
a–e analyses include only subjects with relapsing remitting and secondary progressive disease.
Figure 1 shows the genome-wide trend association results. As expected, the strongest association signal observed in this experiment was in the MHC region in chromosome 6p21. Q–Q plots of the association results are shown in Figure 2. P-values without adjustment for gender, DRB1*1501 and site of sample origin, and the corresponding P-values with adjustment, are plotted for all SNPs as a function of the null distribution, showing an excess in the tail of the distribution at P < 1 × 10−3 (without adjustment) and P < 1 × 10−4 (with adjustment). Following adjustment, the most extreme P-values are still smaller than expected, consistent with the presence of true genetic associations. P-values for SNPs in the HLA region are highlighted, showing that without adjustment, the SNPs from this region are concentrated among the low P-values, whereas with adjustment, they are distributed over the full range of P-values. Logistic regression analysis of these case–control data after adjusting for gender, site and DRB1*1501 identified 242 SNPs with P ≤ 1 × 10−4, including 65 within the MHC locus (Supplementary Material, Table S1.1).
Figure 1.
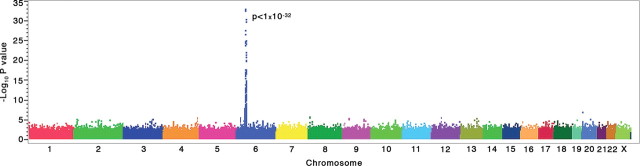
GWA trend test scan. Genome-wide association results for 551 642 SNPs in 978 MS cases and 883 controls using a linear trend test for additive allelic effects on disease penetrance (Cochran–Armitage trend test). As expected, the strongest association signal was located in the MHC region in chromosome 6p21.
Figure 2.

Q–Q plot distribution of the GWAS association results. P-values adjusted for HLA-DRB1*1501 and gender (A, trend test; B, genotypic test) are plotted for all SNPs as a function of the null distribution showing an excess in the tail of the distribution at P < 1 × 10−4. Red line is equal to expectation on H0. Grey/light green, P-values without adjustment for the HLA-DRB1 locus. Black/bright green, P-values with adjustment by including HLA-DRB1 as a covariate in the model. Green (light or bright), SNP loci in HLA region; black/grey, other SNPs.
The top peak SNPs located in the MHC class-II sub-region; rs3129934 (−Log10(P) 32.58) in the intron 3/4 of C6orf10, at the telomeric edge of the class II region, between BTNL2 and NOTCH4, rs9267992 (−Log10(P) 32.00) adjacent (telomeric) to LOC401252, rs9271366 (−Log10(P) 30.00) in the intergenic region between HLA-DRB1 and HLA-DQA1, and rs3129860 adjacent (telomeric) to HLA-DRA (Fig. 3A). It is likely that these signals reflect linkage disequilibrium (LD) with the well-established risk allele HLA-DRB1*1501, although following adjustment for gender and site, and upon conditional analysis for DRB1*1501, there is a residual effect in C6orf10 (−Log10(P) 4.00). The adjusted data also suggest the presence of independent association in the HLA-class I region (Fig. 3B), consistent with recent studies reporting multiple autonomous susceptibility signals in the locus (24,25). The top secondary signals are localized around TRIM 26, TRIM 15 and TRIM 10 members of the tripartite motif (TRIM) gene family. Although their functions are unknown, the presence of a RING domain suggests DNA-binding activity.
Figure 3.
Association and logistic regression analyses of the extended MHC region. Results of allelic tests of association (top panel) and results of subsequent logistic regression analyses conditioning upon HLA-DRB1*1501 (bottom panel).
Results for the non-HLA SNPs showing the strongest evidence of association (threshold of P ≤ 10−5) are shown in Table 2. These include 13 SNPs in eight chromosomes. Of special interest is the identification of Glypican5 (GPC5, 13q32), a heparan sulfate proteoglycan for which an IFNβ-associated pharmacogenomics role in MS was recently suggested (26). Also noteworthy are Tribbles Homolog 2 (TRB2) in 2p25.1-p24.3, a putative autoantigen in uveitis (27), and CUB and sushi multiple domains 1 (CSMD1) in 8p23.2, a complement regulatory protein highly expressed in the CNS (28). It is of interest to note that CMSD1 is located within a genomic region with a remarkable high mutation rate and as a consequence, is highly diverse between humans and non-human primates (29).
Table 2.
Top (non-HLA) markers associated with susceptibility, −Log10(P) ≥5.0
| SNP | Chrom | Position | GeneSymbol | Alleles | Minor allele frequency | Odds ratio | Adj Geno Log P-valuea | Adj Trend Log P-valueb |
|---|---|---|---|---|---|---|---|---|
| rs397020 | 20 | 1153886 | C20orf46 | A/G | 0.484 | 0.711 | 6.097 | 6.102 |
| rs9523762 | 13 | 92129887 | GPC5 | A/G | 0.346 | 1.361 | 5.155 | 5.921 |
| rs1458175 | 12 | 40252128 | PDZRN4 | G/T | 0.486 | 1.342 | 5.102 | 5.770 |
| rs1529316 | 8 | 3815546 | CSMD1 | C/T | 0.468 | 1.356 | 4.921 | 5.699 |
| rs2049306 | 8 | 3819488 | CSMD1 | A/C | 0.495 | 1.362 | 4.796 | 5.585 |
| rs908821 | 3 | 142023416 | SLC25A36 | C/T | 0.285 | 0.732 | 4.854 | 5.538 |
| rs1755289 | 9 | 17928351 | SH3GL2 | C/T | 0.389 | 0.739 | 5.041 | 5.523 |
| rs16914086 | 9 | 98068032 | TBC1D2 | A/G | 0.115 | 0.886 | 5.252 | 1.161 |
| rs651477 | 2 | 119111921 | EN1 | A/G | 0.263 | 1.378 | 4.699 | 5.143 |
| rs7672826 | 4 | 182774844 | MGC45800 | A/G | 0.341 | 1.366 | 4.886 | 5.102 |
| rs1841770 | 3 | 149239384 | ZIC1 | G/T | 0.470 | 0.746 | 4.310 | 5.076 |
| rs1109670 | 2 | 9200636 | DDEF2 | A/C | 0.256 | 1.381 | 4.000 | 5.046 |
| rs7607490 | 2 | 12801718 | TRIB2 | A/G | 0.115 | 1.558 | 4.000 | 5.013 |
aGenotypic test of association (the logistic regression model, adjusted for covariates).
bTrend test for association (the logistic regression model, adjusted for covariates).
Another gene of outstanding interest emerging from this scan is the aminophospholipid transporter (APLT), class I, type 8A, member 1 (ATP8A1), a member of a family of proteins driving uphill transport of ions across membranes. ATP8A1 also transports phosphatidylserine, a phospholipid component of myelin shown to be increased in the experimental autoimmune encepahlomyelitis brain, affecting the fluidity and integrity of the myelin sheath (30,31). In addition, phosphatidylserine on the surface of apoptotic cells serves as molecular addresses for the T-cell immunoglobulin mucin proteins for the effective clearance of apoptotic cells and prevention of autoimmunity (32).
Stratification by gender revealed additional genes of interest (Supplementary Material, Tables S1.2 and 1.3); 112 in the females only group, (39 intersecting with the all group and none with the males only group) and 60 in the males only group (nine intersecting with the all group). Some noteworthy examples include Tankyrase (TNKS) in females, a poly (ADP-ribose) polymerase associated with telomere length control and implicated in the regulation of EBV origin of plasmid replication (33), the prostaglandin receptor EP4 (PTGER4) in females, a gene recently associated with Crohn's disease (34), and the gamma-aminobutyric acid (GABA) A receptor beta 3 (GABRB3) in males, a gene coding for a subunit of a chloride channel that serves as the receptor for the gamma aminobutyric acid, a major inhibitory transmitter in the CNS and associated with Angelman syndrome, Prader–Willi syndrome and autism (35,36).
In order to overcome the potential bias for detection of false-positive associations in large genes with particularly dense SNP coverage, we applied the Šidák correction to the gene-wise minimum P-value. For each statistical and genetic model fitted to the case–control phenotype, values of Pmin without correction and Pmin, corrected, adjusted were obtained for 18 551 genes annotated in the Illumina array (counting different splice variants as different genes). The correlations among these gene-based measures of association are moderately strong (Supplementary Material, Table S2). The genes showing the strongest associations for each measure are, therefore, presented in Supplementary Material, Tables S3.1–3.4. The top MHC secondary signals (TRIM 26, TRIM 15 and TRIM 10) were also detected by the Šidák-corrected minimum P-value analysis of the data (Supplementary Material, Table S3). Outside chromosome 6p21, a group of genes located in chromosome 20p13 displayed robust association, including C20orf46, PSMF1 (Proteasome inhibitor subunit 1), SDCBP2 (Syndecan binding protein 2, or Syntenin) and SNPH (Syntaphylin or Synaptobrevin). The three top non-HLA hits identified in the screening, C20orf46, GPC5 and PCZRN4 (Table 2) survived this correction although GPC5 and PCZRN4 ranked lower than the genes mentioned above (Supplementary Material, Tables S3.1–3.4).
Given the lack of full-scale independent replication, these results were considered with caution. To add confidence to the associations reported here, we compared the results of the present analysis with those reported by the International Multiple Sclerosis Genetics Consortium (IMSGC) (11). SNPs showing support for association in both studies (thresholds P < 10−4) are shown in Table 3. R2 values between the SNPs in the different studies are, however, very low. Nevertheless, the joint association of PARK2 (P = 0.0004), coding for an E3 ubiquitin ligase, is of interest because of a demonstrated role in Parkinson disease (37).
Table 3.
Top (non-HLA) susceptibility markers overlapping between the current and IMSGC studies
| SNP | Chrom | Position | Gene | Alleles | Odds ratio | Adj Geno Log P-valuea | Adj Trend Log P-valuea | PTDT Log P-valueb | HapMap R2 |
|---|---|---|---|---|---|---|---|---|---|
| rs11582200a | 1 | 29810973 | PTPRU | A/C | 1.256 | 4.00 | 2.51 | – | 0.0001 |
| rs4949238b | 1 | 30091689 | PTPRU | A/C | 1.376 | – | – | 3.17 | |
| rs2144208a | 6 | 162020735 | PARK2 | A/G | 0.824 | 4.00 | 1.95 | – | 0.01 |
| rs9458281b | 6 | 161869691 | PARK2 | T/C | 0.706 | – | – | 3.40 | |
| rs10283372a | 8 | 74498045 | RDH10 | C/T | 1.146 | 4.72 | 0.94 | – | 0.02 |
| rs4738350 b | 8 | 74478622 | RDH10 | A/G | 0.469 | – | – | 3.40 | |
| rs2096735a | 11 | 96368652 | JRKL | C/T | 1.151 | 4.00 | 1.27 | – | 0.0001 |
| rs1941861b | 11 | 97548590 | JRKL | A/G | 1.396 | – | – | 3.00 | |
| rs10140464a | 14 | 48280389 | RPS29 | A/G | 0.707 | 4.00 | 4.68 | – | 0.0001 |
| rs10483556b | 14 | 49029992 | RPS29 | G/A | 0.740 | – | – | 2.89 |
aTop SNP in current study.
bTop SNP in IMSGC study (11).
Insofar genes reported by the Consortium and validated by others (11–15), a SNP within CD58 (rs1335532) ranked 16 094th (2.9 percentile) in the current study. The IMSGC SNP (rs12044852) was not represented in the Illumina platform, but according to HapMap data, both SNPs map to intron 10/11 and are in high LD (R2 = 0.93) (Table 4). This indicates high concordance in the results of both studies and suggests that resequencing the flanking exons may identify a functional causative polymorphism in CD58. IL7R ranked 10 167th in this study (1.8 percentile). The exon 6 SNP rs6897932 is represented in both platforms and provides the strongest association in both studies, further supporting a functional role in disease susceptibility (8). The top IL2RA SNP in this study (rs11256497) ranked 7230th (1.3 percentile). Similar to the IMSGC top SNP (rs12722489), it is located in intron 1/2 but they are in relatively low LD (R2 = 0.11). On the other hand, rs12722515 in this study, also in intron 1/2 is in high LD (R2 = 0.93) and confers an OR = 1.21, but ranks 95 421th, suggesting allelic heterogeneity and that the causative variants may be located in the 5′ region of the gene. The best CLEC16A SNP in the current study was rs11074952 in intron 21/22 and ranked 1020 (0.18 percentile). This SNP is in weak LD with the IMSGC SNP rs28087 in intron 22/23 (R2 = 0.24). In this study, the most tightly correlated SNP with the IMSGC is rs42369 (R2 = 0.71), but shows no association ranking 113 789th, indicating the need for additional fine mapping of this intriguing gene. All the top-associated IMSGC SNPs are within the 3.0 percentile of markers in this study. On the other hand, the top EVI5 marker in this study (rs10747446, intron 10/11) is in high LD (R2 = 0.92) with the IMSGC SNP (rs92833128, intron 10/11), but is not significantly associated and ranks 83 341th.
Table 4.
Performance of independently replicated IMSGC (non-HLA) Susceptibility markers in the current study
| SNP | Chrom | Position | Gene | Alleles | Odds ratio | Adj Geno Log P-valuea | Adj Trend Log P-valuea | PTDT Log P-valueb | HapMap R2 |
|---|---|---|---|---|---|---|---|---|---|
| rs10747446a,c | 1 | 92820423 | EVI5 | A/G | 1.00 | 0.99 | 0.12 | – | 0.92 |
| rs10735781b | 1 | 92833128 | EVI5 | C/G | 1.29 | – | – | 3.65 | |
| rs1335532a,c | 1 | 116812999 | CD58 | C/T | 0.78 | 1.53 | 1.78 | – | 0.93 |
| rs12044852b | 1 | 116799821 | CD58 | T/G | 0.68 | – | – | 3.00 | |
| rs6897932a,c | 5 | 35910332 | IL7R | C/T | 1.21 | 1.48 | 2.00 | – | – |
| rs6897932b | 5 | 35910332 | IL7R | T/C | 0.81 | – | – | 2.23 | |
| rs11256497a | 10 | 6127800 | IL2RA | A/G | 1.17 | 1.87 | 2.15 | – | 0.11 |
| rs12722489b | 10 | 6142018 | IL2RA | T/C | 0.74 | – | – | 2.88 | |
| rs12722525c | 10 | 6121236 | IL2RA | G/T | 1.21 | 0.69 | 0.93 | – | 0.93 |
| rs11074952a | 16 | 11126107 | CLEC16A | A/G | 0.80 | 2.59 | 3.10 | – | 0.24 |
| rs28087b | 16 | 11160330 | CLEC16A | G/A | 1.16 | – | – | 1.56 | |
| rs42369a | 16 | 11172113 | CLEC16A | A/G | 0.91 | 0.47 | 0.84 | – | 0.71 |
aTop SNP in current study.
bTop SNP in IMSGC study (11).
cMost tightly correlated SNP in current study with top IMSGC SNP.
SNP rs7321717, located approximately 1240 kb from GPC5, showed a PTD P-value of 0.0004 in the IMSGC study. The metrics of LD between rs9523762 (this study) and rs7321717 (IMSGC) in GPC5 is 0.00014 (R2) and 0.01189 (D′). We followed the GPC5 data by genotyping 3 SNPs in an independent MS group consisting 974 affected individuals, recruited in the US using identical inclusion criteria. This dataset was compared to the control group used in the original screen generating robust evidence of replication when gender and HLA-DRB1*1501 status were included in the logistic regression model (Table 5).
Table 5.
Replication of GPC5
| SNP | Allele, location | Wald Chi-square | Adjusted odds ratio (95% CI)a | Adj Trend Log P-value | Adj Geno Log P-value |
|---|---|---|---|---|---|
| rs553717 | G/A, exon 3 | 1.01 | A/G, OR = 1.12 (0.90–1.39) | 0.50 | 0.22 |
| rs9523762 | G/A, intron 7-8 | 8.34 | A/G, OR = 1.25 (1.07–1.45) | 2.42 | 1.82 |
| rs9516129 | C/T, intron 7-8 | 5.77 | T/C, OR = 1.23 (1.04–1.45) | 1.79 | 1.66 |
aAdditive model, covariates included in the logistic regression model: Gender, HLA-DRB1*1501(presence/absence).
Next, we performed a comprehensive statistical analysis to seek genome-wide associations with phenotypes related to the expression of MS. This analysis was restricted to individuals diagnosed as relapsing remitting or secondary progressive MS (N = 794) to limit the potential confounding influence of disease heterogeneity. Using regression analysis on the combined data from the three sites, and of data from each site separately with subsequent combination of the P-values by the Fisher's method, a number of allelic variants were modestly associated with the different phenotypic traits selected for this study (Supplementary Material, Tables S4–S7). Table 6 displays the top (P < 10−5) markers associated with the following phenotypic endpoints: age of onset, multiple sclerosis severity scale (MSSS), brain parenchymal volume (nBPV) and T2 lesion load.
Table 6.
Top markers associated with the MS phenotype, −Log10(P) ≥5.0
| SNP | Chrom | Position | GeneSymbol | Alleles | Minor allele frequency | Adj Geno Log P-value | Adj Trend Log P-value |
|---|---|---|---|---|---|---|---|
| Age of onset | |||||||
| rs1386330 | 11 | 87459075 | RAB38 | C/T | 0.13 | 5.82 | 6.66 |
| rs7432623 | 3 | 176222035 | NAALADL2 | A/G | 0.18 | 5.72 | 2.52 |
| rs404694 | 16 | 78140299 | MAF | T/G | 0.27 | 5.61 | 0.73 |
| rs17157903 | 7 | 103221987 | RELN | C/T | 0.14 | 4.06 | 5.49 |
| rs2116078 | 8 | 73526543 | KCNB2 | T/G | 0.48 | 5.00 | 5.48 |
| rs1557351 | 18 | 52903312 | WDR7 | C/T | 0.22 | 4.89 | 5.43 |
| rs2842483 | 9 | 76135704 | RFK | T/C | 0.29 | 4.57 | 5.32 |
| rs3804281 | 6 | 41853967 | FRS3 | C/T | 0.46 | 5.31 | 0.72 |
| rs12047808 | 1 | 176200971 | C1orf125 | A/G | 0.13 | 4.82 | 5.24 |
| rs5997184 | 22 | 25873695 | CRYBA4 | C/T | 0.09 | 5.20 | 2.01 |
| rs4704970 | 5 | 155433570 | SGCD | A/G | 0.20 | 3.59 | 5.13 |
| rs1437898 | 2 | 133580362 | FLJ34870 | G/T | 0.40 | 4.36 | 5.09 |
| rs2803418 | 9 | 76138828 | RFK | T/G | 0.26 | 4.37 | 5.08 |
| rs868824 | 7 | 109985872 | IMMP2L | C/T | 0.40 | 3.95 | 5.02 |
| rs7914524 | 10 | 129994857 | MKI67 | C/T | 0.17 | 5.01 | 3.19 |
| MSSS | |||||||
| rs12553535 | 9 | 12902861 | C9orf150 | G/T | 0.24 | 7.26 | 4.51 |
| rs752092 | 15 | 99599457 | CHSY1 | C/T | 0.37 | 5.84 | 1.69 |
| rs12638253 | 3 | 158108793 | FLJ16641 | C/T | 0.47 | 4.19 | 5.68 |
| rs1478091 | 4 | 132148106 | LOC132321 | C/T | 0.06 | 4.53 | 5.68 |
| rs10936043 | 3 | 158179208 | FLJ16641 | A/G | 0.46 | 4.16 | 5.66 |
| rs8043243 | 15 | 99588976 | CHSY1 | C/T | 0.42 | 5.53 | 2.34 |
| rs2028455 | 18 | 46220003 | C18orf24 | A/G | 0.31 | 5.47 | 3.44 |
| rs299175 | 19 | 61005340 | NALP11 | C/T | 0.46 | 4.28 | 5.37 |
| rs10259085 | 7 | 7041671 | C1GALT1 | C/T | 0.46 | 4.49 | 5.37 |
| rs10518025 | 4 | 67893206 | CENPC1 | C/T | 0.14 | 4.85 | 5.35 |
| rs2035213 | 4 | 132145387 | LOC132321 | A/C | 0.06 | 4.23 | 5.33 |
| rs16925027 | 9 | 6968321 | JMJD2C | A/G | 0.24 | 5.33 | 0.66 |
| rs6941421 | 6 | 15197130 | JARID2 | C/T | 0.39 | 4.56 | 5.24 |
| rs7191888 | 16 | 72138559 | C16orf47 | A/G | 0.17 | 4.20 | 5.23 |
| rs180358 | 11 | 116104602 | MGC13125 | G/A | 0.23 | 4.51 | 5.22 |
| rs10243024 | 7 | 115940554 | MET | A/G | 0.23 | 4.58 | 5.21 |
| rs10516537 | 4 | 107985675 | DKK2 | A/C | 0.15 | 5.10 | 3.77 |
| rs337718 | 18 | 67925258 | CBLN2 | T/C | 0.29 | 4.38 | 5.07 |
| rs7253363 | 19 | 11543495 | ACP5 | G/T | 0.05 | 4.66 | 5.05 |
| rs12142240 | 1 | 46459321 | LRRC41 | C/T | 0.29 | 4.52 | 5.00 |
| rs7211577 | 17 | 14055005 | COX10 | A/G | 0.49 | 4.96 | 5.00 |
| Brain parenchymal volume | |||||||
| rs4866550 | 5 | 3361312 | IRX1 | C/T | 0.32 | 6.06 | 2.09 |
| rs10078091 | 5 | 25530762 | CDH10 | A/G | 0.27 | 5.91 | 4.66 |
| rs368380 | 20 | 14762090 | C20orf133 | C/T | 0.33 | 5.73 | 1.47 |
| rs4473631 | 4 | 174876499 | MORF4 | A/C | 0.22 | 5.55 | 0.57 |
| rs1869410 | 2 | 5207954 | SOX11 | C/T | 0.28 | 5.40 | 3.70 |
| rs261902 | 12 | 32367994 | BICD1 | T/C | 0.16 | 4.42 | 5.36 |
| rs11719646 | 3 | 56442731 | CAST1 | A/G | 0.38 | 5.29 | 3.30 |
| rs1354913 | 11 | 89794461 | CHORDC1 | A/G | 0.18 | 5.23 | 0.16 |
| rs13067869 | 3 | 175148379 | NLGN1 | G/T | 0.07 | 5.20 | 4.83 |
| rs9307252 | 4 | 102359544 | PPP3CA | C/T | 0.14 | 5.18 | 4.16 |
| rs9480865 | 6 | 109023266 | FOXO3A | C/T | 0.16 | 4.52 | 5.19 |
| rs9486902 | 6 | 108984745 | FOXO3A | C/T | 0.16 | 4.52 | 5.12 |
| rs1927457 | 10 | 30048669 | SVIL | C/T | 0.31 | 4.69 | 5.11 |
| rs716595 | 10 | 111996476 | MXI1 | A/G | 0.08 | 3.81 | 5.10 |
| rs11957313 | 5 | 169882972 | KCNIP1 | A/G | 0.13 | 3.73 | 5.07 |
| rs9319189 | 13 | 85516099 | SLITRK6 | A/G | 0.35 | 5.01 | 2.54 |
| rs10917727 | 1 | 160028398 | CDCA1 | C/T | 0.47 | 5.00 | 4.28 |
| T2 lesion load | |||||||
| rs12097667 | 1 | 94209 | PLD5 | A/C | 0.15 | 6.38 | 0.73 |
| rs1806468 | 7 | 212245 | KIAA1706 | C/T | 0.11 | 5.65 | 4.13 |
| rs146250 | 6 | 158286 | GPR126 | A/G | 0.09 | 5.55 | 1.66 |
| rs263153 | 6 | 279431 | HIVEP2 | C/A | 0.09 | 5.55 | 1.74 |
| rs6794496 | 3 | 422592 | NPHP3 | C/T | 0.13 | 5.47 | 4.80 |
| rs2602397 | 2 | 277843 | CHRND | C/T | 0.45 | 4.48 | 5.39 |
| rs6899560 | 6 | 429349 | FUT9 | A/G | 0.05 | 5.26 | 2.56 |
| rs2039485 | 14 | 232964 | NUBPL | C/T | 0.22 | 4.63 | 5.22 |
| rs305124 | 4 | 303690 | HIP2 | C/T | 0.07 | 5.20 | 0.68 |
| rs6917747 | 6 | 430506 | IGF2R | A/G | 0.15 | 4.72 | 5.17 |
| rs11666377 | 19 | 74887 | CPAMD8 | C/T | 0.14 | 4.42 | 5.15 |
| rs12202350 | 6 | 98302 | IGF2R | C/T | 0.09 | 5.10 | 4.71 |
| rs6512158 | 19 | 405242 | CPAMD8 | A/C | 0.14 | 4.29 | 5.02 |
In order to explore the potential functionality of the associated SNPs across phenotypes, we conducted a gene ontology (GO) analysis of the most significant markers to assess enrichment in any of the pre-defined functional categories (Biological process, molecular function and cellular component) using a GO tree machine (Vanderbilt University) (Table 7). In this analysis, we included genes containing at least one SNP with a P-value <1 × 10−4 for each of the traits (Supplementary Material, Tables S1.1, 4–7). To avoid redundancy, only the most specific of any given set of nested significant GO categories is reported. Antigen processing and presentation were among the most significantly enriched GO categories for disease susceptibility, together with, surprisingly, CNS development. On the other hand, neural processes including axon guidance and glutamate signaling were over-represented in phenotypes directly related to CNS damage (i.e. T2 lesion load and brain volume, respectively). Cellular mechanisms were primarily represented in time-dependent processes such as MSSS and age of onset.
Table 7.
Gene ontology analysis
| GO (Biological process) | n | Genes (observed) | Genes (expected) | Ratio (O/E) | P-value |
|---|---|---|---|---|---|
| Susceptibility | |||||
| CNS development | 6 | MOG, PARK2, SH3GL2, ZIC1, CHST9, JRKL | 0.52 | 11.5 | 0.00001 |
| Organ morphogenesis | 5 | SPRY2, CITED2, ABLIM1, NPR1, ZIC1 | 0.98 | 5.1 | 0.003 |
| Antigen presentation (MHC class I) | 3 | HLA-A, HLA-B, HLA-G | 0.09 | 33.3 | 0.00009 |
| Antigen presentation (MHC class II) | 3 | HLA-DPB1, HLA-DQA2, HLA-DQB1 | 0.08 | 35.7 | 0.00006 |
| MSSS | |||||
| Protein amino acid N-linked Glycosylation | 2 | FUT8, TM4SF4 | 0.15 | 13.3 | 0.009 |
| Cellular respiration | 2 | ME3, COX10 | 0.15 | 13.3 | 0.009 |
| Embryonic development | 2 | FUT8, KLF4 | 0.1 | 20.0 | 0.005 |
| Age of onset | |||||
| Cell adhesion | 9 | CDH12, DLG1, CNTN6, OPCML, PCDH10, TPBG, PPFIBP1,CASK PSCD1 | 3.3 | 2.8 | 0.005 |
| Signal transduction | 25 | FRS3, RASSF8, PDZD8, CPE, DAPK1, DOCK1, EDNRB, DKK1, RASD2, RAB38, RASGRP3, CNTN6, GRIK1, HTR7, KDR, OR51B6, OR51M1, OR51I1, PDE4D, PDE6A, RGR, VIP, SPSB1, IRS2, PSCD1 | 16.1 | 1.6 | 0.009 |
| CNS development | 4 | CNTN6, GRIK1, PBX1, PCP4 | 0.63 | 6.4 | 0.003 |
| Excretion | 3 | VIP, NPHS2, KCNK5 | 0.22 | 13.6 | 0.001 |
| Brain parenchymal volume | |||||
| Glutamate signaling pathway | 2 | GRIN2A, HOMER2 | 0.07 | 28.6 | 0.002 |
| T2 Lesion load | |||||
| Calcium-mediated signaling | 3 | EGFR, PIP5K3, MCTP2 | 0.11 | 27.2 | 0.0002 |
| G-protein signaling | 3 | DGKG, EDNRB, EGFR | 0.29 | 10.3 | 0.003 |
| Axon guidance | 2 | SLIT2, NRXN1 | 0.12 | 16.7 | 0.007 |
| Hemopoiesis | 3 | JAG1, LRMP, BCL11A | 0.44 | 6.8 | 0.009 |
| Regulation of cell migration | 2 | JAG1, EGFR | 0.13 | 15.4 | 0.008 |
| Amino acid metabolism | 5 | EGFR, MSRA, SLC6A6, UBE1DC1, SLC7A5 | 1.06 | 4.7 | 0.004 |
DISCUSSION
We report the largest GWAS performed so far in a deeply phenotyped MS dataset. The recently reported IMSGC study screened 931 trios with 334 923 SNPs (11). Here we used a case–control design (n = 978 versus 883) with higher SNP density (551 642 SNPs). In contrast to the IMSGC study, which included a second replication stage of top susceptibility hits and candidate genes, the emphasis of this study was to isolate both, disease risk and modifiers. The present scan has identified a number of novel non-HLA candidate genes associated both with susceptibility and with clinical manifestations of the disease. As expected, each significant association has a very modest effect and reflects a small share of the genetic variance affecting MS risk and progression.
Among the most interesting association results is the glypican proteoglycan 5 (GPC5) gene. Glypicans are a class of heparan sulfate proteoglycans bound to the external surface of plasma membranes that play an important role in the control of cell division and growth regulation, but are also implicated in brain pattering, synapse formation, axon regeneration and guidance, and are found in dense networks in active MS plaques, where they may be involved in sequestering pro-inflammatory chemokines (38–42). GPC5 is expressed in neurons (43), and its interaction with growth factors, chemokines and extracellular matrix proteins may affect neuronal growth and repair (44). Recently, we reported GPC5 allelic differences associated with extreme responses to IFNβ in MS patients (26). It is conceivable that a single gene participates in both traits, conferring susceptibility and affecting the phenotype (in this case, a pharmacogenomic response). Alternatively, it is tantalizing to speculate that drug response differences in MS reflect, at least in part, singular etiologies characterized by different genetic backgrounds.
A second gene of interest emerging both from this study and the IMSGC GWAS is PARK2 or Parkin (6q25.2–27), a component of a multiprotein E3 ubiquitin ligase complex that mediates the targeting of substrate proteins for proteasomal degradation. A 4.5 kb transcript is expressed in many human tissues but is abundant in the brain. Mutations in this gene cause a loss of function which may compromise the poly-ubiquitination and proteasomal degradation of specific protein substrates, potentially leading to their deleterious accumulation in autosomal recessive juvenile-onset parkinsonism (37). Parkin mutations also affect mitochondrial function and apoptosis in neuronal cells, processes of importance in MS pathogenesis (45). Interestingly, variations within this gene were found to be associated with susceptibility to leprosy and other infectious diseases (46,47). An important caveat, however, is the very dense coverage of this gene in the Sentrix® HumanHap550 BeadChip (455 SNPs), increasing the possibility for potential type I errors. The signal in PARK2 did not survive the Šidák correction, suggesting the need for a conservative interpretation of its role in MS susceptibility.
The genes that we have associated with the MS phenotype are related to a broad range of cellular functions, but we did not find the expected high representation of genes mediating immunological effector functions. One exception is the association between NALp11 and the severity metric MSSS. NALPs are implicated in the activation of proinflammatory caspases via their involvement in multiprotein complexes called inflammasomes (48). It is striking that RELN (reelin), a gene also implicated in susceptibility to schizophrenia, autism, bipolar disorder, depression, and temporal lobe epilepsy (49–51), shows significant association with age of onset in MS (i.e. first clinical expression of disease). We speculate that the observed effect on age of onset is related to the role of reelin on neuronal survival (52) and layering of neurons in the cerebral cortex and cerebellum, affecting perhaps the threshold of neuronal plasticity required to avoid the clinical manifestation of the neurological damage (53). The associations between NLGN1 (neuroligin 1), HIP2 (huntingtin interacting protein 2) and CDH10 (cadherin 10) with T2 lesion load and brain atrophy, respectively, are consistent with roles for factors facilitating ‘homeostatic’ maintenance of brain function in disease.
Deconstructing the genetic events leading to MS will clarify its pathogenesis and improve opportunities to treat, prevent and cure this disease. This study increases the resolution of the MS genomic map by updating the roster of candidate disease susceptibility and progression loci. The GO summary analysis reflects the compartmentalization of the genetic involvement in the susceptibility versus the neurodegenerative phases of this disease.
MATERIALS AND METHODS
Study participants
The study cohort was ascertained through a prospective multi-center effort initiated in 2003. Three MS clinical centers were involved in patient enrollment and biological specimen collection using identical inclusion criteria, two in Europe (Vrije Universiteit Medical Center, Amsterdam; and University Hospital Basel) and one in the USA (University of California San Francisco). This study recruited patients of northern-European ancestry, preferentially with a relapsing onset of MS, although individuals with all clinical subtypes of the disease participated, including clinically isolated syndrome (CIS), relapsing remitting MS (RRMS), secondary progressive MS (SPMS), primary progressive MS (PPMS) and progressive relapsing MS (PRMS). CIS was defined as the first well-established neurological event lasting ≥48 h, involving optic nerve, spinal cord, brainstem or cerebellum. In CIS patients, the presence of two or more hyperintense lesions on a T2-weighted magnetic resonance imaging (MRI) sequence was also required for enrollment into the study. The diagnosis of RRMS was made utilizing the new International Panel criteria (54,55). SPMS was defined by 6 months of worsening neurological disability not explained by clinical relapse. PPMS was defined both by progressive clinical worsening for more than 12 months from symptom onset without any relapses, and abnormal cerebrospinal fluid as defined by the presence of ≥2 oligoclonal bands or an elevated IgG index. If acute relapses were superimposed on this steadily progressive course, patients were considered to have PRMS. Individuals were excluded if they had experienced a clinical relapse or received treatment with glucocorticosteroids within the previous month of enrollment. The concomitant use of disease modifying therapies for MS was permitted. For all subjects, the expanded disability status scale (56) and MSSS (57) scores were assessed, and brain MRI scans were performed within 2 weeks of entry into the study. In this study, age of onset was defined as the first episode of focal neurological dysfunction suggestive of CNS demyelinating disease (17). This information was obtained via individual recall and verified through review of medical records. The control group consisted of unrelated individuals, primarily spouses/partners, friends and other volunteers. Control subjects were of northern-European ancestry and matched as a group, proportionally with cases according to age (±5 years), gender. A familial history or current diagnosis of MS as well as a relation to another case or control subject were considered exclusionary for this group. Protocols were approved by the Committees on Human Research at all Institutions and informed consent was obtained from all participants prior to participation in the study.
Image acquisition
Brain MRI scans were performed on all cases upon entry into the study in 1.5 (Europe) and 3 (US) Tesla instruments using common sequences and protocols for data acquisition. MRIs were analyzed centrally at the project-MRI Evaluation Center (Basel) without the knowledge of disease subtype, duration or treatment history. Conventional spin echo, T1-weighted images were acquired for 5 min following administration of a single dose (0.1 mm/kg) of contrast agent (TE/TR = 8/467 ms, 256 × 256 × 44 matrix, 240 × 240 × 132 FOV, NEX = 1). Qualitative analysis for the presence of gadolinium enhancement was performed on post-contrast T1-weighted images. Brain lesions were identified and marked by consensus reading on simultaneously viewed T2 long and proton density-weighted images. Additional evaluations were done for new T2 lesions and volume of black holes, volume of T2 and T1 gadolinium enhanced lesions. The volumes were measured using an interactive digital analysis program (AMIRA).
The SIENAX algorithm (an adaptation of SIENA—Structural Image Evaluation using Normalization of Atrophy—for cross-sectional measurements) was used to estimate whole nBPV, normalized for subject head size (58,59). SIENAX extracts brain and skull images from the single structural acquisition. The brain image is registered to a standard space using the skull image to determine the registration scaling. Tissue segmentation, with partial volume estimation, is carried out to calculate total brain volume. SIENAX software are freely available as part of the FMRIB Software Library (www.fmrib.ox.ac.uk/fsl).
Genotyping, quality control and statistical analysis
Genotyping of the complete dataset was performed at the Illumina facilities using the Sentrix® HumanHap550 BeadChip. Sample success rate was 98.2% and the overall call rate was 99.85%. In addition to sample genotyping efficiency, a five-point quality control procedure was implemented before analysis, including assessment of marker allelic frequency, Hardy–Weinberg equilibrium, gender consistency, reproducibility and population genetic structure. Association analysis was carried out on 551 642 SNPs in 978 cases and 883 controls.
DRB1*1501 genotyping was performed using a validated gene-specific TaqMan assay designed to identify, specifically, the presence or absence of DRB1*1501 and/or *1503 alleles. An internal positive control (ß-globin) was included in each well to confirm that the reaction amplified successfully. PCR was carried out in a total volume of 10 µl, containing 20 ng DNA, 1x TaqMan Universal PCR Master Mix (Applied Biosystems), 0.6µM DRB1*1501/1503 specific primers (forward 5′;-ACG TTT CCT GTG GCA GCC TAA-3′, reverse 5′-TGC ACT GTG AAG CTC TCC ACA A-3′), 0.3 µm control primers (forward 5′-ACT GGG CAT GTG GAG ACA GAG AA-3′, reverse 5′-AGG TGA GCC AGG CCA TCA CTA AA-3′), 0.225 µm VIC-labeled DRB1*1501/1503 specific probe (5′-AAC AGC CAG AAG GAC ATC CTG GAG CA-3′) and 0.025 µm 6FAM-labeled control probe (5′-TCT ACC CTT GGA CCC AGA GGT TCT TTG AGT-3′). Amplification was carried out in an ABI PRISM 7900HT Sequence Detection System (Applied Biosystems) with an initial 95°C for 10 min, followed by 50 cycles of 95°C for 15 s and 62°C for 1 min. Results were confirmed with tagging SNP rs3135388 (60).
Two statistical methods were used to account for different models of genetic effect (1): the genotype-based case–control test for testing the dominant/additive/recessive allele effects on disease (61), and (2) a linear trend test for additive allelic effects on disease penetrance [Cochran–Armitage trend test (62)], with an asymptotic χ2 distribution (1 d.f.). In order to address the problem of multiple testing, we implemented the false discovery rate (FDR) method (63). Logistic regression models were used to incorporate multiple covariates in the analysis of the transformed data, including gender, center of sample origin and DRB1*1501 status.
To correct for a potential bias towards detection of false-positive associations in large genes, the Šidák-corrected minimum P-value (64) for each gene, adjusted for LD, was obtained using the formula:
Pmin, corrected = 1−(1−Pmin)Meff, where Pmin is the minimum P-value obtained from the SNPs tested within the gene under consideration, for association with a particular phenotype using a particular statistical and genetic model. Meff is the effective number of SNPs tested within the gene, obtained by the method of Li and Ji (65), which is based on the eigenvalues of the correlation matrix (LD matrix) among the SNPs in the gene (66,67).
If the LD between every pair of SNPs tested within the gene is zero, then Meff is equal to M, the actual number of SNPs tested.
For the genotype–phenotype correlation analysis, four different phenotypes were assessed: T2 lesion load, nBPV, MSSS and age of onset. Covariates included in the model were gender, center of sample origin, age onset, disease duration, treatment status and treatment duration. Transformation of the phenotypic data (cube-root transformation for T2 lesion, square-root transformation for MSSS and log transformation for age of onset) was performed prior to data analysis in order to meet the normality assumption for the regression model. We also performed analyses separately at each site, using the Fisher's method to combine the P-values over sites.
SUPPLEMENTARY MATERIAL
FUNDING
Recruitment of study participants, clinical and MRI assessment and genome scan genotyping was funded by GlaxoSmithKline, of which several authors are employees.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the MS patients and healthy controls who participated in this study. We also thank the IMSGC for access to their GWAS unpublished data.
Conflict of Interest statement. Authors declare no conflict of interest.
REFERENCES
- 1.Hauser S.L., Goodin D.S. Multiple sclerosis and other demyelinating diseases. In: Fauci A.D., Kasper D.L., Braunwald E., Hauser S.L., Longo D.L., Jameson J.L., editors. Harrison's Principle of Internal Medicine. 17th edn. NY: McGraw Hill; 2008. [Google Scholar]
- 2.Compston A., Coles A. Multiple sclerosis. Lancet. 2002;359:1221–1231. doi: 10.1016/S0140-6736(02)08220-X. [DOI] [PubMed] [Google Scholar]
- 3.Pugliatti M., Sotgiu S., Rosati G. The worldwide prevalence of multiple sclerosis. Clin. Neurol. Neurosurg. 2002;104:182–191. doi: 10.1016/s0303-8467(02)00036-7. [DOI] [PubMed] [Google Scholar]
- 4.Hauser S.L., Oksenberg J.R. The neurobiology of multiple sclerosis: genes, inflammation, and neurodegeneration. Neuron. 2006;52:61–76. doi: 10.1016/j.neuron.2006.09.011. [DOI] [PubMed] [Google Scholar]
- 5.Olerup O., Hillert J. HLA class II-associated genetic susceptibility in multiple sclerosis: a critical evaluation. Tissue Antigens. 1991;38:1–15. doi: 10.1111/j.1399-0039.1991.tb02029.x. [DOI] [PubMed] [Google Scholar]
- 6.Teutsch S.M., Booth D.R., Bennetts B.H., Heard R.N., Stewart G.J. Identification of 11 novel and common single nucleotide polymorphisms in the interleukin-7 receptor-alpha gene and their associations with multiple sclerosis. Eur. J. Hum. Genet. 2003;11:509–515. doi: 10.1038/sj.ejhg.5200994. [DOI] [PubMed] [Google Scholar]
- 7.Oksenberg J.R., Barcellos L.F., Cree B.A., Baranzini S.E., Bugawan T.L., Khan O., Lincoln R.R., Swerdlin A., Mignot E., Lin L., et al. Mapping multiple sclerosis susceptibility to the HLA-DR locus in African Americans. Am. J. Hum. Genet. 2004;74:160–167. doi: 10.1086/380997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gregory S.G., Schmidt S., Seth P., Oksenberg J.R., Hart J., Prokop A., Caillier S.J., Ban M., Goris A., Barcellos L.F., et al. Interleukin 7 receptor alpha chain (IL7R) shows allelic and functional association with multiple sclerosis. Nat. Genet. 2007;39:1083–1091. doi: 10.1038/ng2103. [DOI] [PubMed] [Google Scholar]
- 9.Lundmark F., Duvefelt K., Iacobaeus E., Kockum I., Wallstrom E., Khademi M., Oturai A., Ryder L.P., Saarela J., Harbo H.F., et al. Variation in interleukin 7 receptor alpha chain (IL7R) influences risk of multiple sclerosis. Nat. Genet. 2007;39:1108–1113. doi: 10.1038/ng2106. [DOI] [PubMed] [Google Scholar]
- 10.Burton P.R., Clayton D.G., Cardon L.R., Craddock N., Deloukas P., Duncanson A., Kwiatkowski D.P., McCarthy M.I., Ouwehand W.H., Samani N.J., et al. Association scan of 14,500 nonsynonymous SNPs in four diseases identifies autoimmunity variants. Nat. Genet. 2007;39:1329–1337. doi: 10.1038/ng.2007.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.International Multiple Sclerosis Genetics Cosnortium. Risk alleles for multiple sclerosis identified by a genomewide study. N. Engl. J. Med. 2007;357:851–862. doi: 10.1056/NEJMoa073493. [DOI] [PubMed] [Google Scholar]
- 12.Alcina A., Fedetz M., Ndagire D., Fernandez O., Leyva L., Guerrero M., Arnal C., Delgado C., Matesanz F. The T244I variant of the interleukin-7 receptor-alpha gene and multiple sclerosis. Tissue Antigens. 2008;72:158–161. doi: 10.1111/j.1399-0039.2008.01075.x. [DOI] [PubMed] [Google Scholar]
- 13.Hoppenbrouwers I.A., Aulchenko Y.S., Ebers G.C., Ramagopalan S.V., Oostra B.A., van Duijn C.M., Hintzen R.Q. EVI5 is a risk gene for multiple sclerosis. Genes Immun. 2008;9:334–337. doi: 10.1038/gene.2008.22. [DOI] [PubMed] [Google Scholar]
- 14.Rubio J.P., Stankovich J., Field J., Tubridy N., Marriott M., Chapman C., Bahlo M., Perera D., Johnson L.J., Tait B.D., et al. Replication of KIAA0350, IL2RA, RPL5 and CD58 as multiple sclerosis susceptibility genes in Australians. Genes Immun. 2008;9:624–630. doi: 10.1038/gene.2008.59. [DOI] [PubMed] [Google Scholar]
- 15.Weber F., Fontaine B., Cournu-Rebeix I., Kroner A., Knop M., Lutz S., Muller-Sarnowski F., Uhr M., Bettecken T., Kohli M., et al. IL2RA and IL7RA genes confer susceptibility for multiple sclerosis in two independent European populations. Genes Immun. 2008;9:259–263. doi: 10.1038/gene.2008.14. [DOI] [PubMed] [Google Scholar]
- 16.Brassat D., Azais-Vuillemin C., Yaouanq J., Semana G., Reboul J., Cournu I., Mertens C., Edan G., Lyon-Caen O., Clanet M., et al. Familial factors influence disability in MS multiplex families. Neurol. 1999;52:1632–1636. doi: 10.1212/wnl.52.8.1632. [DOI] [PubMed] [Google Scholar]
- 17.Barcellos L.F., Oksenberg J.R., Green A.J., Bucher P., Rimmler J.B., Schmidt S., Garcia M.E., Lincoln R.R., Pericak-Vance M.A., Haines J.L., et al. Genetic basis for clinical expression in multiple sclerosis. Brain. 2002;125:150–158. doi: 10.1093/brain/awf009. [DOI] [PubMed] [Google Scholar]
- 18.Hensiek A.E., Seaman S.R., Barcellos L.F., Oturai A., Eraksoi M., Cocco E., Vecsei L., Stewart G., Dubois B., Bellman-Strobl J., et al. Familial effects on the clinical course of multiple sclerosis. Neurology. 2007;68:376–383. doi: 10.1212/01.wnl.0000252822.53506.46. [DOI] [PubMed] [Google Scholar]
- 19.DeLuca G.C., Ramagopalan S.V., Herrera B.M., Dyment D.A., Lincoln M.R., Montpetit A., Pugliatti M., Barnardo M.C., Risch N.J., Sadovnick A.D., et al. An extremes of outcome strategy provides evidence that multiple sclerosis severity is determined by alleles at the HLA-DRB1 locus. Proc. Natl Acad. Sci. USA. 2007;104:20896–20901. doi: 10.1073/pnas.0707731105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lennon V.A., Wingerchuk D.M., Kryzer T.J., Pittock S.J., Lucchinetti C.F., Fujihara K., Nakashima I., Weinshenker B.G. A serum autoantibody marker of neuromyelitis optica: distinction from multiple sclerosis. Lancet. 2004;364:2106–2112. doi: 10.1016/S0140-6736(04)17551-X. [DOI] [PubMed] [Google Scholar]
- 21.Falush D., Stephens M., Pritchard J.K. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Devlin B., Roeder K., Wassemank L. Genomic control, a new approach to genetic based association studies. Theor. Pop. Biol. 2002;60:155–166. doi: 10.1006/tpbi.2001.1542. [DOI] [PubMed] [Google Scholar]
- 23.Skol A.D., Scott L.J., Abecasis G.R., Boehnke M. Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat. Genet. 2006;38:209–213. doi: 10.1038/ng1706. [DOI] [PubMed] [Google Scholar]
- 24.Burfoot R.K., Jensen C.J., Field J., Stankovich J., Varney M.D., Johnson L.J., Butzkueven H., Booth D., Bahlo M., Tait B.D., et al. SNP mapping and candidate gene sequencing in the class I region of the HLA complex: searching for multiple sclerosis susceptibility genes in Tasmanians. Tissue Antigens. 2008;71:42–50. doi: 10.1111/j.1399-0039.2007.00962.x. [DOI] [PubMed] [Google Scholar]
- 25.Yeo T.W., De Jager P.L., Gregory S.G., Barcellos L.F., Walton A., Goris A., Fenoglio C., Ban M., Taylor C.J., Goodman R.S., et al. A second major histocompatibility complex susceptibility locus for multiple sclerosis. Ann. Neurol. 2007;61:228–236. doi: 10.1002/ana.21063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Byun E., Caillier S.J., Montalban X., Villoslada P., Fernandez O., Brassat D., Comabella M., Wang J., Barcellos L.F., Baranzini S.E., et al. Genome-wide pharmacogenomic analysis of the response to interferon beta therapy in multiple sclerosis. Arch. Neurol. 2008;65:337–344. doi: 10.1001/archneurol.2008.47. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Y., Davis J.L., Li W. Identification of tribbles homolog 2 as an autoantigen in autoimmune uveitis by phage display. Mol. Immunol. 2005;42:1275–1281. doi: 10.1016/j.molimm.2004.11.020. [DOI] [PubMed] [Google Scholar]
- 28.Kraus D.M., Elliott G.S., Chute H., Horan T., Pfenninger K.H., Sanford S.D., Foster S., Scully S., Welcher A.A., Holers V.M. CSMD1 is a novel multiple domain complement-regulatory protein highly expressed in the central nervous system and epithelial tissues. J. Immunol. 2006;176:4419–4430. doi: 10.4049/jimmunol.176.7.4419. [DOI] [PubMed] [Google Scholar]
- 29.Nusbaum C., Mikkelsen T.S., Zody M.C., Asakawa S., Taudien S., Garber M., Kodira C.D., Schueler M.G., Shimizu A., Whittaker C.A., et al. DNA sequence and analysis of human chromosome 8. Nature. 2006;439:331–335. doi: 10.1038/nature04406. [DOI] [PubMed] [Google Scholar]
- 30.Smith M.E. Phagocytosis of myelin in demyelinative disease: a review. Neurochem. Res. 1999;24:261–268. doi: 10.1023/a:1022566121967. [DOI] [PubMed] [Google Scholar]
- 31.Ohler B., Graf K., Bragg R., Lemons T., Coe R., Genain C., Israelachvili J., Husted C. Role of lipid interactions in autoimmune demyelination. Biochim. Biophys. Acta. 2004;1688:10–17. doi: 10.1016/j.bbadis.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 32.Kobayashi N., Karisola P., Pena-Cruz V., Dorfman D.M., Jinushi M., Umetsu S.E., Butte M.J., Nagumo H., Chernova I., Zhu B., et al. TIM-1 and TIM-4 glycoproteins bind phosphatidylserine and mediate uptake of apoptotic cells. Immunity. 2007;27:927–940. doi: 10.1016/j.immuni.2007.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deng Z., Atanasiu C., Zhao K., Marmorstein R., Sbodio J.I., Chi N.W., Lieberman P.M. Inhibition of Epstein-Barr virus OriP function by tankyrase, a telomere-associated poly-ADP ribose polymerase that binds and modifies EBNA1. J. Virol. 2005;79:4640–4650. doi: 10.1128/JVI.79.8.4640-4650.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Libioulle C., Louis E., Hansoul S., Sandor C., Farnir F., Franchimont D., Vermeire S., Dewit O., de Vos M., Dixon A., et al. Novel Crohn disease locus identified by genome-wide association maps to a gene desert on 5p13.1 and modulates expression of PTGER4. PLoS Genet. 2007;3:e58. doi: 10.1371/journal.pgen.0030058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Buxbaum J.D., Silverman J.M., Smith C.J., Greenberg D.A., Kilifarski M., Reichert J., Cook E.H., Jr, Fang Y., Song C.Y., Vitale R. Association between a GABRB3 polymorphism and autism. Mol. Psychiatry. 2002;7:311–316. doi: 10.1038/sj.mp.4001011. [DOI] [PubMed] [Google Scholar]
- 36.Samaco R.C., Hogart A., LaSalle J.M. Epigenetic overlap in autism-spectrum neurodevelopmental disorders: MECP2 deficiency causes reduced expression of UBE3A and GABRB3. Hum. Mol. Genet. 2005;14:483–492. doi: 10.1093/hmg/ddi045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kitada T., Asakawa S., Hattori N., Matsumine H., Yamamura Y., Minoshima S., Yokochi M., Mizuno Y., Shimizu N. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392:605–608. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- 38.Lee J.S., Chien C.B. When sugars guide axons: insights from heparan sulphate proteoglycan mutants. Nat. Rev. Genet. 2004;5:923–935. doi: 10.1038/nrg1490. [DOI] [PubMed] [Google Scholar]
- 39.Van Vactor D., Wall D.P., Johnson K.G. Heparan sulfate proteoglycans and the emergence of neuronal connectivity. Curr. Opin. Neurobiol. 2006;16:40–51. doi: 10.1016/j.conb.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 40.van Horssen J., Bo L., Dijkstra C.D., de Vries H.E. Extensive extracellular matrix depositions in active multiple sclerosis lesions. Neurobiol. Dis. 2006;24:484–491. doi: 10.1016/j.nbd.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 41.Luxardi G., Galli A., Forlani S., Lawson K., Maina F., Dono R. Glypicans are differentially expressed during patterning and neurogenesis of early mouse brain. Biochem. Biophys. Res. Commun. 2007;352:55–60. doi: 10.1016/j.bbrc.2006.10.185. [DOI] [PubMed] [Google Scholar]
- 42.Filmus J., Capurro M., Rast J. Glypicans. Genome Biol. 2008;9:224. doi: 10.1186/gb-2008-9-5-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chernousov M.A., Rothblum K., Stahl R.C., Evans A., Prentiss L., Carey D.J. Glypican-1 and alpha4(V) collagen are required for Schwann cell myelination. J. Neurosci. 2006;26:508–517. doi: 10.1523/JNEUROSCI.2544-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Worapamorn W., Haase H.R., Li H., Bartold P.M. Growth factors and cytokines modulate gene expression of cell-surface proteoglycans in human periodontal ligament cells. J. Cell Physiol. 2001;186:448–456. doi: 10.1002/1097-4652(2001)9999:9999<000::AID-JCP1047>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 45.Kuroda Y., Mitsui T., Kunishige M., Matsumoto T. Parkin affects mitochondrial function and apoptosis in neuronal and myogenic cells. Biochem. Biophys. Res. Commun. 2006;348:787–793. doi: 10.1016/j.bbrc.2006.06.201. [DOI] [PubMed] [Google Scholar]
- 46.Mira M.T., Alcais A., Nguyen V.T., Moraes M.O., Di Flumeri C., Vu H.T., Mai C.P., Nguyen T.H., Nguyen N.B., Pham X.K., et al. Susceptibility to leprosy is associated with PARK2 and PACRG. Nature. 2004;427:636–640. doi: 10.1038/nature02326. [DOI] [PubMed] [Google Scholar]
- 47.Ali S., Vollaard A.M., Widjaja S., Surjadi C., van de Vosse E., van Dissel J.T. PARK2/PACRG polymorphisms and susceptibility to typhoid and paratyphoid fever. Clin. Exp. Immunol. 2006;144:425–431. doi: 10.1111/j.1365-2249.2006.03087.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Petrilli V., Dostert C., Muruve D.A., Tschopp J. The inflammasome: a danger sensing complex triggering innate immunity. Curr. Opin. Immunol. 2007;19:615–622. doi: 10.1016/j.coi.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 49.Shifman S., Johannesson M., Bronstein M., Chen S.X., Collier D.A., Craddock N.J., Kendler K.S., Li T., O'Donovan M., O'Neill F.A., et al. Genome-wide association identifies a common variant in the reelin gene that increases the risk of schizophrenia only in women. PLoS Genet. 2008;4:e28. doi: 10.1371/journal.pgen.0040028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Serajee F.J., Zhong H., Mahbubul Huq A.H. Association of Reelin gene polymorphisms with autism. Genomics. 2006;87:75–83. doi: 10.1016/j.ygeno.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 51.Haas C.A., Dudeck O., Kirsch M., Huszka C., Kann G., Pollak S., Zentner J., Frotscher M. Role for reelin in the development of granule cell dispersion in temporal lobe epilepsy. J. Neurosci. 2002;22:5797–5802. doi: 10.1523/JNEUROSCI.22-14-05797.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ohkubo N., Vitek M.P., Morishima A., Suzuki Y., Miki T., Maeda N., Mitsuda N. Reelin signals survival through Src-family kinases that inactivate BAD activity. J. Neurochem. 2007;103:820–830. doi: 10.1111/j.1471-4159.2007.04804.x. [DOI] [PubMed] [Google Scholar]
- 53.Lee M., Reddy H., Johansen-Berg H., Pendlebury S., Jenkinson M., Smith S., Palace J., Matthews P.M. The motor cortex shows adaptive functional changes to brain injury from multiple sclerosis. Ann. Neurol. 2000;47:606–613. [PubMed] [Google Scholar]
- 54.McDonald W.I., Compston A., Edan G., Goodkin D., Hartung H.P., Lublin F.D., McFarland H.F., Paty D.W., Polman C.H., Reingold S.C., et al. Recommended diagnostic criteria for multiple sclerosis: guidelines from the international panel on the diagnosis of multiple sclerosis. Ann. Neurol. 2001;50:121–127. doi: 10.1002/ana.1032. [DOI] [PubMed] [Google Scholar]
- 55.Polman C.H., Reingold S.C., Edan G., Filippi M., Hartung H.P., Kappos L., Lublin F.D., Metz L.M., McFarland H.F., O'Connor P.W., et al. Diagnostic criteria for multiple sclerosis: 2005 revisions to the ‘McDonald Criteria. Ann. Neurol. 2005;58:840–846. doi: 10.1002/ana.20703. [DOI] [PubMed] [Google Scholar]
- 56.Kurtzke J.F. Rating neurologic impairment in multiple sclerosis: an expanded disability status scale (EDSS) Neurology. 1983;33:1444–1452. doi: 10.1212/wnl.33.11.1444. [DOI] [PubMed] [Google Scholar]
- 57.Roxburgh R.H., Seaman S.R., Masterman T., Hensiek A.E., Sawcer S.J., Vukusic S., Achiti I., Confavreux C., Coustans M., le Page E., et al. Multiple Sclerosis Severity Score: using disability and disease duration to rate disease severity. Neurology. 2005;64:1144–1151. doi: 10.1212/01.WNL.0000156155.19270.F8. [DOI] [PubMed] [Google Scholar]
- 58.Smith S.M., De Stefano N., Jenkinson M., Matthews P.M. Normalized accurate measurement of longitudinal brain change. J. Comput. Assist. Tomogr. 2001;25:466–475. doi: 10.1097/00004728-200105000-00022. [DOI] [PubMed] [Google Scholar]
- 59.Smith S.M., Zhang Y., Jenkinson M., Chen J., Matthews P.M., Federico A., De Stefano N. Accurate, robust, and automated longitudinal and cross-sectional brain change analysis. Neuroimagen. 2002;17:479–489. doi: 10.1006/nimg.2002.1040. [DOI] [PubMed] [Google Scholar]
- 60.de Bakker P.I., McVean G., Sabeti P.C., Miretti M.M., Green T., Marchini J., Ke X., Monsuur A.J., Whittaker P., Delgado M., et al. A high-resolution HLA and SNP haplotype map for disease association studies in the extended human MHC. Nat. Genet. 2006;38:1166–1172. doi: 10.1038/ng1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nielsen R., Palsboll P.J. Single-locus tests of microsatellite evolution: multi-step mutations and constraints on allele size. Mol. Phylogenet. Evol. 1999;11:477–484. doi: 10.1006/mpev.1998.0597. [DOI] [PubMed] [Google Scholar]
- 62.Armitage S.G., Greenberg P.D., Pearl D., Berger D.G., Daston P.G. Predicting intelligence from the Rorschach. J. Consult. Psychol. 1955;19:321–329. doi: 10.1037/h0040299. [DOI] [PubMed] [Google Scholar]
- 63.Benjamini Y., Drai D., Elmer G., Kafkafi N., Golani I. Controlling the false discovery rate in behavior genetics research. Behav. Brain Res. 2001;125:279–284. doi: 10.1016/s0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- 64.Šidák Z. Rectangular confidence regions for the means of multivariate normal distributions. J. Am. Stat. Assoc. 1967;62:626–633. [Google Scholar]
- 65.Li J., Ji L. Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity. 2005;95:221–227. doi: 10.1038/sj.hdy.6800717. [DOI] [PubMed] [Google Scholar]
- 66.Cheverud J.M. A simple correction for multiple comparisons in interval mapping genome scans. Heredity. 2001;87:52–58. doi: 10.1046/j.1365-2540.2001.00901.x. [DOI] [PubMed] [Google Scholar]
- 67.Nyholt D.R. A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am. J. Hum. Genet. 2004;74:765–769. doi: 10.1086/383251. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



