Abstract
Acetaminophen (APAP) overdose is the most frequent cause of drug-induced acute liver failure. Schisandra sphenanthera is a traditional hepato-protective Chinese medicine and Schisandrol B (SolB) is one of its major active constituents. In this study, the protective effect of SolB against APAP-induced acute hepatotoxicity in mice and the involved mechanisms were investigated. Morphological and biochemical assessments clearly demonstrated a protective effect of SolB against APAP-induced liver injury. SolB pretreatment significantly attenuated the increases in alanine aminotransferase and aspartate aminotransferase activity, and prevented elevated hepatic malondialdehyde formation and the depletion of mitochondrial glutathione (GSH) in a dose-dependent manner. SolB also dramatically altered APAP metabolic activation by inhibiting the activities of CYP2E1 and CYP3A11, which was evidenced by significant inhibition of the formation of the oxidized APAP metabolite NAPQI–GSH. A molecular docking model also predicted that SolB had potential to interact with the CYP2E1 and CYP3A4 active sites. In addition, SolB abrogated APAP-induced activation of p53 and p21, and increased expression of liver regeneration and antiapoptotic-related proteins such as cyclin D1 (CCND1), PCNA, and BCL-2. This study demonstrated that SolB exhibited a significant protective effect toward APAP-induced liver injury, potentially through inhibition of CYP-mediated APAP bioactivation and regulation of the p53, p21, CCND1, PCNA, and BCL-2 to promote liver regeneration.
Keywords: acetaminophen, schisandrol B, liver injury, bioactivation, liver regeneration
Acetaminophen (APAP) is the most widely used analgesic and antipyretic agent in the world. Although relatively safe at therapeutic doses, APAP in high doses can cause hepatotoxicity and is recognized as a major cause of acute liver failure in the United States and in many European countries (Blieden et al., 2014; Mitka, 2014). Numerous efforts have been undertaken to elucidate the molecular mechanism of APAP toxicity (Jaeschke et al., 2011; Nelson, 1990). The toxicity is initiated by cytochrome P450-mediated, notably CYP2E1, reactions that convert APAP to an electrophilic metabolite, N-acetyl-p-benzoquinone imine (NAPQI). Subsequently, NAPQI binds to cellular proteins and causes glutathione (GSH)-depletion and oxidative stress that may trigger signaling pathways through mitochondrial toxicity, ultimately resulting in lethal cell injury and liver injury (McGill et al., 2012; Shin et al., 2012). Furthermore, compensational liver regeneration is a vital process for survival after APAP-induced toxic insult (Bajt et al., 2003). It was demonstrated that p53/p21 signaling pathway plays an important role in regulating cell growth, DNA repair, and apoptosis which is related to liver regeneration (Vogelstein et al., 2000), and APAP exposure induces p53 and p21 expression in vitro and in vivo (Chiu et al., 2003; Ray et al., 2001).
Schisandra sphenanthera, the dried ripe fruit of S. sphenanthera Rehd. et Wils, is a traditional Chinese medicine widely used in China, Japan, and Korea for its protective effects in liver, kidney, and heart (Panossian and Wikman, 2008). The whole extract and the components in the extract were shown to possess hepatoprotective effects against viral or chemical hepatitis (Teraoka et al., 2012; Zhu et al., 2000). Most recently, we found that Wuzhi Tablet (a preparation of the ethanol extract of S. sphenanthera) exhibits a significant hepatoprotective effect against APAP-induced liver toxicity (Bi et al., 2013). Schisandrol B (SolB) is one of the most important active components isolated from S. sphenanthera. However, whether SolB exerts a beneficial effect in preventing APAP-induced hepatotoxicity and what mechanisms are involved in remain largely unclear.
This study aimed to explore whether SolB could protect against APAP-induced hepatotoxicity and to determine what molecular pathways were involved in the hepatoprotection. SolB was found to exhibit remarkable dose-dependent hepatoprotection against APAP-induced injury, potentially through inhibition of cytochrome P450 (CYP)-mediated APAP bioactivation and regulation of the p53, p21, cyclin D1 (CCND1), proliferating cell nuclear antigen (PCNA), and B cell lymphoma/lewkmia-2 (BCL-2) to promote liver regeneration.
MATERIALS AND METHODS
Chemicals and reagents
SolB with 98% purity was purchased from Shanghai Winherb Medical Science and Technology Development Co. Ltd. (Shanghai, China). NADPH tetrasodium salt was obtained from AppliChem (Darmstadt, Germany). APAP, l-GSH, loratadine (LOR, 99.4% purity, internal standard), phenacetin (PHE), paracetamol (PAR), chlorzoxazone (CHL), 6-hydroxychlorzoxazone (OHCHL), nifedipine (NIF), dehydronifedipine (DNIF), 4-methylpyrazole (4-ME), ketoconazole (KET), α-naphthoflavone (α-NF), and other chemicals, unless indicated, were obtained from Sigma-Aldrich Chemical Co. (St. Louis, Missouri).
The mouse monoclonal anti-p53 was obtained from Abcam (Cambridge, UK). Rabbit anti-BCL-2 and anti-GAPDH were all purchased from Cell Signaling Technology (Danvers, Massachusetts). The goat polyclonal anti-CYP3A11 and mouse monoclonal anti-p21 were acquired from Santa Cruz Biotechnology (Santa Cruz, California). The rabbit anti-phosphorylated p53, anti-CCND1, anti-PCNA, and anti-cyclin D-dependent kinase 4 (CDK4) were purchased from Sangon Biotechnology (Sangon Tech, Shanghai, China). The rabbit polyclonal anti-CYP2E1 and anti-CYP1A2 antibodies were obtained from Boster Biotechnology (Boster, Wuhan, China). The peroxidase-conjugated antirabbit immunoglobulin G (IgG) and antimouse were purchased from Cell Signaling Technology (Danvers).
Experimental animals and treatment
Male C57BL/6 mice (6–8 weeks old, 20–22 g) were supplied by the Laboratory Animal Service Center at Sun Yat-sen University (Guangzhou, China). The animal room was maintained at 23 ± 1°C with a 12-h light–dark cycle and 55 ± 5% humidity. The mice had free access to standard rodent chow and water. All procedures were in accordance with the Regulations of Experimental Animal Administration issued by the Ministry of Science and Technology of the People’s Republic of China (http://www.most.gov.cn). All animal study protocols were approved by the Ethics Committee on the Care and Use of Laboratory Animals of Sun Yat-sen University.
Mice were randomly divided into six groups: (1) Vehicle, (2) SolB (200 mg/kg/d), (3) APAP (400 mg/kg) + vehicle, (4) APAP (400 mg/kg) + SolB (12.5 mg/kg/d), (5) APAP (400 mg/kg) + SolB (50 mg/kg/d), and (6) APAP (400 mg/kg) + SolB (200 mg/kg/d). The preparation of SolB for oral administration was made by suspending it in 0.5% [w/v] sodium carboxymethyl cellulose (CMC-Na). SolB (6.25, 25, and 100 mg/kg) was gavaged to mice seven times with an interval of 12 h. The control group and APAP group were administered a 0.5% CMC-Na solution. For APAP treatment, APAP was dissolved in a saline solution and administered by intraperitoneal (i.p.) injection at a single dose of 400 mg/kg, and the untreated control group and SolB alone group were injected i.p. with saline solution. For the SolB/APAP group, 15 min after the seventh dose of SolB, APAP was given by i.p. injection. All mice were killed at 1 or 6 h after APAP treatment. Serum samples were collected and the liver was harvested, a portion of the liver was collected in 10% buffered formalin for histology and the remaining tissue was flash frozen in liquid nitrogen and stored at −80°C for further use.
Histological and biochemical assessment
Liver tissues were immediately formalin-fixed, paraffin embedded, cut into sections, and stained with hematoxylin and eosin (H&E) following a standard protocol. H&E-stained liver sections were used to assess liver damage using a LEICA DM5000B Microscope (Leica, Heidelberg, Germany).
APAP-induced liver injury was also evaluated by measuring serum alanine transaminase (ALT) and aspartate transaminase (AST) catalytic activities. ALT and AST levels were determined using a Beckman Synchron CX5 Clinical System and a commercial reagent kit (Kefang Biotech, Guangzhou, China) according to the manufacturer’s protocol.
Total GSH levels in the liver and liver mitochondria extracts were measured using a GSH assay kit (Nanjing Jiancheng Bioengineering Institute, Nanjing, China). Liver mitochondria were isolated by differential centrifugation following the assay kit’s instruction (Sangon Tech).
LC-MS/MS analysis of NAPQI–GSH
The oxidized APAP intermediate NAPQI, formed in mice microsomal incubation system, was trapped by GSH and determined by a previously reported LC-MS/MS method with slight modifications (Tan et al., 2008; Wang et al., 1996). Briefly, the incubation mixture of 150 μl contained 0.5 mM APAP, 5 mM GSH, SolB (0, 2.5, 5, 10, 20, 40, and 80 μM) or positive inhibitor (4-ME at 5 μM for CYP2E1 or KET at 1.1 Μm for CYP3A11), mouse liver microsomes (containing 30 μg protein), 0.4 mM NADPH, and potassium phosphate buffer (50 mM, pH 7.4). The reaction was initiated by adding the NADPH, after a 10-min incubation at 37°C. The reaction was quenched by adding 300 μl ice-cold acetonitrile. The reaction mixtures were centrifuged at 16 000 × g for 15 min and the supernatant was used for analysis of APAP–GSH conjugate formation. The sample was analyzed using Ultimate 3000 UPLC system (Dionex Corporation, Sunnyvale, California) interfaced with triple quadrupole mass spectrometer (TSQ Quantum Access, Thermo Fisher Scientific, Waltham, Massachusetts). Chromatographic separations were performed on Hypurity C18 5 μm column (150 mm × 2.1 mm, Thermo Fisher Scientific). The samples were eluted with 2% acetonitrile (v/v in water) and 0.1% (v/v) formic acid at a flow rate of 0.5 ml/min. Electrospray positive ionization mode (ESI+) was used for NAPQI–GSH analysis. Selective reaction monitoring (SRM) experiments using m/z 457.1–328.1 was performed to profile and quantitate NAPQI-GSH.
Measurement of liver CYPs activity
CYP2E1, CYP1A2, and CYP3A4 (mouse CYP3A11 is a homolog to human CYP3A4) are responsible for APAP oxidation to NAPQI. The effect of SolB on the activity of CYPs was performed using a LC-MS/MS-based “cocktail” incubation approach as previously described with slight modifications (He et al., 2007). Briefly, various concentrations of SolB (2.5, 5, 10, 20, 40, and 80 μM) or positive inhibitor (α-NF at 0.2 μM for CYP1A2, 4-ME at 5 μM for CYP2E1, or KET at 1.1 mM for CYP3A11) and the mixed CYPs probe substrates (PHE for CYP1A2, CHL for CYP2E1, and NIF for CYP3A11) were “cocktail” incubated with mouse liver microsomes. The metabolites (PAR/OHCHL/DNIF) of these CYPs probe substrates in all samples were determined using previously developed LC-MS/MS method.
Molecular docking
Molecular docking analysis was performed by the CDOCKER protocol of Accelrys Discovery Studio 2.5 and Discovery Studio Visualizer 3.5 (Accelrys Software Inc., San Diego, California). The crystal structures of CYP2E1 (PDB: 3E4E) and CYP3A4 (PDB: 3NXU) were adopted as receptor for molecular analysis (Porubsky et al., 2008; Sevrioukova and Poulos, 2010). The active sites of CYPs were defined according to the internal ligand’s binding. The hydrogen positions were optimized with CHARMm force field. The molecules of SolB were selected based on the ability of the chemical structures to generate structure-based pharmacophore, selecting the lowest ranks in the CDOCKER Interaction Energy tactic and visually examining the binding conformation of the docking poses.
Western blot analysis
Western blot analysis was performed as described in a previous report (Chen et al., 2014). Briefly, protein extracted from liver tissue was prepared using RIPA lysis buffer (Biocolors, Shanghai, China) according to the manufacturer’s instructions. Protein concentrations were assayed by bicinchoninic acid (BCA) protein assay (Thermo Scientific, Rockford, Illinois). Every sample was normalized to the 40 µg of protein concentration. Protein extracts were separated in 8%–15% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and electrophoretically transferred onto polyvinylidene fluoride membranes (Millipore, Bedford). After blocked with 5% BSA or 5% nonfat dry milk in Tris-buffered saline, and the membranes were incubated with primary antibody overnight. Specific bands were detected using an electrochemiluminescence (ECL) detection kit (Engreen Biosystem, Beijing, China) and exposed to an X-ray film (GE Healthcare, Piscataway, New Jersey). The intensity of protein bands was analyzed using Quantity One software (Bio-Rad Laboratories, Hercules).
Statistical analysis
Data were expressed as mean ± standard error of the mean (SEM). One-way ANOVA followed by unpaired Student’s t test or Dunnett’s multiple comparison post hoc test was used for statistical analysis of data using GraphPad Prism 5 (GraphPad Software Inc., San Diego, California). P < 0.05 were considered significant.
RESULTS
Protective Effect of SolB Against APAP-Induced Hepatotoxicity
Morphological and histological assessments clearly indicated that treatment with 400 mg/kg APAP for 6 h caused massive hepatotoxicity as evidenced by severe liver cell necrosis (60%–70%) and focal intrahepatic hemorrhage (50%–60%). Pretreatment with SolB at different doses for 3 days before APAP treatment resulted in marked protection against APAP-induced liver injury as indicated by morphological and histological results (Fig. 1A). APAP toxicity was also indicated by serum ALT and AST activities that were markedly increased by APAP treatment (11 900 ± 1270 U/l and 12 600 ± 1700 U/l, respectively) (Figs. 1B and 1C). Pretreatment with SolB at doses of 50 and 200 mg/kg significantly attenuated the increases of ALT activity to 6420 ± 1390 U/l (−46%, P < 0.05) and 3800 ± 515 U/l (−68%, P < 0.001). Consistently, the serum AST levels were significantly attenuated to 6350 ± 1210 U/l (−50%, P < 0.05) and 3790 ± 1410 U/l (−70%, P < 0.01) after pretreatment with SolB at 50 and 200 mg/kg. APAP administration is typically accompanied by a decrease in GSH. Treatment with 400 mg/kg APAP during the first 1 h resulted in a sharp hepatic GSH depletion of about 65% of the control values, whereas SolB pretreatment leaded to a delayed or less GSH depletion of about 15% of the control values, suggesting SolB may inhibit the metabolic activation of APAP. The total hepatic GSH and mitochondrial GSH levels at 6 h after APAP dosing showed similar change trends in the absence or presence of SolB but the GSH depletion was less than that at 1 h may due to a better GSH recovery or a SolB-induced GSH elevation (Figs. 2A–2D). Taken together, these histological and biochemical results clearly demonstrated that SolB could protect against APAP-induced liver injury in a dose-dependent manner.
FIG. 1.

Hepatoprotection of SolB against APAP-induced hepatotoxicity. SolB was gavaged to mice 7 times with an interval of 12 h, and a single dose of APAP (400 mg/kg, i.p.) was given 15 min after the seventh dose of SolB. Mice were sacrificed at 6 h after APAP treatment. A, Histopathological analysis of a representative mice liver samples following H&E staining. (1) Vehicle; (2) SolB (200 mg/kg); (3) APAP (400 mg/kg) + Vehicle; (4) APAP (400 mg/kg) + SolB (200 mg/kg). B and C, Serum ALT and AST activities (n = 6). *P < 0.05, **P < 0.01, and ***P < 0.001 compared with APAP group, #P < 0.05, ##P < 0.01, and ###P < 0.001 compared with vehicle.
FIG. 2.
Change in GSH levels at 1 or 6 h after 400 mg/kg dose of APAP in the absence or presence of SolB (200 mg/kg). A and B, Levels of total and mitochondria GSH at 1 h (n = 6). C and D, Levels of total and mitochondria GSH in livers at 6 h (n = 7). *P < 0.05, **P < 0.01, and ***P < 0.001 compared with APAP group, #P < 0.05, ##P < 0.01, and ###P < 0.001 compared with vehicle.
Effects of SolB on APAP Metabolic Activation
APAP-induced hepatotoxicity is initiated by CYP-mediated bioactivation that converts APAP to NAPQI. To determine whether pretreatment with SolB inhibited APAP bioactivation, liver microsome were incubated with APAP alone and with SolB or CYP inhibitors, then the oxidized APAP intermediate NAPQI was trapped by GSH and NAPQI-GSH was detected by a LC-MS/MS method. The production of NAPQI-GSH after coincubation with SolB was shown in Figures 3A and 3B. It was observed that pretreatment with SolB significantly inhibited the formation of the oxidized APAP metabolite NAPQI in a dose-dependent manner, the levels of NAPQI-GSH were decreased with the increase of SolB. Compared with the control (APAP alone), the positive inhibitors 4-ME for CYP2E1 and KET for CYP3A11 effectively inhibited NAPQI-GSH levels to 14% and 25%; SolB at 5, 10, 20, 40, and 80 μM can significantly inhibit NAPQI-GSH levels to 66%, 43%, 24%, 18%, and 15%, respectively.
FIG. 3.

Effects of SolB on APAP bioactivation. A, Representative and comparative SRM chromatograms of NAPQI-GSH in mouse liver microsomes incubated with APAP and SolB. B, Formation of NAPQI-GSH conjugates in mouse liver microsomes. Data are expressed as means ± SEM (n = 5). *P < 0.05, **P < 0.01, and ***P < 0.001 compared with the control (APAP alone).
Effect of SolB on the Expression and Activity of CYPs Involved in APAP Bioactivation
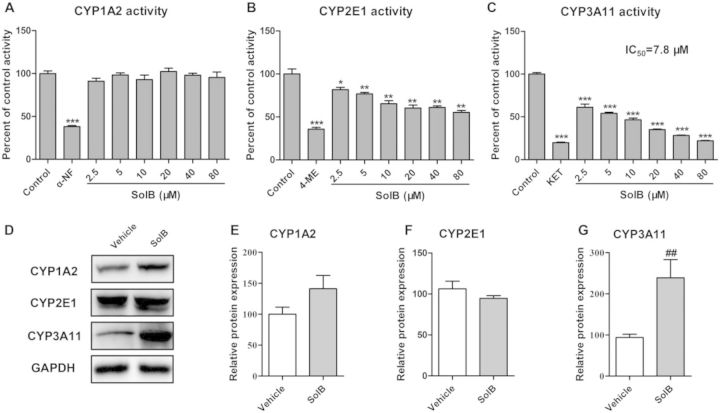
APAP metabolic activation and the formation of NAPQI were mediated by CYPs, thus the effect of SolB on the activity and protein expression of CYPs was determined. The inhibitory effects of positive inhibitors and SolB on CYPs activity were shown in Figures 4A–4C. The typical inhibitors α-NF, 4-ME, and KET effectively inhibited CYP1A2, CYP2E1, and CYP3A11 activities to 38%, 36%, and 20% of the control, respectively. The activities of CYP2E1 and CYP3A11 were markedly inhibited by SolB, whereas the CYP1A2 activity was not significantly inhibited. The activity of CYP2E1 was inhibited by SolB in a dose-dependent manner. CYP3A11 activity was significantly inhibited by SolB in a dose-dependent manner and the IC50 value against CYP3A11 was 7.8 μM. As for the enzyme expression, SolB treatment resulted in an increase in CYP1A2 protein but there was no significant difference compared with the vehicle group; and had no effect on the protein level of CYP2E1. However, SolB can significantly induce the protein expression of CYP3A11 to 2.5-fold compared with the vehical group (Figs. 4D–4G).
FIG. 4.
The effects of SolB on the activity and expression of CYPs. A–C, Inhibitory effects of SolB on CYP1A2, CYP2E1, and CYP3A11 activity were measured by cocktail substrates assay in vitro. Data are expressed as means ± S.E.M. (n = 5). *P < 0.05, **P < 0.01, ***P < 0.001 compared with the control group (D) protein expressions of CYP1A2, CYP2E1, and CYP3A11 were measured by Western blot. E–G, Specific band intensity was quantified, normalized to GAPDH, and expressed as means ± SEM (n = 3). *P < 0.05, **P < 0.01, ***P < 0.001 compared with APAP group, #P < 0.05, ##P < 0.01, and ###P < 0.001 compared with vehicle.
To further validate the inhibitory effect of SolB on CYP2E and CYP3A activities, the molecular binding modes of SolB with CYP2E1 and CYP3A4 were investigated and a computational docking model was established. The binding modes of SolB and 4-ME with CYP2E1 complex were demonstrated (Fig. 5A), and its CDOCKER Interaction Energy were −35.8 and −13.2 kcal/mol, respectively. In particular, hydroxyl of SolB could form a hydrogen bond with Ala438 with distances of 3.9 Å (Fig. 5B). Meanwhile, as shown in Figure 5C, the binding complex of SolB and KET had a stronger CDOCKER Interaction Energy in CYP3A4 (−43.1 and −53.2 kcal/mol), which was in line with the result of enzyme activity. SolB could form two hydrogen bonds with Arg105, with distances of 4.7 and 5.8 Å. Moreover, the benzene rings of SolB formed one potential Pi interaction with Arg105 (Fig. 5D). This docking model predicted that SolB has potential to interact with CYP2E1 and CYP3A4 active sites, which was in line with the result of enzyme activity that the activity of CYP3A11 and CYP2E1 was inhibited by SolB with a dose-dependent manner.
FIG. 5.
Molecular docking (CDOCKER) of SolB, 4-ME, or KET to CYP2E1 or CYP3A4. A, The three-dimensional diagram showed the binding conformation of 4-ME (red), SolB (yellow) with CYP2E1. The heme of CYP2E1 is shown in gray. C, The three-dimensional diagram showed the binding conformation of KET (red), SolB (yellow) with CYP3A4. The heme of CYP3A4 was shown in gray. B and D, The two-dimensional diagrams displayed the docking model of SolB in active site of CYP2E1 and CYP3A4, respectively. The residue colors represent the types of interactions as follows: green (Van der Waals); purple (electrostatic); orange (Pi interaction); and blue arrows (hydrogen bond).
Effect of SolB on the Downstream Signals Involved in Liver Regeneration
The protein expression levels of p-p53, p53, and the downstream genes involved in liver cell proliferation such as p21, CCND1, CDK4, PCNA, and apoptosis protein BCL-2 were also determined. A marked increase in p-p53, p53, and p21 protein expression was observed in the APAP group (3.0-, 2.5-, and 3.3-fold higher than that in vehicle group, respectively), whereas SolB reversed this increase (Figs. 6A–6D). In addition, SolB alone and SolB/APAP cotreatment further enhanced protein levels of the downstream genes such as CCND1, PCNA, and BCL-2. Pretreatment with SolB alone significantly increased the CCND1, PCNA, and BCL-2 protein expression by 3.9-, 4.3-, and 1.4-fold higher than that in vehicle group, whereas SolB/APAP cotreatment significantly increased their expression by 3.1-, 6.9-, and 3.1-fold higher than that in the APAP group. However, down regulation of CDK4 by APAP was not be reversed by SolB. These data indicated that SolB influenced p53 and downstream pathways involved in liver cell proliferation and apoptosis and promotes liver regeneration, which may partially contribute to the hepatoprotective effect of SolB against APAP-induced liver injury.
FIG. 6.
Role of p53 signaling and its downstream proteins in SolB protection against APAP-induced liver toxicity. A, Western blot was used to measure p-p53, p53, p21, CCND1, PCNA, CDK4, and BCL-2 protein expression. B–H, Specific band intensity was quantified, normalized to GAPDH. Data are expressed as means ± SEM (n = 3). *P < 0.05, **P < 0.01, and ***P < 0.001 compared with APAP group, #P < 0.05, ##P < 0.01, and ###P < 0.001 compared with vehicle.
DISCUSSION
Schisandra species have been widely used for the treatment of hepatitis as a well-known traditional Chinese herb (Liu, 1989). In the United States, extracts of Sphenanthera are often combined into multicomponent dietary supplement formulations that are marketed to enhance the function of liver and other organs (Gurley et al., 2012). Recently, Wuzhi Tablet (a preparation of the ethanol extract of S. sphenanthera) was found to have a significant hepatoprotective effect against APAP-induced liver toxicity (Bi et al., 2013). Most recently, it was reported that Schisandrin B, one of the active components isolated from Schisandra, could attenuate APAP-induced hepatotoxicity through heat-shock protein 27 and 70 (Li et al., 2014).
SolB is one of the most important active components isolated from the extract of S. sphenanthera. However, it is not known whether SolB exhibits hepatoprotection against APAP-induced liver injury. Thus, the current study aimed to investigate whether SolB exerts a beneficial effect in preventing APAP-induced hepatotoxicity and what mechanisms are involved. The present data clearly demonstrated that SolB exhibits hepatoprotection against APAP-induced liver injury in a dose-dependent manner, as evidenced by morphological and histological assessment, as well as biochemical data such as AST, ALT, and GSH. Mechanistic studies further indicate that the effect of SolB on CYP-mediated APAP bioactivation and the regulation of signals involved in liver regeneration may contribute to the hepatoprotection of SolB against APAP-induced liver toxicity.
The metabolic activation of APAP is the most proximal event in the toxicity mechanism, thus it is important to realize whether SolB inhibits APAP bioactivation. It has been reported that in the presence of an inhibitor of APAP metabolism, a high overdose APAP may lead to delayed GSH depletion and consequently less toxicity (Saito et al., 2010). In the current study, APAP in the absence of SolB during the first 1 h leaded to hepatic GSH depletion of about 65% of the control values. However, in the presence of SolB, the GSH depletion was delayed or less (depletion of about 15% of the control values), suggesting SolB may inhibit the metabolic activation of APAP and result in a delayed GSH depletion. To confirm whether the protective effect of SolB was correlated with inhibition of APAP bioactivation, the effect of SolB on the formation of NAPQI-GSH was further determined. It was demonstrated that SolB at different concentrations can strongly inhibit the formation of NAPQI-GSH in a dose-dependent manner, directly suggesting SolB can significantly inhibit APAP metabolic activation.
APAP-induced hepatotoxicity is initiated by CYP-mediated conversion of APAP to NAPQI. CYP isoforms such as CYP2E1, CYP3A4 (mouse CYP3A11), and CYP1A2 play the most important role in APAP bioactivation (Manyike et al., 2000; Rumack, 2004). Thus, the effect of SolB on the protein expression and activity of CYP2E1, CYP3A11, and CYP1A2 was further measured. SolB itself induced CYP3A11 and CYP1A2 protein expression but did not affect CYP2E1 expression. Results from the in vitro substrate “cocktail” approach suggest that SolB has no significant inhibitory effect on CYP1A2 activity but SolB exhibits inhibition on CYP2E1 and CYP3A11 enzyme activity which was in line with previous reports (Iwata et al., 2004), and overall impact of SolB on these two CYPs was inhibitory action which has been previously reported and is called net effect (Abel et al., 2008). Most recently, it was reported that Schisandra Lignans Extract, which also contains SolB, possesses complicated inducing and inhibitory effect on CYP3A (Lai et al., 2009). Similarly, induction of CYPs expression but net inhibitory effect on CYPs activity were also observed with other herbs such as St John’s wort (Xie and Kim, 2005). Furthermore, molecular docking analysis with the CDOCKER protocol was used to provide fundamental insights into the mechanism of SolB and CYPs interaction to confirm the inhibitory effect of SolB on CYP2E1 and CYP3A11. The molecular docking results indicated that SolB has distinct high-affinity states with binding sites as a CYP2E1 and CYP3A4 inhibitors, which was in line with the in vitro enzyme activity result. Thus, the net effect of SolB on the CYPs is inhibitory effect on the enzyme activity of CYP2E1 and CYP3A11, which will result in the inhibition of APAP bioactivation and then an important reduction of NAPQI and a decrease in GSH depletion. Therefore, the inhibitory effect of SolB on CYP2E1 and CYP3A11 plays an important role in its inhibition of APAP bioactivation and thus contribute to the hepatoprotective effect of SolB against APAP-induced liver injury.
Liver injury is followed by compensatory liver regeneration, which is a critical determinant of the final outcome of liver injury (Dalhoff et al., 2001; Mehendale, 2005). Liver regeneration is a compensatory regrowth of liver after liver damage. Many genes and signaling pathways, such as cytokines and growth factors, have been identified to initiate or promote this process of liver regrowth. p53 is a tumor suppressor that plays an important role in regulating cell growth, DNA repair, and apoptosis (Shams et al., 2013). Under severe DNA damage by APAP overdose, p53 is activated to inhibit cell proliferation or trigger cell apoptosis (Zhao et al., 2012). p21 is one of the main effectors of p53 that induces cell cycle arrest and senescence in response to triggers such as DNA damage and telomere shortening by inhibiting the activity of cyclin-dependent kinase (CDK)–cyclin complexes and PCNA (Choudhury et al., 2007). The importance of p53/p21 in the regulation of liver regeneration has been studied (Albrecht et al., 1998; Lehmann et al., 2012). In the present study, the protein expression levels of p-p53, p53, p21, and their downstream targets involved in liver cell proliferation, such as CCND1, CDK4, PCNA, and antiapoptotic protein BCL-2, were also determined. The results show a marked increase in p-p53, p53, and p21 protein expression in APAP group, whereas SolB reversed such an increase. In addition, SolB alone and SolB/APAP cotreatment further enhanced the protein level of CCND1, PCNA, and BCL-2, suggesting that uprelation of SolB on CCND1 and PCNA may be involved in affecting cell cycle and proliferation following p53 and p21 signals. On the other hand, upregulation of p53 reduces the expression of antiapoptotic protein BCL-2 to complete the promotion of apoptosis (Weng et al., 2011). In the current study, down-regulation of BCL-2 by APAP and upregulation of BCL-2 by SolB indicate a relevant mechanism for protection of SolB against live cell necrosis. Therefore, regulation of the p53/p21 signaling pathway and upregulation of CCND1, PCNA, and BCL-2 to promote liver regeneration may also contribute to the hepatoprotective effect of SolB against APAP-induced liver injury.
Taken all together, this study reveals a mechanistic understanding of how SolB protects liver from APAP-induced hepatotoxicity, and emphasizes SolB aim at two distinct stages of APAP-induced hepatotoxicity. Stage I is the restrain stage, in which SolB inhibits NAPQI initiated injury via inhibition of CYPs activity. Stage II is the compensatory regeneration stage, in which SolB down-regulates p53/p21 and upregulates CCND1, PCNA, and BCL-2 signaling pathway to facilitate compensatory tissue repair.
In summary, this study clearly demonstrated that SolB can dose-dependently protect against APAP-induced liver injury, potentially through inhibition of CYP-mediated APAP bioactivation and regulation of p53, p21, CCND1, PCNA, and BCL-2 to promote liver regeneration.
Funding
Natural Science Foundation of China (Grant 81373470, 81320108027), the Science and Technology Ministry of China (Grant 2012ZX09506001-004), the Opening Project of Guangdong Provincial Key Laboratory of New Drug Design and Evaluation (Grant 2011A060901014), and the Fundamental Research Funds for the Central Universities (No. 13ykpy08). The authors have declared that there is no conflict of interest.
REFERENCES
- Abel S., Jenkins T. M., Whitlock L. A., Ridgway C. E., Muirhead G. J. (2008). Effects of CYP3A4 inducers with and without CYP3A4 inhibitors on the pharmacokinetics of maraviroc in healthy volunteers. Br. J. Clin. Pharmacol . 65(Suppl. 1), 38–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht J. H., Poon R. Y., Ahonen C. L., Rieland B. M., Deng C., Crary G. S. (1998). Involvement of p21 and p27 in the regulation of CDK activity and cell cycle progression in the regenerating liver. Oncogene 16, 2141–2150. [DOI] [PubMed] [Google Scholar]
- Bajt M. L., Knight T. R., Farhood A., Jaeschke H. (2003). Scavenging peroxynitrite with glutathione promotes regeneration and enhances survival during acetaminophen-induced liver injury in mice. J. Pharmacol. Exp. Ther. 307, 67–73. [DOI] [PubMed] [Google Scholar]
- Bi H., Li F., Krausz K. W., Qu A., Johnson C. H., Gonzalez F. J. (2013). Targeted metabolomics of serum acylcarnitines evaluates hepatoprotective effect of Wuzhi Tablet (Schisandra sphenanthera extract) against acute acetaminophen toxicity. Evid. Based Complement. Alternat. Med. 2013, 985257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blieden M., Paramore L. C., Shah D., Ben-Joseph R. (2014). A perspective on the epidemiology of acetaminophen exposure and toxicity in the United States. Expert Rev. Clin. Pharmacol. 7, 341–348. [DOI] [PubMed] [Google Scholar]
- Chen P., Zeng H., Wang Y., Fan X., Xu C., Deng R., Zhou X., Bi H., Huang M. (2014). Low dose of oleanolic acid protects against lithocholic acid-induced cholestasis in mice: potential involvement of nuclear factor-E2-related factor 2-mediated upregulation of multidrug resistance-associated proteins. Drug Metab. Dispos. 42, 844–852. [DOI] [PubMed] [Google Scholar]
- Chiu H., Gardner C. R., Dambach D. M., Durham S. K., Brittingham J. A., Laskin J. D., Laskin D. L. (2003). Role of tumor necrosis factor receptor 1 (p55) in hepatocyte proliferation during acetaminophen-induced toxicity in mice. Toxicol. Appl. Pharmacol. 193, 218–227. [DOI] [PubMed] [Google Scholar]
- Choudhury A. R., Ju Z., Djojosubroto M. W., Schienke A., Lechel A., Schaetzlein S., Jiang H., Stepczynska A., Wang C., Buer J., et al. (2007). Cdkn1a deletion improves stem cell function and lifespan of mice with dysfunctional telomeres without accelerating cancer formation. Nat. Genet. 39, 99–105. [DOI] [PubMed] [Google Scholar]
- Dalhoff K., Laursen H., Bangert K., Poulsen H. E., Anderson M. E., Grunnet N., Tygstrup N. (2001). Autoprotection in acetaminophen intoxication in rats: the role of liver regeneration. Pharmacol. Toxicol. 88, 135–141. [DOI] [PubMed] [Google Scholar]
- Gurley B. J., Fifer E. K., Gardner Z. (2012). Pharmacokinetic herb–drug interactions (part 2): drug interactions involving popular botanical dietary supplements and their clinical relevance. Planta Med. 78, 1490–1514. [DOI] [PubMed] [Google Scholar]
- He F., Bi H. C., Xie Z. Y., Zuo Z., Li J. K., Li X., Zhao L. Z., Chen X., Huang M. (2007). Rapid determination of six metabolites from multiple cytochrome P450 probe substrates in human liver microsome by liquid chromatography/mass spectrometry: application to high-throughput inhibition screening of terpenoids. Rapid Commun. Mass Spectrom. 21, 635–643. [DOI] [PubMed] [Google Scholar]
- Iwata H., Tezuka Y., Kadota S., Hiratsuka A., Watabe T. (2004). Identification and characterization of potent CYP3A4 inhibitors in Schisandra fruit extract. Drug Metab. Dispos. 32, 1351–1358. [DOI] [PubMed] [Google Scholar]
- Jaeschke H., McGill M. R., Williams C. D., Ramachandran A. (2011). Current issues with acetaminophen hepatotoxicity—a clinically relevant model to test the efficacy of natural products. Life Sci. 88, 737–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai L., Hao H., Wang Q., Zheng C., Zhou F., Liu Y., Wang Y., Yu G., Kang A., Peng Y., et al. (2009). Effects of short-term and long-term pretreatment of Schisandra lignans on regulating hepatic and intestinal CYP3A in rats. Drug Metab. Dispos. 37, 2399–2407. [DOI] [PubMed] [Google Scholar]
- Lehmann K., Tschuor C., Rickenbacher A., Jang J. H., Oberkofler C. E., Tschopp O., Schultze S. M., Raptis D. A., Weber A., Graf R., et al. (2012). Liver failure after extended hepatectomy in mice is mediated by a p21-dependent barrier to liver regeneration. Gastroenterology 143, 1609–1619 e1604. [DOI] [PubMed] [Google Scholar]
- Li L., Zhang T., Zhou L., Zhou L., Xing G., Chen Y., Xin Y. (2014). Schisandrin B attenuates acetaminophen-induced hepatic injury through heat-shock protein 27 and 70 in mice. J. Gastroenterol. Hepatol. 29, 640–647. [DOI] [PubMed] [Google Scholar]
- Liu G. T. (1989). Pharmacological actions and clinical use of fructus schizandrae. Chin. Med. J. (Engl) 102, 740–749. [PubMed] [Google Scholar]
- Manyike P. T., Kharasch E. D., Kalhorn T. F., Slattery J. T. (2000). Contribution of CYP2E1 and CYP3A to acetaminophen reactive metabolite formation. Clin. Pharmacol. Ther. 67, 275–282. [DOI] [PubMed] [Google Scholar]
- McGill M. R., Sharpe M. R., Williams C. D., Taha M., Curry S. C., Jaeschke H. (2012). The mechanism underlying acetaminophen-induced hepatotoxicity in humans and mice involves mitochondrial damage and nuclear DNA fragmentation. J. Clin. Invest. 122, 1574–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehendale H. M. (2005). Tissue repair: an important determinant of final outcome of toxicant-induced injury. Toxicol. Pathol. 33, 41–51. [DOI] [PubMed] [Google Scholar]
- Mitka M. (2014). FDA asks physicians to stop prescribing high-dose acetaminophen products. JAMA 311, 563. [DOI] [PubMed] [Google Scholar]
- Nelson S. D. (1990). Molecular mechanisms of the hepatotoxicity caused by acetaminophen. Semin. Liver Dis. 10, 267–278. [DOI] [PubMed] [Google Scholar]
- Panossian A., Wikman G. (2008). Pharmacology of Schisandra chinensis Bail.: an overview of Russian research and uses in medicine. J. Ethnopharmacol. 118, 183–212. [DOI] [PubMed] [Google Scholar]
- Porubsky P. R., Meneely K. M., Scott E. E. (2008). Structures of human cytochrome P-450 2E1. Insights into the binding of inhibitors and both small molecular weight and fatty acid substrates. J. Biol. Chem. 283, 33698–33707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray S. D., Balasubramanian G., Bagchi D., Reddy C. S. (2001). Ca(2+)-calmodulin antagonist chlorpromazine and poly (ADP-ribose) polymerase modulators 4-aminobenzamide and nicotinamide influence hepatic expression of BCL-XL and P53 and protect against acetaminophen-induced programmed and unprogrammed cell death in mice. Free Radic. Biol. Med. 31, 277–291. [DOI] [PubMed] [Google Scholar]
- Rumack B. H. (2004). Acetaminophen misconceptions. Hepatology 40, 10–15. [DOI] [PubMed] [Google Scholar]
- Saito C., Yan H. M., Artigues A., Villar M. T., Farhood A., Jaeschke H. (2010). Mechanism of protection by metallothionein against acetaminophen hepatotoxicity. Toxicol. Appl. Pharmacol. 242, 182–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevrioukova I. F., Poulos T. L. (2010). Structure and mechanism of the complex between cytochrome P4503A4 and ritonavir. Proc. Natl Acad. Sci. U. S. A. 107, 18422–18427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shams I., Malik A., Manov I., Joel A., Band M., Avivi A. (2013). Transcription pattern of p53-targeted DNA repair genes in the hypoxia-tolerant subterranean mole rat Spalax. J. Mol. Biol. 425, 1111–1118. [DOI] [PubMed] [Google Scholar]
- Shin B. Y., Jin S. H., Cho I. J., Ki S. H. (2012). Nrf2-ARE pathway regulates induction of Sestrin-2 expression. Free Radic. Biol. Med. 53, 834–841. [DOI] [PubMed] [Google Scholar]
- Tan S. C., New L. S., Chan E. C. (2008). Prevention of acetaminophen (APAP)-induced hepatotoxicity by leflunomide via inhibition of APAP biotransformation to N-acetyl-p-benzoquinone imine. Toxicol. Lett. 180, 174–181. [DOI] [PubMed] [Google Scholar]
- Teraoka R., Shimada T., Aburada M. (2012). The molecular mechanisms of the hepatoprotective effect of Gomisin A against oxidative stress and inflammatory response in rats with carbon tetrachloride-induced acute liver injury. Biol. Pharm. Bull. 35, 171–177. [DOI] [PubMed] [Google Scholar]
- Vogelstein B., Lane D., Levine A. J. (2000). Surfing the p53 network. Nature 408, 307–310. [DOI] [PubMed] [Google Scholar]
- Wang E. J., Li Y., Lin M., Chen L., Stein A. P., Reuhl K. R., Yang C. S. (1996). Protective effects of garlic and related organosulfur compounds on acetaminophen-induced hepatotoxicity in mice. Toxicol. Appl. Pharmacol. 136, 146–154. [DOI] [PubMed] [Google Scholar]
- Weng S. Y., Yang C. Y., Li C. C., Sun T. P., Tung S. Y., Yen J. J., Tsai T. F., Chen C. M., Chen S. H., Hsiao M., et al. (2011). Synergism between p53 and Mcl-1 in protecting from hepatic injury, fibrosis and cancer. J. Hepatol. 54, 685–694. [DOI] [PubMed] [Google Scholar]
- Xie H. G., Kim R. B. (2005). St John’s wort-associated drug interactions: short-term inhibition and long-term induction? Clin . Pharmacol. Ther. 78, 19–24. [DOI] [PubMed] [Google Scholar]
- Zhao X., Cong X., Zheng L., Xu L., Yin L., Peng J. (2012). Dioscin, a natural steroid saponin, shows remarkable protective effect against acetaminophen-induced liver damage in vitro and in vivo. Toxicol. Lett. 214, 69–80. [DOI] [PubMed] [Google Scholar]
- Zhu M., Yeung R. Y., Lin K. F., Li R. C. (2000). Improvement of phase I drug metabolism with Schisandra chinensis against CCl4 hepatotoxicity in a rat model. Planta Med. 66, 521–525. [DOI] [PubMed] [Google Scholar]