Abstract
C-reactive protein (CRP) concentration is a heritable systemic marker of inflammation that is associated with cardiovascular disease risk. Genome-wide association studies have identified CRP-associated common variants associated in ∼25 genes. Our aims were to apply exome sequencing to (1) assess whether the candidate loci contain rare coding variants associated with CRP levels and (2) perform an exome-wide search for rare variants in novel genes associated with CRP levels. We exome-sequenced 6050 European-Americans (EAs) and 3109 African-Americans (AAs) from the NHLBI-ESP and the CHARGE consortia, and performed association tests of sequence data with measured CRP levels. In single-variant tests across candidate loci, a novel rare (minor allele frequency = 0.16%) CRP-coding variant (rs77832441-A; p.Thr59Met) was associated with 53% lower mean CRP levels (P = 2.9 × 10−6). We replicated the association of rs77832441 in an exome array analysis of 11 414 EAs (P = 3.0 × 10−15). Despite a strong effect on CRP levels, rs77832441 was not associated with inflammation-related phenotypes including coronary heart disease. We also found evidence for an AA-specific association of APOE-ε2 rs7214 with higher CRP levels. At the exome-wide significance level (P < 5.0 × 10−8), we confirmed associations for reported common variants of HNF1A, CRP, IL6R and TOMM40-APOE. In gene-based tests, a burden of rare/lower frequency variation in CRP in EAs (P ≤ 6.8 × 10−4) and in retinoic acid receptor-related orphan receptor α (RORA) in AAs (P = 1.7 × 10−3) were associated with CRP levels at the candidate gene level (P < 2.0 × 10−3). This inquiry did not elucidate novel genes, but instead demonstrated that variants distributed across the allele frequency spectrum within candidate genes contribute to CRP levels.
INTRODUCTION
C-reactive protein (CRP) is an acute-phase protein reactant produced by the liver in response to proinflammatory stimuli. CRP is a sensitive, but nonspecific heritable biomarker of systemic inflammation that is associated with a variety of inflammation-mediated diseases (1–5). Particular attention has been focused on characterizing the association between CRP and cardiovascular disease (CVD). Prospective epidemiologic studies suggest that basal CRP levels are predictive of risk of future CVD (2,6–8), though the degree of association is dependent on levels of other conventional vascular risk factors (5,9). Current consensus recommendations support the clinical use of CRP to predict CVD risk among a subset of asymptomatic adults and in the selection of statin therapy (10). Despite routine clinical use of CRP levels, data from Mendelian randomization studies suggest that CRP is unlikely to be causally related to CVD (11–17).
Genome-wide association studies (GWASs) have sought to characterize genetic determinates of CRP levels. This approach has been successful at identifying ∼25 loci associated with CRP levels among individuals of European (17–22), Asian (23–25) and African (26,27) descent (Supplementary Material, Table S1). To further interrogate known loci and to search for novel loci associated with CRP levels, we applied exome sequencing, which captures sequence variation in the protein-coding portion of the genome. Exome sequencing has proved useful for identifying rare causal variants for several Mendelian disorders (28–32). Furthermore, recent studies illustrate the application of exome sequencing to identify variation underlying complex traits (33–38).
In this study, we apply exome sequencing to a large sample of European-American (EA) and African-American (AA) ascertained through seven population-based cohorts [Atherosclerosis Risk in Communities (ARIC), Coronary Artery Risk Development in Young Adults (CARDIA), Cardiovascular Health Study (CHS), Framingham Heart Study (FHS), Jackson Heart Study (JHS), Multi-Ethnic Study of Atherosclerosis (MESA) and the Women's Health Initiative (WHI)] that compose the National Heart, Lung, and Blood Institute (NHLBI) Exome Sequencing Project (ESP) and the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) consortium. Our specific aims were to (1) assess whether known CRP loci harboring common variants also contain rare coding variants associated with CRP levels and (2) perform an exome-wide search for rare variants in novel genes associated with CRP levels. Follow-up HumanExome BeadChip genotyping data from an independent sample derived from the WHI and JHS cohorts were used as replication for discovery findings.
RESULTS
Participant characteristics
Race-stratified characteristics of discovery and validation cohorts are summarized in Supplementary Material, Table S2. Overall, compared with EA, AA had a greater proportion of women, higher prevalence of hypertension and type 2 diabetes, higher body mass index (BMI) and higher median CRP levels.
Single-variant test results
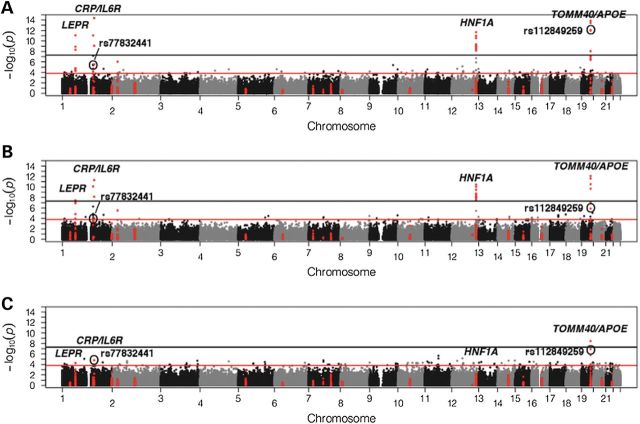
Summary information for uniquely annotated variants included in the ESP–CHARGE meta-analyses is detailed in Supplementary Material, Table S3. The meta-analyzed exome-wide single-variant association results for CRP levels in EA, AA and combined races are summarized in Figure 1 and Supplementary Material, Figure S1A. Overall results for significant association signals were consistent with no significant between-study heterogeneity (Tables 1 and 2).
Figure 1.
Manhattan plots of −log10(P-values) from single-variant analyses. (A) Combined race, (B) EA and (C) AA. Variants in the 25 candidate loci identified through CRP GWAS are highlighted in red, candidate loci with variant reaching exome-wide significance are labeled in italics and rare variants rs77832441 and rs112849259 are also labeled.
Table 1.
Exome-wide significant single-variant associations with CRP (P < 5.00 × 10−08) for common coding variants
| Gene (chromosome: GRCh37 coordinate) | rs# (function; coded/non-coded allele) | Previous report (GWAS variant if different than reported, LD measures) | Sample (N) | MAF | β (SE) | P-value | Het. I2 (P) |
|---|---|---|---|---|---|---|---|
| LEPR (1:66102257) | rs1805096 (Syn; G/A) | Reiner et al. (26) | EA (6050) | 0.37 | −0.11 (0.019) | 3.56 × 10−08 | 70.4 (0.012) |
| AA (3109) | 0.46 | −0.11 (0.027) | 5.37 × 10−05 | 0 (1) | |||
| Combined (9159) | 0.40 | −0.11 (0.016) | 8.34 × 10−12 | 56.7 (0.04) | |||
| IL6R (1:154426970) | rs2228145 (nSyn; C/A) | Curocichin et al. (39) | EA (6050) | 0.40 | −0.12 (0.019) | 7.81 × 10−11 | 0 (0.93) |
| AA (3109) | 0.14 | −0.089 (0.040) | 2.62 × 10−02 | 0 (0.38) | |||
| Combined (9159) | 0.31 | −0.12 (0.017) | 8.81 × 10−12 | 0 (0.86) | |||
| HNF1A (12:121435342) | rs2259820 (Syn; T/C) | Reiner et al. (19) (rs2464196, r2 = 1, D′ = 1)a | EA (6050) | 0.31 | −0.12 (0.020) | 1.83 × 10−09 | 0 (0.69) |
| AA (3109) | 0.12 | −0.067 (0.040) | 9.62 × 10−02 | 0 (0.96) | |||
| Combined (9159) | 0.25 | −0.11 (0.018) | 9.06 × 10−10 | 0 (0.72) | |||
| HNF1A (12:121435427) | rs2464196 (nSyn; A/G) | Reiner et al. (19) | EA (6050) | 0.32 | −0.12 (0.020) | 9.29 × 10−09 | 0 (0.71) |
| AA (3109) | 0.12 | −0.07 (0.040) | 8.07 × 10−02 | 0 (0.80) | |||
| Combined (9159) | 0.25 | −0.11 (0.018) | 3.27 × 10−09 | 0 (0.78) | |||
| HNF1A (12:121416622) | rs1169289 (Syn; G/C) | Kong and Lee (25) (rs2393791, r2 = 0.83, D′ = 0.93)a | EA (6050) | 0.46 | −0.12 (0.019) | 8.98 × 10−11 | 8.5 (0.34) |
| AA (3109) | 0.34 | −0.06 (0.028) | 2.16 × 10−02 | 0 (1) | |||
| Combined (9159) | 0.42 | −0.10 (0.016) | 2.64 × 10−11 | 20.9 (0.28) | |||
| HNF1A (12:121416650) | rs1169288 (nSyn; C/A) | Curocichin et al. (39) | EA (6050) | 0.34 | −0.11 (0.020) | 9.52 × 10−09 | 0 (0.73) |
| AA (3109) | 0.12 | −0.08 (0.041) | 4.04 × 10−02 | 0 (1) | |||
| Combined (9159) | 0.26 | −0.11 (0.018) | 1.36 × 10−09 | 0 (0.90) | |||
| TOMM40 (19:45395714) | rs157581 (Syn; C/T) | Middleburg et al. (40) (rs2075650, r2 = 0.58, D′ = 1)a | EA (6050) | 0.21 | −0.16 (0.023) | 2.42 × 10−12 | 0 (0.68) |
| AA (3109) | 0.47 | −0.09 (0.027) | 5.07 × 10−04 | 0 (0.88) | |||
| Combined (9159) | 0.30 | −0.13 (0.018) | 3.73 × 10−14 | 7.9 (0.37) | |||
| TOMM40 (19:45396144) | rs11556505 (Syn; T/C) | Middleburg et al. (40) (rs2075650, r2 = 1, D′ = 1)a | EA (6050) | 0.14 | −0.18 (0.027) | 2.86 × 10−11 | 0 (0.84) |
| AA (3109) | 0.12 | −0.02 (0.040) | 6.33 × 10−01 | 0 (0.43) | |||
| Combined (9159) | 0.13 | −0.13 (0.023) | 8.62 × 10−09 | 61 (0.025) | |||
| APOE (19:45411941) | rs429358 (nSyn; C/T) | Chasman et al. (41) | EA (1832)b | 0.11 | −0.31 (0.058) | 7.03 × 10−08 | 0 (1) |
| AA (1528)b | 0.19 | −0.24 (0.049) | 1.52 × 10−06 | 0 (1) | |||
| Combined (3360)b | 0.14 | −0.27 (0.038) | 8.05 × 10−13 | 0 (0.82) |
nSyn, nonsynonymous; syn, synonymous; MAF, minor allele frequency; Het. I2, heterogeneity; EA, European-American; AA, African-American; SE, standard error; (P), P-value; β, beta; SNP, single-nucleotide polymorphism; GRCh37, Genome Reference Consortium Human Build 37; rs#, reference SNP ID number.
aD′ and r2 values are based on Broad SNAP proxy search using CEU 1000 Genomes Pilot 1 data.
bThe reduced sample size for rs429358 is explained by the fact the variant passed quality control in ESP, but not in CHARGE.
Table 2.
Single-variant associations of rare and low frequency coding variants with CRP levels
| Gene (GRCh37 coordinate) | rs# (function; coded/non-coded allele) | Previously report | Sample (N) | MAF | β (SE) | P-value | Het. I2 (P) |
|---|---|---|---|---|---|---|---|
| CRP (1:159683438) | rs1800947 (Syn; G/C) | Ridker et al. (2) | EA (6050) | 0.0607 | −0.27 (0.039) | 4.82 × 10−12 | 0 (0.56) |
| AA (3109) | 0.0101 | −0.58 (0.134) | 1.54 × 10−05 | 0 (0.77) | |||
| Combined (9159) | 0.0435 | −0.30 (0.038) | 4.22 × 10−15 | 28.6 (0.22) | |||
| CRP (1:159683814) | rs77832441 (nSyn; A/G) | Not reported | EA (6050) | 0.0022 | −0.75 (0.197) | 1.39 × 10−04 | 0 (0.44) |
| AA (3109) | 0.0005 | −2.06 (0.605) | 6.65 × 10−04 | 0 (0.82) | |||
| Combined (9159) | 0.0016 | −0.88 (0.187) | 2.90 × 10−06 | 28.4 (0.22) | |||
| Replication EA (11 414) | 0.0031 | −0.90 (0.11) | 3.00 × 10−15 | – | |||
| Discovery + replication (14 573) | 0.0034 | – | 3.86 × 10−16 | – | |||
| TOMM40 (19:45397307) | rs112849259 (Syn; C/T) | Not reported | EA (6050) | 0.0266 | −0.28 (0.058) | 1.28 × 10−06 | 14.1 (0.32) |
| AA (3109) | 0.0391 | −0.37 (0.069) | 1.13 × 10−07 | 0 (0.39) | |||
| Combined (9159) | 0.0308 | −0.32 (0.044) | 1.06 × 10−12 | 16.2 (0.31) |
nSyn, nonsynonymous; syn, synonymous; MAF, minor allele frequency; Het. I2, heterogeneity; EA, European-American; AA, African-American; SE, standard error; (P), P-value; β, beta; SNP, single-nucleotide polymorphism; GRCh37, Genome Reference Consortium Human Build 37; rs#, reference SNP ID number.
Confirmation of common coding variants associated with CRP levels
Nine coding variants from four distinct chromosomal regions reached exome-wide significance in the combined sample (Table 1). All nine of these significant variants are common [minor allele frequency (MAF) >10% in both EA and AA] and have been previously reported or are in linkage disequilibrium (LD) with known CRP-associated single-nucleotide polymorphisms (SNPs) (2,19,25,26,39–41). The genes and their variants are hepatocyte nuclear factor 1 homeobox A [HNF1A (MIM 142410); rs2464196 (p.Ser487Asn), rs2259820 (p.Leu459=), rs1169288 (p.Ile27Leu) and rs1169289 (p.Leu17=), 12q24.2], interleukin 6 receptor [IL6R (MIM 147880); rs2228145 (p.Asp358Ala), 1q21.31], leptin receptor [LEPR (MIM 601007); rs1805096 (p.Pro1019=), 1p31.3], translocase of outer mitochondrial membrane 40 [TOMM40 (MIM 608061); rs157581 (p.Phe113=), rs11556505 (p.Phe131=), 19q13.32] and apolipoprotein E [APOE (MIM 107741); rs429358 (p.Cys130Arg), 19q13.32]. These significant hits in HNF1A, IL6R, LEPR, TOMM40 and APOE correspond to four independent association signals. Consistent with the results of prior GWAS meta-analyses, common intronic variants in CRP, LEPR, HNF1A and TOMM40 were also associated with CRP levels at an exome-wide significance level (Supplementary Material, Table S4). Supplementary Material, Table S5 summarizes effect estimates for all variants reaching significance by the analytic group (ESP, CHARGE-ARIC, CHARGE-CHS or CHARGE-FHS).
Lower frequency variation associated with CRP levels
Two lower frequency synonymous variants (TOMM40 rs112849259, p.Asp209=, MAF = 3.1% and CRP rs1800947, p.Leu184=, MAF = 4.4%) reached exome-wide significance in the combined sample of EA and AA participants (Table 2). The CRP rs1800947 synonymous variant has previously been associated with lower CRP levels in EA, independently of the more common CRP-lowering haplotype at the chromosome 1q23 CRP locus (12,20,42). TOMM40 rs112849259 has not previously been reported as associated with CRP levels. Since rs112849259 is not present on the Exome Array, we were unable to assess its association with CRP in our genotype-based validation sample.
In addition to the CRP rs1800947 synonymous variant, we identified a novel association of a rare nonsynonymous CRP variant rs77832441 (Thr59Met; MAF = 0.16%) with lower CRP levels in the combined discovery sample [A allele, beta (β) standard error (SE) = −0.88 (0.19); P = 2.90 × 10−6) that was significant at the candidate gene level (significance threshold P < 1.87 × 10−4). We subsequently validated this finding among 11 414 independent EA Exome Array samples, where the A allele (MAF = 0.31%) was strongly associated with CRP levels (β (SE) = −0.90 (0.11), P = 3.00 × 10−15). The combined discovery and replication P-value was 3.86 × 10−16.
Additional characterization of CRP p.Thr59Met rare variant association signal
To demonstrate that the CRP missense variant rs77832441 p.Thr59Met association signal is independent of other common and lower frequency CRP locus SNPs, we performed conditional regression analyses using either the ESP discovery exome-sequenced samples or the genotype-based Exome Array validation samples. As shown in Supplementary Material, Table S6, the association of rs77832441 p.Thr59Met with CRP levels was independent of CRP variants known to be associated with CRP through prior GWAS (rs1417938, rs3093059 and rs1800947). Furthermore, assessment of LD in the 1000 Genomes European ancestry panel structure showed very weak LD between rs77832441 and common CRP variants (Supplementary Material, Table S7).
The rs77832441-A allele encoding the threonine-to-methionine amino acid substitution could reduce CRP levels by affecting mRNA splicing or stability, reducing CRP synthesis or altering CRP monomer subunit structure or the ability for CRP monomers to associate into native circulating pentamers. The p.Thr59Met missense variant is located at residue 41 of the mature CRP polypeptide. According to the three-dimensional X-ray crystallographic structure of CRP (43), this amino acid lies at the end of a β-sheet (residues 32–41) proximate to a 3/10 alpha helical domain (residues 43–45). The 40–42 region of the CRP protein is involved in interprotomer interactions with residues 115–123 on the adjoining monomer (Supplementary Material, Fig. S2). Therefore, the non-conservative threonine-to-methionine amino acid change [which is predicted to be functional by in silico protein conservation algorithms (44,45)] has the potential to reduce CRP pentamerization or reduce native pentameric CRP stability.
To follow up on this association, we sought to test whether other inflammation-related phenotypes were associated with the CRP-lowering A-allele of rs77832441 p.Thr59Met. In data from 14 727 coronary heart disease (CHD) cases and 30 232 controls from the Myocardial Infarction Genetics Exome Array Consortium, we failed to demonstrate an association between rs77832441 and a decreased risk of CHD [odds ratio (95% confidence interval (CI) ) = 0.99 (0.65–1.51); P = 0.96] despite having an estimated 80% power to detect an odds ratio of 0.67 or less. Furthermore, using data from the EA replication sample, we were unable to demonstrate any association between the CRP rs77832441 variant and incident ischemic stroke [Ncases = 2378, Ncontrols = 16 736; odds ratio (95% CI) = 1.28 (0.76–2.14); P = 0.35], systolic blood pressure [N = 19 111; β (SE) = −0.48 (1.98) mmHg; P = 0.81], waist-to-hip ratio [N = 19 111; β (SE) = −0.0014 (0.0071); P = 0.84] and BMI [N = 19 111; β (SE) = 0.044 (0.54) kg/m2; P = 0.93]. Finally, test for the association of rs77832441 with type 2 diabetes in 6474 cases and 6370 controls from EA participants included in the Type 2 Diabetes Genetic Exploration by Next-generation sequencing in Ethnic Samples Consortium failed to identify an association of the p.Thr59Met variant on type 2 diabetes risk [odds ratio (95% CI) = 0.72 (0.22–2.37); P = 0.59].
Additional analysis of common coding variants at the chromosome 19q13
Two common synonymous variants in TOMM40 (rs157581-C allele, MAF = 30%; and rs11556505-T allele, MAF = 13%) were associated with a ∼12% lower mean CRP level (P ≤ 8.62 × 10−9) in the combined EA and AA single-variant meta-analysis. On the basis of previous studies (39,46), we hypothesized that these TOMM40 common variants and/or the low frequency (rs112849259) variant may be in LD with the functional APOE ε2, ε3 and ε4 alleles. We therefore assessed the relationship between the common and low frequency TOMM40 variants to the APOE ε4 allele-defining variant rs429358 p.Cys130Arg by performing conditional analysis using the ESP exome sequencing data. Conditional analysis in EA and AA demonstrated that all three TOMM40 synonymous CRP-lowering variants (rs157581, rs11556505 and rs112849259) are conditionally dependent on the APOE ε4 allele-defining SNP rs429358 (Table 3).
Table 3.
Conditional analysis of TOMM40 synonymous variants on the APOE ε4-defining missense variant rs429358 in the ESP discovery sample
| rs# (function) | MAF (EA/AA/combined) | Sample (N) | 1000 Genomes pairwise r2 (rs112849259, rs157581, rs11556505) | Without adjustment for rs429358 |
With adjustment for rs429358 |
||
|---|---|---|---|---|---|---|---|
| β (SE) | P-value | β (SE) | P-value | ||||
| rs112849259 (Synonymous) | 0.026/0.039/0.031 | EA (1832) | CEU (1, 0.07, 0.005) | −0.40 (0.11) | 1.29 × 10−06 | −017 (0.12) | 0.13 |
| AA (1528) | YRI (rs112849259 NA in YRI) | −0.30 (0.10) | 2.22 × 10−03 | −0.15 (0.11) | 0.15 | ||
| rs157581 (Synonymous) | 0.21/0.47/0.30 | EA (1832) | CEU (0.07, 1, 0.58) | −0.16 (0.044) | 2.15 × 10−04 | −0.030 (0.058) | 0.60 |
| AA (1528) | YRI (NA, 1, 0.11) | −0.089 (0.038) | 1.99 × 10−02 | −0.025 (0.043) | 0.56 | ||
| rs11556505 (Synonymous) | 0.14/0.12/0.13 | EA (1832) | CEU (0.005, 0.583, 1) | −0.18 (0.052) | 6.77 × 10−04 | −0.014 (0.068) | 0.84 |
| AA (1528) | YRI (NA, 0.11, 1) | −0.051 (0.056) | 0.37 | −0.032 (0.057) | 0.57 | ||
MAF, minor allele frequency; Het. I2, heterogeneity; EA, European-American; AA, African-American; SE, standard error; (P), P-value; β, beta; rs#, reference SNP ID number.
The APOE ε2 allele-defining variant rs7412 p.Arg176Cys failed quality control due to poor sequencing coverage in both ESP and CHARGE discovery samples; however, rs7412 genotype was available in 13 794 individuals from our validation sample genotyped on the Exome Array. Association analyses using the Exome Array samples suggested that the rs7412 minor allele (T) was associated with higher CRP levels in 2379 AA [β (SE) = 0.17 (0.05), P = 6.79 × 10−4] from WHI; however, there was no evidence of association of rs7412 with CRP in 11 404 EA [β (SE) = 0. 018 (0.024), P = 0.46]. We subsequently validated the rs7412 association in an independent sample of 2201 AA from JHS with Exome Array genotype data [β (SE) = 0.20 (0.057); P = 4.44 × 10−4]. The combined meta-analysis results for the association of rs7412 in an Exome Array validation sample with CRP (N = 4609) were β (SE) = 0.18 (0.037); P = 1.09 × 10−6. Using allele dosage for the APOE ε4 variant rs429358 imputed from 1000 Genomes in WHI (imputation Rsq = 0.64), we demonstrated in the combined AA Exome Array validation sample (N = 4211) that the association of APOE ε2-defining variant rs7412 with higher CRP levels (P = 3.74 × 10−5) and the association of APOE ε4 allele-defining variant rs429358 with lower CRP levels (P = 4.78 × 10−9) were conditionally independent.
Gene-based test results
Results for collapsing approach (T1) and sequence kernel association test (SKAT) gene-based tests are summarized in Supplementary Material, Figures S1B and S1C, respectively. None of the genes reached exome-wide significance (significance threshold P < 2.5 × 10−6) in EA or AA alone. In the race-combined sample, only rare variation in CRP was associated with CRP levels (T1 P = 2.36 × 10−6) at an exome-wide significance level.
Among the 25 candidate genes from GWAS (significance threshold P < 2.00 × 10−3), the most significant finding was that rare variation in the CRP locus was associated with CRP levels in EA with both the T1 test (P = 6.80 × 10−4) and SKAT (P = 1.71 × 10−4; Supplementary Material, Tables S8 and S9). Results for both tests of EA on the Exome Array robustly replicated the CRP gene-based association (PSKAT = 3.21 × 10−15 ; PT1 = 2.54 × 10−14). Removal of the rare CRP variant rs77832441 (Thr59Met) from the EA T1 and SKAT tests eliminated significant signal for the gene-based test (PSKAT = 0.96; PT1 = 0.77), suggesting that the CRP gene-based association signal is driven solely by rs77832441. Gene-based results for CRP in AA were less significant (PSKAT = 0.06; PT1 = 1.05 × 10−3), only reaching candidate gene level of significance in the T1 test.
Gene-based testing with SKAT additionally showed that rare putatively deleterious variation in the retinoic acid receptor-related orphan receptor α [RORA (MIM 600825)] locus was associated with CRP levels in AA (P = 1.73 × 10−3), but not in EA (P = 0.83). Using HumanExome BeadChip genotype data from an independent African-American validation sample (P = 0.06), the gene-based test did not reach a statistical significance. However, only one RORA rare variant was shared between exome sequencing discovery and HumanExome BeadChip validation platform, which may explain our inability to replicate the finding.
DISCUSSION
By combining exome sequence data from two large consortia, we have identified and validated a novel association between lower CRP levels and the rs7732441 CRP missense variant. Though strongly associated with CRP levels, rs7732441 was not associated with other inflammation-mediated phenotypes including CHD, stroke, systolic blood pressure, waist-to-hip ratio and BMI. In addition to the previously described CRP-lowering association signal attributable to the canonical APOE ε4 allele, we demonstrate a second, independent association signal from the APOE ε2 allele that is specific to AA. Finally, a targeted gene-based analysis of known CRP-associated genes suggested possible associations between rare coding variants in RORA (in AA) on CRP levels.
In this study, we did not identify any novel genes through aggregated gene-based tests. Meta-analysis of cohort-specific results in this study was carried out in skatMeta, which limited available gene-based methodologies to SKAT and burden tests. Methods development for rare variant association analyses is an active area of research (47). Future efforts may extend available options for meta-analysis, which may be capable of identifying additional rare variant association signals in our dataset.
Previous studies suggest that tens of thousands of samples may be necessary to be sufficiently powered to detect associations between rare variants and complex traits (48,49). Though we uncover associations between CRP levels and rare-to-low frequency variants of larger effect, we recognize that larger sample sizes will be required to detect associations of rare coding variants of more moderate effect sizes. Power calculations based on our discovery sample size of ∼6000 EAs suggest we had 80% power to detect a variant with ∼2 mg/l effect on CRP at 3% MAF but considerably less power to detect the same effect size at MAF ≤ 1% (Supplementary Material, Fig. S3). Beyond limited power, factors such as heterogeneity of study sampling design (inclusive of both extreme sampling for other phenotypes and randomly selected individuals) or exome sequencing platform, and minor differences in variant calling and quality control procedures between ESP and CHARGE consortia, may have limited our ability to detect novel rare variants associated with CRP levels and/or limit generalizability of our findings.
CRP
Previous studies have identified and replicated associations between CRP levels and variants in the CRP locus in populations of European (17,18,20), African (26,27), Asian (17,23,24,39) and Hispanic ancestry (26). These GWAS have identified three independently associated common alleles (rs33116653, rs12093699 and rs1205) and a single low frequency synonymous variant (rs1800947) in the CRP gene associated with lower CRP levels in EA (Supplementary Material, Table S10). GWAS and candidate gene studies (12,42) have identified an additional AA-specific allele (rs16827466) associated with higher CRP levels.
Extending these previous findings, we identify a novel rare CRP variant (rs77832441) that is both strongly associated with CRP level in single-variant tests and drives the gene-based results for CRP. On the basis of conditional analyses (Supplementary Material, Table S6) and LD estimates (Supplementary Material, Table S7), this variant appears to represent an independent signal from known GWAS CRP SNPs. Despite the magnitude of its effect, rs77832441 only contributed 0.5% of the overall natural log(CRP) phenotypic variance. Our results extend the existence of allelic heterogeneity at the CRP locus and suggest that rare coding variation in the CRP gene contributes to CRP phenotypic variation.
Functional prediction algorithms taking into account conservation and protein structure, SIFT (45) and Polyphen-2 (44) predict the p.Thr59Met variant to be deleterious [SIFT = 0.03 (damaging) and Polyphen-2 = 1.0 (probably damaging)]. Similarly, crystal structures of CRP indicate that the variant could disrupt the site involved in monomer subunit interactions (43), which may reduce CRP pentamer formation or stability. In vitro, the monomeric and native pentameric isoforms of CRP exhibit distinct physicochemical and inflammatory properties (50–52), which have potential implications for the role of CRP in atherothrombotic disorders.
It remains possible that, since CRP levels were measured by immunoassay, the p.Thr59Met missense variant may alter an epitope recognized by the monoclonal antibody used for CRP capture or detection, leading to artificially low CRP values. We investigated the possibility that the p.Thr59Met amino acid substitution interferes with CRP detection by examining CRP measurements among a subset of 3442 CARDIA participants for whom both polyclonal and monoclonal antibody assays were performed at study Year 15. We hypothesized that the polyclonal assay would be less susceptible to artifactual bias due to the amino acid substitution at residue 59. Comparison of the results failed to demonstrate a large difference in CRP effect size between monoclonal and polyclonal assays in a subset of the discovery sample (Supplementary Material, Fig. S4). CRP levels detected by monoclonal antibody and polyclonal antibody assays were highly correlated in this sample for p.Thr59Met carriers (r = 0.97) and non-carriers (r = 0.94). These results suggest that p.Thr59Met may confer a true biological reduction in circulating CRP levels, though further studies are warranted to fully characterize the functional significance of this variant.
Despite the large, likely direct effect of rs77832441 on mean CRP levels, investigation of the relationship of the identified rare variant to other inflammation-mediated phenotypes failed to demonstrate an association. Similar to previous studies (11–17,53–55), these findings provide additional evidence that CRP levels may not elicit direct effects on CHD, stroke, systolic blood pressure, hip-to-waist ratio, type 2 diabetes and BMI. While CRP is a robust marker of CHD risk (2,6–8), Mendelian randomization studies have consistently failed to demonstrate that CRP is causally related to CHD (11–17). Our finding of a lack of association between rare, putatively deleterious rs77832441 and CHD provides additional support that CRP is not in the causal pathway for CHD.
TOMM40-APOE
19q13 is a gene-dense region including TOMM40, APOE and apolipoprotein C-I (APOC1; MIM 107710). Variants in all three of these genes have been previously associated with CRP levels through GWAS or candidate gene analyses (17,26,39–41,46). In our exome sequencing analysis, the APOE ε4-tagging missense variant (rs429358) and three synonymous TOMM40 variants were associated with CRP levels (rs157581, rs11556505 and rs112849259). Evidence from previous study and 1000 Genomes sequencing suggest that rs157581 and rs11556505 are in moderate-to-strong LD with rs429358, the variant that defines the APOE ε4 allele (56,57). Similarly, we show in our dataset that the CRP-associated TOMM40 variants (rs157581, rs11556505 and rs112849259) do not represent an independent signal from the APOE rs429358 variant. Furthermore, we identify and replicate an additional AA-specific rs429358-independent novel association between APOE ε2-tagging variant rs7412 and higher CRP level.
APOE is a major constituent of lipoproteins, and the APOE ε2 and ε4 alleles are important genetic determinants of CVD and Alzheimer's disease (58,59). The p.Arg158Cys missense variant encoded by ε2 exhibits reduced affinity for the low-density lipoprotein receptor (LDLR) and reduced clearance of apoE-containing triglyceride-rich lipoprotein particles (such as very low-density lipoprotein), and is associated with lower low-density lipoprotein (LDL) cholesterol levels (41) and type III hyperlipoproteinemia (59). In contrast, the p.Cys112Arg variant encoded by ε4 exhibits more rapid clearance of apoE-containing triglyceride-rich lipoproteins due to increased LDLR affinity and is associated with increased LDL-cholesterol (LDL-c) levels and increased cardiovascular risk (41,58,60,61). In addition to its role in cholesterol transport, APOE has anti-atherogenic and anti-inflammatory effects in experimental systems (59,62). However, the mechanisms for these pleiotropic effects as well as the paradoxical association of ε2 with lower LDL-c/higher CRP levels and ε4 with higher LDL-c/lower CRP levels are not well understood.
RORA
RORA encodes a nuclear receptor with strong homology to the retinoic acid receptor. Data from knockout RORA mice models suggest the importance of RORA in regulating immune and inflammatory responses, atherosclerosis susceptibility and ischemia-induced angiogenesis (63,64). Previous GWAS indicate that common variation in RORA is associated with CRP (18) and liver enzyme levels (65,66) at a genome-wide significance level in EA. The previous GWAS meta-analysis of CRP identified a common variant rs340029-T allele as associated with increased CRP levels (18); however, GWAS in AA and Hispanic Americans found no association between rs340029 and CRP levels (26). Further characterization of rare variation of this locus through sequencing efforts is warranted to follow up on our discovery finding of a burden of rare variants at the locus.
Summary
Overall, our results suggest that variants distributed across the allele frequency spectrum within biological candidate genes identified by GWAS contribute to CRP levels. As suggested by other studies, robustly associating rare coding variants with modest effects on complex traits will require extremely large sample sizes (26,48). Collaborative efforts involving meta-analysis of exome sequence or exome BeadChip genotype data will be necessary to amass the large sample sizes required to identify additional rare coding variants contributing to the phenotypic variance of CRP levels.
MATERIALS AND METHODS
Study subjects and CRP measurements
Our discovery sample consisted of exome sequence data from 3360 individuals from the NHLBI-ESP and 5799 individuals from the CHARGE project with valid CRP measures. In total, these 9159 participants included 6050 EA and 3109 AA sampled from seven population-based cohorts: ARIC (N = 4827), CARDIA (N = 190), CHS (N = 946), FHS (N = 1144), JHS (N = 346), MESA (N = 399) and the WHI (N = 1307) as part of the NHLBI- ESP and an independent sample from three of the same population-based cohorts (ARIC, FHS and CHS) as part of the CHARGE consortium (67). CRP levels were measured by high-sensitivity immunoassay in all seven cohorts. Detailed descriptions of each of the seven cohorts and the techniques used to measure circulating CRP levels are provided in previous publications (68–73) and summarized in Supplementary Material, Table S11. Clinical information was collected through self-report and in-person examination. All participants provided written informed consent as approved by local human subjects committees.
Sampling design
The CHARGE cohorts' study participants included here were selected as part of a large random cohort sample or for extreme values for at least one of the following phenotypes: age at menopause, electrocardiogram QT interval, fasting blood glucose, fibrinogen level, renal function, Stamler–Kannel-like extremes of risk factors selected by principle components (PCs) and waist-to-hip ratio. The sampling design of ESP included disease phenotypes (early onset myocardial infarction and ischemic stroke), and several quantitative cardiovascular risk factors that were sampled on the basis of phenotypic extremes (blood pressure, BMI and LDL-c), as well as a deeply phenotyped random sample. Due to the extreme sampling of phenotypic extremes, we adjusted for sampling design to minimize bias.
Exome sequencing and variant calling
In ESP, the processes of library construction, exome capture, sequencing and mapping were performed as previously described (26,37,74). Sequencing was performed at the University of Washington (UW) and the Broad Institute of MIT/Harvard (Broad). Briefly, exome capture was performed using Roche Nimblegen SeqCap EZ or Agilent SureSelect Human All Exon 50 Mb. Paired-end sequencing (2 × 76 bp) was performed on Illumina GAII and HiSeq instruments. Single-nucleotide variants (SNVs) were called using a maximum-likelihood approach (75) implemented in the UMAKE pipeline at the University of Michigan, which allowed all samples to be analyzed simultaneously, both for variant calling and filtering. Binary Alignment/Map (BAM) (76) files summarizing Burrows–Wheeler Alignment (BWA) (75) alignments generated at the UW and the Broad were used as an input. These BAM files summarized alignments mapped to the Genome Reference Consortium Human Build 37 (GRCh37), refined by duplicate removal, recalibration and indel re-alignment using the Genome Analysis ToolKit (GATK) (77). We excluded all reads that were not confidently mapped (Phred-scaled mapping quality <20) from further analysis. Mean depth was 127× in targeted regions. We then computed genotype likelihoods for exome targeted regions and 50 flanking bases, accounting for per base alignment quality using SAMtools (76). Variable sites and their allele frequencies were identified using a maximum-likelihood model, implemented in glfMultiples (78). These analyses assumed a uniform prior probability of polymorphism at each site. The final call-set was performed on 6823 samples (referred to as the ESP6800 call-set).
In CHARGE, DNA samples were constructed into Illumina paired-end pre-capture libraries according to the manufacturer's protocol. The complete protocol and oligonucleotide sequences are accessible from the Human Genome Sequencing Center (HGSC) website. Either four or six pre-capture libraries were pooled together and then hybridized to Nimblegen exome capture array [HGSC VCRome 2.1 design (79); (42 Mb, NimbleGen)] and sequenced in paired-end mode in a single lane on the Illumina HiSeq 2000 platform. Illumina sequence analysis was performed using the HGSC Mercury analysis pipeline. Pooled samples were de-multiplexed using the Consensus Assessment of Sequence and VAriation (CASAVA) software. Reads were then mapped to the GRCh37 human reference sequence using BWA producing BAM files. Aligned reads were then recalibrated using GATK (77) along with BAM sorting, duplicate read marking and realignment near indels. The Atlas2 (80) suite was used to call variants and produce high-quality variant call files (VCF) (81). The VCF includes a filter indicating variants with apparent strand-bias, low allele fraction, low coverage or low quality to produce a high-quality variant list. Specifically, the poor quality fields included variants with a posterior probability of less than0.95, <3 variant reads, variant read ratio <0.1, >99% variant reads in a single-strand direction, total coverage <6 and homozygous reference alleles with less than 6× coverage.
Quality control
ESP used a support vector machine classifier to separate likely true-positive and false-positive variant sites, applying a series of variant-level filtering steps. Variant-level quality metrics included allelic balance (the proportional representation of each allele in likely heterozygotes), base quality distribution for sites supporting the reference and alternate alleles, and the distribution of supporting evidence between strands and sequencing cycle, among others. These steps were followed by quality control on individual samples within each study. We used as the positive training set variants identified by dbSNP (82) or 1000 Genomes (57), and we used variants that failed multiple filters as the negative training set. We found this method to be effective at removing sequencing artifacts while preserving good quality data, as indicated by the transition–transversion (ti–tv) ratio for previously known and newly identified variant sites, the proportion of high-frequency variants overlapping with dbSNP, and the ratio of synonymous to nonsynonymous variants, as well as attempts at the validation of a subset of sites. We excluded variants with read depth >500, variants with >2 observed alleles in CHARGE and any genotype containing a copy of the less frequent alternate allele in ESP, or missing rates ≥10% for ESP and >20% for CHARGE, and HWE P-value of <5 × 10−8 and <5 × 10−6 for ESP and CHARGE, respectively.
In ESP, samples with discrepant self-reported race and ancestry derived from principal component analysis (PCA) performed on exome sequencing data in PLINK (83) as well as ancestry outliers by PCA were removed. Samples having very low concordance (<90%) with previously obtained SNP array data were considered likely sample swaps and were also dropped from further analysis. In CHARGE, within each cohort, a sample was excluded if it fell beyond 6 standard deviations of any of four selected measures that were calculated by the cohort and ancestry group: number of singletons, heterozygote to homozygote ratio, mean depth or ti–tv ratio.
Variant annotation
To facilitate meta-analysis between CHARGE and ESP, we created a combined variant annotation file including all quality-controlled variant sites observed in either study: 2 706 877 variants in CHARGE and 1 907 911 in ESP6800. We first annotated variants in the two studies separately using an in-house pipeline built on ANNOVAR (84) and dbNSFP v2.0 (85) according to the reference genome GRCh37 and National Center for Biotechnology Information RefSeq. The majority of the exonic variants were annotated to a unique gene and functional category. A variant, however, can be annotated to multiple genes by ANNOVAR and can have more than one of the following functional categories: stop-gain, stop-loss, splicing, nonsynonymous, noncoding RNA splicing, synonymous, exonic, 5′ untranslated region (UTR5), 3′ untranslated region (UTR3), noncoding RNA exonic, upstream, intronic, noncoding RNA intronic, downstream and intergenic, where the first five categories are considered ‘functional’ variants to be included in the rare variant burden tests. We then merged the CHARGE and ESP annotated variant lists to ensure that a variant that was present in both studies has the same reference allele and functional annotation. The combined CHARGE-ESP SNP info file that was used in the skatMeta package included a total of 3 494 971 unique autosomal sites present in either or both ESP and CHARGE.
Association analyses
Samples with very high CRP values (>100 mg/l) were excluded from analyses and measured CRP values below the lower limit of detection were replaced with the assay lower limit value, leaving 3109 AA and 6050 EA available for association testing. CRP values were naturally log (ln) transformed to normalize the distribution of CRP levels. We performed two types of tests, single variant (common and lower frequency variants) and gene burden (rare and lower frequency variants only), as detailed further below and summarized in Supplementary Material, Table S12. Cohort-level analyses were carried out using the R (86) skatMeta package. In the CHARGE cohorts (ARIC, FHS and CHS), the ‘skatCohort’ or ‘skatFamcohort’ functions were used to create datasets for meta-analysis. In ESP, the six cohorts were pooled into a single dataset and were analyzed using the ‘skatCohort’ function.
Meta-analyses of the ‘skatCohort’ and ‘skatFamcohort’ results were conducted at two independent sites to ensure concordance in findings. Both race-stratified meta-analysis considering a single ethnicity, and combined meta-analyses including both ethnicities were carried out because most CRP-associated loci identified to date have shown consistent patterns of association between EA and AA (26). All meta-analyses were conducted in the skatMeta package using the ‘singlesnpMeta’ function for single-variant meta-analyses, and the ‘burdenMeta’ and ‘skatMeta’ functions for gene-level meta-analyses. We considered only variants on autosomal chromosomes in all analyses. Confirmatory analyses in METAL (87) evaluate between-study heterogeneity of significant results using the heterogeneity I2 metric. Based on adjustment for 23 tests of significant CRP-associated SNV in the discovery sample, heterogeneity was considered to be statistically significant (a P-value of <2.17 × 10−3).
Single-variant tests
Using the skatMeta, we ran race-stratified study-specific objects (ESP, CHARGE-ARIC, CHARGE-FHS and CHARGE-CHS) for downstream meta-analyses. Within each meta-analysis group (EA, AA and combined) and for each variant site with five or more minor alleles detected, we tested for association with CRP levels via linear regression with an additive genetic model. We included as covariates, race-specific PCs as needed, age, sex and BMI. In the ESP samples, a dummy variable correcting for the sampling procedure, cohort and capture target was included in the model.
Gene burden tests
Using the skatMeta package, we ran two different types of gene-level tests. The first was adopted from the T1 collapsing approach (15). For the T1 tests, we only considered polymorphic variants having a within race MAF of ≤0.01 that was calculated from the entire ESP–CHARGE call-set. The second gene-level test was adopted from the SKAT approach (47). For the SKAT tests, we only considered variants having an MAF of ≤0.05 that was calculated from the entire ESP–CHARGE call-set. Both T1 and SKAT tests considered only SNVs annotated as nonsynonymous, stop-gain, stop-loss, noncoding RNA splice or splice variants in the shared ESP/CHARGE annotation file. Burden tests considered only considered variants that passed quality control in either or both ESP and CHARGE. All gene-level tests were adjusted for age, sex, BMI and principal components (as needed). In addition, the ESP analyses included a dummy variable correcting for the sampling procedure, cohort and capture target.
Significance thresholds
For the analysis of rare variants (MAF <0.05) in the 25 known CRP-associated genes, we evaluated 267 coding variants and thus, we used a Bonferroni-corrected threshold of P < 1.87 × 10−4 to declare significance at the single-variant level. In the burden test of candidate genes, we corrected for the number of candidate genes surveyed (N = 25) to assign the threshold of significance at P < 2.0 × 10−3. In the exome-wide exploratory analyses, an association was deemed to be statistically significant at P≤5.0 × 10−8 for single variants (the standard GWAS common variant criteria for assessing significance based on a million test), which is a stringent threshold considering that we only tested ∼640 000 variant sites. Significance was assessed at P < 2.5 × 10−6 for the gene-based rare variant association tests based on a correction for 19 230 genes. Supplementary Material, Table S12 summarizes the significance thresholds for all the reported tests.
Validation of CRP association signals in additional samples
Significant association findings from the discovery sample were followed up in an independent sample of 13 794 participants of the WHI with CRP measurement available [11 414 EA participants and a sub-sample of 2380 AA participants from the WHI-SHARe project (26)] and 2201 AA from JHS. Replication samples were independent from samples used in the ESP and CHARGE exome sequencing discovery sample. The JHS and WHI validation samples were genotyped using the Illumina HumanExome v1.0 BeadChip at Broad Institute or at the Translational Genomics Research Institute (Phoenix, AZ), respectively. Genotype calls were assigned using GenomeStudio v2010.3. We removed samples with call-rates <98%, SNPs with call-rates <95% or HWE P-values less than 5 × 10−6. We checked concordance of genotype calls across hundreds of duplicated samples and SNPs with concordance rates <99% were excluded from analysis. High-sensitivity CRP was measured on with the use of a latex-particle enhanced immunoturbidimetric assay. Consistent with the discovery analysis, only samples with CRP values <100 mg/l were included in the replication analyses. Single-variant and gene burden tests for association tests were performed as described above.
Investigation of the association between rs77832441 and inflammation-related phenotypes
Samples from the WHI EA replication with available Illumina Human Exome Array data were used to investigate the association between rs77832441 and inflammation-mediated phenotypes stroke, systolic blood pressure, waist-to-hip ratio and BMI. To investigate the association with CHD, we used EA samples from the Myocardial Infarction Genetics Exome Array Consortium with available Illumina HumanExome BeadChip data (N CHD cases = 14 727, N controls = 30 232). Availability of EA samples from 6474 type 2 diabetes cases and 6370 controls from the Type 2 Diabetes Genetic Exploration by Next-generation sequencing in Ethnic Samples Consortium with available exome sequence data permitted us to evaluate whether rs77832441-A allele was associated with case status. Linear or logistic regression was used to assess association between the predictor (rs77832441—per A-allele) and the continuous or binary inflammation-mediated phenotype with adjustment for covariates as needed. Statistical significance was assessed at P < 0.05.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
ACKNOWLEDGEMENTS
The authors wish to acknowledge the support of the National Heart, Lung, and Blood Institute (NHLBI) and the contributions of the research institutions, study investigators, field staff and study participants in creating this resource for biomedical research. Funding for GO ESP was provided by NHLBI grants RC2 HL-103010 (HeartGO), RC2 HL-102923 (LungGO) and RC2 HL-102924 (WHISP). The exome sequencing was performed through NHLBI grants RC2 HL-102925 (BroadGO) and RC2 HL-102926 (SeattleGO). Funding support for ‘Building on GWAS for NHLBI-diseases: the US CHARGE consortium’ was provided by the NIH through the American Recovery and Reinvestment Act of 2009 (ARRA) (5RC2HL102419). Data for ‘Building on GWAS for NHLBI-diseases: the US CHARGE consortium’ were provided by Eric Boerwinkle on behalf of the Atherosclerosis Risk in Communities (ARIC) Study, L. Adrienne Cupples, principal investigator for the Framingham Heart Study, and Bruce Psaty, principal investigator for the Cardiovascular Health Study. Sequencing was carried out at the Baylor Genome Center (U54 HG003273). The ARIC Study is carried out as a collaborative study supported by National Heart, Lung, and Blood Institute (NHLBI) contracts (HHSN268201100005C, HHSN268201100006C, HHSN268201100007C, HHSN268201100008C, HHSN268201100009C, HHSN2682011000010C, HHSN2682011000011C and HHSN2682011000012C), R01HL087641, R01HL59367 and R01HL086694. The authors thank the staff and participants of the ARIC study for their important contributions. The Framingham Heart Study is conducted and supported by the NHLBI in collaboration with Boston University (Contract No. N01-HC-25195), and its contract with Affymetrix, Inc., for genome-wide genotyping services (contract no. N02-HL-6-4278), for quality control by Framingham Heart Study investigators using genotypes in the SNP Health Association Resource (SHARe) project. A portion of this research was conducted using the Linux Cluster for Genetic Analysis (LinGA) computing resources at Boston University Medical Campus. This CHS research was supported by contracts HHSN268201200036C, HHSN268200800007C, N01 HC55222, N01HC85079, N01HC85080, N01HC85081, N01HC85082, N01HC85083, N01HC85086 and grants HL080295, HL087652, HL105756 from the National Heart, Lung, and Blood Institute (NHLBI) with additional contribution from National Institute of Neurological Disorders and Stroke (NINDS). Additional support was provided through AG023629 from the National Institutes on Aging (NIA). A full list of CHS principal investigators and institutions can be found at CHS-NHLBI.org. Supported in part by grant R25CA094880 from the National Cancer Institute and by R01HL071862 from NHLBI. The Type 2 Diabetes Genetic Exploration by Next-generation sequencing in multi-Ethnic Samples (T2D-GENES) project was supported by NIH grant U01DK085526.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Dehghan A., Kardys I., de Maat M.P., Uitterlinden A.G., Sijbrands E.J., Bootsma A.H., Stijnen T., Hofman A., Schram M.T., Witteman J.C. Genetic variation, C-reactive protein levels, and incidence of diabetes. Diabetes. 2007;56:872–878. doi: 10.2337/db06-0922. [DOI] [PubMed] [Google Scholar]
- 2.Ridker P.M. Clinical application of C-reactive protein for cardiovascular disease detection and prevention. Circulation. 2003;107:363–369. doi: 10.1161/01.cir.0000053730.47739.3c. [DOI] [PubMed] [Google Scholar]
- 3.Sesso H.D., Buring J.E., Rifai N., Blake G.J., Gaziano J.M., Ridker P.M. C-reactive protein and the risk of developing hypertension. J. Am. Med. Assoc. 2003;290:2945–2951. doi: 10.1001/jama.290.22.2945. [DOI] [PubMed] [Google Scholar]
- 4.Erlinger T.P., Platz E.A., Rifai N., Helzlsouer K.J. C-reactive protein and the risk of incident colorectal cancer. J. Am. Med. Assoc. 2004;291:585–590. doi: 10.1001/jama.291.5.585. [DOI] [PubMed] [Google Scholar]
- 5.Kaptoge S., Di Angelantonio E., Lowe G., Pepys M.B., Thompson S.G., Collins R., Danesh J. C-reactive protein concentration and risk of coronary heart disease, stroke, and mortality: an individual participant meta-analysis. Lancet. 2010;375:132–140. doi: 10.1016/S0140-6736(09)61717-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ridker P.M., Hennekens C.H., Buring J.E., Rifai N. C-reactive protein and other markers of inflammation in the prediction of cardiovascular disease in women. N. Engl. J. Med. 2000;342:836–843. doi: 10.1056/NEJM200003233421202. [DOI] [PubMed] [Google Scholar]
- 7.Ridker P.M., Rifai N., Rose L., Buring J.E., Cook N.R. Comparison of C-reactive protein and low-density lipoprotein cholesterol levels in the prediction of first cardiovascular events. N. Engl. J. Med. 2002;347:1557–1565. doi: 10.1056/NEJMoa021993. [DOI] [PubMed] [Google Scholar]
- 8.Tracy R.P., Lemaitre R.N., Psaty B.M., Ives D.G., Evans R.W., Cushman M., Meilahn E.N., Kuller L.H. Relationship of C-reactive protein to risk of cardiovascular disease in the elderly. Results from the Cardiovascular Health Study and the Rural Health Promotion Project. Arterioscler. Thromb. Vasc. Biol. 1997;17:1121–1127. doi: 10.1161/01.atv.17.6.1121. [DOI] [PubMed] [Google Scholar]
- 9.Miller M., Zhan M., Havas S. High attributable risk of elevated C-reactive protein level to conventional coronary heart disease risk factors: the Third National Health and Nutrition Examination Survey. Arch. Intern. Med. 2005;165:2063–2068. doi: 10.1001/archinte.165.18.2063. [DOI] [PubMed] [Google Scholar]
- 10.Greenland P., Alpert J.S., Beller G.A., Benjamin E.J., Budoff M.J., Fayad Z.A., Foster E., Hlatky M.A., Hodgson J.M., Kushner F.G., et al. 2010 ACCF/AHA guideline for assessment of cardiovascular risk in asymptomatic adults: executive summary: a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines. Circulation. 2010;122:2748–2764. doi: 10.1161/CIR.0b013e3182051bab. [DOI] [PubMed] [Google Scholar]
- 11.Kardys I., de Maat M.P., Uitterlinden A.G., Hofman A., Witteman J.C. C-reactive protein gene haplotypes and risk of coronary heart disease: the Rotterdam Study. Eur. Heart J. 2006;27:1331–1337. doi: 10.1093/eurheartj/ehl018. [DOI] [PubMed] [Google Scholar]
- 12.Lange L.A., Carlson C.S., Hindorff L.A., Lange E.M., Walston J., Durda J.P., Cushman M., Bis J.C., Zeng D., Lin D., et al. Association of polymorphisms in the CRP gene with circulating C-reactive protein levels and cardiovascular events. J. Am. Med. Assoc. 2006;296:2703–2711. doi: 10.1001/jama.296.22.2703. [DOI] [PubMed] [Google Scholar]
- 13.Casas J.P., Shah T., Cooper J., Hawe E., McMahon A.D., Gaffney D., Packard C.J., O'Reilly D.S., Juhan-Vague I., Yudkin J.S., et al. Insight into the nature of the CRP-coronary event association using Mendelian randomization. Int. J. Epidemiol. 2006;35:922–931. doi: 10.1093/ije/dyl041. [DOI] [PubMed] [Google Scholar]
- 14.Pai J.K., Mukamal K.J., Rexrode K.M., Rimm E.B. C-reactive protein (CRP) gene polymorphisms, CRP levels, and risk of incident coronary heart disease in two nested case-control studies. PLoS ONE. 2008;3:e1395. doi: 10.1371/journal.pone.0001395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lawlor D.A., Harbord R.M., Timpson N.J., Lowe G.D., Rumley A., Gaunt T.R., Baker I., Yarnell J.W., Kivimaki M., Kumari M., et al. The association of C-reactive protein and CRP genotype with coronary heart disease: findings from five studies with 4,610 cases amongst 18,637 participants. PLoS ONE. 2008;3:e3011. doi: 10.1371/journal.pone.0003011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zacho J., Tybjaerg-Hansen A., Jensen J.S., Grande P., Sillesen H., Nordestgaard B.G. Genetically elevated C-reactive protein and ischemic vascular disease. N. Engl. J. Med. 2008;359:1897–1908. doi: 10.1056/NEJMoa0707402. [DOI] [PubMed] [Google Scholar]
- 17.Elliott P., Chambers J.C., Zhang W., Clarke R., Hopewell J.C., Peden J.F., Erdmann J., Braund P., Engert J.C., Bennett D., et al. Genetic loci associated with C-reactive protein levels and risk of coronary heart disease. J. Am. Med. Assoc. 2009;302:37–48. doi: 10.1001/jama.2009.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dehghan A., Dupuis J., Barbalic M., Bis J.C., Eiriksdottir G., Lu C., Pellikka N., Wallaschofski H., Kettunen J., Henneman P., et al. Meta-analysis of genome-wide association studies in >80 000 subjects identifies multiple loci for C-reactive protein levels. Circulation. 2011;123:731–738. doi: 10.1161/CIRCULATIONAHA.110.948570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reiner A.P., Barber M.J., Guan Y., Ridker P.M., Lange L.A., Chasman D.I., Walston J.D., Cooper G.M., Jenny N.S., Rieder M.J., et al. Polymorphisms of the HNF1A gene encoding hepatocyte nuclear factor-1 alpha are associated with C-reactive protein. Am. J. Hum. Genet. 2008;82:1193–1201. doi: 10.1016/j.ajhg.2008.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ridker P.M., Pare G., Parker A., Zee R.Y., Danik J.S., Buring J.E., Kwiatkowski D., Cook N.R., Miletich J.P., Chasman D.I. Loci related to metabolic-syndrome pathways including LEPR,HNF1A, IL6R, and GCKR associate with plasma C-reactive protein: the Women's Genome Health Study. Am. J. Hum. Genet. 2008;82:1185–1192. doi: 10.1016/j.ajhg.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sabatti C., Service S.K., Hartikainen A.L., Pouta A., Ripatti S., Brodsky J., Jones C.G., Zaitlen N.A., Varilo T., Kaakinen M., et al. Genome-wide association analysis of metabolic traits in a birth cohort from a founder population. Nat. Genet. 2009;41:35–46. doi: 10.1038/ng.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Benjamin E.J., Dupuis J., Larson M.G., Lunetta K.L., Booth S.L., Govindaraju D.R., Kathiresan S., Keaney J.F., Jr, Keyes M.J., Lin J.P., et al. Genome-wide association with select biomarker traits in the Framingham Heart Study. BMC Med. Genet. 2007;8(Suppl 1):S11. doi: 10.1186/1471-2350-8-S1-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu Y., McDade T.W., Kuzawa C.W., Borja J., Li Y., Adair L.S., Mohlke K.L., Lange L.A. Genome-wide association with C-reactive protein levels in CLHNS: evidence for the CRP and HNF1A loci and their interaction with exposure to a pathogenic environment. Inflammation. 2012;35:574–583. doi: 10.1007/s10753-011-9348-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Okada Y., Takahashi A., Ohmiya H., Kumasaka N., Kamatani Y., Hosono N., Tsunoda T., Matsuda K., Tanaka T., Kubo M., et al. Genome-wide association study for C-reactive protein levels identified pleiotropic associations in the IL6 locus. Hum. Mol. Genet. 2011;20:1224–1231. doi: 10.1093/hmg/ddq551. [DOI] [PubMed] [Google Scholar]
- 25.Kong M., Lee C. Genetic associations with C-reactive protein level and white blood cell count in the KARE study. Int. J. Immunogenet. 2013;40:120–125. doi: 10.1111/j.1744-313X.2012.01141.x. [DOI] [PubMed] [Google Scholar]
- 26.Reiner A.P., Beleza S., Franceschini N., Auer P.L., Robinson J.G., Kooperberg C., Peters U., Tang H. Genome-wide association and population genetic analysis of C-reactive protein in African American and Hispanic American women. Am. J. Hum. Genet. 2012;91:502–512. doi: 10.1016/j.ajhg.2012.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Doumatey A.P., Chen G., Tekola Ayele F., Zhou J., Erdos M., Shriner D., Huang H., Adeleye J., Balogun W., Fasanmade O., et al. C-reactive protein (CRP) promoter polymorphisms influence circulating CRP levels in a genome-wide association study of African Americans. Hum. Mol. Genet. 2012;21:3063–3072. doi: 10.1093/hmg/dds133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Choi M., Scholl U.I., Ji W., Liu T., Tikhonova I.R., Zumbo P., Nayir A., Bakkaloglu A., Ozen S., Sanjad S., et al. Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proc. Natl. Acad. Sci. USA. 2009;106:19096–19101. doi: 10.1073/pnas.0910672106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ng S.B., Buckingham K.J., Lee C., Bigham A.W., Tabor H.K., Dent K.M., Huff C.D., Shannon P.T., Jabs E.W., Nickerson D.A., et al. Exome sequencing identifies the cause of a Mendelian disorder. Nat. Genet. 2010;42:30–35. doi: 10.1038/ng.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ng S.B., Turner E.H., Robertson P.D., Flygare S.D., Bigham A.W., Lee C., Shaffer T., Wong M., Bhattacharjee A., Eichler E.E., et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461:272–276. doi: 10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bilguvar K., Ozturk A.K., Louvi A., Kwan K.Y., Choi M., Tatli B., Yalnizoglu D., Tuysuz B., Caglayan A.O., Gokben S., et al. Whole-exome sequencing identifies recessive WDR62 mutations in severe brain malformations. Nature. 2010;467:207–210. doi: 10.1038/nature09327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ng S.B., Bigham A.W., Buckingham K.J., Hannibal M.C., McMillin M.J., Gildersleeve H.I., Beck A.E., Tabor H.K., Cooper G.M., Mefford H.C., et al. Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome. Nat. Genet. 2010;42:790–793. doi: 10.1038/ng.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sanders S.J., Murtha M.T., Gupta A.R., Murdoch J.D., Raubeson M.J., Willsey A.J., Ercan-Sencicek A.G., DiLullo N.M., Parikshak N.N., Stein J.L., et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012;485:237–241. doi: 10.1038/nature10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O'Roak B.J., Deriziotis P., Lee C., Vives L., Schwartz J.J., Girirajan S., Karakoc E., Mackenzie A.P., Ng S.B., Baker C., et al. Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations. Nat. Genet. 2011;43:585–589. doi: 10.1038/ng.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guerreiro R.J., Lohmann E., Kinsella E., Bras J.M., Luu N., Gurunlian N., Dursun B., Bilgic B., Santana I., Hanagasi H., et al. Exome sequencing reveals an unexpected genetic cause of disease: NOTCH3 mutation in a Turkish family with Alzheimer's disease. Neurobiol. Aging. 2012;33:1008.e1017–e1023. doi: 10.1016/j.neurobiolaging.2011.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Albrechtsen A., Grarup N., Li Y., Sparso T., Tian G., Cao H., Jiang T., Kim S.Y., Korneliussen T., Li Q., et al. Exome sequencing-driven discovery of coding polymorphisms associated with common metabolic phenotypes. Diabetologia. 2013;56:298–310. doi: 10.1007/s00125-012-2756-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lange L.A., Hu Y., Zhang H., Xue C., Schmidt E.M., Tang Z.Z., Bizon C., Lange E.M., Smith J.D., Turner E.H., et al. Whole-exome sequencing identifies rare and low-frequency coding variants associated with LDL cholesterol. Am. J. Hum. Genet. 2014;94:233–245. doi: 10.1016/j.ajhg.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peloso G.M., Auer P.L., Bis J.C., Voorman A., Morrison A.C., Stitziel N.O., Brody J.A., Khetarpal S.A., Crosby J.R., Fornage M., et al. Association of low-frequency and rare coding-sequence variants with blood lipids and coronary heart disease in 56,000 whites and blacks. Am. J. Hum. Genet. 2014;94:223–232. doi: 10.1016/j.ajhg.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Curocichin G., Wu Y., McDade T.W., Kuzawa C.W., Borja J.B., Qin L., Lange E.M., Adair L.S., Lange L.A., Mohlke K.L. Single-nucleotide polymorphisms at five loci are associated with C-reactive protein levels in a cohort of Filipino young adults. J. Hum. Genet. 2011;56:823–827. doi: 10.1038/jhg.2011.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Middelberg R.P., Ferreira M.A., Henders A.K., Heath A.C., Madden P.A., Montgomery G.W., Martin N.G., Whitfield J.B. Genetic variants in LPL, OASL and TOMM40/APOE-C1-C2-C4 genes are associated with multiple cardiovascular-related traits. BMC Med. Genet. 2011;12:123. doi: 10.1186/1471-2350-12-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chasman D.I., Kozlowski P., Zee R.Y., Kwiatkowski D.J., Ridker P.M. Qualitative and quantitative effects of APOE genetic variation on plasma C-reactive protein, LDL-cholesterol, and apoE protein. Genes Immun. 2006;7:211–219. doi: 10.1038/sj.gene.6364289. [DOI] [PubMed] [Google Scholar]
- 42.Carlson C.S., Aldred S.F., Lee P.K., Tracy R.P., Schwartz S.M., Rieder M., Liu K., Williams O.D., Iribarren C., Lewis E.C., et al. Polymorphisms within the C-reactive protein (CRP) promoter region are associated with plasma CRP levels. Am. J. Hum. Genet. 2005;77:64–77. doi: 10.1086/431366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shrive A.K., Cheetham G.M., Holden D., Myles D.A., Turnell W.G., Volanakis J.E., Pepys M.B., Bloomer A.C., Greenhough T.J. Three dimensional structure of human C-reactive protein. Nat. Struct. Biol. 1996;3:346–354. doi: 10.1038/nsb0496-346. [DOI] [PubMed] [Google Scholar]
- 44.Adzhubei I., Jordan D.M., Sunyaev S.R. Predicting functional effect of human missense mutations using PolyPhen-2. Curr. Protoc. Hum. Genet. 2013 doi: 10.1002/0471142905.hg0720s76. doi:10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kumar P., Henikoff S., Ng P.C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 46.Judson R., Brain C., Dain B., Windemuth A., Ruano G., Reed C. New and confirmatory evidence of an association between APOE genotype and baseline C-reactive protein in dyslipidemic individuals. Atherosclerosis. 2004;177:345–351. doi: 10.1016/j.atherosclerosis.2004.07.012. [DOI] [PubMed] [Google Scholar]
- 47.Lee S., Emond M.J., Bamshad M.J., Barnes K.C., Rieder M.J., Nickerson D.A., Christiani D.C., Wurfel M.M., Lin X. Optimal unified approach for rare-variant association testing with application to small-sample case-control whole-exome sequencing studies. Am. J. Hum. Genet. 2012;91:224–237. doi: 10.1016/j.ajhg.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kiezun A., Garimella K., Do R., Stitziel N.O., Neale B.M., McLaren P.J., Gupta N., Sklar P., Sullivan P.F., Moran J.L., et al. Exome sequencing and the genetic basis of complex traits. Nat. Genet. 2012;44:623–630. doi: 10.1038/ng.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kryukov G.V., Shpunt A., Stamatoyannopoulos J.A., Sunyaev S.R. Power of deep, all-exon resequencing for discovery of human trait genes. Proc. Natl. Acad. Sci. USA. 2009;106:3871–3876. doi: 10.1073/pnas.0812824106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Boncler M., Watala C. Regulation of cell function by isoforms of C-reactive protein: a comparative analysis. Acta Biochim. Pol. 2009;56:17–31. [PubMed] [Google Scholar]
- 51.Eisenhardt S.U., Thiele J.R., Bannasch H., Stark G.B., Peter K. C-reactive protein: how conformational changes influence inflammatory properties. Cell Cycle. 2009;8:3885–3892. doi: 10.4161/cc.8.23.10068. [DOI] [PubMed] [Google Scholar]
- 52.Thiele J.R., Habersberger J., Braig D., Schmidt Y., Goerendt K., Maurer V., Bannasch H., Scheichl A., Woollard K.J., von Dobschutz E., et al. Dissociation of pentameric to monomeric C-reactive protein localizes and aggravates inflammation: in vivo proof of a powerful proinflammatory mechanism and a new anti-inflammatory strategy. Circulation. 2014;130:35–50. doi: 10.1161/CIRCULATIONAHA.113.007124. [DOI] [PubMed] [Google Scholar]
- 53.Davey Smith G., Lawlor D.A., Harbord R., Timpson N., Rumley A., Lowe G.D., Day I.N., Ebrahim S. Association of C-reactive protein with blood pressure and hypertension: life course confounding and Mendelian randomization tests of causality. Arterioscler. Thromb. Vasc. Biol. 2005;25:1051–1056. doi: 10.1161/01.ATV.0000160351.95181.d0. [DOI] [PubMed] [Google Scholar]
- 54.Timpson N.J., Lawlor D.A., Harbord R.M., Gaunt T.R., Day I.N., Palmer L.J., Hattersley A.T., Ebrahim S., Lowe G.D., Rumley A., et al. C-reactive protein and its role in metabolic syndrome: Mendelian Randomisation study. Lancet. 2005;366:1954–1959. doi: 10.1016/S0140-6736(05)67786-0. [DOI] [PubMed] [Google Scholar]
- 55.Timpson N.J., Nordestgaard B.G., Harbord R.M., Zacho J., Frayling T.M., Tybjaerg-Hansen A., Smith G.D. C-reactive protein levels and body mass index: elucidating direction of causation through reciprocal Mendelian randomization. Int. J. Obes. (Lond.) 2011;35:300–308. doi: 10.1038/ijo.2010.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yu C.E., Seltman H., Peskind E.R., Galloway N., Zhou P.X., Rosenthal E., Wijsman E.M., Tsuang D.W., Devlin B., Schellenberg G.D. Comprehensive analysis of APOE and selected proximate markers for late-onset Alzheimer's disease: patterns of linkage disequilibrium and disease/marker association. Genomics. 2007;89:655–665. doi: 10.1016/j.ygeno.2007.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Abecasis G.R., Auton A., Brooks L.D., DePristo M.A., Durbin R.M., Handsaker R.E., Kang H.M., Marth G.T., McVean G.A. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bennet A.M., Di Angelantonio E., Ye Z., Wensley F., Dahlin A., Ahlbom A., Keavney B., Collins R., Wiman B., de Faire U., et al. Association of apolipoprotein E genotypes with lipid levels and coronary risk. J. Am. Med. Assoc. 2007;298:1300–1311. doi: 10.1001/jama.298.11.1300. [DOI] [PubMed] [Google Scholar]
- 59.Mahley R.W., Rall S.C., Jr Apolipoprotein E: far more than a lipid transport protein. Annu. Rev. Genomics Hum. Genet. 2000;1:507–537. doi: 10.1146/annurev.genom.1.1.507. [DOI] [PubMed] [Google Scholar]
- 60.Rasmussen-Torvik L.J., Pacheco J.A., Wilke R.A., Thompson W.K., Ritchie M.D., Kho A.N., Muthalagu A., Hayes M.G., Armstrong L.L., Scheftner D.A., et al. High density GWAS for LDL cholesterol in African Americans using electronic medical records reveals a strong protective variant in APOE. Clin. Transl. Sci. 2012;5:394–399. doi: 10.1111/j.1752-8062.2012.00446.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sanna S., Li B., Mulas A., Sidore C., Kang H.M., Jackson A.U., Piras M.G., Usala G., Maninchedda G., Sassu A., et al. Fine mapping of five loci associated with low-density lipoprotein cholesterol detects variants that double the explained heritability. PLoS Genet. 2011;7:e1002198. doi: 10.1371/journal.pgen.1002198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Curtiss L.K. ApoE in atherosclerosis: a protein with multiple hats. Arterioscler. Thromb. Vasc. Biol. 2000;20:1852–1853. doi: 10.1161/01.atv.20.8.1852. [DOI] [PubMed] [Google Scholar]
- 63.Besnard S., Silvestre J.S., Duriez M., Bakouche J., Lemaigre-Dubreuil Y., Mariani J., Levy B.I., Tedgui A. Increased ischemia-induced angiogenesis in the staggerer mouse, a mutant of the nuclear receptor Roralpha. Circ. Res. 2001;89:1209–1215. doi: 10.1161/hh2401.101755. [DOI] [PubMed] [Google Scholar]
- 64.Chauvet C., Vanhoutteghem A., Duhem C., Saint-Auret G., Bois-Joyeux B., Djian P., Staels B., Danan J.L. Control of gene expression by the retinoic acid-related orphan receptor alpha in HepG2 human hepatoma cells. PLoS ONE. 2011;6:e22545. doi: 10.1371/journal.pone.0022545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Middelberg R.P., Benyamin B., de Moor M.H., Warrington N.M., Gordon S., Henders A.K., Medland S.E., Nyholt D.R., de Geus E.J., Hottenga J.J., et al. Loci affecting gamma-glutamyl transferase in adults and adolescents show age × SNP interaction and cardiometabolic disease associations. Hum. Mol. Genet. 2012;21:446–455. doi: 10.1093/hmg/ddr478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chambers J.C., Zhang W., Sehmi J., Li X., Wass M.N., Van der Harst P., Holm H., Sanna S., Kavousi M., Baumeister S.E., et al. Genome-wide association study identifies loci influencing concentrations of liver enzymes in plasma. Nat. Genet. 2011;43:1131–1138. doi: 10.1038/ng.970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Psaty B.M., Sitlani C. The Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium as a model of collaborative science. Epidemiology. 2013;24:346–348. doi: 10.1097/EDE.0b013e31828b2cbb. [DOI] [PubMed] [Google Scholar]
- 68.Friedman G.D., Cutter G.R., Donahue R.P., Hughes G.H., Hulley S.B., Jacobs D.R., Jr, Liu K., Savage P.J. CARDIA: study design, recruitment, and some characteristics of the examined subjects. J. Clin. Epidemiol. 1988;41:1105–1116. doi: 10.1016/0895-4356(88)90080-7. [DOI] [PubMed] [Google Scholar]
- 69.Fried L.P., Borhani N.O., Enright P., Furberg C.D., Gardin J.M., Kronmal R.A., Kuller L.H., Manolio T.A., Mittelmark M.B., Newman A., et al. The Cardiovascular Health Study: design and rationale. Ann. Epidemiol. 1991;1:263–276. doi: 10.1016/1047-2797(91)90005-w. [DOI] [PubMed] [Google Scholar]
- 70.Dawber T.R., Meadors G.F., Moore F.E., Jr Epidemiological approaches to heart disease: the Framingham Study. Am. J. Public Health Nations Health. 1951;41:279–281. doi: 10.2105/ajph.41.3.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sempos C.T., Bild D.E., Manolio T.A. Overview of the Jackson Heart Study: a study of cardiovascular diseases in African American men and women. Am. J. Med. Sci. 1999;317:142–146. doi: 10.1097/00000441-199903000-00002. [DOI] [PubMed] [Google Scholar]
- 72.Bild D.E., Bluemke D.A., Burke G.L., Detrano R., Diez Roux A.V., Folsom A.R., Greenland P., Jacob D.R., Jr, Kronmal R., Liu K., et al. Multi-ethnic study of atherosclerosis: objectives and design. Am. J. Epidemiol. 2002;156:871–881. doi: 10.1093/aje/kwf113. [DOI] [PubMed] [Google Scholar]
- 73.Design of the Women's Health Initiative clinical trial and observational study. The Women's Health Initiative Study Group. Control Clin. Trials. 1998;19:61–109. doi: 10.1016/s0197-2456(97)00078-0. [DOI] [PubMed] [Google Scholar]
- 74.Tennessen J.A., Bigham A.W., O'Connor T.D., Fu W., Kenny E.E., Gravel S., McGee S., Do R., Liu X., Jun G., et al. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science. 2012;337:64–69. doi: 10.1126/science.1219240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li H., Ruan J., Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bainbridge M.N., Wang M., Wu Y., Newsham I., Muzny D.M., Jefferies J.L., Albert T.J., Burgess D.L., Gibbs R.A. Targeted enrichment beyond the consensus coding DNA sequence exome reveals exons with higher variant densities. Genome Biol. 2011;12:R68. doi: 10.1186/gb-2011-12-7-r68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Challis D., Yu J., Evani U.S., Jackson A.R., Paithankar S., Coarfa C., Milosavljevic A., Gibbs R.A., Yu F. An integrative variant analysis suite for whole exome next-generation sequencing data. BMC Bioinformatics. 2012;13:8. doi: 10.1186/1471-2105-13-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Danecek P., Auton A., Abecasis G., Albers C.A., Banks E., DePristo M.A., Handsaker R.E., Lunter G., Marth G.T., Sherry S.T., et al. The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sherry S.T., Ward M.H., Kholodov M., Baker J., Phan L., Smigielski E.M., Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang K., Li M., Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Liu X., Jian X., Boerwinkle E. dbNSFP v2.0: a database of human non-synonymous SNVs and their functional predictions and annotations. Hum. Mutat. 2013;34:E2393–E2402. doi: 10.1002/humu.22376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.R Development Core Team. R Foundation for Statistical Computing, Vienna, Austria; 2014. R: A Language and Environment for Statistical Computing. [Google Scholar]
- 87.Willer C.J., Li Y., Abecasis G.R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.