Abstract
Iron is an essential component of many important proteins and enzymes, including hemoglobin, which is responsible for carrying oxygen to the cells. African Americans (AAs) have a greater prevalence of iron deficiency compared with European Americans. We conducted genome-wide admixture-mapping and association studies for serum iron, serum ferritin, transferrin saturation (SAT) and total iron binding capacity (TIBC) in 2347 AAs participating in the Jackson Heart Study (JHS). Follow-up replication analyses for JHS iron-trait associated SNPs were conducted in 329 AA participants in the Healthy Aging in Neighborhoods of Diversity across the Life Span study (HANDLS). Higher estimated proportions of global African ancestry were significantly associated with lower levels of iron (P = 2.4 × 10−5), SAT (P = 0.0019) and TIBC (P = 0.042). We observed significant associations (P < 5 × 10−8) between serum TIBC levels and two independent SNPs around TF on chromosome 3, the first report of a genome-wide significant second independent signal in this region, and SNPs near two novel genes: HDGFL1 on chromosome 6 and MAF on chromosome 16. We also observed significant associations between ferritin levels and SNPs near GAB3 on chromosome X. We replicated our two independent associations at TF and our association at GAB3 in HANDLS. Our study provides evidence for both shared and unique genetic risk factors that are associated with iron-related measures in AAs. The top two variants in TF explain 11.2% of the total variation in TIBC levels in AAs after accounting for age, gender, body mass index and background ancestry.
INTRODUCTION
Iron is critical to an array of metabolic functions, such as oxygen transport and oxidative phosphorylation. Normally, small daily losses of iron in the feces and through menstruation are balanced by its regulated intestinal absorption and its recovery from heme after phagocytosis of senescent red blood cells (RBCs) (1). Iron deficiency can cause anemia, while iron overload may lead to increased risk for cardiovascular disease, including cardiomyopathy, diabetes mellitus, arthritis and liver disease (2). Laboratory tests typically used to assess iron transport and storage include serum iron, total iron binding capacity (TIBC), transferrin saturation (SAT) and serum ferritin. Ferritin, the predominant iron storage protein, reflects the cumulative iron stores in the bone marrow and tissues. Transferrin functions in iron transport, and the concentration of transferrin is proportional to the TIBC in serum. Transferrin SAT, calculated as (serum iron/TIBC) × 100, is affected by the rate of iron absorption in the small bowel as well as the sufficiency of tissue iron stores. Genetic factors could affect iron metabolism through gastrointestinal absorption, transport, tissue uptake, storage or remobilization from tissue stores, and thus could have a large impact on the variation of these iron-related measures (2,3).
Genome-wide association studies (GWAS) have identified seven loci associated with these measures in subjects of European descent (P < 5 × 10−8) (4–7), including variants in or near TF-SRPRB, SLC17A1, HFE, HIST1H2BJ, SIK3, PCSK7 and TMPRSS6. Here, we present a genome-wide admixture and association study of serum iron, TIBC, SAT and ferritin among African Americans (AAs) enrolled in the Jackson Heart Study (JHS) and Healthy Aging in Neighborhoods of Diversity across the Life Span study (HANDLS) cohorts.
RESULTS
Descriptive statistics for the JHS and HANDLS participants included in this study are detailed in Table 1. The correlations between these four phenotypes in JHS are shown in Supplementary Material, Table S1.
Table 1.
Descriptive statistics of JHS and HANDLS participants
| JHS |
HANDLS |
|||||
|---|---|---|---|---|---|---|
| Total | Male | Female | Total | Male | Female | |
| Sample size | 2347 | 1012 | 1335 | 329 | 189 | 140 |
| Age (years) | 54.5 ± 12.6 | 53.0 ± 12.8 | 55.7 ± 12.4 | 49.4 ± 8.3 | 49.0 ± 8.7 | 49.9 ± 7.7 |
| BMI (kg/m2) | 31.5 ± 6.9 | 29.9 ± 6.1 | 32.8 ± 7.3 | 28.8 ± 7.6 | 27.1 ± 5.4 | 31.2 ± 9.3 |
| Ferritin (ng/ml) | 134.0 (74.0, 232.0) | 177.5 (110.8, 286.0) | 105 (58.0, 185.5) | 107.5 (54.0, 201.0) | 137.0 (76.0, 257.0) | 71.0 (35.0, 113.0) |
| Iron (μg/dl) | 81.0 (67.0, 98.0) | 86.0 (70.0, 104.0) | 78.0 (64.0, 94.0) | 83.0 (66.2, 103.8) | 86.0 (70.0, 109.0) | 81.0 (64.0, 98.0) |
| SAT (%) | 28.0 (23.0, 35.0) | 30.0 (25.0, 37.0) | 27.0 (22.0, 33.0) | 25.0 (20.2, 31.1) | 26.4 (20.8, 33.5) | 23.7 (19.3, 27.3) |
| TIBC (μg/dl) | 284 (260.0, 314.0) | 280.0 (256.0, 307.0) | 288.0 (263.0, 319.0) | 332.0 (306.0, 372.0) | 326.0 (300.0, 368.0) | 342.5 (316.8, 379.0) |
Note: data are mean ± SE, median (25th, 75th percentiles).
SAT, transferrin saturation; TIBC, total iron binding capacity.
Admixture analyses
A higher level of estimated overall average African ancestry was significantly associated (P < 0.05) with lower levels of TIBC, iron and SAT (Table 2). No individual region had local ancestry significantly associated with the iron measures. Three regions (DZIP1L on chr 3, TRDN on chr 6 and TMCO5B-RYR3 on chr 15) had a LOD score >3 for TIBC and one region (DEFB129-DEFB132 on chr 20) had a LOD score >3 for iron. The plots of the LOD scores across the whole genome are shown in Supplementary Material, Figure S1a–d.
Table 2.
Association between global African ancestry estimate and phenotypes
| Phenotypes | βa | SE | P-value |
|---|---|---|---|
| TIBC | −13.90 | 6.80 | 0.04 |
| log_ferritin | 0.13 | 0.13 | 0.32 |
| log_iron | −0.20 | 0.05 | 2.4E-05 |
| log_SAT | −0.15 | 0.05 | 0.0019 |
aThe predicted change in the iron measure for each one-percentage point increase in estimated global African ancestry.
Summary GWAS results in JHS
One-hundred-fifty-seven SNPs reached genome-wide significance (P < 5 × 10−8) for TIBC (153 SNPs on chromosome 3, 3 SNPs on chromosome 6 and 1 SNP on chromosome 16); five SNPs were significant for ferritin (all on chromosome X) (Supplementary Material, Table S2). No SNPs reached genome-wide significance for serum iron or SAT. Top results for all four traits are listed in Supplementary Material, Tables S3–S6. Manhattan plots and quantile–quantile (Q–Q) plots for the four traits are shown in Supplementary Material, Figures S2–S5. Q–Q plots revealed no substantial evidence for inflated results, due to population stratification, residual relatedness among subjects or experimental outliers.
TIBC GWAS results on chromosome 3
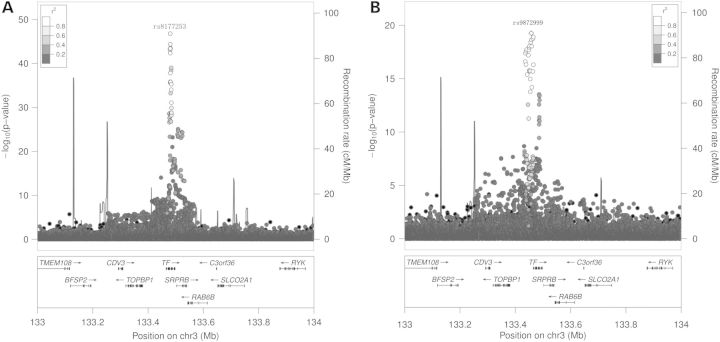
All top SNPs on chromosome 3 clustered within a region spanning <150 kb containing three genes: TOPBP1, TF and SRPRB (Fig. 1A). The strongest signal [rs8177253, P = 1.8 × 10−47, minor allele frequency (MAF) = 0.24] mapped to the TF gene, which encodes transferrin (Table 3). Rs8177253 was also nominally (defined as unadjusted P < 0.05) associated with SAT (P = 3.0 × 10−7). Thirty-six TF region SNPs were genome-wide significant for TIBC levels after multi-SNP analyses, including rs8177253 as a covariate (Supplementary Material, Table S7). The top SNP in the conditional analysis was rs9872999 (P = 5.4 × 10−20, MAF = 0.38), which was not significant (P = 1.0 × 10−6) prior to the adjustment for rs8177253 (Table 3 and Fig. 1B). There was no evidence of an interaction between rs8177253 and rs9872999 (P = 0.11). No SNPs remained genome-wide significant after covariate adjustment for both rs8177253 and rs9872999, although a large number of SNPs remained nominally significant (Supplementary Material, Table S8). SNPs rs8177253 and rs9872999 together explained an estimated 11.2% of the total variance of TIBC after accounting for age, gender, BMI and the first 10 PCs.
Figure 1.
(A) Regional plot of the −log10(P) values for the SNPs in the TF region for TIBC. The X-axis shows the human genome build 19 coordinates (Mb) and the genes in the region. The Y-axis shows the −log10 transformed association P-values of SNPs on the left, and recombination rates in cM per Mb on the right. Different colors of shading indicate the strength of linkage disequilibrium (LD) (r2) between the top SNP and the other SNPs tested in the region. (B) Regional plot of the −log10(P) values for the SNPs in the TF region for TIBC after adjusting for the top SNP rs8177253 in this region. The top SNP was rs9872999 after adjusting for rs8177253.
Table 3.
Top SNPs that reached genome-wide significance (P < 5 × 10−8) in JHS and replication results from HANDLS
| Trait | Chr | Nearest gene | SNP | Pos(hg19) | EA | JHS |
HANDLS |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EAF | N | R2 | β | SE | P | EAF | N | R2 | β | SE | P | ||||||
| TIBC | 3 | TF | rs8177253 | 133480192 | T | 0.24 | 2347 | NA | 19.86 | 1.34 | 1.8E−47 | 0.23 | 329 | 0.98 | 26.72 | 4.91 | 1.1E−07 |
| TIBC | 3 | TF | rs9872999 | 133457514 | T | 0.62 | 2347 | 0.80 | −6.55 | 1.34 | 1.1E−06 | 0.60 | 329 | 0.93 | −14.19 | 4.33 | 0.0012 |
| TIBC | 3 | TF | rs9872999* | 133457514 | T | 0.62 | 2347 | 0.80 | −11.72 | 1.28 | 5.4E−20 | 0.60 | 329 | 0.93 | −19.11 | 4.16 | 6.2E−06 |
| TIBC | 6 | HDGFL1 | rs115923437 | 22678302 | C | 0.06 | 2347 | 0.91 | 14.84 | 2.6 | 1.1E−08 | 0.05 | 329 | 0.96 | −5.28 | 9.93 | 0.60 |
| TIBC | 16 | MAF-DYNLRB2 | rs16951289 | 79790621 | T | 0.07 | 2347 | 0.91 | 13.38 | 2.38 | 2.0E−08 | 0.08 | 329 | 0.97 | −15.95 | 7.92 | 0.04 |
| Log_ferritin | X | GAB3 | rs141555380 | 153906012 | T | 0.14 | 2347 | 0.94 | 0.17 | 0.03 | 1.1E−08 | 0.l3 | 329 | 0.98 | 0.24 | 0.08 | 0.0057 |
*: β, SE, and P were reported for rs9872999 after adjusting for rs8177253.
Chr, chromosome; Pos(hg19), physical position of the SNP according to human genome build version 19; EA, effect allele; EAF, effect allele frequency; β, β coefficients representing the estimated change in the raw or transformed trait value associated with each additional copy of the effect allele; SE, standard error; R2, R2 represents the imputation quality provided. “NA” indicates that the actual genotype data from the Affy 6.0 array were used in the analyses.
Higher local African ancestry at rs8177253 in the TF region was nominally associated with lower TIBC levels (P = 0.0012). The association between TIBC and rs8177253 remained robust after the adjustment for local African ancestry at rs8177253. When stratified by the estimated local number of European versus African chromosomes, the rs8177253–TIBC association was present among 1579 AA who were predicted to carry two African chromosomes (β = 19.64 ± 1.70; P = 8.5 × 10−30) as well as the 768 AA who were predicted to carry at least one European chromosome (β = 19.68 ± 2.26; P = 1.8 × 10−17). The rs9872999–TIBC association also remained significant after adjustment for both rs8177253 and local African ancestry at the genotyped marker nearest its location (data not shown).
TIBC GWAS results on chromosome 6
The three genome-wide significant SNPs on chromosome 6 mapped near the HDGFL1 gene (Supplementary Material, Fig. S6). The strongest signal (rs115923437, P = 1.1 × 10−8) mapped ∼100 Kb distal to HDGFL1, which is a gene that encodes hepatoma-derived growth factor-like 1 and is associated with glycosylated hemoglobin level, and ∼3.5 Mb proximal to the known iron gene HFE. After conditioning on rs115923437, the remaining SNPs within 1 Mb of the SNP no longer showed strong evidence for association (all P > 1 × 10−4). SNP rs115923437 explained 1.3% of the total variance of TIBC after accounting for the covariates.
Local African ancestry near rs115923437 was also nominally associated with TIBC, although this time higher local African ancestry was associated with higher TIBC levels (P = 0.0083). The associations between TIBC and rs115923437 remained significant after the adjustment for local African ancestry at the genotyped marker nearest its location. When stratified by the estimated local number of European versus African chromosomes, the rs115923437–TIBC association was present among the 1569 AA who were predicted to carry two African chromosomes (β = −14.11 ± 2.95; P = 2.0 × 10−6), as well as the 778 AA who were predicted to carry at least one European chromosome (β = −15.42 ± 5.78; P = 0.0070).
TIBC GWAS results on chromosome 16
A single SNP (rs16951289, P = 2.0 × 10−8) on chromosome 16, an intronic variant in uncharacterized gene LOC102467146 that is ∼150 Kb distal to the MAF gene (Supplementary Material, Fig. S7), reached genome-wide significance. MAF encodes v-maf musculoaponeurotic fibrosarcoma oncogene homolog. Local African ancestry in this region was not associated with TIBC (P = 0.97) and did not modify the evidence for association with rs16951289. Rs16951289 explained 1.2% of the total variance of TIBC.
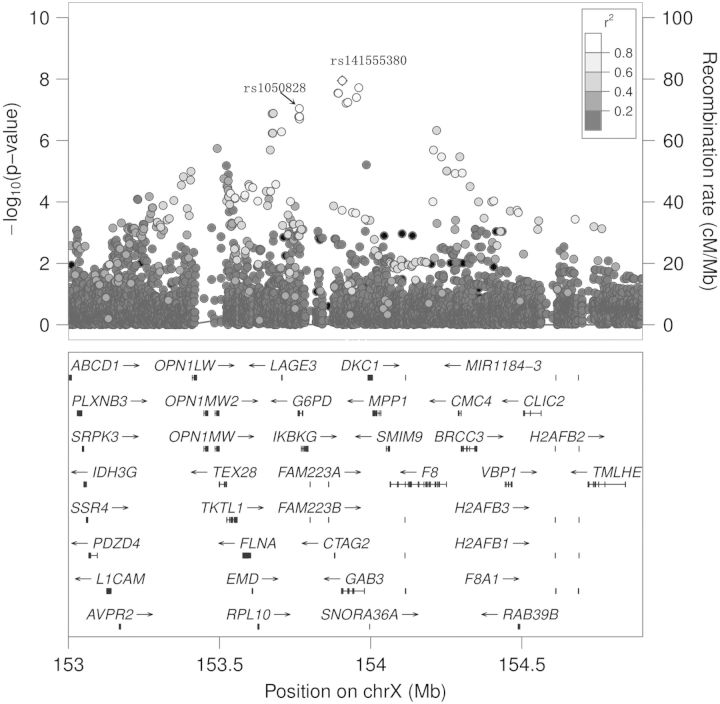
Ferritin GWAS results on chromosome X
Five SNPs on chromosome X near the GAB3 gene reached genome-wide significance for association with ferritin levels (Fig. 2). The strongest signal (rs141555380, P = 1.1 × 10−8) mapped to the UTR-3 region of the GAB3 gene. GAB3 encodes GRB2-associated binding protein 3, which is involved in several growth factor and cytokine signaling pathways. After conditioning on rs141555380, the remaining SNPs within 1 Mb of rs141555380 no longer showed significant evidence for association (all P > 1 × 10−4). The effect estimates for carriers of the rs141555380 minor allele in stratified analysis were similar for hemizygous males and homozygous females (hemizygous males: β = 0.17 ± 0.04, P = 5.05 × 10−6; homozygous females: β = 0.14 ± 0.05, P = 0.002). SNP rs141555380 explained 1.2% of the total variance of ferritin after accounting for age, gender, BMI and the first 10 PCs.
Figure 2.
Regional plot of the −log10(P) values for the SNPs in the GAB3 region for log_ferritin.
Menopause status-adjusted analyses of association with ferritin
Association analysis for serum ferritin was conducted by including menopausal status as an additional covariate. Menopausal status was significantly associated with serum ferritin levels P = 5.3 × 10−17; however, the effect size and significance of SNP-ferritin associations did not change considerably after adjusting for menopausal status. The top SNP rs141555380 (P = 1.1 × 10−8) continued to be the most significant SNP after adjusting for menopause status (P = 1.4 × 10−8). The effect sizes and P-values of top SNPs (P < 10−7) before and after adjusting for menopause status are shown in Supplementary Material, Table S9. This result is consistent with previous findings that the effects of variation in menstrual blood loss, although significant, were small when compared with the genetic effects that influence the iron reserves (8).
Replication results in HANDLS
Four regions contained SNPs that reached genome-wide significance in JHS, including one region, the chromosome 3 TF region, which contained a second significant SNP (rs9872999) after covariate adjustment for the top SNP (rs8177253). Five SNPs (rs8177253, rs9872999, chromosome 6 top SNP rs115923437 and chromosome 16 top SNP rs16951289 for TIBC; chromosome X top SNP rs141555380 for ferritin) were selected and tested for association in HANDLS to determine whether the associations could be replicated (Table 3). Rs9872999 was tested before and after adjustment for rs8177253. Both associations in the TF region with TIBC were replicated (P = 1.1 × 10−7 for rs8177253; P = 0.0012 for rs9872999 before adjusting for rs8177253). As with JHS, the association between rs9872999 and TIBC became more significant after adjusting for the primary signal at rs8177253 (P = 6.2 × 10−6). The association between the GAB3 region SNP rs141555380 and ferritin also replicated (P = 5.7 × 10−3). The associations between TIBC and SNPs rs115923437 and rs16951289 did not replicate in HANDLS; rs16951289 was nominally significant (P = 0.04), but the direction of the effect went in the opposite direction to that observed in JHS.
Prior European GWAS-established signals at P < 5 × 10−8 that are replicated in JHS at P < 0.05
GWAS have reported five regions to contain SNPs to be significantly associated (P < 5 × 10−8) with at least one of the following iron-related traits: iron, ferritin, SAT and transferrin, including TF-SRPRB (rs3811647, rs1830084), SLC17A1 (rs17342717), HFE (rs1799945 [H63D] and rs1800562 [C282Y]), HIST1H2BJ (rs13194491) and TMPRSS6 (rs855791 [V736A] and rs4820268) in subjects of European descent (Table 4). Iron, ferritin and SAT are directly reported in the current study, while TIBC is proportional to transferrin [TIBC (μmol/L) = 25.1 × transferrin (g/L)]. All five regions contained a SNP, either the index SNP in the prior report or a SNP in strong linkage disequilibrium (defined as a SNP with estimated r2 > 0.8 with the index SNP based on CEU 1000 Genomes subjects) with the index SNP, that was nominally associated with at least one corresponding iron measure in JHS at P < 0.05. For HFE rs1800562, only the associations with ferritin and transferrin were replicated, but not the associations with iron and SAT. For HIST1H2BJ rs13194491 and TMPRSS6 rs855791, the associations with the index SNPs were not replicated, but nearby SNPs, which are estimated to be in high LD with the index SNPs in European populations, did reach nominal significance in JHS, suggesting a narrowing of the candidate regions for the causal variants if the causal variants are the same for the two populations.
Table 4.
Replication of signals established in prior GWAS including subjects of European descent
| Trait | Chr | Index SNP result in JHSa |
Most significant SNP in JHSb |
Nearest gene | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Index SNP | Pos(hg19) | EA | EAF | β | SE | P | Most significant SNP | Pos(hg19) | EA | EAF | β | SE | P | LD r2 | |||
| TIBC | 3 | rs3811647*(7) | 133484029 | A | 0.22 | 17.55 | 1.43 | 9.7E−35 | rs8177253 | 133480192 | T | 0.24 | 19.86 | 1.34 | 1.8E−47 | 1 | TF |
| TIBC | 3 | rs1830084(7) | 133508464 | T | 0.16 | 16.84 | 1.60 | 2.2E−25 | rs1830084 | 133508464 | T | 0.16 | 16.84 | 1.60 | 2.2E−25 | 1 | TF |
| log_ferritin | 6 | rs17342717*(4) | 25821770 | T | 0.02 | 0.24 | 0.10 | 0.02 | rs78273613* | 25866075 | G | 0.02 | 0.26 | 0.10 | 0.01 | 0.88 | SLC17A1 |
| log_iron | 6 | rs1799945*(5) | 26091179 | G | 0.04 | 0.08 | 0.03 | 7.8E−04 | rs129128 | 26125342 | C | 0.03 | 0.09 | 0.02 | 1.0E−04 | 0.92 | HFE |
| log_iron | 6 | rs1800562(7) | 26093141 | A | 0.01 | −0.03 | 0.04 | 0.37 | rs1800562 | 26093141 | A | 0.01 | −0.03 | 0.04 | 0.37 | 1 | HFE |
| log_ferritin | 6 | rs1800562(4) | 26093141 | A | 0.01 | 0.20 | 0.10 | 0.05 | rs1800562 | 26093141 | A | 0.01 | 0.20 | 0.10 | 0.05 | 1 | HFE |
| TIBC | 6 | rs1800562(7) | 26093141 | A | 0.01 | −24.63 | 5.13 | 1.7E−06 | rs1800562 | 26093141 | A | 0.01 | −24.63 | 5.13 | 1.7E−06 | 1 | HFE |
| log_sat | 6 | rs1800562(7) | 26093141 | A | 0.01 | 0.05 | 0.04 | 0.14 | rs1800562 | 26093141 | A | 0.01 | 0.05 | 0.04 | 0.14 | 1 | HFE |
| log_sat | 6 | rs13194491*(7) | 27037080 | T | 0.02 | 0.02 | 0.05 | 0.75 | rs35657082* | 27067657 | T | 0.01 | 0.25 | 0.11 | 0.02 | 0.91 | HIST1H2BJ |
| log_iron | 22 | rs855791*(11) | 37462936 | G | 0.83 | 0.03 | 0.01 | 0.08 | rs2072860* | 37470604 | A | 0.73 | 0.03 | 0.01 | 7.7E−03 | 0.90 | TMPRSS6 |
| log_sat | 22 | rs855791*(11) | 37462936 | G | 0.83 | 0.02 | 0.01 | 0.15 | rs4820268* | 37469591 | A | 0.73 | 0.02 | 0.01 | 0.03 | 0.90 | TMPRSS6 |
| log_iron | 22 | rs4820268*(5) | 37469591 | A | 0.73 | 0.03 | 0.01 | 8.4E−03 | rs9610638* | 35775614 | C | 0.80 | 0.05 | 0.02 | 3.5E−03 | 0.83 | TMPRSS6 |
*Imputed.
() besides “Index SNP” contains the citation to the initial association study for each individual variant.
aIndex SNP: Index SNP that was reported to be significantly (P < 5 × 10−8) associated with iron traits in prior GWAS in subjects of European descent.
bMost significant SNP: the proxy for index SNP (LD r2 > 0.8 in CEU 1000G with the index SNP) with the smallest association P-value in JHS.
LD r2: r2 with index SNP in CEU 1000G subjects.
DISCUSSION
We conducted genome-wide admixture and association studies for iron-related phenotypes, including serum iron, serum ferritin, SAT and TIBC in 2347 AAs participating in the JHS. We observed significant associations between SNPs around TF, a well-established region on chromosome 3, and two novel regions: near HDGFL1 on chromosome 6 and MAF on chromosome 16, and TIBC levels. Conditional analyses revealed a second significant SNP associated with TIBC in the TF region that was independent of the top SNP from the unconditional analyses. We also observed significant associations between SNPs around GAB3, a novel region on chromosome X, and ferritin levels. The two independent associations for TIBC at TF and the association for ferritin at GAB3 were successfully replicated in HANDLS. The associations between the JHS index SNPs near HDGFL1 and MAF were not replicated. The available sample size in HANDLS (N = 329) was considerably smaller than for JHS. If we assume the index variants at HDGFL1 and MAF each explain an estimated 1.3% of the total variance of TIBC, then we only had power of 0.34 to observe these associations at P < 0.01 in HANDLS. Thus, larger replication studies will be necessary to make more decisive statements regarding the overall evidence of these other regions. We did not observe any significant SNP associations for the other iron measures, although we were able to replicate associations from previous studies based on subjects of European descent using a less stringent significance threshold (P < 0.05).
The estimated average (“global”) proportion of African ancestry was significantly associated with lower levels of TIBC, serum iron and SAT—which are entirely consistent with previous findings reporting lower levels of these same measures, on average, in AAs compared with European Americans (9,10). Subjects with higher levels of global African ancestry were observed to have higher levels of ferritin, also consistent with prior reported differences between individuals of European and African ancestry (9,10), although this result was not statistically significant. These results implicate novel genetic risk factors in AAs and underscore the importance of studying this population for genetic risk factors that uniquely/disproportionately impact them. Local ancestry was not significantly associated with any iron measures, although a couple of regions containing our GWAS top results were nominally significant.
Variants in and around TF have been observed to be associated with serum ferritin levels (7), transferrin levels (5–7), serum transferrin saturation (11) and serum levels of carbohydrate-deficient transferrin (12) in subjects of European descent. In JHS, the top SNP at TF, rs8177253, was associated with TIBC (P = 1.8 × 10−47) and nominally associated with SAT (P = 3.0 × 10−7). SNP rs8177253 is located in an intronic region of TF and is in high LD (r2 = 1, D′ = 1 in 1000 Genomes CEU) with GWAS index SNP rs3811647 previously reported to be associated with transferrin in subjects of European and Australian descent (7,9) [rs3811647 is also associated with TIBC (P = 9.7 × 10−35) and nominally associated with SAT (P = 2.2 × 10−6) in JHS]. After conditioning on rs8177253, all SNPs, including rs3811647, reported to be significantly associated with TIBC in previous studies became non-significant in JHS.
Our study is the first study to report a second significant independently associated SNP in the TF region for TIBC. Interestingly, this second signal (index SNP rs9872999 which maps to an intergenic region ∼10 Kb proximal to TF) only became significant at the genome-wide level after conditioning on our top TF region SNP rs8177253. Rs8177253 and rs9872999 are in modest LD in 1000 Genomes YRI subjects (r2 = 0.07, D′ = 0.58) and in stronger LD in 1000 Genomes CEU subjects (r2 = 0.27, D′ = 0.71). The allele associated with an increase in TIBC for rs8177253 is preferentially on the same haplotype with the allele associated with a decrease in TIBC for rs9872999. Thus, the mean effects for rs9872999 are shrunk toward the null when not factoring in genotype for rs8177253. There is no evidence for an interaction between these SNPs on TIBC levels; thus the results indicate a second independent signal in the TF region. The MAF for rs9872999 is 0.39 in 1000 Genomes YRI subjects and 0.50 in 1000 Genomes CEU subjects. It is unclear if this second signal is specific to AAs, as results from conditional analyses in other populations have not been reported. Conditional analyses would likely be especially important for detecting significant mean effects for rs9872999 in populations of European descent given the stronger LD between the two SNPs in this population.
Variants in HDGFL1 have been reported previously to be associated with glycosylated hemoglobin levels (P = 2.4 × 10−5) in European type 1 diabetic subjects (13) and nominally associated (P < 0.05) with levels of VLDL, LDL, Apolipoprotein C, HDL and carotid artery disease (14–16). In recent years, there has been considerable interest in the possibility that excessive tissue iron stores may contribute to the pathogenesis of both diabetes and ischemic heart disease (3). MAF, which encodes v-maf musculoaponeurotic fibrosarcoma oncogene homolog, appears to be important in early development. Mutations in MAF have been reported to co-segregate with cerulean congenital cataracts (17) and juvenile-onset pulverulent cataract (18) in human pedigrees. We can find no evidence in the literature suggesting a direct connection between MAF and iron metabolism. However, there is some evidence of pleiotropy for iron metabolism and cataracts, namely hereditary hyperferritinemia cataract syndrome, which is an inherited syndrome caused by a mutation within the L-ferritin gene and characterized by early-onset cataracts and elevated serum ferritin (19).
A cluster of SNPs near GAB3 on chromosome X were significantly associated with ferritin (top SNP rs141555380, MAF = 0.14, P = 1.1 × 10−8) and nominally associated with SAT (rs141555380, P = 0.037). Although no prior studies observed any connection between this gene and iron metabolism, another gene ∼0.2 Mb upstream of GAB3, G6PD, plays a critical role in iron metabolism. G6PD deficiency may cause acute hemolysis or severe chronic non-spherocytic hemolytic anemia. Increases in serum ferritin levels have been observed in G6PD-deficient patients (20,21), which is possibly due to both a shortened life span and increased break down of erythrocytes in G6PD-deficient patients. A functional missense variant in G6PD, rs1050828 (MAF = 0.13, leading to a Val68Met amino acid substitution), was also associated with ferritin but narrowly missed genome-wide significance (P = 9.1 × 10−8). Strong LD exists between this functional variant at G6PD and rs141555380 (R2 = 0.91, D′ = 1 in 1000 Genomes Project participants of African descent), and the association between rs141555380 and ferritin disappears after adjustment for rs1050828 (P = 0.55). Rs1050828 and nearby rs762516 (two SNPs in LD: R2 = 0.68, D′ = 1.0 in HapMap YRI) have been shown to be significantly associated with multiple erythrocyte traits in AAs, including hematocrit, hemoglobin, RBC count, mean corpuscular volume (MCV) and red cell distribution width (RDW) in previous GWAS or candidate gene studies (22,23). In JHS, rs1050828 is significantly associated with MCV (P = 1.7 × 10−8), RBC count (P = 9.9 × 10−16) and RDW (P = 8.9 × 10−21) and nominally associated with hematocrit (P = 8.7 × 10−7) and hemoglobin (7.1 × 10−8). Ferritin levels are correlated with levels of hematocrit (r = 0.25), hemoglobin (r = 0.27), MCV (r = 0.089), RBC (r = 0.15) and RDW (r = −0.13). The associations for both hematocrit (P = 2.9 × 10−8) and hemoglobin (P = 4.3 × 10−10) both became genome-wide significant after additional covariate adjustment for ferritin. Similarly, the association between rs1050828 and ferritin also became genome-wide significant after adjustment for hematocrit (7.2 × 10−9) and hemoglobin (1.3 × 10−9), but evidence for association with ferritin decreased after covariate adjustment for MCV (1.1 × 10−4), RBC (5.2 × 10−6) and RDW (7.2 × 10−3). Since G6DP has been reported to play an important role in hemolysis and affects the levels of erythrocyte traits, the signal we observed at this region may help explain the relationship between hemolysis and iron metabolism. This variant is also implicated in malaria resistance, and the A- form of G6PD deficiency in Africa is under strong natural selection from the preferential protection it provides to hemizygous males and homozygous females against life-threatening malaria (24). This natural selection of G6PD deficiency in African descent may help explain some of the marked differences in iron measures among ethnic groups.
In summary, we report that global genetic admixture is an important predictor of iron measures in AAs, further implicating the importance of unique genetic effect alleles in the AA population. We observed SNPs in or near three genes, TF, HDGFL1 and MAF, which were significantly associated with TIBC in JHS, and SNPs near GAB3 that were significantly associated with ferritin. We identified a novel second independently associated SNP in the TF region for TIBC that was only identified after conditioning on the top SNP in the region. The two TF signals and the GAB3 signal were replicated in a small independent AA sample from HANDLS. Larger replication samples will be necessary to draw firm conclusions regarding the associations for the other loci. The TF region is known to be associated with various serum iron-related measures in subjects of European descent; we now show similar associations in AA. While the G6DP-GAB3 region is known to be associated with multiple erythrocyte traits in AA, this is the first time it has been reported to be significantly associated with ferritin, a specific iron-related measure. We have also nominally replicated four other established loci from other populations in our AA samples. Future fine-mapping studies, including rare and uncommon variants, and functional studies should be undertaken to better characterize these and other loci and to identify the functional variants directly influencing iron levels in AA.
MATERIALS AND METHODS
Study subjects
Discovery stage
The JHS is a longitudinal, population-based cohort designed to identify risk factors for the development of cardiovascular disease, diabetes, obesity, chronic kidney disease and stroke in more than 5000 AAs from the Jackson, metropolitan area (25). The design, recruitment and initial characterization of this study have been described previously (26). The JHS participants for the current study included 1012 AA males and 1335 AA females with available iron-related measures and genome-wide genotype data.
Replication stage
The HANDLS is a community-based, longitudinal epidemiological study that aims to examine the influences of race and socioeconomic status on the development of age-related diseases in African and European Americans from the city of Baltimore. The study consists of 2200 AAs and 1522 European Americans aged 30–64 years. The design, recruitment and initial characterization of this study have been described previously (27). The HANDLS participants for the current study included 329 AAs with iron measures and genome-wide genotype data.
JHS and HANDLS participants provided written informed consent. The study protocols and consent forms for these studies were approved by the responsible research ethics committees and institutional research boards.
Phenotypes
All phenotype measures came from blood samples that were collected from fasting blood during the baseline examination, which occurred during 2000–2004 for JHS and 2004–2009 for HANDLS. Iron measures included total levels of serum iron (μg/dl), serum ferritin (ng/ml), TIBC (μg/dl) and SAT (%). Serum iron in JHS participants was measured by the FerroZine colorimetric assay (Roche), standardized to NIST traceable iron standards and calibrated against control sera from the manufacturer. TIBC was determined by colorimetric (FerroZine) measurement of iron that remains unbound after addition of a known amount of iron to the serum. The assay was standardized and calibrated as for serum iron. Ferritin was measured by an immunoturbidimetric assay (Roche) based on agglutination of anti-ferritin-latex conjugates, standardized with human spleen ferritin and calibrated against a standard protein solution provided by the manufacturer. For HANDLS, serum iron and TIBC were measured using standard clinical laboratory spectrophotometric assays. Serum ferritin was measured using chemiluminescence assays. For both studies, SAT was calculated as: (serum iron/TIBC)*100%.
Participants were excluded if they were taking iron supplements, or not fasting at time of blood draw or if they had known chronic infectious or inflammatory disease, or residual cancer. Additional exclusions included hematocrit <35%, hemoglobin <11 g/dl, mean red cell volume >100 fl, white blood cell count >11 000 per mm3, platelet count >400 000 per mm3, C-reactive protein >3 standard deviations above the mean or transferrin saturation <15% (indicates iron deficiency likely due to blood loss).
Genotyping and imputation
A total of 3030 JHS participants were genotyped using the Affymetrix Genome-Wide Human SNP Array 6.0. 874 712 SNPs, with a call rate >0.95, MAF >0.01 and genotype distributions consistent with the Hardy–Weinberg equilibrium (HWE, P > 1 × 10−5) were included for further analysis. Following pre-phasing using MACH 1.0.18 software (28), 38 million SNPs, excluding SNPs monomorphic in CEU/YRI, were imputed using minimac (29) based on 1000 Genome Project phase I reference samples (November 2010, Version 3). Analyses were limited to the ∼17 million imputed SNPs with estimated imputation quality of r2 >0.3.
A total of 1024 HANDLS participants were genotyped using the Illumina 1 m SNP array, including 329 AAs with iron measures. SNPs with HWE P > 1 × 10−7, MAF > 0.01 and call rate > 95% were included for further analysis. 2 939 993 SNPs were imputed using MACH (28) and Minimac (29) software based on combined reference haplotype data from HapMap Phase 2 CEU + YRI samples that includes monomorphic SNPs in either of the two constituent populations (release 22, build 36.3). Chromosome X variants were imputed based on 1000 Genomes Project EUR + AFR + AMR + ASN reference samples. Only index variants demonstrating significant evidence for association in JHS (P < 5 × 10−8) were subsequently analyzed for the relevant iron phenotype in HANDLS.
Statistical analyses
To assess the impact of genetic admixture on iron measures within the AA population, we first estimated the genome-wide average of African ancestry for each JHS participant (“global ancestry”). We used the software ADMIXTURE (30) with K = 2 clusters and tested, using linear models implemented in R, whether this estimated global ancestry proportion was associated with each of our iron measures after covariate adjustment for age, sex and BMI. Values of serum ferritin, total iron and SAT were natural log-transformed to achieve approximate normality of residuals.
ANCESTRYMAP (31) was used to estimate local ancestry (probabilities of whether an individual has 0, 1 or 2 alleles of European ancestry) at 738 831 autosomal SNPs across the genome, for each participant in JHS, as previously described (32). In brief, local ancestry was inferred using a hidden Markov model based on the genotypes from a panel of densely spaced markers differentiated in frequency between African and European populations. To assess whether there were any regions where local ancestry was associated with iron-related measures, we performed admixture mapping across the whole genome by regressing each of our iron measures on the local ancestry estimates at each SNP location, including covariate adjustment for age, sex, BMI and estimated global ancestry. The conventionally reported LOD score, defined as the log, base 10, ratio of the maximum likelihood of the data under a local-ancestry-associated model divided by the likelihood of the data under the null model (with no local ancestry predictor), was computed at each SNP location. For regions showing association of increased African ancestry with higher levels of iron measures, the LOD scores were assigned positive values, and for regions showing association of increased African ancestry with lower level of iron measures, the LOD scores were assigned negative values. The LOD scores were plotted across the genome, and a LOD score of 5 was assumed to be the threshold of statistical significance (31).
Individual genotyped and imputed SNPs were tested for association using multivariable linear regression models in PLINK (33) and MACH2QTL v.1.08 (28), respectively; adjusted for age, sex, BMI and 10 principal components that we constructed using the software EIGENSOFT (34) to model background ancestry. A second level of covariate adjustment for log-ferritin additionally included self-reported menopausal status, which was available on a large subset of JHS subjects and has been shown previously to be a strong predictor of serum ferritin levels (8,35). We assumed an additive mode of inheritance and reported β coefficients representing the estimated change in the raw or transformed trait value associated with each additional copy of the effect allele. For chromosome X SNPs, hemizygous males were modeled so that males with the minor allele had the same value as females homozygous for the minor allele. We used a significance threshold of P = 5 × 10−8 to maintain an overall type 1 error rate of ∼5% for each phenotype.
Manhattan plots were made to illustrate the association results across the genome and Q–Q plots were made to assess any systematic inflation of the regression test statistics across the genome. In regions demonstrating significant evidence for association, we examined multivariable regression models that included the genotype data of the most strongly associated SNP as a covariate to assess whether there was any evidence for multiple independently associated SNPs in a particular region. If a second signal also reached genome-wide significance after conditioning on the top variant, multivariable regression models were repeated to include the genotypes of both SNPs as covariates. The relevant SNP–SNP interactions were also tested. Region-specific (“locus-zoom”) plots were made to show the magnitude of association between all SNPs and the relevant iron phenotype as well as the LD between each SNP in the region and the most strongly associated SNP (36). Finally, to control for possible confounding between SNP genotype and local ancestry in any observed iron trait-SNP associations, we identified the genetic position of the most strongly associated SNP, selected the local ancestry estimate at the location closest to that SNP (either the SNP itself if genotyped or the closest genotyped SNP) and examined multivariable regression models as described above, but now including estimated local ancestry proportion as an additional covariate.
SNPs that reached genome-wide significance in JHS were selected for testing in the replication study of HANDLS. At each associated region, only the single SNP with the most significant P-value (index SNP) was selected to avoid over adjustment for multiple testing. For the TF region, where conditional analyses revealed a second independently associated SNP with TIBC, the second independent SNP was also included in the replication analyses. A GWAS result was considered replicated if the effect in the replication was in the same direction as in the discovery stage, and if the association in the replication stage was statistically significant after Bonferroni correction adjusting for the number of SNPs tested.
Ethnic differences in iron-related measures have also been observed between subjects of European and African descent (9). Thus, we compared our association results in JHS with established variants from GWASs in populations of European descent in order to assess the importance of these same variants in an AA population. For each prior GWAS-established SNP, we identified and tested all genotyped or imputed proxy SNPs in JHS that were estimated to be in high LD (r2 > 0.8 in CEU based on 1000 Genomes data) with the GWAS index SNPs for association with the reported iron phenotype.
SUPPLEMENTARY MATERIAL
FUNDING
JHS is supported by the National Heart, Lung, and Blood Institute and the National Institute on Minority Health and Health Disparities, with additional support from the National Institute on Biomedical Imaging and Bioengineering (grant numbers HHSN268201300046C, HHSN268201300047C, HHSN268201300048C, HHSN268201300049C, HHSN268201300050C). HANDLS is supported by the Intramural Research Program of the National Institute of Health, National Institute on Aging and the National Center on Minority Health and Health Disparities (project # Z01-AG000513 and human subjects protocol # 2009-149). E.M.L., Y.L. and Q.D. are partially supported by R01HG006703.
Supplementary Material
ACKNOWLEDGEMENTS
The authors express their gratitude to JHS and HANDLS participants who made this research possible.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Andrews N.C., Schmidt P.J. Iron homeostasis. Annu. Rev. Physiol. 2007;69:69–85. doi: 10.1146/annurev.physiol.69.031905.164337. [DOI] [PubMed] [Google Scholar]
- 2.Njajou O.T., Alizadeh B.Z., Aulchenko Y., Zillikens M.C., Pols H.A., Oostra B.A., Swinkels D.W., van Duijn C.M. Heritability of serum iron, ferritin and transferrin saturation in a genetically isolated population, the Erasmus Rucphen Family (ERF) Study. Hum. Hered. 2006;61:222–228. doi: 10.1159/000094777. [DOI] [PubMed] [Google Scholar]
- 3.Wilson J.G., Lindquist J.H., Grambow S.C., Crook E.D., Maher J.F. Potential role of increased iron stores in diabetes. Am. J. Med. Sci. 2003;325:332–339. doi: 10.1097/00000441-200306000-00004. [DOI] [PubMed] [Google Scholar]
- 4.Oexle K., Ried J.S., Hicks A.A., Tanaka T., Hayward C., Bruegel M., Gogele M., Lichtner P., Muller-Myhsok B., Doring A., et al. Novel association to the proprotein convertase PCSK7 gene locus revealed by analysing soluble transferrin receptor (sTfR) levels. Hum. Mol. Genet. 2011;20:1042–1047. doi: 10.1093/hmg/ddq538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pichler I., Minelli C., Sanna S., Tanaka T., Schwienbacher C., Naitza S., Porcu E., Pattaro C., Busonero F., Zanon A., et al. Identification of a common variant in the TFR2 gene implicated in the physiological regulation of serum iron levels. Hum. Mol. Genet. 2011;20:1232–1240. doi: 10.1093/hmg/ddq552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Traglia M., Girelli D., Biino G., Campostrini N., Corbella M., Sala C., Masciullo C., Vigano F., Buetti I., Pistis G., et al. Association of HFE and TMPRSS6 genetic variants with iron and erythrocyte parameters is only in part dependent on serum hepcidin concentrations. J. Med. Genet. 2011;48:629–634. doi: 10.1136/jmedgenet-2011-100061. [DOI] [PubMed] [Google Scholar]
- 7.Benyamin B., McRae A.F., Zhu G., Gordon S., Henders A.K., Palotie A., Peltonen L., Martin N.G., Montgomery G.W., Whitfield J.B., et al. Variants in TF and HFE explain approximately 40% of genetic variation in serum-transferrin levels. Am. J. Hum. Genet. 2009;84:60–65. doi: 10.1016/j.ajhg.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Whitfield J.B., Treloar S., Zhu G., Powell L.W., Martin N.G. Relative importance of female-specific and non-female-specific effects on variation in iron stores between women. Br. J. Haematol. 2003;120:860–866. doi: 10.1046/j.1365-2141.2003.04224.x. [DOI] [PubMed] [Google Scholar]
- 9.McLaren C.E., McLachlan S., Garner C.P., Vulpe C.D., Gordeuk V.R., Eckfeldt J.H., Adams P.C., Acton R.T., Murray J.A., Leiendecker-Foster C., et al. Associations between single nucleotide polymorphisms in iron-related genes and iron status in multiethnic populations. PLoS ONE. 2012;7:e38339. doi: 10.1371/journal.pone.0038339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beutler E., West C. Hematologic differences between African-Americans and whites: the roles of iron deficiency and α-thalassemia on hemoglobin levels and mean corpuscular volume. Blood. 2005;106:740–745. doi: 10.1182/blood-2005-02-0713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Benyamin B., Ferreira M.A., Willemsen G., Gordon S., Middelberg R.P., McEvoy B.P., Hottenga J.J., Henders A.K., Campbell M.J., Wallace L., et al. Common variants in TMPRSS6 are associated with iron status and erythrocyte volume. Nat. Genet. 2009;41:1173–1175. doi: 10.1038/ng.456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kutalik Z., Benyamin B., Bergmann S., Mooser V., Waeber G., Montgomery G.W., Martin N.G., Madden P.A., Heath A.C., Beckmann J.S., et al. Genome-wide association study identifies two loci strongly affecting transferrin glycosylation. Hum. Mol. Genet. 2011;20:3710–3717. doi: 10.1093/hmg/ddr272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paterson A.D., Waggott D., Boright A.P., Hosseini S.M., Shen E., Sylvestre M.P., Wong I., Bharaj B., Cleary P.A., Lachin J.M., et al. A genome-wide association study identifies a novel major locus for glycemic control in type 1 diabetes, as measured by both A1C and glucose. Diabetes. 2010;59:539–549. doi: 10.2337/db09-0653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang Q., Kathiresan S., Lin J.P., Tofler G.H., O'Donnell C.J. Genome-wide association and linkage analyses of hemostatic factors and hematological phenotypes in the Framingham Heart Study. BMC Med. Genet. 2007;8(Suppl. 1):S12. doi: 10.1186/1471-2350-8-S1-S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O'Donnell C.J., Cupples L.A., D'Agostino R.B., Fox C.S., Hoffmann U., Hwang S.J., Ingellson E., Liu C., Murabito J.M., Polak J.F., et al. Genome-wide association study for subclinical atherosclerosis in major arterial territories in the NHLBI's Framingham Heart Study. BMC Med. Genet. 2007;8(Suppl. 1):S4. doi: 10.1186/1471-2350-8-S1-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lowe J.K., Maller J.B., Pe'er I., Neale B.M., Salit J., Kenny E.E., Shea J.L., Burkhardt R., Smith J.G., Ji W., et al. Genome-wide association studies in an isolated founder population from the Pacific Island of Kosrae. PLoS Genet. 2009;5:e1000365. doi: 10.1371/journal.pgen.1000365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vanita V., Singh D., Robinson P.N., Sperling K., Singh J.R. A novel mutation in the DNA-binding domain of MAF at 16q23.1 associated with autosomal dominant "cerulean cataract" in an Indian family. Am. J. Med. Genet. A. 2006;140:558–566. doi: 10.1002/ajmg.a.31126. [DOI] [PubMed] [Google Scholar]
- 18.Jamieson R.V., Perveen R., Kerr B., Carette M., Yardley J., Heon E., Wirth M.G., van Heyningen V., Donnai D., Munier F., et al. Domain disruption and mutation of the bZIP transcription factor, MAF, associated with cataract, ocular anterior segment dysgenesis and coloboma. Hum. Mol. Genet. 2002;11:33–42. doi: 10.1093/hmg/11.1.33. [DOI] [PubMed] [Google Scholar]
- 19.Christiansen G., Mohney B.G. Hereditary hyperferritinemia-cataract syndrome. J. AAPOS. 2007;11:294–296. doi: 10.1016/j.jaapos.2007.01.126. [DOI] [PubMed] [Google Scholar]
- 20.Suga Y., Nagita A., Takesako R., Tanaka I., Kobayashi K., Hirai M., Matsuoka H. A new glucose-6-phosphate dehydrogenase deficiency variant, G6PD Mizushima, showing increases in serum ferritin and cytosol leucine aminopeptidase levels. J. Pediatr. Hematol. Oncol. 2011;33:15–17. doi: 10.1097/MPH.0b013e3181f53da3. [DOI] [PubMed] [Google Scholar]
- 21.Wong C.T., Saha N. Haemoglobin, serum iron, transferrin, ferritin concentrations and total iron-binding capacity in erythrocyte glucose-6-phosphate dehydrogenase deficiency. Trop. Geogr. Med. 1987;39:350–353. [PubMed] [Google Scholar]
- 22.Chen Z., Tang H., Qayyum R., Schick U.M., Nalls M.A., Handsaker R., Li J., Lu Y., Yanek L.R., Keating B., et al. Genome-wide association analysis of red blood cell traits in African Americans: the COGENT Network. Hum. Mol. Genet. 2013;22:2529–2538. doi: 10.1093/hmg/ddt087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lo K.S., Wilson J.G., Lange L.A., Folsom A.R., Galarneau G., Ganesh S.K., Grant S.F., Keating B.J., McCarroll S.A., Mohler E.R., 3rd, et al. Genetic association analysis highlights new loci that modulate hematological trait variation in Caucasians and African Americans. Hum. Genet. 2011;129:307–317. doi: 10.1007/s00439-010-0925-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guindo A., Fairhurst R.M., Doumbo O.K., Wellems T.E., Diallo D.A. X-linked G6PD deficiency protects hemizygous males but not heterozygous females against severe malaria. PLoS Med. 2007;4:e66. doi: 10.1371/journal.pmed.0040066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taylor H.A., Jr The Jackson Heart Study: an overview. Ethn. Dis. 2005;15 S6-1– S6-3. [PubMed] [Google Scholar]
- 26.Wilson J.G., Rotimi C.N., Ekunwe L., Royal C.D., Crump M.E., Wyatt S.B., Steffes M.W., Adeyemo A., Zhou J., Taylor H.A., Jr., et al. Study design for genetic analysis in the Jackson Heart Study. Ethn. Dis. 2005;15 S6-30– S6-37. [PubMed] [Google Scholar]
- 27.Evans M.K., Lepkowski J.M., Powe N.R., LaVeist T., Kuczmarski M.F., Zonderman A.B. Healthy aging in neighborhoods of diversity across the life span (HANDLS): overcoming barriers to implementing a longitudinal, epidemiologic, urban study of health, race, and socioeconomic status. Ethn. Dis. 2010;20:267–275. [PMC free article] [PubMed] [Google Scholar]
- 28.Li Y., Willer C.J., Ding J., Scheet P., Abecasis G.R. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet. Epidemiol. 2010;34:816–834. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Howie B., Fuchsberger C., Stephens M., Marchini J., Abecasis G.R. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat. Genet. 2012;44:955–959. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alexander D.H., Novembre J., Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Patterson N., Hattangadi N., Lane B., Lohmueller K.E., Hafler D.A., Oksenberg J.R., Hauser S.L., Smith M.W., O'Brien S.J., Altshuler D., et al. Methods for high-density admixture mapping of disease genes. Am. J. Hum. Genet. 2004;74:979–1000. doi: 10.1086/420871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deo R.C., Reich D., Tandon A., Akylbekova E., Patterson N., Waliszewska A., Kathiresan S., Sarpong D., Taylor H.A., Jr, Wilson J.G. Genetic differences between the determinants of lipid profile phenotypes in African and European Americans: the Jackson Heart Study. PLoS Genet. 2009;5:e1000342. doi: 10.1371/journal.pgen.1000342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 35.Cade J.E., Moreton J.A., O'Hara B., Greenwood D.C., Moor J., Burley V.J., Kukalizch K., Bishop D.T., Worwood M. Diet and genetic factors associated with iron status in middle-aged women. Am. J. Clin. Nutr. 2005;82:813–820. doi: 10.1093/ajcn/82.4.813. [DOI] [PubMed] [Google Scholar]
- 36.Pruim R.J., Welch R.P., Sanna S., Teslovich T.M., Chines P.S., Gliedt T.P., Boehnke M., Abecasis G.R., Willer C.J. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.