Abstract
Cav1.2 (α1C) and Cav1.3 (α1D) L-type Ca channels are co-expressed in the heart. To date, there are no pharmacological or biophysical tools to separate α1D from α1C Ca currents (ICa-L) in cardiomyocytes. Here, we established a physiological model to study α1D ICa-L in native myocytes using RNA interference. Transfection of rat neonatal cardiomyocytes (RNC) with α1C specific siRNA resulted in low silencing efficiency (50–60%) at the mRNA and protein levels. The use of lentivirus shRNA resulted in 100% transfection efficiency and 92% silencing of the α1C gene by real-time PCR and Western blot. Electrophysiological experiments showed that the total ICa-L was similarly reduced by 80% in lentivirus transfected cells. Both biochemical and functional data demonstrated high transfection and silencing efficiency in the cardiomyocytes using lentiviral shRNA. This novel approach allows for the assessments of the roles of α1C and α1D Ca channels in native myocytes and could be used to examine their roles in physiological and pathological settings.
Keywords: L-type Ca channel, Silencing, Rat neonatal cardiomyocytes, Cardiac, Lentivirus
Introduction
Ca influx through the L-type Ca channels modulates numerous cellular physiological processes ranging from muscle contraction, neuronal excitability, immune regulation, to gene expression [1]. Activation of L-type Ca channels results in the increase in cytosolic Ca thus triggering different cellular events under physiological and pathological settings.
Cardiac L-type Ca channels are complexes of multisubunits composed of α, β2, and α2δ subunits [2,3]. Two types of L-type Ca channels are expressed in the heart: Cav1.2 (α1C) and Cav1.3 (α1D) and both contribute to the total ICa-L. Cav1.2 or α1C is ubiquitously expressed in the cardiovascular system and mediates cardiac excitation–contraction coupling [3]. Cav1.3 or α1D is exclusively restricted to the supraventricular tissue of the adult with higher expression in the sino-atrial node (SAN) and the atrio-ventricular node (AVN) [4]. Up until recently, α1D Ca channel has been thought to be of neuroendocrine origin but recent data confirmed its expression in the heart [4,5].
Electrophysiologically, α1D ICa-L differs from α1C ICa-L in that α1D ICa-L activates at a more negative membrane potentials [6]. Thus, because of its unique localization in the conduction tissue and the range of activation potential, it has been suggested that α1D ICa-L is essential for normal cardiac pacemaker activity and for impulse propagation to the AVN [5,7,8].
Transgenic mice were generated for both Cav1.2 and Cav1.3. α1D knockout (KO) mice are viable and exhibit sinus bradycardia and different degrees of AV blocks [4,5,7,8]. However, deletion of the α1C gene, the predominant form in cardiomyocytes, results in embryonic lethality at day E14.5 [9]. The study of α1D ICa-L has been limited to expression systems such as tsA201 cells because of the lack of efficient pharmacological blockers and biophysical methods to separate α1D ICa-L from α1C ICa-L.
To study α1D Ca channel in a physiological native environment, RNA interference (RNAi) was used to silence the α1C gene and test the effectiveness of RNAi silencing of Cav1.2 in primary rat neonatal cardiomyocyte in culture.
Methods
Animal protocol
All experiments were performed in accordance with animal studies subcommittee regulations at VA New York Harbor Health-care System and all procedures related to animal use conform to the NIH guidelines (Guide for the Care and Use of Laboratory Animals, NIH Publication 86-23).
Preparation of primary neonatal rat cardiomyocyte cultures
Hearts were removed from 1- to 3-day-old Sprague–Dawley rats and ventricles were minced and subjected to consecutive digestion with 0.75 mg/ml trypsin and 20 μg/ml DNase I in CBFHH buffer (137 mM NaCl, 5.4 mM KCl, 0.8 mM MgSO4, 5.6 mM dextrose, 0.4 mM KH2PO4, 0.3 mM Na2HPO4, 20.1 mM HEPES, pH 7.4). Cell suspensions were centrifuged at 1500 rpm for 10 min. The pellets were resuspended in Dulbecco’s Modified Eagle’s Medium (DMEM) containing 10% bovine serum, 0.1 mM bromodeoxyuridine, 1% penicillin–streptomycin, and 1.5 mM vitamin B12. Cell suspensions were filtered through a cell strainer and preplated for 30 min at 37 °C to remove fibroblasts. Cardiac myocytes (3.1 × 106) remaining in suspension were added to 35 mm plates coated with 1% gelatin and incubated overnight in the above medium at 37 °C in humidified air with 5% CO2. Cultures were incubated overnight in serum-free DMEM and then maintained in serum-free DMEM containing 10 μg/ml insulin and 10 μg/ml transferrin at 37 °C in humidified air with 9% CO2 for 3 days prior to exposure to experimental manipulations in serum-free DMEM.
Design and construction of lentiviral siRNA
The sequence of the siRNA specifically targeting the Cav1.2 gene was either purchased from Ambion (Silencer Pre-designed siRNA) siRNA ID # 199812 and 199811 or designed through siRNA Target Finder (Ambion, Austin, TX). The oligonucleotides were selected by Blast homology search among cDNA sequences of target genes from various species including mouse, rat and human. The oligo’s designed were synthesized from IDT with a 5′ phosphate and PAGE purified. The oligo format is the following:
Sense Oligo: 5′ T-(19nt)-TTCAAGAGA)-(91nt)-TTTTTTC-3′ [Sense siRNA-Loop-Antisense siRNA-Stop]. Anti-sense oligo: Compliment of sense but with additional nucleotides at 5′ end generating an XhoI site.
Lentiviral vector cloning
The sense and anti-sense oligo’s were resuspended in water at 60 pmol/μl concentration and annealed in annealing buffer: 100 nM K-acetate, 30 nM HEPES–KOH, pH 7.4, 2 mM Mg-acetate by incubating at 95 °C for 4 min, 70 °C for 10 min, then decreasing the temperature to 4 °C slowly (0.1 °C/min). The annealed oligo was subcloned into XhoI/HpaI site in pLentiLox 3.7 (pLL3.7) vector which encodes the CMV promoter driven eGFP (enhanced green fluorescent protein) marker as internal control. The resulting lentiviral siRNA vector was confirmed by restriction enzyme digestion with XbaI/NotI showing 60 bp band shift on a 2% agarose gel when compared to the parental vector pLL3.7. The constructed vector was also confirmed by DNA sequencing with FLAP primer (5′-CAGTGCAGGGGAAAGAATAGTAGAC-3′).
Lentivirus packaging
Packaging, purification, and titer determination of the lentivirus were performed as described previously [10,11]. The Recombinant viruses were produced by Ca phosphate transfection of HEK293T cells using standard protocols [12]. Briefly, 293T cells were cultured in Dulbecco’s modified Eagles’s medium supplemented with 10% fetal bovine serum (FBS) and penicillin/streptomycin (100 U/ml). At around 70% confluency in a 10 cm culture dish, the cells were co-transfected with the lentiviral vector (10 μg), and the lentiviral packaging vectors pRSV-REV (2 μg), pMDLg/RRE (5 μg), and the vesicular stomatitis virus G glycoprotein (VSVG) expression vector pMD2G (3 μg). The viruses were collected from the culture supernatant 48 h post-transfection, concentrated by ultra-centrifugation for 2 h at 25,000 rpm, and resuspended in phosphate-buffered saline (PBS). Titers were determined by infecting 293T cells with serial dilutions of concentrated lentivirus and counting eGFP positive cells after 48 h under fluorescent microscopy.
Lentiviral transduction in rat neonatal cardiomyocytes
At day 2–3 post-culture, the cells were incubated with lentivirus at various “multiplicity of infection” for 3, 4, and 5 days, respectively. Polybrene (10 μg/ml) was added to increase the efficiency of lentiviral transduction.
Cell sorting using fluorescence-activated cell sorting (FACS)
For sorting by means of FACS, rat neonatal cardiomyocytes were plated and the next day transfected using Lipofectamine 2000 with a combination of α1C siRNA and cy3-labeled GAPDH siRNA [13]. After 48 h, the cells were harvested and FACS sorting of cy3 fluorescent cells was performed at the FACS core facility with a FACS-DIVA sorter. Cy3-positive cells with strong signals in the FL-1 channel were collected and plated on 10 cm dishes. One day after cell sorting, the cells were collected for experiments.
Isolation of RNA and real-time RT-PCR
Total RNA was extracted using the TRIZOL. With this method, we obtained minimal DNA in our RNA samples. RNA was quantified by spectrophotometry at 260 nM and the ratio of absorbance at 260 nm to that of 280 nm was >1.8 for all samples. Degradation of RNA was monitored by the observation of appropriate 28S to 18S rRNA ratios as determined by ethidium bromide staining of agarose gels. Aliquots of RNA were stored in RNase free water at −20 °C. For real-time RT-PCR, first the RNA samples were reverse transcribed using the Ambion reverse transcription Retroscript kit without heat denaturation. Real-time RT-PCR was performed using the Taqman Universal PCR Master Mix (Applied Biosystems) and Pre-designed and labeled primer/Taqman probe (Applied Biosystems) using 18S ribosomal RNA as internal control [14,15]. The reaction solution was assembled in a volume of 50 μl containing the Taqman universal PCR master mix, α1D specific forward and reverse primers (final concentration 300 nM each), the TaqMan probe (200 nM) and the cDNA mixture. The quantity of the α1D cDNA of interest in a certain cDNA mixture was normalized to that of 18S rRNA.
Protein extraction and Western blots
Cells were collected at 48, 72, and 96 h post-transfection. Total protein was prepared as previously described [16]. Briefly, rat neonatal cardiomyocytes were lysed in RIPA buffer (50 mM Tris–Cl, pH 7.4, 150 mM NaCl, 1% NP40, 0.25% Na-deoxycholate, 1 mM PMSF) complete with protease inhibitors (in mmol/l: Leupeptin 0.1 and phenylmethylsulfony fluoride 0.3) and kept for 30 min on ice with vortexing every 5 min. The supernatant was collected following centrifugation at 14,000 rpm for 15 min at 4 °C. The protein concentration was determined in triplicate by a Bio-Rad DC protein assay Kit. The same amount of membrane proteins (50 μg) were loaded for each lane of standard 4–12% SDS–polyacrylamide gels as previously described. After electrophoresis, proteins were transferred to a nitrocellulose membrane. Membranes were blocked in PBS containing 0.1% Tween and 5% non-fat milk. Membranes were incubated overnight at 4 °C in 1:200 primary anti-α1C Ca channel antibody (sigma and Calbiochem). The immunoblots were developed with horseradish peroxidase-labeled goat anti-rabbit IgG (Chemicon) in PBS-Tween as a secondary antibody (1:2000) for 1 h, followed by detection by ECL (Amersham). The density of protein bands was quantified by NIH image software.
Electrophysiological recording of L-type Ca current native cells
Solutions
The composition of external solutions to record whole cell L-type Ca current, ICa-L in (mM) was: NaCl 132, CsCl 5.4, CaCl2 1.8, MgCl2 1.8, NaH2PO4 0.6, 4-amino-pyridine 5, HEPES 10, dextrose 5, Na-pyruvate 5, pH 7.4. Patch electrodes were filled with control internal solution containing (in mM): CsCl 139.8, K-EGTA 10, MgCl2 4, CaCl2 0.062, Na2-creatine phosphate 5, HEPES 10, Na2 ATP 3.1, Na2GTP 0.42, adjusted to pH 7.1 with KOH.
Electrophysiology
The whole cell patch-clamp technique was used [16]. To record L-type ICa-L, all K currents were blocked with intracellular and extracellular Cs2+ and 4-amino-pyridine. The fast Na and T-type Ca currents were blocked by a prepulse to −50 mV from a holding potential of −80 mV every 10 s. The junction potential was always compensated and was smaller than 5 mV. Membrane currents were recorded using Axopatch 200B patch-clamp amplifier (Axon Instruments).
Data analysis
Statistical comparisons were evaluated using either paired or unpaired Student’s t-test, as appropriate. Data are presented as means ± SE. A value of P < 0.05 is considered significant.
Results
Chemically synthesized siRNA were unable to effectively silence Cav1.2
To establish a system for the characterization of Cav1.3 Ca channel in rat neonatal cardiomyocytes, chemically synthesized siRNA against the Cav1.2 gene were purchased from Ambion (siRNA #1 and siRNA#2) and were tested for efficacy. Lipid based reagent Lipofectamine 2000 (Invitrogen) was chosen as a choice for the delivery of the siRNA into the RNC. Different concentrations of siRNA #1 and #2 ranging from 10 to 100 nM were delivered at day 2–3 following the culture and plating of RNC. Forty-eight to 72 h later, the RNC were collected and analyzed using real-time PCR and Western blotting for the residual mRNA and protein content compared to scrambled siRNA controls. Fig. 1A shows the mRNA level for Cav1.2 using siRNA #1 and #2 at concentrations of 50 and 100 nM. Real-time PCR detected only limited silencing of Cav1.2 using siRNA#2 (up to 30%). In contrast siRNA#1 at 100 nM silenced approximately 50% of the Cav1.2 mRNA. Decrease in Cav1.2 protein (45%) was consistent with the decrease in the mRNA as shown in Fig. 1B and C.
Fig. 1.

Effect of Cav1.2 siRNA on Cav1.2 mRNA and protein. siRNA #1 and #2 purchased from Ambion were tested using lipofectamine as a transfection reagent on RNC. Fifty and 100 nM concentration were used from each of the above mentioned siRNA’s. Forty-eight to 72 h post-transfection, RNA was extracted using TRIZOL reagent and subsequently reverse transcribed using Ambion retroscript kit. cDNA from control and silenced were assayed using real-time PCR using Taqman intron spanning primers (Applied Biosystem). siRNA #1 achieved the highest silencing effect reaching merely 50% (A). Similarly, proteins were extracted using RIPA buffer and resolved on 7.5% SDS–PAGE (B). The proteins were analyzed by densitometry (C) and the levels were comparable to the real-time PCR data shown in A.
Optimizing transfection efficiency by cotransfecting with human cy3-labeled GAPDH siRNA
The level of silencing achieved wih lipofectamine transfection as a delivery method for siRNA was low (50%) despite several attempts for optimization. We next used siRNA for GAPDH labeled with cy3 in the transfection experiments. Fig. 2A shows that the maximum achievable level of transfection was around 55% in the RNC. In order to collect only the enriched transfected cells, we cotransfected siRNA #1 along with human GAPDH cy3 labeled siRNA. The fluorescent assay cell sorter was used to sort out the fluorescent cells from the non-fluorescent cells (Fig. 2B). The isolated and transfected RNC were either replated or directly used to extract mRNA and tested for the level of silencing using real-time PCR. The reduction of Cav1.2 mRNA reached 65% with a residual 35% mRNA level remaining (Fig. 2C).
Fig. 2.
Testing and optimizing transfection efficiency using cy3-labeled GAPDH siRNA. To measure the transfection efficiency, human GAPDH siRNA labeled with cy3 was co-transfected with siRNA #1 and immunofluorescent images were captured and analyzed for transfection efficiency. A shows 50–55% transfection efficiency. B shows the sorted cells co-transfected with cy3 labeled siRNA along with siRNA #1. Cells were sorted out using FACS, and tested using real-time PCR for the knockdown effect. C shows that the level of silencing reached 65% compared to control.
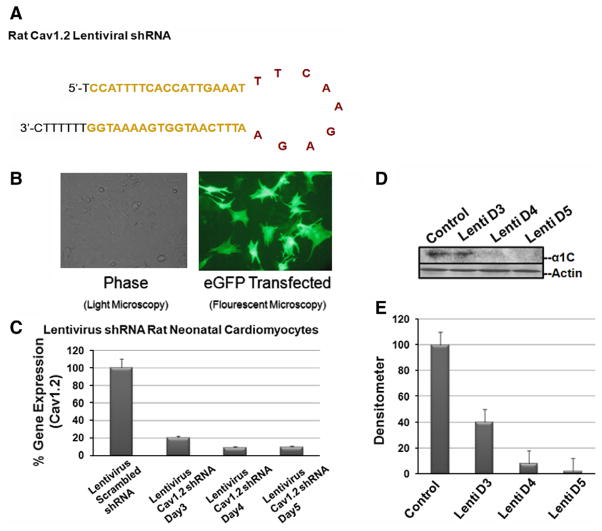
Lentivirus delivered shRNA construct of siRNA #1 efficiently transfected RNC
To achieve higher transfection levels, the lentivirus model was tested. Lentivirus shRNA was constructed from siRNA #1 as shown in Fig. 3A. Restriction sites were added to ends of the sense and anti-sense oligo’s which contained a loop and a termination signal (Fig. 3A). The oligo’s were inserted into pLL3.7 driven under a U6 promoter and contained GFP reporter under CMV promoter to detect the transfection efficiency simultaneously. The level of transfection achieved by the lentivirus construct was almost 100% as shown by immunoflourescent images in Fig. 3B. Next we sought to demonstrate the silencing of Cav1.2 gene. Fig. 3C, D, and E represent the mRNA and protein levels after silencing using different multiplicity of infection at day 3, 4, and 5. At day 4 and 5, more than 90% of Cav1.2 mRNA and more than 95% of the protein were silenced.
Fig. 3.
Lentivirus construct and silencing effects in RNC. A shows the complete shRNA sequence of both sense and anti-sense complementary to each other along with the loop, restriction sites at both ends, and the terminal signal. B shows an immunofluorescent image for the reporter GFP expressed under a CMV promoter in RNC. The level of transfection approached the 100% mark. The level of mRNA after silencing is shown in C using real-time PCR. At day 4 and 5, more than 90% of the Cav1.2 mRNA was abolished. Similarly, D and E show the protein level using Western blotting and the corresponding densitometry, respectively.
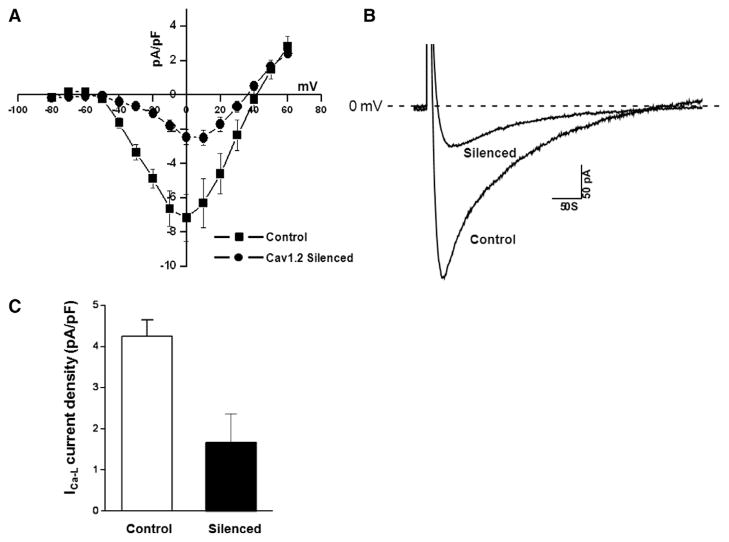
Residual L-type Ca current after Cav1.2 silencing
We examined functional consequences of the Cav1.2 silencing using the whole cell patch-clamp technique. Recording were obtained from GFP expressing RNC at days 4 and 5 as evidence for successful transfection. Fig. 4A and B show typical current recordings from the control and the transfected cells. Fig. 4A shows a current– voltage relationship of ICa-L density (7.1 ± 0.8 pA/pF at 0 mV) in control cells and following the silencing with the lentivirus/Cav1.2 construct (ICa-L density was reduced to 2.5 ± 0.6 pA/pF at 0 mV). Fig. 4B shows representative ICa-L traces elicited from a holding potential of −80 mV to a prepulse to −50 mV followed by voltage steps to 0 mV. The silenced current represents the residual Cav1.2 contribution (minimal) as well as Cav1.3 to the total ICa-L. Thus, we demonstrated a successful model for silencing Cav1.2 at both the molecular level (mRNA and protein) and at the functional level.
Fig. 4.
Functional evidence of knockdown of Cav1.2 using the whole cell patch-clamp recordings. A shows a current–voltage relationship from control and silenced RNC. The current density decreased from from 7.1 pA/pF during control (n = 8) to 2.5 pA/pF in the silenced RNC (n = 6). B shows selected current traces from a control and a silenced cell. C shows the averaged data from the patch-clamp experiments. The residual current is a combination of Cav1.3 and the residual Cav1.2 after silencing.
Discussion
This study is first to establish a model for efficient post-transcriptional silencing of a 240 kDa Ca channel protein (Cav1.2) that is ubiquitously and highly expressed throughout the heart. The results demonstrated a successful shRNA mediated selective silencing of Cav1.2 in primary cultured rat neonatal cardiomyocytes. Chemically synthesized siRNA had limited effects on the silencing of Cav1.2 Ca channel mostly due to difficulty in delivering the siRNA in primary cells. Using FACS sorting, we were able to increase the selection of transfected cells which achieved 65% silencing. When the lentivirus was used as a delivery method, 100% transfection efficiency and 92% silencing at the mRNA and protein level was achieved. To confirm these biochemical results, we functionally recorded the residual Cav1.2 current and showed that the total ICa-L decreased to a 20% level representing the combined residual Cav1.2 (which is minimal post-silencing) and Cav1.3 (which is the main contributor in the silenced cells).
RNAi is a sequence specific post-transcriptional silencing of any gene of interest [17,18]. A limitation for the use of siRNA in primary cardiomyocytes is the low transfection efficiency that can be achieved with difficult delivery. Very few reports published the effective silencing of voltage-gated channels in primary cardiomyocytes [19,20]; none involving voltage gated L-type Ca channels. To achieve high efficiency of transfection, we generated a lentiviral vector carrying shRNA for Cav1.2. Driven under a U6 promoter, the shRNA is subsequently processed into siRNA in the cells and mediated the gene specific silencing.
Several reports show that the α1D deletion causes sinus bradycardia and various degrees of AV-block [4,7,8]. This eludes to the critical role that Cav1.3 Ca channel plays in the diastolic depolarization of the SAN [7]. Cav1.3 Ca channel is expressed in the SAN, AVN and atria, but not in the ventricles of adult hearts [4]. We and others recently demonstrated that Cav1.3 Ca channel could play an important role in cardiac arrhythmias such as atrial fibrillation and heart block [16,21]. The challenge is that both Cav1.2 and Cav1.3 Ca channels contribute to the total ICa-L [9] in the native cardiomyocytes, both are sensitive to the Ca channel blockers such as dihydropurydines and there is no biophysical method to functionally differentiate between Cav1.2 and Cav1.3 Ca channels. The use of transgenic mouse models has been limited due to embryonic lethality at E14.5 of the Cav1.2 knockout mouse [9]. Therefore, the study and characterization of post-natal Cav1.3 by Cav1.2 gene deletion is not possible.
The novel model reported in this study will allow for the investigation and characterization of the Cav1.3 Ca channels in physiological and pathological settings thereby elucidating the Cav1.3 role in cardiac arrhythmias such as atrial fibrillation.
Acknowledgments
This study was supported by NIH R01 HL-077494 and VA MERIT grants to Dr. Boutjdir and VA MREP grant to Dr. Qu.
Footnotes
Conflict of interest
No conflict to disclose.
References
- 1.Reuter H. Ion channels in cardiac cell membranes. Annu Rev Physiol. 1984;46:473–484. doi: 10.1146/annurev.ph.46.030184.002353. [DOI] [PubMed] [Google Scholar]
- 2.Mikami A, Imoto K, Tanabe T, Niidome T, Mori Y, Takeshima H, Narumiya S, Numa S. Primary structure and functional expression of the cardiac dihydropyridine-sensitive calcium channel. Nature. 1989;340:230–233. doi: 10.1038/340230a0. [DOI] [PubMed] [Google Scholar]
- 3.Catterall WA, Perez-Reyes E, Snutch TP, Striessnig J International Union of Pharmacology. XLVIII. Nomenclature and structure–function relationships of voltage-gated calcium channels. Pharmacol Rev. 2005;57:411–425. doi: 10.1124/pr.57.4.5. [DOI] [PubMed] [Google Scholar]
- 4.Mangoni ME, Couette B, Bourinet E, Platzer J, Reimer D, Striessnig J, Nargeot J. Functional role of L-type Cav1.3 Ca2+ channels in cardiac pacemaker activity. Proc Natl Acad Sci USA. 2003;100:5543–5548. doi: 10.1073/pnas.0935295100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang Z, Xu Y, Song H, Rodriguez J, Tuteja D, Namkung Y, Shin HS, Chiamvimonvat N. Functional Roles of Ca(v)1.3 (alpha(1D)) calcium channel in sinoatrial nodes: insight gained using gene-targeted null mutant mice. Circ Res. 2002;90:981–987. doi: 10.1161/01.res.0000018003.14304.e2. [DOI] [PubMed] [Google Scholar]
- 6.Koschak A, Reimer D, Huber I, Grabner M, Glossmann H, Engel J, Striessnig J. Alpha 1D (Cav1.3) subunits can form l-type Ca2+ channels activating at negative voltages. J Biol Chem. 2001;276:22100–22106. doi: 10.1074/jbc.M101469200. [DOI] [PubMed] [Google Scholar]
- 7.Matthes J, Yildirim L, Wietzorrek G, Reimer D, Striessnig J, Herzig S. Disturbed atrio-ventricular conduction and normal contractile function in isolated hearts from Cav1.3-knockout mice. Naunyn Schmiedebergs Arch Pharmacol. 2004;369:554–562. doi: 10.1007/s00210-004-0940-7. [DOI] [PubMed] [Google Scholar]
- 8.Platzer J, Engel J, Schrott-Fischer A, Stephan K, Bova S, Chen H, Zheng H, Striessnig J. Congenital deafness and sinoatrial node dysfunction in mice lacking class D L-type Ca2+ channels. Cell. 2000;102:89–97. doi: 10.1016/s0092-8674(00)00013-1. [DOI] [PubMed] [Google Scholar]
- 9.Seisenberger C, Specht V, Welling A, Platzer J, Pfeifer A, Kuhbandner S, Striessnig J, Klugbauer N, Feil R, Hofmann F. Functional embryonic cardiomyocytes after disruption of the L-type alpha1C (Cav1.2) calcium channel gene in the mouse. J Biol Chem. 2000;275:39193–39199. doi: 10.1074/jbc.M006467200. [DOI] [PubMed] [Google Scholar]
- 10.Rubinson DA, Dillon CP, Kwiatkowski AV, Sievers C, Yang L, Kopinja J, Rooney DL, Zhang M, Ihrig MM, McManus MT, Gertler FB, Scott ML, Van Parijs L. A lentivirus-based system to functionally silence genes in primary mammalian cells, stem cells and transgenic mice by RNA interference. Nat Genet. 2003;33:401–406. doi: 10.1038/ng1117. [DOI] [PubMed] [Google Scholar]
- 11.Follenzi A, Naldini L. Generation of HIV-1 derived lentiviral vectors. Methods Enzymol. 2002;346:454–465. doi: 10.1016/s0076-6879(02)46071-5. [DOI] [PubMed] [Google Scholar]
- 12.Qu Y, Karnabi E, Chahine M, Vassalle M, Boutjdir M. Expression of skeletal muscle Na(V)1.4 Na channel isoform in canine cardiac Purkinje myocytes. Biochem Biophys Res Commun. 2007;355:28–33. doi: 10.1016/j.bbrc.2007.01.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen AA, Derfus AM, Khetani SR, Bhatia SN. Quantum dots to monitor RNAi delivery and improve gene silencing. Nucleic Acids Res. 2005;33:e190. doi: 10.1093/nar/gni188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Razeghi P, Essop MF, Huss JM, Abbasi S, Manga N, Taegtmeyer H. Hypoxia-induced switches of myosin heavy chain iso-gene expression in rat heart. Biochem Biophys Res Commun. 2003;303:1024–1027. doi: 10.1016/s0006-291x(03)00478-9. [DOI] [PubMed] [Google Scholar]
- 15.Xu X, Best PM. Postnatal changes in T-type calcium current density in rat atrial myocytes. J Physiol. 1992;454:657–672. doi: 10.1113/jphysiol.1992.sp019285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qu Y, Baroudi G, Yue Y, Boutjdir M. Novel molecular mechanism involving alpha1D (Cav1.3) L-type calcium channel in autoimmune-associated sinus bradycardia. Circulation. 2005;111:3034–3041. doi: 10.1161/CIRCULATIONAHA.104.517326. [DOI] [PubMed] [Google Scholar]
- 17.Mello CC, Conte D., Jr Revealing the world of RNA interference. Nature. 2004;431:338–342. doi: 10.1038/nature02872. [DOI] [PubMed] [Google Scholar]
- 18.Bass BL. RNA interference. The short answer. Nature. 2001;411:428–429. doi: 10.1038/35078175. [DOI] [PubMed] [Google Scholar]
- 19.Cotella D, Jost N, Darna M, Radicke S, Ravens U, Wettwer E. Silencing the cardiac potassium channel Kv4.3 by RNA interference in a CHO expression system. Biochem Biophys Res Commun. 2005;330:555–560. doi: 10.1016/j.bbrc.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 20.Mikami M, Yang J. Short hairpin RNA-mediated selective knockdown of NaV1.8 tetrodotoxin-resistant voltage-gated sodium channel in dorsal root ganglion neurons. Anesthesiology. 2005;103:828–836. doi: 10.1097/00000542-200510000-00022. [DOI] [PubMed] [Google Scholar]
- 21.Mancarella S, Yue Y, Karnabi E, Qu Y, El-Sherif N, Boutjdir M. Impaired Ca2+ homeostasis is associated with atrial fibrillation in the alpha1D L-type Ca2+ channel KO mouse. Am J Physiol Heart Circ Physiol. 2008;295:H2017–H2024. doi: 10.1152/ajpheart.00537.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]