Abstract
Patterns of colonization and diversification on islands provide valuable insights into evolutionary processes. Due to their unique geographic position and well known history, the Galapagos Islands are an important model system for evolutionary studies. Here we investigate the evolutionary history of a winged grasshopper genus to infer its origin and pattern of colonization in the Galapagos archipelago. The grasshopper genus Sphingonotus has radiated extensively in the Palaearctic and many species are endemic to islands. In the New World, the genus is largely replaced by the genus Trimerotropis. Oddly, in the Caribbean and on the Galapagos archipelago, two species of Sphingonotus are found, which has led to the suggestion that these might be the result of anthropogenic translocations from Europe. Here, we test this hypothesis using mitochondrial and nuclear DNA sequences from a broad sample of Sphingonotini and Trimerotropini species from the Old World and New World. The genetic data show two distinct genetic clusters representing the New World Trimerotropini and the Old World Sphingonotini. However, the Sphingonotus species from Galapagos and the Caribbean split basally within the Old World Sphingonotini lineage. The Galapagos and Caribbean species appear to be related to Old World taxa, but are not the result of recent anthropogenic translocations as revealed by divergence time estimates. Distinct genetic lineages occur on the four investigated Galapagos Islands, with deep splits among them compared to their relatives from the Palaearctic. A scenario of a past wider distribution of Sphingonotus in the New World with subsequent extinction on the mainland and replacement by Trimerotropis might explain the disjunct distribution.
Introduction
Oceanic archipelagos are natural laboratories for studying evolutionary processes [1]. The Galapagos archipelago, in particular, has provided significant insight into our current understanding of speciation [2–4]. Its remote location far off the coast of Ecuador and its well-known geologic history [5] provide a unique opportunity to study colonization and subsequent radiation processes. The islands in their current state developed less than 5 million years ago [5, 6, 7]. The ages of the central and western islands, however, are much younger and range between 0.5 and 2.5 my [8]. This variation in island age might influence patterns of divergence within the archipelago.
The location of the archipelago has determined the general colonization source: phylogeographic studies have shown that most animal and plant species endemic to the Galapagos Islands originated in South America and radiated after one or multiple colonization events [9–13]. Subsequent ‘island hopping’ led to further differentiation among island lineages [7, 13]. Hence, most organisms found in the Galapagos archipelago belong to Neotropic groups and very few studies have shown a direct relationship of Galapagos endemics to any Old World taxon (e.g. the Hemipteran Nezara viridula [14, 15]). However, for many Galapagos endemics no closely related taxa occur in the Old World. In the rare instance that Galapagos species have both New and Old World relatives, phylogeographic studies often neglect the Old World as possible colonization source.
Representatives of the grasshopper genus Sphingonotus Fieber, 1852 provide a rare case of Galapagos endemics for which representatives can be found in both the New and the Old World [16, 17]. This genus is among the most species-rich grasshopper genera worldwide [18, 19]. Its main centres of species richness and endemism are the Mediterranean, central and eastern Asia, but a limited number of species have been described from Australia, South Africa, the Caribbean and Galapagos [17, 20–22]. The genus contains many endemics with very limited geographic distributions [18], often endemic to islands [20, 22]. In North and South America the genus is replaced by the ecologically similar genus Trimerotropis Stål, 1873 [19, 23]. Both genera were until recently grouped in the same tribe (Sphingonotini) [23]. However, it has been demonstrated that they split some 35 million years ago [23]. The presence of Sphingonotus species in the Caribbean (Sphingontous haitensis (Saussure, 1861)) and Galapagos (Sphingonotus fuscoirroratus (Stål, 1861)) is puzzling as these archipelagos are far off the main distribution [24]. Only two other Sphingonotus species have been recorded from the New World, Sphingonotus brasilianus Saussure, 1888 and Sphingonotus punensis Dirsh, 1969. The types of S. brasilianus are lost (NHMW pers. com.) [25] and the description of the species is insufficient to judge the status of the species. Hence, we consider it as nomen dubium. Sphingonotus punensis from Puna Island close to the Ecuadorian coast is morphologically very similar to S. fuscoirroratus [26, 27] and thought to belong to the same species group. However, only a single female of the species is known [26]. Sphingonotus fuscoirroratus itself has a complex history. Originally two species (S. trinesiotis Snodgrass, 1902, S. tetranesiotis Snodgrass, 1902) with several subspecies were described from the Galapagos Islands [28], which later were synonymised [29]. This synonymy was subsequently confirmed by morphological analyses, including inner genitalia, as the island populations could not be separated [26]. Similarly, S. haitensis was originally split in three species (S. haitensis, S. jamaicensis Saussure, 1884, S. cubensis Saussure, 1884). However, currently, only a single species with two subspecies is considered valid [16]. Interestingly, both taxa have been connected to the European species Sphingonotus caerulans in the past due to extremely similar phallic structures [26] and on the basis of the outer morphology [16].
To study the reasons for this disjunct distribution pattern across both continents, we test three hypotheses using a wide geographic sampling and DNA sequences of two mitochondrial genes and a nuclear gene fragment. (i) The taxonomic assignment of the Caribbean and Galapagos species might be wrong and these species may be related to the New World genus Trimerotropis. (ii) It has been suggested that the occurrence of Sphingonotus in the Caribbean is the result of recent anthropogenic translocation of a European species [16]. (iii) Alternatively, their presence may be the result of ancient long-distance colonization from the Old World and may be the relict of a formerly wider distribution.
Results
We sequenced a 651 bp long fragment of the Cytochrome Oxidase I (COI) gene for a total of 104 specimens. The alignment for the NADH Dehydrogenase subunit 5 (ND5) fragment consisted of 955 bp and 104 sequences. For the nuclear Histone 3 (H3) gene fragment 293 bp were sequenced for the same set of taxa (Table 1). The ND5 alignment had 401 variable sites (42.0%), 317 of which were parsimony informative. The COI alignment had 225 variable sites (34.6%), 200 of which were parsimony informative. H3 had 26 variable sites (8.9%), 18 of which were parsimony informative.
Table 1. Overview of all samples used for molecular analyses; given are sampling location, GPS-coordinates, date of sampling and respective Genbank accession numbers.
| ID | Tribe | Genus | Species | Country | County/Island/City | Collector | Genbank accessions | ||
|---|---|---|---|---|---|---|---|---|---|
| COI | ND5 | H3 | |||||||
| T76 | Chortophagini | Chortophaga | viridifasciata | USA, Texas | McLennan Co. | MH | JQ513034 | JQ513132 | JQ513175 |
| T112 | Cibolacrini | Cibolacris | parviceps | USA, Texas | Brewster Co. | MH | JQ513033 | JQ513133 | JQ513176 |
| T25 | Trimerotropini | Circotettix | maculatus | USA, California | Mono Co. | D. Ferguson | JQ513041 | JQ513134 | JQ513177 |
| T26 | Trimerotropini | Circotettix | maculatus | USA, California | Mono Co. | D. Ferguson | JQ513045 | JQ513135 | JQ513178 |
| T10 | Trimerotropini | Circotettix | rabula | USA, New Mexico | Sandoval Co. | D. Ferguson | JQ286519 | JQ286651 | JQ286578 |
| T108 | Trimerotropini | Circotettix | rabula | USA, Montana | Yellowstone Co. | R.D. Scott | JQ513044 | JQ513136 | JQ513179 |
| T9 | Trimerotropini | Circotettix | rabula | USA, New Mexico | Sandoval Co. | D. Ferguson | JQ286518 | JQ286650 | JQ286577 |
| T150 | Trimerotropini | Circotettix | stenometopus | USA, California | Glenn Co. | D. Ferguson | JQ513039 | JQ513137 | JQ513180 |
| T23 | Trimerotropini | Circotettix | undulatus | USA, California | Mono Co. | D. Ferguson | JQ513043 | JQ513138 | JQ513181 |
| T24 | Trimerotropini | Circotettix | undulatus | USA, California | Mono Co. | D. Ferguson | JQ513042 | JQ513139 | JQ513182 |
| T15 | Trimerotropini | Conozoa | texana | USA, New Mexico | Valencia Co. | D. Ferguson | JQ286500 | JQ286632 | JQ286567 |
| K379 | Sphingonotini | Leptopternis | maculatus | Tunisia | Ouesslatia | AH | JQ513074 | JQ513140 | JQ513183 |
| K473 | Sphingonotini | Sphingoderus | carinatus | Tunisia | Bou Hedma | AH | KJ923334 | KJ923393 | KP201145 |
| K315 | Sphingonotini | Sphingonotus | caerulans | France | Vergières / Crau | AH | JQ513068 | JQ513142 | JQ513185 |
| K608 | Sphingonotini | Sphingonotus | caerulans | Finland | Hanko Taktom | AH | JQ513067 | JQ513143 | JQ513186 |
| K613 | Sphingonotini | Sphingonotus | caerulans | Italy | Affi | S. Lötters | KJ923335 | KJ923394 | KP201146 |
| K512 | Sphingonotini | Sphingonotus | canariensis | Cape Verde | Maio | M. Lecoq | JQ513077 | JQ513144 | JQ513187 |
| K403 | Sphingonotini | Sphingonotus | candidus | Italy | Sardinia | Y. Görzig | JQ513066 | JQ513145 | JQ513188 |
| K262 | Sphingonotini | Sphingonotus | corsicus | France | Corse | F. Pahlmann | KJ923336 | EU266719 | KP201147 |
| K90 | Sphingonotini | Sphingonotus | femoralis | Niger | Tabourax | T. McNary | JQ513065 | JQ513146 | JQ513189 |
| K383 | Sphingonotini | Sphingonotus | finotianus | Tunisia | Enfida | AH | JQ513073 | JQ513147 | JQ513190 |
| K456 | Sphingonotini | Sphingonotus | fuerteventurae | Spain | Canary Islands, Fuerteventurae | AH, MH | JQ513071 | JQ513148 | JQ513191 |
| K424 | Sphingonotini | Sphingonotus | fuscoirroratus | Ecuador | Galapagos Islands, Floreana | D. Otte | KJ923337 | KJ923395 | KJ923386 |
| K631 | Sphingonotini | Sphingonotus | fuscoirroratus | Ecuador | Galapagos Islands, San Cristobal | D. Otte | KJ923338 | KJ923396 | KP201148 |
| K632 | Sphingonotini | Sphingonotus | fuscoirroratus | Ecuador | Galapagos Islands, Santa Cruz | D. Otte | KJ923339 | KP201198 | KP201149 |
| T166 | Sphingonotini | Sphingonotus | fuscoirroratus | Ecuador | Galapagos Islands, San Cristobal | D. Otte | KJ923340 | KP201199 | KP201150 |
| T167 | Sphingonotini | Sphingonotus | fuscoirroratus | Ecuador | Galapagos Islands, San Cristobal | D. Otte | KJ923341 | KP201200 | KP201151 |
| T169 | Sphingonotini | Sphingonotus | fuscoirroratus | Ecuador | Galapagos Islands, Floreana | D. Otte | KJ923343 | KJ923397 | KP201152 |
| T170 | Sphingonotini | Sphingonotus | fuscoirroratus | Ecuador | Galapagos Islands, Santa Cruz | D. Otte | KJ923344 | KJ923398 | KP201153 |
| T171 | Sphingonotini | Sphingonotus | fuscoirroratus | Ecuador | Galapagos Islands, Santa Cruz | D. Otte | KJ923345 | KJ923399 | KJ923387 |
| T172 | Sphingonotini | Sphingonotus | fuscoirroratus | Ecuador | Galapagos Islands, Santa Fe | D. Otte | KJ923346 | KJ923400 | KJ923388 |
| T54 | Sphingonotini | Sphingonotus | fuscoirroratus | Ecuador | Galapagos Islands, Santa Fe | D. Otte | KJ923349 | KJ923401 | KP201154 |
| T56 | Sphingonotini | Sphingonotus | fuscoirroratus | Ecuador | Galapagos Islands, Floreana | D. Otte | KJ923350 | KJ923403 | KP201155 |
| T66 | Sphingonotini | Sphingonotus | fuscoirroratus | Ecuador | Galapagos Islands, Floreana | D. Otte | KJ923351 | KJ923404 | KP201156 |
| K14 | Sphingonotini | Sphingonotus | guanchus | Spain | Canary Islands, Gran Canary | AH | JQ513064 | EU266743 | JQ513192 |
| K638 | Sphingonotini | Sphingonotus | guanchus | Spain | Canary Islands, Gran Canary | R. Bland | JQ513063 | JQ513149 | JQ513193 |
| T178 | Sphingonotini | Sphingonotus | haitensis | Dominican Republic | Prov. Independencia | A. Hilario | KP201141 | KJ923405 | KP201157 |
| T179 | Sphingonotini | Sphingonotus | haitensis | Dominican Republic | Prov. San Christobal | A. Hilario | KJ923354 | KJ923406 | KJ923390 |
| T180 | Sphingonotini | Sphingonotus | haitensis | Dominican Republic | Prov. San Christobal | A. Hilario | KJ923355 | KJ923407 | KJ923391 |
| T184 | Sphingonotini | Sphingonotus | haitensis | Dominican Republic | Prov. San Juan | H. Takizawa | KP201142 | KJ923408 | KP201158 |
| T39 | Sphingonotini | Sphingonotus | haitensis | Dominican Republic | Prov. Peravia | D. Perez, B. Hierro | KJ923356 | KJ923409 | KP201159 |
| T40 | Sphingonotini | Sphingonotus | haitensis | Dominican Republic | Prov. Pedernales | D. Perez, B. Hierro, R. Bastardo | KJ923357 | KJ923410 | KP201160 |
| T41 | Sphingonotini | Sphingonotus | haitensis | Dominican Republic | Prov. Pedernales | D. Perez, B. Hierro, R. Bastardo | KJ923358 | KJ923411 | KP201161 |
| K651 | Sphingonotini | Sphingonotus | maroccanus | Morocco | Ameskrout | MH | JQ513075 | JQ513150 | JQ513194 |
| K616 | Sphingonotini | Sphingonotus | ningsianus | China | unknown | unknown | JQ513060 | JQ513151 | JQ513195 |
| K470 | Sphingonotini | Sphingonotus | octofasciatus | Tunisia | Gafsa | AH | JQ513058 | JQ513152 | JQ513196 |
| K351 | Sphingonotini | Sphingonotus | rubescens | Spain | Canary Islands, Fuerteventurae | AH, MH | JQ513069 | JQ513153 | JQ513197 |
| K510 | Sphingonotini | Sphingonotus | rubescens | Cape Verde | Fopo | M. Lecoq | JQ513070 | JQ513154 | JQ513198 |
| K5 | Sphingonotini | Sphingonotus | rugosus | Spain | Canary Islands, Lanzarote | AH | KJ923359 | EU266739 | KP201162 |
| K150 | Sphingonotini | Sphingonotus | savignyi | Spain | Canary Islands, Gran Canary | AH | JQ513076 | JQ513155 | JQ513199 |
| K214 | Sphingonotini | Sphingonotus | scabriculus | Namibia | Otjiu | W. Schuett | JQ513061 | JQ513156 | JQ513200 |
| K615 | Sphingonotini | Sphingonotus | tsinlingensis | China | unknown | unknown | JQ513059 | JQ513157 | JQ513201 |
| K227 | Sphingonotini | Thalpomena | caerulescens | Morocco | Irhil-n’-Isemsiden | AH | JQ513057 | JQ513158 | JQ513203 |
| K641 | Sphingonotini | Thalpomena | viridipennis | Morocco | Imouzzer | MH, JCH | JQ513056 | JQ513159 | JQ513204 |
| T27 | Trimerotropini | Trimerotropis | californica | USA, New Mexico | Socorro Co. | D. Ferguson | KJ923360 | KJ923412 | KP201163 |
| T28 | Trimerotropini | Trimerotropis | californica | USA, New Mexico | Socorro Co. | D. Ferguson | JQ513048 | JQ513160 | JQ513205 |
| T21 | Trimerotropini | Trimerotropis | cincta | USA, New Mexico | Sandoval Co. | D. Ferguson | KJ923361 | KJ923413 | KP201164 |
| T22 | Trimerotropini | Trimerotropis | cincta | USA, New Mexico | Sandoval Co. | D. Ferguson | KJ923362 | KJ923414 | KP201165 |
| T17 | Trimerotropini | Trimerotropis | cyaneipennis | USA, New Mexico | Valencia Co. | D. Ferguson | JQ513040 | JQ513161 | JQ513206 |
| T18 | Trimerotropini | Trimerotropis | cyaneipennis | USA, New Mexico | Valencia Co. | D. Ferguson | KJ923363 | KJ923415 | KP201166 |
| T3 | Trimerotropini | Trimerotropis | cyaneipennis | USA, Arizona | Mojave Co. | D. Ferguson | KJ923364 | KJ923416 | KP201167 |
| T4 | Trimerotropini | Trimerotropis | cyaneipennis | USA, Arizona | Mojave Co. | D. Ferguson | KJ923365 | KJ923417 | KP201168 |
| T104 | Trimerotropini | Trimerotropis | pallidipennis | USA, Montana | Big Horn Co. | R.D. Scott | JQ286536 | JQ286668 | KP201169 |
| T105 | Trimerotropini | Trimerotropis | pallidipennis | USA, Montana | Big Horn Co. | R.D. Scott | JQ286539 | JQ286671 | JQ286598 |
| T109 | Trimerotropini | Trimerotropis | latifasciata | USA, Montana | Blaine Co. | R.D. Scott | KJ923366 | KJ923418 | KP201170 |
| T110 | Trimerotropini | Trimerotropis | latifasciata | USA, Montana | Blaine Co. | R.D. Scott | JQ513047 | JQ513163 | JQ513208 |
| T111 | Trimerotropini | Trimerotropis | latifasciata | USA, Montana | Blaine Co. | R.D. Scott | KJ923367 | KJ923419 | KP201171 |
| T1 | Trimerotropini | Trimerotropis | maritima | USA, Texas | McLennan Co. | MH, PDD | JQ286498 | JQ286630 | JQ286565 |
| T2 | Trimerotropini | Trimerotropis | maritima | USA, Texas | McLennan Co. | MH, PDD | KJ923368 | KJ923420 | KP201172 |
| T52 | Trimerotropini | Trimerotropis | maritima | USA, Texas | Bosque Co. | MH, PDD | JQ286497 | JQ286629 | JQ286564 |
| T86 | Trimerotropini | Trimerotropis | maritima | USA, Texas | Brewster Co. | MH | KJ923369 | KJ923421 | KP201173 |
| T29 | Trimerotropini | Trimerotropis | melanoptera | USA, New Mexico | Valencia Co. | D. Ferguson | KJ923370 | KJ923422 | KP201174 |
| T30 | Trimerotropini | Trimerotropis | melanoptera | USA, New Mexico | Valencia Co. | D. Ferguson | KJ923371 | KJ923423 | KP201175 |
| T14 | Trimerotropini | Trimerotropis | modesta | USA, Arizona | Coconino Co. | D. Ferguson | KJ923372 | KJ923425 | KP201176 |
| T57 | Trimerotropini | Trimerotropis | modesta | USA, Arizona | Cochise Co. | D.R. Swanson | KJ923373 | KP201201 | KP201177 |
| T58 | Trimerotropini | Trimerotropis | modesta | USA, Arizona | Cochise Co. | D.R. Swanson | KJ923374 | KP201202 | KP201178 |
| T152 | Trimerotropini | Trimerotropis | occidentalis | USA, California | Glenn Co. | D. Ferguson | KJ923375 | KJ923426 | KP201179 |
| T153 | Trimerotropini | Trimerotropis | occidentalis | USA, California | Glenn Co. | D. Ferguson | KJ923376 | KP201203 | KP201180 |
| T116 | Trimerotropini | Trimerotropis | ochraceipennis | Chile | Coquimbe | J. Pizarro | JQ286549 | JQ286681 | JQ286607 |
| T117 | Trimerotropini | Trimerotropis | ochraceipennis | Chile | Coquimbe | J. Pizarro | JQ286547 | JQ286679 | KP201181 |
| T118 | Trimerotropini | Trimerotropis | ochraceipennis | Chile | Coquimbe | J. Pizarro | JQ286546 | JQ286678 | KP201182 |
| T119 | Trimerotropini | Trimerotropis | ochraceipennis | Chile | Coquimbe | J. Pizarro | JQ286548 | JQ286680 | JQ286606 |
| T128 | Trimerotropini | Trimerotropis | ochraceipennis | Chile | Coquimbe | J. Pizarro | KJ923377 | JQ286688 | JQ286622 |
| T130 | Trimerotropini | Trimerotropis | pallidipennis | USA, Texas | Brewster Co. | MH | KP201143 | JQ286690 | KP201183 |
| T140 | Trimerotropini | Trimerotropis | pallidipennis | Mexico | El Coptal | D. Salas | JQ286533 | JQ286665 | KP201184 |
| T141 | Trimerotropini | Trimerotropis | pallidipennis | Mexico | Marquez | D. Salas | JQ286527 | JQ286659 | KP201185 |
| T144 | Trimerotropini | Trimerotropis | pallidipennis | Mexico | El Coptal | D. Salas | JQ286562 | KP201204 | KP201186 |
| T156 | Trimerotropini | Trimerotropis | pallidipennis | Mexico | Salamanca | D. Salas | JQ286522 | JQ286654 | JQ286581 |
| T162 | Trimerotropini | Trimerotropis | pallidipennis | Mexico | Salamanca | D. Salas | JQ286537 | JQ286669 | JQ286596 |
| T163 | Trimerotropini | Trimerotropis | pallidipennis | Mexico | Salamanca | D. Salas | JQ286535 | JQ286667 | JQ286594 |
| T124 | Trimerotropini | Trimerotropis | pistrinaria | USA, Texas | Whitney Co. | MH | KJ923379 | KJ923427 | KP201187 |
| T31 | Trimerotropini | Trimerotropis | pistrinaria | USA, New Mexico | Valencia Co. | D. Ferguson | JQ513046 | JQ513165 | JQ513210 |
| T19 | Trimerotropini | Trimerotropis | pseudofasciata | USA, Utah | Tooele Co. | D. Ferguson | KJ923381 | KJ923428 | KP201188 |
| T20 | Trimerotropini | Trimerotropis | pseudofasciata | USA, Utah | Tooele Co. | D. Ferguson | KJ923382 | KJ923429 | KP201189 |
| T132 | Trimerotropini | Trimerotropis | saxatilis | USA, Texas | Hill Co. | M. Hanitzsch | JQ286503 | JQ286635 | JQ286570 |
| T133 | Trimerotropini | Trimerotropis | saxatilis | USA, Texas | Hill Co. | M. Hanitzsch | JQ286502 | JQ286634 | KP201190 |
| T154 | Trimerotropini | Trimerotropis | saxatilis | USA, Missouri | unknown | A. Templeton | KJ923383 | KJ923430 | KP201191 |
| T155 | Trimerotropini | Trimerotropis | saxatilis | USA, Missouri | unknown | A. Templeton | KJ923384 | KJ923431 | KP201192 |
| T68 | Trimerotropini | Trimerotropis | sp | Argentina | Mendoza Prov. | V. Confalonieri | JQ286552 | JQ286684 | KP201193 |
| T69 | Trimerotropini | Trimerotropis | sp | Argentina | Mendoza Prov. | V. Confalonieri | JQ286555 | JQ286687 | KP201194 |
| T70 | Trimerotropini | Trimerotropis | sp | Argentina | San Luis Prov. | V. Confalonieri | JQ286554 | JQ286686 | KP201195 |
| T71 | Trimerotropini | Trimerotropis | sp | Argentina | San Luis Prov. | V. Confalonieri | JQ286553 | JQ286685 | JQ286611 |
| T11 | Trimerotropini | Trimerotropis | verruculata suffusa | USA, New Mexico | Sandoval Co. | D. Ferguson | KP201144 | KJ923432 | KP201196 |
| T12 | Trimerotropini | Trimerotropis | verruculata suffusa | USA, New Mexico | Sandoval Co. | D. Ferguson | KJ923385 | KP201205 | KP201197 |
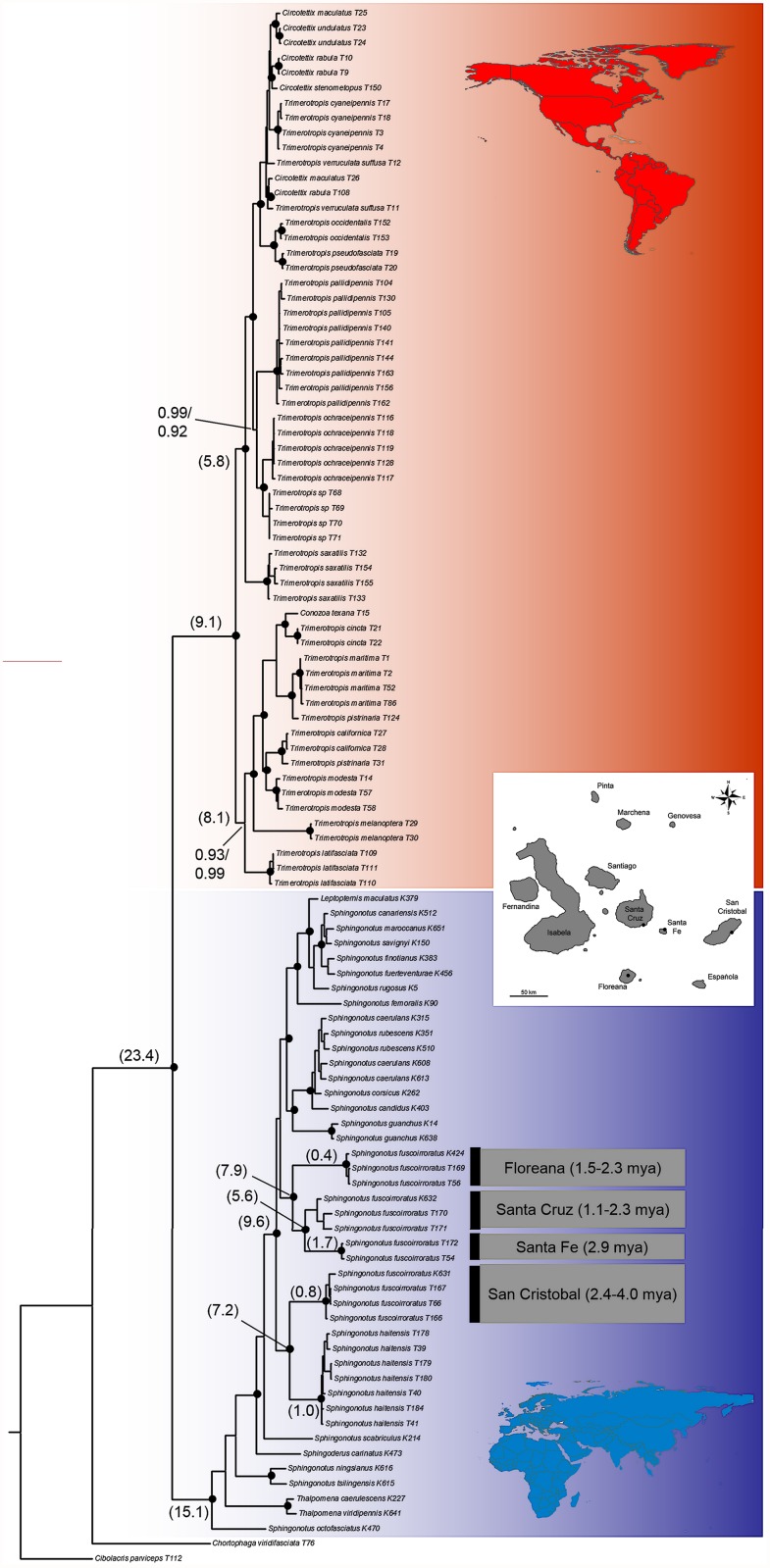
We used two different phylogenetic reconstruction methods, MrBayes and BEAST, which both yielded similar groupings: a major split with high posterior probabilities (pp = 1 for both methods) was identified separating the New World Trimerotropini and the Old World Sphingonotini (Fig. 1). Within the Trimerotropini two groups were detected with high confidence (pp = 1 for both methods) corresponding to the chromosomal groups defined by White [30–32] and previously confirmed by Husemann and colleagues [23]. Further, within the Trimerotropini most species for which multiple individuals were sequenced were monophyletic, besides Trimerotropis pistrinaria Saussure, 1884 and some species of the genus Circotettix Scudder, 1876. Sphingonotus haitensis from the Dominicanian Republic and S. fuscoirroratus from four Galapagos Islands grouped within the Sphingonotini. Within the Sphingonotini S. octofasciatus (Serville, 1838), the genus Thalpomena Saussure, 1884 and the Sphingonotus species from China split basally from the other species in the group. The next split separates Sphingoderus carinatus (Saussure, 1888) from a group consisting of all other Sphingonotus species including S. haitensis and S. fuscoirroratus. The first taxon splitting off in this group is S. scabriculus Stål, 1876 from South Africa followed by the New World Sphingonotus species; Sphingonotus fuscoirroratus from San Cristobal groups together with S. haitensis in both analyses with high support (pp ≥ 0.99). The S. fuscoirroratus lineages from the other three islands form a second monophyletic group with the lineages from Santa Fe and Santa Cruz being sister clades. However, S. fuscoirroratus is not monophyletic in either analysis. The remaining Sphingonotus species from Eurasia and Africa branch off subsequently.
Fig 1. Phylogenetic tree resulting from Bayesian analysis of the combined data set of three genes.
Red color indicates the New World Trimerotropini, blue are the Old World Sphingonotini. Black circles represent posterior probabilities ≥ 0.95 in both analyses. Numbers are posterior probabilities below 0.95 for at least one of the analyses (upper value from BEAST analysis, lower value from MrBayes analysis). The numbers in parentheses represent the divergence time estimates derived from the BEAST analysis. Only the values for main branches of interest are shown and no intraspecific values are presented. Estimates of minimum and maximum emergence times of the studied islands in parentheses next to island names were taken from Geist and colleagues [5].
Both RASP analyses (S-DIVA and Bayes-Lagrange) yielded similar results suggesting an African origin for the Sphingonotini as a whole and a wider distribution (Africa and Galapagos) for the ancestral taxa of the New World Sphingonotus species (S1 Fig.). The molecular clock analyses dated the divergence between the two major clades (Trimerotropini from the New World and Sphingonotini from the Old World) at approximately 23.4 million years ago. The onset of the Trimerotropini radiation was dated at 9.1 million years ago. The Sphingonotini radiation was dated to be older with an age of 15.1 my. The clade including S. fuscoirroratus from San Cristobal and S. haitensis was dated to approximately 9.6 mya whereas the split between the San Cristobal lineage and S. haitensis was dated at 7.2 mya; the radiation of the second S. fuscoirroratus clade started about 7.9 mya. However, the confidence intervals around the estimates were large (S2 Fig.) and hence the results should be only taken as rough guidelines rather than hard evidence.
Discussion
Most oceanic islands are colonized from the closest mainland [7, 33]. For the Galapagos Islands, this means that the common source for most colonizers is the South American mainland, which is ~1000 km away from the archipelago. Our analyses, however, clearly support an Old World origin of the Neotropic Sphingonotus species. The species from Galapagos and the Caribbean Islands group within the Sphingonotini with high support. In addition, the branch lengths of each island population are rather long, which supports the original designation of each island population as a distinct species or subspecies [28] despite limited phenotypic divergence [26].
Phylogeography of the New World Sphingonotus species
The inferred phylogeny interpreted against the background of contemporary species distributions lets us argue that (i) grasshoppers of the tribe Sphingonotini are mainly distributed in the Old World. However, (ii) the focal species found in the Neotropics, i.e. on the Galapagos Islands and in the Caribbean, belong to the Sphingonotini rather than to the Trimerotropini, which is the predominant tribe in the New World. Hence, our analyses reject our first hypothesis that the taxonomic assignment of the Caribbean and Galapagos species to the tribe Sphingonotini is wrong. Rather our data support the hypothesis that the Caribbean (i.e. Atlantic) and the Galapagos Archipelago (i.e. Pacific) species are members of the Sphingonotini.
It has been suggested that the occurrence of Sphingonotus on Galapagos might be the result of a recent introduction from Europe [16]. This hypothesis can be rejected as well, since the species represent rather old lineages within the genus and are much older than most Old World species and diverged prior to any potential introduction date. While the dating is very crude the resulting age estimates are more likely an underestimate than an overestimate; the divergence between the two major clades (Trimerotropini from the New World and Sphingonotini from the Old World) was here estimated at approximately 24.4 million years ago. This dating estimate is more recent (yet both estimates have overlapping 95% HPD) than the estimate derived from a more comprehensive study which dated the split between the clades at about 35 mya [23]. The same split was dated even further back (~55 mya) by a study by Chapco & Contreras [34]. The estimate derived here is therefore a minimum estimate of the age with the lineages likely being much older. The ages of the Galapagos endemics with more than 7 mya at the basis of the lineages predate the origin of the islands.
The observed relationships may be explained by long-distance dispersal via the mainland leading to the colonization of the islands with subsequent extinction on the mainland. One might even speculate that the Sphingonotini might have colonized the American continent (e.g. [23]) and later been displaced by Trimerotropini, except for the oceanic island populations. This is supported by the high age of the islands endemics predating the ages of the islands. Alternatively, the New World Sphingonotus species might have reached the islands via rare long-distance, trans-Atlantic dispersal events. The first colonization step was then likely to the Caribbean, which is supported by the phylogeny. A reasonable number of studies have shown trans-Atlantic dispersal of a variety of animal and plant taxa [35–38]. For example, a study by Carranza and colleagues [39] showed a case of long-distance dispersal, where Tarentola Geckos invaded the Caribbean from Africa [39]; South America has been colonized by Hemidactylus Geckos from Africa [40], and the Americas were colonized from Africa by the grasshopper genus Schistocerca [36].
The Galapagos lineages of Sphingonotus appear to be older than many of the islands and hence a previous mainland distribution with subsequent extinction appears more likely. A continental extinction of the genus would also explain the lack of monophyly of the New World Sphingonotini. However, with our data we are not able to support with confidence either of the following hypotheses: (1) the Sphingonotini had a wider New World distribution which has been largely replaced by the Trimerotropini except for relict occurrences of Sphingonotus on the archipelagos or (ii) the Sphingonotini of the Galapagos archipelago and Hispaniola are the result of trans-Atlantic colonization.
Island colonization and differentiation
In the past, Sphingonotus fuscoirroratus from Galapagos had been divided into two species with several subspecies [28]. Subsequently, these taxa were synonymised as only limited morphological variation between island lineages was found [26]. Our analyses suggest that each island indeed has its own distinct genetic lineage which supports the original species or subspecies status. The extent of genetic divergence of the island populations suggests that no or very little gene flow between islands exists.
Generally, inter-island radiations are typical for the Galapagos as a result of the large distance to the mainland and the relatively high distances between most islands. This can partly be confirmed here (at least for four islands). Similar radiations on the Galapagos are known for mockingbirds (Nesomimus) [10], tenebrionid beetles [41], iguanas (Conolophus) [42], and the Galapagos lava lizards [43]. The lack of monophyly of S. fuscoirroratus due to the position of the San Cristobal lineage might be caused by insufficient resolution of the data or by extinction of true sister species on the American continent. However, another explanation might be that this island was colonized independently from the others as has been shown for the Canary Islands as well [22]. However, this hypothesis would require the assumption that both lineages converged substantially in morphology when adapting to the island habitats.
Conclusion
Our analyses support that the Galapagos endemic S. fuscoirroratus and the Caribbean endemic S. haitensis indeed belong to the tribe Sphingonotini and we therefore reject the hypothesis that these species had been wrongly assigned to the Sphingonotini. The colonization is rather ancient which allows us to reject the hypothesis that the studied species were the result of anthropogenic translocation. However, we cannot infer with certainty if the populations are relicts of a previously more widespread distribution or the result of long-distance, trans-Atlantic dispersal. In demonstrating a close phylogenetic relationship of Galapagos endemic species to Old World taxa, this study highlights the need to include geographically distantly distributed taxa in phylogeographic studies. Following the deep genetic splits detectable for our samples from Galapagos Islands, we assume that at least three to four distinct Sphingonotus species exist on the archipelago. It is likely that further genetic lineages are present on other islands that had not been studied here in concert with the original designation as species and subspecies [28].
Material and Methods
Study species
Grasshoppers of the genus Sphingonotus are widely distributed across major parts of the Palaearctic and Palaeotropic regions. A supposedly close relative, the genus Trimerotropis, can be found exclusively in the Nearctic and Neotropic region [16, 17]. The genera Trimerotropis and Sphingonotus show strong morphological similarities; however, representatives of Trimerotropis are mostly larger [16, 24]. Both genera had been grouped in the tribe Sphingonotini for many decades, but recently the genus Trimerotropis was re-assigned to the previously erected Trimerotropini [23, 44]. Both genera are species-rich with 142 species for Sphingonotus and 52 Species for Trimerotropis [17].
Sampling
In total, 104 individuals belonging to 44 species from four continents were included in the analyses (Table 1). Specimens were collected by hand or netted and subsequently frozen or stored in ethanol. Many samples were obtained from museums or colleagues. None of the collected species are protected and no sampling was performed on protected land aside from the Galapagos. Sampling activities on Galapagos were performed by D. Otte (ANSP, Philadelphia) and S. B. Peck (Carleton University, Ottawa, Canada) under permission of the Galapagos National Park (F. Cepeda, A. Izurieta and E. Cruz, Superintendents, Department of Forestry, Ministry of Agriculture, Republic of Ecuador). The Gomphocerinae Cibolacris parviceps and the Oedipodinae Chortophaga viridifasciata served as outgroups in all analyses. Details about all individuals collected and used for this study are given in Table 1.
Molecular analyses
Genomic DNA was extracted from dried or ethanol preserved hind leg muscle tissue using the Qiagen DNeasy Blood and Tissue Kit (Qiagen, Inc., Valencia, CA) following the manufacturer’s protocol for tissue samples. We amplified two mitochondrial and one nuclear gene fragment using a standard PCR protocol. Primers for the mitochondrial NADH Dehydrogenase subunit 5 (ND5) were obtained from Su and colleagues [45] and for COI from Husemann and colleagues [23]. The primers for Histone 3 (H3) were taken from Colgan and colleagues [46]. PCR reactions were performed using the following setup: 36.6 μl of diH2O, 6 μl of 10 x PCR buffer (reaction concentration 1x), 4.8 μl of dNTP mixture (0.2 μM each), 0.6 μl of DyNAzyme DNA Polymerase (1.2 U, Finnzymes, USA), 3 μl of each primer (0.5 μM, Integrated DNA technologies, USA) and 6 μl of DNA template adding up to a total volume of 60 μl. Amplification conditions were as follows: 94°C for 3 min, followed by 30 cycles of 94°C for 1 min denaturation, 48–57°C 1 min annealing and 72°C for 2 min elongation, with a final elongation step at 72°C for 10 min.
PCR products were visualized on a 1% agarose gel stained with Gel Red (0.1x, Biotium, USA and purified using Solid-phase Reversible Immobilization (SPRI) [47] with carboxylated magnetic beads (Bangs Laboratories, USA) and a 96-Ring SPRIplate (Agencourt, USA). The purified PCR products were sequenced at the Yale Sequencing Facility (New Haven, CT, USA). All sequences were deposited in Genbank; accession numbers are given in Table 1.
Phylogenetic analyses
Sequences were inspected, trimmed and aligned using the MAFFT algorithm in Geneious 5.0.3 [48]. Further we used sequences from previous studies [18–20, 22, 23]. All genes were subsequently analyzed as combined data set. In a first step we identified the best partitioning scheme treating codon positions separately and determined the most suitable substitution models using PartitionFinder v.1.1.1 [49]. We performed two runs of PartitionFinder, one including the models implemented in MrBayes and one including the models implemented in BEAST. We then analyzed the concatenated partitioned data set with MrBayes v.3.1.2 [50]. We ran MrBayes for 50 million generations sampling every 5000 generations. A burn-in of 25% of trees was discarded before constructing a consensus tree. In addition we used BEAST v. 1.8.0 [51] to analyze the data in a supertree framework. The input file for BEAST was setup with BEAUti v. 1.8.0 (implemented in the BEAST package). We used the partitioning scheme from PartitionFinder to link the substitution models. The clock models were linked for mitochondrial genes. The trees were linked for all data. We used the Yule prior as recommended for analyses at species and genus levels and ran the analyses for 100 million iterations sampling every 10,000 iterations. The log-files were checked in Tracer v.1.5 [52] to check for convergence. A burn-in of 1000 trees was discarded before generating a consensus tree. All trees were visualized using FigTree v.1.3.1 [53].
In addition we obtained coarse estimates of divergence dates by applying a molecular clock approach. We used published substitution rates of 0.0113 for ND5 [23] and 0.01 for COI estimating the rate for H3 and applied a strict clock in BEAST v.1.8.0 [51]. No better calibration was possible as no suitable fossil data is available and using island ages as calibration points appeared inappropriate considering that we intended to estimate the divergence times of island lineages. The analysis was run for 100 million generations sampling every 10,000 generations. Trees were summarized with TreeAnnotator and visualized with FigTree.
In a last step we obtained evidence for the origin of the Galapagos taxa by using statistical DIVA and Bayes-Lagrange analyses as implemented in RASP v.3.0 [54]. We used the trees generated by our BEAST run as input and defined the geographic areas as follows: A—N America, B—Africa (including Cape Verde), C—Europe (including the Canary Islands), D—Galapagos Islands, E—Caribbean, F—Asia, G—S America. The maximum areas per node were set as 2.
Supporting Information
We used the trees generated by our BEAST run as input and defined the geographic areas as follows: A—N America, B—Africa (including Cape Verde), C—Europe (including the Canary Islands), D—Galapagos Islands, E—Caribbean, F—Asia, G—S America. The maximum areas per node were set as 2. Values represent posterior probabilities.
(DOC)
We used published substitution rates of 0.0113 for ND5 (Husemann et al. 2012) and 0.01 for COI estimating the rate for H3 and applied a strict clock. The analysis was run for 100 million generations sampling every 10,000 generations. Trees were summarized with TreeAnnotator and visualized with FigTree. Numbers are divergence times in million years. The bars represent the 95% HPDs of age estimates.
(DOC)
Acknowledgments
We would like to thank two anonymous reviewers, David Lightfoot and the editor for valuable comments on the previous versions of the manuscript. We are grateful to D. Ferguson, D.E. Perez, A. Hilario, B. Hierro, R. Bastardo, V. Confalonieri, M.M. Cigliano, D. Salas, J. Pizzaro, R.D. Scott, M. Lecoq, S. Lötters, F. Pahlmann, W. Schuett, D.R. Swanson, A. Templeton, M. Hanitzsch, T. McNary, R. Bland, and Y. Görzig for providing samples for our study. Fieldwork on the Galapagos islands was facilitated by logistical support from the Charles Darwin Research Station, Isla Santa Cruz (G. Reck, D. Evans, C. Blanton, Directors). This work was supported by the German Research Foundation (DFG) and the Technische Universität München within the funding program Open Access Publishing.
Data Availability
All sequence files are available from the NCBI Genbank database (accession number(s) are provided in Table 1).
Funding Statement
The analyses were financed by a small grant of the Orthopterists’ Society to MH, and an URSA grant from Baylor University to PDD and SN. This work was supported by the German Research Foundation (DFG) and the Technische Universität München within the funding programme Open Access Publishing. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. MacArthur RH, Wilson EO (1967) The theory of island biogeography. Princeton: Princeton University Press. [Google Scholar]
- 2.Darwin C (1859) On the origin of species by means of natural selection, or the preservation of favoured races in the struggle for life. Nature 1st ed. [PMC free article] [PubMed]
- 3. Grant PR, Grant BR, Smith JN, Abbott IJ, Abbott LK (1976) Darwin’s finches: population variation and natural selection. Proc Nat Acad Sci 73: 257–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Grant PR, Grant BR (2011) How and why species multiply: the radiation of Darwin’s finches. Princeton: Princeton University Press. [Google Scholar]
- 5. Geist DJ, Snell H, Snell H, Goddard C, Kurz MD (2014) A paleogeographic model of the Galapagos Islands and biogeographical and evolutionary implications In: Harpp KS, Mittelstaedt E, d’Ozouville N, Graham DW, editors. The Galapagos: A natural laboratory fort he earth sciences. New Jersey: John Wiley & Sons. [Google Scholar]
- 6. Rassmann K (1997) Evolutionary age of the Galápagos Iguanas Predates the age of the present Galápagos islands. Mol Phyl Evol 7: 158–172. [DOI] [PubMed] [Google Scholar]
- 7. Parent CE, Caccone A, Petren K (2008) Colonization and diversification of Galápagos terrestrial fauna: a phylogenetic and biogeographical synthesis. Phil Trans Roy Soc B 363: 3347–3361. 10.1098/rstb.2008.0118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. White WM, McBirney AR, Duncan RA (1993) Petrology and geochemistry of the Galapagos Islands: portrait of a pathological mantle plume. J Geophys Res 98: 19533–19563. [Google Scholar]
- 9. Sequira AS, Lanterià AA, Scataglini MA, Confalonieri VA, Farell BD (2000) Are flightless Galapaganus weevils older than the Galápagos Islands they inhabit? Heredity 85: 20–29. [DOI] [PubMed] [Google Scholar]
- 10. Arbogast BS, Drovetski SV, Curry RL, Boag PT, Seutin G et al. (2006) The origin and diversification of Galápagos Mockingbirds. Evolution 60: 370–382. [PubMed] [Google Scholar]
- 11. Bollmer JL, Kimball RT, Whiteman NK, Sarasola JH, Parker PG (2006) Phylogeography of the Galapagos Hawk (Buteo galapagoensis): A recent arrival to the Galapagos Islands. Mol Phyl Evol 39: 237–247. [DOI] [PubMed] [Google Scholar]
- 12. Parent CE, Crespi BJ (2006) Sequential colonization and diversification of Galapagos endemic land snail Genus Bulimulus (Gastropoda, Stylommatophora). Evolution 60: 2311–2328. [PubMed] [Google Scholar]
- 13. Poulakakis N, Russello M, Geist D, Caccone A (2012) Unravelling the peculiarities of island life: vicariance, dispersal and the diversification of the extinct and extant giant Galapagos turtles. Mol Ecol 21: 160–173. 10.1111/j.1365-294X.2011.05370.x [DOI] [PubMed] [Google Scholar]
- 14. Kavar T, Pavlovcic P, Susnik S, Meglic V, Virant-Doverlet M (2006) Genetic differentiation of geographically separate populations of the southern green stink bug Nezara viridula (Hemiptera: Pentatomidae). Bull Entomol Res 96: 117–128. [DOI] [PubMed] [Google Scholar]
- 15. Pavlovcic P, Kavar T, Meglic V, Virant-Doberlet M (2008) Genetic population structure and range colonization of Nezara viridula . B Insectol 61: 191–192. [Google Scholar]
- 16. Otte D (1984) Acrididae: Oedipodinae. North American Grasshoppers. Cambridge: Harvard University Press. [Google Scholar]
- 17.Eades DC, Otte D, Cigliano MM, Braun H (2014) http://orthoptera.speciesfile.org, accessed March 2014.
- 18. Husemann M, Llucia-Pomares D, Hochkirch A (2013) A review of the Iberian Sphingonotini with description of two novel species (Orthoptera: Acrididae: Oedipodinae). Zool J Linn Soc 168: 29–60. [Google Scholar]
- 19. Husemann M, Guzman NV, Danley PD, Cigliano MM, Confalonieri VA (2013) Biogeography of Trimerotropis pallidipennis (Acrididae: Oedipodinae): deep divergence across the Americas. J Biogeogr 40: 261–273. [Google Scholar]
- 20. Hochkirch A, Husemann M (2008) A review of the Canarian Sphingonotini with description of a new species from Fuerteventura (Orthoptera: Acrididae: Oedipodinae). Zool Stud 47: 495–506. [Google Scholar]
- 21. Peck S (1996) Diversity and distribution of the orthopteroid insects of the Galápagos Islands, Ecuador. Can J Zool 74: 1497–1510. [Google Scholar]
- 22. Husemann M, Deppermann J, Hochkirch A (2014) Multiple independent colonization of the Canary Islands by the winged grasshopper genus Sphingonotus Fieber, 1852. Mol Phyl Evol 81: 174–181. 10.1016/j.ympev.2014.09.017 [DOI] [PubMed] [Google Scholar]
- 23. Husemann M, Namkung S, Habel JC, Danley PD, Hochkirch A (2012) Phylogenetic analyses of band-winged grasshoppers (Orthoptera, Acrididae, Oedipodinae) reveal convergence of wing morphology. Zool Scripta 41: 515–526. [Google Scholar]
- 24. Mishchenko L (1936) Revision of palaearctic species of the genus Sphingonotus Fieber (Orth. Acrid.). Eos, Revista española de Entomología (Eos) 12: 65–282. [Google Scholar]
- 25. Hollier J (2012) An annotated list of the Orthoptera (Insecta) species described by Henri de Saussure, with an account of the primary type material housed in the Museum d’histoire naturelle de Geneve, Part 2: The Acrididae: Oedipodinae. Rev Suisse Zool 119: 215–260. [Google Scholar]
- 26. Dirsh VM (1969) Acridoidea of the Galapagos Islands (Orthoptera). Bull Brit Mus Entomol 23: 25–51. [Google Scholar]
- 27. Buzzetti FM, Carotti G (2008) Annotated list of the Caelifera of Ecuador (Insecta: Orthoptera). Biodiversity of South America, I, Memoirs on Biodiversity, 1 2008: 39–66. [Google Scholar]
- 28. Snodgrass RE (1902) Papers from the Hopkins Stanford Galapagos expedition, 1898–1899. viii. Entomological Results (7) Schistocerca, Sphingonotus and Halmenus . Proc Wash Acad Sci 4: 411–454. [Google Scholar]
- 29. Hebard M (1920) Expedition of the California Academy of Sciences to the Galapagos Islands, 1905–106. Proc Cal Acad Sci 2: 311–346. [Google Scholar]
- 30. White MJD (1948). A cytological survey of wild populations of Trimerotropis and Circotettix. (Orthoptera, Acrididae). I. The chromosomes of twelve species. Genetics 34: 537–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. White MJD (1950). A cytological survey of wild populations of Trimerotropis and Circotettix. (Orthoptera, Acrididae). II. Racial differentiation in T. sparsa . Genetics 36: 31–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. White MJD (1973). Animal cytology and evolution. 3rd edition Cambridge University Press. [Google Scholar]
- 33. Juan C, Emerson BC, Oromí P, Hewitt GM (2000) Colonization and diversification: towards a phylogeographic synthesis for the Canary Islands. Trends Ecol Evol 15: 104–109. [DOI] [PubMed] [Google Scholar]
- 34. Chapco W, Contreras D (2011) Subfamilies Acridinae, Gomphocerinae and Oedipodinae are “Fussy Sets”: A proposal for a common African origin. J Orthop Res 20: 173–190. [Google Scholar]
- 35. de Queiroz A (2004) The resurrection of oceanic dispersal in historical biogeography. Trends Ecol Evol 20: 68–73. [DOI] [PubMed] [Google Scholar]
- 36. Lovejoy NR, Mullen SP, Sword GA, Chapman RF, Harrison RG (2006) Ancient trans-Atlantic flight explains locust biogeography: molecular phylogenetics of Schistocerca . Proc Roy Soc B 273: 767–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Clayton JW, Soltis PS, Soltis DE (2009) Recent long-distance dispersal overshadows ancient biogeographical patterns in a pantropical angiosperm family (Simaroubaceae: Sapindales). Syst Biol 58: 395–410. 10.1093/sysbio/syp041 [DOI] [PubMed] [Google Scholar]
- 38. Michalak I, Zhang L-B, Renner SS (2010) Trans-Atlantic, trans-Pacific and trans-Indian Ocean dispersal in the small Gondwanan Laurales family Hernandiaceae. J Biogeogr 37: 1214–1226. 10.3899/jrheum.090988 [DOI] [PubMed] [Google Scholar]
- 39. Carranza S, Arnold EN, Mateo JA, López-Jurado LF (2000) Long-distance colonization and radiation in gekkonid lizards, Tarentola (Reptilia: Gekkonidae), revealed by mitochondrial DNA sequences. Proc Roy Soc B 267: 637–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Carranza S, Arnold EN (2003) Investigating the origin of transoceanic distributions: mtDNA shows Mabuya lizards (Reptilia, Scincidae) crossed the Atlantic twice. Syst Biodiv 1: 275–282. [Google Scholar]
- 41. Finston TL, Peck S (1995) Population structure and gene flow in Stomion: a species swarm of flightless beetles of the Galápagos Islands. Heredity 75: 390–397. [Google Scholar]
- 42. Gentile G, Fabiani A, Marquez C, Snell HL, Snell HM, et al. (2009) An overlooked pink species of land iguana in the Galapagos. Proc Nat Acad Sci 106: 507–511. 10.1073/pnas.0806339106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jordan MA, Snell HL (2008) Historical fragmentation of islands and genetic drift in populations of Galápagos lava lizards (Microlophus albemarlensis complex). Mol Ecol 17: 1224–1237. 10.1111/j.1365-294X.2007.03658.x [DOI] [PubMed] [Google Scholar]
- 44. Blatchley WS (1920) Orthoptera of Northeastern America. Indianapolis: The Nature Publishing Company. [Google Scholar]
- 45. Su Z-H, Tominaga O, Okamoto M, Osawa S (1998) Origin and diversification of hind-wingless Damaster ground beetles within the Japanese Islands as deduced from mitochondrial ND5 gene sequences (Coleoptera, Carabidae). Mol Biol Evol 15: 1025–1039. [DOI] [PubMed] [Google Scholar]
- 46. Colgan DJ, McLauchlan A, Wilson GDF, Livingston SP, Edgecombe GD, et al. (1998) Histone H3 and U2 snRNA DNA sequences and arthropod molecular evolution. Aust J Zool 46: 419–438. [Google Scholar]
- 47. DeAngelis MM, Wang DG, Hawkins TL (1995) Solid-phase reversible immobilization for the isolation of PCR products. Nuc Acids Res 23: 4742–4743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Drummond AJ, Ashton B, Buxton S, Cheung M, Cooper A et al. (2011) Geneious v5.4. Available: http://www.geneious.com/.
- 49. Lanfear R, Calcott B, Ho SYW, Guindon S (2012) PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol Biol Evol 29: 1695–1701. 10.1093/molbev/mss020 [DOI] [PubMed] [Google Scholar]
- 50. Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- 51. Drummond AJ, Suchard MA, Xie D, Rambaut A (2012) Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol 29: 1969–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rambaut A, Drummond AJ (2009) Tracer v1.5. Available: http://beast.bio.ed.ac.uk/Tracer.
- 53.Rambaut A (2010) FigTree 1.3.1. 2010 Available: http://tree.bio.ed.ac.uk/software/figtree/
- 54. Yu Y, Harris AJ, He XJ (2010). S-DIVA (statistical dispersal-vicariance analysis): a tool for inferring biogeographic histories. Mol Phyl Evol 56: 848–850. 10.1016/j.ympev.2010.04.011 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
We used the trees generated by our BEAST run as input and defined the geographic areas as follows: A—N America, B—Africa (including Cape Verde), C—Europe (including the Canary Islands), D—Galapagos Islands, E—Caribbean, F—Asia, G—S America. The maximum areas per node were set as 2. Values represent posterior probabilities.
(DOC)
We used published substitution rates of 0.0113 for ND5 (Husemann et al. 2012) and 0.01 for COI estimating the rate for H3 and applied a strict clock. The analysis was run for 100 million generations sampling every 10,000 generations. Trees were summarized with TreeAnnotator and visualized with FigTree. Numbers are divergence times in million years. The bars represent the 95% HPDs of age estimates.
(DOC)
Data Availability Statement
All sequence files are available from the NCBI Genbank database (accession number(s) are provided in Table 1).