Abstract
Animal study findings have revealed that individual fat depots are not functionally equivalent and have different embryonic origins depending on the anatomic location. Mouse bone regeneration studies have also shown that it is essential to match the Hox code of transplanted cells and host tissues to achieve correct repair. However, subcutaneous fat depots from any donor site are often used in autologous fat grafting. Our study was thus carried out to determine the embryonic origins of human facial (chin) and limb (knee) fat depots and whether they had similar features and molecular matching patterns. Paired chin and knee fat depots were harvested from 11 subjects and gene expression profiles were determined by DNA microarray analyses. Adipose-derived stromal cells (ASCs) from both sites were isolated and analyzed for their capacity to proliferate, form clones, and differentiate. Chin and knee fat depots expressed a different HOX code and could have different embryonic origins. ASCs displayed a different phenotype, with chin-ASCs having the potential to differentiate into brown-like adipocytes, whereas knee-ASCs differentiated into white adipocytes. These results highlighted different features for these two fat sites and indicated that donor site selection might be an important factor to be considered when applying adipose tissue in cell-based therapies.
1. Introduction
Autologous fat grafting remains the gold standard therapy for soft tissue defects, correction or, augmentation in reconstructive and plastic surgery. Autologous fat grafting is particularly useful after tumor removal, for breast reconstruction surgery after mastectomy, to repair extensive facial deformities caused by injury, illness, or congenital abnormalities. With some variations, the technique consists of three stages: fat harvesting from any donor site, processing of the aspirate, and reimplantation at a site that differs from the donor site [1]. Enriching fat grafts with adipose-derived-stromal cells (ASCs) dramatically enhances fat graft survival [2]. A commercial device for ASC isolation and reinjection preparation has recently received FDA approval, illustrating the increased use of adipose tissues and ASCs for a broad range of cell-based therapies [3]. The treatment's main advantages are that autologous fat is easy to obtain in large quantities in various parts of the body with minimum morbidity for patients. However, there are still controversies and unresolved questions regarding autologous fat grafting due to the unpredictability of postoperative outcomes. The main disadvantages of this technique are variable engraftment and resorption rates, microcalcification, and cyst formation due to fat necrosis [4]. The differentiation of ASCs toward an undesirable cell type after grafting cannot be ruled out. Indeed, clinical results are unpredictable, without any clue about the reasons underlying the reconstruction success or failure. Greater cellular and molecular knowledge regarding adipose tissues is essential to improve postoperative outcomes.
Two functionally different types of adipose tissue—brown and white—coexist in mammals. Both are involved in the energy balance but have opposite functions. White adipose tissue is mainly involved in energy storage and mobilization in the form of triglycerides. In contrast, brown adipose tissue burns fat and is specialized in energy expenditure. In this key thermogenic organ, brown adipocytes convert nutrients into heat by uncoupling respiration from ATP synthesis. This process is mediated by the brown adipocyte-specific uncoupling protein (UCP)1 and is stimulated through a PKA-dependent pathway [5]. Human brown adipose tissue represents a minor fraction of adipose tissue, is scattered throughout the body, and disappears from most areas with age, persisting only around deeper organs [6]. White adipose tissue is more abundant, dispersed throughout the body, and functional differences between fat depots have been reported. Individual white adipose tissues are not equivalent, in terms of ASC abundance, proliferation, and differentiation [7, 8]. Recent published data, including ours, demonstrated different embryonic origins for adipocytes in rodents [9, 10]. Contrary to the previous belief that adipocytes derive from mesoderm only, we have shown a neuroectoderm origin for cephalic adipocytes [11]. It has been proposed that embryonic origins could play a critical role in differences observed between fat depots.
Despite the heterogeneity in fat depots, it is still considered that any adipose tissue site, such as abdomen, knee, and hip, could be used as a fat depot donor site for facial transplants. The donor site is usually selected by the surgeon based on the quantity of fat needed or on the patient's preference. The question regarding the best donor site for grafting to a heterotopic site remains unclear because no scientific studies have adequately addressed this issue.
Homeobox (Hox) genes encode transcription factors determining the positional identity along the anterior-posterior body axis of animal embryos [12], and recent studies revealed that they also have prominent roles in adult cells [13]. The Hox code, that is, the Hox gene expression profile, has been shown to play a critical role in stem cell positional identity. More importantly, this positional identity is retained after transplantation [13]. Leucht et al. illustrated the critical role of Hox code, showing that transplanting tibia-derived Hox-positive osteogenic stem cells into a Hox-negative site in mandible leads to aberrant bone regeneration [14]. The influence of the Hox code has also been highlighted in wound healing [15, 16]. Overall, these animal model studies have highlighted that a Hox code mismatch can prevent grafted stem cells from participating in tissue regeneration. The HOX code matching between donor and host fats remains to be determined.
Therefore, the cell sources of adipose tissues from different body sites that are used for transplantation at heterotopic sites may not be equivalent. Molecular characterization of tissues from donor and host fat sites that are used for autologous fat transplantation in reconstructive medicine is crucial. In the present study, we compared two human fat sites—limb and face—as a first step to investigate the therapeutic properties of fat according to its anatomical localization.
2. Materials and Methods
2.1. RNA Preparation
Chin and knee paired fat depots were collected from 11 Caucasians women (age 64 ± 13 years, BMI 21.9 ± 1.5 Kg/m2) who underwent elective liposuction procedures, after obtaining their informed consent. Fat tissues were lysed by the addition of TRIzol reagent (Invitrogen, Life Technologies Corporation, Carlsbad, CA, USA) and RNAs were purified on RNeasy Plus Universal columns (Qiagen, France), according to the manufacturer's instructions. Concentrations of total RNA samples were first evaluated using a Nanodrop spectrophotometer (Thermo Scientific, Waltham, MA, USA) and RNA samples were run in an RNA nanochip in a 2100 Bioanalyzer System (Agilent Technologies, Santa Clara, CA, USA) to verify the RNA sample integrity. All samples displayed good integrity with a measured RNA integrity number (RIN) > 6.
2.2. Whole-Genome Transcriptome Analysis
The Affymetrix analysis was performed with 11 paired samples of chin and knee fat tissues.
Affymetrix Probe Synthesis, Hybridization, Scanning, and Quality Controls. cRNA labelling was achieved starting from 300–500 ng of total RNA from samples. RNA was converted to double-stranded cDNA with a T7-(dT) primer. cDNA was in vitro transcribed to biotinylated complementary RNA (cRNA) by incorporating biotin-CTP and biotin UTP using the Affymetrix IVT labelling kit. 15 µg of biotin-labelled RNA were fragmented to 200 bp size by incubation in fragmentation buffer containing 200 mM Tris-acetate pH 8.2, 500 mM potassium acetate, and 500 mM magnesium acetate for 35 min at 94°C prior to hybridization. For the Affymetrix probe hybridization step, fragmented biotinylated RNA was assessed for relative length on an Agilent 2100 bioanalyzer and hybridized on Affymetrix U133 Plus 2.0 chips for 16 h at 45°C, washed and stained on an Affymetrix fluidic station, and scanned using an Affymetrix genechip scanner. Affymetrix RMA software was used to generate the cell files and to control the data quality prior to the data analysis. These controls included: back_spikes, Housekeeping genes, pm_mean, Mad_residual_mean, Rle_mean, and Box Plot. Validation of all chips was achieved using the principal component analysis (PCA) which is a mathematical procedure based on an orthogonal transformation to convert a set of observations of possibly correlated variables into a set of values of uncorrelated variables called principal components.
Normalization of the Affymetrix Data. All 22 chips were normalized with the Array Studio software package (OmicSoft Corporation, USA) using the three-step robust multichip average (RMA) method: background adjustment, quantile normalization, and finally summarization [17].
Only Affymetrix identifiers (IDs) with expression level ≥26 in at least 6 samples in at least one condition were selected for statistical analysis, resulting in data for 29131 out of 54675 IDs present on the HG_U133_Plus_2 chip.
2.3. Isolation of Adipose-Stromal Cells and Gene Expression Analysis by Real-Time PCR
Adipose-stromal cells (ASCs) were isolated from 9 paired chin-knee adipose tissues and expanded as previously described [18, 19]. Briefly, fat pads were dissociated with 200 mg/ml collagenase and the stromal vascular fraction (SVF) was separated from the adipocyte fraction by low speed centrifugation. Then, the SVF was seeded onto tissue culture plates and cells were maintained in low DMEM supplemented with 10% fetal bovine serum. After reaching 80% confluence, cells were dissociated, split, and expanded in the presence of 2.5 ng/ml FGF2. This passage was designated passage 1. RNAs were purified on RNesay columns (Qiagen, France). Real-time PCR assays were run on a StepOnePlus system (Applied Biosystems, Life Technologies, France). Transcript expression levels were evaluated using the comparative CT method (2-deltaCT). Delta Ct values were used when “expression relative to TBP” is indicated. TBP was used for sample normalization.
2.4. ASC Proliferation, Clonogenic and Differentiation Assays
Chin- and knee-ASCs were split every 4 days. The estimated population doubling time was 30 hours, and analyses were carried out between passage 2 and passage 4.
For proliferation tests, cells were plated on 24-well pates at 5000 cells/well in proliferation medium (DMEM medium supplemented with 10% FCS). Cell numbers were counted at different times after plating using a particle counter (Coulter Beckman).
For the fibroblastoid colony-forming unit assays, cells were plated at 1000-cell density in a 10 mm diameter culture dish in proliferation culture medium. Ten days later, cells were fixed with phosphate-buffer containing 4% formaldehyde and then stained with crystal violet 0.1% before counting.
For adipocyte differentiation, cells were plated on 24-well plates at 100 000 cells/well in proliferation medium. When cells reached confluence, culture medium was replaced by adipogenic differentiation medium composed of proliferation medium supplemented with dexamethasone (0.25 µM), isobutylmethylxanthine (500 µM), human recombinant insulin (0.34 µM), triiodothyronine (0.2 nM), and rosiglitazone (1 µM). The adipogenic differentiation medium was changed twice a week. After 10 days, cells were fixed with Oil red O [19] or lyzed for RNA preparation.
2.5. UCP1 Protein Expression in Differentiated ASCs
ASC-differentiated cells were harvested at day 16 after adipogenic induction. Proteins were extracted and 100 µg protein per condition was separated by 15% SDS-polyacrylamide gel electrophoresis. Proteins were then transferred to polyvinylidene difluoride membrane. The membrane was first incubated with an antibody against UCP1 (Calbiochem) and was then incubated with a secondary antibody rabbit IgG-conjugated horseradish peroxidase (Santa Cruz Biotechnology, Santa Cruz, CA, USA). The membranes were treated with the chemiluminescent HRP substrate (Millipore) according to the manufacturer's instructions. Tubulin was detected as a control with tubulin antibody (Santa Cruz Biotechnology, Santa Cruz, CA, USA).
2.6. Triglyceride Accumulation and Lipolysis Assays
ASCs were induced to undergo differentiation as described above. At day 16 after induction, cells were harvested and the intracellular triglyceride concentration was measured using the Triglyceride Colorimetric Assay Kit (Cayman Chemical) according to the manufacturer's instructions. For lipolysis assays, cells were treated with 10 µM forskolin for 4 h and the secreted glycerol concentration was measured in culture supernatant using the Free Glycerol Reagent Kit (Sigma-Aldrich) according to the manufacturer's instructions.
2.7. Statistics Analysis
Statistical analysis of the normalized Affymetrix data: for the knee versus chin comparison, a two-sided paired t-test was applied using Array Studio software. The Benjamini-Hochberg procedure was used for multiple testing corrections [20]. False discovery rate (FDR) control is a statistical method used in multiple hypotheses testing to correct for multiple comparisons. InStat3 software and a nonparametric unpaired test (Mann-Whitney) were used for statistical analysis of real-time PCR data. P values are indicated on each figure.
3. Results and Discussion
3.1. Large-Scale Affymetrix Transcriptomics Revealed Differential Expression of Pax3 and HOX Genes in Human Chin and Knee Fat Depots
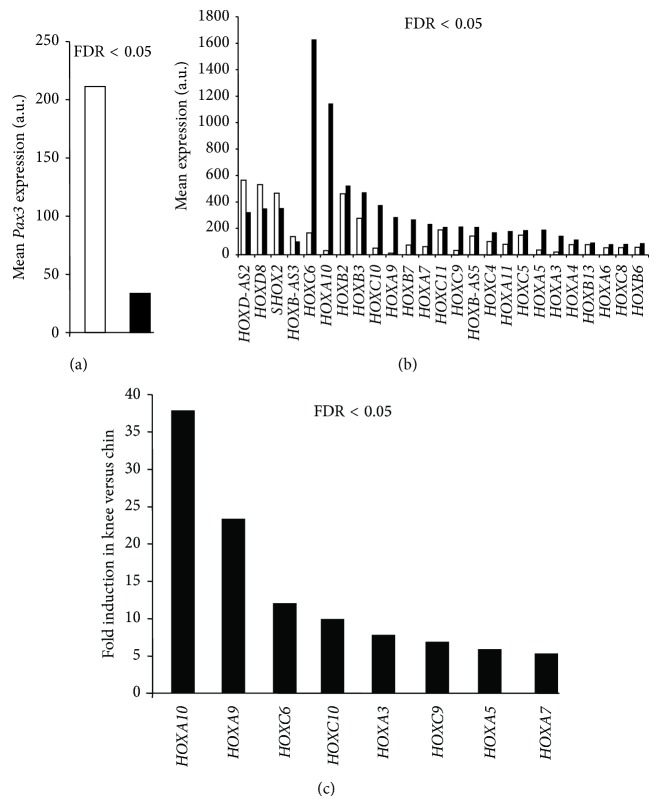
A large-scale transcriptomic analysis was performed using Affymetrix technology to compare the transcriptome from knee versus chin adipose tissues. A total of 346 genes (determined from the 486 modulated Ids) were found to be differentially expressed with a fold < 2 and a FDR value <0.05 (all differentially modulated ids available on demand). These 346 differentially expressed genes were equally distributed between upregulated and downregulated genes (181 up- and 165 downregulated genes). Among the TOP50 upregulated genes were mainly found homeobox genes and inflammatory-related genes. By contrast, most of the genes found in the TOP50 downregulated genes belonged to the “muscle function” (Table 1). Interestingly, microarray analysis revealed that Pax3 expression was enhanced in chin fat compared to knee fat (Figure 1(a)). Lineage tracing experiments in mice allow establishing a neuroectodermic/neural crest and mesodermic embryonic origins for adipose tissues located in the face and trunk, respectively. As lineage tracing is impossible in humans, only specific marker expression could be used to propose an embryonic origin of human samples. Pax3 expression was shown to be associated with neural crest development in humans and to be maintained in adult progenies [21]. Pax3 thus represents a unique neural crest marker in adult humans. Therefore, microarray analysis data strongly suggest that chin fat, but not knee fat, originates from the neural crest. This hypothesis is in agreement with what we observed in mice [11]. HOX gene expression profiles were then compared in paired adipose tissue samples. Thirty-eight HOX genes were detected on Affimetrix chips and 25 of them displayed significant differential expression between both sites (Figure 1(b)). Among those, 4 HOX genes (HOXD-AS2, HOXD8, SHOX2, and HOXB-AS3) were 1.4- to 1.8-fold more expressed in chin fat than in knee fat. In contrast, 21 HOX genes were preferentially expressed in knee fat depots, with the 8 HOX genes, HOXA10, HOXA9, HOXC6, HOXC10, HOXA3, HOXC9, HOXA5, and HOXA7 being the most differentially expressed (Figure 1(c)). The whole genome transcriptomic analysis therefore revealed that chin and knee fat depots expressed a different HOX code. This HOX status in human chin and knee fats resembles the Hox code identified in mouse mandibular and tibial osteogenic progenitors [14]. It should be noted that the Hox code mismatch was responsible for aberrant tibial progenitor differentiation when transplanted into the face.
Table 1.
TOP50 of the down- and upregulated genes between knee and chin adipose tissue depots from the large scale transcriptomic experiment.
| Gene symbol | Gene title | Knee versus chin FDR | Fold change |
|---|---|---|---|
| ACTA1 | Actin, alpha 1, skeletal muscle | 0,0006 | −69,62 |
| MYH2 | Myosin, heavy chain 2, skeletal muscle, adult | 0,0004 | −65,45 |
| KBTBD10 | Kelch repeat and BTB (POZ) domain containing 10 | 0,0016 | −62,65 |
| MYL1 | Myosin, light chain 1, alkali; skeletal, fast | 0,0026 | −31,4 |
| CKM | Creatine kinase, muscle | 0,0026 | −29,47 |
| MYBPC1 | Myosin binding protein C, slow type | 0,0027 | −28,89 |
| MB | Myoglobin | 0,0025 | −25,84 |
| MYL2 | Myosin, light chain 2, regulatory, cardiac, slow | 0,0029 | −24,41 |
| SLN | Sarcolipin | 0,0052 | −22,39 |
| CSRP3 | Cysteine and glycine-rich protein 3 (cardiac LIM protein) | 0,0034 | −18,46 |
| NEB | Nebulin | 0,0037 | −17,84 |
| MYOT | Myotilin | 0,0046 | −16,77 |
| TTN | Titin | 0,0069 | −16,52 |
| SMPX | Small muscle protein, X-linked | 0,0094 | −14,54 |
| COL6A6 | Collagen, type VI, alpha 6 | 0,0004 | −14,47 |
| KBTBD10 | Kelch repeat and BTB (POZ) domain containing 10 | 0,0093 | −14,24 |
| TNNC2 | Troponin C type 2 (fast) | 0,0043 | −13,86 |
| TNNI2 | Troponin I type 2 (skeletal, fast) | 0,0076 | −13,15 |
| TNNC1 | Troponin C type 1 (slow) | 0,0053 | −12,35 |
| TNNT1 | Troponin T type 1 (skeletal, slow) | 0,0051 | −12,08 |
| XIRP2 | Xin actin-binding repeat containing 2 | 0,0086 | −11,91 |
| TPM1 | Tropomyosin 1 (alpha) | 0,0092 | −11,73 |
| ENO3 | Enolase 3 (beta, muscle) | 0,0059 | −11,71 |
| PDLIM5 | PDZ and LIM domain 5 | 0,0085 | −10,49 |
| ASB5 | Ankyrin repeat and SOCS box-containing 5 | 0,0142 | −10,38 |
| MYLPF | Myosin light chain, phosphorylatable, fast skeletal muscle | 0,0092 | −9,51 |
| CADM2 | Cell adhesion molecule 2 | 5,51E − 05 | −9,22 |
| MYH6 | Myosin, heavy chain 6, cardiac muscle, alpha///myosin | 0,0105 | −8,65 |
| CA3 | Carbonic anhydrase III, muscle specific | 0,0006 | −8,27 |
| MYBPC2 | Myosin binding protein C, fast type | 0,0097 | −8,06 |
| MYOZ1 | Myozenin 1 | 0,012 | −7,9 |
| ACTN2 | Actinin, alpha 2 | 0,0155 | −7,86 |
| TRDN | Triadin | 0,0075 | −7,71 |
| CADM2 | Cell adhesion molecule 2 | 0,0003 | −6,45 |
| CYR61 | Cysteine-rich, angiogenic inducer, 61 | 0,0002 | −6,29 |
| PAX3 | Paired box 3 | 0,0001 | −6,26 |
| TTN | Titin | 0,0317 | −6,11 |
| CADM2 | Cell adhesion molecule 2 | 0,0003 | −5,98 |
| AMPD1 | Adenosine monophosphate deaminase 1 | 0,0243 | −5,97 |
| LDB3 | LIM domain binding 3 | 0,0143 | −5,96 |
| LMOD2 | Leiomodin 2 (cardiac) | 0,0218 | −5,94 |
| TPM3 | Tropomyosin 3 | 0,0139 | −5,9 |
| APOBEC2 | Apolipoprotein B mRNA editing enzyme | 0,0126 | −5,8 |
| C8orf22 | Chromosome 8 open reading frame 22 | 0,008 | −5,79 |
| RBM24 | RNA binding motif protein 24 | 0,0006 | −5,74 |
| TTN | Titin | 0,0171 | −5,62 |
| PDLIM5 | PDZ and LIM domain 5 | 0,0325 | −5,57 |
| CYR61 | Cysteine-rich, angiogenic inducer, 61 | 0,0002 | −5,45 |
| CMYA5 | Cardiomyopathy associated 5 | 0,0152 | −5,31 |
| PKIA | Protein kinase (cAMP-dependent, catalytic) inhibitor alpha | 0,0038 | −5,31 |
| PCDH7 | Protocadherin 7 | 0,0008 | −5,23 |
| HSPB3 | Heat shock 27kDa protein 3 | 0,0235 | −5,19 |
| PPBP | Proplatelet basic protein (chemokine (C-X-C motif) ligand 7) | 0,0153 | −5,14 |
| MYH1 | Myosin, heavy chain 1, skeletal muscle, adult | 0,0276 | −5,11 |
| NRAP | Nebulin-related anchoring protein | 0,0129 | −5,08 |
| POPDC3 | Popeye domain containing 3 | 0,0033 | −5,03 |
| MYOZ2 | Myozenin 2 | 0,0209 | −4,99 |
| HOXA10 | Homeobox A10 | 1,40E − 09 | 37,88 |
| HOXA9 | Homeobox A9 | 9,16E − 08 | 23,43 |
| HOXA9 | Homeobox A9 | 8,09E − 07 | 13,38 |
| HOXC6 | Homeobox C6 | 1,93E − 07 | 9,92 |
| TNC | Tenascin C | 0,0005 | 8,26 |
| HOXC10 | Homeobox C10 | 3,47E − 06 | 7,8 |
| IGH@///IGHG1/// | Immunoglobulin heavy locus | 0,0021 | 7,59 |
| IGJ | Immunoglobulin J polypeptide | 0,0083 | 7,41 |
| HOXA10 | Homeobox A10 | 3,47E − 06 | 7,37 |
| CHI3L1 | Chitinase 3-like 1 (cartilage glycoprotein-39) | 0,0008 | 7,09 |
| SAA1///SAA2 | Serum amyloid A1///serum amyloid A2 | 0,0004 | 7,04 |
| HOXA3 | Homeobox A3 | 5,60E − 05 | 6,88 |
| HOXC9 | Homeobox C9 | 1,32E − 06 | 5,87 |
| CYAT1///IGLV1-44 | Cyclosporin A transporter 1/ | 0,0076 | 5,83 |
| ASPN | Asporin | 1,32E − 06 | 5,76 |
| HOXA5 | Homeobox A5 | 9,27E − 05 | 5,28 |
| ASPN | Asporin | 4,94E − 06 | 5,2 |
| CHI3L1 | Chitinase 3-like 1 (cartilage glycoprotein-39) | 0,0021 | 4,82 |
| SAA1///SAA2 | Serum amyloid A1///serum amyloid A2 | 0,0011 | 4,75 |
| ATP8B4 | ATPase, class I, type 8B, member 4 | 2,99E − 06 | 4,61 |
| IGL@ | Immunoglobulin lambda locus | 0,0058 | 4,6 |
| CSN1S1 | Casein alpha s1 | 0,0143 | 4,04 |
| CYAT1///IGLV1-44 | Cyclosporin A transporter 1 | 0,008 | 3,96 |
| PRG4 | Proteoglycan 4 | 0,001 | 3,95 |
| IGH@///IGHA1 | Immunoglobulin heavy locus | 0,0172 | 3,75 |
| HOXA7 | Homeobox A7 | 5,44E − 05 | 3,69 |
| HOXB7 | Homeobox B7 | 2,43E − 05 | 3,63 |
| IGLV1-44 | Immunoglobulin lambda variable 1-44 | 0,013 | 3,6 |
| ALDH1A3 | Aldehyde dehydrogenase 1 family, member A3 | 0,0002 | 3,54 |
| FGFBP2 | Fibroblast growth factor binding protein 2 | 0,0013 | 3,54 |
| ZIC1 | Zic family member 1 (odd-paired homolog, Drosophila) | 0,0054 | 3,5 |
| CP | Ceruloplasmin (ferroxidase) | 0,0003 | 3,43 |
| IGK@///IGKC | Immunoglobulin kappa locus t | 0,0132 | 3,39 |
| IGK@///IGKC | Immunoglobulin kappa locus///immunoglobulin kappa constant | 0,012 | 3,37 |
| ISM1 | Isthmin 1 homolog (zebrafish) | 0,0006 | 3,36 |
| PHLDA2 | Pleckstrin homology-like domain, family A, member 2 | 0,0011 | 3,34 |
| NPY1R | Neuropeptide Y receptor Y1 | 0,0017 | 3,31 |
| EVI2A | Ecotropic viral integration site 2A | 0,0014 | 3,29 |
| HOXB7 | Homeobox B7 | 2,86E − 05 | 3,29 |
| FCER1A | Fc fragment of IgE, high affinity I, receptor for alpha polypeptide | 0,0012 | 3,28 |
| MSR1 | Macrophage scavenger receptor 1 | 0,0002 | 3,23 |
| CNTN4 | Contactin 4 | 0,0021 | 3,23 |
| IGKC | Immunoglobulin kappa constant | 0,01 | 3,21 |
| PLA2G7 | Phospholipase A2, group VII (platelet-activating factor acetylhydrolase, plasma) | 0,0076 | 3,21 |
| MS4A7 | Membrane-spanning 4 domains, subfamily A, member 7 | 0,0004 | 3,19 |
| RNASE6 | Ribonuclease, RNase A family, k6 | 0,0003 | 3,18 |
| IGK@///IGKC | Immunoglobulin kappa locus///immunoglobulin kappa constant | 0,0184 | 3,18 |
| CRISPLD1 | Cysteine-rich secretory protein LCCL domain containing 1 | 0,0009 | 3,17 |
| CP | Ceruloplasmin (ferroxidase) | 0,0004 | 3,17 |
| CHI3L2 | Chitinase 3-like 2 | 0,0033 | 3,12 |
| CLEC7A | C-type lectin domain family 7, member A | 0,0205 | 3,1 |
| LYZ | Lysozyme | 0,0123 | 3,07 |
| BGN | Biglycan | 0,0005 | 3,06 |
| LYZ | Lysozyme | 0,0083 | 2,94 |
| MARCH1 | Membrane-associated ring finger (C3HC4) 1 | 0,0002 | 2,92 |
| IGK@///IGKC///LOC100291464 | Immunoglobulin kappa locus///immunoglobulin kappa constant///similar to hCG26659 | 0,0172 | 2,91 |
| SAMSN1 | SAM domain, SH3 domain, and nuclear localization signals 1 | 0,0013 | 2,91 |
| ANKDD1A | Ankyrin repeat and death domain containing 1A | 0,0006 | 2,88 |
Figure 1.
Pax3 and HOX gene expression in paired chin and knee fat depots from large-scale Affymetrix transcriptomics. (a) Mean expression of Pax3 in chin (open bar) and in knee (black bar) fat depots (n = 11 paired). (b) Mean expression of HOX genes in chin (open bars) and knee (black bars) (n = 11 paired). (c) HOX genes with the highest differential expression are presented.
3.2. Differential Pax3 and HOX Gene Expression Was Conserved in Adipose-Stromal Cells Derived from Chin and Knee Fat Depots
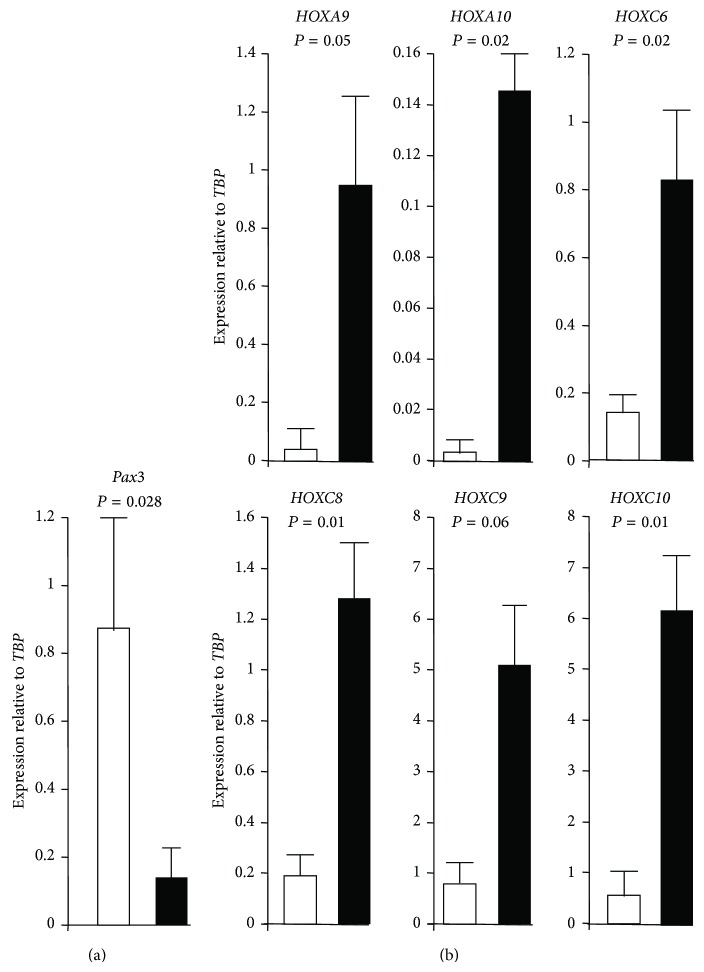
As adipose tissues contain several cell types, such as adipose-stromal cells (ASCs), adipocytes, endothelial cells, and immune cells, we wondered whether chin and knee molecular signatures revealed by microarray data were conserved in ASCs and derived adipocytes. For that purpose ASCs were isolated from the stromal vascular fraction of paired chin and knee fat samples. The skeletal muscle signature of chin fat depot was not detectable neither in undifferentiated ASCs nor in their differentiated progenies (not shown), indicating that other cell types in chin fat biopsies could be responsible for muscle gene expression. In contrast, Pax3 and HOX differential expression was conserved in ASCs. Pax3 expression was analysed by real-time PCR and its gene expression was normalized according to TATA box binding protein (TBP) gene expression, that is, a housekeeping gene used as reference gene. As shown in Figure 2(a), Pax3 was expressed significantly higher in chin ASCs than in knee ASCs (6.2-fold), supporting the neuroectoderm origin of chin-ASCs.
Figure 2.
Expression of Pax3 and HOX genes in adipose-stromal cells derived from chin and knee fat: ASCs were isolated from chin (open bars) and knee (black bars) paired fats and were expanded ex vivo. (a) Pax3 expression was quantified by real-time PCR and normalized with TBP housekeeping gene. Values are means ± SEM. P is indicated (n = 4 paired depots). (b) Expression of indicated HOX genes was quantified by real-time PCR and normalized with TBP housekeeping gene. Values are means ± SEM. P are indicated (n = 3 paired depots).
It has been previously reported that fat from different depots displays distinct developmental gene expression profiles, including Hox genes [10, 22, 23], but limb and facial fat were not included in these analyses. We performed real-time PCR to monitor the expression of 6 HOX genes that displayed the highest differential expression between knee and chin according to the microarray data. As shown in Figure 2(b), HOX genes were expressed at different levels. More importantly, HOX gene expression was significantly enhanced in knee ASCs compared to chin ASCs (more than 20-fold for HOXA9 and HOXA10 and between 5- and 10-fold for HOXC6, HOXC8, HOXC, and HOXC10). These data are in line with the HOX expression revealed by the Affimetrix analysis from whole fat depots (compare Figures 1(c) and 2(b)), indicating that the HOX code of the whole fat depot reflected the HOX code of ASCs. The HOX code thus distinguished ASCs from facial and limb depots. Overall, these results strongly suggest that chin and knee fats could have different functional features.
3.3. Properties of Chin- and Knee-ASCs and Adipocyte Progenies
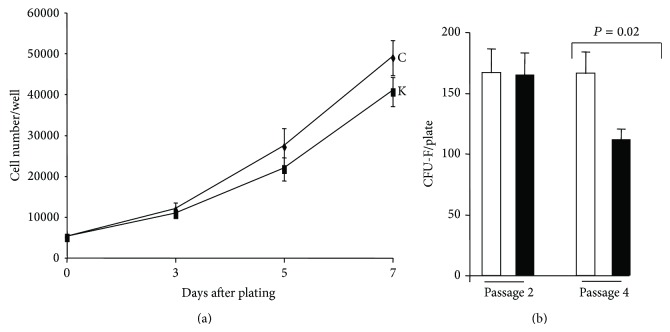
ASCs and their progenies were characterized in order to determine if there are functional differences between chin and knee fats. We hence compared ASC proliferation, fibroblastoid colony-forming units (CFU-F), and differentiation. Cells were plated at low cell density and allowed to proliferate for 7 days. As shown in Figure 3, the proliferation curve of both ASCs was similar. In contrast, a significant difference in CFU-F potential was observed after cell expansion. Indeed, at passage 2, the CFU-F frequency was identical for both ASCs (16%, which is in the same range as previously reported for other human fat depots [3]). However, after 2 more passages, the CFU-F potential decreased for knee ASCs, whereas it remained unchanged for chin ASCs (Figure 3(b)). These results indicated that both fat depots contained ASCs that could be expanded but that chin ASCs had a higher self-renewal potential, at least ex vivo.
Figure 3.
Proliferation and clonogenic potentials of chin and knee ASCs. (a) ASCs isolated from chin (C) and knee (K) were plated at 5000 cells/well in 24-well plates and cell number was quantified at the indicated times. Values are means ± SEM. (n = 8 paired depots). (b) Chin-ASCs (open bars) and knee-ASCs (black bars) were plated at a clonal cell density and maintained in proliferative medium. The numbers of colonies were assessed after 10 days of culture. Values are means ± SEM. P is indicated. Experiments were performed at cell passage 2 (n = 7 paired depots) and at passage 4 (n = 6 paired depots).
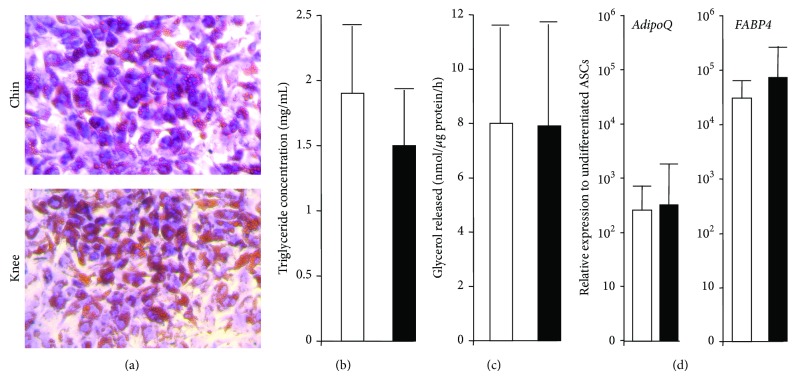
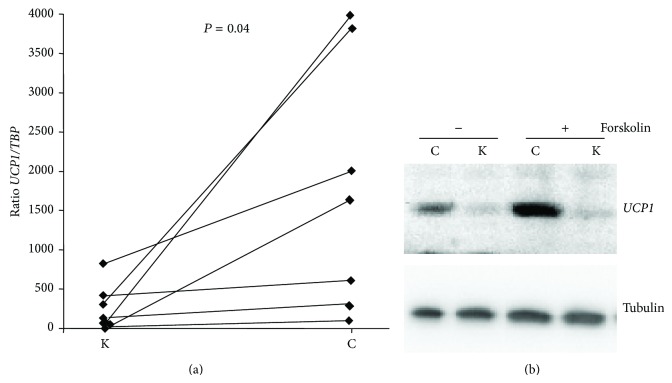
Adipocytes generated from ASCs of both depots were then characterized. Adipocytes of both sites accumulated intracellular lipid droplets to a similar extent after 10 days of differentiation, as qualitatively determined by Oil red O staining (Figure 4(a)). Quantitative tests then revealed that adipocytes derived from chin and knee ASCs displayed a similar intracellular triglyceride concentration and lipolysis activity (Figures 4(b) and 4(c)). Expression of adipocyte-specific genes, such as Adiponectin (AdipoQ) and FABP4, was similar for adipocytes generated from both ASCs, supporting at the molecular level that chin- and knee-ASCs have a similar adipogenic capacity (Figure 4(d)). However, a major difference appeared regarding the adipocyte phenotype. As shown in Figure 5(a), the brown adipocyte marker UCP1 was highly enriched in chin adipocytes compared to paired knee adipocytes. UCP1 differential expression was confirmed at the protein level (Figure 5(b)). Interestingly, UCP1 expression increased in chin adipocytes upon PKA pathway activation via forskolin treatment (Figure 5(b)), as expected for functional brown adipocytes. Overall, our data indicated that chin host site contained brown or brown-like adipocytes, whereas the knee donor site contained white adipocytes. The presence of functional brown and brown-like adipocytes in human neck has been previously reported [24], and we have observed UCP1 gene expression in human cheek (not shown). The human face thus contains cells having the potential to enable the dissipation of excessive caloric intake. The tissue regeneration and facial rejuvenation consequences of transplanting white adipocytes in a fat environment with different metabolic properties remain to be investigated.
Figure 4.
Adipocyte differentiation of chin- and knee-ASCs. ASCs were induced to undergo differentiation for 10 days. (a) Cells were stained with Oil Red O for lipid droplets. (b) Cells were harvested and analyzed for their triglyceride content. (c) Cells were stimulated with forskolin for 4 h and then glycerol release was determined. (d) RNAs were prepared and differentiation levels were assessed by the expression of the indicated genes. Expression of each gene was normalized to TBP and then relative to the expression in undifferentiated cells. Values are means ± SEM. P is indicated (n = 7 paired depots). Open bars: chin; black bars: knee.
Figure 5.
UCP1 expression in adipocytes generated from chin ASCs. ASCs derived from chin (C) and knee (K) were induced to undergo differentiation for 10 days. (a) RNAs were prepared and UCP1 expression was determined by real-time PCR. n = 7 paired depots. Corresponding data points of knee and chin paired depots are connected with lines. (b) Cells were stimulated (+) or not (−) with forskolin for 4 h and then proteins were prepared. Expression of UCP1 was assessed by Western-blotting. Tubulin was used as loading control.
4. Conclusions
Our results strongly suggest that ASCs present in knee and chin fats derived from different embryonic origins. They did not express the same HOX code and displayed different adipocyte phenotypes. The consequences of these differences on autologous fat grafting outcomes remain to be investigated in detail. It should be noted that mesenchymal stem cells from different embryonic origins have been found to display different tissue regeneration capacities [25] and that Hox code matching was confirmed to be critical for successful tissue regeneration in mouse [14]. Our data highlighted that donor site selection might be an important factor to be considered for fat grafting. Matching Pax3 and/or HOX code and/or UCP1 expression between host and donor sites could improve postoperative outcomes.
Acknowledgments
The authors would like to thank Sophie Déret for her expertise in BioInformatics for Affymetrix data analysis and the “stem cells and differentiation” team for helpful discussions. This work was supported by the French Government (National Research Agency, ANR) through the “Investments for the Future” LABEX SIGNALIFE: Program reference no. ANR-11-LABX-0028-01 and the National Research Agency against HIV (ANRS, Grant AO 2013-1 CSS3).
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
Authors' Contribution
Bérengère Chignon-Sicard and Christian Dani contributed equally to this work.
References
- 1.Coleman S. R. Long-term survival of fat transplants: controlled demonstrations. Aesthetic Plastic Surgery. 1995;19(5):421–425. doi: 10.1007/bf00453875. [DOI] [PubMed] [Google Scholar]
- 2.Kølle S.-F. T., Fischer-Nielsen A., Mathiasen A. B., et al. Enrichment of autologous fat grafts with ex-vivo expanded adipose tissue-derived stem cells for graft survival: a randomised placebo-controlled trial. The Lancet. 2013;382(9898):1113–1120. doi: 10.1016/s0140-6736(13)61410-5. [DOI] [PubMed] [Google Scholar]
- 3.Bourin P., Bunnell B. A., Casteilla L., et al. Stromal cells from the adipose tissue-derived stromal vascular fraction and culture expanded adipose tissue-derived stromal/stem cells: a joint statement of the International Federation for Adipose Therapeutics and Science (IFATS) and the International Society for Cellular Therapy (ISCT) Cytotherapy. 2013;15(6):641–648. doi: 10.1016/j.jcyt.2013.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yoshimura K., Sato K., Aoi N., et al. Cell-assisted lipotransfer for facial lipoatrophy: efficacy of clinical use of adipose-derived stem cells. Dermatologic Surgery. 2008;34(9):1178–1185. doi: 10.1111/j.1524-4725.2008.34256.x. [DOI] [PubMed] [Google Scholar]
- 5.Fredriksson J. M., Thonberg H., Ohlson K. B. E., Ohba K.-I., Cannon B., Nedergaard J. Analysis of inhibition by H89 of UCP1 gene expression and thermogenesis indicates protein kinase A mediation of β3-adrenergic signalling rather than β3-adrenoceptor antagonism by H89. Biochimica et Biophysica Acta—Molecular Cell Research. 2001;1538(2-3):206–217. doi: 10.1016/s0167-4889(01)00070-2. [DOI] [PubMed] [Google Scholar]
- 6.Enerbäck S. Human brown adipose tissue. Cell Metabolism. 2010;11(4):248–252. doi: 10.1016/j.cmet.2010.03.008. [DOI] [PubMed] [Google Scholar]
- 7.Tchkonia T., Giorgadze N., Pirtskhalava T., et al. Fat depot origin affects adipogenesis in primary cultured and cloned human preadipocytes. The American Journal of Physiology—Regulatory Integrative and Comparative Physiology. 2002;282(5):R1286–R1296. doi: 10.1152/ajpregu.00653.2001. [DOI] [PubMed] [Google Scholar]
- 8.Tchkonia T., Tchoukalova Y. D., Giorgadze N., et al. Abundance of two human preadipocyte subtypes with distinct capacities for replication, adipogenesis, and apoptosis varies among fat depots. American Journal of Physiology—Endocrinology and Metabolism. 2005;288(1):E267–E277. doi: 10.1152/ajpendo.00265.2004. [DOI] [PubMed] [Google Scholar]
- 9.Billon N., Dani C. Developmental origins of the adipocyte lineage: new insights from genetics and genomics studies. Stem Cell Reviews and Reports. 2012;8(1):55–66. doi: 10.1007/s12015-011-9242-x. [DOI] [PubMed] [Google Scholar]
- 10.Gesta S., Blüher M., Yamamoto Y., et al. Evidence for a role of developmental genes in the origin of obesity and body fat distribution. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(17):6676–6681. doi: 10.1073/pnas.0601752103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Billon N., Iannarelli P., Monteiro M. C., et al. The generation of adipocytes by the neural crest. Development. 2007;134(12):2283–2292. doi: 10.1242/dev.002642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krumlauf R. Hox genes in vertebrate development. Cell. 1994;78(2):191–201. doi: 10.1016/0092-8674(94)90290-9. [DOI] [PubMed] [Google Scholar]
- 13.Wang K. C., Helms J. A., Chang H. Y. Regeneration, repair and remembering identity: the three Rs of Hox gene expression. Trends in Cell Biology. 2009;19(6):268–275. doi: 10.1016/j.tcb.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leucht P., Kim J.-B., Amasha R., James A. W., Girod S., Helms J. A. Embryonic origin and Hox status determine progenitor cell fate during adult bone regeneration. Development. 2008;135(17):2845–2854. doi: 10.1242/dev.023788. [DOI] [PubMed] [Google Scholar]
- 15.Creuzet S., Couly G., Vincent C., le Douarin N. M. Negative effect of Hox gene expression on the development of the neural crest-derived facial skeleton. Development. 2002;129(18):4301–4313. doi: 10.1242/dev.129.18.4301. [DOI] [PubMed] [Google Scholar]
- 16.White P., Thomas D. W., Fong S., et al. Deletion of the homeobox gene PRX-2 affects fetal but not adult fibroblast wound healing responses. Journal of Investigative Dermatology. 2003;120(1):135–144. doi: 10.1046/j.1523-1747.2003.12015.x. [DOI] [PubMed] [Google Scholar]
- 17.Bolstad B. M., Irizarry R. A., Åstrand M., Speed T. P. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19(2):185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 18.Rodriguez A.-M., Pisani D., Dechesne C. A., et al. Transplantation of a multipotent cell population from human adipose tissue induces dystrophin expression in the immunocompetent mdx mouse. The Journal of Experimental Medicine. 2005;201(9):1397–1405. doi: 10.1084/jem.20042224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wdziekonski B., Mohsen-Kanson T., Villageois P., Dani C. The generation and the manipulation of human multipotent adipose-derived stem cells. Methods in Molecular Biology. 2011;702:419–427. doi: 10.1007/978-1-61737-960-4_30. [DOI] [PubMed] [Google Scholar]
- 20.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B: Methodological. 1995;57(1):289–300. [Google Scholar]
- 21.Betters E., Liu Y., Kjaeldgaard A., Sundström E., García-Castro M. I. Analysis of early human neural crest development. Developmental Biology. 2010;344(2):578–592. doi: 10.1016/j.ydbio.2010.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tchkonia T., Lenburg M., Thomou T., et al. Identification of depot-specific human fat cell progenitors through distinct expression profiles and developmental gene patterns. American Journal of Physiology—Endocrinology and Metabolism. 2007;292(1):E298–E307. doi: 10.1152/ajpendo.00202.2006. [DOI] [PubMed] [Google Scholar]
- 23.Cantile M., Procino A., D'Armiento M., Cindolo L., Cillo C. HOX gene network is involved in the transcriptional regulation of in vivo human adipogenesis. Journal of Cellular Physiology. 2003;194(2):225–236. doi: 10.1002/jcp.10210. [DOI] [PubMed] [Google Scholar]
- 24.Cypess A. M., White A. P., Vernochet C., et al. Anatomical localization, gene expression profiling and functional characterization of adult human neck brown fat. Nature Medicine. 2013;19(5):635–639. doi: 10.1038/nm.3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Uccelli A., Moretta L., Pistoia V. Mesenchymal stem cells in health and disease. Nature Reviews Immunology. 2008;8(9):726–736. doi: 10.1038/nri2395. [DOI] [PubMed] [Google Scholar]