Abstract
Objective. The aim of this study was to investigate the prognostic value of MicroRNA-210 (miR-210) expression in patients with non-small-cell lung cancer (NSCLC). Methods. We examined the miR-210 expression of samples of 80 patients, who underwent surgical resection at Fukushima Medical University from 2004 to 2007, by using quantitative RT-PCR. The relationship between miR-210 expression and clinicopathological factors as well as histological subtype was statistically analyzed. Results. miR-210 expression showed an inverse correlation with disease-free and overall survival in patients with NSCLC. Significant correlations were found between miR-210 expression and lymph node metastasis, late disease stages, and poor prognosis in patients with adenocarcinoma. Multivariate Cox analysis indicated that miR-210 expression was an independent prognostic factor for disease-free survival in patients with adenocarcinoma. Conclusions. We showed that miR-210 may be a prognostic biomarker for patients with NSCLC, especially for those with lung adenocarcinoma.
1. Introduction
Hypoxia is a common feature of pathological conditions such as tissue ischemia and inflammation, as well as of the microenvironment of solid tumors [1]. Many cellular responses to hypoxia are thought to be mediated through changes in targeted gene expression. One major mechanism mediating cellular responses to hypoxia is the pathway of hypoxia inducible factor-1 (HIF-1) [2]. HIF-1 is a member of the basic helix-loop-helix/Per-Arnt-Sim (bHLH-PAS) family of proteins and binds to hypoxia-response elements (HRE) in the promoters of target genes. HIF-1 consists of an alpha (HIF-1α) and a beta (HIF-1β) subunit and activates the expression of at least 150 genes, which encode proteins that regulate cell metabolism, cell cycle, proliferation, apoptosis, autophagy, erythropoiesis, immune reactions, cytokine production, and angiogenesis as well as many other functions [3]. HIF-1β is a non-oxygen-responsive nuclear protein. In contrast, HIF-1α is highly inducible by hypoxia [4].
In human cancers, HIF-1α is overexpressed as a result of intratumoral hypoxia and of genetic alterations affecting crucial oncogenes and tumor suppressor genes [3]. HIF-1α overexpression has been associated with increased patient mortality in many different human cancers [3]. Similarly, HIF-1α overexpression has been reported at both the protein [5, 6] and the mRNA [7, 8] level in non-small-cell lung cancer (NSCLC) patients with poor prognosis. In preclinical studies, inhibition of HIF-1α activity has marked effects on tumor growth; inhibitors of HIF-1α have therefore attracted much attention as new therapeutic agents for patients with advanced malignancies, and several clinical studies have been performed [3]. Research has shown that HIF-1α antagonists, such as EZN-2968 and PX-478, inhibit tumor cell proliferation in vitro and in vivo [9, 10].
miRNAs have emerged as a new class of noncoding genes that are involved in the regulation of cell proliferation, differentiation, and viability [11]. miRNAs are single-stranded small RNA molecules of approximately 22 nucleotides that silence the expression of target genes either through mRNA degradation or suppression of transcription [12–14]. The miRNAs that are regulated by hypoxia were examined in a 2007 study in which miR-210 was identified as the most consistently and robustly induced miRNA in hypoxic cells and tissues [15]. miR-210 expression is frequently elevated in a variety of cancers [15], including lung cancer [16–20]. miR-210 is regulated by both HIF-1α [21–23] and HIF-2α [24], and a recent study demonstrated that HIF-1α directly binds to an HRE on the proximal miR-210 promoter [23]. miR-210 plays numerous crucial roles in the cellular response to hypoxia, such as in apoptosis [15, 25], angiogenesis [26], cell cycle regulation [24, 27], DNA damage repair [22], mitochondrial metabolism [28, 29], and tumor growth [19]. Furthermore, miR-210 is also involved in stem cell biology [30]. Thus, miR-210 is thought to have essential roles in tumorigenesis along with HIF-1α.
It has been reported that miR-210 overexpression is correlated with poor prognosis in breast [21, 31], pancreatic [32], and head and neck cancer patients [31]. Recently, two systematic reviews and a meta-analysis confirmed that miR-210 is useful for prediction of the survival of patients with various tumors, especially breast cancers [33, 34]. However, these two studies did not include the outcome of patients with lung cancer. Therefore, the prognostic impact of miR-210 in patients with lung cancer remains unclear. Within this context, we analyzed miR-210 expression in NSCLC patient samples, and showed that it could be a prognostic biomarker, especially for patients with adenocarcinoma.
2. Materials and Methods
2.1. Patient and Tissue Samples
In total, 80 snap-frozen NSCLC and 30 matched normal adjacent lung tissue samples were evaluated for miR-210 expression. These consecutive samples were obtained from patients who underwent surgical resection at the Department of Regenerative Surgery, Fukushima Medical University, Fukushima, Japan, from January 2004 to December 2007. The clinical characteristics of the 80 patients included in this study were typical of the characteristics of resected NSCLC reported by the Japan Lung Cancer Society (2004) with respect to age, sex, histology, and pathological stage [35]. None of the patients had received any previous anticancer treatment. Ethical approval for analysis of samples and patient notes was obtained from the local research ethics committee. Tumor types and stages were determined according to the 7th edition of Union for International Cancer Control TNM classification. At the time of surgery, all tissue samples were immediately frozen in liquid nitrogen and stored at −80°C until assay. All samples were analyzed histologically to assess the amount of tumor component (at least 70% tumor cells) and the quality of the material (i.e., absence of necrosis). These 80 cases consisted of 34 female and 46 male patients with a median age of 69 years (range: 51–85), of which 54 were stage I cases, 12 were stage II, and 14 were stage III. The median observation period was 74.5 months (range: 5–117), and the five-year survival rate was 62.4%. Full patient clinicopathological characteristics are provided in Table 1.
Table 1.
Relationship between miR-210 expression and clinicopathological factors.
| Clinicopathological factor | Patient number | miR-210 miRNA expression | P | |
|---|---|---|---|---|
| High group | Low group | |||
| Gender | 0.258 | |||
| Male | 46 | 26 | 20 | |
| Female | 34 | 14 | 20 | |
| Age | 0.222 | |||
| ≤65 | 24 | 9 | 15 | |
| ≥66 | 56 | 31 | 25 | |
| Tumor size | 0.82 | |||
| <30 mm | 48 | 23 | 25 | |
| ≥30 mm | 32 | 17 | 15 | |
| T factor | 0.481 | |||
| T1-T2 | 61 | 34 | 37 | |
| T3-T4 | 9 | 6 | 3 | |
| Lymph node | 0.293 | |||
| Negative | 61 | 28 | 33 | |
| Positive | 19 | 12 | 7 | |
| P-stage | 0.232 | |||
| Stage I | 54 | 24 | 30 | |
| Stages II-III | 26 | 16 | 10 | |
| ly factor | 0.502 | |||
| Negative | 41 | 19 | 22 | |
| Positive | 39 | 21 | 17 | |
| v factor | 0.359 | |||
| Negative | 43 | 22 | 27 | |
| Positive | 27 | 18 | 13 | |
| Relapse | 0.007* | |||
| With | 37 | 25 | 12 | |
| Without | 43 | 15 | 28 | |
Relapse was defined as first evidence of radiographic metastatic disease after surgery.
* P < 0.05.
2.2. RNA Extraction and Quantitative Real-Time PCR
TaqMan-based quantitative real-time polymerase chain reaction (qRT-PCR) was applied to assess miRNA expression levels in tissue samples. RNA was extracted from snap-frozen lung tumor samples or normal lung tissue using the miRVana miRNA Isolation Kit (Ambion, Austin TX, USA). The quantity and quality of the extracted RNA were determined spectrophotometrically by measurement of absorbance at 260 and 280 nm using a DU530 UV-VIS spectrophotometer (Beckman Coulter, Fullerton, CA, USA). Samples with a 260/280 ratio of 1.80 or greater were used for analysis. miRNA-cDNA was synthesized from 5 ng of total RNA using microRNA-specific primers and the TaqMan MicroRNA Reverse Transcription kit (Applied Biosystems, Foster City, CA, USA). The TaqMan MicroRNA assays for miR-210 (ID: 000512) and RNU6B (ID: 001093) were purchased from Applied Biosystems. Real-time PCR was performed in triplicate using a StepOnePlus Real-Time PCR system (Applied Biosystems). For miRNA assays, each PCR reaction contained 1.33 μL reverse transcription product, 2×TaqMan Universal Master Mix, and 1 μL TaqMan MicroRNA assay. The 20 μL reactions were incubated in a 96-well optical plate at 95°C for 10 minutes, followed by 40 cycles at 95°C for 15 seconds, and 60°C for 60 seconds. Changes in miRNA expression between treatment and controls were determined using the 2−ΔΔCt method [36], and results were normalized against RNU6B expression levels. For lung cancer samples, the controls consisted of the median of 30 normal lung tissues.
2.3. Statistical Analysis
Correlations between the status of miR-210 expression and clinical characteristics were assessed using Student's t-test, Pearson and Spearman's rank test, or the Mann-Whitney U test. Kaplan-Meier survival analysis was performed by applying the long-rank test to miR-210 expression and was stratified by median values and quartiles. Disease-specific overall survival (referred to as overall survival hereafter) was defined as the time from surgery to last follow-up or time of NSCLC-specific death. Disease-free survival was defined as the time from surgery to the time of first evidence of radiographic metastatic disease. Univariate and multivariate analyses were performed using the Cox proportional hazard model. All statistical analyses were performed using SPSS 17 software (SPSS Inc., Chicago, IL, USA), and P values of <0.05 were considered significant.
3. Results
3.1. Relationship between miR-210 Expression and Clinical Characteristics in Patients with NSCLC
To examine whether miR-210 expression correlates with clinical characteristics in patients with NSCLC, we analyzed miR-210 expression of NSCLC samples using qRT-PCR. For each sample, the data were normalized using RNU6B as a reference. The fold-change in miR-210 expression for each NSCLC sample was calculated by comparison with the median of 30 normal control samples. Patients were divided into two subgroups according to high or low miR-210 expression levels. Patients with higher than the median expression level of miR-210 were defined as the high group of miR-210 expression. No significant association between the status of miR-210 expression and clinical characteristics such as sex, age, tumor size, histology, T factor, lymph node status, pathological stage, ly factor, or v factor was observed. However, we found that the status of miR-210 expression was significantly correlated with disease relapse (P = 0.007, Pearson and Spearman's rank test; Table 1).
3.2. miR-210 Expression Is a Prognostic Factor for Disease-Free Survival and Overall Survival of NSCLC Patients
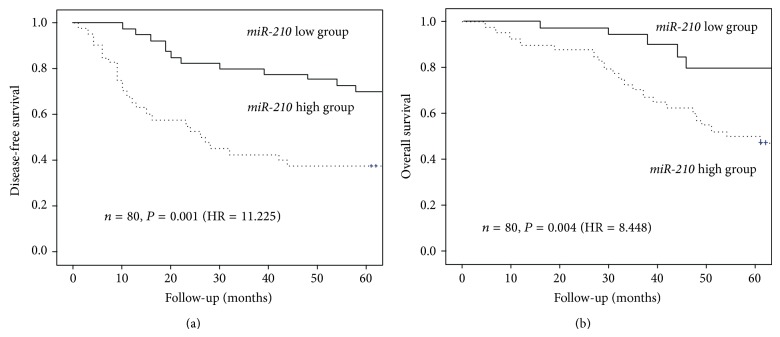
To confirm the correlation between miR-210 and prognosis, we analyzed the relationship between miR-210 expression and patient survival by performing Kaplan-Meier survival analysis, applying the long-rank test to miR-210 expression. The miR-210-high group showed significantly shorter disease-free survival than the miR-210-low group (log rank chi-square = 11.225, P = 0.001; Figure 1(a)). Five-year disease-free survival was 37.5% in the miR-210-high group and 70.0% in the miR-210-low group. Furthermore, the miR-210-high group showed significantly shorter overall survival than the miR-210-low group. Five-year overall survival was 47.5% in the miR-210-high group and 77.5% in the miR-210-low group (log rank chi-square = 8.448, P = 0.004; Figure 1(b)).
Figure 1.
Kaplan-Meier curves of survival of patients with NSCLC (n = 80). Kaplan-Meier curves of (a) disease-free and (b) overall survival of 80 patients with NSCLC, stratified according to miR-210 levels. Expression levels were stratified by the median value; follow-up was limited to 60 months.
3.3. miR-210 Expression Is Significantly Associated with Important Clinicopathological Factors in Patients with Lung Adenocarcinoma
Although we found that miR-210 expression is a prognostic factor for disease-free survival and overall survival in NSCLC patient samples; the status of miR-210 expression was not significantly correlated with important clinicopathological factors. We therefore analyzed the correlation of each histological type with miR-210 expression and clinical characteristics. In 62 patient samples with adenocarcinoma, miR-210 expression was significantly correlated with lymph node status, pathological stage, ly factor, v factor, and disease relapse (P = 0.018, P = 0.003, P = 0.0009, P = 0.044, and P = 0.002, resp., Mann-Whitney U test), whereas, in 18 patient samples with squamous cell carcinoma, it was not significantly correlated with any clinical characteristic (Table 2).
Table 2.
Relationship between miR-210 expression and clinical characteristics.
| Clinical characteristics | miR-210 expression | |||
|---|---|---|---|---|
| Ad cases | P | Sq cases | P | |
| Gender | 0.086 | 1.000 | ||
| Male | 2.32 ± 2.89 | 2.88 ± 2.00 | ||
| Female | 1.45 ± 1.67 | 2.93 ± 0.88 | ||
| Age | 0.029* | — | ||
| ≤65 | 1.18 ± 1.71 | — | ||
| ≥66 | 2.17 ± 2.60 | 2.87 ± 1.89 | ||
| Tumor size | 0.402 | 1.000 | ||
| <30 mm | 1.84 ± 1.95 | 2.87 ± 1.62 | ||
| ≥30 mm | 1.87 ± 2.98 | 3.25 ± 2.14 | ||
| Histology | — | — | ||
| Adeno | 1.86 ± 2.40 | — | ||
| Squamous | — | 2.87 ± 1.89 | ||
| T factor | 0.417 | 1.000 | ||
| T1-T2 | 1.82 ± 2.48 | 2.88 ± 1.65 | ||
| T3-T4 | 2.34 ± 1.70 | 3.37 ± 4.40 | ||
| Lymph node | 0.034* | — | ||
| Negative | 1.44 ± 2.01 | 2.87 ± 1.89 | ||
| Positive | 2.34 ± 2.97 | — | ||
| P-stage | 0.019* | 1.000 | ||
| Stage I | 1.37 ± 2.02 | 2.88 ± 1.65 | ||
| Stages II-III | 2.62 ± 2.77 | 3.37 ± 4.40 | ||
| ly factor | 0.025* | 0.620 | ||
| Negative | 1.23 ± 1.44 | 3.30 ± 2.07 | ||
| Positive | 2.24 ± 2.84 | 2.09 ± 1.23 | ||
| v factor | 0.097 | 1.000 | ||
| Negative | 1.75 ± 1.97 | 2.12 ± 1.84 | ||
| Positive | 2.34 ± 2.98 | 3.01 ± 1.86 | ||
| Relapse | 0.007* | 1.000 | ||
| With | 2.72 ± 2.63 | 2.88 ± 1.96 | ||
| Without | 1.35 ± 1.75 | 2.87 ± 1.84 | ||
Ad: adenocarcinoma.
Sq: squamous cell carcinoma.
Relapse was defined as first evidence of radiographic metastatic disease after surgery.
* P < 0.05.
3.4. miR-210 Expression Is a Prognostic Factor for Disease-Free and Overall Survival in Patients with Lung Adenocarcinoma
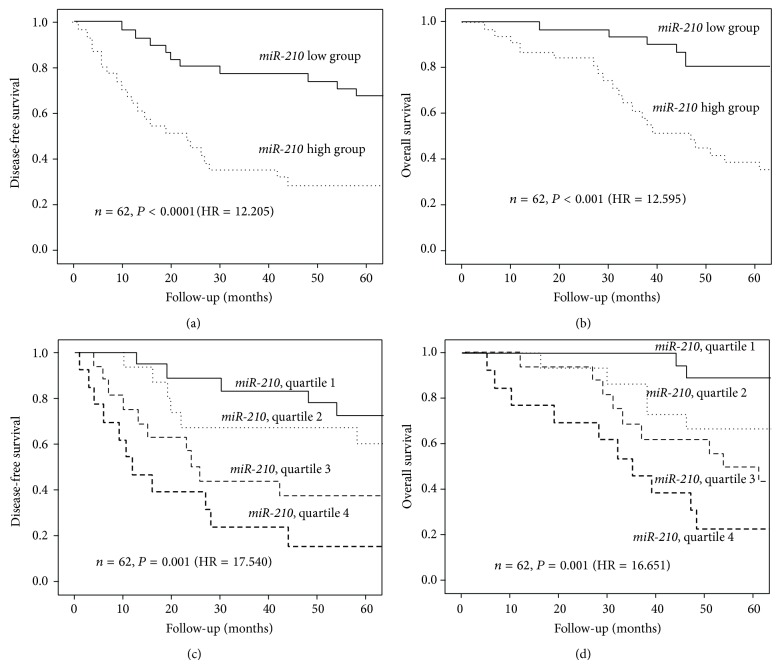
Because miR-210 expression in adenocarcinoma patient samples was correlated with many important clinicopathological factors, we hypothesized that miR-210 expression would be more closely correlated with prognosis in adenocarcinoma patient samples than in NSCLC patient samples. To test this hypothesis, Kaplan-Meier survival analysis was performed by applying the log-rank test to miR-210 expression of 62 patient samples with adenocarcinoma. miR-210 expression was a strong adverse prognostic factor for disease-free and overall survival when considered as a binary variable divided by median value (log rank chi-square = 12.205, P < 0.001; Figure 2(a), log rank chi-square = 12.595, P < 0.001; Figure 2(b), resp.). However, in the 18 patient samples with squamous cell carcinoma, no significant correlation between miR-210 expression and disease-free or overall survival was observed. In the 62 lung adenocarcinoma patient samples, these relationships were also statistically significant when the patients were divided into quartiles on the basis of miR-210 expression levels; miR-210 expression was an adverse prognostic factor for disease-free and overall survival (log rank chi-square = 17.540, P < 0.001; Figure 2(c), log rank chi-square = 16.651, P = 0.001; Figure 2(d), resp.).
Figure 2.
Kaplan-Meier survival curves for patients with adenocarcinoma (n = 62). Kaplan-Meier curves were constructed of the following: (a) disease-free and (b) overall survival of 62 patients with adenocarcinoma stratified according to miR-210 levels. Expression levels were stratified by the median value; follow-up was limited to 60 months. (c) Disease-free and (d) overall survival of patients with adenocarcinoma stratified according to miR-210 levels. Expression levels were stratified by quartiles; follow-up was limited to 60 months.
3.5. Multivariate Analysis Indicated That miR-210 Expression in Adenocarcinoma Patient Samples Is an Independent Prognostic Factor for Disease-Free Survival
We performed further studies of diagnostic factors of disease-free survival and overall survival, specifically in adenocarcinoma patients, using univariate and multivariate analysis. These data are summarized in Table 3. In a Cox univariate analysis, sex, tumor size, T factor, lymph node metastases, p-stage, ly factor, v factor, and miR-210 expression were significantly correlated with disease-free survival (P = 0.024, P = 0.034, P = 0.036, P < 0.0001, P < 0.0001, P = 0.001, P = 0.004, and P = 0.001, resp.), whereas only age was not correlated with disease-free survival. Furthermore, a Cox multivariate analysis defined sex, ly factor, and miR-210 expression as independent prognostic factors for disease-free survival (P = 0.008, P = 0.021, and P = 0.020, resp.). Moreover, in a Cox univariate analysis, sex, tumor size, T factor, lymph node metastases, p-stage, ly factor, v factor, and miR-210 expression were also significantly correlated with overall survival (P = 0.005, P = 0.044, P = 0.018, P = 0.001, P < 0.0001, P = 0.004, P = 0.032, and P = 0.001, resp.). Furthermore, Cox multivariate analysis showed that the usefulness of miR-210 as a prognostic factor for overall survival was marginal (P = 0.057).
Table 3.
Disease-free (a) and overall survival (b) of 62 adenocarcinoma patients according to clinical characteristics and miR-210 expression, analyzed by Cox proportional regression.
(a) Disease-free survival
| Variables | Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|---|
| HR (95% CI) | P | HR (95% CI) | P | ||
| Gender | Male/female | 2.263 (1.113–4.601) | 0.024* | 3.600 (1.392–9.309) | 0.008* |
| Age | ≥66/≤65 | 0.860 (0.427–1.730) | 0.672 | ||
| Tumor size | ≥33/<33 | 2.130 (1.052–4.735) | 0.034* | 2.310 (0.819–6.518) | 0.114 |
| T factor | T1/T2–T4 | 2.232 (1.052–4.735) | 0.036* | 0.469 (0.134–1.645) | 0.237 |
| Lymph node | +/− | 3.754 (1.857–7.590) | <0.0001* | 1.038 (0.300–3.589) | 0.953 |
| P-stage | I/II-III | 4.969 (2.396–10.304) | <0.0001* | 3.270 (0.894–11.964) | 0.073 |
| ly factor | +/− | 4.078 (1.822–9.128) | 0.001* | 4.027 (1.229–13.193) | 0.021* |
| v factor | +/− | 2.768 (1.376–5.566) | 0.004* | 0.994 (0.412–2.398) | 0.989 |
| miR-210 | High/Low | 0.284 (0.134–0.604) | 0.001* | 0.355 (0.148–0.847) | 0.02* |
(b) Overall survival
| Variables | Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|---|
| HR (95% CI) | P | HR (95% CI) | P | ||
| Gender | Male/female | 3.218 (1.434–7.222) | 0.005* | 2.151 (1.889–14.047) | 0.001* |
| Age | ≥66/≤65 | 1.011 (0.473–2.162) | 0.977 | ||
| Tumor size | ≥33/<33 | 2.173 (1.019–4.630) | 0.044* | 1.725 (0.618–4.811) | 0.298 |
| T factor | T1/T2–T4 | 2.988 (1.204–7.415) | 0.018* | 0.801 (0.211–3.039) | 0.744 |
| Lymph node | +/− | 3.453 (1.614–7.391) | 0.001* | 1.858 (0.499–6.918) | 0.356 |
| P-stage | I/II-III | 4.533 (2.027–10.138) | <0.0001* | 1.492 (0.380–5.853) | 0.567 |
| ly factor | +/− | 3.865 (1.554–9.615) | 0.004* | 3.387 (0.894–12.836) | 0.073 |
| v factor | +/− | 2.303 (1.075–4.937) | 0.032* | 0.602 (0.226–1.607) | 0.311 |
| miR-210 | High/low | 0.235 (0.099–0.561) | 0.001* | 0.361 (0.127–1.031) | 0.057 |
CI: confidence interval.
* P < 0.05.
4. Discussion
By univariate and multivariate analyses our study provides clear evidence that upregulation of miR-210 is a prognostic factor in patients with lung adenocarcinoma and is correlated with important clinicopathological factors including nodal involvement, pathological stage, lymphatic vessel invasion, and cancer relapse.
Recently, a meta-analysis of human lung cancer microRNA expression profiling studies that compared cancer tissues with normal tissues showed that the top two most consistently reported upregulated microRNAs were miR-210 and miR-21 [37]. In addition, systematic reviews and meta-analyses of two studies confirmed that upregulation of miR-210 is predictive of poor survival of patients with various tumors, especially breast cancers [33, 34]. However, these two systematic reviews did not include the outcome of patients with lung cancer. Thus, the prognostic impact of miR-210 in patients with lung cancer remained unclear.
Recently, Eilertsen et al. reported a large-scale study of the prognostic role of miR-210 in NSCLC [38]. In that study, upregulation of miR-210 expression was a positive prognostic factor for disease-free survival in 335 NSCLC patients. This result is not consistent with our findings. One reason for the differences between these results might be the different methods used, since our study assessed miR-210 expression using qRT-PCR, whereas the previous study used in situ hybridization. However, other previous studies strongly suggest that high expression of miR-210 could be a biomarker of bad prognosis in lung cancer. Puisségur et al. reported that miR-210 was significantly elevated in patients with advanced disease such as stages II-III disease compared with stage I A disease (n = 20) [39]. Li et al. reported that miR-210 was significantly elevated in patients with stages III-IV disease compared with stages I-II disease (n = 60) [40]. Furthermore, in the majority of the previous studies, miR-210 upregulation was significantly correlated with poor prognosis in patients with various cancers such as breast cancer [21], pancreatic cancer [32], head and neck cancer [31], colorectal cancer [41], and glioblastoma [42], though not renal cancer [43]. Our data are consistent with these previous important findings. Further study is still needed to clarify the clinical impact of miR-210 as a prognostic factor in patients with NSCLC.
In vitro functional studies regarding miR-210 function in cancer progression provide even further contradictory results. For example, Zhang et al. found that miR-210 inhibits MNT, an antagonist of c-MYC, and promotes cell proliferation in transformed cells such as colon and cervical cancer cells [24]. However, Giannakakis et al. found that miR-210 acts as a tumor suppressor by inhibiting cell proliferation via E2F3 regulation in ovarian cancer cell lines [27]. It is unclear how to reconcile these paradoxical findings that miR-210 acts primarily as a positive or a negative regulator of proliferation.
To understand these conflicting findings, we hypothesized that miR-210 may play various roles depending on the cancer type or histological subtype in which it is expressed. In the present study, we first analyzed the correlation of miR-210 expression in NSCLC patient samples with each histological subtype of NSCLC and with the clinical characteristics of patients with each subtype. We then focused on miR-210 expression in samples with histology specific for adenocarcinoma. In patients with adenocarcinoma, we clearly showed that miR-210 expression was strongly associated with important clinical parameters such as age, lymph node metastasis, pathological stage, ly factor, v factor, and relapse, while, in patients with squamous cell carcinoma, miR-210 was not associated with any clinical characteristic. In our study, squamous cell carcinoma showed high levels of baseline miR-210 expression compared with adenocarcinoma. The uniformly high expression levels of miR-210 in most squamous cell carcinomas meant that a prognostic impact of miR-210 on squamous cell carcinoma could not be determined. However, miR-210 could be a biomarker of adenocarcinoma because adenocarcinomas showed varying levels of miR-210 expression.
5. Conclusions
In conclusion, this study demonstrated for the first time that miR-210 was correlated with poor prognosis in patients with NSCLC, especially in lung adenocarcinoma. This evidence could contribute to biomarker studies in patients with lung adenocarcinoma. Because this was an exploratory study and the sample size was very small, our results warrant further investigation and require independent validation. In particular, miR-210 levels in the plasma may have special prognostic significance since miRNA has been shown to circulate in various body fluids in remarkably stable forms, resulting in the identification of novel noninvasive biomarkers for the diagnosis and prognosis of various cancers and other diseases [44, 45]. Based on these evidences, the prognostic signature of miR-210 in the plasma of NSCLC patients should be examined in future studies.
Acknowledgments
This study was supported by the Department of Regenerative Surgery, Fukushima Medical University. The authors gratefully thank Yuka Kimura, Kaoru Ogoshi, Yukiko Kikuta, and Hidemi I for excellent technical assistant and data acquisition and analysis.
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
References
- 1.Keith B., Simon M. C. Hypoxia-inducible factors, stem cells, and cancer. Cell. 2007;129(3):465–472. doi: 10.1016/j.cell.2007.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Semenza G. L., Wang G. L. A nuclear factor induced by hypoxia via de novo protein synthesis binds to the human erythropoietin gene enhancer at a site required for transcriptional activation. Molecular and Cellular Biology. 1992;12(12):5447–5454. doi: 10.1128/mcb.12.12.5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Semenza G. L. Targeting HIF-1 for cancer therapy. Nature Reviews Cancer. 2003;3(10):721–732. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- 4.Weidemann A., Johnson R. S. Biology of HIF-1α . Cell Death and Differentiation. 2008;15(4):621–627. doi: 10.1038/cdd.2008.12. [DOI] [PubMed] [Google Scholar]
- 5.Giatromanolaki A., Koukourakis M. I., Sivridis E., et al. Relation of hypoxia inducible factor 1α and 2α in operable non-small cell lung cancer to angiogenic/molecular profile of tumours and survival. British Journal of Cancer. 2001;85(6):881–890. doi: 10.1054/bjoc.2001.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Swinson D. E. B., Jones J. L., Cox G., Richardson D., Harris A. L., O'Byrne K. J. Hypoxia-inducible factor-1α in non small cell lung cancer: relation to growth factor, protease and apoptosis pathways. International Journal of Cancer. 2004;111(1):43–50. doi: 10.1002/ijc.20052. [DOI] [PubMed] [Google Scholar]
- 7.Lau S. K., Boutros P. C., Pintilie M., et al. Three-gene prognostic classifier for early-stage non-small-cell lung cancer. Journal of Clinical Oncology. 2007;25(35):5562–5569. doi: 10.1200/JCO.2007.12.0352. [DOI] [PubMed] [Google Scholar]
- 8.Yohena T., Yoshino I., Takenaka T., et al. Upregulation of hypoxia-inducible factor-1αmRNA and its clinical significance in non-small cell lung cancer. Journal of Thoracic Oncology. 2009;4(3):284–290. doi: 10.1097/JTO.0b013e31819852d5. [DOI] [PubMed] [Google Scholar]
- 9.Greenberger L. M., Horak I. D., Filpula D., et al. A RNA antagonist of hypoxia-inducible factor-1α, EZN-2968, inhibits tumor cell growth. Molecular Cancer Therapeutics. 2008;7(11):3598–3608. doi: 10.1158/1535-7163.mct-08-0510. [DOI] [PubMed] [Google Scholar]
- 10.Jacoby J. J., Erez B., Korshunova M. V., et al. Treatment with hif-1α antagonist PX-478 inhibits progression and spread of orthotopic human small cell lung cancer and lung adenocarcinoma in mice. Journal of Thoracic Oncology. 2010;5(7):940–949. doi: 10.1097/JTO.0b013e3181dc211f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bartel D. P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 12.Lagos-Quintana M., Rauhut R., Lendeckel W., Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294(5543):853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 13.Lau N. C., Lim L. P., Weinstein E. G., Bartel D. P. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans . Science. 2001;294(5543):858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 14.Lee R. C., Ambros V. An extensive class of small RNAs in Caenorhabditis elegans . Science. 2001;294(5543):862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 15.Kulshreshtha R., Ferracin M., Wojcik S. E., et al. A microRNA signature of hypoxia. Molecular and Cellular Biology. 2007;27(5):1859–1867. doi: 10.1128/MCB.01395-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cho W. C. S., Chow A. S. C., Au J. S. K. Restoration of tumour suppressor hsa-miR-145 inhibits cancer cell growth in lung adenocarcinoma patients with epidermal growth factor receptor mutation. European Journal of Cancer. 2009;45(12):2197–2206. doi: 10.1016/j.ejca.2009.04.039. [DOI] [PubMed] [Google Scholar]
- 17.Miko E., Czimmerer Z., Cśanky E., et al. Differentially expressed micrornas in small cell lung cancer. Experimental Lung Research. 2009;35(8):646–664. doi: 10.3109/01902140902822312. [DOI] [PubMed] [Google Scholar]
- 18.Raponi M., Dossey L., Jatkoe T., et al. MicroRNA classifiers for predicting prognosis of squamous cell lung cancer. Cancer Research. 2009;69(14):5776–5783. doi: 10.1158/0008-5472.CAN-09-0587. [DOI] [PubMed] [Google Scholar]
- 19.Volinia S., Calin G. A., Liu C.-G., et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(7):2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yanaihara N., Caplen N., Bowman E., et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9(3):189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 21.Camps C., Buffa F. M., Colella S., et al. Hsa-miR-210 is induced by hypoxia and is an independent prognostic factor in breast cancer. Clinical Cancer Research. 2008;14(5):1340–1348. doi: 10.1158/1078-0432.CCR-07-1755. [DOI] [PubMed] [Google Scholar]
- 22.Crosby M. E., Kulshreshtha R., Ivan M., Glazer P. M. MicroRNA regulation of DNA repair gene expression in hypoxic stress. Cancer Research. 2009;69(3):1221–1229. doi: 10.1158/0008-5472.CAN-08-2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang X., Ding L., Bennewith K. L., et al. Hypoxia-inducible mir-210 regulates normoxic gene expression involved in tumor initiation. Molecular Cell. 2009;35(6):856–867. doi: 10.1016/j.molcel.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Z., Sun H., Dai H., et al. MicroRNA miR-210 modulates cellular response to hypoxia through the MYC antagonist MNT. Cell Cycle. 2009;8(17):2756–2768. doi: 10.4161/cc.8.17.9387. [DOI] [PubMed] [Google Scholar]
- 25.Cheng A. M., Byrom M. W., Shelton J., Ford L. P. Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Research. 2005;33(4):1290–1297. doi: 10.1093/nar/gki200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fasanaro P., D'Alessandra Y., Di Stefano V., et al. MicroRNA-210 modulates endothelial cell response to hypoxia and inhibits the receptor tyrosine kinase ligand Ephrin-A3. The Journal of Biological Chemistry. 2008;283(23):15878–15883. doi: 10.1074/jbc.m800731200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Giannakakis A., Sandaltzopoulos R., Greshock J., et al. miR-210 links hypoxia with cell cycle regulation and is deleted in human epithelial ovarian cancer. Cancer Biology and Therapy. 2008;7(2):255–264. doi: 10.4161/cbt.7.2.5297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chan S. Y., Zhang Y.-Y., Hemann C., Mahoney C. E., Zweier J. L., Loscalzo J. MicroRNA-210 controls mitochondrial metabolism during hypoxia by repressing the iron-sulfur cluster assembly proteins ISCU1/2. Cell Metabolism. 2009;10(4):273–284. doi: 10.1016/j.cmet.2009.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen Z., Li Y., Zhang H., Huang P., Luthra R. Hypoxia-regulated microRNA-210 modulates mitochondrial function and decreases ISCU and COX10 expression. Oncogene. 2010;29(30):4362–4368. doi: 10.1038/onc.2010.193. [DOI] [PubMed] [Google Scholar]
- 30.Kim H. W., Haider H. K., Jiang S., Ashraf M. Ischemic preconditioning augments survival of stem cells via miR-210 expression by targeting caspase-8-associated protein 2. The Journal of Biological Chemistry. 2009;284(48):33161–33168. doi: 10.1074/jbc.m109.020925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gee H. E., Camps C., Buffa F. M., et al. hsa-mir-210 is a marker of tumor hypoxia and a prognostic factor in head and neck cancer. Cancer. 2010;116(9):2148–2158. doi: 10.1002/cncr.25009. [DOI] [PubMed] [Google Scholar]
- 32.Greither T., Grochola L. F., Udelnow A., Lautenschläger C., Würl P., Taubert H. Elevated expression of microRNAs 155, 203, 210 and 222 in pancreatic tumors is associated with poorer survival. International Journal of Cancer. 2010;126(1):73–80. doi: 10.1002/ijc.24687. [DOI] [PubMed] [Google Scholar]
- 33.Li M., Ma X., Zhang B., Huang J., Liu L., Wei Y. Prognostic role of MicroRNA-210 in various carcinomas: a systematic review and meta-analysis. Disease Markers. 2014;2014:10. doi: 10.1155/2014/106197.106197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang J., Zhao J., Shi M., et al. Elevated expression of miR-210 predicts poor survival of cancer patients: a systematic review and meta-analysis. PLoS ONE. 2014;9(2) doi: 10.1371/journal.pone.0089223.e89223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sawabata N., Miyaoka E., Asamura H., et al. Japanese lung cancer registry study of 11,663 surgical cases in 2004: demographic and prognosis changes over decade. Journal of Thoracic Oncology. 2011;6(7):1229–1235. doi: 10.1097/jto.0b013e318219aae2. [DOI] [PubMed] [Google Scholar]
- 36.Livak K. J., Schmittgen T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 37.Guan P., Yin Z., Li X., Wu W., Zhou B. Meta-analysis of human lung cancer microRNA expression profiling studies comparing cancer tissues with normal tissues. Journal of Experimental and Clinical Cancer Research. 2012;31(1, article 54) doi: 10.1186/1756-9966-31-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eilertsen M., Andersen S., Al-Saad S., et al. Positive prognostic impact of miR-210 in non-small cell lung cancer. Lung Cancer. 2014;83(2):272–278. doi: 10.1016/j.lungcan.2013.11.005. [DOI] [PubMed] [Google Scholar]
- 39.Puisségur M.-P., Mazure N. M., Bertero T., et al. miR-210 is overexpressed in late stages of lung cancer and mediates mitochondrial alterations associated with modulation of HIF-1 activity. Cell Death and Differentiation. 2011;18(3):465–478. doi: 10.1038/cdd.2010.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li Z.-H., Zhang H., Yang Z.-G., Wen G.-Q., Cui Y.-B., Shao G.-G. Prognostic significance of serum microRNA-210 levels in nonsmall-cell lung cancer. The Journal of International Medical Research. 2013;41(5):1437–1444. doi: 10.1177/0300060513497560. [DOI] [PubMed] [Google Scholar]
- 41.Qu A., Du L., Yang Y., et al. Hypoxia-inducible MiR-210 is an independent prognostic factor and contributes to metastasis in colorectal cancer. PLoS ONE. 2014;9(3) doi: 10.1371/journal.pone.0090952.e90952 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 42.Qiu S., Lin S., Hu D., Feng Y., Tan Y., Peng Y. Interactions of miR-323/miR-326/miR-329 and miR-130a/miR-155/miR-210 as prognostic indicators for clinical outcome of glioblastoma patients. Journal of Translational Medicine. 2013;11(1, article 10) doi: 10.1186/1479-5876-11-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McCormick R. I., Blick C., Ragoussis J., et al. miR-210 is a target of hypoxia-inducible factors 1 and 2 in renal cancer, regulates ISCU and correlates with good prognosis. British Journal of Cancer. 2013;108(5):1133–1142. doi: 10.1038/bjc.2013.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen X., Ba Y., Ma L., et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Research. 2008;18(10):997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 45.Wang J., Chen J., Chang P., et al. MicroRNAs in plasma of pancreatic ductal adenocarcinoma patients as novel blood-based biomarkers of disease. Cancer Prevention Research. 2009;2(9):807–813. doi: 10.1158/1940-6207.capr-09-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]