Abstract
Determining the genetic diversity of M. tuberculosis strains allows identification of the distinct Mycobacterium tuberculosis genotypes responsible for tuberculosis in different regions. Several studies have reported the genetic diversity of M. tuberculosis strains in Mexico, but little information is available from the state of Jalisco. Therefore, the aim of this study was to determine the genetic diversity of Mycobacterium tuberculosis clinical isolates from Western Mexico. Sixty-eight M. tuberculosis isolates were tested for susceptibility to first-line drugs using manual Mycobacteria Growth Indicator Tube method and genotyped using spoligotyping and IS6110-restriction fragment length polymorphism (RFLP) pattern analyses. Forty-seven (69.1%) isolates were grouped into 10 clusters and 21 isolates displayed single patterns by spoligotyping. Three of the 21 single patterns corresponded to orphan patterns in the SITVITWEB database, and 1 new type that contained 2 isolates was created. The most prevalent lineages were T (38.2%), Haarlem (17.7%), LAM (17.7%), X (7.4%), S (5.9%), EAI (1.5%) and Beijing (1.5%). Six (12.8%) of the clustered isolates were MDR, and type 406 of the Beijing family was among the MDR isolates. Seventeen (26.2%) isolates were grouped into 8 clusters and 48 isolates displayed single patterns by IS6110-RFLP. Combination of IS6110-RFLP and spoligotyping reduced the clustering rate to 20.0%. The results show that T, Haarlem, and LAM are predominant lineages among clinical isolates of M. tuberculosis in Guadalajara, Mexico. Clustering rates indicated low transmission of MDR strains. We detected a rare Beijing genotype, SIT406, which was a highly resistant strain. This is the first report of this Beijing genotype in Latin America.
Introduction
It is estimated that a third of the world’s population is infected with Mycobacterium tuberculosis and over 9 million new tuberculosis (TB) cases are diagnosed annually [1]. In Mexico, more than 2000 people die every year from TB, and the rate is 16.8 cases per 100,000 in 2010. In the western state of Jalisco, the TB rate was 13 per 100,000 [2]. Multidrug-resistant strains of M. tuberculosis (MDR-TB) resistant to isoniazid (INH) and rifampicin (RIF) are a challenge for clinicians. Patients with MDR-TB are significantly more difficult to treat and mortality is higher than in drug-susceptible cases [3].
Genotyping has been used to study TB transmission [4,5]. One genotyping technique is restriction fragment length polymorphism (RFLP) analysis, where the distribution and number of copies of an insertion sequence, IS6110, in a chromosome is monitored, and this event varies among different strains. Spoligotyping is based on polymorphisms in the direct repeat (DR) locus of the M. tuberculosis genome. Identification of identical genotypes from different patients generally indicates recent infection by a common source [6], although this greatly depends on the genotyping method used. Spoligotyping has gained international approval as a robust, fast, and reproducible typing method suitable for epidemiological research [7].
Several studies have reported the genetic diversity of M. tuberculosis strains in Mexico, but little information is available from the state of Jalisco [8–17]. Previously we evaluated phenotypic and genotypic resistance of M. tuberculosis isolates from Guadalajara, Jalisco [18]. Therefore, the aim of this study was to analyze the genetic diversity of M. tuberculosis clinical isolates from patients with pulmonary TB in Guadalajara, Jalisco. Shared types, clade predominance, and clustering rates of both susceptible and MDR isolates were determined.
Methods
Ethics Statement
The local ethics committee (Comité de Ética y Comité de Investigación, Facultad de Medicina y Hospital “Dr José Eleuterio González”, Universidad Autónoma de Nuevo León, Mexico) approved the study and written informed consent to participate in this study was obtained from each patient.
Clinical specimens and isolates
The Hospital Civil de Guadalajara, Fray Antonio Alcalde, admitted more than 240 patients with TB between 2005 and 2011, mostly patients from Jalisco and the surrounding Western states. From September 2010 to November 2011, specimens (n = 351) were referred to the collaborative mycobacteriology laboratory for further testing. All samples were decontaminated by the Petroff method [19] and cultured on Löwestein-Jensen slants at 37°C. Smears were made and stained using the Ziehl-Neelsen method and were examined for the presence of acid-fast bacilli. The cultures were examined twice a week and their growth rates and colony morphologies were recorded. Isolates were identified by traditional biochemical methods including niacin production and nitrate reduction tests. One isolate per patient was assayed.
DNA isolation and species identification
Mycobacteria were first subjected to enzymatic lysis with lysozyme and proteinase K. DNA was then obtained using a phenol extraction and ethanol precipitation [20]. Species identification was performed using multiplex PCR amplification of the cfp32 (Rv0577), RD9 (Rv2073c) and RD12 (Rv3120) genes [21,22].
Drug susceptibility testing
Drug susceptibilities were tested in all M. tuberculosis isolates using the manual Mycobacteria Growth Indicator Tube (MGIT) method. Critical concentrations of STR (0.8 μg/mL), INH (0.1 μg/mL), RIF (1.0 μg/mL), and EMB (3.5 μg/mL) were used according to the manufacturer’s instructions (Becton Dickinson and Co., Sparks Glencoe, MD) [23]. Fluorescence at 365nm indicating microbial growth was detected with a UV light from a transilluminator. The susceptible H37Ra strain was used as a control.
Spoligotyping analysis
Spoligotyping was performed as described previously [5] using a commercially available kit (Isogen Life Science, De Meern, Netherlands) according to the manufacturer’s instructions. Spoligotype results were compared to the SITVITWEB database (available at http://www.pasteur-guadeloupe.fr:8081/SITVIT_ONLINE/). At the time of the study, SITVITWEB contained 62,582 clinical isolates with 153 countries of origin [7]. A dendrogram of spoligotype patterns was generated with the unweighted pair group method and the neighbor joining algorithm using the online MIRU-VTRN plus application [24].
IS6110-RFLP analysis
IS6110-RFLP analysis was performed according to the international standard method [4]. Comparison of the results was done using the IBM SPSS statistical software for Windows version 20.0 (SPSS Inc, Chicago, IL, USA). Isolates with clustered IS6110-RFLP banding patterns containing more than five bands were considered related. A dendrogram was generated with the neighbor joining algorithm using the Jaccard coefficient.
Results
Patients and strains
Clinical samples from 351 patients with pulmonary TB were collected and processed. From these, 68 M. tuberculosis isolates were obtained. Patients were predominantly male (53/68, 77.9%). The median age was 43 years (ranging 1 to 85 years).
Shared types, clades, and clusters of M. tuberculosis isolates
From the 68 isolates, 31 different spoligotype patterns were detected. Unique spoligotype patterns were detected in 3 strains. They did not match any preexisting patterns in the SITVITWEB database and were considered orphans (Table 1). A total of 27 SITs (including 63 isolates) matched pre-existing SITs in the SITVITWEB database, and one new SIT containing 2 isolates was created in this study (Table 2).
Table 1. Description of orphan strains found in this study.
| Strain ID | Sex | Age | Spoligotype Description | Octal Code | Drug Resistance |
|---|---|---|---|---|---|
| MEX47 | Male | 26 | □□□□□□□□□□□□■■■■■□□■■■■□□□□□□□□□□□□□□□□■■■■ | 000076360000071 | Susceptible |
| MEX224 | Female | 38 | ■■■■■■■■□□■■■□■■■■■■■■■■■■■■■■■■□□□□■□■■■■■ | 776357777760571 | RIF Resistant |
| MEX257 | Male | 56 | ■■■■■■□□□□□□□■■■■■■■■■■■■■■■■■■■□□□□■■■■■■■ | 770037777760771 | Susceptible |
Orphan strains had spoligotype patterns that did not match a preexisting pattern in the SITVITWEB database.
Table 2. Description of shared types found in this study.
| SIT a | Spoligotype Description | Octal Code | Clade b | n (%) |
|---|---|---|---|---|
| 2 | □□□□□□□□□□□□□□□□□□□□□□□□■□□□□□□■□□□□■■■■■■■ | 000000004020771 | H2 | 1 (1.5) |
| 17 | ■■□■■■■■■■■■□■■■■■■■□□□□■■■■■■■■□□□□■■■■■■■ | 677737607760771 | LAM2 | 4 (5.9) |
| 19 | ■■□■■■■■■■■■■■■■■■■□□■■■■■■■□□□□■□■■■■■■■■■ | 677777477413771 | EAI2-Manila | 1 (1.5) |
| 20 | ■■□■■■■■■■■■■■■■■■■■□□□□■■■■■■■■□□□□■■■■■■■ | 677777607760771 | LAM1 | 1 (1.5) |
| 34 | ■■■■■■■■□□□□■■■■■■■■■■■■■■■■■■■■□□□□■■■■■■■ | 776377777760771 | S | 2 (2.9) |
| 42 | ■■■■■■■■■■■■■■■■■■■■□□□□■■■■■■■■□□□□■■■■■■■ | 777777607760771 | LAM9 | 5 (7.4) |
| 47 | ■■■■■■■■■■■■■■■■■■■■■■■■□□□□□□□■□□□□■■■■■■■ | 777777774020771 | H1 | 2 (2.9) |
| 50 | ■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■□■□□□□■■■■■■■ | 777777777720771 | H3 | 6 (8.8) |
| 51 | ■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■□□□□■■■□□□□ | 777777777760700 | T1 | 2 (2.9) |
| 53 | ■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■□□□□■■■■■■■ | 777777777760771 | T1 | 17 (25) |
| 92 | ■■■□□□□□□□□□■■■■■□■■■■■■■■■■■■■■□□□□■■■■■■■ | 700076777760771 | X3 | 1 (1.5) |
| 137 | ■■■■■■■■■■■■■■■■■□■■■■■■■■■■■■■■□□□□■■□□□□■ | 777776777760601 | X2 | 3 (4.4) |
| 232 | ■■■■■■■■■■■■■■■■■■■■■■□□□□□□□□□□□□□□□■■■■■■ | 777777760000171 | Unk | 1 (1.5) |
| 335 | ■■■■■■■■■■■■■■■■■■■■□□□□■■■■■■□■□□□□■■■■■■■ | 777777607720771 | Unk | 1 (1.5) |
| 371 | ■■■■■■■■■■■■■■■■■■□□□□□□■■■■■■□■□□□□■■■■■■■ | 777777007720771 | H3 | 1 (1.5) |
| 406 | □□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□□■■■□■■■ | 000000000000731 | Beijing | 1 (1.5) |
| 469 | □□□■■■■■■■■■■■■■■■■■□□□□■■■■■■■■□□□□■■■■■■■ | 077777607760771 | LAM9 | 1 (1.5) |
| 478 | ■■□□□■■■■■■■■■■■■□■■■■■■■■■■■■■■□□□□■■□□□□■ | 617776777760601 | X2 | 1 (1.5) |
| 535 | ■■■■■■■■■■■■■■■■■■■■■□□□■■■■■■■■□□□□■■■■■■■ | 777777707760771 | T1 | 1 (1.5) |
| 732 | ■■■■■■■■■■■■■■□□■■■■■■■■■■■■■■■■□□□□■■■■■■■ | 777763777760771 | T1 | 1 (1.5) |
| 1215 | ■■□■■■■■■■■■□■■■□■■■□□□□■■■■■■■■□□□□■■■■■■■ | 677735607760771 | LAM2 | 1 (1.5) |
| 1225 | ■■■■■■■■□□■■■■■■■■■■■□□□□□□□□■■■□□□□■■■■■■■ | 776377700160771 | S | 1 (1.5) |
| 1231 | ■■■■■■■■■■■■■■■■□□□□□■□■■■■■■■■■□□□□■■■■■■■ | 777774057760771 | T5 | 4 (5.9) |
| 1356 | ■■■■■■■■□□■■■■■■■■■■■■■■■■■■■■■■□□□□■■■■□■■ | 776377777760751 | S | 1 (1.5) |
| 1686 | ■■■■■■■■■■■■■□□□□□□□□□□□■□□□□□□■□□□□■■■■■■■ | 777740004020771 | H1 | 1 (1.5) |
| 2007 | ■■■■■■■■■■■■□■■■■■■■□■■■■■■■■■■■□□□□■■■■■■■ | 777737677760771 | T3 | 1 (1.5) |
| 2212 | ■■■■■■■■■■■■■■■■■■■□□□□□■□□■■■□■□□□□■□■■■■■ | 777777404720571 | H3 | 1 (1.5) |
| 3434* | ■■■■■■■□□■■■■□□□□□□□□□□■□■■■■□■□□□□□■■■■■■■ | 774740005720771 | Unk | 2 (2.9) |
aIn the SITVITWEB database, the spoligo (shared) international type (SIT) numbers designate spoligotypes shared by two or more patient isolates.
*SIT3434 is a newly created shared type due to two strains belonging to a new pattern within this study.
bClade designations according to the SITVITWEB database: Beijing clade, East African-Indian (EAI) clade and 9 sublineages, Haarlem (H) clade and 3 sublineages, Latin American-Mediterranean (LAM) clade and 12 sublineages, the ancestral “Manu” family and 3 sublineages, the S clade, the IS6110-low-banding X clade and 3 sublineages, and an ill-defined T clade with 5 sublineages. Unk, unknown patterns within any of the major clades described in the database.
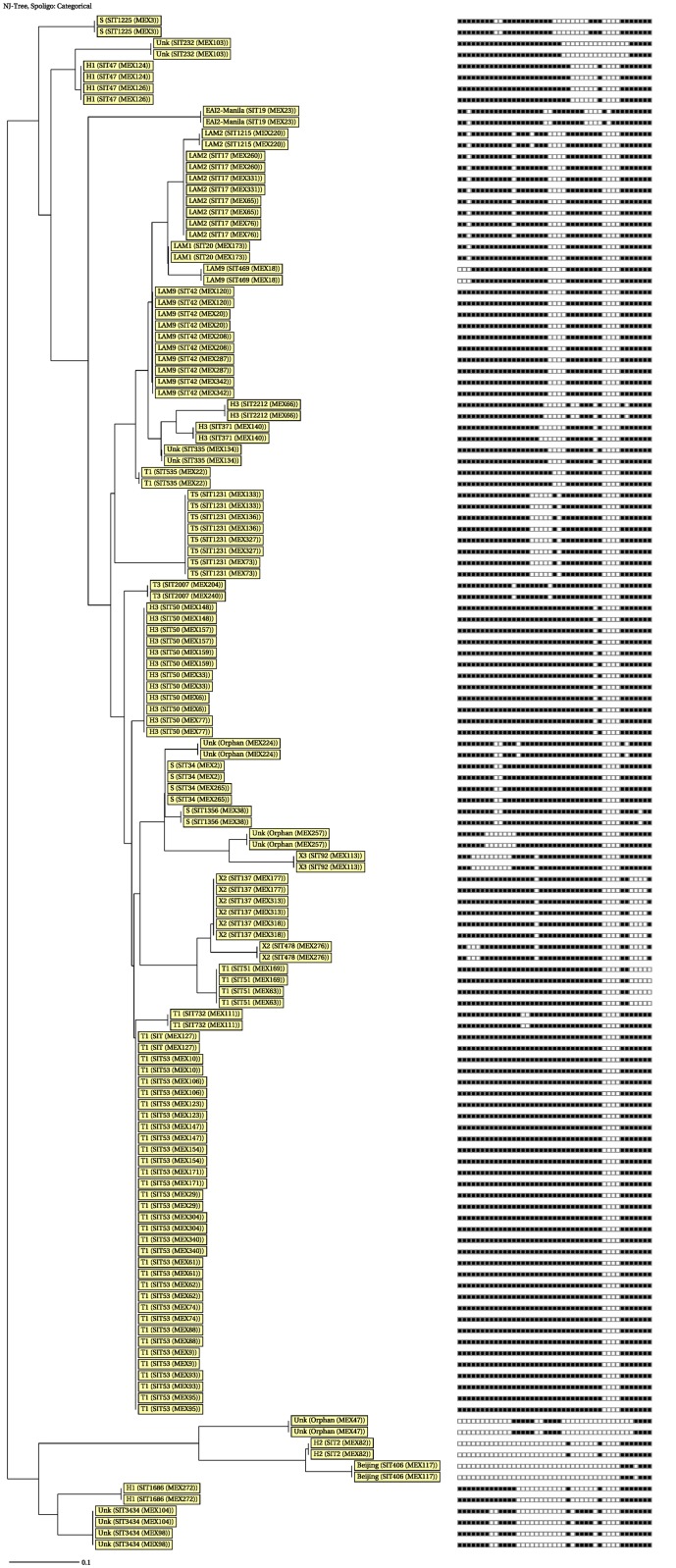
Clustering was detected in 47 isolates (69.1%) (Fig. 1) including SIT53 (T1 of ill-defined T clade, n = 17, 25.0%), SIT50 (H3 of Haarlem clade, n = 6, 8.8%), SIT42 (LAM9 of the Latin American-Mediterranean clade, n = 5, 7.4%), SIT17 (LAM2, n = 4, 5.9%), SIT1231 (T5, n = 4, 5.9%), SIT137 (X2 of the IS6110-low-banding X clade, n = 3, 4.4%), SIT34 (the S clade, n = 2, 2.9%), SIT47 (H1, n = 2, 2.9%), SIT51 (T1, n = 2, 2.9%), and SIT3434 (Unknown clade, n = 2, 2.9%), which was created in this study. Single patterns were observed in 18 isolates including SIT2 (H2), SIT19 (EAI2-Manila), SIT20 (LAM1), SIT92 (X3), SIT232 (Unk), SIT335 (Unk), SIT371 (H3), SIT406 (Beijing), SIT469 (LAM9), SIT478 (X2), SIT535 (T1), SIT732 (T1), SIT1215 (LAM2), SIT1225 (S), SIT1356 (S), SIT1686 (H1), SIT2007 (T3) and SIT2212 (H3).
Fig 1. Spoligotype tree of M. tuberculosis isolates (n = 68).
Lineage, Spoligotype International Type (SIT) as defined in the SITVITWEB proprietary database of the Pasteur Institute of Guadeloupe, and the strain identifications are shown.
IS6110-RFLP patterns of M. tuberculosis isolates
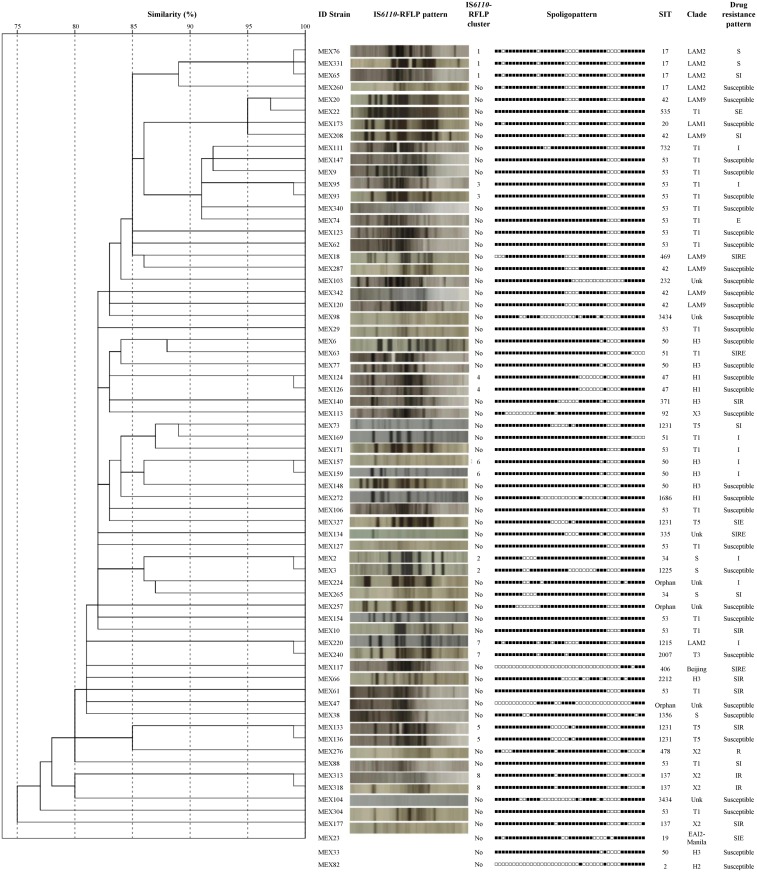
IS6110-RFLP patterns of three isolates (MEX23, MEX33 and MEX82) were not obtained despite three attempts. The remaining 65 isolates were typified by IS6110-RFLP typing (Fig. 2). We identified 56 fingerprint patterns, 8 were clustered including 17 isolates with a clustering rate of 26.2% and the remaining 48 had single patterns. The number of IS6110 copies varied from 1 to 14; 13.8% (n = 9) of the isolates were low-copy-number isolates having less than six copies of IS6110. Combination of IS6110-RFLP and spoligotyping reduced the number of clustered isolates to 13 and the clustering rate to 20.0%. Isolates lacking the IS6110 element were not observed.
Fig 2. IS6110-RFLP clustering patterns, spoligopatterns and drug resistance profiles of M. tuberculosis isolates.
Drug resistance profile indicates resistance to I: isoniazid; R: rifampicin; S: streptomycin and E: ethambutol.
Drug resistant M. tuberculosis isolates
Drug resistance was detected in 33 (48.5%) isolates. Among the 13 MDR isolates, SIT53 (n = 3) and SIT137 (n = 3) were clustered by spoligotyping and only 2 isolates (MEX313 and MEX318) were clustered by IS6110-RFLP (Fig. 2). The single pattern isolates SIT335 (Unk), SIT371 (H3), SIT406 (Beijing), SIT469 (LAM9) and SIT2212 (H3) were also MDR.
Discussion
Determining the genetic diversity of M. tuberculosis strains allows identification of the distinct M. tuberculosis genotypes responsible for TB in different regions. Genetic variation in M. tuberculosis strains contributes to the variability in disease presentation, frequency of transmission, and treatment outcomes [25,26]. Knowing these factors is important for appropriate treatment and control of TB.
The most predominant clades found in our study were the ill-defined T clade (SIT53), the Haarlem clade (SIT50), the Latin American-Mediterranean clade (SIT42) and the IS6110-low-banding X clade (SIT137). These results are similar to those reported in several studies from across the country [8–17]. And considering that SIT42 evolved originally from SIT53, this group is very dominant in this region [27].
We detected high clustering rates of spoligotypes (69.1%) consistent with previous reports in Mexico [9,11–13]. However, despite the single spoligopattern presented, four isolates (MEX2, MEX3, MEX220 and MEX240) were in 2 clusters according to IS6110-RFLP. Thus, combination of IS6110-RFLP and spoligotyping reduced the clustering rate to 20.0%. This indicates that spoligotyping alone is not discriminatory enough to determine epidemiological links and for transmission analysis, due to the high rate of homoplasy (independent mutational events that result in the loss of the same spacer) exhibited by the spacers used in spoligotyping [28–30].
The low clustering rate detected in this study indicates low transmission of M. tuberculosis clones. Most of the clustered isolates, regardless of genotypic family, were drug susceptible, also indicating low transmission of MDR isolates. In addition, changes in the drug resistance pattern of the isolates in one cluster were not accompanied by changes of the IS6110-RFLP pattern (clusters 1, 2, 3, 5 and 7). This is in accordance with previous studies where the rate of change of IS6110 fingerprint patterns in drug-resistant isolates seems not to differ from that in drug-susceptible isolates, thus indicating a high degree of stability of IS6110 patterns [31–33].
Although a high number of previously reported spoligotypes were identified in Mexico [12,13,17], 19.1% of our isolates showed SIT (335, 371, 406, 469, 1231, 1686, 2007, 2212 and 3434) which were previously unreported in the country.
M. tuberculosis isolates with no IS6110 elements are rare [34] and we did not find them in Jalisco. In Mexico, there is only one report in Mexico City, constituting as high as 20% of the isolates studied [13]. It will be important to determine the ancestry of these patients, since zero IS6110 M. tuberculosis isolates have been found elsewhere in Mexico.
Some M. tuberculosis strains of the Beijing genotype have been often associated with MDR phenotype and higher transmission compared to other genotypes [26,35]. The Beijing genotype is principally identified by the deletion of spacers 1–34 and the presence of at least 3 of the 9 spacers between positions 35 and 43 of the DR locus [36–38]. To date, 27 Beijing family genotypes have been reported in the STVIT2 international database (including 6,148 strains from 71 countries). SIT1 was the classical Beijing genotype in the SITVITWEB database at the time this study was conducted. Although these strains are prevalent throughout Asia, they are found worldwide [39]. The SIT1 genotype has been reported in several states in Mexico including Puebla [14], Jalisco [9], Baja California, Sinaloa, Veracruz [15] and San Luis Potosi [16]. Both drug-susceptible and resistant strains have been documented.
In this study, we detected an MDR strain with the Beijing genotype and a SIT406 pattern. To our knowledge, this is the first report of Beijing SIT406 in Mexico or Latin America. At the time of this comparison, SIT406 represented a total of 14 strains in the SITVITWEB database, which corresponded to 0.2% (14/6,148) of all Beijing strains, in contrast to SIT1, which corresponded to 94.3% (5,800/6,148) of all Beijing strains. According to the available entries from the database, the distribution of SIT406 strains is limited to the United States (64.3%), Belgium, India, Italy, Japan, and Tunisia (7.1% each).
Little is known about the differences between disease caused by the rare Beijing and classical Beijing genotypes. In Latin America only two rare Beijing strains have been reported in Mexico (SIT269 and SIT941) [7] and one case was reported in Colombia (SIT190) [40]. TB treatment outcomes are associated with MDR phenotypes rather than Beijing genotypes [41]. However, most studies of Beijing strains only include SIT1 strains, and the behavior of other Beijing family genotypes is undefined. Spoligotyping is commonly used in high-income countries and data from countries such as Mexico is limited.
One limitation of this work is the lack of use of mycobacterium interspersed repetitive unit-variable number of tandem repeat (MIRU-VNTR) as a genotyping method for M. tuberculosis isolates. It is well known that in the M. tuberculosis epidemiology, MIRU-VNTR and spoligotyping are increasingly being used as first line methods for their ease, discriminatory power, and digital analysis of generated data [6] compared with the gold standard IS6110 typing method. Nonetheless, MIRU-VNTR genotyping is commonly used in high-income countries and availability in countries such as Mexico is limited. Incidentally, it has been reported a reduced discriminatory power of 24-loci MIRU-VNTR compared to IS6110 typing especially in Beijing lineages [42,43]. In these cases, 24-loci MIRU-VNTR typing is inadequate for accurately identifying transmission chains of Beijing strains, and thus needs to be complemented either by IS6110 typing or combinations of MIRU-VNTR loci more variable in Beijing strains [43]. Contrariwise, MIRU-VNTR allows a higher discrimination of the low-copy-number isolates, which in our case corresponded to 13.8% of the isolates studied. Therefore, an expansion of this work is required to further analyze this isolates using MIRU-VNTR, both to allow the inclusion of a greater sample number and to further describe the Beijing genotype detected in this study.
In summary, T, Haarlem, and LAM were predominant lineages in clinical isolates of M. tuberculosis in Guadalajara, Jalisco. Transmission of MDR strains was low in Guadalajara. However, some isolated cases of TB were caused by the rare MDR Beijing genotype. The report of an MDR strain with a SIT406 pattern indicated additional effort should be placed on the search for these genotypes. The status of Beijing strains in Mexico needs to be monitored to determine whether they are expanding.
Acknowledgments
VBG is a COFAA and EDI scholarship recipient of the Instituto Politecnico Nacional. We thank Sergio Lozano-Rodríguez, M.D., from the Hospital Universitario for his review of the manuscript.
Data Availability
All relevant data are within the paper.
Funding Statement
The authors have no support or funding to report.
References
- 1. World Health Organization Global Tuberculosis Report 2012 WHO/HTM/TB/2011.16. Geneva, Switzerland: WHO, 2012. [Google Scholar]
- 2.Secretaria de Salud (Mexico) (2012) Perfil Epidemiológico de la Tuberculosis en México. Sistema Nacional de Vigilancia Epidemiológica, Direccion General de Epidemiología, Secretaría de Salud.
- 3. Zazueta-Beltran J, Leon-Sicairos C, Canizalez-Roman A (2009) Drug resistant Mycobacterium tuberculosis in Mexico. J Infect Dev Ctries 3: 162–168. [DOI] [PubMed] [Google Scholar]
- 4. van Embden JD, Cave MD, Crawford JT, Dale JW, Eisenach KD, et al. (1993) Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J Clin Microbiol 31: 406–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kamerbeek J, Schouls L, Kolk A, van Agterveld M, van Soolingen D, et al. (1997) Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J Clin Microbiol 35: 907–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Barnes PF, Cave MD (2003) Molecular epidemiology of tuberculosis. N Engl J Med 349: 1149–1156. [DOI] [PubMed] [Google Scholar]
- 7. Demay C, Liens B, Burguiere T, Hill V, Couvin D, et al. (2012) SITVITWEB—a publicly available international multimarker database for studying Mycobacterium tuberculosis genetic diversity and molecular epidemiology. Infect Genet Evol 12: 755–766. 10.1016/j.meegid.2012.02.004 [DOI] [PubMed] [Google Scholar]
- 8. Yang ZH, Rendon A, Flores A, Medina R, Ijaz K, et al. (2001) A clinic-based molecular epidemiologic study of tuberculosis in Monterrey, Mexico. Int J Tuberc Lung Dis 5: 313–320. [PubMed] [Google Scholar]
- 9. Macias-Parra M, Kumate Rodriguez J, Arredondo Garcia JL, Lopez-Vidal Y, Castanon-Arreola M, et al. (2011) Mycobacterium tuberculosis Complex Genotype Diversity and Drug Resistance Profiles in a Pediatric Population in Mexico. Tuberc Res Treat 2011: 239042 10.1155/2011/239042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Quitugua TN, Seaworth BJ, Weis SE, Taylor JP, Gillette JS, et al. (2002) Transmission of drug-resistant tuberculosis in Texas and Mexico. J Clin Microbiol 40: 2716–2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Molina-Torres CA, Moreno-Torres E, Ocampo-Candiani J, Rendon A, Blackwood K, et al. (2010) Mycobacterium tuberculosis spoligotypes in Monterrey, Mexico. J Clin Microbiol 48: 448–455. 10.1128/JCM.01894-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nava-Aguilera E, Lopez-Vidal Y, Harris E, Morales-Perez A, Mitchell S, et al. (2011) Clustering of Mycobacterium tuberculosis cases in Acapulco: Spoligotyping and risk factors. Clin Dev Immunol 2011: 408375 10.1155/2011/408375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lopez-Alvarez R, Badillo-Lopez C, Cerna-Cortes JF, Castillo-Ramirez I, Rivera-Gutierrez S, et al. (2010) First insights into the genetic diversity of Mycobacterium tuberculosis isolates from HIV-infected Mexican patients and mutations causing multidrug resistance. BMC Microbiol 10: 82 10.1186/1471-2180-10-82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Martinez-Gamboa A, Ponce-de-Leon A, Galindo-Fraga A, Bobadilla-del-Valle M, Kato-Maeda M, et al. (2008) Molecular analysis of Mycobacterium tuberculosis strains with an intact pks15/1 gene in a rural community of Mexico. Arch Med Res 39: 809–814. 10.1016/j.arcmed.2008.08.006 [DOI] [PubMed] [Google Scholar]
- 15. Martinez-Guarneros A, Rastogi N, Couvin D, Escobar-Gutierrez A, Rossi LM, et al. (2013) Genetic diversity among multidrug-resistant Mycobacterium tuberculosis strains in Mexico. Infect Genet Evol 14: 434–443. 10.1016/j.meegid.2012.12.024 [DOI] [PubMed] [Google Scholar]
- 16. Lopez-Rocha E, Juarez-Alvarez J, Riego-Ruiz L, Enciso-Moreno L, Ortega-Aguilar F, et al. (2013) Genetic diversity of the Mycobacterium tuberculosis Complex in San Luis Potosi, Mexico. BMC Res Notes 6: 172 10.1186/1756-0500-6-172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Flores-Trevino S, Mendoza-Olazaran S, Garza-Gonzalez E (2014) Drug resistance and molecular epidemiology of Mycobacterium tuberculosis in Mexico: a systematic review. Salud Publica Mex 56: 63–77. [DOI] [PubMed] [Google Scholar]
- 18. Flores-Trevino S, Morfin-Otero R, Rodriguez-Noriega E, Gonzalez-Diaz E, Perez-Gomez HR, et al. (2014) Characterization of phenotypic and genotypic drug resistance patterns of Mycobacterium tuberculosis isolates from a city in Mexico. Enferm Infecc Microbiol Clin. 10.1016/j.eimc.2014.10.020 [DOI] [PubMed] [Google Scholar]
- 19. Petroff SA (1915) A New and Rapid Method for the Isolation and Cultivation of Tubercle Bacilli Directly from the Sputum and Feces. J Exp Med 21: 38–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Belisle JT, Mahaffey SB, Hill PJ (2009) Isolation of mycobacterium species genomic DNA. Methods Mol Biol 465: 1–12. 10.1007/978-1-59745-207-6_1 [DOI] [PubMed] [Google Scholar]
- 21. Huard RC, Lazzarini LC, Butler WR, van Soolingen D, Ho JL (2003) PCR-based method to differentiate the subspecies of the Mycobacterium tuberculosis complex on the basis of genomic deletions. J Clin Microbiol 41: 1637–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nakajima C, Rahim Z, Fukushima Y, Sugawara I, van der Zanden AG, et al. (2010) Identification of Mycobacterium tuberculosis clinical isolates in Bangladesh by a species distinguishable multiplex PCR. BMC Infect Dis 10: 118 10.1186/1471-2334-10-118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Palomino JC, Traore H, Fissette K, Portaels F (1999) Evaluation of Mycobacteria Growth Indicator Tube (MGIT) for drug susceptibility testing of Mycobacterium tuberculosis. Int J Tuberc Lung Dis 3: 344–348. [PubMed] [Google Scholar]
- 24. Weniger T, Krawczyk J, Supply P, Niemann S, Harmsen D (2010) MIRU-VNTRplus: a web tool for polyphasic genotyping of Mycobacterium tuberculosis complex bacteria. Nucleic Acids Res 38: W326–331. 10.1093/nar/gkq351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Aguilar D, Hanekom M, Mata D, Gey van Pittius NC, van Helden PD, et al. (2010) Mycobacterium tuberculosis strains with the Beijing genotype demonstrate variability in virulence associated with transmission. Tuberculosis (Edinb) 90: 319–325. 10.1016/j.tube.2010.08.004 [DOI] [PubMed] [Google Scholar]
- 26. Hanekom M, Gey van Pittius NC, McEvoy C, Victor TC, Van Helden PD, et al. (2011) Mycobacterium tuberculosis Beijing genotype: a template for success. Tuberculosis (Edinb) 91: 510–523. 10.1016/j.tube.2011.07.005 [DOI] [PubMed] [Google Scholar]
- 27. Realpe T, Correa N, Rozo JC, Ferro BE, Gomez V, et al. (2014) Population structure among mycobacterium tuberculosis isolates from pulmonary tuberculosis patients in Colombia. PLoS One 9: e93848 10.1371/journal.pone.0093848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fenner L, Malla B, Ninet B, Dubuis O, Stucki D, et al. (2011) "Pseudo-Beijing": evidence for convergent evolution in the direct repeat region of Mycobacterium tuberculosis. PLoS One 6: e24737 10.1371/journal.pone.0024737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kato-Maeda M, Metcalfe JZ, Flores L (2011) Genotyping of Mycobacterium tuberculosis: application in epidemiologic studies. Future Microbiol 6: 203–216. 10.2217/fmb.10.165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Comas I, Homolka S, Niemann S, Gagneux S (2009) Genotyping of genetically monomorphic bacteria: DNA sequencing in Mycobacterium tuberculosis highlights the limitations of current methodologies. PLoS One 4: e7815 10.1371/journal.pone.0007815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Niemann S, Rusch-Gerdes S, Richter E, Thielen H, Heykes-Uden H, et al. (2000) Stability of IS6110 restriction fragment length polymorphism patterns of Mycobacterium tuberculosis strains in actual chains of transmission. J Clin Microbiol 38: 2563–2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Niemann S, Richter E, Rusch-Gerdes S (1999) Stability of IS6110 restriction fragment length polymorphism patterns of multidrug-resistant Mycobacterium tuberculosis strains. J Clin Microbiol 37: 3078; author reply 3078–3079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Niemann S, Richter E, Rusch-Gerdes S (1999) Stability of Mycobacterium tuberculosis IS6110 restriction fragment length polymorphism patterns and spoligotypes determined by analyzing serial isolates from patients with drug-resistant tuberculosis. J Clin Microbiol 37: 409–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lok KH, Benjamin WH Jr., Kimerling ME, Pruitt V, Lathan M, et al. (2002) Molecular differentiation of Mycobacterium tuberculosis strains without IS6110 insertions. Emerg Infect Dis 8: 1310–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Parwati I, van Crevel R, van Soolingen D (2010) Possible underlying mechanisms for successful emergence of the Mycobacterium tuberculosis Beijing genotype strains. Lancet Infect Dis 10: 103–111. 10.1016/S1473-3099(09)70330-5 [DOI] [PubMed] [Google Scholar]
- 36. van Soolingen D, Qian L, de Haas PE, Douglas JT, Traore H, et al. (1995) Predominance of a single genotype of Mycobacterium tuberculosis in countries of east Asia. J Clin Microbiol 33: 3234–3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kremer K, Glynn JR, Lillebaek T, Niemann S, Kurepina NE, et al. (2004) Definition of the Beijing/W lineage of Mycobacterium tuberculosis on the basis of genetic markers. J Clin Microbiol 42: 4040–4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tsolaki AG, Gagneux S, Pym AS, Goguet de la Salmoniere YO, Kreiswirth BN, et al. (2005) Genomic deletions classify the Beijing/W strains as a distinct genetic lineage of Mycobacterium tuberculosis. J Clin Microbiol 43: 3185–3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bifani PJ, Mathema B, Kurepina NE, Kreiswirth BN (2002) Global dissemination of the Mycobacterium tuberculosis W-Beijing family strains. Trends Microbiol 10: 45–52. [DOI] [PubMed] [Google Scholar]
- 40. Murcia MI, Manotas M, Jimenez YJ, Hernandez J, Cortes MI, et al. (2010) First case of multidrug-resistant tuberculosis caused by a rare “Beijing-like” genotype of Mycobacterium tuberculosis in Bogota, Colombia. Infect Genet Evol 10: 678–681. 10.1016/j.meegid.2010.03.010 [DOI] [PubMed] [Google Scholar]
- 41. Buu TN, Huyen MN, van Soolingen D, Lan NT, Quy HT, et al. (2010) The Mycobacterium tuberculosis Beijing genotype does not affect tuberculosis treatment failure in Vietnam. Clinical Infectious Diseases 51: 879–886. 10.1086/656410 [DOI] [PubMed] [Google Scholar]
- 42. Hanekom M, van der Spuy GD, Gey van Pittius NC, McEvoy CR, Hoek KG, et al. (2008) Discordance between mycobacterial interspersed repetitive-unit-variable-number tandem-repeat typing and IS6110 restriction fragment length polymorphism genotyping for analysis of Mycobacterium tuberculosis Beijing strains in a setting of high incidence of tuberculosis. J Clin Microbiol 46: 3338–3345. 10.1128/JCM.00770-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Aleksic E, Merker M, Cox H, Reiher B, Sekawi Z, et al. (2013) First molecular epidemiology study of Mycobacterium tuberculosis in Kiribati. PLoS One 8: e55423 10.1371/journal.pone.0055423 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.