Abstract
Myeloid malignancies consist of acute myeloid leukemia (AML), myelodysplastic syndromes (MDS) and myeloproliferative neoplasm (MPN). The latter two diseases have preleukemic features and frequently evolve to AML. As with solid tumors, multiple mutations are required for leukemogenesis. A decade ago, these gene alterations were subdivided into two categories: class I mutations stimulating cell growth or inhibiting apoptosis; and class II mutations that hamper differentiation of hematopoietic cells. In mouse models, class I mutations such as the Bcr-Abl fusion kinase induce MPN by themselves and some class II mutations such as Runx1 mutations induce MDS. Combinations of class I and class II mutations induce AML in a variety of mouse models. Thus, it was postulated that hematopoietic cells whose differentiation is blocked by class II mutations would autonomously proliferate with class I mutations leading to the development of leukemia. Recent progress in high-speed sequencing has enabled efficient identification of novel mutations in a variety of molecules including epigenetic factors, splicing factors, signaling molecules and proteins in the cohesin complex; most of these are not categorized as either class I or class II mutations. The functional consequences of these mutations are now being extensively investigated. In this article, we will review the molecular basis of hematological malignancies, focusing on mouse models and the interfaces between these models and clinical findings, and revisit the classical class I/II hypothesis.
Keywords: acute myeloid leukemia, myelodysplastic syndromes, myeloproliferative neoplasm, epigenetic factors, splicing, mutations
Introduction
The first gene alteration identified in hematologic malignancies was the Bcr-Abl fusion kinase caused by the Ph1 chromosome t(9:22) in patients with chronic myeloid leukemia (CML).1) Bcr-Abl was detected in most CML patients, and it was thought that the specific chromosomal translocation was the sole cause of the disease. In fact, Bcr-Abl induced CML-like diseases in transgenic mice as well as in mouse bone marrow transplant (BMT) models where bone marrow cells transduced with Bcr-Abl are transplanted to irradiated mice.2,3) In addition to Bcr-Abl, many fusion genes caused by chromosomal translocations were identified, some of which were specific to particular types of leukemia; Runx1-ETO for AML-M2 and PML-RARa for AML-M3 (acute promyelocytic leukemia:APL). However, it was experimentally shown that these fusion genes were not by themselves able to efficiently induce the corresponding leukemia in mouse models.4,5) In addition, more than two gene alterations — including fusion genes, deletions and point mutations — were identified in clinical samples of AML patients, suggesting that multiple mutations are required for leukemogenesis, as it is the case with solid tumors.
Two-step model of leukemogenesis
The hypothesis that multiple mutations are required for leukemogenesis was experimentally proven for the first time by a group in the USA using transgenic mice of PML-RARa. Only 15–20% of the transgenic mice developed AML-M3-like leukemias with long latencies. However, retrovirus-mediated transduction of FLT3-ITD (internal tandem duplication), a constitutively active mutant of tyrosine kinase receptor FLT3, to bone marrow cells of PML-RARa transgenic mice induced AML-M3-like disease in all of the transplanted mice with short latencies.5) The same group also demonstrated that FLT3-ITD by itself was capable of inducing MPN in mice.6) Based on these results, a “two-step model of leukemogenesis” was proposed.7) In this model, leukemia-associated gene alterations were divided into two groups: class I mutations included activating mutations of tyrosine kinase receptors such as FLT3-ITD and oncogenes such as N-Ras as well as inactivation mutations of tumor suppressors including p53, inducing cell proliferation or inhibiting apoptosis. On the other hand, class II mutations include inactivating mutations of transcription factors such as AML1/Runx1 and chromosome modification enzymes such as MLL that hamper cell differentiation (Table 1). It was conceivable that hematopoietic cells whose differentiation is blocked by class II mutations would proliferate with class I mutations, leading to development of leukemia. In fact, combination of class I and class II mutations has been shown to induce AML in a variety of mouse models.8–14)
Table 1.
Classification of mutations found in hematological malignancies
| Class I mutations (induce proliferation or inhibit apoptosis) |
| FLT3-ITD, cKit, PDGFR, TEL-PDGFR, FGFR |
| JAK2, JAK3, Bcr-Abl |
| N-Ras, K-Ras |
| p53, NF1 |
| Class II mutations (inhibit differentiation or induce immortalization) |
| Runx 1/AML1, PU1, C/EBPα |
| MLL fusions, PML-RARα, AMΛ1–ETO |
Among the gene alterations, C/EBPα is unique because two distinct C/EBPα mutations seem to play either class I or class II-like roles in inducing leukemia. C/EBPα is a transcription factor involved in the differentiation of granulocytes as well as adipocytes. On one hand, C/EBPα also has some features of a tumor suppressor; C/EBPα knockout increases the number of hematopoietic stem cells and enhances their proliferation.15) Mutations of C/EBPα are found in 10% of AML patients without chromosomal translocations. There are two types of mutations: C-terminal mutations disrupt the DNA binding activity while N-terminal mutations disrupt full-length C/EBPα, leaving an N-terminal truncated form of C/EBPα which is a naturally occurring dominant-negative form.16) Both mutations could inhibit the functions of the wild-type C/EBPα. Interestingly, C-terminal and N-terminal mutations were frequently found in the different alleles of the C/EBPα gene in the same patient. That is, 70% of AML patients harboring a C/EBPα mutation on one allele carried a distinct mutation on the other allele,17) suggesting co-operability of N- and C-terminal mutations of C/EBPα in leukemogenesis. In fact, this has been experimentally proven using a knock-in mouse18) or a BMT model.19) In the BMT model, we demonstrated that the C-terminal mutant but not the N-terminal mutant of C/EBPα efficiently blocked the differentiation of hematopoietic cells and collaborated with a representative class I mutation FLT3-ITD in inducing acute leukemia with very short latencies. This result indicated that the C-terminal mutant plays class II-like roles in inducing leukemia.
Myeloproliferative neoplasm (MPN) is caused by class I mutations
As stated above, one of the class I mutations Bcr-Abl induced chronic myelogeneous leukemia (CML)-like disease in transgenic mice as well as BMT models. Later, another class I mutation, an activating mutation of a tyrosine kinase JAK2 was identified in MPN including polycytemia vera (PV), essential thrombocytosis (ET) and myelofibrosis (MF).20–23) Like Bcr-Abl, the mutant JAK2 JAK2-V617F induced MPN including PV or ET in the transgenic mouse models.24,25) It seems that the expression levels of JAK2-V617F determines the phenotypes of the disease. In MPN patients, in addition to the JAK2 mutation, mutations of MPL, Lnk and Calreticulin (CALR) have been identified which are exclusive with the JAK2 mutation.26–29) MPL mutations induced constitutive activation of signaling pathways like JAK2-V617F and induced MPN in mouce models.26) How the CALR mutation induces MPN is not clear at present; however it has been reported that the CALR mutation induced constitutive activation of transcription factor STAT5, a major downstream target of JAK2, and autonomous proliferation of an IL-3-dependent cell line Ba/F3.29) Some other class I mutations such as activating mutations of Ras and fusion kinase FIP1L1-PDGFRα was reported to induce MPN in mouse models30–32) although it is not known if these mutations induce MPN in humans. These results indicated that class I mutations alone could induce MPN by inducing proliferation and inhibiting apoptosis of hematopoietic cells.
Myelodysplastic syndromes (MDS) is caused by class II mutations
MDS is characterized by pancytopenia, myelodysplasia, increased apoptosis and frequent progression to leukemia. MDS is a heterogenous disease, associated with a variety of chromosomal abnormalities including 5q-, 20q-, 7q-, and inv(3)(q21;q26). Until quite recently, point mutations or frame shift mutations had been identified only in a restricted number of genes including Runx1/AML1,33) C/EBPα34) and Ras35) in MDS patients. Ras mutations, categorized as class I mutations, are known to be associated with progression of MDS to AML (MDS/overt leukemia). In the same context, C/EBPα mutations are found in AML patients more frequently than in MDS patients, and it was reported that an N-terminal C/EBPα mutation was identified in a patient progressing to acute leukemia.36) These results implicated that the N-terminal C/EBPα mutation as well as Ras mutations played a role as a class I mutation in the progression of MDS to AML. This is consistent with our results that the C-terminal mutation plays class II-like roles and collaborates with the N-terminal mutant of C/EBPa in leukemogenesis.19) However, it is not clear at present whether C-terminal mutations of C/EBPα could induce MDS in patients. In our BMT model, C-terminal mutations of C/EBPα, which are believed to play class II-like roles in leukemogenesis, never gave rise to MDS-like symptoms in the transplanted mice,19) but induced AML in the transplanted mice with long latencies.37)
There have been several MDS models in mice. Among them, one established by Lin et al. well recapitulated human MDS. They established transgenic mice expressing a fusion gene, NUP98-HOXD13, caused by t(2;11)38) and found that these mice displayed pancytopenia, ineffective hematopoiesis with increased apoptosis, myelodysplasia, and frequent progression to acute leukemia — all common features of human MDS. The NUP98-HOXD13 fusion is found in patients with MDS as well as those with AML, and it has been recently reported that the combination of FLT3-ITD and NUP98-HOXD13 induced AML in a mouse model.39) These results suggested that NUP98-HOXD13 played class II-like roles in MDS pathogenesis and induced AML in concert with a class I mutation, FLT3-ITD.
It was also reported that either heterozygote mice for nucleophosmin 1 (NPM1) or overexpression of Evi1 induced MDS-like symptoms. NPM1 is a nucleolar protein with an important role in ribosome biogenesis and is frequently mutated in AML patients, losing its nucleolar localization and gaining cytosol localization.40) It is known that point mutations of NPM1 frequently co-exist with a representative class I mutation FLT3-ITD in AML patients, implying that the NPM1 mutations play class II-like roles. Nonetheless, it has been suggested that the NPM1 mutant plays rather class I-like roles; the NPM1 mutant localizing in the cytoplasma inhibits PTEN or Bax, and induces the survival or proliferation of the cells, playing rather class I like roles.41,42) At present, it remains elusive how NPM1 mutations contribute to MDS pathogenesis and leukemogenesis.
Whether Evi1 overexpression plays class II-like roles is also not clear. In our hands, overexpression of Evi1 rather induced AML with a long latency in the transplanted mice in concert with C/EBP β overexpression caused by retroviral integration.43) In addition, Evi1 represses PTEN, leading to the activation of the PI3K-Akt pathway,44) suggesting that overexpression of Evi1 rather plays a class I-like role. Consistent with the class I/class II hypothesis, in our BMT model, a class II mutation Runx1 mutant D171N (Runx1-D171N) induced MDS-like symptoms and that co-expression of Evi1 with the mutant Runx1 induced AML in the transplanted mice,45) again suggesting that Evi1 overexpression plays class I-like roles. In fact, one patient out of fifteen with MDS/MPN harboring Runx1 mutations was found to express a high amount of Evi1.46) This patient’s MDS cells harbored D171G mutation and evolved to AML 10 months after the onset of MDS. Thus, our mouse model well recapitulated the disease phenotype and course of this patient. Expression profile has revealed that a substantial number of MDS patients with Runx1 mutations displayed Bmi1 overexpression.46) Indeed, co-expression of Runx1-D171N and Bmi1 induced AML with shorter latencies when compared with MDS or MDS/AML induced by the Runx1 mutant alone. Bmi1 is a component of polycomb 1 complex (PRC1) and plays important roles in inhibiting the expression of tumor suppressor genes such as p16 and p19. In fact, mouse MDS/AML induced by Runx1-D171N and Bmi1 displayed reduced levels of expression of p16 and p19. Thus, overexpression of Bmi1 would play a class I-like role by inducing cell cycling via suppression of cell cycle inhibitors p16 and p19. In summary, experimental results as well as clinical observations suggest that class II mutations induce MDS and that acquisition of additional class I mutation(s) leads to the progression of MDS to MDS/AML, although how Bmi1 or Evi1 is upregulated remains elusive.
The class I/class II hypothesis cannot explain the whole story
MLL1 is a methyltransferase of histone H3K4, forms a variety of fusion genes with more than 60 genes47,48) in leukemia and was originally categorized as a class II mutation. However, most MLL-fusions are able to induce acute leukemia by itself in mouse models although there are several reports that MLL fusions collaborate with class I mutations in inducing acute leukemia.8–11) In addition, it is known that MLL fusions play critical roles in inducing the expression of posterior HOXA genes and Meis 1 through recruiting Menin and LEDGF to these genes.49) Recently, a histone 3 lysine 79 (H3K79) methyltransferase DOT1L has been implicated in MLL-AF9-induced leukemogenesis; DOT1L induces the expression of MLL-AF9 target genes through H3K79me2.50) These results indicate that MLL fusions are not typical class II mutations. Instead, MLL fusions should be categorized to a different entity together with some of the mutations recently identified in a variety of epigenetics-related genes as described later.
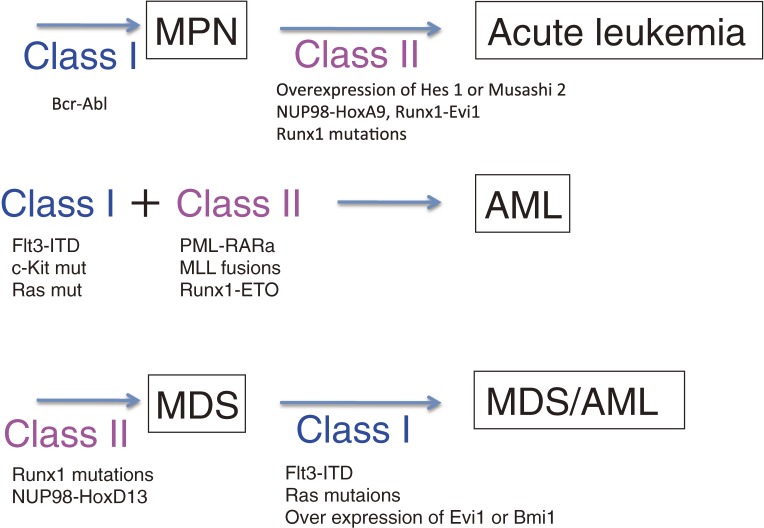
The hypothesis for the etiology of hematopoietic malignancies shown in Fig. 1 seemed to be consistent with the clinical observations. However, this is a simplified model, and there are obviously exceptions such as double ph1 in which two identical class I mutations (Bcr-Abl) induce blast crisis in CML, progression of MPN to AML. In addition, as described later in this review, recent technical progress in high-speed sequencing and SNP (single nucleotide polymorphism) microarray has identified a number of gene mutations in patients with hematological malignancies that are not categorized as either class I or class II mutations.
Figure 1.
Class I/class II hypothesis for leukemogenesis. Class I mutations Bcr-Abl or JAK2-V617F induce CML or MPN, respectively, and class II mutations such as Runx1 mutations, Nup98-HoxA9 and Runx1-Evi1 play crucial roles in inducing blast crisis of CML. Hes1 overexpression may also play roles as a class II mutation by suppressing C/EBPa. On the other hand, class II mutations such as Runx1 mutations induce MDS. Class I mutations including FLT3-ITD and activating mutants of N-Ras may play roles in the progression of MDS to leukemia. Overexpression of Evi1 or Bmi1 may play class I-like roles by repressing PTEN or p16/p19, respectively.
Novel mutations identified in hematological malignancies
With the recent progress in SNP microarray and high-speed sequencing, mutations of a variety of molecules have been identified in myeloid malignancies, including AML, MDS and MPN.51–53) Most of the newly identified mutations are not categorized as typical class I or class II mutations and include mutations of splicing factors, epigenetic factors, signaling proteins and molecules of the cohesin complex (Table 2). In addition, many patients with AML as well as MPN and MDS harbor more than two mutations. These findings have lead some researchers to propose to divide these mutations to four classes including mutations inducing clonal dominance of hemopoietic stem cells (Runx1 and TET2), those promoting a pathway to leading towards either to primary or secondary leukemia (ASXL1 and NPM1), those associated with signaling and proliferation (CBL, FLT3, JAK2, Ras), and those associated with acute phase (IDH1/2, WT1).54) Hoffmeister has proposed 5 classes of mutations including mutations in epigenetic molecules (class III), adhesion molecules (class IV) and DNA repair/RNA splicing (class V) in addition to classical class I and II mutations.55) However, these classifications were not based on the experimental results but were mostly dependent on speculation. Therefore, its biological significance awaits verification.
Table 2.
Novel mutations identified in hematological malignancies
| Spliceosomal proteins: |
| SF3B1, SRSF2, U2AF1, ZRSRS2, SF3A1, U2AF65, SF1 |
| Epigenetic factors: |
| TET2, IDH1/IDH2, ASXL1/ASXL2, EZH2, DNMT3A, EED, Suz12 |
| Signaling proteins: |
| CBL, Calreticulin (CALR), SETBP1, GNAS, RHOA |
| Cohesin complex and related proteins: |
| STAG2, SMC1A, SMC3, RAD21, CTCF1 |
The novel mutations are found in a wide range of myeloid malignancies including AML, MDS, and MPN. Some mutations are specific to certain types of diseases. For instance, splicesome mutations are found in MDS more frequently than de novo AML and MPN.56) Moreover, among splicesome mutations, SF3B1 mutations are closely associated with a specific type of MDS, RARS (refractory anemia with ringed sideroblast); 57–75% of RARS patients harbor SF3B1 mutations.57,58) On the other hand, SRSF2 mutations are closely associated with CMML; 28.4% of patients with CMML harbor SRSF2 mutations.58) EZH2 mutations are more frequently found in MDS patients and are rare in de novo AML patients.51,59) In addition, there are some frequent combinations of mutations in AML. Currently, the biological meanings of these novel mutations and their combinations are being extensively studied in relation to the pathogenesis of myeloid malignancies as described below.
Functional consequences of newly found mutations: loss-of-function mutations
The pathological meanings of the newly identified gene mutations are now being extensively sought using knockout mice, knock-in mice, and mouse BMT models. Some of these mutations are thought to be loss-of-function mutations; thus knockout mice have been used for the investigation of the functional consequences of these mutations. Gene deprivation of TET2 (TET2 knockout) expanded hematopoietic stem cells (HSCs) and progenitors, eventually leading to myeloproliferation, MPN and CMML,60,61) suggesting a class I-like feature of TET2 deprivation. Conditional knockout of the polycomb-related genes EZH2 or ASXL1 has been shown to induce MDS-like symptoms in mice.62,63) EZH2 is a component of PRC2 and is a histone methyltransferase responsible for tri-methylation of lysine 27 of histone H3 (H3K27me3), a representative repressive mark of histone modification. ASXL1 binds EZH2 and other members of polycomb complex 2 (PRC2) and is required for PRC2-mediated gene repression.64) However, gene deprivation of EZH2 did not induce derepression of HoxA9 expression unlike that of ASXL1 despite the fact that EZH2 and ASXL1 collaborate in repressing gene expression through EZH2-mediated H3K27me3. Although the reason for this discrepancy are not yet clarified, an EZH2 homologue EZH1 may compensate for some functions of EZH2 in the absence of EZH2, but may not function in the absence of ASXL1. Alternatively, ASXL1 may have PRC2-independent functions as well. In fact, ASXL1-KO reduced H3K4me3 a representative positive mark in addition to H3K27me3 although the underlying molecular mechanism remains elusive.65) In relation to this, it is known that ASXL1 plays dual roles in inhibiting and activating the transcription.66,67)
Gene deletion of EED, another component of PRC2 complex required for EZH2-mediated H3K27me3, has been reported to induce pancytopenia and increased apoptosis via derepression of p16 and p19 tumor suppressors as well as several EZH2 target genes HoxC4, Id2, Sox7 and Noxa.68) In a conditional knockout mouse, hematopoietic cells quickly declined after the deprivation of EED. Therefore, the loss-of-function mutation of EED must be accompanied with other gene mutations that would rescue the survival of hematopoietic cells. As for the mutations of the molecules involved in splicing machinery, it has been suggested that these mutations either induce genomic instability or abnormal splicing of specific genes57,58) although it is not clear whether they are loss-of-function mutations or gain-of-function mutations. Recently, it has been reported that U2AF1 mutations induces missplicing of specific genes69) but the relationship between the missplicing of specific genes and MDS pathogenesis remains to be clarified.
Thus, some of the newly found mutations induce MPN or MDS but not AML in mouse models, suggesting that multiple mutations would be required for AML to develop. In fact, the ASXL1 mutation or deletion and the activated form of N-Ras (N-Ras-G12V) have been shown to co-operate in inducing leukemia.64,70) On the other hand, some mutations by themselves do not seem to induce hematological malignancies in mice, suggesting the possibility that the number of the mutations required for MDS or MPN pathogenesis may vary depending on the kinds of the mutations.
Functional consequences of newly found mutations: gain-of-function or dominant-negative mutations
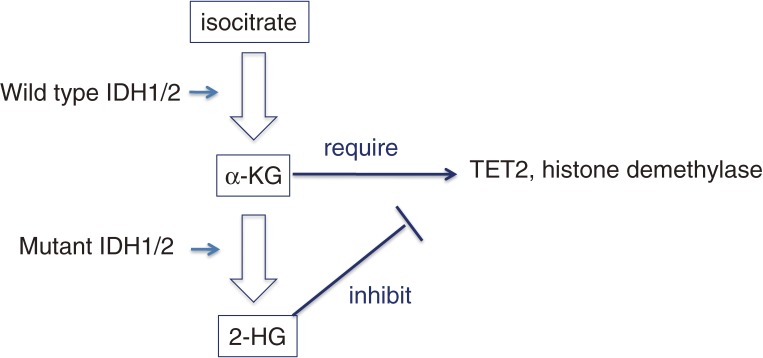
Some of the newly identified mutations are gain-of-function or dominant-negative mutations. First, IDH1/2 (isocitrate dehydrogenase 1/2) mutations are gain-of-function mutations; wide type IDH1/2 converts isocitrate to α-ketoglutarate, and the mutant IDH1/2 converts α-ketoglutarate (αKG) to 2-hydroglutarate (2HG), which inhibits TET2 as well as αKG-dependent dioxygenases and histone demethylases (Fig. 2).71) In addition, IDH1/2 mutations and TET2 mutations are mutually exclusive and both mutations induce the hypermethylation phenotype,72) identifying an unexpected link between IDH1/2 mutations and TET2. The most frequent mutant, IDH1-R132H, alone did not induce hematological malignancies in a mouse BMT model; however, it did collaborate with HoxA9 in accelerating the induction of MPN-like disease in the transplanted mice.73) Knock-in of the same mutant resulted in increased hemopoietic progenitors, but again did not induce MPN or AML.74) Similarly, transgenic expression of IDH2-R140Q resulted in an increased number of hemopoietic progenitor cells and aberrant hematopoiesis in vivo, but did not induce any hematological malignancies. Interestingly, IDH2-R140Q was found to collaborate with HoxA9/Meis1 or FLT3-ITD in inducing leukemia.75) In summary, IDH1/2 mutants increase the number of progenitor cells, do not induce MPN or AML on their own, but collaborate with class I-like mutations in inducing MPN or AML.
Figure 2.
An example of gain-of-function mutations.
Second, mutations of SETBP1 have been identified in a variety of myeloid malignancies.76–79) The mutations are restricted to the SKI-homologous region, which retards degradation of SETBP1. The resulting increase in SETBP1 expression stabilizes the nuclear oncoprotein SET, which is an inhibitor of PP2A, leading to the suppression of this tumor suppressor and the activation of the intracellular signaling pathways, including the PI3K-Akt pathway. It has also been reported that SETBP1 directly activates transcription of HoxA9 and HoxA10.80) HoxA9 is known to confer a growth advantage for hemopoietic stem cells.81) In addition, as described below, SETBPT-mutant (SETBPI-MT) collaborates with ASXLT-MT in inducing AML in the BMT model.82) These results imply that the SETBP1 mutations, or overexpression of SETBP1 have class I-like features.
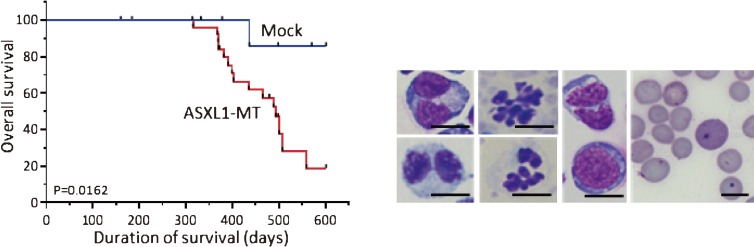
Third, mutations of ASXL1 in patients with myeloid malignancies are almost exclusively found in the 5′ end of the last exon. Most are frame-shift mutations, which are thought to produce C-terminal truncated forms of ASXL1.83,84) In addition, the mutations are always heterozygous. These facts suggested that ASXL1 mutations are dominant-negative or gain-of-function mutations.85) Consistent with this, we have shown that bone marrow cells transduced with the C-terminal truncated ASXL1 mutant (ASXL1-MT) induced MDS-like symptoms in the transplanted mice (Fig. 3).70) Concerning the underlying molecular mechanisms, ASXL1-MT inhibits EZH2-mediated trimethylation of lysine 27 of histone H3 (H3K27me3), derepressing posterior Hox genes, including HoxA9, as well as oncogenic miRNA miR125a. miR125a targets the expression of Clec5a/MDL1 which is required for granulocytic differentiation.70) In fact, the ASXL1-MT inhibition of G-SCF-induced granulocytic differentiation of 32D cells was rescued by Clec5a expression. Given that ASXL1-MT inhibited differentiation of hematopoietic cells, ASXL1-MT may play class II-like roles.70) Therefore, it is reasonable that ASXL1-MT collaborates with N-Ras-G12V or SETBPI-MT,82) a class I mutation, in inducing leukemia.
Figure 3.
Expression of the ASXL1 mutant with C-terminal truncation induced MDS-like diseases in mouse BMT model. Left panel: After transplantation of mouse bone marrow cells transduced with the ASXL1-MT, the transplanted mice developed MDS-like with long latencies. Right panel: These mice displayed morphological abnormalities of myeloid cells typical for MDS including Pelger-Huet anomaly, and hypersegmented nuclei of neutrophil, and polychromasia, Howel-Jolly body and anisopoikilocytosis of erythrocytes.
Fourth, a mutant of DNA methyltransferase DNMT3A-R882H induced CMML with thrombocytosis in a BMT model through aberrant DNA methylation and increased expression of several genes including MPL, the thrombopoietin receptor.86) DNMT3A-R882H mutant lacks the DNA methyl transferase activity but still binds the wild-type DNMT3A and DNMT3B, suggesting that R882H is a dominant-negative mutant.87) In addition, interestingly, DNMT3A-R882H could form a complex with cyclin-dependent kinase 1 (CDK1) with higher capacity than wild type DNMT3A, leading to increased CDK1 protein and enhanced cell cycle progression, suggesting a gain-of-function as well.86) Although further characterization is required, it is possible that DNMT3A-R882H plays class I-like roles.
Finally, the recently found mutations of calreticulin (CALR) always result in +1 frame shift, which results in the expression of the common novel C-terminus. Although how CALR mutations contribute to the pathogenesis of MPN remain elusive at present, these mutations have been shown to induce the activation of STAT5.29) In addition, CALR mutations are exclusive with activating mutations of JAK2 and MPL, indicating that the CALR mutation is a classical class I mutation. In addition to these five examples described above, mutations in splicing machinery (U2AF1, SF3B1) are thought to be gain-of-function mutations limited to particular residues that are conserved among species.69)
In summary, functional studies of the newly identified mutations have just begun, and it is clear from the initial reports that newly identified mutations are also involved in multi-step leukemogenesis. Further research will be required to reveal the interactions among various mutations, which would like lylead to a new hypothesis for leukemogenesis.
Are multiple mutations required for pathogenesis of MDS and MPN?
It is now well accepted that multiple gene alterations are required for leukemogenesis. Are multiple mutations required for the etiologies of MDS and MPN as well? Both MDS and MPN are sets of heterogenous diseases. As described above, there are many mouse models of MDS and MPN. In most models, a single mutant gene, such as the ASXL1 mutant, Runx1 mutant or Nup98-HoxD13 fusion gene for MDS and Bcr-Abl or JAK2V617F for MPN, was able to induce the disease by itself, with some cases progressing to overt leukemia.38,45,70) These results suggest that MDS and MPN could be induced by a single mutation or chromosomal translocation. Consistent with this idea, patients with early phase MDS, MDS-RA, harbor one or a little more than 1 mutation on average.88) On the other hand, more than two mutations are identified in most patients with progressed MDS (MDS-RAEB: RA with excess blasts) and MDS/AML.88) It is not clear at present if multiple gene alterations are required for the development of MDS or MPN, but there are some frequent combinations — such as ASXL1 and SETBP1 or N-Ras76,78) or RUNX1 and STAG2 or ASXL1 for MDS89) — suggesting that some of these combinations functionally collaborate in MDS etiology. However, among these combinations, ASXL1-MT collaborates with the N-Ras mutant70) as well as the SETBP1 mutation82) in inducing AML rather than MDS, indicating that the Ras mutant or the SETBP1 mutant is required for the progression of MDS to AML but not to pathogenesis of MDS. Our result thus indicates that ASXL1-MT induced MDS as a class II-like mutation and co-operated with class I mutations in inducing acute leukemia. Based on these results, we assume that one driver mutation (class II-like) would in some cases be enough to induce MDS, and it is possible that the mutations of TET2/DNMT3A as well as splicesomal proteins inhibit the expression of important genes, leading to the loss-of-function mutations, or induce genome instability, leading to causative mutations such as ASXL1 or Runx1. In relation to this, Abdel-Wahab and colleagues have demonstrated that expression of an SRSF2 mutant in a knock-in mouse model suppressed the normal hematopoiesis in a week or so, by inhibiting E2H2 expression (personal communication). On one hand, ASXL1 mutations are frequently associated with mutations of Runx1 or EZH2. Although these mutations seemed to induce MDS in mouse models by themselves, it requires long latencies, from several months to 2 years, suggesting the possibility that additional mutations incorporated in the course of the experiment contribute to MDS pathogenesis, which is now under investigation.
Concerning MPN, it is possible that both survival and proliferative signals are required for pathogenesis. Bcr-Abl and JAK2V617F could deliver both signals simultaneously by activating multiple signaling pathways. Another important factor may be where the mutations are incorporated; Hematopoietic stem cells (HSC) has self-renewal ability while committed progenitors CMP (common myeloid progenitor) and GMP (granulocyte macrophage progenitor) do not. Indeed, Bcr-Abl can transform HSC but not CMP or GMP.90) Functional meanings of the combination of different mutations have to be further investigated for MPN pathogenesis. Further complexity comes from the situations where several independent clones exist in some patients, one of which could become dominant in the late phase of the disease.91,92) Intriguingly, in some patients with MPN, one of the subclones without JAK2V617F evolves to acute leukemia.92)
Progression of MDS and MPN to acute leukemia
It is well known that MDS and MPN frequently evolve to acute leukemia. In particular, CML eventually evolves to acute leukemia (blast crisis) in most patients after a long chronic phase. Blast crisis (CML-BC) is almost always associated with additionally acquired gene alterations including double Ph1, NUP98-HoxA9 caused by t(7;11), Runx1-Evi1 caused by t(3;21), and Runx1 mutations,93,94) indicating that additional gene alterations are required for CML progression. Although the underlying mechanisms for the up-regulation remains elusive, up-regulation of Musashi 2 or Hes1 has been shown to play a critical role in CML-BC by inhibiting the differentiation of CML cells.95,96) It is conceivable that overexpression of Musashi 2 or Hes1 plays class II-like roles and co-operates with BCR-ABL a class I mutation in inducing CML-BC (Fig. 1). Unlike CML, probabilities of leukemic transformation of other MPNs are relatively low (few %).92) It has been recently reported that the presence of two or more mutations reduced overall survival and increased the risk of transformation of MPN into AML.92) Interestingly, the number of mutations did not significantly change between early and late MPN samples, and most somatic mutations were present when the diagnosis of MPN was made. These results suggest that unlike CML, other MPNs (PV, ET and CMF) already harbor the set of mutations at diagnosis, that the diseases do not induce genome instability, and that leukemic transformation is predetermined by the combination of mutations at diagnosis. For example, somatic mutations in TP53 and TET2 at diagnosis were reported to be associated with the risk for leukemic transformation. Thus, information about the mutations at diagnosis would be clinically important in choosing therapeutic strategies, including stem cell transplantation.
The situations are completely different in MDS; the number of mutations seems to increase with the disease progression.88,97) Interestingly, the additionally acquired mutations in patients with MDS-RAEB and those with MDS/AML are mostly class I mutations, including FLT3-ITD, cKit-D816 mutations, and N-Ras mutations.97) Recently, high-speed sequencing has shown that Runx1 and SMC3 mutations could also be acquired in some patients in the progression of MDS to AML in addition to class I mutations such as WT1 and PTPN11.91) Among gene alterations found in MDS, mutations of the genes involved in DNA methylation and mRNA splicing — including TET2, IDH2, DNMT3a, SF3B1, and U2AF1 — are thought to be present in the early phase of the disease. On the other hand, mutations of molecules involved in signaling, such as N-Ras and c-Kit, are thought to be acquired in the late phase.89,97) As discussed above, the preceding mutations in the molecules involved in DNA modification and RNA splicing might induce genome instability, leading to acquisition of additional or causative mutations in the later phase of the disease. The fact that TET2/DNMT3a and splicesome mutations are found in MDS patients more frequently than in MPN patients is consistent with the above findings that MDS patients tend to acquire additional mutations during the course of the disease, leading to the leukemic transformation.
Conclusion
In this article, we have reviewed the relationship between hematopoietic malignancies and the mutations identified in various molecules. In principle, all mutations will contribute to the pathogenesis through altering the gene expression, either down-regulation or up-regulation. In particular, mutations in epigenetic factors and splicing machinery could lead to changes in the expression levels of a wide variety of genes. So it is possible that the disregulated expressions of multiple genes, caused by a mutation of one protein, would collaborate in inducing hematological malignancies.
Newly identified mutations have been seen as distinct from “classical” class I or class II mutations. However, some of them could be categorized as either class I or class II mutation. For example, as we demonstrated, the truncated mutant of ASXL1 (ASXL1-MT) blocked granulocytic differentiation, induced an MDS-like disease in the transplanted mice and collaborated with the active mutant of Ras (class I mutation) in inducing leukemia,70) indicating that ASXL1-MT is equivalent to the “classical” class II mutation. SETBP1 mutations protect SETBP1 from protein degradation and increase its expression levels, leading to PP2A inhibition and activation of the PI3K-Akt pathway, which induces cell cycle progression, suggesting that this mutation belongs to class I.76,78) Intriguingly, we have recently found that SETBPI-MT is found almost exclusively in advanced MDS patients harboring ASXLI-MT, and that the combination of the SETBP1 mutant and ASXL1-MT, and that quickly induced AML in the transplanted mice.82) These results match well with the original hypothesis (Fig. 1). However, many examples do not match the class I/class II hypothesis. For example, the IDH1 mutants increased the HSC number, suggesting the class I-like feature of the IDH1 mutant, but did not induce any disease on their own.72–74) Combination of the IDH1 mutant and HoxA9 accelerated MPN in mice.75) Together, the IDH1 mutant could be a “weak” class I mutation. On the other hand, the combination of the IDH1 mutant and either HoxA9/Meis1 or FLT3-ITD induced AML in mouse models,75) suggesting that the IDH1 mutant also has a class II-like feature.
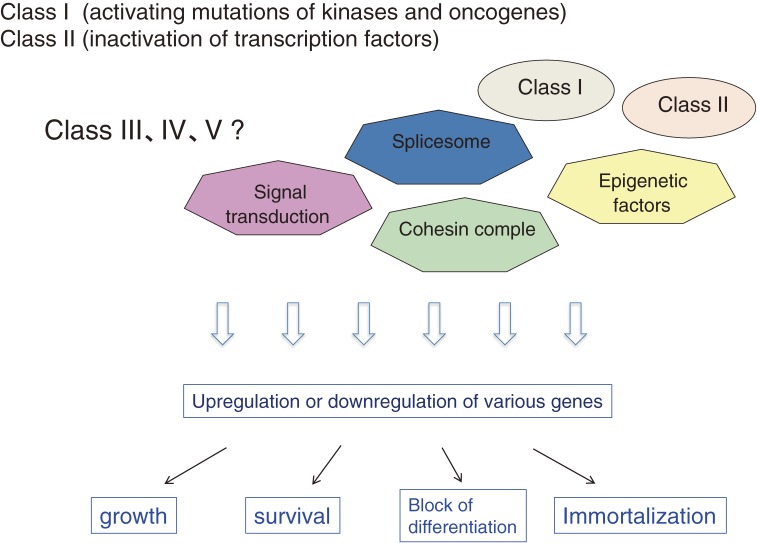
Thus, it would be difficult to classify the mutations by two simple cell fates (induction of proliferation and inhibition of differentiation) caused by the mutations. To understand the molecular basis of hematological malignancies, we might do better to classify the cell fate into 4 categories, including proliferation, cell survival, block of differentiation, and immortalization. These cell fates are determined by the changes of gene expression profiles caused by a variety of mutations identified in hematological malignancies (Fig. 4). In addition, the disease phenotypes will be determined by the combinations of the cell fate changes. For example, the combination of proliferation and immortalization will lead to the development of MPN, and the combination of either cell survival or immortalization and block of differentiation may lead to the development of MDS. In addition, we also have to take into account the cell origin of mutations; hematopoietic stem cell but not progenitors already have self-renewability and may not need the immortalization process. Recent progress in gene set enrichment analysis and gene ontology analysis will greatly contribute to our ability to correlate the changes in cell fate with those in gene expression profiles of hematological malignancies, eventually leading to the comprehensive understanding of the molecular basis of these diseases.
Figure 4.
Molecular basis of hematological malignancies: working hypothesis. Disease phenotypes are determined by the disregulated expression of various genes caused by a variety of gene mutations. We assume that biological outcomes caused by the disregulated expression are determined by but are more important than the kind of mutant genes when we think about the molecular basis of hematological malignancies. “Proliferation” is caused by strong class I mutations while “survival” may be caused by weak class I mutations. “Immortalization” is caused by class II mutations while differentiation block may be caused by weak class II mutations.
Acknowledgment
We thank Dr. Dovie Wylie for her excellent work to edit the manuscript. This work was supported by Grants-in-Aid for Scientific Research on Innovative Areas and Grants-in-Aid for Scientific Research (A) from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan.
Profile
Toshio Kitamura was born in Osaka in 1956, and graduated Faculty of Medicine, University of Tokyo in 1981. After 2 years training as a physician (internal medicine), he started his career as a scientist in retrovirology and hematology in National Cancer Center in Japan. After 2 years research training, he returned to the Third Department of Internal Medicine, University of Tokyo (Professor Fumimaro Takaku), and started a project for identification of cytokine receptors while working as a clinical hematologist. He established a cytokine-dependent cell line TF-1, which is now widely used in the field, and using TF-1, formed a hypothesis that IL-3 and GM-CSF receptors share a common subunit. He then moved to California in 1989 and joined Atsushi Miyajima’s lab in DNAX Research Institute as a postdoctoral fellow, where he isolated human cDNAs for IL-3 and GM-CSF receptors and presented the first example for the shared subunit of cytokine receptors. He also established a retrovirus-mediated expression cloning method, and started a small laboratory in DNAX Research Institute working on the cytokine signal transduction. In 1996, he returned to Japan to establish a new laboratory (donation lab) in the Institute of Medical Science, The University of Tokyo (IMSUT), and was promoted to a professor of the division of Cellular Therapy. In IMSUT, he has performed several projects using the retrovirus-mediated high-efficiency gene transfer and expression cloning, and made a number of achievements in a variety of fields including hematology (molecular basis of hematological malignancies), cell biology (cell differentiation, cell division and cellular signaling) and immunology (allergy and paired IG receptors). In addition to the scientific achievements, he widely distributed the efficient retrovirus system (pMX vectors and PLAT packaging cell lines) developed by his group to the scientific community. The retrovirus system that his group established is now used in more than 5,000 laboratories in the world.
References
- 1).Bartram C.R., de Klein A., Hagemeijer A., van Agthoven T., Geurts van Kessel A., Bootsma D., Grosveld G., Ferguson-Smith M.A., Davies T., Stone M., Heisterkamp N., Stephenson J.R., Groffen J. (1983) Translocation of c-ab1 oncogene correlates with the presence of a Philadelphia chromosome in chronic myelocytic leukaemia. Nature 306, 277–280. [DOI] [PubMed] [Google Scholar]
- 2).Wong S., Witte O.N. (2001) Modeling Philadelphia chromosome positive leukemias. Oncogene 20, 5644–5659. [DOI] [PubMed] [Google Scholar]
- 3).Van Etten R.A. (2001) Retroviral transduction models of Ph+ leukemia: advantages and limitations for modeling human hematological malignancies in mice. Blood Cells Mol. Dis. 27, 201–205. [DOI] [PubMed] [Google Scholar]
- 4).Grisolano J.L., Wesselschmidt R.L., Pelicci P.G., Ley T.J. (1997) Altered myeloid development and acute leukemia in transgenic mice expressing PML-RAR alpha under control of cathepsin G regulatory sequences. Blood 89, 376–387. [PubMed] [Google Scholar]
- 5).Kelly L.M., Kutok J.L., Williams I.R., Boulton C.L., Amaral S.M., Curley D.P., Ley T.J., Gilliland D.G. (2002) PML/RARalpha and FLT3-ITD induce an APL-like disease in a mouse model. Proc. Natl. Acad. Sci. U.S.A. 99, 8283–8288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6).Kelly L.M., Liu Q., Kutok J.L., Williams I.R., Boulton C.L., Gilliland D.G. (2002) FLT3 internal tandem duplication mutations associated with human acute myeloid leukemias induce myeloproliferative disease in a murine bone marrow transplant model. Blood 99, 310–318. [DOI] [PubMed] [Google Scholar]
- 7).Gilliland D.G., Griffin J.D. (2002) The roles of FLT3 in hematopoiesis and leukemia. Blood 100, 1532–1542. [DOI] [PubMed] [Google Scholar]
- 8).Stubbs M.C., Kim Y.M., Krivtsov A.V., Wright R.D., Feng Z., Agarwal J., Kung A.L., Armstrong S.A. (2008) MLL-AF9 and FLT3 cooperation in acute myelogenous leukemia: development of a model for rapid therapeutic assessment. Leukemia 22, 66–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9).Moriya K., Suzuki M., Watanabe Y., Takahashi T., Aoki Y., Uchiyama T., Kumaki S., Sasahara Y., Minegishi M., Kure S., Tsuchiya S., Sugamura K., Ishii N. (2012) Development of a multi-step leukemogenesis model of MLL-rearranged leukemia using humanized mice. PLoS ONE 7, e37892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10).Ono R., Kumagai H., Nakajima H., Hishiya A., Taki T., Horikawa K., Takatsu K., Satoh T., Hayashi Y., Kitamura T., Nosaka T. (2009) Mixed-lineage-leukemia (MLL) fusion protein collaborates with Ras to induce acute leukemia through aberrant Hox expression and Raf activation. Leukemia 23, 2197–2209. [DOI] [PubMed] [Google Scholar]
- 11).Ono R., Nakajima H., Ozaki K., Kumagai H., Kawashima T., Taki T., Kitamura T., Hayashi Y., Nosaka T. (2005) Dimerization of MLL fusion proteins and FLT3 activation synergize to induce multiple-lineage leukemogenesis. J. Clin. Invest. 115, 919–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12).Nick H.J., Kim H.G., Chang C.W., Harris K.W., Reddy V., Klug C.A. (2012) Distinct classes of c-Kit-activating mutations differ in their ability to promote RUNX1-ETO-associated acute myeloid leukemia. Blood 119, 1522–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13).Kagiyama Y., Kitaura J., Togami K., Uchida T., Inoue D., Matsukawa T., Izawa K., Kawabata K.C., Komeno Y., Oki T., Nakahara F., Sato K., Aburatani H., Kitamura T. (2012) Upregulation of CD200R1 in lineage-negative leukemic cells is characteristic of AML1-ETO-positive leukemia in mice. Int. J. Hematol. 96, 638–648. [DOI] [PubMed] [Google Scholar]
- 14).Gilliland D.G. (2001) Hematologic malignancies. Curr. Opin. Hematol. 8, 189–191. [DOI] [PubMed] [Google Scholar]
- 15).Ye M., Zhang H., Amabile G., Yang H., Staber P.B., Zhang P., Levantini E., Alberich-Jorda M., Zhang J., Kawasaki A., Tenen D.G. (2013) C/EBPa controls acquisition and maintenance of adult haematopoietic stem cell quiescence. Nat. Cell Biol. 15, 385–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16).Nerlov C. (2004) C/EBPalpha mutations in acute myeloid leukaemias. Nat. Rev. Cancer 4, 394–400. [DOI] [PubMed] [Google Scholar]
- 17).Pabst T., Mueller B.U. (2007) Transcriptional dysregulation during myeloid transformation in AML. Oncogene 26, 6829–6837. [DOI] [PubMed] [Google Scholar]
- 18).Bereshchenko O., Mancini E., Moore S., Bilbao D., Mansson R., Luc S., Grover A., Jacobsen S.E., Bryder D., Nerlov C. (2009) Hematopoietic stem cell expansion precedes the generation of committed myeloid leukemia-initiating cells in C/EBPalpha mutant AML. Cancer Cell 16, 390–400. [DOI] [PubMed] [Google Scholar]
- 19).Kato N., Kitaura J., Doki N., Komeno Y., Watanabe-Okochi N., Togami K., Nakahara F., Oki T., Enomoto Y., Fukuchi Y., Nakajima H., Harada Y., Harada H., Kitamura T. (2011) Two types of C/EBPalpha mutations play distinct but collaborative roles in leukemogenesis: lessons from clinical data and BMT models. Blood 117, 221–233. [DOI] [PubMed] [Google Scholar]
- 20).Levine R.L., Wadleigh M., Cools J., Ebert B.L., Wernig G., Huntly B.J., Boggon T.J., Wlodarska I., Clark J.J., Moore S., Adelsperger J., Koo S., Lee J.C., Gabriel S., Mercher T., D’Andrea A., Frohling S., Dohner K., Marynen P., Vandenberghe P., Mesa R.A., Tefferi A., Griffin J.D., Eck M.J., Sellers W.R., Meyerson M., Golub T.R., Lee S.J., Gilliland D.G. (2005) Activating mutation in the tyrosine kinase JAK2 in polycythemia vera, essential thrombocythemia, and myeloid metaplasia with myelofibrosis. Cancer Cell 7, 387–397. [DOI] [PubMed] [Google Scholar]
- 21).James C., Ugo V., Le Couedic J.P., Staerk J., Delhommeau F., Lacout C., Garcon L., Raslova H., Berger R., Bennaceur-Griscelli A., Villeval J.L., Constantinescu S.N., Casadevall N., Vainchenker W. (2005) A unique clonal JAK2 mutation leading to constitutive signalling causes polycythaemia vera. Nature 434, 1144–1148. [DOI] [PubMed] [Google Scholar]
- 22).Baxter E.J., Scott L.M., Campbell P.J., East C., Fourouclas N., Swanton S., Vassiliou G.S., Bench A.J., Boyd E.M., Curtin N., Scott M.A., Erber W.N., Green A.R. (2005) Acquired mutation of the tyrosine kinase JAK2 in human myeloproliferative disorders. Lancet 365, 1054–1061. [DOI] [PubMed] [Google Scholar]
- 23).Kralovics R., Passamonti F., Buser A.S., Teo S.S., Tiedt R., Passweg J.R., Tichelli A., Cazzola M., Skoda R.C. (2005) A gain-of-function mutation of JAK2 in myeloproliferative disorders. N. Engl. J. Med. 352, 1779–1790. [DOI] [PubMed] [Google Scholar]
- 24).Tiedt R., Hao-Shen H., Sobas M.A., Looser R., Dirnhofer S., Schwaller J., Skoda R.C. (2008) Ratio of mutant JAK2-V617F to wild-type Jak2 determines the MPD phenotypes in transgenic mice. Blood 111, 3931–3940. [DOI] [PubMed] [Google Scholar]
- 25).Shide K., Kameda T., Markovtsov V., Shimoda H.K., Tonkin E., Fang S., Liu C., Gelman M., Lang W., Romero J., McLaughlin J., Bhamidipati S., Clough J., Low C., Reitsma A., Siu S., Pine P., Park G., Torneros A., Duan M., Singh R., Payan D.G., Matsunaga T., Hitoshi Y., Shimoda K. (2011) R723, a selective JAK2 inhibitor, effectively treats JAK2V617F-induced murine myeloproliferative neoplasm. Blood 117, 6866–6875. [DOI] [PubMed] [Google Scholar]
- 26).Pikman Y., Lee B.H., Mercher T., McDowell E., Ebert B.L., Gozo M., Cuker A., Wernig G., Moore S., Galinsky I., DeAngelo D.J., Clark J.J., Lee S.J., Golub T.R., Wadleigh M., Gilliland D.G., Levine R.L. (2006) MPLW515L is a novel somatic activating mutation in myelofibrosis with myeloid metaplasia. PLoS Med. 3, e270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27).Lasho T.L., Pardanani A., Tefferi A. (2010) LNK mutations in JAK2 mutation-negative erythrocytosis. N. Engl. J. Med. 363, 1189–1190. [DOI] [PubMed] [Google Scholar]
- 28).Nangalia J., Massie C.E., Baxter E.J., Nice F.L., Gundem G., Wedge D.C., Avezov E., Li J., Kollmann K., Kent D.G., Aziz A., Godfrey A.L., Hinton J., Martincorena I., Van Loo P., Jones A.V., Guglielmelli P., Tarpey P., Harding H.P., Fitzpatrick J.D., Goudie C.T., Ortmann C.A., Loughran S.J., Raine K., Jones D.R., Butler A.P., Teague J.W., O’Meara S., McLaren S., Bianchi M., Silber Y., Dimitropoulou D., Bloxham D., Mudie L., Maddison M., Robinson B., Keohane C., Maclean C., Hill K., Orchard K., Tauro S., Du M.Q., Greaves M., Bowen D., Huntly B.J., Harrison C.N., Cross N.C., Ron D., Vannucchi A.M., Papaemmanuil E., Campbell P.J., Green A.R. (2013) Somatic CALR mutations in myeloproliferative neoplasms with nonmutated JAK2. N. Engl. J. Med. 369, 2391–2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29).Klampfl T., Gisslinger H., Harutyunyan A.S., Nivarthi H., Rumi E., Milosevic J.D., Them N.C., Berg T., Gisslinger B., Pietra D., Chen D., Vladimer G.I., Bagienski K., Milanesi C., Casetti I.C., Sant’Antonio E., Ferretti V., Elena C., Schischlik F., Cleary C., Six M., Schalling M., Schonegger A., Bock C., Malcovati L., Pascutto C., Superti-Furga G., Cazzola M., Kralovics R. (2013) Somatic mutations of calreticulin in myeloproliferative neoplasms. N. Engl. J. Med. 369, 2379–2390. [DOI] [PubMed] [Google Scholar]
- 30).MacKenzie K.L., Dolnikov A., Millington M., Shounan Y., Symonds G. (1999) Mutant N-ras induces myeloproliferative disorders and apoptosis in bone marrow repopulated mice. Blood 93, 2043–2056. [PubMed] [Google Scholar]
- 31).Cools J., Stover E.H., Boulton C.L., Gotlib J., Legare R.D., Amaral S.M., Curley D.P., Duclos N., Rowan R., Kutok J.L., Lee B.H., Williams I.R., Coutre S.E., Stone R.M., DeAngelo D.J., Marynen P., Manley P.W., Meyer T., Fabbro D., Neuberg D., Weisberg E., Griffin J.D., Gilliland D.G. (2003) PKC412 overcomes resistance to imatinib in a murine model of FIP1L1-PDGFRalpha-induced myeloproliferative disease. Cancer Cell 3, 459–469. [DOI] [PubMed] [Google Scholar]
- 32).Uchida T., Kitaura J., Nakahara F., Togami K., Inoue D., Maehara A., Nishimura K., Kawabata K.C., Doki N., Kakihana K., Yoshioka K., Izawa K., Oki T., Sada A., Harada Y., Ohashi K., Katayama Y., Matsui T., Harada H., Kitamura T. (2014) Hes1 up-regulation contributes to the development of FIP1L1-PDGFRA-positive leukemia in blast crisis. Exp. Hematol. 42, 369–379. [DOI] [PubMed] [Google Scholar]
- 33).Harada H., Harada Y., Tanaka H., Kimura A., Inaba T. (2003) Implications of somatic mutations in the AML1 gene in radiation-associated and therapy-related myelodysplastic syndrome/acute myeloid leukemia. Blood 101, 673–680. [DOI] [PubMed] [Google Scholar]
- 34).Gombart A.F., Hofmann W.K., Kawano S., Takeuchi S., Krug U., Kwok S.H., Larsen R.J., Asou H., Miller C.W., Hoelzer D., Koeffler H.P. (2002) Mutations in the gene encoding the transcription factor CCAAT/enhancer binding protein alpha in myelodysplastic syndromes and acute myeloid leukemias. Blood 99, 1332–1340. [DOI] [PubMed] [Google Scholar]
- 35).Hirai H., Kobayashi Y., Mano H., Hagiwara K., Maru Y., Omine M., Mizoguchi H., Nishida J., Takaku F. (1987) A point mutation at codon 13 of the N-ras oncogene in myelodysplastic syndrome. Nature 327, 430–432. [DOI] [PubMed] [Google Scholar]
- 36).Kaeferstein A., Krug U., Tiesmeier J., Aivado M., Faulhaber M., Stadler M., Krauter J., Germing U., Hofmann W.K., Koeffler H.P., Ganser A., Verbeek W. (2003) The emergence of a C/EBPalpha mutation in the clonal evolution of MDS towards secondary AML. Leukemia 17, 343–349. [DOI] [PubMed] [Google Scholar]
- 37).Togami K., Kitaura J., Uchida T., Inoue D., Nishimura K., Kawabata K.C., Nagase R., Horikawa S., Izawa K., Fukuyama T., Nakahara F., Oki T., Harada Y., Harada H., Aburatani H., Kitamura T. (2014) C-terminal mutant of C/EBPα (C/EBPα-Cm) down-regulates M-CSF receptor which is a potent accelerator in the progression of AML with C/EBPα-Cm B. Exp. Hematol. (in press). [DOI] [PubMed] [Google Scholar]
- 38).Lin Y.W., Slape C., Zhang Z., Aplan P.D. (2005) NUP98-HOXD13 transgenic mice develop a highly penetrant, severe myelodysplastic syndrome that progresses to acute leukemia. Blood 106, 287–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39).Greenblatt S., Li L., Slape C., Nguyen B., Novak R., Duffield A., Huso D., Desiderio S., Borowitz M.J., Aplan P., Small D. (2012) Knock-in of a FLT3/ITD mutation cooperates with a NUP98-HOXD13 fusion to generate acute myeloid leukemia in a mouse model. Blood 119, 2883–2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40).Falini B., Martelli M.P., Bolli N., Sportoletti P., Liso A., Tiacci E., Haferlach T. (2011) Acute myeloid leukemia with mutated nucleophosmin (NPM1): is it a distinct entity? Blood 117, 1109–1120. [DOI] [PubMed] [Google Scholar]
- 41).Noguera N.I., Song M.S., Divona M., Catalano G., Calvo K.L., Garcia F., Ottone T., Florenzano F., Faraoni I., Battistini L., Colombo E., Amadori S., Pandolfi P.P., Lo-Coco F. (2013) Nucleophosmin/B26 regulates PTEN through interaction with HAUSP in acute myeloid leukemia. Leukemia 27, 1037–1043. [DOI] [PubMed] [Google Scholar]
- 42).Wang Z., Gall J.M., Bonegio R., Havasi A., Illanes K., Schwartz J.H., Borkan S.C. (2013) Nucleophosmin, a critical Bax cofactor in ischemia-induced cell death. Mol. Cell. Biol. 33, 1916–1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43).Watanabe-Okochi N., Yoshimi A., Sato T., Ikeda T., Kumano K., Taoka K., Satoh Y., Shinohara A., Tsuruta T., Masuda A., Yokota H., Yatomi Y., Takahashi K., Kitaura J., Kitamura T., Kurokawa M. (2013) The shortest isoform of C/EBPbeta, liver inhibitory protein (LIP), collaborates with Evi1 to induce AML in a mouse BMT model. Blood 121, 4142–4155. [DOI] [PubMed] [Google Scholar]
- 44).Yoshimi A., Goyama S., Watanabe-Okochi N., Yoshiki Y., Nannya Y., Nitta E., Arai S., Sato T., Shimabe M., Nakagawa M., Imai Y., Kitamura T., Kurokawa M. (2011) Evi1 represses PTEN expression and activates PI3K/AKT/mTOR via interactions with polycomb proteins. Blood 117, 3617–3628. [DOI] [PubMed] [Google Scholar]
- 45).Watanabe-Okochi N., Kitaura J., Ono R., Harada H., Harada Y., Komeno Y., Nakajima H., Nosaka T., Inaba T., Kitamura T. (2008) AML1 mutations induced MDS and MDS/AML in a mouse BMT model. Blood 111, 4297–4308. [DOI] [PubMed] [Google Scholar]
- 46).Harada Y., Inoue D., Ding Y., Imagawa J., Doki N., Matsui H., Yahata T., Matsushita H., Ando K., Sashida G., Iwama A., Kitamura T., Harada H. (2013) RUNX1/AML1 mutant collaborates with BMI1 overexpression in the development of human and murine myelodysplastic syndromes. Blood 121, 3434–3446. [DOI] [PubMed] [Google Scholar]
- 47).Liedtke M., Cleary M.L. (2009) Therapeutic targeting of MLL. Blood 113, 6061–6068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48).Krivtsov A.V., Armstrong S.A. (2007) MLL translocations, histone modifications and leukaemia stem-cell development. Nat. Rev. Cancer 7, 823–833. [DOI] [PubMed] [Google Scholar]
- 49).Yokoyama A., Cleary M.L. (2008) Menin critically links MLL proteins with LEDGF on cancer-associated target genes. Cancer Cell 14, 36–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50).Bernt K.M., Zhu N., Sinha A.U., Vempati S., Faber J., Krivtsov A.V., Feng Z., Punt N., Daigle A., Bullinger L., Pollock R.M., Richon V.M., Kung A.L., Armstrong S.A. (2011) MLL-rearranged leukemia is dependent on aberrant H3K79 methylation by DOT1L. Cancer Cell 20, 66–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51).The Cancer Genome Atlas Research Network (2013) Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 368, 2059–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52).Raza A., Galili N. (2012) The genetic basis of phenotypic heterogeneity in myelodysplastic syndromes. Nat. Rev. Cancer 12, 849–859. [DOI] [PubMed] [Google Scholar]
- 53).Milosevic J.D., Kralovics R. (2013) Genetic and epigenetic alterations of myeloproliferative disorders. Int. J. Hematol. 97, 183–197. [DOI] [PubMed] [Google Scholar]
- 54).Rocquain J., Carbuccia N., Trouplin V., Raynaud S., Murati A., Nezri M., Tadrist Z., Olschwang S., Vey N., Birnbaum D., Gelsi-Boyer V., Mozziconacci M.J. (2010) Combined mutations of ASXL1, CBL, FLT3, IDH1, IDH2, JAK2, KRAS, NPM1, NRAS, RUNX1, TET2 and WT1 genes in myelodysplastic syndromes and acute myeloid leukemias. BMC Cancer 10, 401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55).Thiede C. (2012) Mutant DNMT3A: teaming up to transform. Blood 119, 5615–5617. [DOI] [PubMed] [Google Scholar]
- 56).Abdel-Wahab O., Levine R. (2011) The spliceosome as an indicted conspirator in myeloid malignancies. Cancer Cell 20, 420–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57).Papaemmanuil E., Cazzola M., Boultwood J., Malcovati L., Vyas P., Bowen D., Pellagatti A., Wainscoat J.S., Hellstrom-Lindberg E., Gambacorti-Passerini C., Godfrey A.L., Rapado I., Cvejic A., Rance R., McGee C., Ellis P., Mudie L.J., Stephens P.J., McLaren S., Massie C.E., Tarpey P.S., Varela I., Nik-Zainal S., Davies H.R., Shlien A., Jones D., Raine K., Hinton J., Butler A.P., Teague J.W., Baxter E.J., Score J., Galli A., Della Porta M.G., Travaglino E., Groves M., Tauro S., Munshi N.C., Anderson K.C., El-Naggar A., Fischer A., Mustonen V., Warren A.J., Cross N.C., Green A.R., Futreal P.A., Stratton M.R., Campbell P.J. (2011) Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. N. Engl. J. Med. 365, 1384–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58).Yoshida K., Sanada M., Shiraishi Y., Nowak D., Nagata Y., Yamamoto R., Sato Y., Sato-Otsubo A., Kon A., Nagasaki M., Chalkidis G., Suzuki Y., Shiosaka M., Kawahata R., Yamaguchi T., Otsu M., Obara N., Sakata-Yanagimoto M., Ishiyama K., Mori H., Nolte F., Hofmann W.K., Miyawaki S., Sugano S., Haferlach C., Koeffler H.P., Shih L.Y., Haferlach T., Chiba S., Nakauchi H., Miyano S., Ogawa S. (2011) Frequent pathway mutations of splicing machinery in myelodysplasia. Nature 478, 64–69. [DOI] [PubMed] [Google Scholar]
- 59).Ernst T., Chase A.J., Score J., Hidalgo-Curtis C.E., Bryant C., Jones A.V., Waghorn K., Zoi K., Ross F.M., Reiter A., Hochhaus A., Drexler H.G., Duncombe A., Cervantes F., Oscier D., Boultwood J., Grand F.H., Cross N.C. (2010) Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nat. Genet. 42, 722–726. [DOI] [PubMed] [Google Scholar]
- 60).Quivoron C., Couronne L., Della Valle V., Lopez C.K., Plo I., Wagner-Ballon O., Do Cruzeiro M., Delhommeau F., Arnulf B., Stern M.H., Godley L., Opolon P., Tilly H., Solary E., Duffourd Y., Dessen P., Merle-Beral H., Nguyen-Khac F., Fontenay M., Vainchenker W., Bastard C., Mercher T., Bernard O.A. (2011) TET2 inactivation results in pleiotropic hematopoietic abnormalities in mouse and is a recurrent event during human lymphomagenesis. Cancer Cell 20, 25–38. [DOI] [PubMed] [Google Scholar]
- 61).Moran-Crusio K., Reavie L., Shih A., Abdel-Wahab O., Ndiaye-Lobry D., Lobry C., Figueroa M.E., Vasanthakumar A., Patel J., Zhao X., Perna F., Pandey S., Madzo J., Song C., Dai Q., He C., Ibrahim S., Beran M., Zavadil J., Nimer S.D., Melnick A., Godley L.A., Aifantis I., Levine R.L. (2011) Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell 20, 11–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62).Muto T., Sashida G., Oshima M., Wendt G.R., Mochizuki-Kashio M., Nagata Y., Sanada M., Miyagi S., Saraya A., Kamio A., Nagae G., Nakaseko C., Yokote K., Shimoda K., Koseki H., Suzuki Y., Sugano S., Aburatani H., Ogawa S., Iwama A. (2013) Concurrent loss of Ezh2 and Tet2 cooperates in the pathogenesis of myelodysplastic disorders. J. Exp. Med. 210, 2627–2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63).Abdel-Wahab O., Gao J., Adli M., Dey A., Trimarchi T., Chung Y.R., Kuscu C., Hricik T., Ndiaye-Lobry D., Lafave L.M., Koche R., Shih A.H., Guryanova O.A., Kim E., Li S., Pandey S., Shin J.Y., Telis L., Liu J., Bhatt P.K., Monette S., Zhao X., Mason C.E., Park C.Y., Bernstein B.E., Aifantis I., Levine R.L. (2013) Deletion of Asxl1 results in myelodysplasia and severe developmental defects in vivo. J. Exp. Med. 210, 2641–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64).Abdel-Wahab O., Adli M., LaFave L.M., Gao J., Hricik T., Shih A.H., Pandey S., Patel J.P., Chung Y.R., Koche R., Perna F., Zhao X., Taylor J.E., Park C.Y., Carroll M., Melnick A., Nimer S.D., Jaffe J.D., Aifantis I., Bernstein B.E., Levine R.L. (2012) ASXL1 mutations promote myeloid transformation through loss of PRC2-mediated gene repression. Cancer Cell 22, 180–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65).Wang J., Li Z., He Y., Pan F., Chen S., Rhodes S., Nguyen L., Yuan J., Jiang L., Yang X., Weeks O., Liu Z., Zhou J., Ni H., Cai C.L., Xu M., Yang F.C. (2014) Loss of Asxl1 leads to myelodysplastic syndrome-like disease in mice. Blood 123, 541–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66).Cho Y.S., Kim E.J., Park U.H., Sin H.S., Um S.J. (2006) Additional sex comb-like 1 (ASXL1), in cooperation with SRC-1, acts as a ligand-dependent coactivator for retinoic acid receptor. J. Biol. Chem. 281, 17588–17598. [DOI] [PubMed] [Google Scholar]
- 67).Fisher C.L., Lee I., Bloyer S., Bozza S., Chevalier J., Dahl A., Bodner C., Helgason C.D., Hess J.L., Humphries R.K., Brock H.W. (2010) Additional sex combs-like 1 belongs to the enhancer of trithorax and polycomb group and genetically interacts with Cbx2 in mice. Dev. Biol. 337, 9–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68).Xie H., Xu J., Hsu J.H., Nguyen M., Fujiwara Y., Peng C., Orkin S.H. (2014) Polycomb repressive complex 2 regulates normal hematopoietic stem cell function in a developmental-stage-specific manner. Cell Stem Cell 14, 68–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69).Przychodzen B., Jerez A., Guinta K., Sekeres M.A., Padgett R., Maciejewski J.P., Makishima H. (2013) Patterns of missplicing due to somatic U2AF1 mutations in myeloid neoplasms. Blood 122, 999–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70).Inoue D., Kitaura J., Togami K., Nishimura K., Enomoto Y., Uchida T., Kagiyama Y., Kawabata K.C., Nakahara F., Izawa K., Oki T., Maehara A., Isobe M., Tsuchiya A., Harada Y., Harada H., Ochiya T., Aburatani H., Kimura H., Thol F., Heuser M., Levine R.L., Abdel-Wahab O., Kitamura T. (2013) Myelodysplastic syndromes are induced by histone methylation-altering ASXL1 mutations. J. Clin. Invest. 123, 4627–4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71).Xu W., Yang H., Liu Y., Yang Y., Wang P., Kim S.H., Ito S., Yang C., Wang P., Xiao M.T., Liu L.X., Jiang W.Q., Liu J., Zhang J.Y., Wang B., Frye S., Zhang Y., Xu Y.H., Lei Q.Y., Guan K.L., Zhao S.M., Xiong Y. (2011) Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer Cell 19, 17–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72).Figueroa M.E., Abdel-Wahab O., Lu C., Ward P.S., Patel J., Shih A., Li Y., Bhagwat N., Vasanthakumar A., Fernandez H.F., Tallman M.S., Sun Z., Wolniak K., Peeters J.K., Liu W., Choe S.E., Fantin V.R., Paietta E., Lowenberg B., Licht J.D., Godley L.A., Delwel R., Valk P.J., Thompson C.B., Levine R.L., Melnick A. (2010) Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell 18, 553–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73).Chaturvedi A., Araujo Cruz M.M., Jyotsana N., Sharma A., Yun H., Gorlich K., Wichmann M., Schwarzer A., Preller M., Thol F., Meyer J., Haemmerle R., Struys E.A., Jansen E.E., Modlich U., Li Z., Sly L.M., Geffers R., Lindner R., Manstein D.J., Lehmann U., Krauter J., Ganser A., Heuser M. (2013) Mutant IDH1 promotes leukemogenesis in vivo and can be specifically targeted in human AML. Blood 122, 2877–2887. [DOI] [PubMed] [Google Scholar]
- 74).Sasaki M., Knobbe C.B., Munger J.C., Lind E.F., Brenner D., Brustle A., Harris I.S., Holmes R., Wakeham A., Haight J., You-Ten A., Li W.Y., Schalm S., Su S.M., Virtanen C., Reifenberger G., Ohashi P.S., Barber D.L., Figueroa M.E., Melnick A., Zuniga-Pflucker J.C., Mak T.W. (2012) IDH1(R132H) mutation increases murine haematopoietic progenitors and alters epigenetics. Nature 488, 656–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75).Kats L.M., Reschke M., Taulli R., Pozdnyakova O., Burgess K., Bhargava P., Straley K., Karnik R., Meissner A., Small D., Su S.M., Yen K., Zhang J., Pandolfi P.P. (2014) Proto-Oncogenic Role of Mutant IDH2 in Leukemia Initiation and Maintenance. Cell Stem Cell 14, 329–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76).Makishima H., Yoshida K., Nguyen N., Przychodzen B., Sanada M., Okuno Y., Ng K.P., Gudmundsson K.O., Vishwakarma B.A., Jerez A., Gomez-Segui I., Takahashi M., Shiraishi Y., Nagata Y., Guinta K., Mori H., Sekeres M.A., Chiba K., Tanaka H., Muramatsu H., Sakaguchi H., Paquette R.L., McDevitt M.A., Kojima S., Saunthararajah Y., Miyano S., Shih L.Y., Du Y., Ogawa S., Maciejewski J.P. (2013) Somatic SETBP1 mutations in myeloid malignancies. Nat. Genet. 45, 942–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77).Thol F., Suchanek K.J., Koenecke C., Stadler M., Platzbecker U., Thiede C., Schroeder T., Kobbe G., Kade S., Loffeld P., Banihosseini S., Bug G., Ottmann O., Hofmann W.K., Krauter J., Kroger N., Ganser A., Heuser M. (2013) SETBP1 mutation analysis in 944 patients with MDS and AML. Leukemia 27, 2072–2075. [DOI] [PubMed] [Google Scholar]
- 78).Piazza R., Valletta S., Winkelmann N., Redaelli S., Spinelli R., Pirola A., Antolini L., Mologni L., Donadoni C., Papaemmanuil E., Schnittger S., Kim D.W., Boultwood J., Rossi F., Gaipa G., De Martini G.P., di Celle P.F., Jang H.G., Fantin V., Bignell G.R., Magistroni V., Haferlach T., Pogliani E.M., Campbell P.J., Chase A.J., Tapper W.J., Cross N.C., Gambacorti-Passerini C. (2013) Recurrent SETBP1 mutations in atypical chronic myeloid leukemia. Nat. Genet. 45, 18–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79).Sakaguchi H., Okuno Y., Muramatsu H., Yoshida K., Shiraishi Y., Takahashi M., Kon A., Sanada M., Chiba K., Tanaka H., Makishima H., Wang X., Xu Y., Doisaki S., Hama A., Nakanishi K., Takahashi Y., Yoshida N., Maciejewski J.P., Miyano S., Ogawa S., Kojima S. (2013) Exome sequencing identifies secondary mutations of SETBP1 and JAK3 in juvenile myelomonocytic leukemia. Nat. Genet. 45, 937–941. [DOI] [PubMed] [Google Scholar]
- 80).Oakley K., Han Y., Vishwakarma B.A., Chu S., Bhatia R., Gudmundsson K.O., Keller J., Chen X., Vasko V., Jenkins N.A., Copeland N.G., Du Y. (2012) Setbp1 promotes the self-renewal of murine myeloid progenitors via activation of Hoxa9 and Hoxa10. Blood 119, 6099–6108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81).Thorsteinsdottir U., Mamo A., Kroon E., Jerome L., Bijl J., Lawrence H.J., Humphries K., Sauvageau G. (2002) Overexpression of the myeloid leukemia-associated Hoxa9 gene in bone marrow cells induces stem cell expansion. Blood 99, 121–129. [DOI] [PubMed] [Google Scholar]
- 82).Inoue D., Kitaura J., Matsui H., Hou H.-A., Chou W.-C., Nagamachi A., Kawabata K.C., Togami K., Nagase R., Horikawa S., Saika M., Micol J.-P., Hayashi Y., Harada Y., Harada H., Inaba T., Tien H.-F., Abdel-Wahab O., Kitamura T. (2014) SETBP1 mutations drive leukemic transformation in ASXL1-murtated MDS. Leukemia (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83).Gelsi-Boyer V., Trouplin V., Adelaide J., Bonansea J., Cervera N., Carbuccia N., Lagarde A., Prebet T., Nezri M., Sainty D., Olschwang S., Xerri L., Chaffanet M., Mozziconacci M.J., Vey N., Birnbaum D. (2009) Mutations of polycomb-associated gene ASXL1 in myelodysplastic syndromes and chronic myelomonocytic leukaemia. Br. J. Haematol. 145, 788–800. [DOI] [PubMed] [Google Scholar]
- 84).Thol F., Friesen I., Damm F., Yun H., Weissinger E.M., Krauter J., Wagner K., Chaturvedi A., Sharma A., Wichmann M., Gohring G., Schumann C., Bug G., Ottmann O., Hofmann W.K., Schlegelberger B., Heuser M., Ganser A. (2011) Prognostic significance of ASXL1 mutations in patients with myelodysplastic syndromes. J. Clin. Oncol. 29, 2499–2506. [DOI] [PubMed] [Google Scholar]
- 85).Nikoloski G., van der Reijden B.A., Jansen J.H. (2012) Mutations in epigenetic regulators in myelodysplastic syndromes. Int. J. Hematol. 95, 8–16. [DOI] [PubMed] [Google Scholar]
- 86).Xu J., Wang Y.Y., Dai Y.J., Zhang W., Zhang W.N., Xiong S.M., Gu Z.H., Wang K.K., Zeng R., Chen Z., Chen S.J. (2014) DNMT3A Arg882 mutation drives chronic myelomonocytic leukemia through disturbing gene expression/DNA methylation in hematopoietic cells. Proc. Natl. Acad. Sci. U.S.A. 111, 2620–2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87).Kim S.J., Zhao H., Hardikar S., Singh A.K., Goodell M.A., Chen T. (2013) A DNMT3A mutation common in AML exhibits dominant-negative effects in murine ES cells. Blood 122, 4086–4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88).Haferlach T., Nagata Y., Grossmann V., Okuno Y., Bacher U., Nagae G., Schnittger S., Sanada M., Kon A., Alpermann T., Yoshida K., Roller A., Nadarajah N., Shiraishi Y., Shiozawa Y., Chiba K., Tanaka H., Koeffler H.P., Klein H.U., Dugas M., Aburatani H., Kohlmann A., Miyano S., Haferlach C., Kern W., Ogawa S. (2014) Landscape of genetic lesions in 944 patients with myelodysplastic syndromes. Leukemia 28, 241–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89).Papaemmanuil E., Gerstung M., Malcovati L., Tauro S., Gundem G., Van Loo P., Yoon C.J., Ellis P., Wedge D.C., Pellagatti A., Shlien A., Groves M.J., Forbes S.A., Raine K., Hinton J., Mudie L.J., McLaren S., Hardy C., Latimer C., Della Porta M.G., O’Meara S., Ambaglio I., Galli A., Butler A.P., Walldin G., Teague J.W., Quek L., Sternberg A., Gambacorti-Passerini C., Cross N.C., Green A.R., Boultwood J., Vyas P., Hellstrom-Lindberg E., Bowen D., Cazzola M., Stratton M.R., Campbell P.J. (2013) Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood 122, 3616–3627; quiz 3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90).Huntly B.J., Shigematsu H., Deguchi K., Lee B.H., Mizuno S., Duclos N., Rowan R., Amaral S., Curley D., Williams I.R., Akashi K., Gilliland D.G. (2004) MOZ-TIF2, but not BCR-ABL, confers properties of leukemic stem cells to committed murine hematopoietic progenitors. Cancer Cell 6, 587–596. [DOI] [PubMed] [Google Scholar]
- 91).Walter M.J., Shen D., Ding L., Shao J., Koboldt D.C., Chen K., Larson D.E., McLellan M.D., Dooling D., Abbott R., Fulton R., Magrini V., Schmidt H., Kalicki-Veizer J., O’Laughlin M., Fan X., Grillot M., Witowski S., Heath S., Frater J.L., Eades W., Tomasson M., Westervelt P., DiPersio J.F., Link D.C., Mardis E.R., Ley T.J., Wilson R.K., Graubert T.A. (2012) Clonal architecture of secondary acute myeloid leukemia. N. Engl. J. Med. 366, 1090–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92).Lundberg P., Karow A., Nienhold R., Looser R., Hao-Shen H., Nissen I., Girsberger S., Lehmann T., Passweg J., Stern M., Beisel C., Kralovics R., Skoda R.C. (2014) Clonal evolution and clinical correlates of somatic mutations in myeloproliferative neoplasms. Blood 123, 2220–2228. [DOI] [PubMed] [Google Scholar]
- 93).Calabretta B., Perrotti D. (2004) The biology of CML blast crisis. Blood 103, 4010–4022. [DOI] [PubMed] [Google Scholar]
- 94).Perrotti D., Jamieson C., Goldman J., Skorski T. (2010) Chronic myeloid leukemia: mechanisms of blastic transformation. J. Clin. Invest. 120, 2254–2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95).Ito T., Kwon H.Y., Zimdahl B., Congdon K.L., Blum J., Lento W.E., Zhao C., Lagoo A., Gerrard G., Foroni L., Goldman J., Goh H., Kim S.H., Kim D.W., Chuah C., Oehler V.G., Radich J.P., Jordan C.T., Reya T. (2010) Regulation of myeloid leukaemia by the cell-fate determinant Musashi. Nature 466, 765–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96).Nakahara F., Sakata-Yanagimoto M., Komeno Y., Kato N., Uchida T., Haraguchi K., Kumano K., Harada Y., Harada H., Kitaura J., Ogawa S., Kurokawa M., Kitamura T., Chiba S. (2010) Hes1 immortalizes committed progenitors and plays a role in blast crisis transition in chronic myelogenous leukemia. Blood 115, 2872–2881. [DOI] [PubMed] [Google Scholar]
- 97).Bacher U., Haferlach T., Kern W., Haferlach C., Schnittger S. (2007) A comparative study of molecular mutations in 381 patients with myelodysplastic syndrome and in 4130 patients with acute myeloid leukemia. Haematologica 92, 744–752. [DOI] [PubMed] [Google Scholar]