Background: Deconjugation of polyubiquitin is an essential step in preparing substrates for proteolysis by the 26S proteasome.
Results: Proteasome-associated DUBs, Rpn11 and Ubp6, process long Lys11- or Lys63-linked polyUb more efficiently than Lys48 linkages.
Conclusion: 26S proteasomes can completely disassemble a mixed/branched polyUb conjugate.
Significance: These observations call into question what constitutes an efficient signal for proteasome targeting versus proteolysis.
Keywords: Cysteine Protease, Deubiquitination, Metalloprotease, Proteasome, Ubiquitin, Rpn11, Ubp6, Polyubiquitin
Abstract
Protein homeostasis is largely dependent on proteolysis by the ubiquitin-proteasome system. Diverse polyubiquitin modifications are reported to target cellular proteins to the proteasome. At the proteasome, deubiquitination is an essential preprocessing event that contributes to degradation efficiency. We characterized the specificities of two proteasome-associated deubiquitinases (DUBs), Rpn11 and Ubp6, and explored their impact on overall proteasome DUB activity. This was accomplished by constructing a panel of well defined ubiquitin (Ub) conjugates, including homogeneous linkages of varying lengths as well as a heterogeneously modified target. Rpn11 and Ubp6 processed Lys11 and Lys63 linkages with comparable efficiencies that increased with chain length. In contrast, processing of Lys48 linkages by proteasome was inversely correlated to chain length. Fluorescently labeled tetra-Ub chains revealed endo-chain preference for Ubp6 acting on Lys48 and random action for Rpn11. Proteasomes were more efficient at deconjugating identical substrates than their constituent DUBs by roughly 2 orders of magnitude. Incorporation into proteasomes significantly enhanced enzymatic efficiency of Rpn11, due in part to alleviation of the autoinhibitory role of its C terminus. The broad specificity of Rpn11 could explain how proteasomes were more effective at disassembling a heterogeneously modified conjugate compared with homogeneous Lys48-linked chains. The reduced ability to disassemble homogeneous Lys48-linked chains longer than 4 Ub units may prolong residency time on the proteasome.
Introduction
In eukaryotic cells, versatile polyubiquitin (polyUb)5 modifications direct protein-protein interactions to numerous cellular functions, including DNA repair, chromatin dynamics, mRNA export, membrane trafficking, and proteasome-mediated degradation (1–4). The polyUb signal is initiated with attachment of ubiquitin (Ub) to a target protein by a cascade of three ubiquitinating enzymes (E1, E2, and E3). The target protein is selected by a dedicated E3 Ub ligase that catalyzes conjugation of a lysine ϵ-amine on the target to the C terminus of Ub. Subsequently, any of eight positions on the first Ub (an ϵ-amine at Lys6, Lys11, Lys27, Lys29, Lys33, Lys48, or Lys63 or the N terminus at Met1) can be covalently attached to additional Ub molecules (5–7). The resulting Ub-Ub linkage is typically directed by an E2 conjugating enzyme; for example, E2-25K (Ube2K) forms chains through Lys48, whereas Ubc13 (Ube2N) paired with Uev1a (Ube2v1) forms chains through Lys63 (8). Each linkage results in a unique three-dimensional conformation of the polyUb chain (supplemental Fig. S1; for comprehensive reviews, see Refs. 2 5, and 6). In turn, polyUb receptors can differentiate between these structurally distinct signals, allowing for divergent outcomes. Modification by polyUb is reversible through the action of deubiquitinases (DUBs), dedicated enzymes that can hydrolyze after the C-terminal Gly76 of Ub to remove covalently attached adducts or other Ub units. Through their combined action, numerous DUBs (∼90 in the human genome, classified into five different families; for comprehensive reviews, see Refs. 9 and 10) sculpt the heterogeneous Ub landscape. Thus, some DUBs display broad specificity, whereas others are relatively selective for substrate or for linkage type; some can completely disassemble a chain, whereas others amputate an entire polyUb chain “en bloc” from the substrate (10–12). All linkage types have been detected in vivo. Lys48 and Lys63 linkages and, to a lesser extent, Lys11 account for the bulk of linkage types (13, 14), although relative levels may change in response to specific conditions. Lys63 linkages have been reported to serve in primarily regulatory non-degradative processes, such as DNA repair and membrane trafficking, whereas Lys48 is the canonical signal that targets substrates to the proteasome (3, 4, 15, 16). Lys11 linkages alone or branched with Lys48 linkages are thought to serve as a degradation signal enhancer of anaphase-promoting complex substrates during cell (17, 18) or on ERAD pathways (19).
The 26S proteasome is a 2.5-MDa ATP-dependent protease, made up of a proteolytic 20S core particle capped by one or two 19S regulatory particles (RP) abutting the core particle (20, 21). The 19S RP can be further subdivided in two major subcomplexes: base and lid. The base contains six AAA-ATPases, Rpt1–6, along with non-ATPases Rpn1, -2, -10, and -13. The lid is composed of six PCI (proteasome, COP9, initiation factor-3) domain subunits, Rpn3, -5, -6, -7, -9, and -12, and two MPN (Mpr1-Pad1-N-terminal) domain subunits, Rpn8 and Rpn11.
Several DUBs associate with the 19S RP, where they participate in preparing substrates for degradation and rescue Ub from a similar fate (22, 23). Ubp6/USP14 and Rpn11/PSMD14 (yeast/human, respectively) are the primary DUBs studied in this context. UCH37/UCHL5, a third proteasome-associated DUB, is not found in all eukaryotes (24). Ubp6 is a cysteine protease that transiently associates with the proteasome by docking to the Rpn1 subunit in the 19S RP (25, 26). In contrast, Rpn11, a MPN+/JAMM zinc metalloprotease (23, 27), is an integral component of the 19S RP. Recent crystal structures of Rpn11 MPN+ domain in combination with cryo-EM position Rpn11 at the center of the 19S RP (28–30). Further analysis by a cryo-EM study has hinted that the position of Rpn11 in the proteasome differs by 18 Å between the free and substrate-engaged forms, pointing to a dramatic reaction-induced conformational change (31). Rpn11 and Ubp6 activities within the proteasome are synchronized to ATP consumption by neighboring Rpt ATPases and substrate degradation, and therefore complicate understanding of their respective contributions (23, 31–34). So far, Ubp6/USP14 and UCH37 are thought to trim polyUb chains from the distal end (11, 25), gradually decreasing affinity of the conjugate as the chain shortens, whereas Rpn11 amputates entire chains at the proximal side coupled to ATP-dependent degradation (11, 35). Alternatively, Rpn11 has also been demonstrated to disassemble Lys63-linked unconjugated polyUb chains (36). Deciphering DUB action of both Ubp6 and Rpn11 is key to understanding how they influence residency time of polyUb substrates at the proteasome (32, 33, 37–40).
Our main objective was to understand the properties associated with each proteasomal DUB uninfluenced by the many other interlinked ATP-dependent processes of the proteasome. With this in mind, we deliberately studied their enzymatic activities in isolation and in complex by utilizing non-degradable substrates: a panel of polyUb conjugates with defined properties, chain length and linkage type. With this experimental setup, we found that the overall DUB activity of proteasome primarily reflects properties of Rpn11. When incorporated into proteasome, Rpn11 is activated over 100-fold compared with unattached subcomplexes. Due to the broad specificity of Rpn11, proteasomes were able to completely remove all Ub from a model substrate Ubch5b-Ubn, which included diverse Ub modifications on multiple lysines. In summary, proteasomes coordinate multiple DUBs and polyUb receptors to act on a wide range of substrates for both degradative and non-degradative outcomes.
EXPERIMENTAL PROCEDURES
Purification of Yeast Proteasome
The 26S proteasome was purified from wild type Saccharomyces cerevisiae strain using conventional chromatography as described previously (20). The 26S proteasomes were purified from wild type or isogenetic Δubp6 Saccharomyces cerevisiae strains using conventional chromatography as described previously (20). Because Ubp6 is a labile proteasome-associated protein (37, 41, 42), equimolar levels of Ubp6 were added to purified 26S proteasomes to guarantee stoichiometric levels. Full incorporation of Ubp6 into 26S proteasomes was validated in each case (Fig. 1B). Intact 26S proteasomes were confirmed by non-denaturing PAGE and peptidase activity with Suc-LLVY-AMC as described (20).
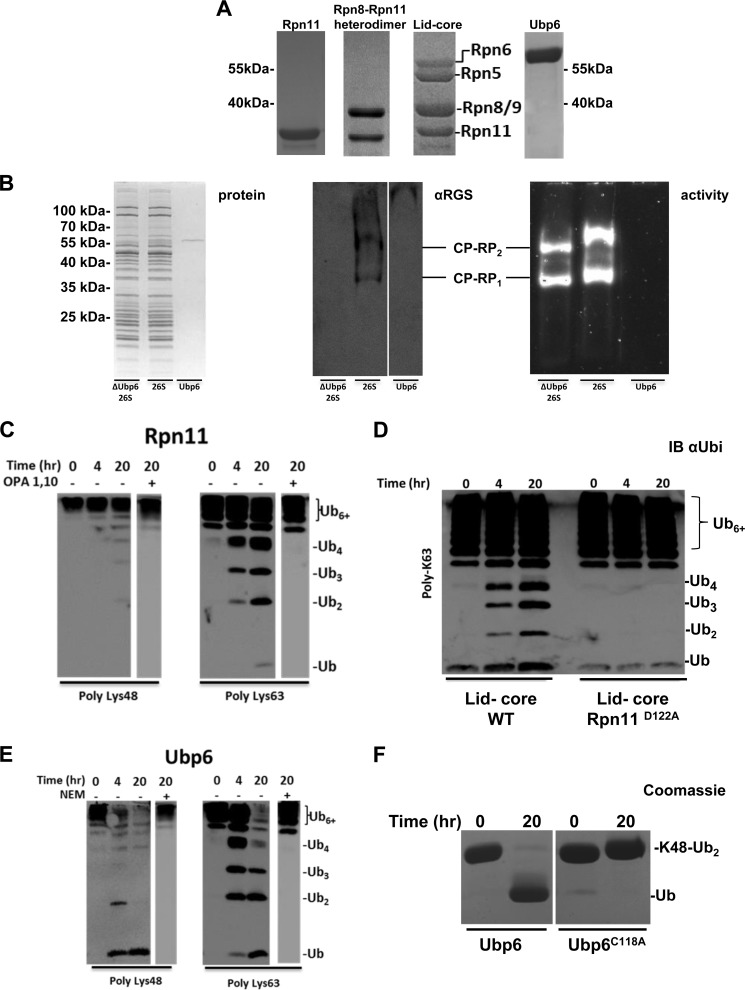
FIGURE 1.
A, purification of recombinant Rpn11, Rpn8·Rpn11 heterodimer, lid-core, and Ubp6 evaluated by Coomassie-stained SDS-PAGE. B, SDS-PAGE reveals proper subunit composition of proteasome with and without Ubp6 (left). Native gel was immunoblotted for His6-Ubp6 to ensure incorporation into proteasome (middle). Gel shifts in native PAGE indicate the additional association of Ubp6 to active ΔUbp6 26S proteasome (right). C, activity of Rpn11 in complete isolation on Lys48-Ub6+ (left) and Lys63-Ub6+ (right); the end point of the reaction in the presence of 10 mm 1,10-phenanthroline (OPA) is shown in the right lanes. D, deubiquitination of Lys63-Ub6+ by WT lid-core compared with Rpn11D122A lid-core is monitored using anti-Ub (IB αUbi). E, deubiquitination of Lys48-Ub6+ (left) or Lys63-Ub6+ (right) by Ubp6 with Ub6+ chains in the presence of 10 mm N-ethylmaleimide (NEM) thiol modifier. F, Lys48-Ub2 serves as control to demonstrate that Ubp6C118A is deactivated.
Plasmid Design
Full-length coding sequence of S. cerevisiae Ubp6 and Rpn11 was amplified from the genomic DNA and cloned into the pQE30 expression vector (Qiagen) with an N-terminal RGS-His6 tag. The lid-core plasmid containing Rpn5, -6, -8, -9, and -11-His6 in pETDuet1 was kindly provided by Prof. Andreas Martin (University of California, Berkeley) (43). The heterodimer Rpn8·Rpn11-His6 was amplified from the lid-core plasmid and cloned to pET28b expression vector (Novagen). Truncated Rpn8(1–186)·Rpn11(1–229)-His6 heterodimer was created by replacing full-length Rpn8 with truncated Rpn8(1–186) on the heterodimer plasmid and then replacing Rpn11 with truncated Rpn11(1–229). The Rpn11D122A mutated lid-core and Ubp6C118A were prepared using site-directed mutagenesis.
Preparation of Proteasomal Deubiquitinases and Subcomplexes
pQE30 plasmids were transformed to Escherichia coli M15 cells, whereas pET28b and pETDuet1 plasmids were transformed to BL-21 Rosetta 2 cells (Novagen). To ensure that Rpn11 was contained in all complexes, Rpn11 was N-terminally His6-tagged following a published methodology (43). Proteins were expressed in LB medium at 37 °C supplemented with 60 μg/ml ampicillin or 50 μg/ml kanamycin and induced with 0.5 mm isopropyl β-d-1-thiogalactopyranoside at A600 of ∼0.7. Following induction, 2-liter cultures were grown overnight at 16 °C, harvested, and stored at −80 °C. Cells were lysed using a French press, the lysates were clarified by centrifugation at 18,000 rpm for 30 min at 4 °C, and the supernatant was loaded onto a 10-ml His-Trap (GE Healthcare) column that was pre-equilibrated with 50 mm Tris, pH 7.4, 300 mm KCl, 5% glycerol, 10 mm imidazole in order to isolate recombinant His6-Ubp6 and His6-Rpn11. A solution of 50 mm pH 7.4, 100 mm NaCl, 100 mm KCl, 5% glycerol, 10 mm imidazole was used for Rpn11 subcomplexes. Proteins were eluted in the same buffer with 280 mm imidazole. Following elution, fractions containing proteins of interest were pooled and dialyzed in PBS, pH 7.4, buffer for His6-Ubp6 and His6-Rpn11, whereas Rpn11 subcomplexes were dialyzed into 50 mm, pH 7.4, 100 mm NaCl, 100 mm KCl, 5% glycerol. To obtain the highest purity and isolate stoichiometric Rpn11-containing subcomplexes, gel filtration was performed with a Sephacryl 400 column (GE Healthcare), and for monomeric proteins, it was performed with a Superdex 200 column (GE Healthcare). After confirming the purity with SDS-PAGE, all enzymes were aliquoted and stored at −80 °C.
Purification and Assembly of Fluorescently Labeled Tetra-Ub Chains
Codon-optimized ORF of human Ub in pETm60 vector was kindly provided by Dr. V. Dötsch (Goethe University, Frankfurt am Main) (44). Ub and variants (UbS20C, UbS20C,K63R, and UbK63R) were expressed in E. coli BL21 Rosetta 2 cells (Novagen), lysed, and processed as described previously (45). Following precipitation with perchloric acid and centrifugation, the pH of the Ub-containing supernatant was adjusted to neutral and concentrated using an Ultra-15 filter, 3 kDa cut-off (Millipore). Gel filtration (HiLoad 26/60 Superdex, 75 pg; GE Healthcare) in 50 mm Tris, pH 8, was performed to eliminate reaming impurities. Cysteine variants of Ub were fluorescently labeled (Alexa Fluor 488 C5-maleimide; UbS20CA488, UbS20C,K63RA488) as described previously (46). Uba1 in pET21d was kindly provided by Prof. C. Wolberger (Johns Hopkins, Howard Hughes Medical Institute, Baltimore, MD) and produced as described (47). Hrd1 and Ubc7 were expressed and purified following a published protocol (46). Ubc13 and Uev1a in pGEX were expressed in E. coli BL21 Rosetta 2 cells. GST fusion proteins were purified by affinity chromatography using a 10-ml GSTrap column (GE Healthcare) according to the manufacturer's instructions.
In vitro ubiquitination reactions for preparative assembly of fluorescently labeled Lys48-linked Ub chains included 0.2 μm E1 (Uba1), 6 μm Hrd1, 2 μm Ubc7, 1.485 mm Ub, and 15 μm UbS20CA488 in 9 mm ATP, 0.9 mm DTT, 4.5 mm MgCl2, and 17.5 mm HEPES, pH 7.5. Lys63-linked Ub chains included 1 μm E1 (Uba1), 8 μm GST-Ubc13, 8 μm GST-Uev1a, 1.2 mm Ub, 12 μm UbS20CA488, 0.8 mm UbK63R, and 8 μm UbS20C,K63RA488 in 20 mm ATP, 0.9 mm DTT, 9 mm MgCl2, and 40 mm Tris, pH 7.5. Reactions were performed at 30 °C for 18 h in the dark. At the end point, each reaction was centrifuged at 20,000 × g for 30 min at 4 °C, and tetra-Ub species were isolated using gel filtration (HiLoad 26/60 Superdex 75 pg, GE Healthcare) in PBS, pH 7.4, and then concentrated using an Ultra-15, 10 kDa cut-off (Millipore).
Preparation of Dimeric Ub, Ub4, Ub6+, and Ubch5b-Ubn Conjugates
Monomeric Ub mutants, E2 conjugating enzymes, and human E1 were obtained recombinantly as described (8, 48). Enzymatically synthesized Lys11-, Lys48-, and Lys63-linked Ub dimers were assembled by combining a proximally blocked Ub mutant (UbD77 or Ub-His6) in combination with a distally blocked Lys to Arg Ub variant as described (48, 49). Lys11-linked dimers were obtained from a reaction containing 10 mg of each Ub-His6 and UbK11R,K63R, 500 nm Uba1, 30 μm Ube2s, 5 mm tris(2-carboxyethyl)phosphine, and 15 mm ATP in a volume of 2 ml with a 50 mm Tris, pH 8.0, buffer incubated at 30 °C for 20 h. Lys48-linked dimers were obtained in a similar reaction with E2-25K as the sole E2 and UbD77 and UbK48R,K63R monomers. In a similar fashion, reactions to generate Lys63-linked dimers contained a 20 μm concentration of each Ubc13 and Uev1a with UbD77 and UbK48R/K63R monomers. Following the completion of each reaction, 10 ml of cation buffer A (50 mm ammonium acetate, pH 4.5) was added, the solution was centrifuged at 13,000 rpm for 10 min to remove precipitated E1 and E2 enzymes, and the supernatant was slowly injected onto a 5 ml HiTrap SP column (GE Healthcare) at 0.2 ml/min. The polyUb species were eluted with cation buffer B (50 mm ammonium acetate, 1 m NaCl, pH 4.5); exchanged into PBS, pH 7.4, buffer; and concentrated to a final volume of 1 ml. Monomeric and dimeric Ub species were separated on a Superdex 75 size exclusion column (GE Healthcare) in PBS, pH 7.4, buffer with a flow rate of 0.35 ml/min. Fractions containing pure dimers were detected using SDS-PAGE. Non-fluorescent Lys48-Ub4 and Lys63-Ub4 were prepared as described above, without Ub cysteine variants.
High molecular weight Lys48-Ub6+ and Lys63-Ub6+ chains were created using reaction conditions identical to the dimers with 30 mg of wild type Ub as the only monomer. Following the Superdex 75 column, fractions containing the desired chain lengths were detected by SDS-PAGE and pooled. The autoubiquitinating properties of the E2 Ub-conjugating enzyme, Ubch5b, were exploited to generate Ubch5b-Ubn conjugates under similar reaction conditions using 2 mm of human wild type Ub, 5 mm DTT, 15 mm ATP, and 75 μm Ubch5b-His6. Ubch5b-Ubn conjugates were purified as described (50). DTT was used in place of tris(2-carboxyethyl)phosphine to minimize the accumulation of thioester-linked Ubs on the active site cysteine of Ubch5b. Desired oligomeric states of Ubc5b-Ubn were detected using 15% SDS-PAGE; exchanged into PBS, pH 7.4, without DTT; and stored at −20 °C.
In Vitro Deubiquitination Reactions of Fluorescently Labeled Tetra-Ub Chains
In vitro deubiquitination time course experiments containing 25 μm Ubp6, Rpn8·Rpn11 heterodimer, or lid-core were incubated with 25 μm Lys48-Ub4 or Lys63-Ub4 fluorescent substrates in PBS, pH 7.4. Both tetra-Ub substrates included 1% A488-labeled Ub. All deubiquitination reactions were performed at 30 °C in the dark. Samples were taken at given time points and analyzed by 18% SDS-PAGE. Gels were analyzed using fluorescence scanning on a TyphoonTM FLA 9500 scanner (GE Healthcare) employing emission filter BPB1 (530DF20) with excitation at 473 nm and Coomassie staining. Fluorescence intensity was quantified using ImageQuant TL software (GE Healthcare).
Gel-based Deubiquitination Assays
All cleavage assays were performed in vitro. 50 nm 26S proteasome was incubated with a 500 nm concentration of either Lys48- or Lys63-Ub6+, whereas 1 μm lid-core, Rpn8·Rpn11 heterodimer, or Ubp6 was incubated with 10 μm Ub6+. 10 μm isolated Rpn11 was incubated with 100 μm Ub6+ in order to observe activity. All reactions were incubated at 30 °C. Samples of the deubiquitination assay were taken at the indicated points, mixed with 5× protein loading dye, boiled at 95 °C for 5 min, and kept at room temperature until analysis by 15% SDS-PAGE. Gels were visualized with Coomassie staining or transferred and immunoblotted with anti-ubiquitin antibody polyclonal rabbit anti-Ub Dako (z0458) in a 1:1,000 dilution. For detection of Lys11, Lys48, and Lys63 linkages, rabbit monoclonal Lys11 (Millipore catalog no. 2021885), Lys48 (Millipore catalog no. 2197314), and Lys63 (Millipore catalog no. 2063204) antibodies were used in a 1:1,000 dilution. IgG goat anti-rabbit HRP conjugate (Bio-Rad catalog no. 170-6515) was used as the secondary antibody in a 1:50,000 dilution. Gel bands were quantified using ImageJ analysis. Enzymatic efficiency in units of nm−1 × min−1 was determined using Equation 1.
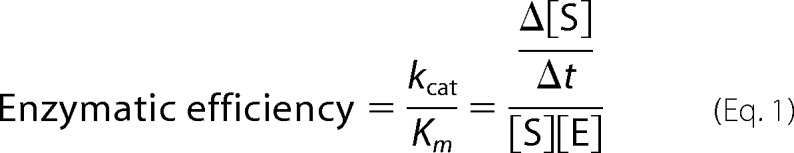 |
All reactions were repeated in triplicate. Average values with S.D. are summarized in Table 1.
TABLE 1.
Summary of deubiquitination efficiency (nm−1 × min−1) for proteasome and associated DUBs
Enzymatic efficiency of polyUb chain disassembly by deubiquitination enzymes in this study was calculated using Equation 1 (see “Experimental Procedures”). Disassembly efficiency of Ub2 substrate was determined by quantifying the Ub2 gel band intensity. Values for Ub4, Ub6+, and UbcH5b-Ubn were determined by following the decrease in signal intensity of residual substrate relative to the entire signal generated by Ub in all of its forms.
| Ub2 |
Ub4 |
Ub6+ |
Ubch5b-Ubn (heterogeneous) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Lys11 | Lys48 | Lys63 | Lys48 | Lys63 | Lys11 | Lys48 | Lys63 | ||
| Rpn8 + 11 | 0.12 ± 0.03 | 0.07 ± 0.01 | 0.05 ± 0.02 | 0.046 ± 0.00 | 0.07 ± 0.01 | 0.8 ± 0.2 | 0 | 0.61 ± 0.11 | 0.54 ± 0.06 |
| Lid-core | 0.22 ± 0.01 | 0.12 ± 0.05 | 0.13 ± 0.03 | 0.06 ± 0.01 | 0.13 ± 0.02 | 2.9 ± 0.4 | 0 | 2.04 ± 0.34 | 0.76 ± 0.06 |
| Ubp6 | 0.20 ± 0.02 | 0.28 ± 0.04 | 0.14 ± 0.04 | 0.15 ± 0.01 | 0.42 ± 0.14 | 3.5 ± 0.3 | 1.17 ± 0.12 | 2.49 ± 0.08 | 0.66 ± 0.15 |
| ΔUbp6 26S | 38.29 ± 1.09 | 25.15 ± 4.4 | 19.55 ± 2.31 | 5.81 ± 0.42 | 124.08 ± 0.2 | 100.1 ± 13.1 | 6 ± 1 | 94.2 ± 2.6 | 81 ± 15 |
| 26S | 36.88 ± 3.26 | 21.82 ± 1.36 | 16.58 ± 4.74 | 4.63 ± 0.21 | 141.8 ± 5.9 | 136.1 ± 12.5 | 7.73 ± 1.59 | 80.1 ± 6.5 | 86.7 ± 14.3 |
Mathematical Analysis of Fluorescent Tetra-Ub Disassembly Intermediates
At each time point of tetra-Ub disassembly, the fluorescent signal for each Ub species was quantified. Disassembly was analyzed by fitting simultaneously the intensities of the gel bands corresponding to Ub4, Ub3, Ub2, and Ub1 over the time course to Equations 2–5) and treating the rate constants as fitting parameters. Here the tetramer is initially cleaved at an exo position (rate constant k4→3) or the endo (k4→2), as schematically illustrated in supplemental Fig. S2. The trimer is cleaved to a dimer (k3→2), and finally, the dimer is converted to monomeric Ub (k2→1). Each rate constant was determined numerically, and the results are summarized in (supplemental Fig. S2).
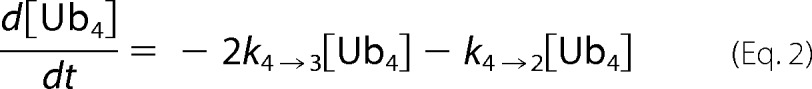 |
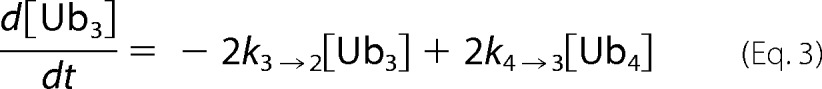 |
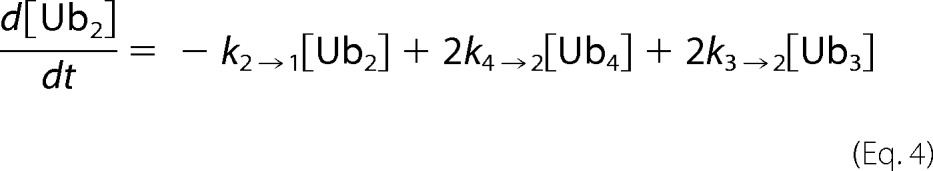 |
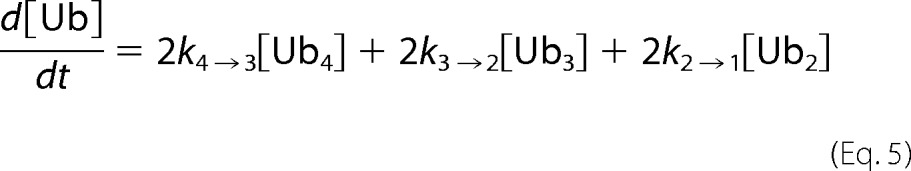 |
MS/MS Analysis of Ubch5b-Ubn Modifications
The Ub linkages on the Ubch5b-Ubn substrate were determined by tryptic digestion in solution followed by LC-MS/MS analysis. Ubch5b-Ubn in PBS buffer was reduced by 10 mm DTT and reacted with 40 mm iodoacetamide (at 25 °C) and trypsinized (modified trypsin (Promega)) at a 1:100 enzyme/substrate ratio for 18 h at 37 °C. The resulting tryptic peptides were desalted and resolved by reverse-phase chromatography on 0.075 × 200-mm fused silica capillaries (J&W) packed with Reprosil reversed phase material (Dr. Maisch GmbH, Ammerbuch-Entringen, Germany). The peptides were eluted with linear 65-min gradients of 5–45% and 15 min at 95% acetonitrile with 0.1% formic acid in water at flow rates of 0.25 μl/min. Mass spectrometry was performed by “hybrid” mass spectrometer (Orbitrap, Thermo) in a positive mode using a repetitively full MS scan followed by collision-induced dissociation of the seven most dominant ions selected from the first MS scan. The mass spectrometry data were analyzed using Trans Proteomic Pipeline (TPP) version 4.6.3 (51). TPP-processed centroid fragment peak lists in mzML format were searched against a database composed of S. cerevisiae yeast and E. coli proteins (Uniprot) and human Ub supplemented with their corresponding decoy sequences (as described on the Matrix Science Web site). The database searches were performed using X! Tandem with the k-score plugin through TPP. Search parameters include trypsin cleavage specificity with two missed cleavage sites; cysteine carbamidomethyl as a fixed modification; and lysine ubiquitination, methionine oxidation, and protein N-terminal acetylation as variable modifications. Peptide tolerance and MS/MS tolerance were set at 20 ppm and 0.8 Da, respectively. X! Tandem refinement included semistyle cleavages and variable lysine GG modification. Peptide and protein lists were generated following Peptide Prophet and Protein Prophet analysis using a protein false discovery rate of <1%.
RESULTS
Proteasome-associated DUBs Are Active in Isolation
To assess the individual properties of proteasome-associated DUBs, we prepared isolated enzymes to compare their activity with that of 26S proteasome holoenzyme (Fig. 1, A and B). Recent studies have demonstrated that a truncated Rpn8·Rpn11 heterodimer as well as an Rpn8·Rpn11 fusion used for crystallization maintain catalytic activity (28, 29). For in depth investigation of Rpn11 activity, we recombinantly purified full-length Rpn11, an Rpn8·Rpn11 “heterodimer,” and another subcomplex containing Rpn5, -6, -8, -9, and -11 (referred to as “lid-core”), which has been demonstrated to be a stable complex (43, 52, 53). We remind the reader that Rpn8 is an inactive paralogue, of Rpn11; both are adjacently located at the center of the 19S RP (28–30, 52). The composition of each preparation was confirmed by SDS-PAGE and MS analysis (Fig. 1A and supplemental Table S1). Purified proteasomes from yeast (54, 55) were used to compare the activity of these enzymes in isolated subcomplexes with that of intact proteasome holoenzymes. We found that Ubp6, a non-essential subunit transiently associated with proteasome (37, 41, 42), easily dissociated during purification, leading to inconsistent levels of Ubp6. Therefore, to ensure full incorporation (verified by gel shift (41)), recombinant Ubp6 was added back at a 1:1 molar ratio (Fig. 1B).
Given that both Lys48 and Lys63 linkages are processed by the proteasome (56, 57), all recombinant enzyme preparations were tested on polymeric Lys48 and Lys63 chains representative of proteasome targeting signals. In complete isolation as a standalone enzyme, Rpn11 was competent to process polymeric Lys63 linkages (Fig. 1C). Despite not having been previously documented, this property is consistent with other members of the MPN+/JAMM metalloprotease family, such as AMSH and BRCC36 (58, 59), which have strict specificity for Lys63 linkages. Lending further support to the idea Rpn11 as a member of the MPN+/JAMM metalloprotease, the zinc ion chelator 1,10-phenanthroline (OPA) abolished all measurable DUB activity (Fig. 1C, right), reaffirming that Rpn11 is a MPN/JAMM metalloprotease family member. To demonstrate that Rpn11 was solely responsible for DUB activity in the lid-core, we introduced the D122A substitution (37) to the active enzyme, which resulted in ablation of DUB activity (Fig. 1D). In contrast to Rpn11, significant levels of Ubp6 have been documented unassociated from proteasomes (26, 42). And indeed, Ubp6 efficiently disassembled both Lys48 and Lys63 polyUb chains uncomplexed, on its own (Fig. 1E). This DUB activity was arrested by N-ethylmaleimide (NEM), a general inhibitor of cysteine proteases. Moreover, substitution of the active site cysteine C118A rendered the enzyme inactive (Fig. 1F).
Ubiquitin Dimers Are Processed by 26S Proteasome but Are Poor Substrates for Associated DUBs
The ease of synthesis and homogeneity of dimeric Ub facilitates direct comparison of linkage types. We initiated our investigation of proteasome-associated DUBs by synthesizing the most abundant linkage types: Lys11, Lys48, and Lys63. The Km of dimeric substrates for Rpn11 has been reported to be in the hundreds of μm (29), which makes it impractical to assess conditions where the substrate concentration exceeds Km. Therefore, by working well below the Km, we calculated the apparent rate of conversion of dimer to monomer and extracted the bimolecular rate constant (nm−1 × min−1) of enzymatic efficiency. This analysis allowed the direct quantitative comparison of different enzymes for an array of substrates.
All preparations of proteasome-associated DUBs had measurable activity for all three dimers (Fig. 2A). Both Rpn11 containing complexes showed a significant preference for Lys11 linkages; however, the lid-core converted each substrate at approximately double the efficiency (Table 1). From this analysis, the lid-core and Rpn8·Rpn11 heterodimer exhibited similar linkage preferences, with efficiency for Lys11 approximately twice that for Lys48 and Lys63 (Table 1). Regardless of linkage type, the larger lid-core complex had enhanced activity demonstrated by the greater efficiency. In contrast, under the identical conditions, the preferred linkage for Ubp6 was Lys48 (Fig. 2A and Table 1).
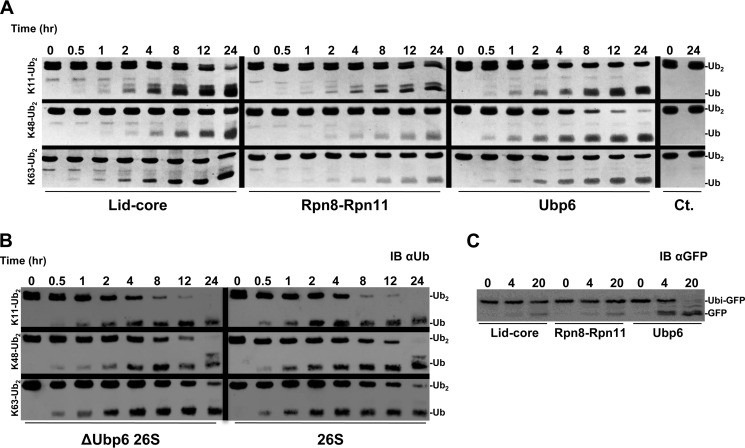
FIGURE 2.
A, Lys11-Ub2, Lys48-Ub2, and Lys63-Ub2 were incubated with the indicated enzyme, sampled over a time course, and visualized by Coomassie-stained SDS-PAGE to determine linkage preference. B, the same Ub2 substrates were incubated with ΔUbp6 proteasome or wild type and visualized with anti-Ub (IB αUb). C, Ub-GFP was incubated with the indicated enzyme and analyzed with anti-GFP (IB αGFP).
It is notable that DUBs intimately associated with the proteasome process a Lys11 linkage on par with Lys48 and Lys63. This led us to test whether the preference of the whole proteasome reflects those of its constituent DUBs. Disassembly of Lys11 was remarkably faster than of the other linkage types by 26S holoenzymes (Fig. 2B). With an increase in more than 2 orders of magnitude, the enzymatic efficiency of proteasome dwarfs its isolated constituents (Table 1). We attribute the bulk of this activity to the presence of Rpn11 in the proteasome because proteasomes lacking Ubp6 had comparable activity (Fig. 2B and Table 1). Processing of Lys11 is a new undocumented property of Rpn11 beyond the characteristic Lys63 cleavage activity of other MPN+/JAMM family members.
To reconcile the documented activity of Ubp6 on model substrates containing a single Ub domain (37, 60–62) with our current results using dimeric Ub, we assessed how each proteasomal DUB processed linear Ub fusions lacking an isopeptide bond, using the proven Ub-GFP substrate (37, 63, 64). Ubp6 deconjugated nearly all of the Ub form GFP, whereas neither Rpn11 complex at equimolar concentration was capable of efficiently hydrolyzing the peptide bond within the same time frame (Fig. 2C). The potency of Ubp6 to remove a substrate-attached Ub may explain one of the phenotypes associated with loss of Ubp6 function: low levels of cellular Ub caused by degradation of Ub at the proteasome along with its conjugate (65, 66). The results thus far point to Rpn11 accounting for the majority of Lys11 and Lys63 processing at the proteasome.
Patterns of Lys48-Ub4 and Lys63-Ub4
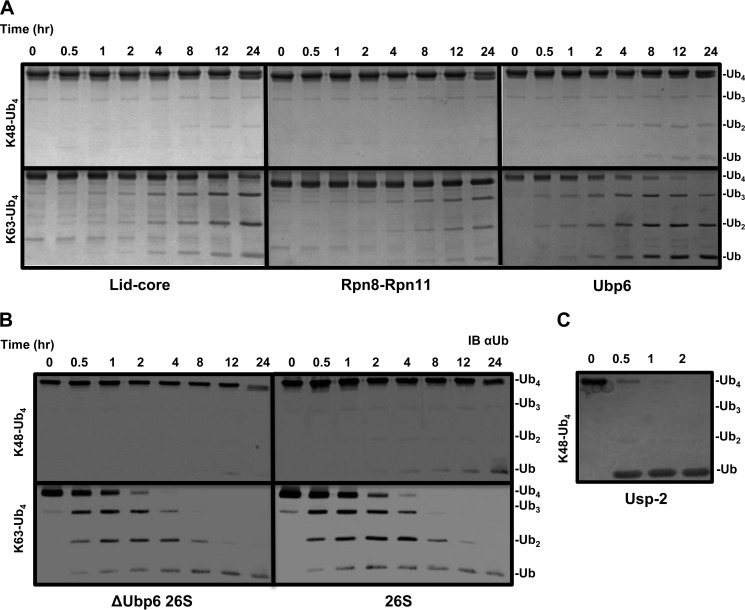
Use of dimeric Ub substrates for DUBs is informative regarding linkage specificity yet probably does not represent natural proteasome targeting signals, which often contain longer Ub polymers (67). Tetramers can provide information on the position preference (i.e. endo/exo chain cleavage) within a polymer as well as general effect of length on activity outcome. Homogeneously linked Lys48 or Lys63 tetramers were obtained in high purity and used to assess DUB action associated with proteasome. Both complexes of Rpn11 continued the trend demonstrating Lys63 linkage preference over Lys48 (Fig. 3A). Surprisingly, with tetramers, Ubp6 also exhibited preference for Lys63 linkages in contrast to dimeric substrates, where Lys48 was processed with a greater efficiency (Fig. 3A and Table 1). The resistance of Lys48-Ub4 to disassembly by individual DUBs was carried over to the proteasome, which again showed a dramatic preference for Lys63-Ub4. As was the case with processing of dimers, Ubp6 did not greatly contribute to the total DUB activity of the proteasome (Fig. 3B). Nonetheless, proteasome holoenzymes were 2 orders of magnitude more efficient in processing and therefore were able to fully convert the Lys63-Ub4 substrate within the time frame of the reaction. Increased Lys63 efficiency and the minimal effect of Ubp6 are in agreement with early reports that Rpn11 is activated when incorporated in the proteasome. USP2, a DUB unrelated to proteasome function was able to fully convert Lys48-Ub4 to monomeric units, highlighting that this substrate is not problematic for dedicated enzymes (Fig. 3C).
FIGURE 3.
A and B, Lys48-Ub4 and Lys63-Ub4 were systematically incubated with lid-core, Rpn8·Rpn11 heterodimer, and Ubp6 and visualized by Coomassie staining (A) or incubated with ΔUbp6 proteasome or wild type and visualized by anti-Ub (IB αUb) (B). C, USP2 was incubated with Lys48-Ub4 to ensure complete disassembly to monomeric Ub.
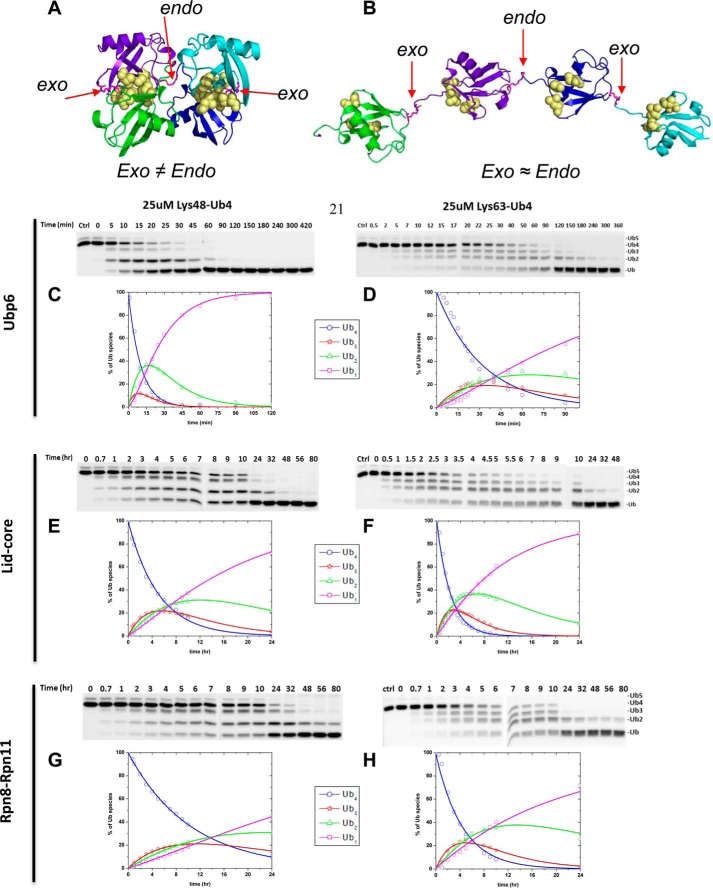
Despite having potentially more binding sites on a per mole basis, cleavage of Lys48-Ub4 by proteasomal DUBs occurred at a lower efficiency than Lys48-Ub2. Given the unique spatial environments within Ub polymers, individual isopeptide bonds are probably recognized differently (Fig. 4A and B). To address DUB preference for linkages within a chain, we designed a set of homogeneously linked Lys48 or Lys63 tetra-Ub chains in which a randomly positioned Ub unit was labeled with Alexa 488 on position 20 using the UbS20C mutant. Concentrations of DUBs were increased relative to the previous reactions, in order to collect information on all reaction intermediates. For this reason, this specific experimental setup was not used to extract enzymatic efficiency but was subject to separate kinetic analysis. Changes in concentrations of initial tetra-Ub substrate as well as the mono-, di-, and tri-Ub cleavage products were quantified independently by in-gel fluorescence. The total fluorescent signal emanating from all Ub species remained constant throughout the time course for the reaction, indicating no loss of material and supporting use of this approach for monitoring reaction progression. Nevertheless, the relative contribution of Ub intermediates did differ for certain enzyme substrate pairs. Ubp6 acting on Lys48-Ub4 stood out by initially generating one primary intermediate; Lys48-Ub2 accumulated until the Ub4 substrate was depleted, and only then was Lys48-Ub2 fully converted into the final product, mono-Ub (Fig. 4C, green curve). The low abundance of Ub3 as an intermediate suggests that Ubp6 favors the endo isopeptide bond in Ub4. Such behavior was not observed for Ubp6 with Lys63-Ub4 (Fig. 4D). Likewise, Rpn11 in either complex equally generated all three Ub intermediates initially, suggesting random action on the three isopeptide bonds in Ub4 (Fig. 4, E–H). As a note, concentration of Ub intermediates depends on both rate of generation from longer chains and conversion into shorter ones, hence the unique profile of tri- and di-Ub intermediates in each reaction.
FIGURE 4.
A, the tetrameric form of Lys48-Ub4 adopts a highly compact structural conformation at physiological conditions (100). B, in contrast, the extended conformation of tetrameric Lys63-Ub4 similarly exposes all Lys63 linkages (101). The positions of the isopeptide bonds are indicated with red arrows. The Leu8, Ile44, and Val70 hydrophobic patch of Ub is represented as yellow spheres. Ub4 disassembly was quantified by in-gel fluorescence. Ubp6 (C and D), lid-core (E and F), and Rpn8·Rpn11 heterodimer (G and H) were incubated with Alexa Fluor 488-labeled Lys48-Ub4 chains (left) or Lys63-Ub4 (right). The percentage of each Ub species calculated from contribution to total fluorescence is displayed graphically. Experimental data points are plotted as symbols along with the curves representing the results of mathematical analysis (Equations 2–5); the results for Ub4 species are shown in blue, Ub3 results are in red, Ub2 results are in green, and Ub1 results are in magenta. The total florescence signal integrated from contributions of mono-, di-, tri-, and tetra-Ub remained constant throughout the time course with less than 5% S.D.
We expanded on the advantages offered by the fluorescent labeling, which enabled a more accurate quantification of changes in each intermediate concentration. Therefore, we were not limited to monitoring solely depletion of initial substrate and could apply our mathematical models to deconvolute parallel reactions. We wrote the reaction progression equations to describe the pattern of all intermediates displayed in panels. Trend lines derived from fitting parallel reactions in Equations 2–5 were overlaid on the experimental data points (Fig. 4, C–H). Supporting our initial observation for Ubp6 acting on Lys48-Ub4, the rate constant for the endo reaction (conversion of tetramer to dimer (k4→2)) was more than twice that for each possible exo reaction (k4→3) (supplemental Fig. S2). Interestingly, Lys48-Ub3 was processed (k3→2) nearly equivalently to the preferred bond cleavage in the tetramer. In the case of Lys63-Ub4, rate constants for endo (k4→2) and exo (k4→3) cleavage were equivalent, indicating that Ubp6 lacks a preference for either site. Using the same analysis, we found that Rpn11 acts on all isopeptide linkages within a chain with equal preference; thus, we conclude that Rpn11 disassembles the chain from random position. The preference Rpn11 for Lys63 over Lys48 linkages was maintained regardless of chain length. With Ubp6, fluorophore labeling on position UbS20C slightly enhanced processing of Lys48-Ub4, presumably due to alterations in the compact conformation. However, the overall pattern of intermediates was retained (Figs. 3A and 4C).
Extended Lys11- or Lys63-linked Ub Polymers Are Preferred over Lys48 as Substrates for Disassembly by 26S Proteasome and Associated DUBs
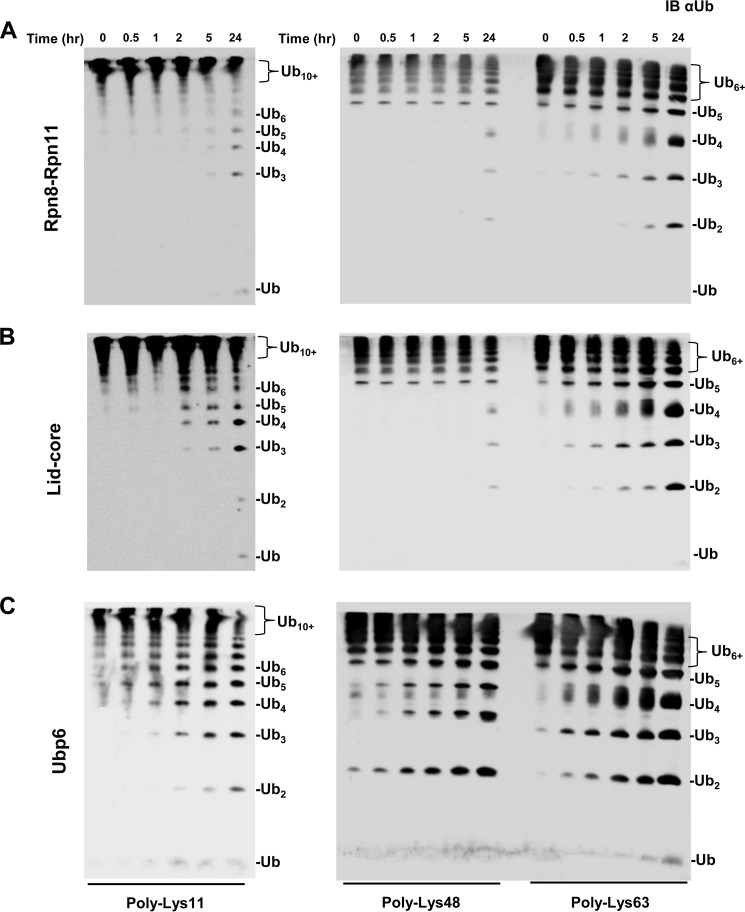
The fact that tetrameric Lys48-linked Ub chains posed a unique challenge for disassembly relative to shorter dimeric or trimeric chains encouraged us to investigate further chain length on DUB activity. For this purpose, we examined the disassembly of longer (Ub6+) homogeneously linked chains, which we refer to herein as Lys11-Ub6+, Lys48-Ub6+, and Lys63-Ub6+. Processing efficiency was calculated based on the depletion of the starting polymeric substrate at initial time points to avoid the complicated treatment of multiple reactions as the reaction progressed. Both Lys11-Ub6+ and Lys63-Ub6+ were disassembled by Rpn11-containing complexes with an efficiency exceeding that of tetramers or dimers (Table 1). Rpn11-related DUB activity revealed an interesting trend: increased efficiency with chain length for Lys11 and Lys63 polyUb and an inverse relationship for Lys48 linkages. In fact, disassembly efficiency by either Rpn11 subcomplex for Lys48-Ub6+ was not calculated due to slow progression (Fig. 5A and B). Once again, the lid-core exhibited a higher efficiency than the Rpn8·Rpn11 heterodimer (Table 1). Ubp6 was able to process both Lys11- and Lys63-linked Ub6+ conjugates with a similar efficiency to the lid-core and could even act on long Lys48 chains (Fig. 5C and Table 1). However, with these longer substrates, Lys48-linked Ub6+ was not the preferred linkage for Ubp6 (Table 1).
FIGURE 5.
Panel of homogeneously linked Ub6+ chains Lys11-Ub6+ (left), Lys48-Ub6+ (center), and Lys63-Ub6+ (right) subjected to Rpn8·Rpn11 heterodimer (A), lid-core (B), and Ubp6 (C). Products were visualized by immunoblotting with anti-Ub (IB αUb).
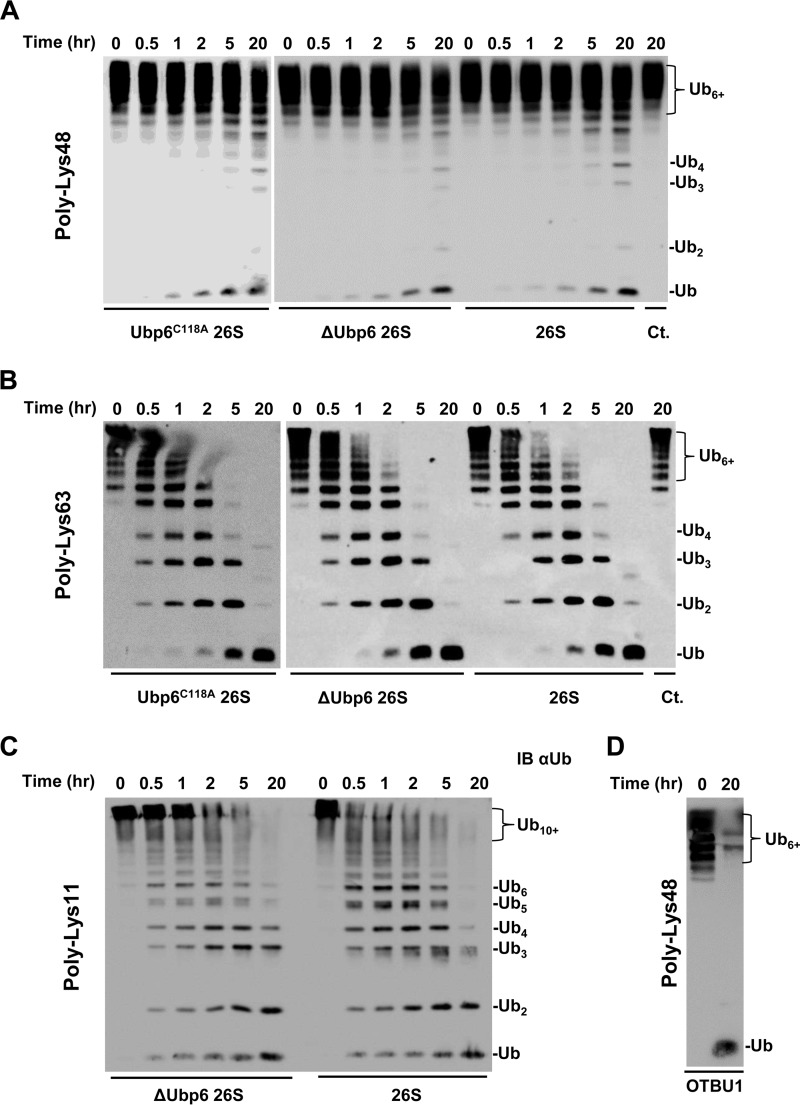
Because polyUb is considered the canonical signal for targeting substrates to the proteasome (3, 68), we continued to assess 26S proteasome action on Ub6+ of each linkage type. Enzymatic reactions were performed at 0.5 μm substrate concentration, intentionally below the reported Km for tetra-Ub (56) in order to extract bimolecular rate constants from initial rates at non saturating conditions. The proteasome was competent to disassemble Lys11-Ub6+ and Lys63-Ub6+, in contrast to Lys48-Ub6+ which was remarkably resilient to proteasome action (Fig. 6, A–C, and Table 1). The highly compact nature of Lys48-Ub6+ (supplemental Fig. S1) may explain the relatively poor processing of this substrate by proteasome-associated DUBs, in contrast to another DUB adapted for Lys48 linkages (Fig. 6D).
FIGURE 6.
A and B, proteasomes purified from ubp6C118A, Δubp6, or wild-type strains were incubated with Lys48-Ub6+ (A) and Lys63-Ub6+ (B). C, Lys11-Ub6+ was assayed with ΔUbp6 and wild-type proteasome. D, full disassembly of Lys48-Ub6+ was achieved by the addition of OTUB1; all blots were visualized using anti-Ub (IB αUb).
With Lys63-Ub6+, we expand on proteasome action for Lys63 linkages previously documented for Lys63-Ub4 (56). Although proteasome holoenzymes processed Ub polymers faster than did the individual DUBs, the overall preference for Lys11 and Lys63 over Lys48 mimicked that of Rpn11 with a dramatic increase in apparent rate constants (Table 1). This completes the trend observed in this study that larger Rpn11 assemblies possess greater rate constants, in particular for Lys11 and Lys63 linkages. To address potential catalytic and non-catalytic contributions of Ubp6 to overall proteasome DUB activity, we compared proteasomes containing active Ubp6 with those incorporating inactive (Ubp6C118A) or no Ubp6. Inactivation of or the absence of Ubp6 had a minimal effect on processing of unanchored Ub6+ of the three linkages tested (Fig. 6, A–C). The cumulative observations from our panel of unanchored polyUb chains did not identify a significant contribution of Ubp6 in the proteasome to DUB activity.
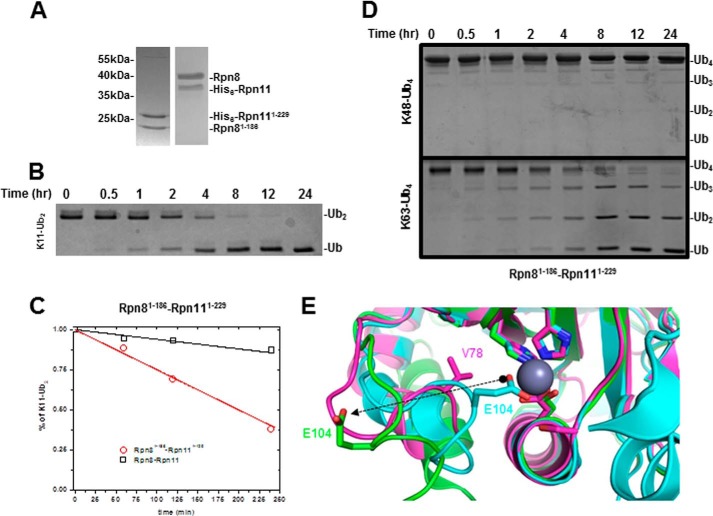
Rpn11 C Terminus Hints at Autoinhibitory Mechanism
Thus far, the bulk of DUB activity measured for proteasome complexes can be attributed to Rpn11. This suggests that Rpn11 is significantly activated when incorporated into proteasome holoenzyme. Activation of CSN5, a structurally related JAMM/MPN+ metalloprotease, is mediated by unblocking the active site zinc ion by repositioning Glu104 (69). The equivalent position in the ins-1 loop of Rpn11 is not conserved, suggesting involvement of additional resides in autoinhibition isolated from the proteasome. The C terminus of Rpn11 has been documented to facilitate the delicate coordination of Rpn11 into the proteasome (70). This encouraged us to test the impact Rpn11 C terminus has on its DUB activity. For this purpose, we obtained the truncated Rpn8(1–186)·Rpn11(1–229) heterodimer spanning the MPN domains (Fig. 7A). As a case in point, we reevaluated Rpn8(1–186)·Rpn11(1–229) with Lys11-Ub2 (Fig. 7B). Direct comparison clearly demonstrates significant activation of truncated Rpn8(1–186)·Rpn11(1–229) compared with the full-length heterodimer (Fig. 7C). Activation also extended to Lys63 chains, yet even with this enhanced activity, truncated Rpn8(1–186)·Rpn11(1–229) still had reduced ability to process Lys48-Ub4 (Fig. 7D). Truncation of the C terminus of CSN5 correlates with a 12-Å transition of Glu104 in the ins-1 loop away from the active site Zn2+ ion (Fig. 7E). Available high resolution structural data position the ins-1 loop of Rpn11 at an intermediate position between the autoinhibited and derepressed forms of CSN5 (Fig. 7E). We speculate that rearrangement of the C terminus of Rpn11 upon incorporation into proteasome is partially emulated by truncation described herein.
FIGURE 7.
A, SDS-PAGE comparing truncated Rpn8(1–186)·Rpn11(1–229) and wild type heterodimer. B, DUB activity of truncated heterodimer was assessed on Lys11-Ub2. C, initial processing of Lys11-Ub2 plotted for truncated (red) and wild-type heterodimer (black). D, truncated heterodimer fails to process Lys48-Ub4 efficiently (top); however, it is competent to process Lys63-Ub4. E, structural alignment of full-length CSN5 (Protein Data Bank entry 4D10; cyan), truncated CSN5(1–257) (Protein Data Bank entry 4F7O; green), and truncated Rpn11(2–239) (Protein Data Bank entry 4O8X; purple) shows classical MPN motif coordinating the active site Zn2+ ion. Glu104 (sticks) undergoes a 12-Å transition between the two forms of CSN5, regulating access to the Zn2+ ion in the process. Val78 (purple sticks) of Rpn11 is the equivalent of Glu104 in CSN5.
Complete Disassembly of a Heterogeneously Ubiquitinated Substrate Conjugate at the Proteasome
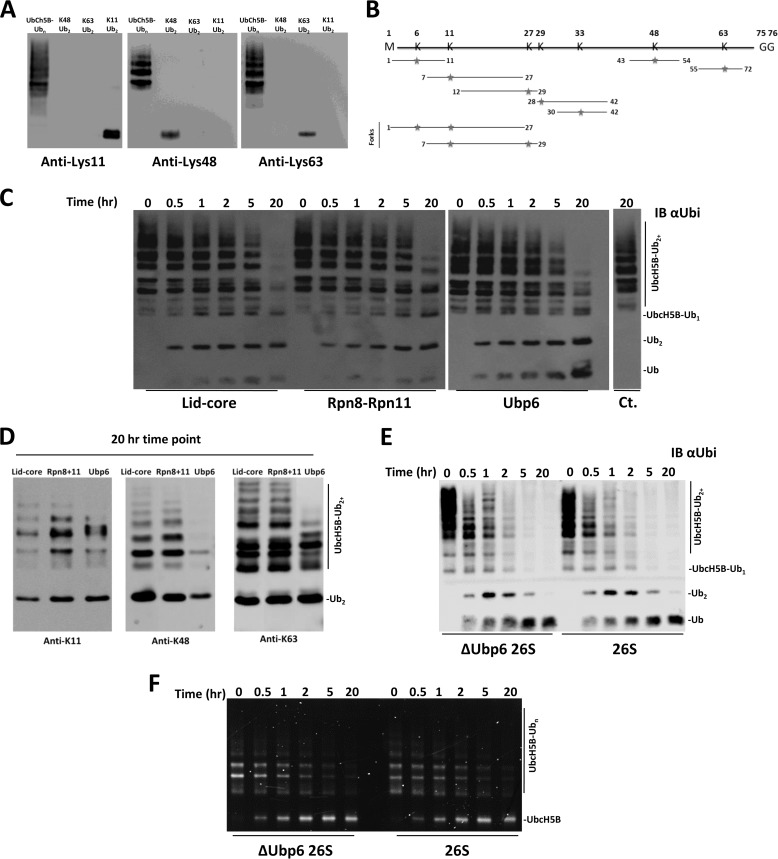
In a cellular environment, proteasomes may encounter more complex substrates heterogeneously modified with multiple Ub linkages rather than unanchored homogeneous chains (7). Moreover, in a conjugate, Ub units distal or proximal to substrate add an extra dimension for deubiquitinating machinery to distinguish. As a representative polymorphic substrate, we utilized the ability of Ubch5b to autoubiquitinate (50, 71) (supplemental Fig. S3). Lys11, Lys48, and Lys63 linkages were found across a broad range of modified Ubch5b using linkage-specific antibodies (Fig. 8A). Closer analysis using tryptic MS/MS identified all isopeptide Ub-Ub linkages and even two forked linkages, Lys6-Lys11 and Lys11-Lys27, simultaneously modifying several lysines on the Ubch5b target (verified on Lys8, Lys128, and Lys133 (50)). Ubch5b-Ubn is thereby established as an extreme example of a heterogeneously modified substrate (Fig. 8B and supplemental Fig. S4). Processing of such a complex substrate by proteasomal DUBs was an intriguing concept to put to an experimental test.
FIGURE 8.
Deconjugation of a heterogeneous polyubiquitinated substrate by 26S proteasomes and associated DUBs. A, Lys11 (left), Lys48 (center), and Lys63 (right) linkage-specific antibodies confirm the presence of all three linkage types in Ubch5b-Ubn. B, tryptic MS/MS of UbcH5B-Ubn identified all seven Ub linkage types, including forked. Tryptic GG sites are indicated with red stars (spectra are shown; supplemental Fig. S4). C, Ubch5b-Ubn was incubated with each indicated enzyme and analyzed with anti-Ub (IB αUbi). D, the 20 h end point for each enzyme was subjected to blotting with Lys11, Lys48, and Lys63 linkage-specific antibodies. E, anti-Ub reveals that all polyUb is fully removed from the Ubch5b-Ubn substrate by both ΔUbp6 and wild-type proteasome. F, a fluorescent label on Ubch5b emphasizes accumulation of free substrate following action of ΔUbp6 and wild type proteasome.
Rpn11 subcomplexes and Ubp6 were capable of deubiquitinating Ubch5b-Ubn (Fig. 8C). Although enzymatic efficiencies at initial time points were similar (Table 1), conversion of Ubch5b-Ubn by Ubp6 was more complete (Fig. 8C). This may reflect a broader range of associated activities for Ubp6 as an isolated enzyme. After prolonged incubation with each DUB, we analyzed the residual short modifications on Ubch5b using linkage specific antibodies (Fig. 8D). Ubp6 eliminated nearly all Lys48 linkages, whereas the lid-core eliminated nearly all of the Lys11 linkages in Ubch5b-Ubn (Fig. 8D). As a side product, accumulation of unanchored dimeric Ub supports the common theme emanating from our current work, that dimers of all linkages are poor substrates compared with longer polyUb chains.
As with homogeneously linked polyUb chains, the proteasome processed Ubch5b-Ubn with an efficiency nearly 100-fold greater than any constituent DUB. Once again, Ubp6 had a minimal effect on overall DUB activity of the proteasome (Fig. 8E and Table 1). When presented simultaneously with multiple linkages on the same substrate, the proteasome experienced no limitation in processing, reflecting diverse capabilities harbored in resident DUBs. To determine the fate of Ubch5b modified with this mixture of Ub linkages, we tracked the substrate with a fluorescent label. Indicating a lack of degradation, a Ub-free form of Ubch5b accumulated as a result of proteasomal deubiquitination (Fig. 8F). Consequently, deubiquitination proceeded to completion, removing all modifications from the substrate. Previous studies have demonstrated that branched polyUb chains can serve as substrate for the proteasome (18, 48), and expanding on this concept, we demonstrate that mixed and branched modifications render a conjugate as an adequate substrate for deubiquitination (Fig. 8, A–F).
DISCUSSION
The 26S proteasome emerges from this study as a broad spectrum multicatalytic DUB, capable of reducing a heterogeneously modified conjugate containing all linkage types down to mono-Ub. Disassembly of polymeric Ub, especially of well defined linkage type or chain length, poses experimental challenges in the synthesis of substrates and in the isolation, characterization, and quantification of generated products. Due to these experimental constraints, DUBs are routinely studied with synthetic model substrates that allow for easy detection of liberated products or for continuous assays. Hence, fluorescently labeled Ub (e.g. Ub-AMC) or substrates modified by a single Ub are prevalent in experimental setups. The current study found that Ub dimers are poor substrates for deconjugation by proteasomes or their associated DUBs. Not only was processing slow, but Ub dimers failed to expose significant differences between linkages. Purified homogeneously linked Ub tetramers or longer poly-Ub6+ proved useful to flush out differences in the processing of linkage types. For instance, the potency of proteasome complexes or their constituent DUBs for disassembling Lys11-linked polyUb became apparent. Longer or more complex chains may provide the three-dimensional spatial information needed for recognition or efficient processing by proteasomes (supplemental Fig. S1).
Regardless of linkage type, processing of chains was generally more efficient than removal of a single conjugated Ub (as in unanchored Ub dimers). However, one notable exception was polyUb linked via Lys48 (e.g. Lys48-Ub4 or Lys48-Ub6+ chains) that were processed with difficulty. The compact conformation of Lys48-linked polyUb longer than 4 units (supplemental Fig. S5) may obstruct access of cleavage sites to certain DUBs (72, 73). Could Lys48-linked tetra-Ub be the optimal “Goldilocks” proteasomal signal (74) precisely because it is rather resilient to disassembly? In this regard, Lys63 linkages are thought to be poor proteasome-targeting signals (56, 57), presumably because they can be disassembled rapidly and are therefore unable to provide the residency time required to commit a substrate for unfolding. Given that we have now uncovered an ability of proteasomes to rapidly disassemble Lys11 linkages, it is unclear how they may serve as a targeting signal (17, 18, 75, 76). Beyond homogeneously linked chains, mixing of linkages into a single chain or onto a single substrate may provide for new signaling properties (77, 78), unexpected when considering information from individual linkages alone. Mixed or branched linkages may disrupt a repetitive configuration of a homogeneous chain, exposing initiation sites for DUBs. This may be one explanation for efficient proteasome processing of the heterogeneously modified Ubch5b-Ubn, although it was abundant in Lys48 linkages. Likewise, a recent study demonstrated that a branched Lys11/Lys48-linked chain had a greater affinity for proteasome receptor S5a and was also deconjugated faster at the proteasome than either homogeneous Lys11- or Lys48-linked chains (18). Nevertheless, linkage branching should be considered on a case-by-case basis, given that some types of forked linkages have been reported to inhibit proteasomal degradation (79), whereas a Lys48/Lys63-linked branched chain had no effect on deconjugation (48).
In separate studies, activities of recombinant Rpn8·Rpn11 or USP14 were demonstrated against short substrates (25, 28, 29). Further expanding on these findings, Ubp6 and Rpn11 cleaved a range of longer Ub conjugates linked via Lys11, Lys48, or Lys63 as well as heterogeneously. Biological roles of Ubp6 or Rpn11 could henceforth be evaluated under the notion that a portion is uncoupled from the proteasome and partakes in biological pathways that do not necessarily involve Lys48 linkages (80–82). Involvement of Rpn11 in Lys63-linked Ub signaling at DNA double-stranded breaks has been documented, but it was unclear whether Rpn11 acted independently of the 20S (83). Other MPN+/JAMM family DUBs are also known to act in Lys63-signaling pathways, such as DNA repair or membrane trafficking (36, 58, 59, 83–85). With the development of functional Rpn11 and Ubp6 assays, their direct involvement in these processes could now be reevaluated also for Lys11 linkages.
Relative to either DUB, proteasome deubiquitination efficiency was enhanced over 100-fold. With a model substrate, Ubch5b-Ubn, a multiubiquitinated globular protein, proteasomes selectively deconjugated rather than proteolyzed the substrate. Under these conditions, removal of Ubp6 had only a marginal effect on overall proteasome DUB activity. One explanation is that proteasome-incorporated Ubp6 remains in a latent form until triggered into action either by substrate fully committed to degradation after initiation of unfolding (60, 86, 87) or by conformational changes upon engagement of RPT ATPases (32). In either scenario, we propose that the primary contribution of Ubp6 is near the end of the proteasome catalytic cycle. In contrast, deconjugating efficiency of proteasome-incorporated Rpn11 was enhanced against the same model substrate. PolyUb receptors on the proteasome (Rpn10 and Rpn13) may compensate for the low inherent affinity of Rpn11 for Ub (29, 56). Rpn10, in particular, is situated adjacent to Rpn11 within the 19S RP (30, 43, 88, 89). Activation of Rpn11 may also be driven through a conformational change upon incorporation into the proteasome involving slight rearrangements of Zn2+-coordinating residues in the active site (Fig. 7E). That the Rpn8·Rpn11 heterodimer was more active as truncated MPN domains rather than as full-length proteins (Fig. 7) points to a role for their C termini in autoinhibition until property incorporated into proteasomes. Indeed, the C termini participate in critical subunit interactions stabilizing mature proteasome (90, 91). Given that lid-core was also less active than Rpn11 lacking its C terminus, the full extent of Rpn11 activation probably involves interactions beyond the lid subcomplex, for instance by synchronization with AAA-ATPases (29). In the cell, proteasomes encounter numerous ubiquitinated proteins that are not necessarily intended for degradation. By removing Lys11 and Lys63 linkages, the preference of Rpn11 may aid in deflecting these substrates from an unwanted fate, acting independently uncoupled from degradation.
The central role of the proteasome in protein homeostasis, as well as its ability to regulate oncogenic factors and toxic proteins, makes it a lead pharmacological target (92). Inhibition of USP14 increased the turnover of oxidized and damaged proteins by the proteasome (60). In addition, the therapeutic importance of inhibiting USP14 and UCHL5 have been demonstrated for controlling progression of acute myeloid leukemia (63, 64) or tumor growth (93). Although its roles in cell viability, life span, and cancers (94–96) make it a promising drug target (62), there are currently no specific inhibitors for the Rpn11 metalloprotease. An ability to study individual proteasome-associated DUBs in isolation should facilitate intensive ongoing drug discovery efforts (61, 63, 93, 97–99) relative to the dynamic, ATP-dependent, multisubunit proteasome complex.
This work was supported, in whole or in part, by National Institutes of Health Grants GM065334 (to D. F.) and GM095755 (to D. F. and M. H. G.). This work was also supported by a USA-Israel Binational Science Foundation grant (to D. F. and M. H. G.) and a Deutsch-Israel Program grant (to T. S. and M. H. G.).

This article contains supplemental Table S1 and Figs. S1–S5.
- polyUb
- polyubiquitin
- Ub
- ubiquitin
- DUB
- deubiquitinase
- RP
- regulatory particle(s)
- AMC
- 7-amino-4-methylcoumarin.
REFERENCES
- 1. Hershko A., Ciechanover A., Varshavsky A. (2000) Basic Medical Research Award: the ubiquitin system. Nat. Med. 6, 1073–1081 [DOI] [PubMed] [Google Scholar]
- 2. Komander D. (2009) The emerging complexity of protein ubiquitination. Biochem. Soc Trans 37, 937–953 [DOI] [PubMed] [Google Scholar]
- 3. Finley D. (2009) Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu. Rev. Biochem. 78, 477–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Glickman M. H., Ciechanover A. (2002) The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol. Rev. 82, 373–428 [DOI] [PubMed] [Google Scholar]
- 5. Ikeda F., Dikic I. (2008) Atypical ubiquitin chains: new molecular signals. EMBO Rep. 9, 536–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pickart C. M., Fushman D. (2004) Polyubiquitin chains: polymeric protein signals. Curr. Opin. Chem. Biol. 8, 610–616 [DOI] [PubMed] [Google Scholar]
- 7. Kravtsova-Ivantsiv Y., Sommer T., Ciechanover A. (2013) The lysine48-based polyubiquitin chain proteasomal signal: not a single child anymore. Angew Chem. Int. Ed. Engl. 52, 192–198 [DOI] [PubMed] [Google Scholar]
- 8. Volk S., Wang M., Pickart C. M. (2005) Chemical and genetic strategies for manipulating polyubiquitin chain structure. Methods Enzymol. 399, 3–20 [DOI] [PubMed] [Google Scholar]
- 9. Reyes-Turcu F. E., Ventii K. H., Wilkinson K. D. (2009) Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu. Rev. Biochem. 78, 363–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Komander D. (2010) Mechanism, specificity and structure of the deubiquitinases. Subcell. Biochem. 54, 69–87 [DOI] [PubMed] [Google Scholar]
- 11. Lee M. J., Lee B. H., Hanna J., King R. W., Finley D. (2011) Trimming of ubiquitin chains by proteasome-associated deubiquitinating enzymes. Mol. Cell Proteomics 10.1074/mcp.R110.003871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Eletr Z. M., Wilkinson K. D. (2014) Regulation of proteolysis by human deubiquitinating enzymes. Biochim. Biophys. Acta 1843, 114–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ziv I., Matiuhin Y., Kirkpatrick D. S., Erpapazoglou Z., Leon S., Pantazopoulou M., Kim W., Gygi S. P., Haguenauer-Tsapis R., Reis N., Glickman M. H., Kleifeld O. (2011) A perturbed ubiquitin landscape distinguishes between ubiquitin in trafficking and in proteolysis. Mol. Cell Proteomics 10, 10.1074/mcp.M111.009753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dammer E. B., Na C. H., Xu P., Seyfried N. T., Duong D. M., Cheng D., Gearing M., Rees H., Lah J. J., Levey A. I., Rush J., Peng J. (2011) Polyubiquitin linkage profiles in three models of proteolytic stress suggest the etiology of Alzheimer disease. J. Biol. Chem. 286, 10457–10465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dikic I., Dötsch V. (2009) Ubiquitin linkages make a difference. Nat. Struct. Mol. Biol. 16, 1209–1210 [DOI] [PubMed] [Google Scholar]
- 16. Kim W., Bennett E. J., Huttlin E. L., Guo A., Li J., Possemato A., Sowa M. E., Rad R., Rush J., Comb M. J., Harper J. W., Gygi S. P. (2011) Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell 44, 325–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Matsumoto M. L., Wickliffe K. E., Dong K. C., Yu C., Bosanac I., Bustos D., Phu L., Kirkpatrick D. S., Hymowitz S. G., Rape M., Kelley R. F., Dixit V. M. (2010) K11-linked polyubiquitination in cell cycle control revealed by a K11 linkage-specific antibody. Mol. Cell 39, 477–484 [DOI] [PubMed] [Google Scholar]
- 18. Meyer H. J., Rape M. (2014) Enhanced protein degradation by branched ubiquitin chains. Cell 157, 910–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Locke M., Toth J. I., Petroski M. D. (2014) Lys11- and Lys48-linked ubiquitin chains interact with p97 during endoplasmic-reticulum-associated degradation. Biochem. J. 459, 205–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Glickman M. H., Rubin D. M., Fried V. A., Finley D. (1998) The regulatory particle of the Saccharomyces cerevisiae proteasome. Mol. Cell. Biol. 18, 3149–3162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tanaka K., Mizushima T., Saeki Y. (2012) The proteasome: molecular machinery and pathophysiological roles. Biol. Chem. 393, 217–234 [DOI] [PubMed] [Google Scholar]
- 22. Ambroggio X. I., Rees D. C., Deshaies R. J. (2004) JAMM: a metalloprotease-like zinc site in the proteasome and signalosome. PLoS Biol. 2, E2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Verma R., Aravind L., Oania R., McDonald W. H., Yates J. R., 3rd, Koonin E. V., Deshaies R. J. (2002) Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science 298, 611–615 [DOI] [PubMed] [Google Scholar]
- 24. Koulich E., Li X., DeMartino G. N. (2008) Relative structural and functional roles of multiple deubiquitylating proteins associated with mammalian 26S proteasome. Mol. Biol. Cell 19, 1072–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hu M., Li P., Song L., Jeffrey P. D., Chenova T. A., Wilkinson K. D., Cohen R. E., Shi Y. (2005) Structure and mechanisms of the proteasome-associated deubiquitinating enzyme USP14. EMBO J. 24, 3747–3756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rosenzweig R., Bronner V., Zhang D., Fushman D., Glickman M. H. (2012) Rpn1 and Rpn2 coordinate ubiquitin processing factors at proteasome. J. Biol. Chem. 287, 14659–14671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Maytal-Kivity V., Reis N., Hofmann K., Glickman M. H. (2002) MPN+, a putative catalytic motif found in a subset of MPN domain proteins from eukaryotes and prokaryotes, is critical for Rpn11 function. BMC Biochem. 3, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pathare G. R., Nagy I., Śledź P., Anderson D. J., Zhou H. J., Pardon E., Steyaert J., Förster F., Bracher A., Baumeister W. (2014) Crystal structure of the proteasomal deubiquitylation module Rpn8-Rpn11. Proc. Natl. Acad. Sci. U.S.A. 111, 2984–2989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Worden E. J., Padovani C., Martin A. (2014) Structure of the Rpn11-Rpn8 dimer reveals mechanisms of substrate deubiquitination during proteasomal degradation. Nat. Struct. Mol. Biol. 21, 220–227 [DOI] [PubMed] [Google Scholar]
- 30. Bhattacharyya S., Yu H., Mim C., Matouschek A. (2014) Regulated protein turnover: snapshots of the proteasome in action. Nat. Rev. Mol. Cell Biol. 15, 122–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Matyskiela M. E., Lander G. C., Martin A. (2013) Conformational switching of the 26S proteasome enables substrate degradation. Nat. Struct. Mol. Biol. 20, 781–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Peth A., Kukushkin N., Bossé M., Goldberg A. L. (2013) Ubiquitinated proteins activate the proteasomal ATPases by binding to Usp14 or Uch37 homologs. J. Biol. Chem. 288, 7781–7790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Peth A., Besche H. C., Goldberg A. L. (2009) Ubiquitinated proteins activate the proteasome by binding to Usp14/Ubp6, which causes 20S gate opening. Mol. Cell 36, 794–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hanna J., Hathaway N. A., Tone Y., Crosas B., Elsasser S., Kirkpatrick D. S., Leggett D. S., Gygi S. P., King R. W., Finley D. (2006) Deubiquitinating enzyme Ubp6 functions noncatalytically to delay proteasomal degradation. Cell 127, 99–111 [DOI] [PubMed] [Google Scholar]
- 35. Yao T., Cohen R. E. (2002) A cryptic protease couples deubiquitination and degradation by the proteasome. Nature 419, 403–407 [DOI] [PubMed] [Google Scholar]
- 36. Cooper E. M., Cutcliffe C., Kristiansen T. Z., Pandey A., Pickart C. M., Cohen R. E. (2009) K63-specific deubiquitination by two JAMM/MPN+ complexes: BRISC-associated Brcc36 and proteasomal Poh1. EMBO J. 28, 621–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Guterman A., Glickman M. H. (2004) Complementary roles for Rpn11 and Ubp6 in deubiquitination and proteolysis by the proteasome. J. Biol. Chem. 279, 1729–1738 [DOI] [PubMed] [Google Scholar]
- 38. Glickman M. H., Adir N. (2004) The proteasome and the delicate balance between destruction and rescue. PLoS Biol. 2, E13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Inobe T., Matouschek A. (2014) Paradigms of protein degradation by the proteasome. Curr. Opin. Struct. Biol. 24, 156–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Peth A., Uchiki T., Goldberg A. L. (2010) ATP-dependent steps in the binding of ubiquitin conjugates to the 26S proteasome that commit to degradation. Mol. Cell 40, 671–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Leggett D. S., Hanna J., Borodovsky A., Crosas B., Schmidt M., Baker R. T., Walz T., Ploegh H., Finley D. (2002) Multiple associated proteins regulate proteasome structure and function. Mol. Cell 10, 495–507 [DOI] [PubMed] [Google Scholar]
- 42. Matiuhin Y., Kirkpatrick D. S., Ziv I., Kim W., Dakshinamurthy A., Kleifeld O., Gygi S. P., Reis N., Glickman M. H. (2008) Extraproteasomal Rpn10 restricts access of the polyubiquitin-binding protein Dsk2 to proteasome. Mol. Cell 32, 415–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lander G. C., Estrin E., Matyskiela M. E., Bashore C., Nogales E., Martin A. (2012) Complete subunit architecture of the proteasome regulatory particle. Nature 482, 186–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rogov V. V., Rozenknop A., Rogova N. Y., Löhr F., Tikole S., Jaravine V., Guntert P., Dikic I., Dötsch V. (2012) A universal expression tag for structural and functional studies of proteins. Chembiochem 13, 959–963 [DOI] [PubMed] [Google Scholar]
- 45. Pickart C. M., Raasi S. (2005) Controlled synthesis of polyubiquitin chains. Methods Enzymol. 399, 21–36 [DOI] [PubMed] [Google Scholar]
- 46. Bagola K., von Delbrück M., Dittmar G., Scheffner M., Ziv I., Glickman M. H., Ciechanover A., Sommer T. (2013) Ubiquitin binding by a CUE domain regulates ubiquitin chain formation by ERAD E3 ligases. Mol. Cell 50, 528–539 [DOI] [PubMed] [Google Scholar]
- 47. Berndsen C. E., Wolberger C. (2011) A spectrophotometric assay for conjugation of ubiquitin and ubiquitin-like proteins. Anal. Biochem. 418, 102–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nakasone M. A., Livnat-Levanon N., Glickman M. H., Cohen R. E., Fushman D. (2013) Mixed-linkage ubiquitin chains send mixed messages. Structure 21, 727–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Castañeda C. A., Kashyap T. R., Nakasone M. A., Krueger S., Fushman D. (2013) Unique structural, dynamical, and functional properties of k11-linked polyubiquitin chains. Structure 21, 1168–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cannon J., Nakasone M., Fushman D., Fenselau C. (2012) Proteomic identification and analysis of K63-linked ubiquitin conjugates. Anal. Chem. 84, 10121–10128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Keller A., Eng J., Zhang N., Li X. J., Aebersold R. (2005) A uniform proteomics MS/MS analysis platform utilizing open XML file formats. Mol. Syst. Biol. 1, 2005.0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Fu H., Reis N., Lee Y., Glickman M. H., Vierstra R. D. (2001) Subunit interaction maps for the regulatory particle of the 26S proteasome and the COP9 signalosome. EMBO J. 20, 7096–7107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lasker K., Förster F., Bohn S., Walzthoeni T., Villa E., Unverdorben P., Beck F., Aebersold R., Sali A., Baumeister W. (2012) Molecular architecture of the 26S proteasome holocomplex determined by an integrative approach. Proc. Natl. Acad. Sci. U.S.A. 109, 1380–1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Glickman M. H., Rubin D. M., Larsen C. N., Finley D. (2000) in Proteasomes: The World of Regultory Proteolysis (Hilt W., Wolf D. H., eds) pp. 71–90, Eurekah.com/Landes Bioscience Publishing Co., Georgetown, TX [Google Scholar]
- 55. Leggett D. S., Glickman M. H., Finley D. (2005) Purification of proteasomes, proteasome subcomplexes, and proteasome-associated proteins from budding yeast. Methods Mol. Biol. 301, 57–70 [DOI] [PubMed] [Google Scholar]
- 56. Jacobson A. D., Zhang N. Y., Xu P., Han K. J., Noone S., Peng J., Liu C. W. (2009) The lysine 48 and lysine 63 ubiquitin conjugates are processed differently by the 26 S proteasome. J. Biol. Chem. 284, 35485–35494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Nathan J. A., Kim H. T., Ting L., Gygi S. P., Goldberg A. L. (2013) Why do cellular proteins linked to K63-polyubiquitin chains not associate with proteasomes? EMBO J. 32, 552–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Cooper E. M., Boeke J. D., Cohen R. E. (2010) Specificity of the BRISC deubiquitinating enzyme is not due to selective binding to Lys63-linked polyubiquitin. J. Biol. Chem. 285, 10344–10352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sato Y., Yoshikawa A., Yamagata A., Mimura H., Yamashita M., Ookata K., Nureki O., Iwai K., Komada M., Fukai S. (2008) Structural basis for specific cleavage of Lys 63-linked polyubiquitin chains. Nature 455, 358–362 [DOI] [PubMed] [Google Scholar]
- 60. Lee B. H., Lee M. J., Park S., Oh D. C., Elsasser S., Chen P. C., Gartner C., Dimova N., Hanna J., Gygi S. P., Wilson S. M., King R. W., Finley D. (2010) Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature 467, 179–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lee B. H., Finley D., King R. W. (2012) A high-throughput screening method for identification of inhibitors of the deubiquitinating enzyme USP14. Curr. Protoc. Chem. Biol. 4, 311–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Krutauz D., Reis N., Nakasone M. A., Siman P., Zhang D., Kirkpatrick D. S., Gygi S. P., Brik A., Fushman D., Glickman M. H. (2014) Extended ubiquitin species are protein-based DUB inhibitors. Nat. Chem. Biol. 10, 664–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. D'Arcy P., Linder S. (2012) Proteasome deubiquitinases as novel targets for cancer therapy. Int. J. Biochem. Cell Biol. 44, 1729–1738 [DOI] [PubMed] [Google Scholar]
- 64. D'Arcy P., Brnjic S., Olofsson M. H., Fryknäs M., Lindsten K., De Cesare M., Perego P., Sadeghi B., Hassan M., Larsson R., Linder S. (2011) Inhibition of proteasome deubiquitinating activity as a new cancer therapy. Nat. Med. 17, 1636–1640 [DOI] [PubMed] [Google Scholar]
- 65. Hanna J., Leggett D. S., Finley D. (2003) Ubiquitin depletion as a key mediator of toxicity by translational inhibitors. Mol. Cell. Biol. 23, 9251–9261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Anderson C., Crimmins S., Wilson J. A., Korbel G. A., Ploegh H. L., Wilson S. M. (2005) Loss of Usp14 results in reduced levels of ubiquitin in ataxia mice. J. Neurochem. 95, 724–731 [DOI] [PubMed] [Google Scholar]
- 67. Shabek N., Herman-Bachinsky Y., Buchsbaum S., Lewinson O., Haj-Yahya M., Hejjaoui M., Lashuel H. A., Sommer T., Brik A., Ciechanover A. (2012) The size of the proteasomal substrate determines whether its degradation will be mediated by mono- or polyubiquitylation. Mol. Cell 48, 87–97 [DOI] [PubMed] [Google Scholar]
- 68. Fushman D., Wilkinson K. D. (2011) Structure and recognition of polyubiquitin chains of different lengths and linkage. F1000 Biol. Rep. 3, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Lingaraju G. M., Bunker R. D., Cavadini S., Hess D., Hassiepen U., Renatus M., Fischer E. S., Thomä N. H. (2014) Crystal structure of the human COP9 signalosome. Nature 512, 161–165 [DOI] [PubMed] [Google Scholar]
- 70. Rinaldi T., Hofmann L., Gambadoro A., Cossard R., Livnat-Levanon N., Glickman M. H., Frontali L., Delahodde A. (2008) Dissection of the carboxyl-terminal domain of the proteasomal subunit Rpn11 in maintenance of mitochondrial structure and function. Mol. Biol. Cell 19, 1022–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Kim H. T., Kim K. P., Lledias F., Kisselev A. F., Scaglione K. M., Skowyra D., Gygi S. P., Goldberg A. L. (2007) Certain pairs of ubiquitin-conjugating enzymes (E2s) and ubiquitin-protein ligases (E3s) synthesize nondegradable forked ubiquitin chains containing all possible isopeptide linkages. J. Biol. Chem. 282, 17375–17386 [DOI] [PubMed] [Google Scholar]
- 72. Varadan R., Walker O., Pickart C., Fushman D. (2002) Structural properties of polyubiquitin chains in solution. J. Mol. Biol. 324, 637–647 [DOI] [PubMed] [Google Scholar]
- 73. Schaefer J. B., Morgan D. O. (2011) Protein-linked ubiquitin chain structure restricts activity of deubiquitinating enzymes. J. Biol. Chem. 286, 45186–45196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Thrower J. S., Hoffman L., Rechsteiner M., Pickart C. M. (2000) Recognition of the polyubiquitin proteolytic signal. EMBO J. 19, 94–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Wickliffe K. E., Williamson A., Meyer H. J., Kelly A., Rape M. (2011) K11-linked ubiquitin chains as novel regulators of cell division. Trends Cell Biol. 21, 656–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Xu P., Duong D. M., Seyfried N. T., Cheng D., Xie Y., Robert J., Rush J., Hochstrasser M., Finley D., Peng J. (2009) Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell 137, 133–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Lin D. Y., Diao J., Zhou D., Chen J. (2011) Biochemical and structural studies of a HECT-like ubiquitin ligase from Escherichia coli O157:H7. J. Biol. Chem. 286, 441–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Ben-Saadon R., Zaaroor D., Ziv T., Ciechanover A. (2006) The polycomb protein Ring1B generates self atypical mixed ubiquitin chains required for its in vitro histone H2A ligase activity. Mol. Cell 24, 701–711 [DOI] [PubMed] [Google Scholar]
- 79. Kim H. T., Kim K. P., Uchiki T., Gygi S. P., Goldberg A. L. (2009) S5a promotes protein degradation by blocking synthesis of nondegradable forked ubiquitin chains. EMBO J. 28, 1867–1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Qin S., Wang Q., Ray A., Wani G., Zhao Q., Bhaumik S. R., Wani A. A. (2009) Sem1p and Ubp6p orchestrate telomeric silencing by modulating histone H2B ubiquitination and H3 acetylation. Nucleic Acids Res. 37, 1843–1853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Wu N., Liu C., Bai C., Han Y. P., Cho W. C., Li Q. (2013) Over-expression of deubiquitinating enzyme USP14 in lung adenocarcinoma promotes proliferation through the accumulation of β-catenin. Int. J. Mol. Sci. 14, 10749–10760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Jung H., Kim B. G., Han W. H., Lee J. H., Cho J. Y., Park W. S., Maurice M. M., Han J. K., Lee M. J., Finley D., Jho E. H. (2013) Deubiquitination of dishevelled by Usp14 is required for Wnt signaling. Oncogenesis 2, e64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Butler L. R., Densham R. M., Jia J., Garvin A. J., Stone H. R., Shah V., Weekes D., Festy F., Beesley J., Morris J. R. (2012) The proteasomal de-ubiquitinating enzyme POH1 promotes the double-strand DNA break response. EMBO J. 31, 3918–3934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Feng L., Wang J., Chen J. (2010) The Lys63-specific deubiquitinating enzyme BRCC36 is regulated by two scaffold proteins localizing in different subcellular compartments. J. Biol. Chem. 285, 30982–30988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Patterson-Fortin J., Shao G., Bretscher H., Messick T. E., Greenberg R. A. (2010) Differential regulation of JAMM domain deubiquitinating enzyme activity within the RAP80 complex. J. Biol. Chem. 285, 30971–30981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Schrader E. K., Harstad K. G., Matouschek A. (2009) Targeting proteins for degradation. Nat. Chem. Biol. 5, 815–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Kraut D. A., Prakash S., Matouschek A. (2007) To degrade or release: ubiquitin-chain remodeling. Trends Cell Biol. 17, 419–421 [DOI] [PubMed] [Google Scholar]
- 88. da Fonseca P. C., He J., Morris E. P. (2012) Molecular model of the human 26S proteasome. Mol. Cell 46, 54–66 [DOI] [PubMed] [Google Scholar]
- 89. Beck F., Unverdorben P., Bohn S., Schweitzer A., Pfeifer G., Sakata E., Nickell S., Plitzko J. M., Villa E., Baumeister W., Förster F. (2012) Near-atomic resolution structural model of the yeast 26S proteasome. Proc. Natl. Acad. Sci. U.S.A. 109, 14870–14875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Chandra A., Chen L., Madura K. (2010) Synthetic lethality of rpn11–1 rpn10Δ is linked to altered proteasome assembly and activity. Curr. Genet. 56, 543–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Rinaldi T., Pick E., Gambadoro A., Zilli S., Maytal-Kivity V., Frontali L., Glickman M. H. (2004) Participation of the proteasomal lid subunit Rpn11 in mitochondrial morphology and function is mapped to a distinct C-terminal domain. Biochem. J. 381, 275–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Goldberg A. L. (2012) Development of proteasome inhibitors as research tools and cancer drugs. J. Cell Biol. 199, 583–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Liu N., Liu C., Li X., Liao S., Song W., Yang C., Zhao C., Huang H., Guan L., Zhang P., Liu S., Hua X., Chen X., Zhou P., Lan X., Yi S., Wang S., Wang X., Dou Q. P., Liu J. (2014) A novel proteasome inhibitor suppresses tumor growth via targeting both 19S proteasome deubiquitinases and 20S proteolytic peptidases. Sci. Rep. 4, 5240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Schmidt M., Finley D. (2014) Regulation of proteasome activity in health and disease. Biochim. Biophys. Acta 1843, 13–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Tonoki A., Kuranaga E., Tomioka T., Hamazaki J., Murata S., Tanaka K., Miura M. (2009) Genetic evidence linking age-dependent attenuation of the 26S proteasome with the aging process. Mol. Cell. Biol. 29, 1095–1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Gallery M., Blank J. L., Lin Y., Gutierrez J. A., Pulido J. C., Rappoli D., Badola S., Rolfe M., Macbeth K. J. (2007) The JAMM motif of human deubiquitinase Poh1 is essential for cell viability. Mol. Cancer Ther. 6, 262–268 [DOI] [PubMed] [Google Scholar]
- 97. Tian Z., D'Arcy P., Wang X., Ray A., Tai Y. T., Hu Y., Carrasco R. D., Richardson P., Linder S., Chauhan D., Anderson K. C. (2014) A novel small molecule inhibitor of deubiquitylating enzyme USP14 and UCHL5 induces apoptosis in multiple myeloma and overcomes bortezomib resistance. Blood 123, 706–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Nag D. K., Finley D. (2012) A small-molecule inhibitor of deubiquitinating enzyme USP14 inhibits Dengue virus replication. Virus Res. 165, 103–106 [DOI] [PubMed] [Google Scholar]
- 99. Kapuria V., Peterson L. F., Fang D., Bornmann W. G., Talpaz M., Donato N. J. (2010) Deubiquitinase inhibition by small-molecule WP1130 triggers aggresome formation and tumor cell apoptosis. Cancer Res. 70, 9265–9276 [DOI] [PubMed] [Google Scholar]
- 100. Eddins M. J., Varadan R., Fushman D., Pickart C. M., Wolberger C. (2007) Crystal structure and solution NMR studies of Lys48-linked tetraubiquitin at neutral pH. J. Mol. Biol. 367, 204–211 [DOI] [PubMed] [Google Scholar]
- 101. Datta A. B., Hura G. L., Wolberger C. (2009) The structure and conformation of Lys63-linked tetraubiquitin. J. Mol. Biol. 392, 1117–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]