Background: Nucleolin is a multifunctional protein, but nucleolin-SUMO is unexploited.
Results: Nucleolin-SUMO at Lys-294 facilitated binding with mRNA substrate gadd45α by maintaining its nuclear localization during cellular response to arsenite exposure.
Conclusion: Nucleolin-SUMO promoted arsenite-induced apoptosis by increasing GADD45α expression.
Significance: We identified a new modification of nucleolin and its contribution to the functional paradigm of nucleolin in mRNA stability regulation.
Keywords: Apoptosis, Metal, Nucleolus, Oxidative Stress, Small Ubiquitin-like Modifier (SUMO)
Abstract
Nucleolin is a ubiquitously expressed protein and participates in many important biological processes, such as cell cycle regulation and ribosomal biogenesis. The activity of nucleolin is regulated by intracellular localization and post-translational modifications, including phosphorylation, methylation, and ADP-ribosylation. Small ubiquitin-like modifier (SUMO) is a category of recently verified forms of post-translational modifications and exerts various effects on the target proteins. In the studies reported here, we discovered SUMOylational modification of human nucleolin protein at Lys-294, which facilitated the mRNA binding property of nucleolin by maintaining its nuclear localization. In response to arsenic exposure, nucleolin-SUMO was induced and promoted its binding with gadd45α mRNA, which increased gadd45α mRNA stability and protein expression, subsequently causing GADD45α-mediated cell death. On the other hand, ectopic expression of Mn-SOD attenuated the arsenite-generated superoxide radical level, abrogated nucleolin-SUMO, and in turn inhibited arsenite-induced apoptosis by reducing GADD45α expression. Collectively, our results for the first time demonstrate that nucleolin-SUMO at K294R plays a critical role in its nucleus sequestration and gadd45α mRNA binding activity. This novel biological function of nucleolin is distinct from its conventional role as a proto-oncogene. Therefore, our findings here not only reveal a new modification of nucleolin protein and its novel functional paradigm in mRNA metabolism but also expand our understanding of the dichotomous roles of nucleolin in terms of cancer development, which are dependent on multiple intracellular conditions and consequently the appropriate regulations of its modifications, including SUMOylation.
Introduction
Ubiquitin and small ubiquitin-like modifier (SUMO)2 represent a category of recently verified forms of protein post-translational modification, which alters protein conformation, activity, localization, and protein-protein interaction (1). In most cases, SUMOylation occurs in the nucleus or at the nuclear periphery and is involved in transcription initiation, RNA processing and metabolism, chromatin remodeling, maintenance of genome stability, and nucleo-cytoplasmic transport (2). Protein SUMOylation also regulates ion channel activity, the metabolic pathway, and cell mobility (2–6). Most of the SUMO-targeted proteins have the consensus sequence of ΦKX(E/D), where Φ is a large hydrophobic amino acid, K is the target lysine, X is any amino acid, and E or D is an acidic residue (7). However, recent proteomics studies reveal that 52% of the identified SUMO-modified proteins do not contain known consensus sequences, suggesting that non-consensus SUMO acceptor sites are more common than originally suspected (8, 9).
There are four isoforms of SUMO that have been characterized in higher eukaryotes. SUMO1–3 are ubiquitously expressed in different types of mammalian cells, whereas SUMO4 only expresses in the kidney and other specific tissues, suggesting a limited biological role for SUMO4 (10). In mammals, SUMO2–3/4 can form a poly-SUMO chain through the internal consensus SUMO sites (ΦKX(E/D)) at its N terminus and is involved in ubiquitin-dependent protein degradation (11). Due to the lack of this motif, SUMO1 usually occurs in mono-form and leads to functional modification of the target proteins (12). However, the existence of polymeric chains of SUMO1 has also been reported in the case of nuclear proteins of RanBP2 (13) and topoisomerase 1 (14), which might be formed by a noncovalent bond between the C-terminal glycine residue of SUMO1 and the ϵ-amino group of an internal lysine residue of another SUMO1 (14).
Considering the broad participation of SUMO target proteins in proliferation, differentiation and development, senescence, and apoptosis, it is not surprising to find the dysfunctional modification of SUMO in various disease states (2), such as neurodegeneration, placental insufficiency disorders, cardiovascular disease, diabetes, and cancer, which are characterized by ischemia, inflammation, and aberrant apoptosis evasion (2, 15–17). Although knowledge of the molecular mechanisms that mediate or execute these pathologic outcomes is expanding, the discovery of a novel individual SUMO protein target is still of great significance in understanding the development of human diseases as well as in pursuing more effective therapeutic intervention methods.
Nucleolin (also known as C23) is a multifunctional nucleus protein, implicated in the fundamental aspects of DNA and RNA metabolisms, such as transcription, ribosome biogenesis, mRNA stability, and translation; therefore, nucleolin closely associates with regulation of cell proliferation and growth (18). As a result, deregulation of nucleolin is observed in several pathologies, including viral infection (19), autoimmune diseases (20), Alzheimer disease (21), Parkinson disease (22), and cancer development (23, 24). Nucleolin is highly conserved in vertebrates, and analysis of the amino acid sequences reveals the presence of three different structure domains (25). The N-terminal domain consists of highly acidic regions separated by basic sequences and contains multiple phosphorylation sites followed by the nuclear localization signal (NLS) (25). The central domain contains four RNA-binding domains (RBDs). The C-terminal domain contains glycine- and arginine-rich repeats (25), which can be dimethylated (26). The activity of nucleolin is usually regulated by intracellular localization and post-translational modification, including phosphorylation, methylation and ADP-ribosylation (25).
Our present studies, for the first time to the best of our knowledge, provide direct evidence for the discovery of SUMOylational modification of nucleolin at Lys-294, which responded rapidly to the arsenic exposure. In addition, nucleolin-SUMO was found to be required for maintaining its nuclear localization and promoting its activity in the mediation of gadd45α mRNA stability, which in turn increased cell apoptosis under the arsenic exposure. This novel biological function of nucleolin is distinct from its conventional proto-oncogenic role. Therefore, our findings here not only reveal a new modification of nucleolin protein and its novel functional paradigm in mRNA biogenesis processes but also expand our understanding regarding the dichotomous roles of nucleolin in term of cancer development. In other words, the specific intracellular conditions, such as oxidative stress versus pro-proliferative signals, might make appropriate regulations of nucleolin modifications, including SUMOylation and phosphorylation, which further determines the final fate of nucleolin function.
MATERIALS AND METHODS
Cell Culture and Reagents
Human embryonic kidney fibroblast 293T cells, 3T3 protocol-immortalized mouse embryonic fibroblasts (MEFs), and their stable transfectants were maintained at 37 °C in a 5% CO2 incubator with Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 2 mm l-glutamine, and 25 μg/ml gentamicin. Mouse epidermal JB6 Cl41 cells and their stable transfectants were cultured in minimum Eagle's medium with 5% FBS. The cultures were detached with trypsin and transferred to new 75-cm2 culture flasks (Fisher) twice a week. FBS was purchased from Invitrogen; minimum Eagle's medium and actinomycin D were from Calbiochem; sodium arsenite was purchased from Aldrich. Hydroethidine was from Molecular Probes (Carlsbad, CA).
Plasmids and Stable Transfection
shRNAs for nucleolin and UBC9 were purchased from Open Biosystems (Huntsville, AL). GFP-Mn-SOD, mitochondrial signal-targeted catalase, and catalase expression plasmids as well as the parental control vector pSV-Zeo construct were generous gifts from Dr. J. Andres Melendez (Center for Immunology and Microbial Disease, Albany Medical College) (27). gadd45α promoter-based luciferase reporter gene vector (−2252 to +1) was a generous gift from Dr. Fei Chen (Pathology and Physiology Research Branch, Health Effects Laboratory Division, National Institute for Occupational Safety and Health) (28). Human GFP-nucleolin construct was kindly provided by Dr. Michael B. Kastan (Comprehensive Cancer Center, St. Jude Children's Research Hospital) (29). Mouse FLAG-nucleolin was kindly provided by Dr. Kenji Kadomatsu (Department of Biochemistry, Nagoya University School of Medicine) (30). The stable transfectants were established by G418 (400 μg/ml), hygromycin (100 μg/ml), or puromycin (2 μg/ml) selection. Stable transfectants were cultured in antibiotic-free medium for at least two passages before experimentation.
Analysis of His-tagged SUMO1 Conjugation Using an Ni2+-NTA Bead Pull-down Assay
Ni2+-NTA beads were used to pull down all His-tagged SUMO1-conjugated proteins. Briefly, the cells were infected with adenovirus expressing His6-tagged SUMO1, and then the cell lysis were subjected to Ni2+-NTA beads to pull down the proteins that are covalently conjugated to SUMO1. The pull-down complexes were subjected to Western blotting to detect the presence of nucleolin-SUMO1. The adenoviruses carrying Dual-His-S-SUMO1/IRES/HA-UBC9 (Ad-SUMO1-WT), Dual-His-S-SUMO1-Q94P/IRES/HA-UBC9 (Ad-SUMO1-Q94P), and Dual-His-S-SUMO1ΔGG/IRES/HA-UBC9 (Ad-SUMO1-ΔGG) as well as the control LacZ virus (Ad-LacZ) were provided by Dr. German Rosas-Acosta (31). Twenty-four hours after the above virus infection of 293T cells, the cell lysates were subjected to an Ni2+-NTA bead pull-down assay. Briefly, the cells were lysed in 3 ml of His lysis buffer and incubated with 60 μl of Ni2+-NTA-agarose beads (Qiagen, Valencia, CA) with rotation at 4 °C overnight. The beads were washed for 5 min for each step at room temperature with 750 μl of each of the following buffers: washing buffer 1, washing buffer 2, washing buffer 3, and washing buffer 4 (4). After the last wash, His6-tagged SUMOylated products were eluted by incubating the beads in 75 μl of elution buffer for 20 min at room temperature. The elutes were analyzed by Western blotting. The densitometric analyses of the SUMOylated nucleolin versus the unmodified nucleolin were performed using ImageQuant version 5.2 software (GE Healthcare). The results shown are representative of three independent experiments.
RNA IP
The indicated cells were cultured in 10-cm dishes and harvested by scraping after washing twice with PBS. Polysome lysis buffer, containing 10 mm HEPES, pH 7.0, 100 mm KCl, 5 mm MgCl2, 25 mm EDTA, 0.5% IGEPAL, 2 mm DTT, 50 units/ml RNase OUTTM, 50 units/ml Superase INTM, 0.2 mg/ml heparin, and complete proteinase inhibitor, was used to lyse the cell pellet. The cell lysis was centrifuged at 14,000 × g for 10 min at 4 °C. The anti-GFP antibody coupled directly to agarose beads (Vector Laboratories, Burlingame, CA) was added into the supernatant and rotated overnight at 4 °C in NET2 buffer containing 50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1 mm MgCl2, 0.05% IGEPAL, 50 units/ml RNase OUT, 50 units/ml Superase IN, 1 mm dithiothreitol, and 30 mm EDTA. The beads were washed three times, resuspended in 100 μl of NET2 and 100 μl of SDS-TE (20 mm Tris-HCl, pH 7.5, 2 mm EDTA, and 2% SDS), and then incubated for 30 min at 55 °C, mixing occasionally. The RNAs in the buffer off the beads were extracted by phenol/chloroform/isoamyl alcohol. RT-PCR was performed to detect the mRNA present in the immune complex.
Gene Reporter Assay
A confluent monolayer of gadd45α promoter-based luciferase reporter stable transfectants was trypsinized, and 8 × 103 viable cells suspended in 100 μl of medium were seeded to each well of 96-well plates. After the cells were treated with arsenite or UVB radiation for different doses and time periods as indicated in the figure legends, the cells were then lysed with 50 μl of lysis buffer, and the luciferase activity was measured using Promega luciferase assay reagent with a luminometer (Wallac 1420 Victor2 multipliable counter system). The results were expressed as GADD45α induction relative to the medium control.
Western Blotting
The cells were washed once with ice-cold PBS and then extracted with SDS-sample buffer. The cell extracts were separated on polyacrylamide-SDS gels, transferred, and probed with each of the antibodies against ATF2, GAPDH, caspase-3, and poly(ADP-ribose) polymerase (Cell Signaling Technology Inc., Danvers, MA); Mn-SOD (Upstate Biotechnology, Inc., Lake Placid, NY); HA, GFP, catalase, GADD45α, and UBC9 (Santa Cruz Biotechnology, Inc.); SUMO1 (Epitomics, Burlingame, CA); and nucleolin and β-actin (Sigma). The protein bands specifically bound to the primary antibodies were detected using an alkaline phosphatase-linked secondary antibody and an ECF Western blotting system (Amersham Biosciences).
RT-PCR
Total RNA was extracted from the cells using TRIzol reagent (Invitrogen). cDNAs were synthesized by the ThermoScriptTM RT-PCR system (Invitrogen). The mRNA amount presented in the cells was measured by semiquantitative RT-PCR. The primers were as follows: for mouse gadd45α, 5′-ATG ACT TTG GAG GAA TTC TCG-3′ and 5′-CAC TGA TCC ATG TAG CGA CTT-3′; for mouse β-actin, 5′-GAC GAT GAT ATT GCC GCA CT-3′ and 5′-GAT ACC ACG CT T GCT CTG AG-3′; for human gadd45α, 5′-CGT TTT GCT GCG AGA ACG AC-3′ and 5′-GAA CCC ATT GAT CCA TGT AG-3′; for human β-actin, 5′-GCG AGA AGA TGA CCC AGA TCA T-3′ and 5′-GCT CAG GAG GAG CAA TGA TCT T-3′. The PCR products were separated on 2% agarose gels and stained with ethidium bromide, and the images were scanned from a UV light. The densitometric analyses of the product bands were conducted using ImageQuant version 5.2 software (GE Healthcare).
Measurement of Superoxide Production
Cells were treated with arsenite for 6 h and then washed thoroughly with PBS. After a 10-min incubation of the cells with PBS containing 10 μm hydroethidine. Then the cells were collected and subjected to fluorescent quantification using a Coulter XL cytometer (Beckman-Coulter Corp., Miami, FL). Ethidium fluorescence was collected using a 610-nm long pass filter.
Cell Death Assay
After the cells were treated with arsenite as indicated in the figure legends, the cell death was analyzed by flow cytometry following propidium iodide staining of the nuclei. Briefly, the cells were fixed in ice-cold 80% ethanol at −20 °C overnight. The fixed cells were permeabilized in buffer containing 100 mm sodium citrate, 0.1% Triton X-100 at room temperature for 15 min as well as RNase A (0.02 mg/ml; Sigma) for 10 min, stained with propidium iodide (50 μg/ml) at 4 °C for at least 1 h, and then analyzed using the Coulter XL cytometer (Beckman-Coulter).
Site-directed Mutagenesis
Site-directed mutagenesis was performed with the QuikChange II XL site-directed mutagenesis kit (Stratagene, La Jolla, CA) according to the manufacturer's protocol. The pairs of primers for mutations of SUMOylation sites in human nucleolin protein are listed as follows: K142R, 5′-GAA TGG CAA GAA TGC CAA GAG GGA AGA CAG TGA TGA A-3′ and 5′-CCT CTT GGC ATT CTT GCC ATT CTT TGC CCC CCT GGC T-3′; K294R, 5′-AGC AGC TCC TGA AGC CA GGA AAC AGA AAG TGG AAG-3′ and 5′-CTT CCA CTT TCT GTT TCC TGG CTT CAG GAG CTG CT-3′; K295R, 5′-CAG CTC CTG AAG CCA AGA GAC AGA AAG TGG AAG GC-3′ and 5′-TCT CTT GGC TTC AGG AGC TGC TTT CTG TTT GGC CA-3′; K333R, 5′-GTA TCA GCG ATG TTT TTG CTA GAA ATG ATC TTG CTG TTG TGG A-3′ and 5′-TCC ACA ACA GCA AGA TCA TTT CTA GCA AAA ACA TCG CTG ATA C-3′; K377R, 5′-GTT TGA AAG TCT TTG GCA ATG AAA TTA GAC TAG AGA AAC CAA AAG G-3′ and 5′-CCT TTT GGT TTC TCT AGT CTA ATT TCA TTG CCA AAG ACT TTC AAA C-3′; K387R, 5′-CAA AAG GAA AAG ACA GTA GGA AAG AGC GAG ATG CGA G-3′ and 5′-CTC GCA TCT CGC TCT TTC CTA CTG TCT TTT CCT TTT G-3′; K437R, 5′-AAA GTA AAG GGA TTG CTT ATA TTG AAT TTA GGA CAG AAG CTG ATG CA-3′ and 5′-TGC ATC AGC TTC TGT CCT AAA TTC AAT ATA AGC AAT CCC TTT ACT TT-3′; K513R, 5′-GGA AGT ATT TGA GAA AGC AAC TTT TAT CAG AGT ACC CCA GAA CCA-3′ and 5′-TGG TTC TGG GGT ACT CTG ATA AAA GTT GCT TTC TCA AAT ACT TCC-3′; K708R, 5′-AAA GAA GAC GAG GTT TGA ATA GCT CGA GCA TGC ATC-3′ and 5′-TAT TCA AAC CTC GTC TTC TTT CCT TGT GGC TTG TGG-3′. The nucleotide sequences of the mutants were confirmed by automatic DNA sequencing.
Nuclear Extract Preparation
Preparation of nuclear extracts was assessed as described previously (32). The indicated stable transfectants were plated into 10-cm culture dishes at 80% confluence. The nuclear proteins were extracted according to the protocol of the Nuclear/Cytosol Fractionation Kit (BioVision Technologies, Mountain View, CA). Equal protein concentrations were determined using a protein quantification assay kit (Bio-Rad). Nuclear extracts were stored at −80 °C until they were used.
Immunofluorescence Microscopy
293T cells grown on 8-well chamber Lab-Tek® slides (Electron Microscopy Sciences, Hatfield, PA) were transfected with the indicated GFP-tagged plasmids for 24 h. The cells were fixed with 3.7% paraformaldehyde for 15 min and then permeabilized with 0.1% Triton X-100 in PBS for 15 min at room temperature. The cells were then blocked with 1% BSA/PBS for 30 min and incubated with or without anti-nucleolin antibody (Sigma) overnight at 4º C. After washing with PBS three times, the slide was incubated with Alexa Fluor 595-conjugated goat anti-rabbit IgG (Invitrogen) for 1 h. Cellular DNA was finally stained with 4′,6-diamidino-2-phenylindole (DAPI) (Molecular Probes, Inc., Eugene, OR). Fluorescence signals were detected on a Leica TCS SP5 confocal microscope or on a Leica AF6000 fluorescence microscope.
Statistical Analysis
The significance of the difference between the treated and untreated groups was determined with Student's t test. The results are expressed as mean ± S.D.
RESULTS
Discovery of Nucleolin Protein SUMOylation
Our recent findings showed that nucleolin regulated cancer microenvironments by controlling HIF-1α expression in a JNK2-dependent manner (33). During these studies, we unexpectedly noticed an obvious slower migrating band on the Western blotting gel when probed with anti-nucleolin antibody in various cell lines, including human embryonic kidney fibroblast 293T cells (Fig. 1A), mouse embryonic fibroblasts (MEFs; Fig. 1B), mouse epidermal JB6 Cl41 cells (Fig. 1C), human fibrosarcoma HT1080 cells, and several other cell lines (data not shown). To identify whether this slower migrating band represents modified nucleolin rather than a reaction with nonspecific proteins, we knocked down nucleolin expression in MEFs using two sets of shRNAs specifically targeting different sequences of mouse nucleolin mRNA (33). Expression of the modified band was attenuated accordingly upon introduction of nucleolin shRNA (Fig. 1D), suggesting that the slower migrating band represents a modified form of nucleolin protein. This undefined nucleolin modification band accounted for 8.92% of the native nucleolin and appeared at the molecular mass of ∼130 kDa, which increased about 20 kDa from the original molecular mass at 110 kDa, quite distinct from any of the reported nucleolin protein modifications, including phosphorylation or methylation (25). To determine the feasibility of SUMOylational modification of nucleolin, we employed the dicistronic expression construct His6-S-SUMO1 Q94P/IRES/HA-UBC9 (SUMO1 Q94P for short), which can elevate global SUMOylation levels of proteins by simultaneously overexpressing E2 SUMO-conjugating enzyme UBC9 and SUMO1 Q94P, the mutant form of SUMO1, which lacks the interaction site with deconjugating enzymes (5, 31). After 293T cells were infected with the recombinant adenoviruses expressing SUMO1 Q94P (Ad-SUMO1 Q94P), the slower migrating protein band of nucleolin was detected much more strongly when compared with that of the cells infected with LacZ control adenovirus (Ad-LacZ; Fig. 1E), supporting our presumption regarding SUMOylational modification of nucleolin. Consistent with the nuclear compartmentation of original nucleolin, the modified form of nucleolin occurred mostly in the nucleus rather than in the cytosolic part (Fig. 1F).
FIGURE 1.
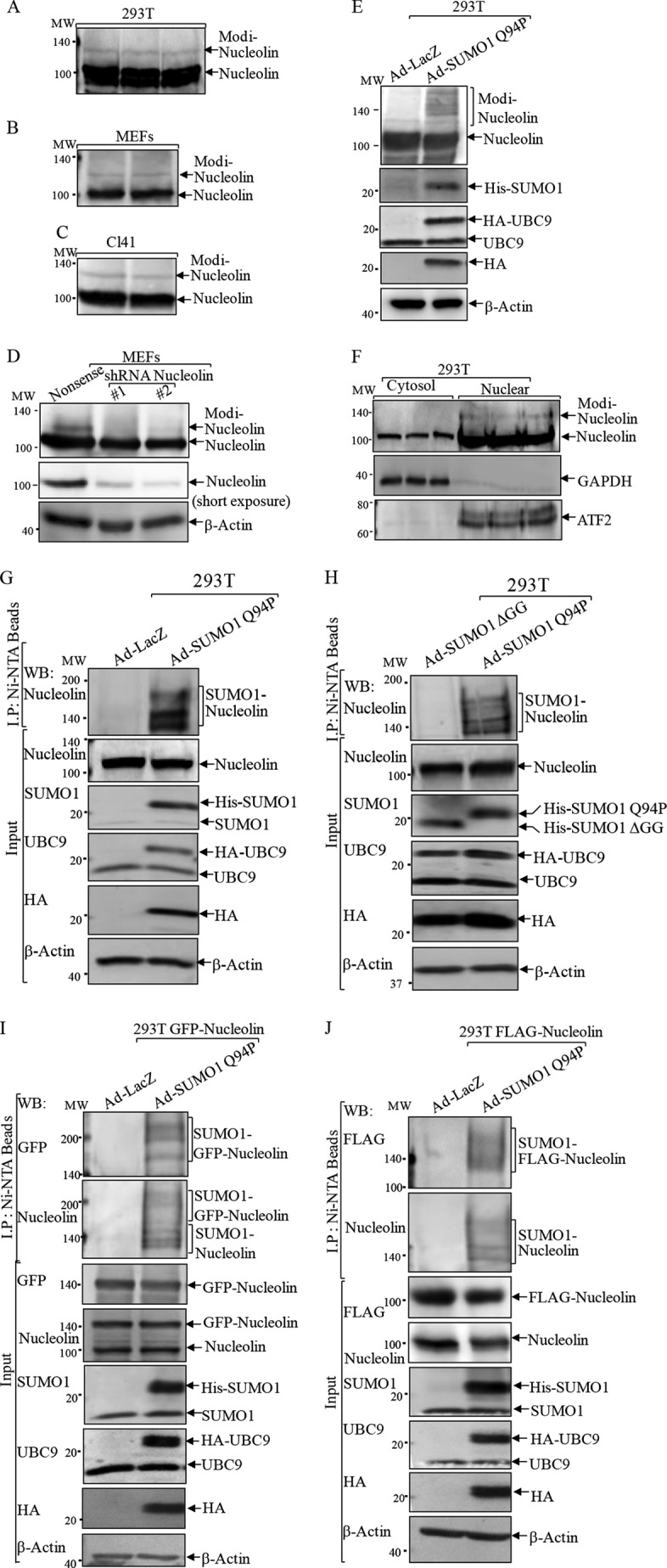
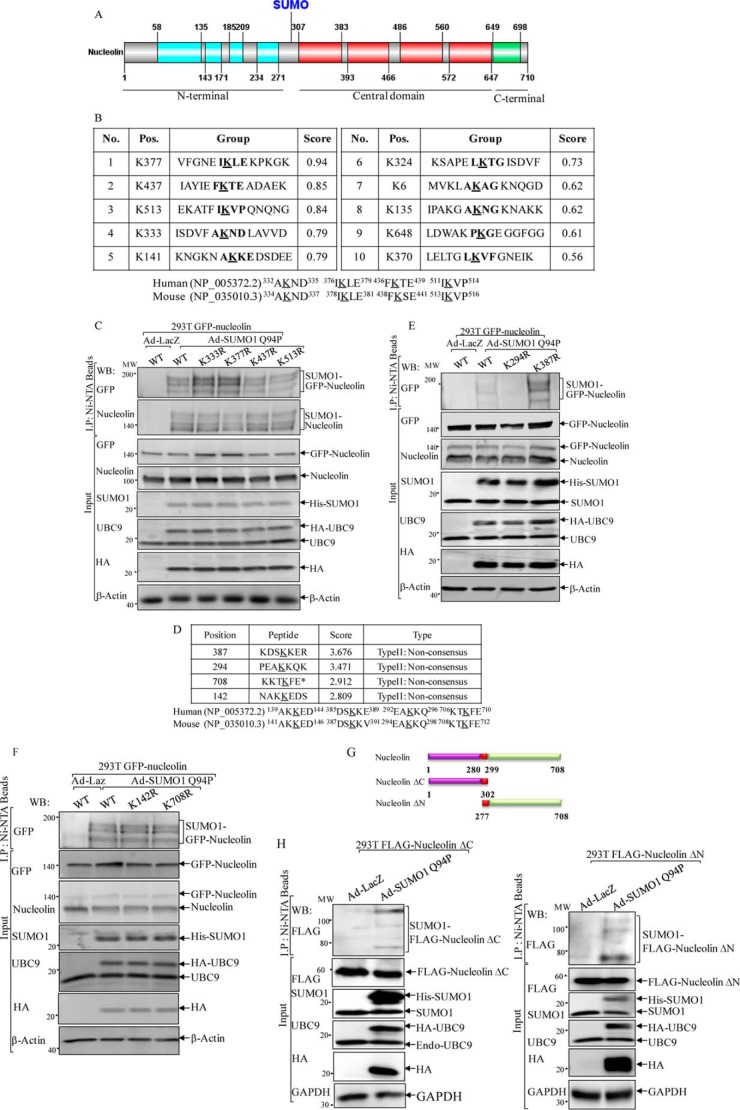
Detection of a novel modified form of nucleolin protein. A–C, cell extracts from 293T cells (A), MEFs (B), and Cl41 cells (C) were subjected to a Western blotting assay for determination of nucleolin protein modification. D, two sets of nucleolin shRNA were stably transfected into MEFs to verify nucleolin modification band. E, 293T cells were infected with adenovirus carrying Dual His6-S-SUMO1 Q94P/IRES/HA-UBC9 construct (Ad-SUMO1 Q94P) or control adenovirus (Ad-LacZ). The cell extracts were subjected to a Western blotting assay to detect nucleolin protein modification. F, cytoplasmic and nuclear extracts that were isolated from 293T cells were subjected to a Western blotting assay (WB) for determination of nucleolin localization. GADPH and ATF2 were used as cytosol and nuclear markers, respectively. G–J, 293T cells transfected with either control vector (G and H) or human GFP-nucleolin (I) or mouse FLAG-nucleolin (J) were infected with Ad-SUMO1 Q94P (G, I, and J) or Ad-SUMO1 ΔGG (H). Ad-LacZ (G, I, and J) was used as a negative control. Ni2+-NTA beads were employed to pull down proteins that were conjugated with His6-tagged SUMO1. A Western blotting assay was carried out for determination of nucleolin SUMOylation.
To provide direct evidence for nucleolin SUMOylation, the above cell lysates (Fig. 1E) were subjected to a pull-down assay using Ni2+-NTA beads to precipitate the proteins that were conjugated with His6-tagged SUMO1. In Fig. 1G, nucleolin protein was present in the pull-down complex in the presence of Ad-SUMO1 Q94P, whereas when 293T cells were infected with a defective mutant SUMO1 that harbors a deletion of the C-terminal two glycine residues (SUMO1-ΔGG), nucleolin was not found in the pull-down complex (Fig. 1H). The findings were further reinforced when either human GFP-tagged nucleolin (Fig. 1I) or mouse FLAG-tagged nucleolin (Fig. 1J) was ectopically expressed in 293T cells and examined by an Ni2+-NTA bead pull-down assay. Thus, our results clearly suggested that nucleolin protein could be SUMOylated inside cells.
SUMOylation of Nucleolin Occurred at Lys-294
The promising consensus SUMOylation sites of human nucleolin protein were predicted by the program Abgent SUMOplotTM. Four lysine residues (Lys-333, -377, -437, and -513) with high scores were considered as a potential SUMOylation consensus site(s), and all sites were present in the RBDs and conserved between humans and mice (Fig. 2, A and B). To validate the predictions, we mutated each of above four lysine residues to arginine, respectively, and then transfected these point-mutated GFP-nucleolin constructs into 293T cells, followed by infection of the cells with Ad-SUMO1 Q94P. The result of a pull-down assay using Ni2+-NTA beads indicated that none of these mutations impaired GFP-nucleolin SUMOylation (Fig. 2C), indicating that nucleolin SUMOylation did not occur at these predicted sites; rather, it might be present at other non-consensus residues.
FIGURE 2.
Identification of human nucleolin SUMOylation at lysine 294. A, schematic representation of functional domains of human nucleolin protein. B, SUMOylation sites of human nucleolin protein were predicted by the program Abgent SUMOplotTM. Four lysine residues (Lys-333, -377, -437, and -513) with high scores are the putative SUMOylation sites, and these sites were conserved between humans and mice. C, 293T cells were transiently transfected with constructs of GFP-nucleolin WT or mutants (K333R, K377R, K437R, or K513R). Twenty-four hours post-transfection, the cells were infected with Ad-LacZ or Ad-SUMO1 Q94P. Cell extracts were used for Ni2+-NTA precipitation, and pulled down proteins were identified by a Western blotting assay (WB). D, the non-consensus SUMO acceptor residues of human nucleolin were predicted by the Site-Specific Predictor of the SUMOsp2.0 software, including Lys-387, -294, -708, and -142. E and F, the prediction was validated by transient transfection with constructs of GFP-nucleolin WT or mutants as indicated into 293T cells. G, schematic representation of various FLAG-mouse nucleolin constructs. H, FLAG-tagged mouse nucleolin plasmids that presented the N terminus and C terminus deletions were transiently transfected into 293T cells, followed by infection with Ad-LacZ or Ad-SUMO1 Q94P. Cell extracts were used for Ni2+-NTA precipitation, and pulled-down proteins were subjected to Western blotting assay as indicated.
It has been suggested that SUMOylational modification on non-consensus sites is a more common event in mammalian cells (34). Therefore, the non-consensus sites were examined in the following studies, and several putative SUMO acceptor residues of human nucleolin were predicted by the Site-Specific Predictor of the SUMOsp2.0 software (35). Four lysine residues, including Lys-387, -294, -708, and -142, were predicted as non-consensus SUMO acceptor sites, which were found to be conserved between humans and mice (Fig. 2D). A lysine to arginine mutagenesis approach was employed once again to determine the putative SUMOylation sites. As shown in Fig. 2, E and F, GFP-nucleolin was detected in the Ni2+-NTA bead pull-down complex from the cells transfected with wild type GFP-nucleolin (WT) and with the K387R, K142R, and K708R mutants, whereas the GFP tag could not be detected in the Ni2+-NTA bead pull-down complex from the cells transfected with GFP-nucleolin K294R mutant, demonstrating that nucleolin SUMOylation occurred at the position of Lys-294, a non-consensus SUMO acceptor site.
It is noteworthy that the Ni2+-NTA bead pull-down results from Ad-SUMO1 Q94P-infected cells clearly demonstrated three SUMO-nucleolin bands around 130, 150, and 180 kDa compared with the unmodified form of nucleolin at 110 kDa (Fig. 1, G and H), whereas the K294R mutation caused loss of all three SUMO-nucleolin bands (Fig. 2, E and F). Therefore, the following possibilities were hypothesized: 1) nucleolin might have multiple SUMO sites, with Lys-294 as the preferably modified residue whose modification facilitates the subsequent conjugation of either monomeric or polymeric SUMO1 at one or two other distinct promising SUMO sites, respectively; 2) nucleolin can be poly-SUMOylationally modified, and the Lys-294 residue was the sole poly-SUMO1 acceptor site. To test these two scenarios, we took advantage of FLAG-tagged mouse nucleolin plasmids (30) that contained a deletion of either the C terminus (amino acids 1–302, ΔC) or the N terminus (amino acids 277–708, ΔN), but both with a reserved NLS (amino acids 280–299) as delineated in Fig. 2G. As shown in Fig. 2H, when either FLAG-nucleolin ΔC or ΔN was overexpressed in 293T cells, SUMO1-nucleolin was nevertheless detected as three obvious bands separated by molecular mass shifts at 20, 40, and 70 kDa, using anti-FLAG antibody. Therefore, it was most likely that nucleolin was SUMOylated by a polymeric SUMO1 chain conjugated to one acceptor site at Lys-294.
Nucleolin SUMOylation Was Responsive to Arsenic Exposure
To determine the category of physiological and/or pathological alteration of nucleolin SUMOylation and its related biological outcomes, the responsiveness of nucleolin-SUMO to oxidative stress was assessed. To this end, GFP-Mn-SOD was overexpressed in 293T cells to reduce the O2⨪ level, and its effect on nucleolin-SUMO was determined by an Ni2+-NTA bead pull-down assay following Ad-SUMO1 Q94P infection of cells (Fig. 3A). The results showed that nucleolin-SUMO was decreased nearly 4-fold upon Mn-SOD overexpression (Fig. 3A), indicating that O2⨪ generation promoted SUMOylation of nucleolin. In contrast, ectopic expression of mitochondrial signal-targeted catalase failed to induce a change of nucleolin SUMOylation under the same experimental conditions (1.15 versus 1.00; Fig. 3B), whereas overexpressing catalase only partially reduced nucleolin SUMOylation (0.76 versus 1.00; Fig. 3B), suggesting that nucleolin SUMOylation was specifically regulated by the intracellular O2⨪ level but not by H2O2.
FIGURE 3.
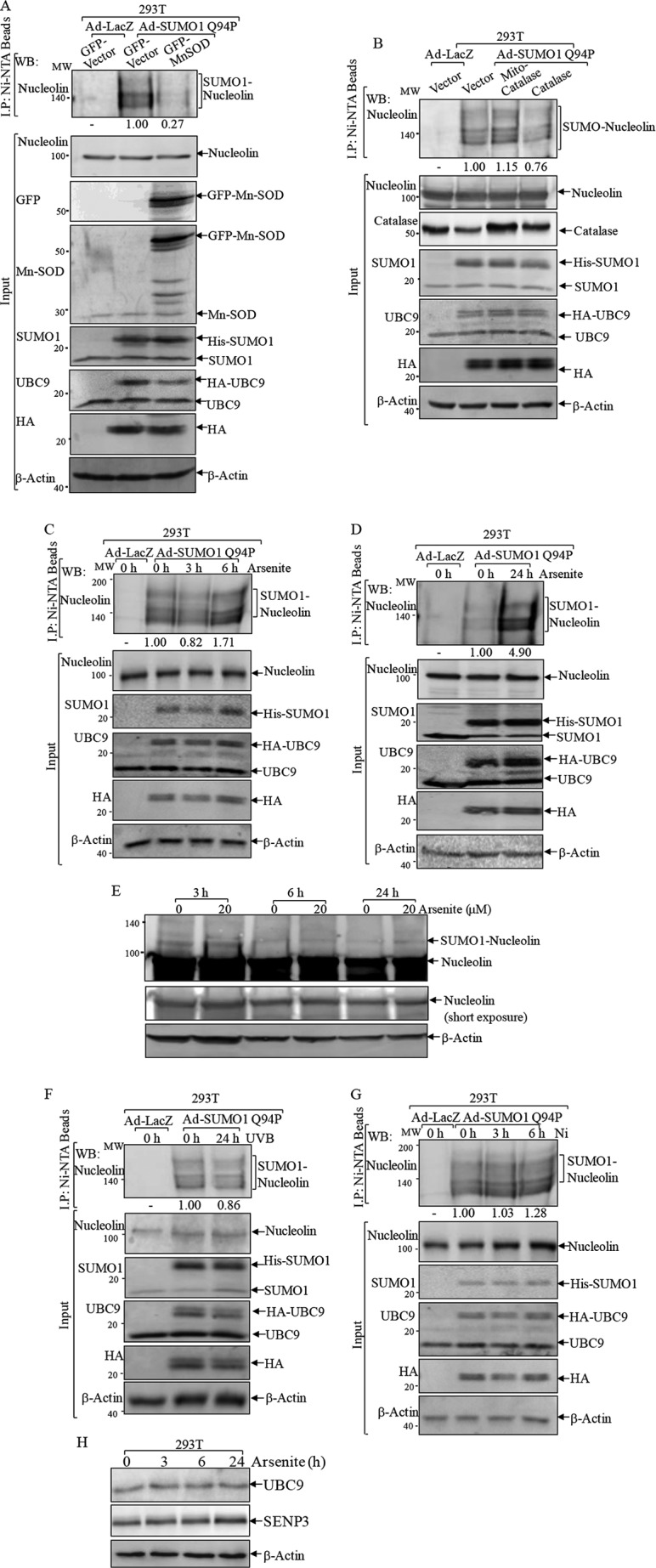
Nucleolin SUMOylation was responsive to oxidative stress. A and B, 293T cells were transiently transfected with GFP-Mn-SOD (A), catalase, or mitochondrial signal-targeted catalase (mito-catalase) (B). Twenty-four hours post-transfection, the cells were infected with Ad-LacZ or Ad-SUMO1 Q94P. Cell extracts were used for Ni2+-NTA precipitation, and pulled-down proteins were subjected to a Western blotting assay (WB). C, D, F, and G, 293T cells infected with Ad-LacZ or Ad-SUMO1 Q94P were treated with 20 μm arsenite for the indicated times (C and D), with UVB (1 kJ/m2) for 24 h (F), or with 0.5 mm nickel chloride for 3 and 6 h (G). An Ni2+-NTA pulled-down assay was performed to detect SUMO-nucleolin. E and H, 293T cells were treated with 20 μm arsenite and subjected to a Western blotting assay to determine expressions of the shifted nucleolin band (E) or UBC9 and SENP3 (H).
Arsenic is a metalloid known as a potent reactive oxygen species producer (33). Arsenite treatment induces SUMOylation and degradation of PML-RARA fusion protein in leukemic cells, which underlies its clinical implications in remissions of acute promyelocytic leukemia (36, 37). Therefore, we used arsenite as an oxidative stress inducer and investigated its potential effect on nucleolin SUMOylation. The result of the Ni2+-NTA bead pull-down assay indicated that acute arsenite exposure for 3 h slightly decreased nucleolin-SUMO, whereas a weak induction was observed as early as 6 h following arsenite treatment (Fig. 3C). When arsenite exposure time was prolonged to 24 h, a robust increase of nucleolin-SUMO was observed (Fig. 3D), indicating that nucleolin-SUMO was induced by prolonged arsenite exposure. This finding was also observed using whole cell lysis to detect the shifted band of nucleolin (Fig. 3E). To discriminate whether nucleolin SUMOylation is responsive to other oxidative stresses, we examined the nucleolin SUMOylation levels following UV and nickel exposure, which are known to induce reactive oxygen species generation (38, 39). As shown in Fig. 3, F and G, both UV and nickel exposure only showed a marginal effect on nucleolin SUMOylation. Therefore, our results demonstrated that nucleolin SUMOylation was responsive to arsenite treatment specifically. In addition, we evaluated the expression levels of UBC9 and SENP3 following arsenite treatment to determine the possible mechanisms underlying arsenite-regulated nucleolin SUMOylation. As shown in Fig. 3H, arsenite treatment did not affect UBC9 expression, whereas it showed a slight up-regulation in SENP3 expression at 24 h postexposure (Fig. 3H), indicating that arsenite might not up-regulate SUMO conjugation or down-regulate deconjugation enzyme expression.
O2⨪-mediated Nucleolin-SUMO Was Critical for Arsenite-induced Cell Death
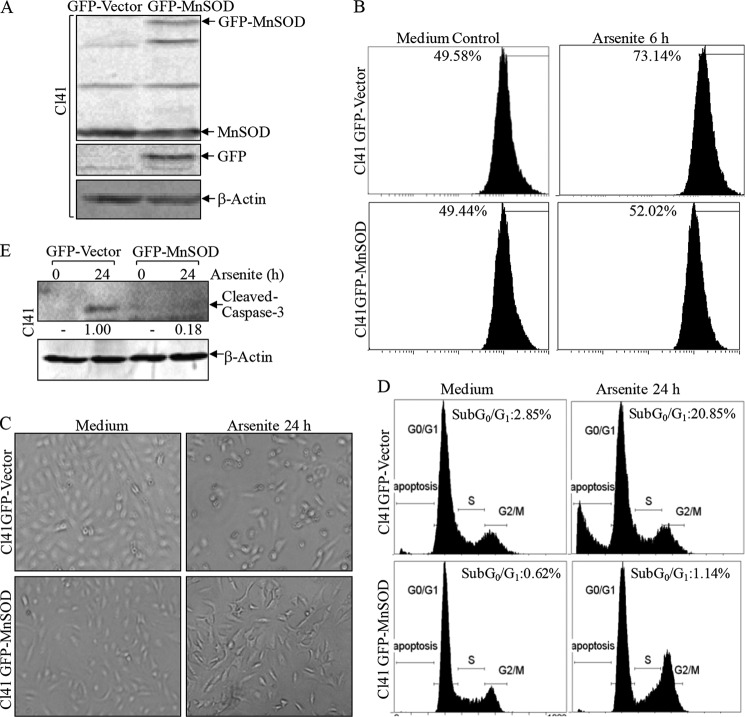
Oxidative stress plays a crucial role in apoptosis (40, 41). In order to define the biological function of redox-responsive nucleolin-SUMO, Mn-SOD was stably transfected into mouse epidermal JB6 Cl41 cells to investigate whether Mn-SOD defends against arsenite toxicity because skin is the primary target organ of arsenite (Fig. 4A). Upon arsenite exposure, the increased O2⨪ generation was markedly scavenged by overexpression of Mn-SOD (52.02 versus 73.14%) assessed by ethidine fluorescent density (Fig. 4B). Moreover, arsenite-induced apoptosis was remarkably reduced by Mn-SOD overexpression determined by morphological change (Fig. 4C), accumulation of sub-G0/G1 phase (20.85 versus 1.14%; Fig. 4D), and caspase-3 cleavage, the hallmark of apoptosis (Fig. 4E). Thus, these results suggested that O2⨪ generation was implicated in arsenite-induced apoptosis.
FIGURE 4.
Mn-SOD inhibited arsenite-induced apoptosis in Cl41 cells. A, GFP-Mn-SOD was transfected into Cl41 cells, and the stable transfectants were identified by a Western blotting assay. B, the indicated cells were treated with 20 μm arsenite for 6 h. The cells were washed thoroughly with PBS and then incubated with PBS containing 10 μm hydroethidine for 10 min. The cells were collected and subjected to fluorescent quantification by cytometry. C–E, the indicated cells were treated with arsenite for 24 h. The morphological images were captured under an inverted microscope (C). Cell death was quantified by flow cytometry after propidium iodide staining (D). The cell extracts were subjected to a Western blotting assay as indicated (E).
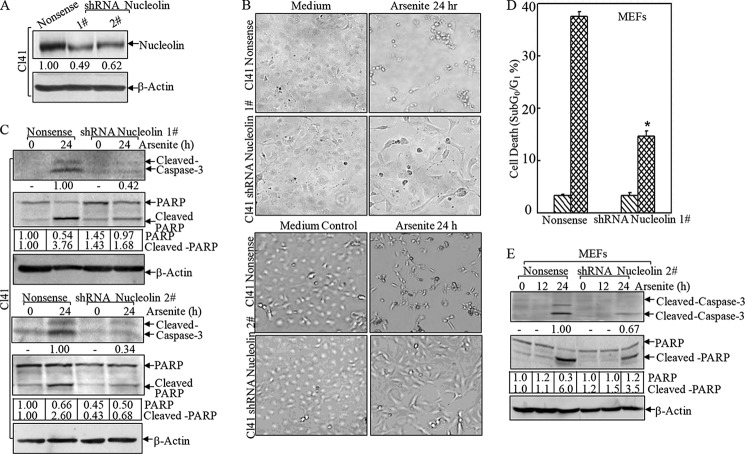
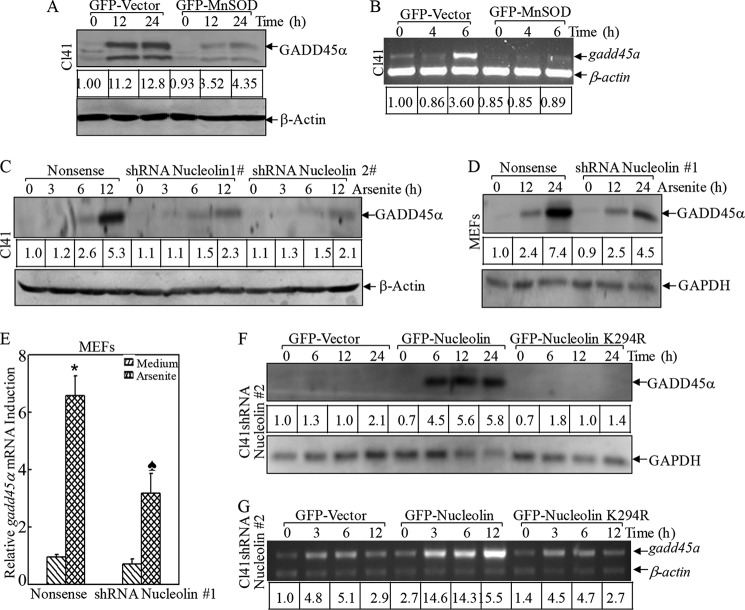
Next we exploited the role of nucleolin in arsenite-associated apoptosis using Cl41 shRNA nucleolin transfectants (Fig. 5A). As shown in Fig. 5, B and C, knocking down nucleolin by both sets of shRNAs in Cl41 cells markedly attenuated arsenite-induced apoptosis, manifested by blockage of typical apoptotic morphological changes (Fig. 5B) and activation of caspase-3 and cleavage of poly(ADP-ribose) polymerase (Fig. 5C). This finding was also reproducible in MEFs (Fig. 5, D and E), indicating that nucleolin exerted a proapoptotic effect due to arsenite exposure in a cell type-independent manner. This newly defined role of nucleolin in promoting cell death under stress conditions was contrary to its conventional function as a proto-oncogene (42). Therefore, these results to some degree suggested that the diverse functions of this protein were relying on various intracellular conditions, such as oxidative stress versus pro-proliferative signals.
FIGURE 5.
Nucleolin promoted arsenite-induced apoptosis. shRNAs targeting mouse nucleolin were stably transfected into Cl41 cells (A–C) and MEFs (D and E). *, significant increase compared with nonsense control transfectants (p < 0.05). The nonsense control and shRNA nucleolin stable transfectants were treated with arsenite, and the morphological images were captured under an inverted microscope (B). Cell death was quantified by flow cytometry (D). The cell extracts were subjected to a Western blotting assay to detect cleavage of caspase-3 and poly(ADP-ribose) polymerase (PARP) (C and E). Error bars, S.D.
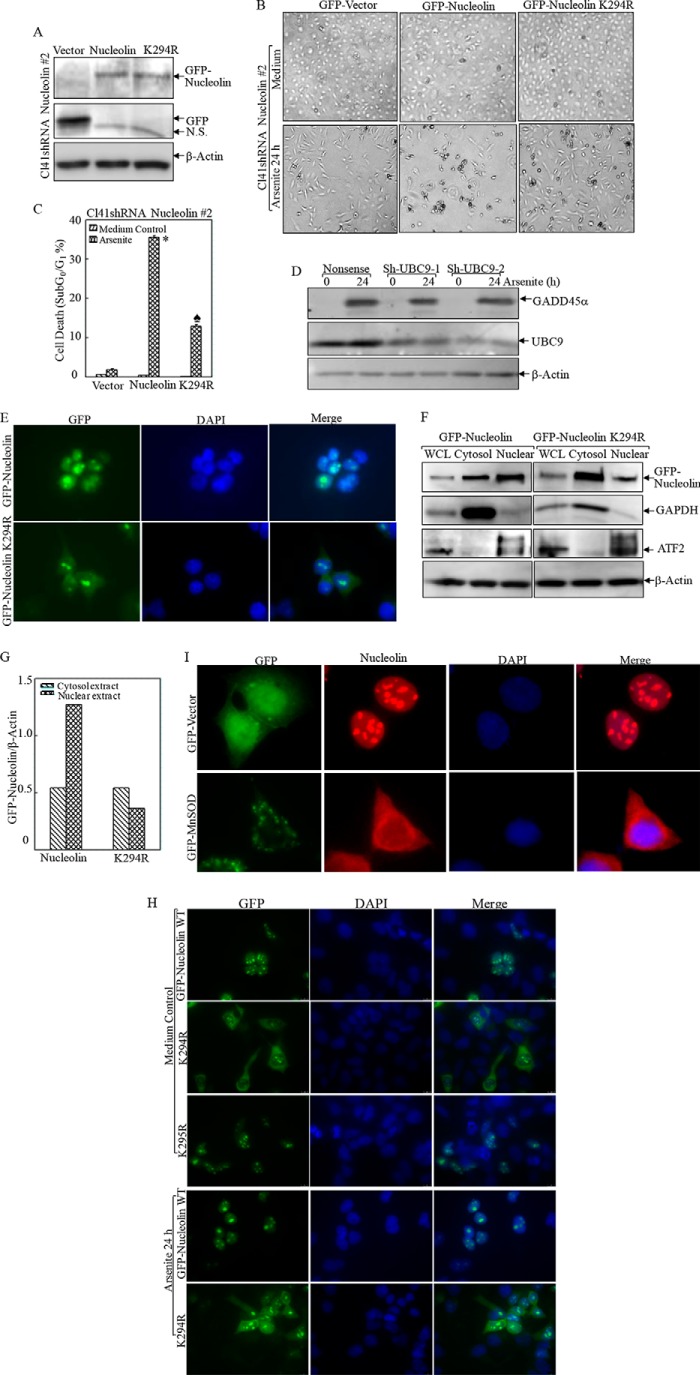
To further assess the biological effect of nucleolin-SUMO in this process, we carried out gain-of-function studies by overexpressing wild type and K294R GFP-nucleolin. For these overexpression experiments, we did not use the parental Cl41 cells because the endogenous nucleolin expression level was so high that the exogenous GFP-nucleolin K294R was incapable of competing with endogenous nucleolin. To overcome this problem, we first knocked down nucleolin in Cl41 cells using nucleolin shRNA 2, which only specifically targeted mouse nucleolin mRNA without affecting human nucleolin mRNA. Then, Cl41 nucleolin shRNA 2 stable transfectants were used to transfect with either human GFP-nucleolin or its K294R mutant in order to minimize the interference of highly expressed endogenous nucleolin (Fig. 6A). Ectopic expression of human GFP-nucleolin in Cl41 nucleolin shRNA 2 transfectants sensitized the apoptotic response compared with that of GFP-vector transfection (35.55 ± 0.42 versus 1.91 ± 0.2%; Fig. 6, B and C), whereas overexpression of SUMOylation-deficient GFP-nucleolin K294R mutant was less effective in restoring cell death (12.88 ± 0.46 versus 35.55 ± 0.42%; Fig. 6, B and C). Our results indicated that SUMOylation at Lys-294 was a functionally privileged form of nucleolin in the mediation of apoptosis due to arsenite exposure. However, we found that knockdown of SUMO conjugation enzyme UBC9 expression by introducing two sets of shRNA only marginally affected GADD45α induction by arsenite (Fig. 6D). Consistent with our observation in Fig. 3G, we anticipated that arsenite might specifically regulate nucleolin SUMO E3 ligase expression or activity rather than affect the global SUMO activation (E1) or conjugation (E2) process. The SUMO E3 ligase, protein inhibitor of activated STAT1 (PIAS1), is reported to mediate the SUMOylation of hairy and enhancer of split 1 (Hes-1), which could transcriptionally regulate GADD45α expression as well as proapoptotic function (43). The role of PIAS1 in the regulation of nucleolin SUMOylation is currently under investigation in our laboratory.
FIGURE 6.
Nucleolin SUMOylation was essential for its nuclear localization and proapoptotic effect following arsenite treatment. A, wild type and K294R mutant human GFP-tagged nucleolin were overexpressed in Cl41 nucleolin shRNA 2 stable transfectants. B and C, the indicated cells were treated with arsenite, and the morphological images were captured under an inverted microscope (B). Cell death was quantified by flow cytometry after propidium iodide staining (C). *, significant increase compared with vector control transfectants (p < 0.05). ♠, significant decrease compared with GFP-nucleolin transfectants (p < 0.05). D, two sets of shRNA UBC9 were stably transfected into MEFs and exposed to arsenite (20 μm) for 24 h. The induction of GADD45α by arsenite exposure was evaluated in the cell extracts obtained from nonsense control and shRNA UBC9 transfectants by Western blotting. E–H, the subcellular distribution of WT or mutant nucleolin was captured by fluorescent microscopy without (E) or with arsenite treatment (H) or extraction of cytosol and nuclear lysis (F). The relative amount of cytosol and nuclear nucleolin protein was quantified by ImageQuant version 5.2 (G). The data shown are representative of three independent experiments. I, the subcellular distribution of nucleolin in the transfectants of GFP-vector and GFP-Mn-SOD was captured by fluorescent microscopy. WCL, whole cell lysis. Error bars, S.D.
Nucleolin SUMOylation Maintained Its Nuclear Localization
SUMOylation is reported to have a significant effect on subcellular localization of nuclear proteins (44, 45). In agreement with this notion, our defined nucleolin-SUMO site Lys-294 was found resident within the NLS region (amino acids 282–301 of human nucleolin) (46). Therefore, we transfected GFP-nucleolin and GFP-nucleolin K294R into 293T cells to determine whether SUMOylation affects nucleolin compartmentation. As shown in Fig. 6E, GFP-nucleolin was found to mainly localize in the nucleolus and to a lesser degree in the nucleoplasm but was barely detected in the cytoplasm. In contrast, GFP-nucleolin K294R was diffusively present in both the cytoplasm and nucleolus as well as in the nucleoplasm. This finding was further extended by extracting cytosol and nuclear cell lysis, which showed that the amount of nucleus-localized GFP-nucleolin was twice that of the cytoplasm-restricted form, whereas GFP-nucleolin K294R distribution in the nucleus was less compared with that in the cytoplasm (Fig. 6, F and G). This distribution pattern was not disturbed by arsenic treatment (Fig. 6H). Considering that our above defined nucleolin SUMOylation site, Lys-294, resides within the NLS region (amino acids 282–301), we doubt that the mutation in Lys-294 would actually affect the localization of nucleolin irrespective of its SUMOylation. To this end, we mutated another lysine residue in NSL, Lys-295, to separate these effects. As shown in Fig. 6H, K295R mutation did not affect nucleolin intracellular distribution. This suggested that Lys-294 was critically required for both nucleolin SUMOylation and nuclear sequestration. Consistently, when nucleolin-SUMO was greatly abrogated by Mn-SOD overexpression (Fig. 3A), the distribution of nucleolin was found in both the nucleus and cytoplasm in comparison with the localization mainly in the nucleolus and nucleoplasm of vector control transfectant (Fig. 6I). Taken together, our results, for the first time, demonstrated that nucleolin SUMOylation at Lys-294 maintains its subcellular localization.
SUMOylation of Nucleolin Increased gadd45α mRNA Stability
Growth arrest and DNA damage-inducible gene 45α (GADD45α) is one of the mediators of arsenite-induced apoptosis, as we found previously (47, 48); therefore, GADD45α induction was investigated to define its involvement in the biological function of SUMO-nucleolin. Consistent with inhibition of the apoptotic response, overexpression of Mn-SOD (Fig. 7, A and B) and knockdown of nucleolin (Fig. 7, C–E) both attenuated arsenite-induced GADD45α expression at the protein as well as the mRNA levels. In addition, ectopic expression of GFP-nucleolin elevated GADD45α induction by arsenite treatment as compared with that in GFP-vector transfectants, whereas expression of GFP-nucleolin K294R failed to elevate its induction (Fig. 7, F and G), suggesting that nucleolin SUMOylation at Lys-294 was crucial for GADD45α induction, which further mediated the cell apoptosis following arsenite exposure.
FIGURE 7.
Nucleolin SUMOylation was crucial for GADD45α expression following arsenite treatment. A, Cl41 GFP-vector control and Mn-SOD stable transfectants were treated with arsenite for 12 and 24 h. The induction of GADD45α protein was compared between the indicated cell lines by a Western blotting assay. B, the above cells were treated with arsenite for 4 and 6 h and then extracted with TRIzol reagent for the total RNA isolation. Gadd45α mRNA was amplified with specific primers by RT-PCR. β-Actin was used as an internal control. C, two sets of shRNA targeting mouse nucleolin were stably transfected into Cl41 cells. The induction of GADD45α protein arsenite was compared between the shRNA nucleolin transfectants and the nonsense control cells by a Western blotting assay. D and E, nucleolin shRNA 1 plasmid was stably transfected into MEFs. The inductions of GADD45α protein and mRNA were analyzed by a Western blotting assay and RT-PCR. *, a significant increase compared with medium control (p < 0.05). ♠, significant decrease compared with nonsense control transfectants (p < 0.05). F and G, wild type and K294R mutant human GFP-tagged nucleolin were overexpressed in Cl41 nucleolin shRNA 2 stable transfectants. The inductions of GADD45α protein and mRNA were analyzed as described above. Error bars, S.D.
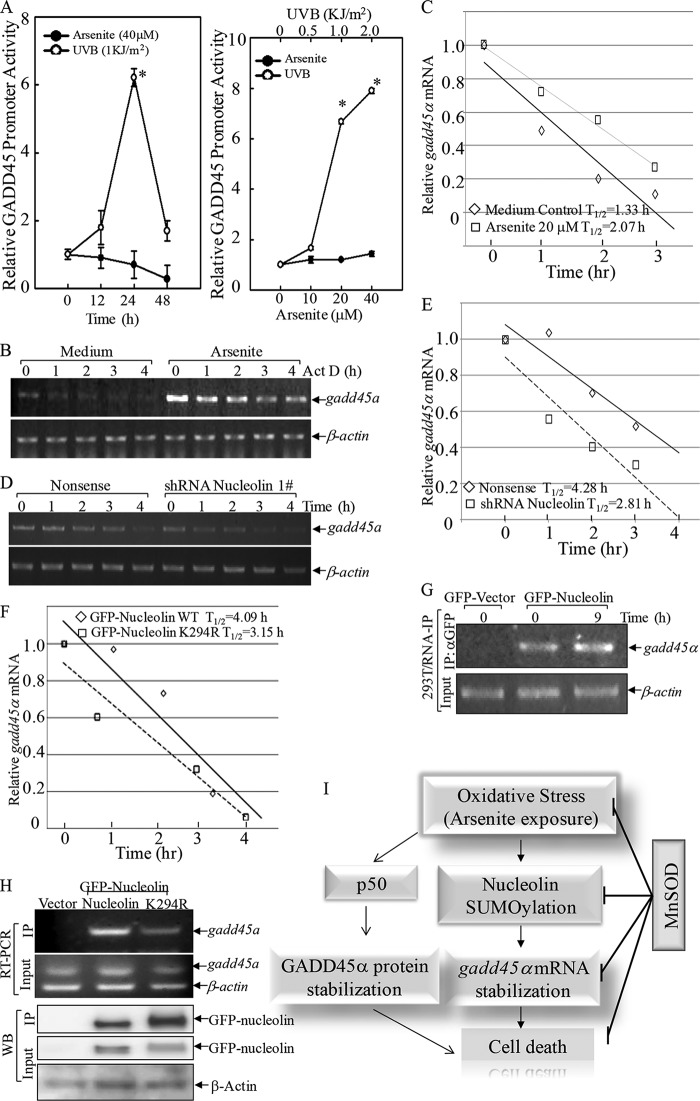
Our current studies further demonstrated that arsenite elevated gadd45α mRNA independent of transcription, because arsenite treatment failed to increase gadd45α promoter-driven luciferase reporter activity, whereas UVB exposure caused a marked induction of gadd45α promoter transcriptional activity (Fig. 8A). This suggested that gadd45α mRNA induction by arsenite might be associated with up-regulation of its mRNA stability. Next, the gadd45α mRNA decay rate was determined following treatment of actinomycin D in the absence or presence of arsenite treatment. As shown in Fig. 8, B and C, half-life (t½) of gadd45α mRNA was about 1.33 h, whereas arsenite treatment attenuated gadd45α mRNA degradation, and its t½ was prolonged to 2.07 h (increased more than 50%), indicating that arsenite treatment increased gadd45α mRNA stability. In addition, knockdown of nucleolin resulted in a reduction of the half-life of gadd45α mRNA compared with that in nonsense transfectants following arsenite treatment (t½ = 2.81 versus 4.28 h, reduced by 34%; Fig. 8, D and E). In addition, we determined the effect of nucleolin SUMOylation on gadd45α mRNA stability by comparing gadd45α mRNA half-life after arsenic treatment. As shown in Fig. 8F, overexpression of GFP-nucleolin K294R reduced gadd45α mRNA half-life by nearly 30% compared with that in GFP-nucleolin WT transfectants (t½ = 3.15 h versus 4.09 h). Moreover, the physical interaction between nucleolin and gadd45α mRNA was further observed in the RNA IP assay, using αGFP-beads to pull down GFP-nucleolin. Arsenite exposure could increase the interaction of nucleolin with gadd45α mRNA (Fig. 8G). Importantly, point mutation of GFP-nucleolin at Lys-294 dramatically reduced binding activity toward gadd45α mRNA in the RNA IP assay (Fig. 8H), indicating that SUMOylation of nucleolin at Lys-294 played a key role in its stabilizing gadd45α mRNA and consequently promoted cell apoptosis (Fig. 8I).
FIGURE 8.
Nucleolin SUMOylation bound with gadd45α mRNA and regulated its stability. A, Cl41 cells stably transfected with a gadd45α promoter-based luciferase reporter were seeded into each well of 96-well plates and then exposed to arsenite or UVB radiation. The cells were then extracted for determination of luciferase activity. The results are presented as gadd45α promoter luciferase activity relative to medium control (relative gadd45α induction). Each bar indicates the mean and S.D. (error bars) of three independent experiments. *, significant increase compared with medium control (p < 0.05). B and C, Cl41 cells were incubated with actinomycin D (Act D) with or without a 6-h arsenite pretreatment. The mRNA degradation rate of gadd45α was analyzed by RT-PCR (B). The densitometric analyses of the product bands were used to calculate the half-life of gadd45α mRNA (t½; C). D–F, shRNA nucleolin and the nonsense control transfectants (D and E) or GFP-nucleolin WT and GFP-nucleolin K294R transfectants (F) were pretreated with arsenite for 6 h, followed by actinomycin D treatment for the indicated times. The degradation rate of gadd45α mRNA of the indicated cell lines treated with arsenite was analyzed by RT-PCR (D), and t½ of gadd45α mRNA was calculated as described above (E and F). G and H, RNA IP was performed using αGFP antibody-conjugated agarose resins to examine the binding of nucleolin to gadd45α mRNA. I, proposed model of cascade activation of the superoxide/nucleolin-SUMO/GADD45α module in promoting oxidative stress-induced cell death. Our previous findings regarding p50-dependent GADD45α induction are also summarized in response to arsenite. WB, Western blot.
DISCUSSION
Nucleolin is a ubiquitously expressed protein involved in multiple important biological processes, such as cell cycle regulation and ribosomal biogenesis (49). Nucleolin possesses four RBDs and reportedly stabilizes multiple RNA molecules (50–52). In the studies reported here, we discovered that human nucleolin was SUMOylationally modified at the Lys-294 residue (for mice, Lys-296), and this SUMOylation played an important role in maintaining its nuclear compartmentation and facilitated its mRNA binding property. We also found that arsenic exposure induced nucleolin SUMOylation and consequently stabilized gadd45α mRNA, through which nucleolin executed a proapoptotic function in arsenic response.
Most eukaryote proteins undergo one or several forms of post-translational modification, such as phosphorylation, glycosylation, and methylation, which provide a variety of functional alterations of the proteins (53). As a multifunctional RNA-binding protein, nucleolin has key functions in DNA/RNA metabolisms, chromatin remodeling, and nucleocytoplasmic transport (54). Nucleolin has three major functional domains (25). The acidic N-terminal domain of nucleolin participates in the transcription of rRNA. Phosphorylation of nucleolin in this region is stimulated by growth factors, hormones, or cell cycle regulator and is correlated with increased rRNA transcription and cell proliferation (25). For instance, phosphorylations of nucleolin by CDK1 ((S/T)PXKK) and casein kinase 2 (X(S/T)XX(E/D)) promote its helicase activity toward RNA/RNA and RNA/DNA duplexes, therefore affecting cell cycle progression, embryonic development, and tumorigenesis (25). Casein kinase II (CKII)-mediated nucleolin phosphorylation leads to its autocleavage into 30- and 72-kDa peptides (55). The specific interaction between nucleolin and nucleic acid significantly relies on the central four RBDs that function redundantly to ensure the strong and specific affinity toward RNAs (25). The C-terminal domain of nucleolin contains high levels of dimethylated arginine, which performs a nonspecific interaction with nucleic acid so as to strengthen the RBD-dependent specific binding of nucleolin to RNA. The structure between the N-terminal and central domains contains the bipartite NLS of nucleolin that executes nucleocytoplasmic translocation (56). Thus, nucleolin also serves as an “adaptor” for karyophilic proteins (such as ribosomal proteins) as well as a specific RNA-binding protein (49). During the translocation through the nuclear pore complex, the NLS of nuclear proteins remains bound to importin-α; therefore, the modifications within NLS might affect the interaction with importin-α, changing protein compartmentation (57). However, until now there has been no report about nucleolin protein modifications within this region except for proteomic predictions of acetylation on Lys-294 (see the PhosphoSitePlus Web site) and phosphorylation on Ser-301 (58). Instead, massive phosphorylation by CDK11 and CKII kinases has been reported to be required for nucleolin localization to the cytoplasm in the developing Xenopus embryo (46). In the current studies, we successfully demonstrated that SUMO1 was conjugated to human nucleolin at Lys-294, a non-consensus SUMO acceptor site within the NLS region. Mutation of this site to arginine impaired nuclear sequestration of nucleolin and attenuated its function in maintaining mRNA stability. It suggested that SUMOylation was the functionally privileged form of this protein that sustained its nuclear localization and mRNA binding property. We also found that this SUMOylation was inducible upon arsenite exposure, whereas ectopic expression of Mn-SOD impaired its SUMOylation as well as its nuclear localization. Moreover, we noticed that mutation of Lys-387, a promising acetylation site (59), increased nucleolin SUMOylation. Lys-387 resides in the central RNA binding domain, and it is likely that mutation of this site to arginine might change nucleolin confirmation, affecting the interaction with either E3 ligase or deconjugation enzymes to increase the nucleolin SUMOylation level.
Under normal physiological conditions, only a small portion of a particular protein pool is SUMOylated (often <5%) (60, 61). This small amount of SUMOylated protein maintains the basal activity or function of these proteins. In terms of nucleolin, we did observe a weak presence of nucleolin SUMOylation without any stimulation, which might account for its steady nuclear sequestration. Meanwhile, the protein SUMO level is dynamically regulated under various stress conditions, including osmotic stress, hypoxia, heat shock, and oxidative stress (36, 37, 48). On one hand, a low or moderate level of oxidative stress inhibits SUMOylation by oxidation of SUMO E1 and E2 enzymes, leading to disulfide bond formation between SAE1 and UBC9 (62). On the other hand, a high level of oxidative stress is reported to inactivate SUMO protease SENP1 by cross-linking disulfide sites at Cys-603 and Cys-613, which increases the global protein SUMO level (63). This is consistent with our observation of the biphasic response of arsenic-induced nucleolin SUMOylation, which showed partial inhibition at an early time point (3 h) and incrementation at a later time point of exposure (24 h).
Although nucleolin is overexpressed in highly proliferating cells, such as cancer and stem cells, it does hamper cell growth under stress conditions caused by heat shock or ionizing radiation (64). After heat shock, nucleolin was found to redistribute from the nucleolus to the nucleoplasm, where it formed a complex with RPA to inhibit DNA replication in a p53-dependent manner (65). Consistently, here we provided experimental evidence supporting the proapoptotic effect of nucleolin through up-regulating gadd45α mRNA stability under lethal oxidative stress conditions initiated by arsenite treatment. GADD45α is an evolutionarily conserved protein that is implicated in cell cycle arrest (66), DNA repair (67), and cell survival and apoptosis (47, 48, 68). GADD45α is induced in response to multiple environmental and physiological stresses, including UV radiation (18), arsenite (28, 47, 48, 69), and inflammatory cytokines (70). Our previous findings showed that GADD45α was one of the crucial mediators of arsenite-induced apoptosis (47, 48). However, the mechanisms underlying GADD45α induction by arsenite varied in various experimental conditions, including transcriptional and post-transcriptional regulations (28, 47, 48, 69). The cell growing conditions seem critical for the gadd45α mRNA induction by arsenite (69). When the cells were in a logarithmically growing condition, arsenite resulted in a time-dependent increase in gadd45α mRNA in the late phase of exposure (47, 69). However, if the cells were growing in the arrested condition, in which the cells reached almost confluence with a high density, gadd45α mRNA was not induced by arsenite (48); rather, it was decreased with prolonged exposure time (69). Our current studies showed that arsenite exposure did increase the gadd45α mRNA level; however, the transcriptional regulation is unlikely to be the main mechanism of arsenite-induced expression of GADD45α. Using a gadd45α promoter-based luciferase reporter, we did not find an obvious increase in gadd45α promoter luciferase transcription activity, whereas UVB radiation induced a profound gadd45α transcription. Consistent with our findings, Chen and co-workers (69) also failed to observe any appreciable transcription of the gadd45α gene in a nuclear run-on assay after exposure of the Beas2B cells to arsenite. Thus, our studies indicated that gadd45α mRNA expression was mainly regulated through increasing its mRNA stability following arsenite exposure.
Regulation of eukaryotic mRNA stability is an important control point in the regulation of gene expression. The rate of mRNA decay is regulated by the interaction of cis-acting elements in the transcripts and sequence-specific RNA-binding proteins. One of the most studied cis-acting elements is the AU-rich element present in the 3′-untranslated region (3′-UTR) of several unstable mRNAs. These sequences are targets of many AU-rich element-binding proteins, some of which induce mRNA degradation, whereas others promote mRNA stabilization (71). Analysis of gadd45α 3′-UTR using a conventional method (72) revealed two distinct positions containing AUUUA sequences, at the sites of nucleotides 1211–1215/1430–1434 in human gadd45α 3′-UTR (and nucleotides 901–905/1146–1150 in mouse gadd45α 3′-UTR). Similar sequences were also found in other higher vertebrates, such as chimpanzee, rat, and bovine. Nucleolin has been found to bind to the mRNA of several important genes, including p53 (29), bcl-2 (73), bcl-xl (52), and hif-1a (33), leading to regulation of mRNA turnover or translation. In our present studies, nucleolin was found to bind to and stabilize the gadd45α mRNA, which promoted GADD45α protein expression and cell death upon arsenite exposure. In addition, we found that the SUMOylation status of nucleolin was induced upon arsenite-initiated oxidative stress. SUMOylated nucleolin mainly localized in the nucleus and augmented its RNA binding property, thereby facilitating gadd45α mRNA stabilization and GADD45α-mediated cell death, whereas overexpressing Mn-SOD scavenged superoxide generation and blocked nucleolin SUMOylation, which destabilized gadd45α mRNA and subsequently led to protection of cell death induced by arsenite exposure.
O2⨪ is one of the major reactive oxygen species that can lead to the formation of the highly reactive hydroxyl radical; therefore, the excess production of O2⨪ may play a role in the progression of apoptosis (40, 41). Many reports observe a decrease in the levels and activities of antioxidants after acute exposure to high dose arsenic, including SOD (74–76). SOD pretreatment has a protective effect against arsenic-induced DNA damage, such as sister chromatin exchanges (77). However, the effect of SOD on arsenic-induced cell death seems contradictory. For the macrophages of CDF1 mice, the cell death induced by sodium arsenite (10 and 20 μm) was significantly reduced by Mn-SOD (78), but the cell death induced by arsenic trioxide (7 μm) could not be reversed by SOD in HeLa cells (79); nor could SOD prevent sodium arsenite-induced cell death in NIH3T3 cells (80). However, the specimens of SOD used in the two latter reports were not clearly elucidated. In our present studies, we overexpressed Mn-SOD in mouse epidermal cell Cl41 and found that the sodium arsenite-induced cell death was obviously blocked via attenuating the nucleolin-SUMO/GADD45α axis.
In summary, we discovered a novel nucleolin protein modification, SUMOylation at Lys-294, and its induction as well as its function in regulation of gadd45α mRNA stability and apoptosis in the cellular response to arsenic exposure, which provides valuable insight into the new face of nucleolin function under arsenic-induced oxidative stress conditions.
Acknowledgments
We thank Dr. J. Andres Melendez for the generous gift of GFP-Mn-SOD plasmid, Dr. Fei Chen for GADD45α promoter-based luciferase reporter gene vector (−2252 to +1), Dr. Michael B. Kastan for the GFP-nucleolin construct, and Dr. Kenji Kadomatsu for mouse FLAG-nucleolin plasmid.
This work was supported, in whole or in part, by National Institutes of Health (NIH), NCI, Grants CA112557, CA177665, and CA165980 and by NIH, NIEHS, Grant ES000260. This work was also supported by National Science Foundation of China Grant C81472945.
- SUMO
- small ubiquitin-like modifier
- SOD
- superoxide dismutase
- NLS
- nuclear localization signal
- RBD
- RNA-binding domain
- NTA
- nitrilotriacetic acid
- IP
- immunoprecipitation
- MEF
- mouse embryo fibroblast
- Ad
- adenovirus.
REFERENCES
- 1. Hay R. T. (2005) SUMO: a history of modification. Mol. Cell 18, 1–12 [DOI] [PubMed] [Google Scholar]
- 2. Anderson D., Cimarosti H., Henley J. (2009) The role of sumoylation in neurodegenerative diseases. in SUMO Regulation of Cellular Processes (Wilson V. G., ed) pp. 233–251, Springer, Dordrecht, Netherlands [Google Scholar]
- 3. Liu J., Zhang D., Luo W., Yu Y., Yu J., Li J., Zhang X., Zhang B., Chen J., Wu X.-R., Rosas-Acosta G., Huang C. (2011) X-linked inhibitor of apoptosis protein (XIAP) mediates cancer cell motility via Rho GDP dissociation inhibitor (RhoGDI)-dependent regulation of the cytoskeleton. J. Biol. Chem. 286, 15630–15640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Liu J., Zhang D., Luo W., Yu J., Li J., Yu Y., Zhang X., Chen J., Wu X.-R., Huang C. (2012) E3 ligase activity of XIAP RING domain is required for XIAP-mediated cancer cell migration, but not for its RhoGDI binding activity. PLoS One 7, e35682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yu J., Zhang D., Liu J., Li J., Yu Y., Wu X.-R., Huang C. (2012) RhoGDI SUMOylation at Lys-138 increases its binding activity to Rho GTPase and its inhibiting cancer cell motility. J. Biol. Chem. 287, 13752–13760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Castillo-Lluva S., Tatham M. H., Jones R. C., Jaffray E. G., Edmondson R. D., Hay R. T., Malliri A. (2010) SUMOylation of the GTPase Rac1 is required for optimal cell migration. Nat. Cell Biol. 12, 1078–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Johnson E. S. (2004) Protein modification by SUMO. Annu. Rev. Biochem. 73, 355–382 [DOI] [PubMed] [Google Scholar]
- 8. Hoshino D., Tomari T., Nagano M., Koshikawa N., Seiki M. (2009) A novel protein associated with membrane-type 1 matrix metalloproteinase binds p27(kip1) and regulates RhoA activation, actin remodeling, and Matrigel invasion. J. Biol. Chem. 284, 27315–27326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Blomster H. A., Imanishi S. Y., Siimes J., Kastu J., Morrice N. A., Eriksson J. E., Sistonen L. (2010) In vivo identification of sumoylation sites by a signature tag and cysteine-targeted affinity purification. J. Biol. Chem. 285, 19324–19329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Deyrieux A. F., Rosas-Acosta G., Ozbun M. A., Wilson V. G. (2007) Sumoylation dynamics during keratinocyte differentiation. J. Cell Sci. 120, 125–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tatham M. H., Geoffroy M.-C., Shen L., Plechanovova A., Hattersley N., Jaffray E. G., Palvimo J. J., Hay R. T. (2008) RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat. Cell Biol. 10, 538–546 [DOI] [PubMed] [Google Scholar]
- 12. Ghioni P., D'Alessandra Y., Mansueto G., Jaffray E., Hay R. T., La Mantia G., Guerrini L. (2005) The protein stability and transcriptional activity of p63α are regulated by SUMO-1 conjugation. Cell Cycle 4, 183–190 [DOI] [PubMed] [Google Scholar]
- 13. Pichler A., Gast A., Seeler J. S., Dejean A., Melchior F. (2002) The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell 108, 109–120 [DOI] [PubMed] [Google Scholar]
- 14. Yang M., Hsu C.-T., Ting C.-Y., Liu L. F., Hwang J. (2006) Assembly of a polymeric chain of SUMO1 on human topoisomerase I in vitro. J. Biol. Chem. 281, 8264–8274 [DOI] [PubMed] [Google Scholar]
- 15. Baczyk D., Drewlo S., Kingdom J. C. P. (2013) Emerging role of SUMOylation in placental pathology. Placenta 34, 606–612 [DOI] [PubMed] [Google Scholar]
- 16. Abe J.-I., Manabe I., Aikawa M., Aikawa E. (2013) Cardiovascular inflammation 2012: reactive oxygen species, SUMOylation, and biomarkers in cardiovascular inflammation. Int. J. Inflam. 2013, 953463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yang W., Sheng H., Homi H. M., Warner D. S., Paschen W. (2008) Cerebral ischemia/stroke and small ubiquitin-like modifier (SUMO) conjugation: a new target for therapeutic intervention? J. Neurochem. 106, 989–999 [DOI] [PubMed] [Google Scholar]
- 18. Lapeyre B., Bourbon H., Amalric F. (1987) Nucleolin, the major nucleolar protein of growing eukaryotic cells: an unusual protein structure revealed by the nucleotide sequence. Proc. Natl. Acad. Sci. U.S.A. 84, 1472–1476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Greco A. (2009) Involvement of the nucleolus in replication of human viruses. Rev. Med. Virol. 19, 201–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Seko Y., Cole S., Kasprzak W., Shapiro B. A., Ragheb J. A. (2006) The role of cytokine mRNA stability in the pathogenesis of autoimmune disease. Autoimmun. Rev. 5, 299–305 [DOI] [PubMed] [Google Scholar]
- 21. Dranovsky A., Vincent I., Gregori L., Schwarzman A., Colflesh D., Enghild J., Strittmatter W., Davies P., Goldgaber D. (2001) Cdc2 phosphorylation of nucleolin demarcates mitotic stages and Alzheimer's disease pathology. Neurobiol. Aging 22, 517–528 [DOI] [PubMed] [Google Scholar]
- 22. Caudle W. M., Kitsou E., Li J., Bradner J., Zhang J. (2009) A role for a novel protein, nucleolin, in Parkinson's disease. Neurosci. Lett. 459, 11–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Storck S., Shukla M., Dimitrov S., Bouvet P. (2007) Functions of the histone chaperone nucleolin in diseases. Subcell. Biochem. 41, 125–144 [DOI] [PubMed] [Google Scholar]
- 24. Grinstein E., Du Y., Santourlidis S., Christ J., Uhrberg M., Wernet P. (2007) Nucleolin Regulates Gene Expression in CD34-positive Hematopoietic Cells. J. Biol. Chem. 282, 12439–12449 [DOI] [PubMed] [Google Scholar]
- 25. Ginisty H., Sicard H., Roger B., Bouvet P. (1999) Structure and functions of nucleolin. J. Cell Sci. 112, 761–772 [DOI] [PubMed] [Google Scholar]
- 26. Tuteja R., Tuteja N. (1998) Nucleolin: a multifunctional major nucleolar phosphoprotein. Crit. Rev. Biochem. Mol. Biol. 33, 407–436 [DOI] [PubMed] [Google Scholar]
- 27. Connor K. M., Hempel N., Nelson K. K., Dabiri G., Gamarra A., Belarmino J., Van De Water L., Mian B. M., Melendez J. A. (2007) Manganese superoxide dismutase enhances the invasive and migratory activity of tumor cells. Cancer Res. 67, 10260–10267 [DOI] [PubMed] [Google Scholar]
- 28. Bower J. J., Leonard S. S., Chen F., Shi X. (2006) As(III) transcriptionally activates the gadd45α gene via the formation of H2O2. Free Radic. Biol. Med. 41, 285–294 [DOI] [PubMed] [Google Scholar]
- 29. Takagi M., Absalon M. J., McLure K. G., Kastan M. B. (2005) Regulation of p53 translation and induction after DNA damage by ribosomal protein L26 and nucleolin. Cell 123, 49–63 [DOI] [PubMed] [Google Scholar]
- 30. Shibata Y., Muramatsu T., Hirai M., Inui T., Kimura T., Saito H., McCormick L. M., Bu G., Kadomatsu K. (2002) Nuclear targeting by the growth factor midkine. Mol. Cell. Biol. 22, 6788–6796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pal S., Rosas J. M., Rosas-Acosta G. (2010) Identification of the non-structural influenza A viral protein NS1A as a bona fide target of the small ubiquitin-like modifier by the use of dicistronic expression constructs. J. Virol. Methods 163, 498–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fang Y., Yu Y., Hou Q., Zheng X., Zhang M., Zhang D., Li J., Wu X.-R., Huang C. (2012) The Chinese herb isolate isorhapontigenin induces apoptosis in human cancer cells by down-regulating overexpression of antiapoptotic protein XIAP. J. Biol. Chem. 287, 35234–35243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhang D., Li J., Zhang M., Gao G., Zuo Z., Yu Y., Zhu L., Gao J., Huang C. (2012) The requirement of c-Jun N-terminal kinase 2 in regulation of hypoxia-inducing factor-1α mRNA stability. J. Biol. Chem. 287, 34361–34371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Blomster H. A., Hietakangas V., Wu J., Kouvonen P., Hautaniemi S., Sistonen L. (2009) Novel proteomics strategy brings insight into the prevalence of SUMO-2 target sites. Mol. Cell. Proteomics 8, 1382–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ren J., Gao X., Jin C., Zhu M., Wang X., Shaw A., Wen L., Yao X., Xue Y. (2009) Systematic study of protein sumoylation: development of a site-specific predictor of SUMOsp2.0. Proteomics 9, 3409–3412 [DOI] [PubMed] [Google Scholar]
- 36. Tempé D., Piechaczyk M., Bossis G. (2008) SUMO under stress. Biochem. Soc. Trans. 36, 874–878 [DOI] [PubMed] [Google Scholar]
- 37. Manza L. L., Codreanu S. G., Stamer S. L., Smith D. L., Wells K. S., Roberts R. L., Liebler D. C. (2004) Global shifts in protein sumoylation in response to electrophile and oxidative stress. Chem. Res. Toxicol. 17, 1706–1715 [DOI] [PubMed] [Google Scholar]
- 38. Ding M., Li J., Leonard S. S., Shi X., Costa M., Castranova V., Vallyathan V., Huang C. (2002) Differential role of hydrogen peroxide in UV-induced signal transduction. Mol. Cell. Biochem. 234, 81–90 [PubMed] [Google Scholar]
- 39. Salnikow K., Gao M., Voitkun V., Huang X., Costa M. (1994) Altered oxidative stress responses in nickel-resistant mammalian cells. Cancer Res. 54, 6407–6412 [PubMed] [Google Scholar]
- 40. Ozben T. (2007) Oxidative stress and apoptosis: impact on cancer therapy. J. Pharm. Sci. 96, 2181–2196 [DOI] [PubMed] [Google Scholar]
- 41. Matés J. M., Segura J. A., Alonso F. J., Márquez J. (2012) Oxidative stress in apoptosis and cancer: an update. Arch. Toxicol. 86, 1649–1665 [DOI] [PubMed] [Google Scholar]
- 42. Kim S. K., Srivastava M. (2003) Stability of nucleolin protein as the basis for the differential expression of nucleolin mRNA and protein during serum starvation. DNA Cell Biol. 22, 171–178 [DOI] [PubMed] [Google Scholar]
- 43. Chiou H.-Y., Liu S.-Y., Lin C.-H., Lee E. H. (2014) Hes-1 SUMOylation by protein inhibitor of activated STAT1 enhances the suppressing effect of Hes-1 on GADD45α expression to increase cell survival. J. Biomed. Sci. 21, 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lin D.-Y., Huang Y.-S., Jeng J.-C., Kuo H.-Y., Chang C.-C., Chao T.-T., Ho C.-C., Chen Y.-C., Lin T.-P., Fang H.-I., Hung C.-C., Suen C.-S., Hwang M.-J., Chang K.-S., Maul G. G., Shih H.-M. (2006) Role of SUMO-interacting motif in Daxx SUMO modification, subnuclear localization, and repression of sumoylated transcription factors. Mol. Cell 24, 341–354 [DOI] [PubMed] [Google Scholar]
- 45. Lin X., Sun B., Liang M., Liang Y.-Y., Gast A., Hildebrand J., Brunicardi F. C., Melchior F., Feng X.-H. (2003) Opposed regulation of corepressor CtBP by SUMOylation and PDZ binding. Mol. Cell 11, 1389–1396 [DOI] [PubMed] [Google Scholar]
- 46. Schwab M. S., Dreyer C. (1997) Protein phosphorylation sites regulate the function of the bipartite NLS of nucleolin. Eur. J. Cell Biol. 73, 287–297 [PubMed] [Google Scholar]
- 47. Zhang D., Song L., Li J., Wu K., Huang C. (2006) Coordination of JNK1 and JNK2 is critical for GADD45α induction and its mediated cell apoptosis in arsenite responses. J. Biol. Chem. 281, 34113–34123 [DOI] [PubMed] [Google Scholar]
- 48. Song L., Li J., Zhang D., Liu Z.-G., Ye J., Zhan Q., Shen H.-M., Whiteman M., Huang C. (2006) IKKβ programs to turn on the GADD45α-MKK4-JNK apoptotic cascade specifically via p50 NF-κB in arsenite response. J. Cell Biol. 175, 607–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Srivastava M., Pollard H. B. (1999) Molecular dissection of nucleolin's role in growth and cell proliferation: new insights. FASEB J. 13, 1911–1922 [PubMed] [Google Scholar]
- 50. Chen C.-Y., Gherzi R., Andersen J. S., Gaietta G., Jürchott K., Royer H.-D., Mann M., Karin M. (2000) Nucleolin and YB-1 are required for JNK-mediated interleukin-2 mRNA stabilization during T-cell activation. Genes Dev. 14, 1236–1248 [PMC free article] [PubMed] [Google Scholar]
- 51. Yang C., Maiguel D. A., Carrier F. (2002) Identification of nucleolin and nucleophosmin as genotoxic stress-responsive RNA-binding proteins. Nucleic Acids Res. 30, 2251–2260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhang J., Tsaprailis G., Bowden G. T. (2008) Nucleolin Stabilizes Bcl-XL Messenger RNA in Response to UVA Irradiation. Cancer Res. 68, 1046–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Jentsch S., Haendler B., Hunter T., Sun H. (2009) The Ubiquitin System in Health and Disease, pp. 1–16, Springer, Berlin [Google Scholar]
- 54. Abdelmohsen K., Gorospe M. (2012) RNA-binding protein nucleolin in disease. RNA Biology 9, 799–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Warrener P., Petryshyn R. (1991) Phosphorylation and proteolytic degradation of nucleolin from 3T3-F442A cells. Biochem. Biophys. Res. Commun. 180, 716–723 [DOI] [PubMed] [Google Scholar]
- 56. Créancier L., Prats H., Zanibellato C., Amalric F., Bugler B. (1993) Determination of the functional domains involved in nucleolar targeting of nucleolin. Mol. Biol. Cell 4, 1239–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Shin I., Rotty J., Wu F. Y., Arteaga C. L. (2005) Phosphorylation of p27Kip1 at Thr-157 interferes with its association with importin α during G1 and prevents nuclear re-entry. J. Biol. Chem. 280, 6055–6063 [DOI] [PubMed] [Google Scholar]
- 58. Ruse C. I., McClatchy D. B., Lu B., Cociorva D., Motoyama A., Park S. K., Yates J. R. (2008) Motif-specific sampling of phosphoproteomes. J. Proteome Res. 7, 2140–2150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hornbeck P. V., Kornhauser J. M., Tkachev S., Zhang B., Skrzypek E., Murray B., Latham V., Sullivan M. (2012) PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res. 40, D261–D270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Schneider Aguirre R., Karpen S. J. (2013) Inflammatory mediators increase SUMOylation of retinoid X receptor α in a c-Jun N-terminal kinase-dependent manner in human hepatocellular carcinoma cells. Mol. Pharmacol. 84, 218–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Da Silva-Ferrada E., Lopitz-Otsoa F., Lang V., Rodríguez M. S., Matthiesen R. (2012) Strategies to identify recognition signals and targets of SUMOylation. Biochem. Res. Int. 2012, 875148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Bossis G., Melchior F. (2006) Regulation of SUMOylation by reversible oxidation of SUMO conjugating enzymes. Mol. Cell 21, 349–357 [DOI] [PubMed] [Google Scholar]
- 63. Xu Z., Lam L. S. M., Lam L. H., Chau S. F., Ng T. B., Au S. W. N. (2008) Molecular basis of the redox regulation of SUMO proteases: a protective mechanism of intermolecular disulfide linkage against irreversible sulfhydryl oxidation. FASEB J. 22, 127–137 [DOI] [PubMed] [Google Scholar]
- 64. Cong R., Das S., Bouvet P. (2011) The multiple properties and functions of nucleolin. in The Nucleolus (Olson M. O. J., ed) pp. 185–212, Springer, New York [Google Scholar]
- 65. Daniely Y., Borowiec J. A. (2000) Formation of a complex between nucleolin and replication protein A after cell stress prevents initiation of DNA replication. J. Cell Biol. 149, 799–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wang X. W., Zhan Q., Coursen J. D., Khan M. A., Kontny H. U., Yu L., Hollander M. C., O'Connor P. M., Fornace A. J., Jr., Harris C. C. (1999) GADD45 induction of a G2/M cell cycle checkpoint. Proc. Natl. Acad. Sci. U.S.A. 96, 3706–3711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Smith M. L., Chen I. T., Zhan Q., Bae I., Chen C. Y., Gilmer T. M., Kastan M. B., O'Connor P. M., Fornace A. J., Jr. (1994) Interaction of the p53-regulated protein Gadd45 with proliferating cell nuclear antigen. Science 266, 1376–1380 [DOI] [PubMed] [Google Scholar]
- 68. Yasui K., Baba A. (2006) Therapeutic potential of superoxide dismutase (SOD) for resolution of inflammation. Inflamm. Res. 55, 359–363 [DOI] [PubMed] [Google Scholar]
- 69. Chang Q., Bhatia D., Zhang Y., Meighan T., Castranova V., Shi X., Chen F. (2007) Incorporation of an internal ribosome entry site-dependent mechanism in arsenic-induced GADD45α expression. Cancer Res. 67, 6146–6154 [DOI] [PubMed] [Google Scholar]
- 70. Hovanessian A. G., Soundaramourty C., El Khoury D., Nondier I., Svab J., Krust B. (2010) Surface expressed nucleolin is constantly induced in tumor cells to mediate calcium-dependent ligand internalization. PLoS One 5, e15787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Bolognani F., Perrone-Bizzozero N. I. (2008) RNA-protein interactions and control of mRNA stability in neurons. J. Neurosci. Res. 86, 481–489 [DOI] [PubMed] [Google Scholar]
- 72. Halees A. S., El-Badrawi R., Khabar K. S. A. (2008) ARED organism: expansion of ARED reveals AU-rich element cluster variations between human and mouse. Nucleic Acids Res. 36, D137–D140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Soundararajan S., Chen W., Spicer E. K., Courtenay-Luck N., Fernandes D. J. (2008) The nucleolin targeting aptamer AS1411 destabilizes Bcl-2 messenger RNA in human breast cancer cells. Cancer Res. 68, 2358–2365 [DOI] [PubMed] [Google Scholar]
- 74. Maiti S., Chatterjee A. (2000) Differential response of cellular antioxidant mechanism of liver and kidney to arsenic exposure and its relation to dietary protein deficiency. Environ. Toxicol. Pharmacol. 8, 227–235 [DOI] [PubMed] [Google Scholar]
- 75. Sinha M., Manna P., Sil P. C. (2008) Protective effect of arjunolic acid against arsenic-induced oxidative stress in mouse brain. J. Biochem. Mol. Toxicol. 22, 15–26 [DOI] [PubMed] [Google Scholar]
- 76. Yang P., He X.-Q., Peng L., Li A.-P., Wang X.-R., Zhou J.-W., Liu Q.-Z. (2007) The role of oxidative stress in hormesis induced by sodium arsenite in human embryo lung fibroblast (HELF) cellular proliferation model. J. Toxicol. Environ. Health A 70, 976–983 [DOI] [PubMed] [Google Scholar]
- 77. Nordenson I., Beckman L. (1991) Is the genotoxic effect of arsenic mediated by oxygen free radicals? Hum. Hered. 41, 71–73 [DOI] [PubMed] [Google Scholar]
- 78. Sakurai T., Kaise T., Matsubara C. (1998) Inorganic and methylated arsenic compounds induce cell death in murine macrophages via different mechanisms. Chem. Res. Toxicol. 11, 273–283 [DOI] [PubMed] [Google Scholar]
- 79. Han Y. H., Kim S. Z., Kim S. H., Park W. H. (2008) Suppression of arsenic trioxide-induced apoptosis in HeLa cells by N-acetylcysteine. Mol. Cells 26, 18–25 [PubMed] [Google Scholar]
- 80. Chen Y.-C., Lin-Shiau S.-Y., Lin J.-K. (1998) Involvement of reactive oxygen species and caspase 3 activation in arsenite-induced apoptosis. J. Cell. Physiol. 177, 324–333 [DOI] [PubMed] [Google Scholar]