Background: Na,K-ATPase is stabilized by phosphatidylserine/cholesterol and is stimulated by neutral phospholipids.
Results: Three specific lipid-Na,K-ATPase interactions are detectable that either (a) stabilize the protein or (b) stimulate or (c) inhibit Na,K-ATPase activity, with distinct kinetic mechanisms.
Conclusion: There are separate binding sites for phosphatidylserine/cholesterol (stabilizing), polyunsaturated phosphatidylethanolamine (stimulatory), and sphingomyelin/cholesterol (inhibitory).
Significance: In physiological conditions, specifically bound lipids may regulate Na,K-ATPase activity.
Keywords: Cholesterol, Membrane Protein, Membrane Transport, Na+/K+-ATPase, Phospholipid, Lipid-Protein Interaction, Specific Lipid Sites
Abstract
The activity of membrane proteins such as Na,K-ATPase depends strongly on the surrounding lipid environment. Interactions can be annular, depending on the physical properties of the membrane, or specific with lipids bound in pockets between transmembrane domains. This paper describes three specific lipid-protein interactions using purified recombinant Na,K-ATPase. (a) Thermal stability of the Na,K-ATPase depends crucially on a specific interaction with 18:0/18:1 phosphatidylserine (1-stearoyl-2-oleoyl-sn-glycero-3-phospho-l-serine; SOPS) and cholesterol, which strongly amplifies stabilization. We show here that cholesterol associates with SOPS, FXYD1, and the α subunit between trans-membrane segments αTM8 and -10 to stabilize the protein. (b) Polyunsaturated neutral lipids stimulate Na,K-ATPase turnover by >60%. A screen of the lipid specificity showed that 18:0/20:4 and 18:0/22:6 phosphatidylethanolamine (PE) are the optimal phospholipids for this effect. (c) Saturated phosphatidylcholine and sphingomyelin, but not saturated phosphatidylserine or PE, inhibit Na,K-ATPase activity by 70–80%. This effect depends strongly on the presence of cholesterol. Analysis of the Na,K-ATPase activity and E1-E2 conformational transitions reveals the kinetic mechanisms of these effects. Both stimulatory and inhibitory lipids poise the conformational equilibrium toward E2, but their detailed mechanisms of action are different. PE accelerates the rate of E1 → E2P but does not affect E2(2K)ATP → E13NaATP, whereas sphingomyelin inhibits the rate of E2(2K)ATP → E13NaATP, with very little effect on E1 → E2P. We discuss these lipid effects in relation to recent crystal structures of Na,K-ATPase and propose that there are three separate sites for the specific lipid interactions, with potential physiological roles to regulate activity and stability of the pump.
Introduction
P-type ATPases are a diverse group of ion-transporting membrane proteins consisting of bacterial ion translocators (type I), plant and fungal H-pumps (type III), phospholipid flippases (type IV), and proteins of unknown function (type V). Type II comprises plasma membrane Ca-pumps, SR Ca-pumps, gastric H,K-ATPase, and the ubiquitous Na,K-ATPase (1, 2). The Na,K-ATPase transports Na+ and K+ ions against their electrochemical gradients across the plasma membrane. For each hydrolyzed ATP, three Na+ ions are pumped out of the cell in exchange for two K+ ions. The sodium pump is active as a heterodimer of one α-subunit and one β-subunit. α harbors ion-binding sites, the ion transport pathway, and three intracellular domains that orchestrate ATP binding and phosphorylation of the catalytic aspartate residue (3). The β-subunit acts as a chaperone and furthermore affects the apparent K+ affinity (4, 5). Although not essential for pump activity, FXYD proteins associate with the Na,K-ATPase and modulate its function in a tissue-specific manner (6).
Transport is achieved by cycling of the enzyme between two principle conformations, E1 and E2 (3, 7), with the conformational transitions E1P(3Na+) → E2P and E2(2K+) → E1 being the rate-limiting steps of the cycle (8, 9). In E1, the affinity for Na+ is high, and the ion binding sites are accessible from the cytoplasm; E2 is open to the extracellular side and has a high affinity for K+ (10). High resolution structures of SERCA, which were obtained for several conformations, revealed that the conformational transitions require substantial movements of TM1–63 as well as rearrangements of the three cytoplasmic domains (11). Until now, Na,K-ATPase has been crystallized in only three conformations either from shark rectal gland or pig kidney: E2·MgF42−·2K+ (12, 13) and E2·MgF42−·2K+·ouabain (14), E2P·ouabain (15, 16), and E1·AlF4−·ADP·3Na+ (17, 18). Several of these high resolution structures revealed the position of associated phospholipid and cholesterol molecules bound to crevices of the transmembrane domain and between subunits. Although the dependence of Na,K-ATPase activity on cholesterol has been known for many years (19), the function of other associated lipids is unclear.
In addition to cholesterol Na,K-ATPase activity depends on phosphatidylserine (20). Besides these specific lipid requirements, it has been proposed that membrane protein functions may be modified by annular lipids responding to physical properties of the membrane (e.g. membrane fluidity, lateral pressure, and hydrophobic thickness) (21). For instance, sodium pump activity was highest in proteoliposomes composed of 22:1 PC in the absence or 18:1 PC in the presence of cholesterol, and the difference was attributed to a contraction of the bilayer thickness in the presence of cholesterol (22). Similarly, in other studies, the optimal chain length for Ca-ATPase was 18:1, and it was shown that the lower rate of ATP hydrolysis in “thinner” or “thicker” membranes follows from a reduced rate of phosphorylation or dephosphorylation, respectively (23, 24). In addition, low resolution x-ray crystallography and molecular dynamic calculations have suggested that Ca-ATPase may adapt structurally to membranes of different hydrophobic thicknesses (25).
It is often difficult to distinguish annular from specific lipid interactions, and the effects of certain lipids differ when comparing proteins reconstituted in liposomes with solubilized proteins. For instance, 22:6 PC or 16:0/22:6 PC reduced Na,K-ATPase turnover in liposomes, whereas polyunsaturated phospholipids stimulate pump activity of Na,K-ATPase in lipid-detergent micelles (26, 27). Use of soluble detergent-lipid-protein mixed micelles removes the ambiguity in interpretation of effects of lipids when studied in membrane or proteoliposome systems and allows specific protein-lipid interactions to be studied systematically. For example, observations of the lipid dependence of recombinant Na,K-ATPase expressed in Pichia pastoris provided strong evidence for a specific interaction of the protein with 18:0/18:1 PS (SOPS) that stabilizes the protein against thermal inactivation. In the presence of SOPS, cholesterol causes strong additional stabilization of the Na,K-ATPase (28). Using a mutant of the thermo-labile α2 isoform (α2VFP), we have recently shown that SOPS is bound between αTM8 and -10 and interacts with the FXYD protein (29). Using the same mutant, we provide here evidence for a direct interaction of cholesterol and SOPS, making the C-terminal helices of the α-subunit, the FXYD protein, and the lipids bound between αTM8 and -10 a stability “hotspot.” Whereas SOPS and cholesterol stabilize the sodium pump but do not per se alter activity, neutral phospholipids increase Na,K-ATPase turnover (27). In this paper, we analyze the specificity of this effect in detail and also describe a third effect of lipids on the Na,K-ATPase, namely the inhibition of pump activity by saturated PC or sphingomyelin in the presence of cholesterol. Finally, we have investigated the mechanisms of the kinetic effects of both activating and inhibitory effects of lipids by analyzing the properties of the Na,K-ATPase activity, including E1-E2 conformational changes detected by steady-state and stopped-flow fluorescence measurements.
EXPERIMENTAL PROCEDURES
Materials
n-Dodecyl-β-d-maltopyranoside (catalogue no. D310) and C12E8 (25% (w/w), catalogue no. 0330) were purchased from Anatrace. All lipids were stored as chloroform solutions and obtained from Avanti Polar Lipids (synthetic PC, PE, and brain SM) or Sigma (cholesterol and chicken egg yolk SM). [γ-32P]Adenosine 5′-triphosphate was purchased from PerkinElmer Life Sciences, and BD Talon metal affinity resin was purchased from Clontech (catalogue no. 635503). FITC was obtained from Sigma. All chemicals used were of the highest available quality.
Recombinant Human Na,K-ATPase Expression and Purification
P. pastoris transformation, yeast growth, membrane preparation, and His tag purification of recombinant human α1β1FXYD1 Na,K-ATPase were done essentially as described previously (27–29).
Detergent-soluble Na,K-ATPase complexes were purified in a mix of 0.3 mg/ml C12E8, 0.05 mg/ml cholesterol, and 0.17 mg/ml PS or 0.07 mg/ml SOPS and a 0.1 mg/ml concentration of the indicated lipid (PC, PE, or SM). For lipid specificity assays, the enzyme was prepared in batch mode, and gravity column purification was used for kinetic measurements.
FITC Labeling of Yeast Membranes
Na,K-ATPase in yeast membranes was labeled with fluorescein 5′-isothiocyanate as described (30) with modifications. Membranes were suspended in 50 mm NaCl, 1 mm EDTA, and 50 mm sodium carbonate, pH 9.2, at 2 mg/ml. FITC was prepared freshly and added to a final concentration of 4 μm. Membranes were then labeled for 45 min at room temperature in the dark with stirring, diluted with cold MOPS/Tris, pH 6.6, and collected by ultracentrifugation.
Recombinant Human FXYD1 Expression, Purification, and Reconstitution
Human FXYD1 (phospholemman (PLM)) was expressed in Escherichia coli CD41 cells from pET28-TevH vector (29). FXYD1 purification was described in detail previously (27). For reconstitution with the Na,K-ATPase, PLM was dialyzed against 500 mm NaCl, 10% glycerol, 0.1 mm DTT, and 25 mm MOPS, pH 7.4, and the His tag was removed by tobacco etch virus protease. The cleaved PLM was then added to the solubilized yeast membranes with the Na,K-ATPase bound to BD Talon beads at a molar ratio of 10:1 (PLM/Na,K-ATPase) and incubated overnight.
Size Exclusion HPLC
Size exclusion HPLC was done using a Superdex 200 column (300 × 10 mm) (Amersham Biosciences). The protein was eluted at 0.5 ml/min in a medium containing 150 mm NaCl, 20 mm MOPS/Tris, pH 7.2, 0.1 mg/ml C12E8 (see Ref. 28).
Blue Native Gel Electrophoresis
Blue native gel electrophoresis was carried out as described previously (31).
Activity Assays
Enzyme activity was measured using PiColorLockTM malachite green assay (Inova Biosciences), detecting free phosphate from ATP hydrolysis in a medium containing 130 mm NaCl, 20 mm KCl, 3 mm MgCl2, 1 mm EGTA, 25 mm histidine, pH 7.4, and 1 mm ATP at 37 °C. For ion titrations, activity was measured at varying concentrations of sodium or potassium, and choline chloride was used in order to maintain ionic strength. Phosphoenzyme was determined as described previously (27).
Equilibrium and Stopped-flow Fluorescence Measurements
Equilibrium fluorescence experiments were carried out in a Varian fluorimeter at room temperature using the following conditions: 8 μg/ml FITC-labeled Na,K-ATPase in 150 mm choline chloride, 10 mm MOPS/Tris, pH 7.4. The excitation wavelength was set to 485 nm and emission to 520 nm with a 5-nm slit width each. For titrations, small aliquots of concentrated RbCl or NaCl were added until no further fluorescence changes were observed. NaCl and KCl titrations with RH421 were performed as described previously (32) with emission and excitation wavelength set to 580 and 680 nm with a 5-nm slit width.
For stopped-flow measurements, an Applied Photophysics SX20 system (for FITC) or a BioLogic SFM 400 instrument (for RH421) was used. Excitation wavelength was 485 nm for FITC and 577 nm for RH421 using a combined xenon/mercury lamp, and fluorescence was measured at emission ≥515 nm for FITC and ≥665 nm for RH421 using cut-off filters. Solutions were mixed 1:1 using ∼120 μl/syringe, and the temperature of the measurements was set to 23 °C. All solutions were buffered to pH 7.2 using MOPS/Tris, and ionic strength was kept constant at 120 mm for all measurements using choline chloride.
FITC-labeled Enzyme
For E2(2Rb) → E1·3Na, syringe 1 contained 15 μg/ml FITC-labeled enzyme in 20 mm RbCl and was mixed with 80 mm NaCl in syringe 2.
RH421
For E2(2K)ATP → E1·3NaATP, syringe 1 contained 20 μg/ml of enzyme non-covalently labeled with 200 nm RH421 in 20 mm KCl, 1 mm EDTA and was mixed with 80 mm NaCl, 2 mm ATP, 1 mm EDTA in syringe 2. E1·3Na → E2P, 10 μg/ml Na,K-ATPase non-covalently labeled with 200 nm RH421 in 100 mm NaCl and 4 mm MgCl2 in syringe 1 was mixed with 1 mm ATP in the same solution in syringe 2.
Data Fitting
Data were fitted using KaleidaGraph (Synergy Software). Ion titrations were fitted using the Hill equation,
 |
where n is the Hill coefficient, [ion] is the free concentration of the respective ion, and K½ is the concentration required to obtain the half-maximal fluorescence signal.
Stopped-flow traces were fitted to a monoexponential function,
or double exponential function,
where A is the amplitude of the fluorescence signal, k is the rate of the reaction, and c is the equilibrium fluorescence level after the reaction is complete. All values are expressed as averages of at least three experiments ± S.E.
Lipidomics Analysis
Right-side-out membrane vesicles (ROVs) were prepared from rabbit kidney microsomes as described (33).
Lipid Extraction
10 μl of ROV preparation (64 μg of equivalent protein) was homogenized in 150 mm ammonium bicarbonate buffer. The lipids from ROV samples were extracted according to the procedure described previously (34).
Shotgun Mass Spectrometry
Dried lipid extract was resuspended in 100 μl of chloroform/methanol mixture (1:2, v/v). For the analysis, 10 μl of lipid extract were loaded in a 96-well plate (Eppendorf, Hamburg) and diluted with 20 μl of 0.05% triethylamine in methanol or ammonium acetate 13 mm in isopropyl alcohol for negative ion mode analysis Fourier transform-MS/MS. Cholesterol was subjected to chemical acetylation and analyzed by positive ion mode Fourier transform-MS analysis (35). Data were collected after samples were infused via robotic nanoflow electrospray ionization source Triversa NanoMate (Advion Biosciences, Ithaca, NY) into a hybrid quadrupole Orbitrap tandem mass spectrometer Q Exactive (Thermo Fisher Scientific, Waltham, MA).
Data Processing
Lipids were identified by LipidXplorer software by matching m/z of their monoisotopic peaks to the corresponding elemental composition constraints (36).
RESULTS
As an indication of the state of the Na,K-ATPase preparation used in the current studies, namely detergent-soluble purified human α1β1FXYD1 complexes, Fig. 1A shows an SDS gel, Fig. 1B a blue native gel, and Fig. 1C a size exclusion HPLC run. The data in Fig. 1, A and C, resemble the data in Refs. 27 and 37, except that the preparations are purer, because they were prepared by a column rather than batch procedure, and the α1β1 complexes were also reconstituted with FXYD1 and with different added lipids, as indicated. The SDS-PAGE (Fig. 1A), overloaded with α1β1FXYD1 complexes prepared with SOPS/cholesterol or with added brain PE and sphingomyelin, shows that the protein is largely pure (∼90%). Blue native gels are used to separate hydrophobic membrane proteins and complexes in the native state (31). The blue native gel in Fig. 1B shows that the protein is soluble and pure, and the great majority consists of an α1β1FXYD1 protomer, with apparent molecular mass between 250 and 300 kDa, with a minor amount of the diprotomer (α1β1FXYD1)2, ∼500 kDa.4 Finally, a size exclusion HPLC run (Fig. 1C) confirmed that the preparation consists mainly of the soluble α1β1FXYD1-lipid-detergent protomer, with small amounts of protein aggregates, higher oligomers, and protein-free lipid-detergent micelles (see also Ref. 28). No differences were found for preparations made with different lipids. In relation to the lipid content of the preparations, preliminary shotgun mass spectrometry analyses of the lipids in the preparations did not reveal species other than those added to the preparations.5
FIGURE 1.

Purified human α1β1FXYD1 Na,K-ATPase protomer. A, SDS-PAGE. PS, protein prepared with SOPS/cholesterol; PE, protein prepared with SOPS/cholesterol/brain PE; SM, protein prepared with SOPS/cholesterol/brain SM as described under “Experimental Procedures.” 15 μg of protein was used per lane. B, blue native gel. 15 μg of purified α1β1FXYD1 complex, prepared with SOPS/cholesterol, was applied to the gel and stained with Coomassie Brilliant Blue. C, size exclusion HPLC. 50 μg of protein was prepared in 0.17 mg/ml SOPS, 0.05 mg/ml cholesterol, 0.3 mg/ml C12E8 and applied to the Superdex 200 column.
Lipid Specificity of Stimulation of NaK-ATPase
As shown recently, unsaturated neutral phospholipids (PC or PE, especially 18:2/18:2 PC and 18:2/18:2 PE) stimulate the specific Na,K-ATPase activity of human α1β1 FXYD1 complexes by increasing the molar turnover rate (27, 37). We have now investigated the selectivity of this effect more systematically using a selection of synthetic mono-, di-, and polyunsaturated PCs and PEs (Fig. 2). All samples were prepared in a mix of SOPS, cholesterol, C12E8, and the indicated lipid with the total lipid and detergent concentration kept constant. One result of this screen is that phospholipids with polyunsaturated acyl chains, especially lipids with mixed saturated and polyunsaturated acyl groups, are more effective than mono- or di-unsaturated lipids. For example, 18:0/22:6 PE increased Na,K-ATPase activity by 65%, followed by 18:0/20:4 PE or 18:1/20:4 plasmalogen PE and brain PE (mainly 18:1/20:4 PE), which increased activity by 55%, whereas 18:0/18:2 PE was less effective (30% effect), and 18:0/18:1 PE was a poor activator, increasing activity only by 13%.6 A second result is that PEs tend to be slightly more effective than PCs esterified with the same fatty acids. Compare, for example, 18:0/20:4 and 18:0/22:6 PE and PC and 18:0/18:1 PE, which increased activity by 13%, whereas 18:0/18:1 PC did not affect pump activity. In addition, purification with the symmetrical highly unsaturated 20:4/20:4 or 20:6/20:6 PC was also suboptimal.
FIGURE 2.
Na,K-ATPase activity purified with polyunsaturated PC or PE. Human α1β1FXYD1 complexes were prepared in a mix of 0.07 mg/ml SOPS, 0.05 mg/ml cholesterol, a 0.1 mg/ml concentration of the indicated lipid, and 0.3 mg/ml C12E8. Activities were normalized to the SOPS reference (black). Phospholipids esterified with two identical fatty acids are shown in light gray, and mixed acyl phospholipids are depicted in dark gray. P 18:0–20:4 EP, plasmalogen PE. Error bars, S.E.
Because the α3 and α2 isoforms are expressed strongly in nerves and glial cells (40), the high degree of stimulation of α1 by brain PE raised the question of whether α2 and α3 are also stimulated by polyunsaturated phospholipids. We prepared α1β1FXYD1, α2β1FXYD1, and α3β1FXYD1 without and with different PEs. Fig. 3 shows that all three isoforms were stimulated by polyunsaturated PEs and that there is no significant difference between the isoforms. 18:0/22:6 PE was most effective, followed by 18:0/20:4 PE and brain PE, whereas 18:0/18:2 PE was about half as effective.
FIGURE 3.

Activity of Na,K-ATPase isoforms α1, α2, and α3 purified with polyunsaturated PE. Na,K-ATPase isoforms α1β1, α2β1, and α3β1 were prepared with FXYD1 as in Fig. 1 with the indicated lipid, and activity was normalized to the sample purified with SOPS/cholesterol. Error bars, S.E.
The specific Na,K-ATPase activity of α1β1FXYD1 prepared with SOPS/cholesterol/brain PE was in the range of 30–36 μmol/min/mg protein, similar to that of highly purified native renal Na,K-ATPase (41, 42). Another feature of the preparation made with PE similar to the renal Na,K-ATPase concerns thermostability. Renal Na,K-ATPase is more thermostable in KCl than NaCl (43). In contrast, the purified recombinant protein prepared in the normal way with SOPS/cholesterol is more thermostable in NaCl compared with KCl (38). Fig. 4 shows thermal stability of the α1β1FXYD complex purified with brain PE/SOPS/cholesterol in NaCl (E13Na) or KCl (E2(2K)) compared with that purified only with SOPS/cholesterol. As expected, the standard preparation with SOPS/cholesterol is more stable in NaCl than in KCl. However, when the protein is prepared with SOPS/cholesterol/PE, the overall thermostability is reduced (see also Ref. 27), and now the preparation is more stable in KCl than in NaCl, similar to the renal Na,K-ATPase.
FIGURE 4.

Thermostability of Na,K-ATPase in the presence of NaCl and KCl. Na,K-ATPase was purified with SOPS/cholesterol (circle) or SOPS/cholesterol/Brain PE (square) in the presence of 100 mm KCl (open; stabilization of E2) or NaCl (filled symbols; stabilization of E1) and incubated for the indicated time at 47 °C. Enzyme activity at each time point was normalized to the maximal activity at t0. Error bars, S.E.
Saturated PC and Sphingomyelin Inhibit Na,K-ATPase Activity
The strong stimulation by mixed saturated/polyunsaturated neutral lipids raises the question of how all-saturated lipids affect Na,K-ATPase activity. α1β1FXYD1 was prepared as in Fig. 2 with saturated instead of polyunsaturated lipids and SOPS/cholesterol, and activity was compared with a sample purified only with SOPS/cholesterol (Fig. 5). Noticeably, 18:0/18:0 and 20:0/20:0 PC strongly reduced Na,K-ATPase activity by 75–80%, and PCs with shorter saturated fatty acyl chains (e.g. 14:0/14:0) were less effective. In contrast to unsaturated PEs, which had stimulatory effects similar to those of polyunsaturated PCs, saturated 18:0/18:0 PE was a less effective inhibitor than 18:0/18:0 PC and reduced Na,K-ATPase activity by only 40%. The negatively charged 18:0/18:0 PS was even less effective and reduced specific activity by only 20%. This selectivity pattern is consistent with a specific protein-lipid interaction. The rather specific effect of PC led us to consider whether sphingomyelin has a similar effect because sphingomyelin consists of phosphocholine and ceramide, and its fatty acids are generally more unsaturated than those of phospholipids. As seen in Fig. 5, sphingomyelin from chicken egg and porcine brain was almost as effective as 18:0/18:0 PC and reduced Na,K-ATPase activity by 65 and 71%, respectively. The strong opposing effects of the mixed saturated/polyunsaturated PE and saturated PC or sphingomyelin imply that Na,K-ATPase activity could be regulated over a wide range, depending on the composition of its lipid environment. We have tested this notion by purifying the pump in varying proportions of brain PE and brain SM (Fig. 6, left and right). When the brain PE was replaced by gradually increasing amounts of brain SM, at constant total PE plus SM concentration, the Na,K-ATPase-specific activity decreases in a linear manner. Thus, the relative lipid composition can alter the molar activity of recombinant Na,K-ATPase by almost 10-fold. In order to establish whether the PE and SM compete for the same site, brain SM was increased gradually, at constant total SM +PS, without or with PE (Fig. 6, right). The initial activities without SM were 17 and 26 μmol/min/mg without and with PE, respectively. Increasing concentrations of SM inhibited the activity along virtually identical hyperbolic curves, with fitted Ki values (∼0.02 mg/ml) and maximal degrees of inhibition (∼70%) without or with PE, respectively. Thus, the experiment proves that the SM and PE do not compete for the same binding site.
FIGURE 5.

Inhibitory effects of saturated phospholipids and sphingomyelin. α1β1FXYD1 was purified with SOPS/cholesterol or SOPS/cholesterol and the indicated lipid, and its specific activity was normalized to the SOPS reference. Error bars, S.E.
FIGURE 6.

Activity of recombinant Na,K-ATPase purified in a mixture of brain SM and brain PE. Left, Na,K-ATPase was purified in a mixture of 0.07 mg/ml SOPS, 0.05 mg/ml cholesterol, and a total of 0.1 mg/ml brain PE and sphingomyelin, which were mixed at the indicated ratio. Activity was normalized to the maximal activity when purified with PS/cholesterol/brain PE (right). Error bars, S.E.
Effects of Cholesterol on Stimulation, Inhibition, and Stabilization by Phospholipids
It has been known for many years that cholesterol is required for optimal activity of the Na,K-pump (19), and we have shown previously that cholesterol strongly stabilizes the purified detergent-soluble recombinant Na,K-ATPase when present with SOPS but does not per se alter pump activity (28). The latter feature is seen in the bar graph in Fig. 7 (left two columns). In addition, the graph shows that the raised Na,K-ATPase activity of enzyme purified with 18:0/20:4 PE is unaffected by the presence or absence of cholesterol (columns 3 and 4). However, Fig. 7 shows that 18:0/18:0 PC and brain SM are significantly less efficient inhibitors in the absence of cholesterol, the level of inhibition being only ∼40% compared with ∼70% with cholesterol. The inhibition by SM is also reflected in the turnover rates, estimated as 5809 ± 243 min−1 for SOPS and 3843 ± 169 min−1 for SOPS/brain SM in the absence of cholesterol (34% inhibition) versus 5256 ± 185 and 1747 ± 128 min−1 in the presence of cholesterol (67% inhibition) (Table 1).
FIGURE 7.

Dependence of activation and inhibition on cholesterol. Na,K-ATPase was prepared with SOPS, SOPS/18:0/20:4 PE, SOPS/18:0 PC, or SOPS/brain SM with and without cholesterol. The free detergent concentration was kept constant by varying the concentration of SOPS. Activities were normalized to the SOPS/cholesterol sample. Error bars, S.E.
TABLE 1.
Reduction of turnover rate by brain sphingomyelin and its dependence on cholesterol
| Turnover rate at 37 °C |
||
|---|---|---|
| With cholesterol | Without cholesterol | |
| min−1 | ||
| SOPS | 5256 ± 185 | 5809 ± 243 |
| Brain SM | 1747 ± 128 | 3843 ± 169 |
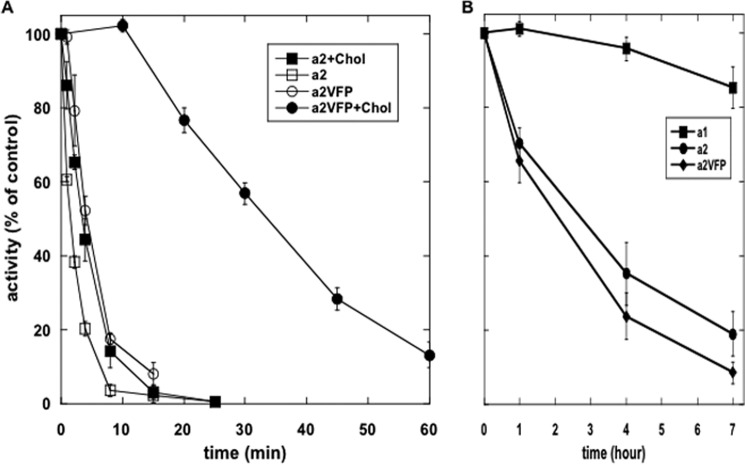
The finding that cholesterol both stabilizes in the presence of SOPS and enhances inhibition by SM or 18:0/18:0 PC suggests that there are more than one binding site. A recent high resolution structure of Na,K-ATPase revealed that cholesterol is bound to at least two sites (17). One cholesterol was observed in the αM8–10 pocket next to the FXYD protein, a location we recently reported to be interacting with SOPS (29) (see also Fig. 12). Is this cholesterol molecule responsible for the stabilization with SOPS? This question has been answered using a triple mutant of the thermo-labile α2 isoform, with three residues mutated to the equivalent α1 residues (A920V (M8), L955F (M9), V981P (M10); human α2 numbering). The thermostability of the purified α2VFPβ1 mutant is greatly increased over thermolabile wild type α2β1 and is close to that of the α1β1 isoform (29). In the experiment in Fig. 8A, wild-type α2β1 and α2VFPβ1 were purified either with SOPS alone or with SOPS/cholesterol and heated at 42 °C for different lengths of time, and the activity was measured. The order of thermal stability was α2VFP-PS/cholesterol ≫ α2VFP-PS = α2-PS/cholesterol > α2-PS (i.e. the α2VFPβ1 mutant stabilized by 2.59 fold without cholesterol but by 8.9-fold in the presence of cholesterol; see Table 2). Thus, the experiment provides clear cut evidence that the stabilizing effect of cholesterol is exerted in the same TM8–10 pocket as that of SOPS.
FIGURE 12.
Hydrophobic cluster surrounding cholesterol in the αTM8–10 pocket. A, ribbon representation of the Na,K-ATPase. α is shown in black, β in cyan, and FXYD2 in magenta. The three associated cholesterol molecules are depicted in white (CHL 3, CHL 2, and CHL 1), and the αTM helices 8 (red), 9 (green), and 10 (orange) are highlighted. B, αTM8–10 crevice harboring a cholesterol molecule. VFP side chains are shown in blue, and residues within 4 Å of cholesterol are represented as sticks. Numbering is for pig α1 and FXYD.
FIGURE 8.
Thermal inactivation of WT α2β1 and α2VFPβ1. A, purified wild type α2β1 and α2VFPβ1 prepared without or with cholesterol were incubated for the indicated time, and activity was calculated as a percentage of that of the untreated t0 sample. Half-times and stabilization factors are summarized in Table 2. B, P. pastoris membranes harboring α1β1, α2β1, and α2VFPβ1 were incubated at 45 °C prior to measuring residual ouabain binding capacity. Error bars, S.E.
TABLE 2.
Effect of cholesterol on thermal inactivation of Na,K-ATPase α2 and α2VFP
| t½ at 47 °C ±S.E. | Ratio α2VFPβ1/α2β1 without/with cholesterol ± S.E. | |
|---|---|---|
| min | ||
| α2β1 | 1.60 ± 0.05 | |
| α2VFPβ1 | 4.14 ± 0.23 | 2.59 ± 0.17 |
| α2β1 + cholesterol | 3.77 ± 0.31 | |
| α2VFPβ1 + cholesterol | 33.61 ± 1.46 | 8.92 ± 2.15 |
Another striking demonstration that cholesterol is required for stabilization of α2VFPβ1 is presented in Fig. 8B. We measured thermal stability of ouabain binding in the membranes of P. pastoris expressing α1, α2, and α2VFP. In contrast to the purified proteins, α2VFP is no more stable than wild-type α2, and both are much more thermolabile than α1. The sterol in yeast membrane is ergosterol rather than cholesterol, and we have shown that ergosterol is a poor stabilizer of the purified recombinant Na,K-ATPase compared with cholesterol (28). Thus, thermolability of α2VFP in the yeast membranes lacking cholesterol is now understandable.
Kinetic Mechanism of PE and Sphingomyelin/Cholesterol Effects
The kinetic mechanism by which brain PE and brain SM mediate stimulation or inhibition was investigated by a combination of steady-state Na,K-ATPase kinetics and fluorescence measurements of conformational changes (Tables 3–5 and Figs. 9 and 10). First, we determined apparent affinities for sodium and potassium and Ki for vanadate in Na,K-ATPase activity assays (Table 3). For the Na,K-ATPase purified with brain PE, in comparison with the SOPS reference, the K0.5K was reduced from 1.47 ± 0.06 to 1.01 ± 0.02 mm; K0.5Na increased from 16.4 ± 0.4 to 25 ± 0.5 mm; and the Ki value for vanadate decreased from 2.2 ± 0.04 to 1.0 ± 0.08 μm. Na,K-ATPase prepared with brain SM showed the same tendency, with the respective values being K0.5K = 1.16 ± 0.06 mm, K0.5Na = 20.3 ± 0.4 mm; and a Ki value for vanadate of 1.3 ± 0.2 μm. The differences in cation K0.5 values and vanadate Ki values are moderate, ranging from 20 to 55%, but are significant and are suggestive of a shift of the conformational equilibrium toward E2. The KmATP of the preparations made with SM (268 ± 12 μm) is significantly higher than for the control with only SOPS (207 ± 3 μm), also indicative of stabilization of the E2 conformation, although no difference was observed for the preparations made with PE (204 ± 5 μm). Finally, no significant difference between the three preparations was observed for inhibition by digoxin.
TABLE 3.
Apparent affinities for sodium, potassium, ATP, vanadate, and digoxin under Na,K-ATPase turnover conditions
| SOPS | Brain PE | Brain SM | |
|---|---|---|---|
| K0.5Na+ (mm) | 16.4 ± 0.4 | 25.0 ± 0.5 | 20.3 ± 0.4 |
| p < 0.01 | p < 0.01 | ||
| K0.5K+ (mm) | 1.47 ± 0.06 | 1.01 ± 0.02 | 1.16 ± 0.06 |
| p < 0.01 | p < 0.05 | ||
| KmATP (μm) | 207 ± 3 | 204 ± 5 | 268 ± 12 |
| p < 0.05 | |||
| Kivanadate (μm) | 2.2 ± 0.04 | 1.0 ± 0.08 | 1.3 ± 0.2 |
| p < 0.01 | p < 0.05 | ||
| Kidigoxin (nm) | 298 ± 20 | 236 ± 23 | 257 ± 28 |
TABLE 4.
Equilibrium titrations with FITC and RH421 labeled enzyme
The differences between the K0.5Na and K0.5K determined by RH421 are not significant.
| SOPS | Brain PE | Brain SM | |
|---|---|---|---|
| FITC | |||
| K0.5Rb+ (mm), E1 → E2(2Rb) | 1.44 ± 0.007 | 0.86 ± 0.06 | 0.58 ± 0.01 |
| p < 0.01 | p < 0.01 | ||
| K0.5Na+ (mm), E2(2Rb) → E1·3Na | 15.5 ± 0.3 | 22.8 ± 1.2 | 24.7 ± 0.8 |
| p < 0.01 | p < 0.01 | ||
| RH421 | |||
| K0.5cyt Na+ (mm) | 7.7 ± 0.5 | 8.7 ± 0.1 | 8.5 ± 0.7 |
| K0.5ex K+ (μm) | 0.58 ± 0.04 | 0.65 ± 0.02 | 0.64 ± 0.06 |
TABLE 5.
Rates of the conformational transitions
| SOPS | Rates | |||
|---|---|---|---|---|
| Brain PE | Brain SM | |||
| s−1 | ||||
| RH421, E1·3Na → E2P, 1 mm ATP | ||||
| k1 | 131 ± 9 | 270 ± 10 | 162 ± 12 | |
| k2 | 8.7 ± 0.9 | 9.7 ± 1 | 8.7 ± 0.5 | |
| RH421, E2(2K) → E1·3Na, 1 mm ATP | 61 ± 2 | 67 ± 2 | 24 ± 2 | |
| FITC, E2(2Rb)→E1·3Na | 0.19 ± 0.001 | 0.19 ± 0.001 | 0.14 ± 0.001 | |
FIGURE 9.

RH421 standard experiment and equilibrium titrations. A, standard experiment showing the principle ion binding and release steps of the Post-Albers cycle (sodium binding, sodium release after transition from E1P-E2P, potassium binding). B, equilibrium titration of sodium binding to the E1 conformation. C, equilibrium titration of potassium binding to the E2P conformation. Error bars, S.E.
FIGURE 10.
Stopped-flow traces of conformational transitions monitored with RH421. A, E1Na → E2P transition fitted to a double exponential function. Na,K-ATPase was prebound with sodium and mixed with ATP. B, Na,K-ATPase was preincubated with potassium and mixed with sodium and ATP, triggering the E2(2K)ATP → E13NaATP transition, which was fitted to a single exponential function.
We have studied E1-E2 conformational transitions using two independent fluorescent probes (Tables 4 and 5 and Figs. 9 and 10). FITC covalently labels Lys-501 of the N-domain of purified native or recombinant Na,K-ATPase and responds to E1-E2(2K) conformational transitions with relatively large fluorescence changes (30, 44). Alternatively, the non-covalent label electrochromic shift dye RH421 allows the recording of ion movements either in membrane proteins incorporated in membrane fragments or the detergent-solubilized recombinant protein complexes (32, 45, 46).
Equilibrium fluorescence titrations (Table 4) of the E1-E2(2Rb) and E2(2Rb)-E13Na transition using FITC-labeled protein revealed that both brain PE and brain SM/cholesterol significantly reduced the K0.5Rb and increased K0.5Na of the E2(2Rb)-E13Na when compared with a sample purified only with SOPS/cholesterol. Equilibrium sodium and potassium titrations of RH421 fluorescence signals provide a measure of intrinsic cation binding affinities, summarized in Fig. 9. Fig. 9A shows the standard RH421 changes of the purified soluble recombinant protein associated with sodium binding to E1 (E1-E1(3Na)) and ATP/Mg (E13Na-E2P) and potassium binding to E2P (E2P-E2(2K)). Fig. 9, B and C, and Table 4 show results of titrations of sodium and potassium binding reactions, and, as seen, neither PE nor SM significantly affected the K0.5Na or K0.5K. Taken together, the lack of effects of the PE and SM on the sodium and potassium binding affinities detected by RH421 but significant and opposite effects on K0.5Rb and K0.5Na detected with FITC-labeled protein are indeed strongly suggestive of stabilization of an E2 state either by PE or by SM compared with SOPS/cholesterol.
At first sight, it seems paradoxical that both the activator (PE) and inhibitory lipid (SM/cholesterol) stabilize an E2 conformation. In principle, stimulation of Na,K-ATPase activity by polyunsaturated PE and inhibitory effects of sphingomyelin must both affect rate-limiting reactions of the Post-Albers cycle. Measurements of the individual conformational transitions, E2(2Rb)ATP-E13NaATP and E13Na-E2P, have revealed how the opposite effects of these lipids can both stabilize the E2 conformation. Fig. 10 and Table 5 show the data for such measurements in stopped-flow experiments using the RH421 dye. When the Na,K-ATPase prebound with sodium and magnesium ions is mixed with ATP in the stopped-flow fluorimeter, the protein undergoes the E1-E2P transition. Similarly, when protein prebound with potassium ions (E2(2K)) is mixed with sodium ions plus ATP, the protein undergoes the E2(2K)ATP-E13NaATP transitions. Note here that the rates of interconversion between the different ion bound states detected by the RH421 signals must reflect the limiting rates of the conformational transitions and not the rates of cation binding or dissociation themselves, which are assumed to be orders of magnitude faster.
Fig. 10 shows traces of the E13Na-E2P and E2(2K)ATP-E1(3Na)ATP interconversions for preparations made with SOPS/cholesterol alone or with brain PE or with SM/cholesterol, and Table 5 shows the data for fitted rate constants. As described previously (47), the E1-E2P transition showed a rapid major phase and minor slow phase. Compared with the SOPS sample (k1 = 131 ± 9 s−1), the rapid phase (k1) was clearly accelerated by brain PE (270 ± 10 s−1) but was unaffected by SM/cholesterol (162 ± 12 s−1) (Fig 10A). The slow phase was unaffected by the different lipids. In contrast, the rate of E2(2K)ATP-E1(3Na)ATP was strongly inhibited by the SM/cholesterol (24 ± 2 s−1) compared with the SOPS sample (61 ± 2 s−1) (61% inhibition) but was unaffected by PE (67 ± 2 s−1) (Fig. 10B). Table 5 also shows the slow rate of E2(2K)-E1(3Na) for the three lipid conditions measured with the FITC-labeled α1β1FXYD1 complex. Here, of course, the fast rate of the transition with ATP cannot be detected because FITC labeling prevents ATP binding (44). The sample made with SM/cholesterol showed some inhibition (26%) compared with those with SOPS/cholesterol without or with PE, which were identical.
In summary, although both PE and SM/cholesterol stabilize the E2 conformation, they do so by different mechanisms. In Na,K-ATPase turnover conditions, PE accelerates the E13Na-E2P conversion, whereas SM/cholesterol inhibits the E2(2K)ATP-E1(3Na)ATP transition and also raises the KmATP.
Lipidomic Analysis of Native Renal Membranes
We have undertaken a full lipidomics analysis of rabbit kidney membranes, including quantities and relative enrichment of the synthetic lipids that produce specific effects on purified proteins, in order to establish whether they could interact with the Na,K-ATPase in a native membrane. As an example, a quantitative shotgun mass spectrometry analysis of lipids extracted from rabbit kidney membrane right-side-out vesicles, enriched with Na,K-ATPase (α1β1FXYD2), is presented in Fig. 11. As shown from this analysis and also less quantitative analyses in the literature (48, 49), PC contains a much lower proportion of polyunsaturated fatty acids than PE (or PE plasmalogen). Because the functional tests showed only moderate selectivity between polyunsaturated PC and PE (Fig. 2), on this basis alone, it is difficult to identify a “physiological ligand.” However, the analysis of acyl chain composition of native membrane lipids points to PE rather than PC, making 18:0/20:4, 18:0/22:6, or a similar polyunsaturated PE species (or perhaps PE plasmalogen) the more likely stimulatory phospholipid in physiological conditions.
FIGURE 11.

Lipid composition of right-side-out membrane vesicles from rabbit kidney. Lipids of right-side-out membrane vesicles from rabbit kidney (64 μg of protein equivalent) were extracted and analyzed by shotgun lipidomics. Molecular lipid species are expressed as pmol/μg of protein content. Cholesterol was estimated at 463.18 ± 1.89 pmol/μg proteins. Lysolipid species were detected as minor lipids and are not shown. Error bars, S.E.
Similarly, as seen in Fig. 11, the native rabbit kidney membrane lipids contain only a small amount and very low proportion of all saturated PC (16:0/16:0 PC) but a relatively large amount and high proportion of 16:0 SM. Thus, in a physiological situation, the inhibitory effect is more likely to be mediated by the SM/cholesterol than by 16:0–16:0 PC/cholesterol.
DISCUSSION
Multiple Specific Lipid Binding Sites in Na,K-ATPase
We discuss here accumulating biochemical and structural evidence for specific phospholipid and cholesterol binding to the Na,K-ATPase and their functional effects. At least three independent specific sites for phospholipids with or without cholesterol can be inferred. Crystal structures of Na,K-ATPase have revealed several bound phospholipids and cholesterol molecules, in three distinct pockets (without evidence for specific functional roles of the observed lipids) (see Refs. 13 and 17). Interestingly, crystal structures of Ca-ATPase prepared with different lipids also reveal three specific lipid binding sites in the pockets analogous to those proposed here for Na,K-ATPase (50). Another recent approach for detection of specific lipid binding to membrane proteins utilizes ion mobility mass spectrometry (51, 52). Indeed, we have recently applied this technique and obtained direct evidence for binding of one SOPS molecule to the α1β1FXYD1 complex (but no additional phospholipids such as might be carried over from the yeast).7 This approach could become invaluable for detailed characterization of specific lipid binding sites on the recombinant Na,K-ATPase inferred from the biochemical approach described here.
Site A, SOPS/Cholesterol
The biochemical evidence for specific binding of SOPS within the αTM8–10 and TMFXYD1 pocket has been described extensively (28, 29, 38). The characteristic of this interaction is that the SOPS stabilizes the protein against thermal or detergent inactivation but does not itself affect the Na,K-ATPase activity, and cholesterol strongly enhances stabilization by SOPS (28). Previously, it was proposed that SOPs and cholesterol interact directly (28), but their location in the protein was not known. As shown here, thermal stabilization of the α2VFP triple mutant described previously (29) is also strongly dependent on the presence of cholesterol together with SOPS, thus locating both cholesterol and SOPS in the same pocket. The fact that the α2VFP mutant is not stabilized in P. pastoris membranes, which lack cholesterol, implies that these residues in αTM8–10 are crucial for the binding of cholesterol. Thus, we propose that the αTM8–10 pocket harbors a cholesterol molecule together with the SOPS molecule, and the two interact to stabilize the protein.
The recent high resolution E1·AlF4−·ADP·3Na+ structure provides some structural insight (17). Fig. 12A shows the whole molecule with three bound cholesterol molecules (CHL1, -2, and -3). The experiment in Fig. 8A allows us to discriminate between them. Fig. 12B shows the immediate vicinity of the cholesterol bound in the TM8–10 pocket (CHL3). This cholesterol interacts mainly with aromatic residues in TM8, -9, and -10 and the FXYD protein, which is typical for a cholesterol binding pocket (53, 54). In addition, two of the residues altered in the α2VFP mutant interact directly or indirectly with the bound CHL3. Pro-978 in TM10 is within van der Waals distance of the cholesterol headgroup, and Val-917 in TM8 interacts indirectly with the cholesterol by a close contact with Trp-981, which forms the direct contact with cholesterol. The third residue altered in α2VFP, Phe-952, does not contact the cholesterol but interacts with the FXYD protein (via Val-40 and Ala-37), which interacts directly with the cholesterol (via Phe-33, Gly-30, and Val-26). We have also shown previously that the FXYD protein interacts with the bound SOPS to further strongly stabilize the protein. In a previous paper (27), based on previously unassigned electron density in the shark rectal gland Na,K-ATPase crystal structure (Protein Data Bank code 2ZXE), we modeled a PS molecule bound near the cytoplasmic surface lysines Lys-252 and Lys-250, in contact with the FXYD protein. One can propose that lipid site A consists of a SOPS molecule bound at this cytoplasmic surface interacting via the acyl chains with the CHL3 bound near the extracellular surface. This particular hypothesis needs to be tested independently, by mutagenesis of Lys-252 and Lys-250, especially because a lipid molecule in this position was seen in the shark rectal gland structure (Protein Data Bank code 2ZXE) but not the pig kidney (Protein Data Bank code 3WGU) structure. In any event, the accumulated evidence supports the notion that there is a stability “hotspot” that includes the bound SOPS, cholesterol in Fig. 12, and the FXYD protein all interacting with each other and the α subunit. It was hypothesized previously that these interactions of the FXYD protein with SOPS and cholesterol and αTM8–10 at the cytoplasmic surface, together with αβ subunit interactions at the extracellular surface (L7-8-β), maintain the topological stability of the TM8-M10, which can be everted upon thermal destabilization (55, 56). An important role of this “hotspot” may be to maintain the unique configuration of sodium site III, comprising residues from TM5, TM6, and TM8, which determines the selectivity and cooperativity of binding of the three sodium ions (17).
Site B, Polyunsaturated Neutral Phospholipids
The structural selectivity for the PE- or PC-dependent stimulation of Na,K-ATPase, shown here to be optimal for 18:0/20:4 and 18:0/22:6 PE, with 18:0/20:4 and 18:0/22:6 PC slightly less effective, provides strong evidence for a specific lipid-protein interaction. As inferred from the lipidomics analysis, 18:0/20:4, 18:0/22:6, or a similar polyunsaturated PE species (or perhaps PE plasmalogen) is the more likely physiologically relevant stimulatory phospholipid.
There are remarkable functional similarities between recombinant Na,K-ATPase purified with SOPS/cholesterol and polyunsaturated PE and purified native rabbit or porcine renal Na,K-ATPase that also make it likely that the pump-PE interaction is specific and corresponds to a physiological interaction. These include similar specific Na,K-ATPase activities (30–35 units min−1), EP levels (∼5 nmol/mg), kinetic properties (K0.5Na, K0.5K, and vanadate and ouabain inhibition; Tables 3 and 4), rates of conformational changes (Table 5), and stability in different conformations (8, 41, 47, 57). The similar effect of the PE on Na,K-ATPase activity of all three isoforms (Fig. 3) also suggests that this pump-PE interaction is a general rather than tissue-specific feature.
A structural model for polyunsaturated phospholipid binding to a crevice between transmembrane helices 2, 4, 6, and 9 was proposed recently (27). This model is in good agreement with the kinetic data described above. It is conceivable that the bound PE accelerates the opening of this pocket during the E1P-E2P transition, leading to the stabilization of the E2 conformation and an overall acceleration of the Post-Albers cycle. A similar mechanism has been derived from structural data of Ca-ATPase. In E2, the headgroup of a PE molecule was identified between TM2, -4, -6, and -9 (58, 59). In E1, this space is occupied by TM2, and the phospholipid is bound more superficially between TM2 and -9 (60). Accordingly, PE was proposed to stabilize the E2 conformation of SERCA (61).
Site C, Sphingomyelin/Cholesterol
The third lipid effect shows strong selectivity for saturated fatty acyl chains and choline headgroups (18:0/18:0 PC or 16:0 or 18:0 SM) causes inhibition of Na,K-ATPase activity. The structural selectivity for inhibition (PC > PE > PS and 22:0 < 20:0 ≈ 18:0 > 16:0 > 14:0, and 18:0/18:0 PC ≈ 18:0 SM or 16:0 SM) indicates a specific lipid-protein interaction, As inferred from the lipidomics analysis of the kidney membranes, the 18:0 SM or 16:0 SM is the more likely inhibitory lipid. Because the inhibition depends strongly on the presence of cholesterol (Fig. 7), unlike stimulation by polyunsaturated PE or PC, and it is well established that cholesterol forms strong interactions with saturated fatty acyl chains, especially of SM (53, 62), the SM/cholesterol is a likely inhibitory pair in native membranes.
The SM/cholesterol is presumed to occupy an independent site C from the activating unsaturated PE, as judged by the lack of competition (Fig. 6), different dependence on cholesterol (Fig. 7), and also different kinetic mechanism, but the location of SM/cholesterol is not known. Because the only lipids identified so far in crystal structures of Na,K-ATPase are PC and cholesterol molecules, possible clues as to the SM/cholesterol site focus on bound cholesterol molecules (see Fig. 12). One putative site (CHL1) is near the transmembrane helix of β and αTM7. Several crystal structures revealed the binding of cholesterol to this site (13, 17). Interestingly, the cholesterol is in close contact with a phospholipid, modeled as PC, showing that this pocket is large enough to accommodate a cholesterol/lipid heterodimer. It was proposed that the cholesterol molecule in this pocket stabilizes the bound ions (13), but it is unclear whether the cholesterol/lipid dimer affects kinetic parameters of Na,K-ATPase. Tyr-40 of the β-subunit is in stacking interaction with the aromatic ring of cholesterol (54), and Tyr-40 and Tyr-44 are important for the stabilization of the E2 conformation of H,K- and Na,K-ATPase (63), indicating that these residues can affect the conformational equilibrium. Neither of two other cholesterol molecules (CHL3 and CHL2) bound in the pocket between TMFXYD, αTM8–10, and βTM is likely to be involved in the inhibitory effects of SM/cholesterol because the C-terminal TM helices of the α-subunit undergo little if any movement during the reaction cycle (11). Because an inhibitory effect of SM/cholesterol could work by preventing otherwise mobile trans-membrane segments from moving, one might also speculate that the SM/cholesterol in site C is bound in the TM2, -4, -6, and -9 pocket. However, this would have to be in a way that does not overlap or compete with the activating PE in site B (e.g. at the extracellular surface as opposed to the activating lipid at the cytoplasmic surface). Obviously, either of the potential locations for the inhibitory site C mentioned here are speculative and require further investigation.
Physiological Significance
Assuming that the specific lipid-protein interactions observed here reflect interactions that can occur in vivo, one can propose that they reflect evolutionary adaptations of the pump to its immediate lipid environment, which provide optimal stability or activity. The following examples illustrate this idea.
Site A
An insight into the possible role of the stabilizing interactions of SOPS/cholesterol can be inferred from the data in supplemental Fig. 1, which shows sequence conservation over many species for the three residues Val-917, Phe-952, and Pro-978 that stabilize the α2VFP mutant. The striking observation is that these three residues are highly conserved in α1 and α3 of warm-blooded (homeothermic) animals, such as mammals and birds, whereas α2 almost exclusively contains ALV, which makes it unstable to thermal inactivation. In contrast, the same three residues are much less conserved in cold-blooded (poikilothermic) animals, such as fish, amphibians, reptiles, crustaceans, molluscs, annelids, hydrozoan, and insects. Thus, one can propose that the Na,K-ATPase of the poikilothermic animals is stable enough at the given ambient and body temperatures (in general much lower than in homeothermic animals) and did not need to evolve to a more stable form. In contrast, for mammals and birds, the “all purpose” α1 isoform evolved to be optimally stable at the raised body temperature of, ∼37 °C and thus reduced its cellular turnover. In contrast, α2, which has a more distinct regulatory physiological function in mammals, may turn over more quickly and can tolerate a lower stability than α1 even with bound cholesterol (see Ref. 29 for a discussion).
Site B
In a series of papers, it was shown that the molecular turnover rate for membrane-bound Na,K-ATPase of various birds and mammals, over a wide range of basal metabolic rates, is strongly positively correlated with the fraction of polyunsaturated lipids in the membranes, specifically in the PE fraction (64–66). This phenomenon was explained by assuming that an increased fraction of polyunsaturated acyl chains affects lipid packing densities, and the pump turnover rate increases in response to this change in physical state of the bilayer. However, enrichment of 18:1 PC or 18:1/cholesterol proteolipsomes with 16:0/22:6 PS or 22:6 PC was found to inhibit Na,K-ATPase activity (26). Thus, an increased molecular Na,K-ATPase turnover rate associated with increased fractions of polyunsaturated PE might be attributable to the specific stimulatory interaction described in this paper. In other words, the increased turnover rate associated with the specific PE interactions may explain specific adaptation to the requirement for faster Na,K-pumping in animals with a higher basal metabolic rate.
Site C
In principle, the inhibitory effect of SM/cholesterol, independent of the PE interaction, could imply the possibility of a pool of inactive or non-pumping Na,K-ATPase under physiological conditions. For example, it was proposed that 50% of Na,K-ATPase in LLC-PK1 cells is non-pumping and acts as non-canonical receptor for cardiotonic steroids triggering a protein kinase cascade (67). This non-pumping sodium pump was proposed to reside in caveolae, a membrane microdomain enriched in cholesterol and sphingomyelin (68). Such an inactive pool of Na,K-ATPase in the SM/cholesterol-rich caveolar environment could, in principle, be involved in cardiac glycoside signaling or as a temporally silenced reserve that can be activated if necessary (e.g. by transport along the cytoskeleton into an environment that contains a higher content of polyunsaturated phospholipids). On the other hand, this notion must be considered speculative because no significant differences in Na,K-ATPase activity were found in a study that compared kinetic properties of renal non-caveolar and caveolar Na,K-ATPase (69).
The almost 10-fold difference in enzyme activity of Na,K-ATPase purified with PE or SM raises the question of whether Na,K-ATPase subspecies with different activities coexist in a cell. Of possible relevance, Klodos and Post (70) proposed a functional heterogeneity model in which Na,K-ATPase molecules are randomly distributed in at least two different membrane phases in which the kinetic properties of these subgroups differ accordingly.
Conclusion
It is remarkable that three different phospholipid and cholesterol binding pockets are observed in the crystal structures of Na,K-ATPase (13, 15, 17, 18), and three separate functional effects have now been detected: stabilization by phosphatidylserine/cholesterol, stimulation by polyunsaturated phosphatidylethanolamine, and inhibition by sphingomyelin/cholesterol. Of course, the inference that these three effects are mediated within three separate lipid binding sites, A, B, and C, must now be tested in detail by mutating the putative phospholipid/cholesterol binding residues and by ion mobility mass spectrometry. Nevertheless, direct and specific phospholipid and cholesterol interactions that determine both stability and activity of the Na,K-ATPase seem to represent a new concept or paradigm for lipid-protein interactions of the pump.
Acknowledgment
We thank Prof. Amnon Horovitz (Weizmann Institute) for the use of a SX20 stopped-flow fluorimeter.

This article contains supplemental Fig. 1.
The apparent molecular mass of a soluble detergent-lipid-protein protomer includes the protein mass of α1β1FXYD1 (α, 112.3 kDa; β, ∼43.5 kDa (37); FXYD1, 8.6 kDa (38)) plus bound detergent (0.5–1 mg/mg of protein) (37) and annular lipid (∼30 molecules/per molecule (39) and is predicted to be ∼260–330 kDa. The apparent mass range in Fig. 1B is close to this estimate and is too low to represent a diprotomer.
S. Ayciriex, A. Katz, S. J. D. Karlish, and A. Shevchenko, unpublished results.
The concentration of PC or PE used in all of these preparations (0.1 mg/ml) was sufficient to give maximal effects.
M. Habeck, S. Karlish, and M. Sharon, unpublished results.
- TM
- transmembrane helix
- PS
- phosphatidylserine
- PC
- phosphatidylcholine
- PE
- phosphatidylethanolamine
- SM
- sphingomyelin
- SOPS
- 1-stearoyl-2-oleoyl-sn-glycero-3-phospho-l-serine
- C12E8
- octaethylene glycol monododecyl ether
- RH421
- N-(4-sulfobutyl)-4-(4-(p-(dipentylamino)phenyl)butadienyl)pyridinium inner salt
- PLM
- phospholemman
- ROV
- right-side-out membrane vesicle.
REFERENCES
- 1. Palmgren M. G., Nissen P. (2011) P-type ATPases. Annu. Rev. Biophys. 40, 243–266 [DOI] [PubMed] [Google Scholar]
- 2. Kühlbrandt W. (2004) Biology, structure and mechanism of P-type ATPases. Nat. Rev. Mol. Cell Biol. 5, 282–295 [DOI] [PubMed] [Google Scholar]
- 3. Jorgensen P. L., Hakansson K. O., Karlish S. J. (2003) Structure and mechanism of Na,K-ATPase: functional sites and their interactions. Annu. Rev. Physiol. 65, 817–849 [DOI] [PubMed] [Google Scholar]
- 4. Geering K. (2001) The functional role of β subunits in oligomeric P-type ATPases. J. Bioenerg. Biomembr. 33, 425–438 [DOI] [PubMed] [Google Scholar]
- 5. Geering K. (2008) Functional roles of Na,K-ATPase subunits. Curr. Opin. Nephrol. Hypertens. 17, 526–532 [DOI] [PubMed] [Google Scholar]
- 6. Garty H., Karlish S. J. (2006) Role of FXYD proteins in ion transport. Annu. Rev. Physiol. 68, 431–459 [DOI] [PubMed] [Google Scholar]
- 7. Post R. L., Hegyvary C., Kume S. (1972) Activation by adenosine triphosphate in the phosphorylation kinetics of sodium and potassium ion transport adenosine triphosphatase. J. Biol. Chem. 247, 6530–6540 [PubMed] [Google Scholar]
- 8. Karlish S. J., Yates D. W. (1978) Tryptophan fluorescence of (Na+ + K+)-ATPase as a tool for study of the enzyme mechanism. Biochim. Biophys. Acta 527, 115–130 [DOI] [PubMed] [Google Scholar]
- 9. Forbush B., 3rd, Klodos I. (1991) Rate-limiting steps in Na translocation by the Na/K pump. Soc. Gen. Physiol. Ser. 46, 210–225 [PubMed] [Google Scholar]
- 10. Apell H. J. (2004) How do P-type ATPases transport ions? Bioelectrochemistry 63, 149–156 [DOI] [PubMed] [Google Scholar]
- 11. Toyoshima C. (2009) How Ca2+-ATPase pumps ions across the sarcoplasmic reticulum membrane. Biochim. Biophys. Acta 1793, 941–946 [DOI] [PubMed] [Google Scholar]
- 12. Morth J. P., Pedersen B. P., Toustrup-Jensen M. S., Sørensen T. L., Petersen J., Andersen J. P., Vilsen B., Nissen P. (2007) Crystal structure of the sodium-potassium pump. Nature 450, 1043–1049 [DOI] [PubMed] [Google Scholar]
- 13. Shinoda T., Ogawa H., Cornelius F., Toyoshima C. (2009) Crystal structure of the sodium-potassium pump at 2.4 A resolution. Nature 459, 446–450 [DOI] [PubMed] [Google Scholar]
- 14. Ogawa H., Shinoda T., Cornelius F., Toyoshima C. (2009) Crystal structure of the sodium-potassium pump (Na+,K+-ATPase) with bound potassium and ouabain. Proc. Natl. Acad. Sci. U.S.A. 106, 13742–13747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Laursen M., Yatime L., Nissen P., Fedosova N. U. (2013) Crystal structure of the high-affinity Na+,K+-ATPase-ouabain complex with Mg2+ bound in the cation binding site. Proc. Natl. Acad. Sci. U.S.A. 110, 10958–10963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yatime L., Laursen M., Morth J. P., Esmann M., Nissen P., Fedosova N. U. (2011) Structural insights into the high affinity binding of cardiotonic steroids to the Na+,K+-ATPase. J. Struct. Biol. 174, 296–306 [DOI] [PubMed] [Google Scholar]
- 17. Kanai R., Ogawa H., Vilsen B., Cornelius F., Toyoshima C. (2013) Crystal structure of a Na+-bound Na+,K+-ATPase preceding the E1P state. Nature 502, 201–206 [DOI] [PubMed] [Google Scholar]
- 18. Nyblom M., Poulsen H., Gourdon P., Reinhard L., Andersson M., Lindahl E., Fedosova N., Nissen P. (2013) Crystal structure of Na+,K+-ATPase in the Na+-bound state. Science 342, 123–127 [DOI] [PubMed] [Google Scholar]
- 19. Cornelius F. (1995) Cholesterol modulation of molecular activity of reconstituted shark Na+,K+-ATPase. Biochim. Biophys. Acta 1235, 205–212 [DOI] [PubMed] [Google Scholar]
- 20. Wheeler K. P., Whittam R. (1970) The involvement of phosphatidylserine in adenosine triphosphatase activity of the sodium pump. J. Physiol. 207, 303–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lee A. G. (2004) How lipids affect the activities of integral membrane proteins. Biochim. Biophys. Acta 1666, 62–87 [DOI] [PubMed] [Google Scholar]
- 22. Cornelius F. (2001) Modulation of Na,K-ATPase and Na-ATPase activity by phospholipids and cholesterol. I. Steady-state kinetics. Biochemistry 40, 8842–8851 [DOI] [PubMed] [Google Scholar]
- 23. Lee A. G. (2011) Lipid-protein interactions. Biochem. Soc. Trans. 39, 761–766 [DOI] [PubMed] [Google Scholar]
- 24. Starling A. P., East J. M., Lee A. G. (1995) Effects of phospholipid fatty acyl chain length on phosphorylation and dephosphorylation of the Ca2+-ATPase. Biochem. J. 310, 875–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sonntag Y., Musgaard M., Olesen C., Schiøtt B., Møller J. V., Nissen P., Thøgersen L. (2011) Mutual adaptation of a membrane protein and its lipid bilayer during conformational changes. Nat. Commun. 2, 304. [DOI] [PubMed] [Google Scholar]
- 26. Cornelius F. (2008) Cholesterol-dependent interaction of polyunsaturated phospholipids with Na,K-ATPase. Biochemistry 47, 1652–1658 [DOI] [PubMed] [Google Scholar]
- 27. Haviv H., Habeck M., Kanai R., Toyoshima C., Karlish S. J. (2013) Neutral phospholipids stimulate Na,K-ATPase activity: a specific lipid-protein interaction. J. Biol. Chem. 288, 10073–10081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Haviv H., Cohen E., Lifshitz Y., Tal D. M., Goldshleger R., Karlish S. J. (2007) Stabilization of Na+,K+-ATPase purified from Pichia pastoris membranes by specific interactions with lipids. Biochemistry 46, 12855–12867 [DOI] [PubMed] [Google Scholar]
- 29. Kapri-Pardes E., Katz A., Haviv H., Mahmmoud Y., Ilan M., Khalfin-Penigel I., Carmeli S., Yarden O., Karlish S. J. (2011) Stabilization of the α2 isoform of Na,K-ATPase by mutations in a phospholipid binding pocket. J. Biol. Chem. 286, 42888–42899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Belogus T., Haviv H., Karlish S. J. (2009) Neutralization of the charge on Asp-369 of Na+,K+-ATPase triggers E1 ↔ E2 conformational changes. J. Biol. Chem. 284, 31038–31051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wittig I, Braun HP., Schägger H. (2006) Blue native PAGE. Nat. Protoc. 1, 418–428 [DOI] [PubMed] [Google Scholar]
- 32. Habeck M., Cirri E., Katz A., Karlish S. J., Apell H. J. (2009) Investigation of electrogenic partial reactions in detergent-solubilized Na,K-ATPase. Biochemistry 48, 9147–9155 [DOI] [PubMed] [Google Scholar]
- 33. Jørgensen P. L. (1988) Purification of Na+,K+-ATPase: enzyme sources, preparative problems, and preparation from mammalian kidney. Methods Enzymol. 156, 29–43 [DOI] [PubMed] [Google Scholar]
- 34. Carvalho M., Sampaio J. L., Palm W., Brankatschk M., Eaton S., Shevchenko A. (2012) Effects of diet and development on the Drosophila lipidome. Mol. Syst. Biol. 8, 600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Liebisch G., Binder M., Schifferer R., Langmann T., Schulz B., Schmitz G. (2006) High throughput quantification of cholesterol and cholesteryl ester by electrospray ionization tandem mass spectrometry (ESI-MS/MS). Biochim. Biophys. Acta 1761, 121–128 [DOI] [PubMed] [Google Scholar]
- 36. Herzog R., Schwudke D., Schuhmann K., Sampaio J. L., Bornstein S. R., Schroeder M., Shevchenko A. (2011) A novel informatics concept for high-throughput shotgun lipidomics based on the molecular fragmentation query language. Genome Biol. 12, R8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cohen E., Goldshleger R., Shainskaya A., Tal D. M., Ebel C., le Maire M., Karlish S. J. (2005) Purification of Na+,K+-ATPase expressed in Pichia pastoris reveals an essential role of phospholipid-protein interactions. J. Biol. Chem. 280, 16610–16618 [DOI] [PubMed] [Google Scholar]
- 38. Mishra N. K., Peleg Y., Cirri E., Belogus T., Lifshitz Y., Voelker D. R., Apell H. J., Garty H., Karlish S. J. (2011) FXYD proteins stabilize Na,K-ATPase: amplification of specific phosphatidylserine-protein interactions. J. Biol. Chem. 286, 9699–9712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Esmann M., Marsh D. (2006) Lipid-protein interactions with the Na,K-ATPase. Chem. Phys. Lipids 141, 94–104 [DOI] [PubMed] [Google Scholar]
- 40. Blanco G., Mercer R. W. (1998) Isozymes of the Na-K-ATPase: heterogeneity in structure, diversity in function. Am. J. Physiol. 275, F633–F650 [DOI] [PubMed] [Google Scholar]
- 41. Jorgensen P. L. (1974) Purification and characterization of (Na+ plus K+)-ATPase. 3. Purification from the outer medulla of mammalian kidney after selective removal of membrane components by sodium dodecylsulphate. Biochim. Biophys. Acta 356, 36–52 [DOI] [PubMed] [Google Scholar]
- 42. Jørgensen P. L. (1988) Overview: structural basis for coupling of E1-E2 transitions in α β-units of renal Na,K-ATPase to Na,K-translocation. Prog. Clin. Biol. Res. 268A, 19–38 [PubMed] [Google Scholar]
- 43. Jørgensen P. L., Andersen J. P. (1986) Thermoinactivation and aggregation of α β units in soluble and membrane-bound (Na,K)-ATPase. Biochemistry 25, 2889–2897 [DOI] [PubMed] [Google Scholar]
- 44. Karlish S. J. (1980) Characterization of conformational changes in (Na,K) ATPase labeled with fluorescein at the active site. J. Bioenerg. Biomembr. 12, 111–136 [DOI] [PubMed] [Google Scholar]
- 45. Bühler R., Stürmer W., Apell H. J., Läuger P. (1991) Charge translocation by the Na,K-pump. I. Kinetics of local field changes studied by time-resolved fluorescence measurements. J. Membr. Biol. 121, 141–161 [DOI] [PubMed] [Google Scholar]
- 46. Cirri E., Katz A., Mishra N. K., Belogus T., Lifshitz Y., Garty H., Karlish S. J., Apell H. J. (2011) Phospholemman (FXYD1) raises the affinity of the human α1β1 isoform of Na,K-ATPase for Na ions. Biochemistry 50, 3736–3748 [DOI] [PubMed] [Google Scholar]
- 47. Lupfert C., Grell E., Pintschovius V., Apell H. J., Cornelius F., Clarke R. J. (2001) Rate limitation of the Na+,K+-ATPase pump cycle. Biophys. J. 81, 2069–2081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yabuuchi H., O'Brien J. S. (1968) Positional distribution of fatty acids in glycerophosphatides of bovine gray matter. J. Lipid Res. 9, 65–67 [PubMed] [Google Scholar]
- 49. Nealon J. R., Blanksby S. J., Mitchell T. W., Else P. L. (2008) Systematic differences in membrane acyl composition associated with varying body mass in mammals occur in all phospholipid classes: an analysis of kidney and brain. J. Exp. Biol. 211, 3195–3204 [DOI] [PubMed] [Google Scholar]
- 50. Drachmann N. D., Olesen C., Møller J. V., Guo Z., Nissen P., Bublitz M. (2014) Comparing crystal structures of Ca2+-ATPase in the presence of different lipids. FEBS J. 281, 4249–4262 [DOI] [PubMed] [Google Scholar]
- 51. Barrera N. P., Zhou M., Robinson C. V. (2013) The role of lipids in defining membrane protein interactions: insights from mass spectrometry. Trends Cell Biol. 23, 1–8 [DOI] [PubMed] [Google Scholar]
- 52. Laganowsky A., Reading E., Allison T. M., Ulmschneider M. B., Degiacomi M. T., Baldwin A. J., Robinson C. V. (2014) Membrane proteins bind lipids selectively to modulate their structure and function. Nature 510, 172–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Fantini J., Barrantes F. J. (2013) How cholesterol interacts with membrane proteins: an exploration of cholesterol-binding sites including CRAC, CARC, and tilted domains. Front. Physiol. 4, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Adamian L., Naveed H., Liang J. (2011) Lipid-binding surfaces of membrane proteins: evidence from evolutionary and structural analysis. Biochim. Biophys. Acta 1808, 1092–1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Donnet C., Arystarkhova E., Sweadner K. J. (2001) Thermal denaturation of the Na,K-ATPase provides evidence for α-α oligomeric interaction and γ subunit association with the C-terminal domain. J. Biol. Chem. 276, 7357–7365 [DOI] [PubMed] [Google Scholar]
- 56. Goldshleger R., Tal D. M., Karlish S. J. (1995) Topology of the α-subunit of Na,K-ATPase based on proteolysis: lability of the topological organization. Biochemistry 34, 8668–8679 [DOI] [PubMed] [Google Scholar]
- 57. Jørgensen P. L., Andersen J. P. (1988) Structural basis for E1-E2 conformational transitions in Na,K-pump and Ca-pump proteins. J. Membr. Biol. 103, 95–120 [DOI] [PubMed] [Google Scholar]
- 58. Toyoshima C., Nomura H., Tsuda T. (2004) Lumenal gating mechanism revealed in calcium pump crystal structures with phosphate analogues. Nature 432, 361–368 [DOI] [PubMed] [Google Scholar]
- 59. Winther A. M., Liu H., Sonntag Y., Olesen C., le Maire M., Soehoel H., Olsen C. E., Christensen S. B., Nissen P., Møller J. V. (2010) Critical roles of hydrophobicity and orientation of side chains for inactivation of sarcoplasmic reticulum Ca2+-ATPase with thapsigargin and thapsigargin analogs. J. Biol. Chem. 285, 28883–28892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Toyoshima C., Yonekura S., Tsueda J., Iwasawa S. (2011) Trinitrophenyl derivatives bind differently from parent adenine nucleotides to Ca2+-ATPase in the absence of Ca2+. Proc. Natl. Acad. Sci. U.S.A. 108, 1833–1838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Obara K., Miyashita N., Xu C., Toyoshima I., Sugita Y., Inesi G., Toyoshima C. (2005) Structural role of countertransport revealed in Ca2+ pump crystal structure in the absence of Ca2+. Proc. Natl. Acad. Sci. U.S.A. 102, 14489–14496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Song Y., Kenworthy A. K., Sanders C. R. (2014) Cholesterol as a co-solvent and a ligand for membrane proteins. Protein Sci. 23, 1–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Dürr K. L., Tavraz N. N., Dempski R. E., Bamberg E., Friedrich T. (2009) Functional significance of E2 state stabilization by specific α/β-subunit interactions of Na,K- and H,K-ATPase. J. Biol. Chem. 284, 3842–3854 [DOI] [PubMed] [Google Scholar]
- 64. Else P. L., Wu B. J., Storlien L. H., Hulbert A. J. (2003) Molecular activity of Na+,K+-ATPase relates to the packing of membrane lipids. Ann. N.Y. Acad. Sci. 986, 525–526 [DOI] [PubMed] [Google Scholar]
- 65. Turner N., Haga K. L., Else P. L., Hulbert A. J. (2006) Scaling of Na+,K+-ATPase molecular activity and membrane fatty acid composition in mammalian and avian hearts. Physiol. Biochem. Zool. 79, 522–533 [DOI] [PubMed] [Google Scholar]
- 66. Turner N., Hulbert A. J., Else P. L. (2003) Molecular activity of sodium pumps in the kidney of mammals and birds. Ann. N.Y. Acad. Sci. 986, 606–607 [DOI] [PubMed] [Google Scholar]
- 67. Liang M., Tian J., Liu L., Pierre S., Liu J., Shapiro J., Xie Z. J. (2007) Identification of a pool of non-pumping Na/K-ATPase. J. Biol. Chem. 282, 10585–10593 [DOI] [PubMed] [Google Scholar]
- 68. Anderson R. G. (1998) The caveolae membrane system. Annu. Rev. Biochem. 67, 199–225 [DOI] [PubMed] [Google Scholar]
- 69. Liu L., Ivanov A. V., Gable M. E., Jolivel F., Morrill G. A., Askari A. (2011) Comparative properties of caveolar and noncaveolar preparations of kidney Na+/K+-ATPase. Biochemistry 50, 8664–8673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Post R. L., Klodos I. (1996) Interpretation of extraordinary kinetics of Na+-K+-ATPase by a phase change. Am. J. Physiol. 271, C1415–C1423 [DOI] [PubMed] [Google Scholar]