Background: Retbindin is a retina-specific protein with unknown function.
Results: Retbindin is expressed and secreted by rods into the extracellular environment and is capable of binding riboflavin.
Conclusion: Retbindin may be involved in retinal acquisition of flavins.
Significance: This work assigns a function to retbindin, a rod-specific protein that may be essential for the high metabolic rate of the retina.
Keywords: Extracellular Matrix, Flavoprotein, Photoreceptor, Retina, Riboflavin, Retbindin
Abstract
Retbindin is a novel retina-specific protein of unknown function. In this study, we have used various approaches to evaluate protein expression, localization, biochemical properties, and function. We find that retbindin is secreted by the rod photoreceptors into the inter-photoreceptor matrix where it is maintained via electrostatic forces. Retbindin is predominantly localized at the interface between photoreceptors and retinal pigment epithelium microvilli, a region critical for retinal function and homeostasis. Interestingly, although it is associated with photoreceptor outer segments, retbindin's expression is not dependent on their presence. In vitro, retbindin is capable of binding riboflavin, thus implicating the protein as a metabolite carrier between the retina and the retinal pigment epithelium. Altogether, our data show that retbindin is a novel photoreceptor-specific protein with a unique localization and function. We hypothesize that retbindin is an excellent candidate for binding retinal flavins and possibly participating in their transport from the extracellular space to the photoreceptors. Further investigations are warranted to determine the exact function of retbindin in retinal homeostasis and disease.
Introduction
Although the retina is composed of multiple cell types, photoreceptors are the light-responsive cells that initiate phototransduction. Phototransduction begins in a modified cilium known as the outer segment (OS),2 which is present on both rod and cone photoreceptors. Because of the complex and energy-intensive nature of light perception, photoreceptors display an elevated energy metabolism unlike any other cell type (1–3). This is evident by the high consumption of glucose and oxygen in the retina (4, 5). Photoreceptors acquire their nutrients and oxygen from the retinal pigment epithelium (RPE), which transports nutrients from the choroidal blood supply into the retina (7).
A dynamic interface exists between the photoreceptor OSs and the RPE known as the inter-photoreceptor matrix (IPM) (8). This matrix is responsible for metabolite acquisition and exchange between the photoreceptors and RPE (8–10). Among the many nutrients transported through the IPM, two important metabolites are the riboflavin cofactor derivatives, flavin adenine dinucleotide (FAD) and flavin adenine mononucleotide (FMN). FAD and FMN are functionally equivalent as redox co-enzymes and are highly concentrated in the retina relative to blood (∼50 pmol/mg total protein versus ∼2.5 in the retina and blood, respectively) (11, 12). How the retina concentrates such high levels of flavins is currently unknown, but the high level of metabolism in the retina would predict that large concentrations of these nutrients would be required there. FAD is needed for the citric acid cycle and fatty acid oxidation (13–15), which are important for photoreceptor metabolism and function. Flavins are usually tightly bound to flavoproteins, and unbound flavins are known to undergo photoreduction to produce free radicals that can cause lipid peroxidation (16–18), to which the photoreceptor OSs are particularly sensitive (19). Thus, flavoproteins are a critical part of maintaining homeostasis in the retina. Given the high concentration of flavins in the retina, it makes sense that the tissue would contain processes for transporting flavins from the RPE to the IPM and then to the photoreceptors, but such systems are not yet described.
An expressed sequence tag analysis of an un-amplified human retinal cDNA library identified a novel, highly expressed transcript whose translated peptide shares 27% sequence homology to the riboflavin-binding protein (RBP), a member of the folate receptor superfamily. RBP is specific to chicken oviduct and is known to be involved in the deposition of riboflavin in the chicken egg (20) but with no known mammalian homologs. The newly identified gene, now known as retbindin (Retb), is located on human chromosome 19 (21).
Regulation of flavins in the metabolically active retina is critical, but little is known about retinal flavoproteins. The homology between RBP and Retb suggested that Retb could be a retinal flavoprotein, so here we elected to further investigate the biochemical properties of Retb. We provide evidence that Retb is a mammalian- and neural retina-specific gene with a single transcript and a predicted protein tertiary structure similar to that of RBP. Biochemical analysis revealed that Retb is a secreted peripheral membrane protein and a component of the retinal IPM. In the mouse retina, Retb localized specifically in the photoreceptor cell layer, with predominant localization near the tip of the rod outer segments. Furthermore, heterologously expressed Retb is capable of binding riboflavin, suggesting it may play a key role in the regulation of retinal flavins.
EXPERIMENTAL PROCEDURES
In Silico Analysis
Retb and RBP sequences were obtained from the National Center for Biotechnology Information database (21). All known sequences were aligned using Clustal Omega software (22). STRAP software was used to generate a phylogenic tree based on species and sequence divergence (23). The Phyre2 server was used to generate predicted tertiary structures based on protein sequence (24).
Animals
All experiments involving mice were approved by the local Institutional Animal Care and Use Committee and adhered to the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health and the Association for Research in Vision and Ophthalmology Resolution on the Use of Animals in Research. All mice were negative for the rd8 allele. C57BL/6J mice were purchased from the Jackson Laboratory (Bar Harbor, ME). P30 wild-type and Rd/Rd mice were euthanized using CO2 asphyxiation, and then the neural retina and PECS were harvested. For light-induced protein translocation, mice were placed in a light box (7,000 lux) for 1 h, and dark-adapted mice were kept in a dark box for 4 h.
Bovine and Human Ocular Tissues
Enucleated bovine eyes were obtained from Country Home Meat Co. (Edmond, OK). Eyes were dissected, and the neural retina and RPE were harvested separately and frozen in liquid nitrogen. A 56-year-old Caucasian phakic donor eye and a 61-year-old Caucasian normal donor eye were obtained from the Illinois Eye Bank, Chicago, IL. A 4-mm trephine was used to separate the macula from the peripheral neural retina; also the RPE behind the macula was separated from the RPE in the periphery. Another normal 72-year-old Caucasian male eye was dissected, and the neural retina and the RPE were separated. Samples were frozen in liquid nitrogen and stored at −80 °C until used.
Antibody Characterization
The anti-Retb antibody was produced by 21st Century Biologicals (Marlboro, MA) against a peptide corresponding to mouse Retb amino acids 115–131. The antiserum was tested at various concentrations against Retb protein recombinantly expressed in vitro (Escherichia coli and COS-7). Full-length Retb cDNA was inserted into a pTriEx-4 vector backbone (EMD Millipore, Billerica, MA). This construct was transformed into DE3 pLacI-competent cells (EMD Millipore). Cultures were grown overnight at 30 °C. Cultures were sonicated on ice five times for 30 s each. Following the manufacturer's protocols, the protein was isolated using nickel-nitrilotriacetic acid beads (Thermo, Waltham, MA). Retb was eluted from the beads using 200 mm imidazole. Samples were analyzed via immunoblots as described under “Preparation of Total Retinal/RPE Extracts and Immunoblotting.” COS-7 cells were transfected with the full-length Retb cDNA in a pK vector backbone. ∼1 million cells were transfected with 0.6 μg of DNA via Lipofectamine 2000 according to the manufacturer's protocols (Invitrogen). These cells were analyzed as outlined under “Preparation of Total Retinal/RPE Extracts and Immunoblotting.” A corresponding peptide (amino acids 115–131) was used in a competition assay to demonstrate the Retb specificity of the antibody. In this experiment, anti-Retb antibody was incubated overnight with 50 μg of the peptide used for immunization. This antibody/peptide solution was diluted in either 1× TBST, 1% Tween 20 (details below) or immunofluorescence (IF) blocking solution (details below) and used during the primary antibody incubation step at a Retb antibody final concentration of 1:500. Peptide competition removed all antibody labeling, and no band was observed in mock-transfected cells indicating the specificity of the antibody to retbindin.
Immunofluorescence
Eyes were processed, sectioned, and stained as described previously (25). Briefly, whole eyes were placed in Davidson's fixative (32% ethanol, 11% acetic acid, 2% formaldehyde) overnight, washed in 1× PBS, and then stored in 70% ethanol. Eyes were dehydrated, embedded in paraffin, and sectioned at 10 μm. Sections were blocked in 2.5% donkey serum, 0.5% Triton X-100, and 1% fish gelatin in 1× PBS (pH 7.2) (blocking solution) for 30 min at room temperature. Anti-Retb antibody (1:500, described above) was then applied to the slides for 30 min at room temperature in blocking solution. The following primary antibodies were applied in conjunction with Retb: rhodopsin 1D4 (1:1,000, a generous gift from Dr. Robert Molday); S-opsin (1:500, Santa Cruz Biotechnology, Santa Cruz, CA); retinal degeneration slow (Rds-CT, 1:1,000 (26)); ezrin (1:50, Abcam, Cambridge, MA); and arrestin (1:500, Santa Cruz Biotechnology, sc-166383). Fluorescently labeled secondary antibodies (1:500, Invitrogen) were applied for 1.5 h at room temperature. Fluorescently tagged wheat germ agglutinin (WGA) and peanut agglutinin (PNA) (1:500, Invitrogen) were applied with the secondary antibodies. Slides were mounted with ProLong Gold antifade with DAPI (Invitrogen), and images were captured using a Hamamatsu C-4742 camera on an Olympus BX62 upright spinning disk confocal microscope equipped with UplanSapo objectives (Olympus, Tokyo, Japan). Images were analyzed using Slidebook version 4 and Adobe Photoshop software licensed to the University of Oklahoma Health Sciences Center.
Relative Quantitative Real Time-PCR
Relative quantitative real time-PCR was performed as described previously (27). Briefly, RNA was extracted from the various tissues shown in Fig. 2 using TRIzol (Invitrogen). cDNA was generated by first strand synthesis using an oligo(dT) primer and Superscript III reverse transcriptase (Invitrogen). Quantitative real time-PCR was performed in triplicate on each sample using a C1000 Thermal Cycler (Bio-Rad). Three separate amplicons were used for retbindin corresponding to the full-length sequence of exon 1a, exon 1b, and exon 6. The primer pairs are shown in Table 1. Relative expression of Retb amplicons was normalized to hypoxanthine-guanine phosphoribosyltransferase as a housekeeping gene and plotted. Three independent quantitative real time-PCR experiments were performed on cDNA samples generated from three separate animals. Melt curve analysis and semi-quantitative RT-PCR were used to verify proper amplicon generation and to ensure the absence of primer dimers. Slope of the amplification curve was used to measure the efficiency of the reaction at 104%, which falls within our predefined limits set at 110 and 90%.
FIGURE 2.
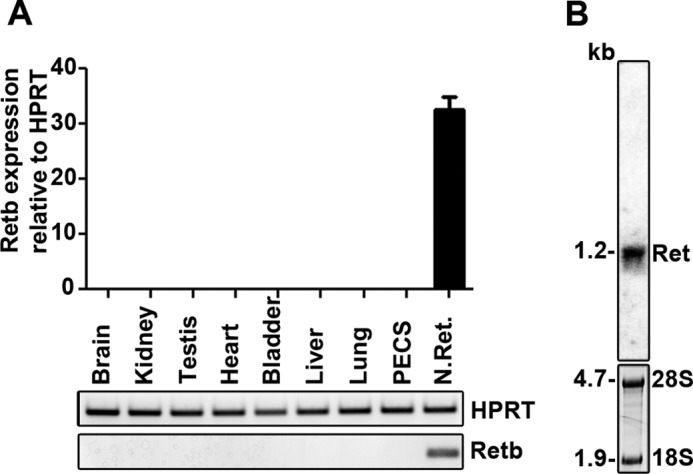
Retb is specific to the neural retina. A, Retb quantitative RT-PCR results on cDNA prepared from total RNA isolated from P30 mouse brain, kidney, testis, heart, bladder, liver, lung, PECS (RPE, choroid, and sclera), and neural retina. Sequences of the primers are listed in Table 1. Results are plotted relative to Hprt and show retina-specific expression of the Retb gene. B, Northern blot of total retinal RNA isolated from P30 wild-type mice and probed with radiolabeled Retb full-length cDNA. 28 S and 18 S ribosomal RNAs are marked for size reference and loading control. Shown is a single Retb transcript of ∼1.2 kb in size.
TABLE 1.
Retb primer sequences for quantitative RT-PCR
| Exon 1a | |
|---|---|
| Forward | 5′-TACACTCGAGCTTGTAGATATAAATGCACCAGTCCC-3′ |
| Reverse | 5′-TACATCTAGACACTGCCACTCACCTGTTAGCT-3′ |
| Exon 1b | |
| Forward | 5′-TACACCTCGAGTGAGAACCGAGAGGTGACCG-3′ |
| Reverse | 3′-TACATCTAGACCACGGATTAAAACACCGATCC-3′ |
| Exon 6 | |
| Forward | 5′-TACAGCCCACTAGGGCCTTAACTC-3′ |
| Reverse | 5′-TACAGTACCGCGGAGATGGAGAT-3′ |
Preparation of Total Retinal/RPE Extracts and Immunoblotting
Where not specified elsewhere, tissue extracts for immunoblotting were prepared from mouse, bovine, and human tissues by homogenization with a handheld motor and pestle tip (VWR, Radnor, PA) in a 1× PBS (pH 7.2) solution containing 1% Triton X-100 and complete protease inhibitor mixture (Roche Applied Science) and incubated on a nutator for 1 h at 4 °C. The insoluble material was pelleted using centrifugation at 4,000 × g, and the supernatants were incubated in reducing Laemmli buffer for 1 h at room temperature. The samples were then separated on a 10% SDS-polyacrylamide gel and transferred to PVDF membranes. Samples under nonreducing conditions had N-ethylmaleimide added to a final concentration of 200 mm. Membranes were blocked for 30 min at room temperature in 5% milk, 1× TBST, 1% Tween 20. Anti-Retb antibody (1:500) in 1× TBST, 1% Tween 20 was added to the membrane and then incubated with shaking for 30 min at room temperature. Blots were then washed three times for 10 min each in 1× TBST, 1% Tween 20. Goat-anti rabbit conjugated to horseradish peroxidase (1:25,000) (KPL, Gaithersburg, MD) was added to the membrane and incubated with shaking for 30 min at room temperature. The blot was washed for 10 min in 1× TBST, 1% Tween 20 three times. The following primary antibodies were used under the same conditions after the membrane was allowed to dry overnight: rhodopsin 1D4 (1:1,000); RPE65 (1:500, Abcam ab-105366); Rds-CT (1:1,000 (26)); GAPDH (1:1,000, Genetex, Irvine, CA GT329); IRBP (1:10,000 (28)); and anti-FLAG (1:1000) (Cell Signaling, Boston, MA). Blots were probed with β-actin-HRP antibody (1:100,000, Sigma) as a loading control.
PNGase Treatment
Intact P30 mouse neural retinas were used to prepare retinal protein lysates as described above. Following the manufacturer's protocols, retinal protein lysates were incubated either with or without PNGase F (New England Biolabs, Ipswich, MA) under reducing conditions overnight. The following morning, samples were analyzed via immunoblotting as described above.
Separation of Soluble IPM from Insoluble IPM and Retinal Cells
Intact P30 mouse neural retinas were incubated on ice in 1× PBS (pH 7.2) with 1× protease inhibitors for 30 min without agitation. The insoluble IPM and retinal cells were then separated from the soluble interphotoreceptor matrix by centrifugation at 750 × g for 10 min. The supernatant was considered to contain soluble components of the IPM, although the pellet, which was resuspended in 1× PBS, was considered insoluble IPM and retinal cells. The samples were analyzed via immunoblotting as described above.
Preparation of Retinal Membranes
P30 mouse neural retinas or bovine/human retinas were homogenized with a handheld motor and pestle tip in an isotonic 100 mm Tris buffer (pH 7) containing 1× protease inhibitors. The soluble and membranous portions were separated by centrifugation at 50,000 × g for 30 min in a Sorvall Discovery M150 ultracentrifuge (Thermo Scientific) equipped with a fixed angle rotor (Sorvall no. S55S-1009). The pellet (membrane fraction) contains retinal membranes, organelles, cytoskeletal components, and insoluble extracellular matrix. The supernatant represents soluble extracellular matrix and cytosolic components. The pellet was resuspended in the same isotonic Tris buffer and analyzed via immunoblotting as described above.
Release of Peripheral Membrane Proteins with Na2CO3
The membrane pellet from the above section was resuspended in a 100 mm Tris buffer (pH 10) containing 100 mm Na2CO3 and 1× protease inhibitors. The pellet was incubated for 30 min at 4 °C. The Na2CO3-soluble and -insoluble portions were separated by centrifugation at 50,000 × g for 30 min. The pellet (Na2CO3 insoluble) was resuspended in the 100 mm Na2CO3 buffer and analyzed via immunoblotting as described above.
Release of Peripheral Membrane Proteins from Intact Neural Retinas with NaCl
Intact P30 mouse neural retinas were incubated at 4 °C in 1× PBS (pH 7.2) containing 1 m NaCl and 1× protease inhibitors for 30 min without agitation. The insoluble IPM and retinal cells were then separated from the NaCl-soluble extracellular matrix by centrifugation at 750 × g for 10 min. The pellet was resuspended in the same buffer, and the samples were analyzed via immunoblotting as described above.
In Vitro Binding Assay
COS-7 cells were grown to 90% confluency in 6-well plates in DMEM made in-house without riboflavin. Cells were transfected with Retb, rbp, Enox2 (ecto-NOX disulfide-thiol exchanger 2), or backbone only vectors using Lipofectamine 2000 (Invitrogen). All constructs included a pK backbone with a CMV promoter and N-terminal FLAG tag. After 48 h of growth, the media were exchanged with DMEM containing 0.075 mm riboflavin and the cells were incubated at 37 °C for 1 h. After incubation, the cells were washed four times with 1× PBS, then scraped and collected in 1× PBS. The cells were homogenized with a handheld motor and pestle tip and centrifuged at 30,000 × g for 15 min to separate the membranous fraction (contains membranes, organelles, cytoskeletal components, and insoluble extracellular matrix). The pellet was resuspended in 100 mm Tris buffer (pH 10) containing 100 mm Na2CO3 and 1× protease inhibitors to release peripheral membrane proteins (Retb and RBP), and the absorbance was measured at 340 nm. The resuspended pellet was then incubated at 4 °C for 30 min and centrifuged again at 30,000 × g for 15 min to separate the Na2CO3-insoluble fraction from the soluble. The pellet was resuspended in 100 mm Na2CO3 buffer, and the absorbance was measured again and immunoblotted as described above. Relative absorbance was measured as 340 nm absorbance divided by the ratio of FLAG to GAPDH signal. We chose 340 nm wavelength of light as riboflavin absorbs light strongly at 340 nm (16), and we chose to use a concentration of 0.075 mm because it is within the linear range of a standard curve for riboflavin concentration (data not shown). The experiment was repeated five independent times, and samples in each iteration were run in triplicate. Two-way analysis of variance followed by Bonferroni pairwise comparative statistical analysis was performed using GraphPad Prism 5 (GraphPad Software, La Jolla, CA).
Immunocytochemistry
Coverslips were placed on the bottom of 6-well plates and cells were grown and transfected as described in “In Vitro Binding Assay.” After the media were removed and cells were washed with PBS, a 4% paraformaldehyde solution was applied to the cells on the coverslips for 15 min at room temperature to fix the cells. After fixation, the experiments were carried out as described above with the primary anti-FLAG antibody (1:1,000) (Cell Signaling) overnight at 4 °C. Secondary antibody and DiI membrane marker (1:500, Invitrogen) were applied at room temperature for 1.5 h prior to mounting and imaging as described in “Immunofluorescence.”
RESULTS
Retb Exhibits Similarities to Riboflavin-binding Protein
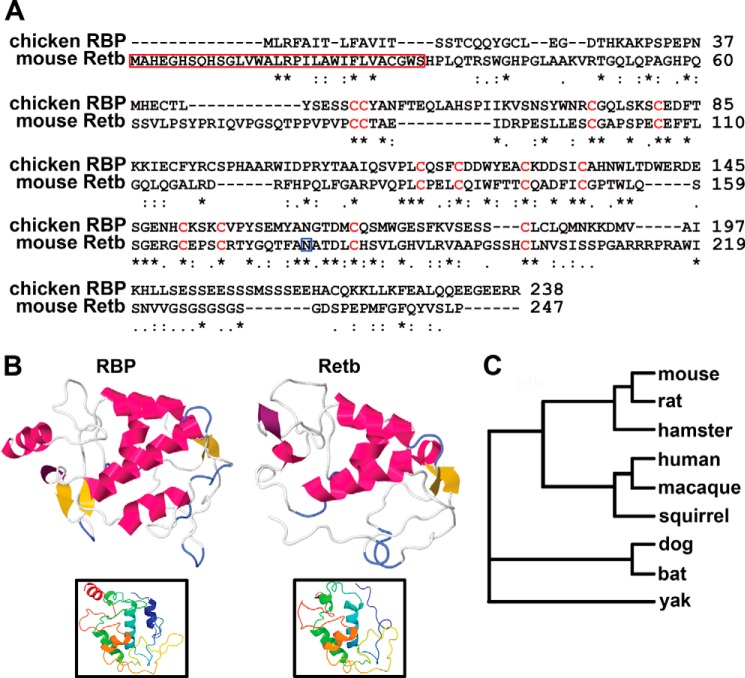
According to NCBI, the mouse Retb coding sequence contains 744 nucleotides (21). This is predicted to be translated into a 247-amino acid protein with a molecular mass of 26.6 kDa. The amino acid sequence can be accessed at the NCBI Protein Database, accession number _659178.1 (21). Amino acids 1–31 constitute a potential signal peptide (predicted to be cleaved) (Fig. 1A, red box), and there is a predicted N-linked glycosylation site at residue 178 (Fig. 1A, blue box) (21). Retb has no transmembrane domains, amphipathic loops, or covalently linked membrane anchoring domains (29). BLAST analysis reveals only one similar sequence, the aforementioned RBP from chicken oviduct (at 27% sequence identity over 135 residues) (20). Clustal Omega alignment of these two peptide sequences shows many conserved and similar residues between the two proteins (Fig. 1A). Of particular importance are 12 cysteine residues that are necessary for riboflavin binding to RBP (30, 31). These cysteines are conserved in Retb (Fig. 1A, red) and are also present in other members of the folate receptor superfamily (21). Phyre2 tertiary structure prediction software (24) shows striking similarities between RBP and Retb, particularly in the riboflavin binding fold (Fig. 1B). This structure exists at comparable positions within the two peptide sequences (Fig. 1B, insets). The Retb gene is only present in mammals, and an expanded analysis of Clustal Omega alignments for some potential Retb orthologs showed that across species a core of 11 cysteines and the potential signal sequence are conserved. The phylogenic tree in Fig. 1C illustrates the evolutionary divergence of Retb protein sequences. The maintenance of the gene through multiple speciation events supports assignment of an important biological function to Retb. These in silico data suggest that Retb may function similarly to RBP. To determine whether these predictions are accurate, we next characterized the gene and protein in mice.
FIGURE 1.
Retb has significant sequence homology to riboflavin-binding protein. A, Clustal Omega alignment of chicken RBP and mouse Retb showing conserved residues indicated by (*), strongly similar by (:), and weakly similar by (.). Conserved cysteines are shown in red. Red box indicates potential cleaved signal sequence, and blue box points to potential N-linked glycosylation site. B, Phyre2 server tertiary structure prediction of RBP and Retb showing helices in pink, β-sheets in yellow, and turns in purple. Insets represent the N-to-C tertiary structure as a rainbow (blue to red). C, STRAP phylogenic tree of the Retb sequence is displayed showing similarity across mammalian species.
Retb Is Expressed Exclusively in the Photoreceptor Cell Layer of the Neural Retina
To determine the tissue distribution of Retb, reverse-transcribed total RNA was analyzed by quantitative RT-PCR utilizing an amplicon corresponding to exon 6. The presence of this amplicon exclusively in the neural retina (Fig. 2A) confirms the retina-specific expression of Retb in mice (20). This was further confirmed with two sets of primers corresponding to exons 1a and 1b (data not shown). Northern blot analysis using a full-length Retb cDNA probe and total RNA isolated from mouse neural retinas revealed a single Retb transcript ∼1.2 kb in size (Fig. 2B), consistent with the predicted message size.
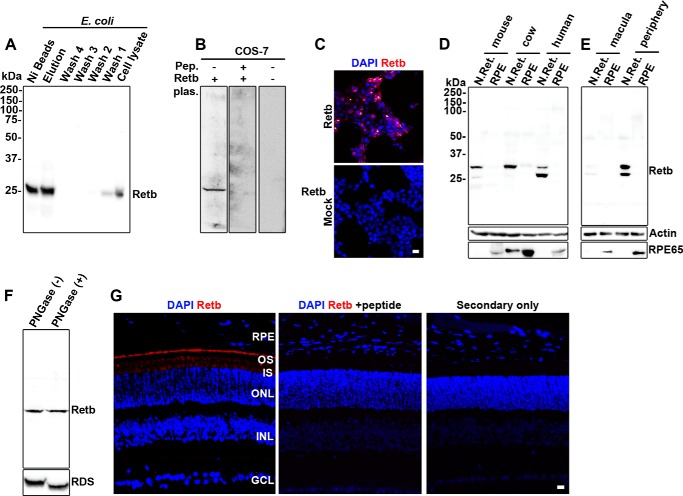
To analyze Retb protein, we generated a polyclonal anti-peptide antibody against Retb amino acids 115–131. On immunoblots, this antibody detected a single band at the predicted size when either recombinant His-tagged Retb was purified from E. coli (Fig. 3A, first two lanes) or when total E. coli extracts were run on the gel (Fig. 3A, last lane). Importantly, this result was recapitulated in mammalian cells; the antibody recognized a single, correctly sized band on immunoblots of lysates from COS-7 cells transfected with a Retb plasmid but not on blots that were incubated with a peptide corresponding to Retb (115–131) or on blots from mock-transfected cells (Fig. 3B). The antibody was also effective in immunofluorescence (Fig. 3C), and as a result of these positive characterizations, it is used throughout.
FIGURE 3.
Retb protein is found in the periphery of the neural retina but not the macula or RPE. A–C, specificity of Retb polyclonal antibody was assessed against recombinantly expressed Retb. A, immunoblots of His-tagged full-length Retb eluted from nickel-nitrilotriacetic acid beads using imidazole exhibited a single band of predicted size. B and C, COS-7 cells were transiently transfected with Retb plasmid (or Mock). B, immunoblot showing that Retb antibody signal was completely competed out using 50 μg of the peptide (Pep.) used to generate the antibody. Plasm, plasmid. C, immunocytochemistry showing Retb labeling (red) in COS-7 cells transfected with Retb at ×10, although mock-transfected cells showed no signal (bottom panel) indicating specificity of the antibody to the expressed protein. D, immunoblot of mouse, bovine, and human neural retina (N.Ret.) and RPE (PECS for mice and RPE for human and bovine) protein extracts probed with anti-Retb antibody. A single band of ∼30 kDa is shown in all samples, and a lower band at varying intensities is shown in human and mouse retinal extracts. E, immunoblot of human macula and peripheral neural retina and the corresponding RPE blotted with the anti-Retb antibody. Retb is preferentially expressed in the peripheral human retina than in the macula. F, immunoblot of PNGase F-treated and -untreated P30 neural retinal extracts probed with anti-Retb or anti-Rds antibodies. G, representative IF using the anti-Retb antibody in red (left panel), anti-Retb antibody competed out by preincubation with peptide (middle panel), and secondary antibody only as control (right panel). Nuclei were counterstained with DAPI (blue). OS, outer segment; IS, inner segment; ONL, outer nuclear layer; INL, inner nuclear layer; GCL, ganglion cell layer. Scale bars, 10 μm.
Immunoblot analysis of retinal lysates using this anti-Retb antibody showed one or two bands at ∼30 kDa (Fig. 3D). A single band was observed in bovine neural retinal extracts, although two bands with different intensities were observed in human neural retinal extracts and with inverse intensities in mouse neural retinal extracts (Fig. 3, D and E). Consistent with the previous finding (Fig. 2, A and B), we found Retb protein highly enriched in neural retina. Interestingly, Retb was highly expressed in extracts prepared from the peripheral human retina, with very little detectable Retb in the human macula (Fig. 3E). The lower band is the size predicted from virtual translation of the Retb transcript. To determine whether the larger protein observed in the mouse neural retina results from N-linked glycosylation (as predicted from sequence analysis), we performed PNGase F treatment on mouse neural retinal extracts and probed subsequent immunoblots for Retb and Rds (a known N-glycosylated protein). As expected, PNGase F treatment shifted the Rds band toward lower molecular mass, but no shift in size was observed for Retb (Fig. 3F). This result suggests that the higher molecular weight Retb band is either the result of alternative splicing or yet to be determined post-translational modifications (20, 32).
IF labeling revealed that Retb labeling is present in the OS and inner segment layer with marked localization at the interface between the OS/RPE (Fig. 3G, left panel). To confirm the specificity of the signal localization, we fully competed out the antibody via co-incubation with the peptide that was used to generate the antibody (Fig. 3G, middle panel). No signal was detected in secondary only control experiments (Fig. 3G, right panel). Although the IF data in Fig. 3G do not clearly distinguish between photoreceptor OS and RPE, our biochemical data (Fig. 3, D and E) show that the RPE does not express Retb.
Retb Is Located at the OS/RPE Interface
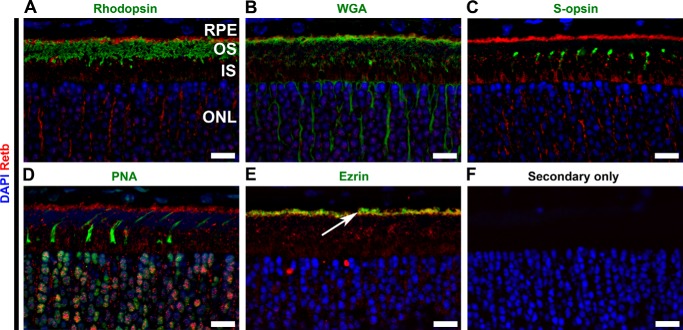
To further study the subcellular localization of Retb, we co-labeled retinal sections with Retb (Fig. 4, red) and the following markers in green: rhodopsin (rod OS, Fig. 4A), WGA (rod extracellular matrix, Fig. 4B), S-opsin (cone OS, Fig. 4C), PNA (cone extracellular matrix, Fig. 4D), and ezrin (RPE microvilli, Fig. 4E). Although Retb is present in the inner segment layer, it is primarily concentrated at the tips of rod OSs as evident from its apical location relative to rhodopsin (Fig. 4A) and WGA (Fig. 4B). We also observe nonuniform distribution of Retb around the inner segments, likely attributed to the fact that these images are single planes from a deconvolved image stack and that Retb may be localized in individual domains/clusters within the extracellular space. No co-localization (Fig. 4A) is observed between Retb and rhodopsin, suggesting Retb is not likely to be a disc component. In contrast, the co-localization between Retb and WGA (Fig. 4B) suggests Retb could be part of the extracellular matrix surrounding rods, an hypothesis consistent with the predicted secretory signal found in the Retb sequence. Retb was not observed close to S-opsin (Fig. 4C) or PNA (Fig. 4D), suggesting that it is not present in/around cone photoreceptors nor is it likely to be a component of the cone extracellular matrix, in agreement with the absence of Retb in human macula (Fig. 3E). Interestingly, Retb exhibited co-localization with ezrin (Fig. 4E, white arrow), with the majority of Retb residing basal to ezrin (i.e. toward the photoreceptors) at the RPE microvilli (Fig. 4E). Again, no signal was detected in secondary only control (Fig. 4F) clearly demonstrating the specificity of Retb staining.
FIGURE 4.
Retb is localized at the interface between the OSs and RPE microvilli. Representative single confocal images of P30 retinal cross-sections were immunolabeled for Retb (red) and different markers (green) for photoreceptor, RPE, and IPM. Nuclei are counterstained with DAPI. A, IF images co-labeled with anti-Retb (red) and anti-rhodopsin (green). Retb is primarily concentrated at the tips of the rod OSs and is apical to rhodopsin. B, IF images co-labeled with Retb (red) and WGA (green). Retb co-localized with WGA. C, IF images co-labeled with Retb (red) and S-opsin (green). No co-localization was seen between Retb and S-opsin. D, IF images co-labeled with Retb (red) and PNA (green). No co-localization was seen between Retb and PNA. E, IF images co-labeled with Retb (red) and ezrin (green). Retb co-localized with ezrin and located basal to the RPE microvilli (indicated by an arrow). F, secondary antibody alone as a control. Nuclei are counterstained with DAPI (blue). Scale bars, 10 μm. The distribution of Retb around the inner segments seems nonuniform, which may be partly due to the fact that those images represent single planes from a deconvolved image stack and that Retb may be localized in individual domains/clusters within the extracellular space. OS, outer segment; IS, inner segment; ONL, outer nuclear layer.
Co-labeling for Retb and arrestin on sections from dark-adapted mouse eyes showed that Retb in the inner segment layer does not co-localize with arrestin (Fig. 5A), again suggestive of an extracellular localization. Furthermore, Retb (unlike arrestin) does not translocate in a light-dependent manner. This localization shows that Retb is concentrated in the area of the outer retina corresponding to the OS/RPE interface but is not in the RPE cell bodies (consistent with lack of biochemical detection in RPE extracts). Combined, these data suggest that Retb is likely in the IPM.
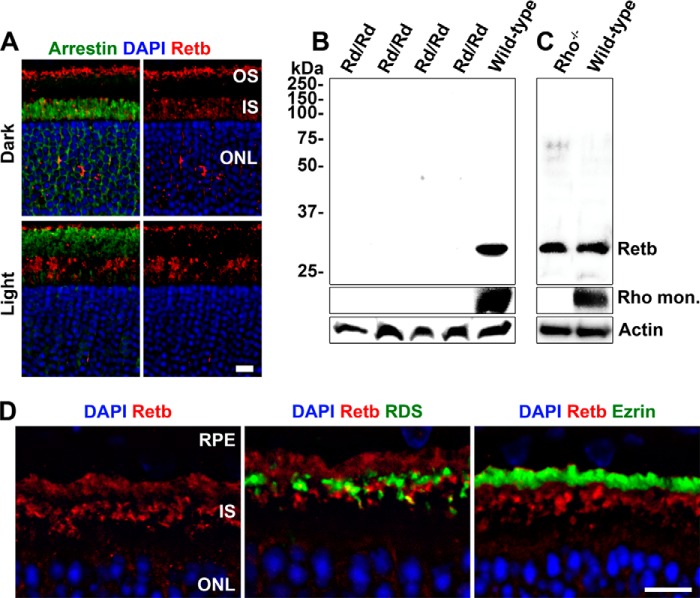
FIGURE 5.
Retb is produced by photoreceptors but not dependent on the presence of OSs. A, IF images of WT retinal sections co-labeled with Retb (red) and arrestin (green) (left panels) or Retb alone (right panels) in retinas taken from dark-adapted (top panels) and light-adapted (bottom panels) mice. B, immunoblot detection of Retb protein in P30 wild-type and Rd/Rd neural retinal extracts. No Retb was detected in any of the four different Rd/Rd retinal extracts. C, immunoblot of extracts from wild-type and Rho−/− neural retinas at P21, probed with anti-Retb antibody. Retb is detected in neural retinal extracts from both Rho−/− and wild-type animals. D, IF of Retb (red) in P21 Rho−/− retina (left panel), for Rds (middle panel) or ezrin (right panel) in green, and nuclei counterstained with DAPI. Scale bars, 10 μm. OS, outer segment; IS, inner segment; ONL, outer nuclear layer.
IF analysis strongly suggests that Retb is a rod photoreceptor-specific protein localized at the interface between the rod OSs and RPE microvilli. To further confirm this finding, we used degenerative models to determine whether Retb is actually produced by rod photoreceptors. We began with Rd/Rd neural retinas (also known as rd1) in which a naturally occurring mutation in the Pde6b gene leads to rapid photoreceptor degeneration. At P30, the Rd/Rd retina is devoid of rods, although some cones remain intact (33). Fig. 5B shows that Retb is not present in the Rd/Rd P30 neural retinas, indicating that it is produced exclusively by rod photoreceptors.
Because Retb is localized to the OS/RPE interface, we next asked whether its expression and/or localization were dependent on the presence of rod OSs. We utilized P21 rhodopsin knock-out (Rho−/−) neural retinas. At this time, the rod photoreceptor cells are present, but no elaborated OSs are formed (34). Instead, these cells produce tiny OS-like structures at the apical end of the connecting cilia. They have no discs, but Rds and other OS proteins are present (34, 35). Consistent with the presence of rod cells in Rho−/− retinas, Retb protein is detected in retinal extracts from this model (Fig. 5C). Retb filled the subretinal space (Fig. 5D, left panel) around the OS-like nubs (labeled with Rds in green, Fig. 5D, middle panel). Interestingly, in this model Retb does not co-localize with the RPE microvilli (Fig. 5D, right panel shows co-labeling with ezrin in green). This finding suggests proper OSs are necessary for the restricted localization of Retb to the tip of OS/RPE interface in the wild-type retina.
Retb Is a Member of the IPM
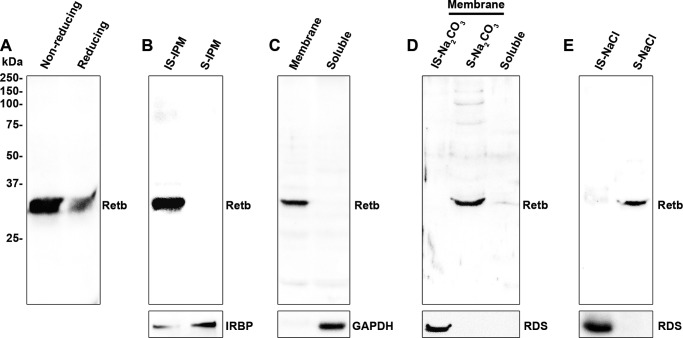
Because of the presence of 12 cysteines in Retb, we first asked whether Retb multimerizes via covalent disulfide linkages to another protein(s). Mouse retinal extracts were prepared under nonreducing or reducing conditions, and subsequent immunoblots were probed for Retb. No size difference was detected between the two samples (Fig. 6A), suggesting that Retb does not form intermolecular disulfide linkages.
FIGURE 6.
Retb is a secreted extracellular peripheral membrane protein. Protein extracts were obtained from P30 wild-type neural retinas under various conditions and probed for Retb or other proteins as indicated in each panel as controls. A, immunoblot of retinal extracts showing Retb under either reducing or nonreducing conditions. B, immunoblot of PBS S-IPM and IS-IPM of wild-type neural retinal extracts showing exclusive localization of Retb in the IS-IPM. IRBP was used as a positive control for the fractionation. C, immunoblot of membrane fraction (also containing insoluble matrix and cellular matter/organelles) versus soluble fraction (soluble IPM components and cytoplasmic components) probed with Retb or the control Gapdh. D, immunoblot of membrane fraction (prepared as in C) treated with 100 mm Na2CO3 followed by centrifugation. Shown are immunoblots of the Na2CO3-insoluble membrane fraction (IS-Na2CO3), the fraction that was Na2CO3-soluble (S-Na2CO3), and the soluble fraction from the initial separation (Soluble) probed for Retb or Rds (control). E, immunoblot of IS-IPM preparation from intact neural retinal extract treated with 1 m NaCl followed by centrifugation to separate insoluble (IS-NaCl) and soluble (S-NaCl) fractions and probed with anti-Retb or Rds (control).
Given our IF data and sequence predictions, we undertook a series of experiments to assess the possibility of Retb being an extracellular protein. We began by separating the soluble IPM (S-IPM) from the insoluble IPM (IS-IPM) fraction by incubating intact mouse neural retinas in 1× PBS on ice for 30 min without agitation. The S-IPM was removed from the IS-IPM by centrifugation at 750 × g for 10 min. The known S-IPM member, Irbp, was used as a positive control for the fractionation and was found highly enriched in the S-IPM fraction. In contrast, Retb was found exclusively in the IS-IPM fraction, suggesting Retb is not a soluble component of the IPM (Fig. 6B).
Next, membrane fractions (containing retinal membranes, organelles, cytoskeletal components, and insoluble IPM) were prepared by homogenizing mouse neural retinas in isotonic buffer followed by centrifugation at 50,000 × g to separate the soluble fraction. Retb was found exclusively in the membranous fraction, although the soluble protein Gapdh (positive control) was exclusively in the soluble fraction (Fig. 6C).
The in silico analysis showed that Retb has no transmembrane domains, no amphipathic loops, and no covalently linked membrane anchoring moieties (21), suggesting it could be a soluble protein. However, our biochemical analyses indicated that Retb was associated with the insoluble/membranous fractions. Thus, we hypothesized that Retb is maintained as a peripheral membrane protein. To test this, we utilized a procedure known to strip peripheral membrane proteins from their associated membranes: incubation in buffer containing Na2CO3 (36). To accomplish this, we resuspended the membranous fraction (isolated as in Fig. 6C) in 100 mm Na2CO3 buffer to detach peripheral membrane proteins. Retb localized exclusively to the Na2CO3-soluble fraction, although the known transmembrane protein Rds remained in the Na2CO3-insoluble fraction (Fig. 6D), supporting the notion that Retb is a peripheral membrane protein.
To determine whether this peripheral membrane attachment is intra- or extracellular, we made an IS-IPM preparation from intact mouse neural retinas and treated it with 1 m NaCl to detach electrostatically linked peripheral membrane proteins (36). Retb localized exclusively to the NaCl-soluble fraction, although the transmembrane protein Rds remained in the NaCl-insoluble fraction (Fig. 6E), suggesting that Retb is localized extracellularly. These biochemical analyses taken together with the IF results in Fig. 4 confirm that Retb is an extracellular peripheral membrane protein maintained in the insoluble IPM by electrostatic forces. Our data place Retb in a unique extracellular environment at the interface of the OS and RPE microvilli where many important photoreceptor processes, such as OS shedding and nutrient/oxygen exchange, take place (9, 37).
Retb Binds Riboflavin in Vitro
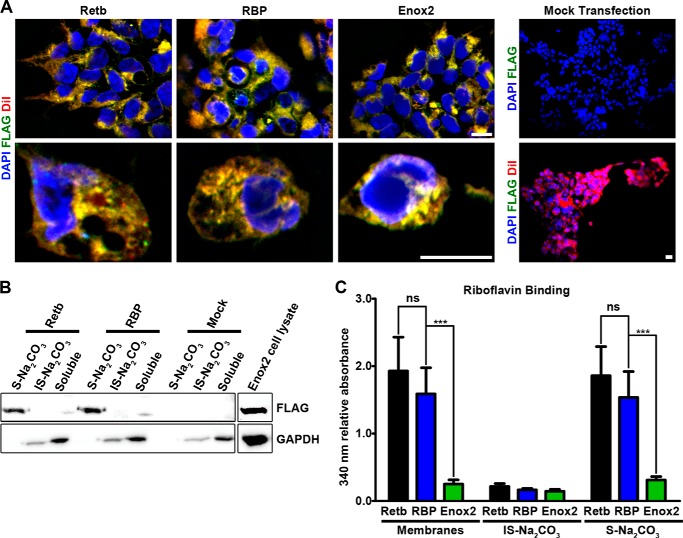
Retb homology to RBP coupled with its localization at the OS/RPE interface make it uniquely positioned to play a role in the transfer of flavins from the RPE to the retina. To determine whether Retb has the ability to bind flavins, we undertook a series of in vitro experiments. COS-7 cells were transfected with vectors containing FLAG-tagged (N-terminal) Retb, rbp, or Enox2. The positive control (rbp) was chosen because it has been shown to bind riboflavin, and Enox2 was chosen as a negative control because it is a cell surface protein that binds a similar ligand (NADH) but not riboflavin (38, 39). To confirm that all three genes were expressed in their respected transfected cells, we performed immunocytochemistry with anti-FLAG antibody and the cell membrane marker DiI (3) and confirmed that the gene products were expressed at the cell membrane (Fig. 7A). To verify that Retb and RBP were positioned at the membrane, we detached the peripheral membrane proteins using 100 mm Na2CO3 buffer as in Fig. 6. Because Enox2 is a transmembrane protein, it is not stripped by Na2CO3, but we did confirm it was expressed in the appropriate cells (Fig. 7B, right panels).
FIGURE 7.
Retb binds riboflavin in vitro. A, immunocytochemistry of Retb-, rbp-, and Enox2-transfected cells at ×60 (upper panels), ×100 (lower panels), secondary only (mock-transfected cells) at ×20 (upper panel), and Dil only at ×20 (lower panel). B, immunoblots of soluble, S-Na2CO3 and IS-Na2CO3 protein extracts from COS-7 cells transfected with FLAG-tagged Retb-, rbp-, Enox2-, or mock-transfected COS-7 cells. Upper blots probed with FLAG antibody, and lower panels probed with Gapdh antibody. C, absorbance of membranes isolated from COS-7 cells incubated in media containing 0.075 mm riboflavin as well as from subsequent separated IS-Na2CO3 and S-Na2CO3 fractions. Absorbance at 340 nm was divided by the ratio of FLAG to Gapdh signal assessed from immunoblots. Results demonstrate that Retb binds riboflavin at levels comparable with RBP. Scale bar, 10 μm.
After 24 h growing in riboflavin-free DMEM, the transfected cells were incubated for 30 min in 1 ml of DMEM supplemented with 0.075 mm riboflavin, washed four times, scraped, and collected in 1× PBS. Following homogenization, the membranous fraction (containing membranes, organelles, cytoskeletal components, and insoluble ECM) was pelleted (as in Fig. 6B), resuspended, and the absorbance measured at 340 nm (the wavelength to measure riboflavin absorbance) (16). Relative riboflavin binding was determined by subtracting the absorbance of mock-transfected cell membranes from the absorbance of transfected cells and then dividing by the ratio of FLAG to Gapdh signal (analyzed via immunoblots). We determined that normalizing the absorbance to the FLAG/Gapdh ratio would control for the amount of transfected protein present in each well. As shown in Fig. 7C, membranes from cells expressing Retb bound riboflavin at levels comparable to that of membranes from RBP-expressing cells (per protein concentration), although negligible amounts were bound by the negative control Enox2. To prove that the binding was due to the presence of Retb or RBP, membrane homogenates were resuspended in 100 mm Na2CO3 buffer to detach Retb and RBP from the membranes (Fig. 7B). This treatment abolished riboflavin binding (Fig. 7C). The binding data and the localization of Retb together with its biochemical characteristics support the notion that Retb is likely to be involved in the process of photoreceptor flavin acquisition and/or utilization.
DISCUSSION
The photoreceptor is a highly organized and specialized neuronal cell type characterized by the expression of a number of cell-specific proteins. These proteins are essential for proper photoreceptor structure, function, and maintenance. Aberrations in these proteins result in an array of retinal degenerative disorders such as retinitis pigmentosa, age-related macular degeneration, or Leber's congenital amaurosis. It was previously reported that Retb is retina-specific (20). In this study, we verified that Retb is specific to the neural retina, and we present new data showing that Retb is a rod-specific protein and is capable of binding riboflavin efficiently as RBP. Our results also showed that Retb is a secreted protein that is localized to the tips of the rod OSs directly adjacent to the RPE microvilli. The placement of Retb at this OS/RPE interface suggests that it may be involved in the processes that occur within the IPM such as OS phagocytosis by the RPE, visual pigment regeneration, metabolite exchange, and neural retina attachment to the RPE. Well controlled regulation of these processes is essential for proper photoreceptor maintenance and phototransduction.
Because of the homology to RBP (Fig. 1) (20), we chose to study the possibility that Retb binds to riboflavin. We confirmed that retbindin is capable of binding riboflavin in an in vitro expression system. The riboflavin binding capabilities and Retb's localization make it an ideal candidate for the in vivo photoreceptor flavin acquisition from the RPE at the apical tips of the OS. The flavin cofactors FAD and FMN play major roles in cells due to their direct involvement in fatty acid oxidation and citric acid cycle. These processes are very important to photoreceptors as they have a high concentration of polyunsaturated fatty acids and high energy consumption (40).
Flavins' role in the retina is emphasized by their high concentration (48 ± 1.7 pmol/mg−1), which is ∼20-fold higher than in the blood (2.57 ± 0.31 pmol/mg−1) (11). Said et al. (41) showed that cultured human RPE cells (ARPE-19) uptake riboflavin via a Ca2+-calmodulin-regulated process. It is not known how the retina in general and specifically the photoreceptor cells acquire flavins. Flavins are key cofactors involved in fatty acid oxidation and citric acid cycle (42). However, these processes alone may not account for the high levels of flavins found in the retina. Despite their high levels in the retina, their concentration must be tightly regulated because riboflavin deficiency (ariboflavinosis) results in photosensitivity and poor dim light vision (43, 44), although excess dietary riboflavin causes photoreceptor cell death via OS lipid peroxidation (45).
Besides their role in energy metabolism, flavins likely play other yet uncharacterized roles in retinal function and/or homeostasis. It is known that flavins are cofactors in many isomerization reactions (42). For instance, FAD is the cofactor used by xanthine oxidase in the conversion of retinol to retinoic acid (46–48). Interestingly, this flavoprotein is localized to cone OSs in the human photoreceptor layer (49). FAD is also present in the blue light cryptochromes, which help mediate circadian rhythm in the inner neural retina (50–52). Despite the absence of Retb from cones, the diversity of these two processes is a good example of how widely flavoproteins are used in the retina.
Whatever the function of flavoproteins in the photoreceptors may be, it is well known that unbound flavins are reduced by light and cause lipid peroxidation (17, 45), to which the OSs are particularly sensitive (19, 53). For instance, lipid peroxidation and its by-products are present in degenerative retinas of patients of age-related macular degeneration (54, 55). The localization and binding data presented in this study point to Retb as a key component of this process. Potentially, Retb binds flavins to protect OSs from light damage, although it or another protein(s) (yet to be discovered) may provide a sufficient amount to the flavoproteins needed for photoreceptor metabolism and function. This may also explain the presence of some Retb around the inner segments but not within them.
In conclusion, we have identified a new IPM-specific protein capable of binding riboflavin in vitro. Our findings on riboflavin binding will need to be verified in vivo to better understand the role of Retb and flavins in the neural retina. Given the localization to the OS/RPE interface, Retb's role should also be investigated in OS phagocytosis by the RPE, visual pigment regeneration, metabolite exchange, and neural retina attachment. Examining Retb in various disease models at various times could reveal more information about IPM processes in which this protein may be involved. Future studies will pinpoint what exact role(s) Retb is playing in the retina. Separately, the specificity of the Retb protein for rod photoreceptors provides an exciting new locus for use in genetic and gene therapy studies. The findings presented here warrant further investigation of this novel and exciting protein. Studies on the function of this protein in normal and diseased states will yield exciting new information about how changes in the IPM can affect photoreceptor function and the treatment of retinal diseases.
Acknowledgments
We thank Dr. Shannon Conley for scientific input and careful reading of the manuscript and Dr. Jody Summers-Rada for providing chicken oviduct RNA.
This work was supported, in whole or in part, by National Institutes of Health Grants R01EY10609 (to M. I. N.), R01EY018137 (to M. R. A.), and P30EY021725 from NEI. This work was also supported by the Foundation Fighting Blindness (to M. I. N. and M. R. A.).
- OS
- outer segment
- RPE
- retinal pigment epithelium
- IPM
- inter-photoreceptor matrix
- WGA
- wheat germ agglutinin
- PNA
- peanut agglutinin
- PNGase F
- peptide:N-glycosidase F
- IF
- immunofluorescence
- S-IPM
- soluble IPM
- IS-IPM
- insoluble IPM
- DiI
- 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate
- PECS
- pigment epithelia/choroid/sclera
- IRBP
- interphotoreceptor retinoid-binding protein.
REFERENCES
- 1. Alder V. A., Ben-Nun J., Cringle S. J. (1990) PO2 profiles and oxygen consumption in cat retina with an occluded retinal circulation. Invest. Ophthalmol. Vis. Sci. 31, 1029–1034 [PubMed] [Google Scholar]
- 2. Braun R. D., Linsenmeier R. A., Goldstick T. K. (1995) Oxygen consumption in the inner and outer retina of the cat. Invest. Ophthalmol. Vis. Sci. 36, 542–554 [PubMed] [Google Scholar]
- 3. Njie-Mbye Y. F., Kulkarni-Chitnis M., Opere C. A., Barrett A., Ohia S. E. (2013) Lipid peroxidation: pathophysiological and pharmacological implications in the eye. Front. Physiol. 4, 366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Winkler B. S. (1981) Glycolytic and oxidative metabolism in relation to retinal function. J. Gen. Physiol. 77, 667–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Futterman S., Kinoshita J. H. (1959) Metabolism of the retina. I. Respiration of cattle retina. J. Biol. Chem. 234, 723–726 [PubMed] [Google Scholar]
- 6. Deleted in proof.
- 7. Sparrow J. R., Hicks D., Hamel C. P. (2010) The retinal pigment epithelium in health and disease. Curr. Mol. Med. 10, 802–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Röhlich P. (1970) The interphotoreceptor matrix: electron microscopic and histochemical observations on the vertebrate retina. Exp. Eye Res. 10, 80–86 [DOI] [PubMed] [Google Scholar]
- 9. Feeney L. (1973) The interphotoreceptor space. II. Histochemistry of the matrix. Dev. Biol. 32, 115–128 [DOI] [PubMed] [Google Scholar]
- 10. Fong S. L., Liou G. I., Landers R. A., Alvarez R. A., Bridges C. D. (1984) Purification and characterization of a retinol-binding glycoprotein synthesized and secreted by bovine neural retina. J. Biol. Chem. 259, 6534–6542 [PubMed] [Google Scholar]
- 11. Batey D. W., Daneshgar K. K., Eckhert C. D. (1992) Flavin levels in the rat retina. Exp. Eye Res. 54, 605–609 [DOI] [PubMed] [Google Scholar]
- 12. Batey D. W., Eckhert C. D. (1991) Analysis of flavins in ocular tissues of the rabbit. Invest. Ophthalmol. Vis. Sci. 32, 1981–1985 [PubMed] [Google Scholar]
- 13. Tomitsuka E., Hirawake H., Goto Y., Taniwaki M., Harada S., Kita K. (2003) Direct evidence for two distinct forms of the flavoprotein subunit of human mitochondrial complex II (succinate-ubiquinone reductase). J. Biochem. 134, 191–195 [DOI] [PubMed] [Google Scholar]
- 14. Crane F. L., Beinert H. (1956) On the mechanism of dehydrogenation of fatty acyl derivatives of coenzyme A. II. The electron-transferring flavoprotein. J. Biol. Chem. 218, 717–731 [PubMed] [Google Scholar]
- 15. Crane F. L., Mii S., Hauge J. G., Green D. E., Beinert H. (1956) On the mechanism of dehydrogenation of fatty acyl derivatives of coenzyme A. I. The general fatty acyl coenzyme A dehydrogenase. J. Biol. Chem. 218, 701–706 [PubMed] [Google Scholar]
- 16. Oster G., Bellin J. S., Holmstrom B. (1962) Photochemistry of riboflavin. Experientia 18, 249–253 [DOI] [PubMed] [Google Scholar]
- 17. Treadwell G. E., Cairns W. L., Metzler D. E. (1968) Photochemical degradation of flavins. V. Chromatographic studies of the products of photolysis of riboflavin. J. Chromatogr. 35, 376–388 [DOI] [PubMed] [Google Scholar]
- 18. Huvaere K., Cardoso D. R., Homem-de-Mello P., Westermann S., Skibsted L. H. (2010) Light-induced oxidation of unsaturated lipids as sensitized by flavins. J. Phys. Chem. B 114, 5583–5593 [DOI] [PubMed] [Google Scholar]
- 19. Wiegand R. D., Giusto N. M., Rapp L. M., Anderson R. E. (1983) Evidence for rod outer segment lipid peroxidation following constant illumination of the rat retina. Invest. Ophthalmol. Vis. Sci. 24, 1433–1435 [PubMed] [Google Scholar]
- 20. Wistow G., Bernstein S. L., Wyatt M. K., Ray S., Behal A., Touchman J. W., Bouffard G., Smith D., Peterson K. (2002) Expressed sequence tag analysis of human retina for the NEIBank Project: retbindin, an abundant, novel retinal cDNA and alternative splicing of other retina-preferred gene transcripts. Mol. Vis. 8, 196–204 [PubMed] [Google Scholar]
- 21. Geer L. Y., Marchler-Bauer A., Geer R. C., Han L., He J., He S., Liu C., Shi W., Bryant S. H. (2010) The NCBI BioSystems database. Nucleic Acids Res. 38, D492–D496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., Higgins D. G. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 [DOI] [PubMed] [Google Scholar]
- 23. Gille C., Robinson P. N. (2006) HotSwap for bioinformatics: a STRAP tutorial. BMC Bioinformatics 7, 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kelley L. A., Sternberg M. J. (2009) Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4, 363–371 [DOI] [PubMed] [Google Scholar]
- 25. Stricker H. M., Ding X. Q., Quiambao A., Fliesler S. J., Naash M. I. (2005) The Cys214→Ser mutation in peripherin/rds causes a loss-of-function phenotype in transgenic mice. Biochem. J. 388, 605–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ding X. Q., Nour M., Ritter L. M., Goldberg A. F., Fliesler S. J., Naash M. I. (2004) The R172W mutation in peripherin/rds causes a cone-rod dystrophy in transgenic mice. Hum. Mol. Genet. 13, 2075–2087 [DOI] [PubMed] [Google Scholar]
- 27. Han Z., Conley S. M., Makkia R. S., Cooper M. J., Naash M. I. (2012) DNA nanoparticle-mediated ABCA4 delivery rescues Stargardt dystrophy in mice. J. Clin. Invest. 122, 3221–3226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Donoso L. A., Gregerson D. S., Smith L., Robertson S., Knospe V., Vrabec T., Kalsow C. M. (1990) S-antigen: preparation and characterization of site-specific monoclonal antibodies. Curr. Eye Res. 9, 343–355 [DOI] [PubMed] [Google Scholar]
- 29. Gasteiger E., Gattiker A., Hoogland C., Ivanyi I., Appel R. D., Bairoch A. (2003) ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31, 3784–3788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hamazume Y., Mega T., Ikenaka T. (1987) Positions of disulfide bonds in riboflavin-binding protein of hen egg white. J. Biochem. 101, 217–223 [DOI] [PubMed] [Google Scholar]
- 31. Monaco H. L. (1997) Crystal structure of chicken riboflavin-binding protein. EMBO J. 16, 1475–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ota T., Suzuki Y., Nishikawa T., Otsuki T., Sugiyama T., Irie R., Wakamatsu A., Hayashi K., Sato H., Nagai K., Kimura K., Makita H., Sekine M., Obayashi M., Nishi T., Shibahara T., Tanaka T., Ishii S., Yamamoto J., Saito K., Kawai Y., Isono Y., Nakamura Y., Nagahari K., Murakami K., Yasuda T., Iwayanagi T., Wagatsuma M., Shiratori A., Sudo H., Hosoiri T., Kaku Y., Kodaira H., Kondo H., Sugawara M., Takahashi M., Kanda K., Yokoi T., Furuya T., Kikkawa E., Omura Y., Abe K., Kamihara K., Katsuta N., Sato K., Tanikawa M., Yamazaki M., Ninomiya K., Ishibashi T., Yamashita H., Murakawa K., Fujimori K., Tanai H., Kimata M., Watanabe M., Hiraoka S., Chiba Y., Ishida S., Ono Y., Takiguchi S., Watanabe S., Yosida M., Hotuta T., Kusano J., Kanehori K., Takahashi-Fujii A., Hara H., Tanase T. O., Nomura Y., Togiya S., Komai F., Hara R., Takeuchi K., Arita M., Imose N., Musashino K., Yuuki H., Oshima A., Sasaki N., Aotsuka S., Yoshikawa Y., Matsunawa H., Ichihara T., Shiohata N., Sano S., Moriya S., Momiyama H., Satoh N., Takami S., Terashima Y., Suzuki O., Nakagawa S., Senoh A., Mizoguchi H., Goto Y., Shimizu F., Wakebe H., Hishigaki H., Watanabe T., Sugiyama A., Takemoto M., Kawakami B., Yamazaki M., Watanabe K., Kumagai A., Itakura S., Fukuzumi Y., Fujimori Y., Komiyama M., Tashiro H., Tanigami A., Fujiwara T., Ono T., Yamada K., Fujii Y., Ozaki K., Hirao M., Ohmori Y., Kawabata A., Hikiji T., Kobatake N., Inagaki H., Ikema Y., Okamoto S., Okitani R., Kawakami T., Noguchi S., Itoh T., Shigeta K., Senba T., Matsumura K., Nakajima Y., Mizuno T., Morinaga M., Sasaki M., Togashi T., Oyama M., Hata H., Watanabe M., Komatsu T., Mizushima-Sugano J., Satoh T., Shirai Y., Takahashi Y., Nakagawa K., Okumura K., Nagase T., Nomura N., Kikuchi H., Masuho Y., Yamashita R., Nakai K., Yada T., Nakamura Y., Ohara O., Isogai T., Sugano S. (2004) Complete sequencing and characterization of 21,243 full-length human cDNAs. Nat. Genet. 36, 40–45 [DOI] [PubMed] [Google Scholar]
- 33. Carter-Dawson L. D., LaVail M. M., Sidman R. L. (1978) Differential effect of the rd mutation on rods and cones in the mouse retina. Invest. Ophthalmol. Vis. Sci. 17, 489–498 [PubMed] [Google Scholar]
- 34. Lem J., Krasnoperova N. V., Calvert P. D., Kosaras B., Cameron D. A., Nicolò M., Makino C. L., Sidman R. L. (1999) Morphological, physiological, and biochemical changes in rhodopsin knockout mice. Proc. Natl. Acad. Sci. U.S.A. 96, 736–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chakraborty D., Conley S. M., Al-Ubaidi M. R., Naash M. I. (2014) Initiation of rod outer segment disc formation requires RDS. PLoS One 9, e98939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Doonan S. (1996) Protein purification protocols. General strategies. Methods Mol. Biol. 59, 1–16 [DOI] [PubMed] [Google Scholar]
- 37. Hollyfield J. G. (1999) Hyaluronan and the functional organization of the interphotoreceptor matrix. Invest. Ophthalmol. Vis. Sci. 40, 2767–2769 [PubMed] [Google Scholar]
- 38. Becvar J., Palmer G. (1982) The binding of flavin derivatives to the riboflavin-binding protein of egg white. A kinetic and thermodynamic study. J. Biol. Chem. 257, 5607–5617 [PubMed] [Google Scholar]
- 39. Morré D. J., Morré D. M. (2003) Cell surface NADH oxidases (ECTO-NOX proteins) with roles in cancer, cellular time-keeping, growth, aging and neurodegenerative diseases. Free Radic. Res. 37, 795–808 [DOI] [PubMed] [Google Scholar]
- 40. Stone J., Maslim J., Valter-Kocsi K., Mervin K., Bowers F., Chu Y., Barnett N., Provis J., Lewis G., Fisher S. K., Bisti S., Gargini C., Cervetto L., Merin S., Peér J. (1999) Mechanisms of photoreceptor death and survival in mammalian retina. Prog. Retin. Eye Res. 18, 689–735 [DOI] [PubMed] [Google Scholar]
- 41. Said H. M., Wang S., Ma T. Y. (2005) Mechanism of riboflavin uptake by cultured human retinal pigment epithelial ARPE-19 cells: possible regulation by an intracellular Ca2+-calmodulin-mediated pathway. J. Physiol. 566, 369–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fraaije M. W., Mattevi A. (2000) Flavoenzymes: diverse catalysts with recurrent features. Trends Biochem. Sci. 25, 126–132 [DOI] [PubMed] [Google Scholar]
- 43. Goldsmith G. A. (1975) in Riboflavin (Rivlin R. S., ed) pp. 221–238, Plenum Press, New York [Google Scholar]
- 44. Kruse H. D., Sydensticker V. P., Sebrell W. H., Cleckley H. M. (1940) Ocular manifestations of ariboflavinosis. Public Health Reports 55, 157–169 [Google Scholar]
- 45. Eckhert C. D., Hsu M. H., Pang N. (1993) Photoreceptor damage following exposure to excess riboflavin. Experientia 49, 1084–1087 [DOI] [PubMed] [Google Scholar]
- 46. Fridovich I., Handler P. (1958) Xanthine oxidase. II. Studies of the active site. J. Biol. Chem. 231, 899–911 [PubMed] [Google Scholar]
- 47. Taibi G., Nicotra C. M. (2007) Xanthine oxidase catalyzes the oxidation of retinol. J. Enzyme Inhib. Med. Chem. 22, 471–476 [DOI] [PubMed] [Google Scholar]
- 48. Taibi G., Paganini A., Gueli M. C., Ampola F., Nicotra C. M. (2001) Xanthine oxidase catalyzes the synthesis of retinoic acid. J. Enzyme Inhib. 16, 275–285 [DOI] [PubMed] [Google Scholar]
- 49. Fox N. E., van Kuijk F. J. (1998) Immunohistochemical localization of xanthine oxidase in human retina. Free Radic. Biol. Med. 24, 900–905 [DOI] [PubMed] [Google Scholar]
- 50. Thompson C. L., Bowes Rickman C., Shaw S. J., Ebright J. N., Kelly U., Sancar A., Rickman D. W. (2003) Expression of the blue-light receptor cryptochrome in the human retina. Invest. Ophthalmol. Vis. Sci. 44, 4515–4521 [DOI] [PubMed] [Google Scholar]
- 51. Thresher R. J., Vitaterna M. H., Miyamoto Y., Kazantsev A., Hsu D. S., Petit C., Selby C. P., Dawut L., Smithies O., Takahashi J. S., Sancar A. (1998) Role of mouse cryptochrome blue-light photoreceptor in circadian photoresponses. Science 282, 1490–1494 [DOI] [PubMed] [Google Scholar]
- 52. Oztürk N., Song S. H., Selby C. P., Sancar A. (2008) Animal type 1 cryptochromes. Analysis of the redox state of the flavin cofactor by site-directed mutagenesis. J. Biol. Chem. 283, 3256–3263 [DOI] [PubMed] [Google Scholar]
- 53. Cairns W. L., Metzler D. E. (1971) Photochemical degradation of flavins. VI. A new photoproduct and its use in studying the photolytic mechanism. J. Am. Chem. Soc. 93, 2772–2777 [DOI] [PubMed] [Google Scholar]
- 54. De La Paz M., Anderson R. E. (1992) Region- and age-dependent variation in susceptibility of the human retina to lipid peroxidation. Invest. Ophthalmol. Vis. Sci. 33, 3497–3499 [PubMed] [Google Scholar]
- 55. Kopitz J., Holz F. G., Kaemmerer E., Schutt F. (2004) Lipids and lipid peroxidation products in the pathogenesis of age-related macular degeneration. Biochimie 86, 825–831 [DOI] [PubMed] [Google Scholar]