Abstract
Peripheral T-cell lymphomas not otherwise specified (PTCLs/NOS) are rare and aggressive tumours whose molecular pathogenesis and diagnosis are still challenging. The microRNA (miRNA) profile of 23 PTCLs/NOS was generated and compared with that of normal T-lymphocytes (CD4+, CD8+, naive, activated). The differentially expressed miRNA signature was compared with the gene expression profile (GEP) of the same neoplasms. The obtained gene patterns were tested in an independent cohort of PTCLs/NOS. The miRNA profile of PTCLs/NOS then was compared with that of 10 angioimmunoblastic T-cell lymphomas (AITLs), 6 anaplastic large-cell lymphomas (ALCLs)/ALK+ and 6 ALCLs/ALK−. Differentially expressed miRNAs were validated in an independent set of 20 PTCLs/NOS, 20 AITLs, 19 ALCLs/ALK− and 15 ALCLs/ALK+. Two hundred and thirty-six miRNAs were found to differentiate PTCLs/NOS from activated T-lymphocytes. To assess which miRNAs impacted on GEP, a multistep analysis was performed, which identified all miRNAs inversely correlated to different potential target genes. One of the most discriminant miRNAs was selected and its expression was found to affect the global GEP of the tumours. Moreover, two sets of miRNAs were identified distinguishing PTCL/NOS from AITL and ALCL/ALK−, respectively. The diagnostic accuracy of this tool was very high (83.54%) and its prognostic value validated.
Introduction
Peripheral T-cell lymphomas (PTCLs) correspond to a heterogeneous group of nodal and extranodal tumours, which account for 10 to 15% of all lymphoid neoplasms. Four subtypes, namely PTCL not otherwise specified (NOS), angioimmunoblastic T-cell lymphoma (AITL) and anaplastic large-cell lymphoma (ALCL) (ALK+ and ALK−), represent about 60% of cases in Europe and the United States of America.1, 2 Particularly, PTCL/NOS is an aggressive disease whose molecular pathogenesis and distinction from AITL and ALCLs are still challenging.3 The latter is however relevant because of possible differences in prognosis4, 5, 6, 7 and therapeutic options.8, 9 Of note, despite the heterogeneity of cytogenetic features,10, 11, 12, 13, 14, 15, 16 PTCLs/NOS share a consistent transcriptional signature, likely indicative of common altered pathways. During the past few years, gene expression profiling (GEP) provided some hints for the distinction among different PTCL entities.17, 18, 19, 20, 21, 22, 23 In this respect, our group recently reported a molecular classifier that might represent an useful tool in the diagnostic workup of nodal PTCLs.7, 24
MicroRNAs (miRNAs) are small noncoding single-stranded RNA molecules (18–22nt) that mainly control gene expression posttranscriptionally by binding the 3′-untranslated region of the target mRNA. This results in the degradation of target mRNA or inhibition of its translation.25 miRNA expression is tissue-specific and changes in miRNA expression have been associated with various human diseases, including cancer.26, 27 Several studies highlighted the role of specific miRNAs in lymphoma biology, in particular miR-101, miR-16, miR-135b and miR-29a; five members of the miR-17–92 cluster have been reported to have a role in the oncogenic ALK signalling in ALCLs,28, 29, 30, 31, 32 and miR-187 was overexpressed in PTCL/NOS associated with high Ki-67 expression.33 Furthermore, expression levels of miR-155 were significantly higher in ALCL/ALK− than in ALCL/ALK+.28 In a recent study, Liu et al.34 showed a signature of 11 miRNAs that differentiated ALCL/ALK− from other PTCLs.
However, the possible impact of miRNA deregulation on the transcriptional profile of PTCL/NOS has not been explored yet, as well as the possible application of miRNA profiling for diagnostic purposes. In the present study, by applying a phase IV diagnostic accuracy study respecting the EBM rules and NCI criteria for biomarkers validation, we performed extensive miRNA profiling of PTCL/NOS, ALCLs, AITL and normal T cells aiming to (1) determine the possible contribution of miRNAs to PTCL/NOS transcriptional profile, and (2) to assess whether differentially expressed miRNAs may improve the differential diagnosis among the four subtypes of nodal PTCL.
Materials and methods
Case collection and study design
We retrieved 174 formalin-fixed, paraffin-embedded tissue (FFPE) nodal PTCLs from the archive of the Haematopathology Unit of Bologna University, including 98 PTCLs/NOS, 30 AITLs and 44 ALCLs (21 ALK+, 23 ALK−).
All the cases had been diagnosed by expert haematopathologists (ES, FB, CA, CASS and SAP) according to the criteria of the WHO classification1 and collected at diagnosis before any treatment. Written informed consent was obtained from all patients for tissue analysis. Patient characteristics are summarized in the Supplementary Table 1.
The cases were used to assess (1) the possible impact of miRNAs on the regulation of PTCL/NOS transcriptome, and (2) to assess specific miRNA signatures useful for the differential diagnosis among PTCL/NOS, AITL and ALCLs.
For the former purpose, 23 PTCLs/NOS were chosen, all provided with GEP6 data and characterized by at least 80% of neoplastic cells as defined based on their aberrant phenotypic profile. Their miRNA profile was compared with that of 12 normal T-lymphocyte samples (corresponding naive CD4+ (n=4), activated CD4+ (n=2), naive CD8+ (n=4) and activated CD8+ (n=2)). A signature based on miRNAs differentially expressed between tumours and normal T-lymphocyte samples (see below) was compared with GEP of the same neoplasms. The obtained gene patterns were then tested in an independent cohort of PTCLs/NOS (n=55), which had been included in a previous GEP analysis.7
For the other aim of the present study, the miRNA profile of the above-mentioned 23 PTCLs/NOS was compared with that of 10 AITLs and 12 ALCLs (6 ALK+ and 6 ALK−). The differentially expressed miRNAs were validated in an independent set of cases (20 PTCLs/NOS, 20 AITLs, 19 ALCLs/ALK− and 15 ALCLs/ALK+) by quantitative reverse transcriptase-PCR (qRT-PCR).
The study was conducted according to the principles of the Declaration of Helsinki after approval of the Internal review Board (Prot. Numb. 001-2011-U-Tess).
CD4+ and CD8+ normal T-lymphocytes isolation
Whole blood samples were collected by venipuncture from six healthy donors. Peripheral blood mononucleated cells were separated by Ficoll gradient (Ficoll-Histopaque; Pharmacia, Uppsala, Sweden). CD4+- and CD8+- naive T cells were isolated from peripheral blood mononucleated cells using magnetic MACS microbeads according to manufacturer's instructions (Miltenyi, Bergisch Gladbach, Germany) The CD4+- and CD8+-naive cell isolation was analysed by flow cytometry (FACS Calibur; Becton-Dickinson, San Diego, CA, USA), and the cell purity was >94% for CD4+/CD45RA+ or CD8+/CD45RA+ cells, respectively. Isolated cells were cultured in RPMI-1640 (Lonza, Basel, Switzerland) with 10% foetal bovine serum (GIBCO, Paisley, UK) and activated with phytohaemagglutinin (5 μg/ml; Sigma, St Louis, MO, USA) plus IL-2 (10 U/ml; Miltenyi) for 4 days.
RNA extraction, microRNA profiling and gene expression profiling
RecoverAll Total Nucleic Acid Isolation Kit (Ambion, Life Technologies, Carlsbad, CA, USA) was used to extract total RNA from FFPE tissues according to the manufacturer's procedure. Total RNA from normal T-lymphocytes subset was extracted with Trizol according to the manufacturer's instructions (Invitrogen, Life Technologies, Carlsbad, CA, USA). RNA was quantified using ND-1000 spectrophotometer running software version 3.0.1 (NanoDrop Technologies Inc., Rockland, DE, USA).
After RNA extraction of samples, microRNA profiling was carried out by using the TaqMan Array Human MicroRNA Card A v.2.0 (Life Technologies, Carlsbad, CA USA). We used the Illumina Whole Genome DASL assay for gene profile generation from FFPE samples, as described previously7, 35, 36 (details are provided in Supplementary File).
miRNA qRT-PCR
Validation of an miRNA signature with potential differential diagnostic value (see above) was carried by qRT-PCR (TaqMan 7900HT; Life Technologies) (details are provided in Supplementary File).
miRNA transfection and GEP
A human PTCL CD30+ cell line (Fe-Pd) was used to perform the ex vivo functional experiments. Transient transfections were performed by nucleofection, using an Amaxa apparatus, program X-01 and solution V (Amaxa, Cologne, Germany). Total RNA from Fe-Pd cell line was extracted with Trizol according to the manufacturer's instructions (Invitrogen, Life Technologies). RNA was quantified using ND-1000 spectrophotometer running software version 3.0.1 (NanoDrop Technologies Inc.).
Gene expression profiling was executed on transfected and untransfected Fe-Pd cells using Gene Chip 2.0 Whole Transcript Assay (Affymetrix, Santa Clara, CA, USA) (details are provided in Supplementary File).
Data analysis
miRNA and gene expression analysis
Based on unsupervised and supervised analyses, which showed that activated T-lymphocytes represent the closest normal counterpart of PTCLs/NOS (see below) differentially expressed miRNAs between PTCL/NOS and activated T-lymphocytes were identified, using a two-tailed Student's t-test with Welch approximation for different variance among groups and adjusted Bonferroni correction for false discovery rate, applying the following filtering criteria: P-value <0.05, and fold change (FC) in absolute value >2 for log2-transformed data.
To identify miRNAs more likely impacting on the transcriptome of PTCL/NOS, we used a three-step bioinformatic approach: (1) miRNAs differentially expressed between PTCL/NOS- and CD4+/CD8+ -activated T-lymphocytes were selected using a two-tailed Student's t-test with Welch approximation for different variance among groups, with the same filtering parameters mentioned above; (2) Spearman's correlation between every differentially expressed miRNA–gene pairs was calculated using a custom R script and only miRNA–gene pairs with negative Spearman's correlation and P-value <0.01 were selected; (3) microRNA Data Integration Portal was used for further filter of miRNA–gene pairs, selecting only miRNA–gene target predictions confirmed by at least 5 out of 12 data sets.37
The miRNA-related genes signature was then tested in 55 PTCLs/NOS, which had been the object of a previous GEP study by hierarchical clustering (HC) and applying a classifier based on a support vector machine algorithm.7 Broad Institute Gene Set Enrichment Analysis (GSEA) software38 was performed to identify significant enrichments in differentially expressed genes signatures, in case of nominal P-values corrected for false discovery rate for enrichment scores <0.05. GSEA was also used to identify KEGG pathways significantly enriched for genes found after filtering steps. DAVID Functional Annotation Bioinformatics Microarray Analysis (http://david.abcc.ncifcrf.gov/) was also used to establish whether specific pathways and biological processes defined according to gene ontology were significantly represented among the deregulated genes.
Classification of PTCL subtypes
Molecular signatures of differentially expressed miRNAs between PTCL/NOS vs AITL, PTCL/NOS vs ALCL/ALK− and ALCL/ALK+ were generated by supervised analysis (two-tailed Student's t-test). Differentially expressed miRNAs were selected according to P-value (<0.05) and FC (⩾2). Stepwise discriminant analysis was performed using IBM SPSS Statistics 20.0 (IBM, Armonk, NY, USA) to identify the minimal number of miRNAs able to classify correctly a given sample in different PTCL subgroups. For each step of the analysis, the miRNA that minimizes Wilks' lambda test probability distribution was selected to enter in the discriminant model and included in the list of discriminant miRNAs if its probability associated with Fisher's exact test was lower than 0.05. Otherwise, the miRNA with probability associated with Fisher's exact test >0.10 were excluded from the list. Stepwise method was run until no further miRNAs could be added or excluded to reduce Wilks' lambda probability distribution. A discriminant function based on linear combination of the expression of discriminant miRNAs was created for each classification step (PTCL/NOS vs AITL and PTCL/NOS vs ALCL/ALK−), to assign a discriminant score for each sample.
To test discriminant function associated to reduced miRNA signatures, each case was then assigned to a specific PTCL subgroup according to the distance between its discriminant score and a cutoff value was calculated for each predicted group. Samples with discriminant score rate <60% were not assigned to a particular PTCL subgroup and were flagged as unclassified. Calculations of sensitivity (ST), specificity (SP), positive predictive value (PPV), negative predictive value (NPV), positive and negative likelihood ratio (LR) and odds ratio were made by CAT maker software (Centre for Evidence Based Medicine, Oxford University, http://www.cebm.net).
Survival analyses
Clinical information and complete follow-up were available for 65/119 cases for which miRNA profile and miRNA qRT-PCR were performed. Overall survival (OS) was calculated from the time of diagnosis to death or last follow-up. Statistical analyses were carried out by IBM SPSS Statistics 20.0. Survival data were analysed with the Kaplan–Meier estimator method.39 The limit of significance for all analyses was defined as P<0.05 for the log-rank Mantle–Cox test.
More data analysis details and any associated reference are available in the Supplementary File.
Results
Identification of normal T-lymphocyte-specific miRNA signature and its expression in PTCL/NOS samples
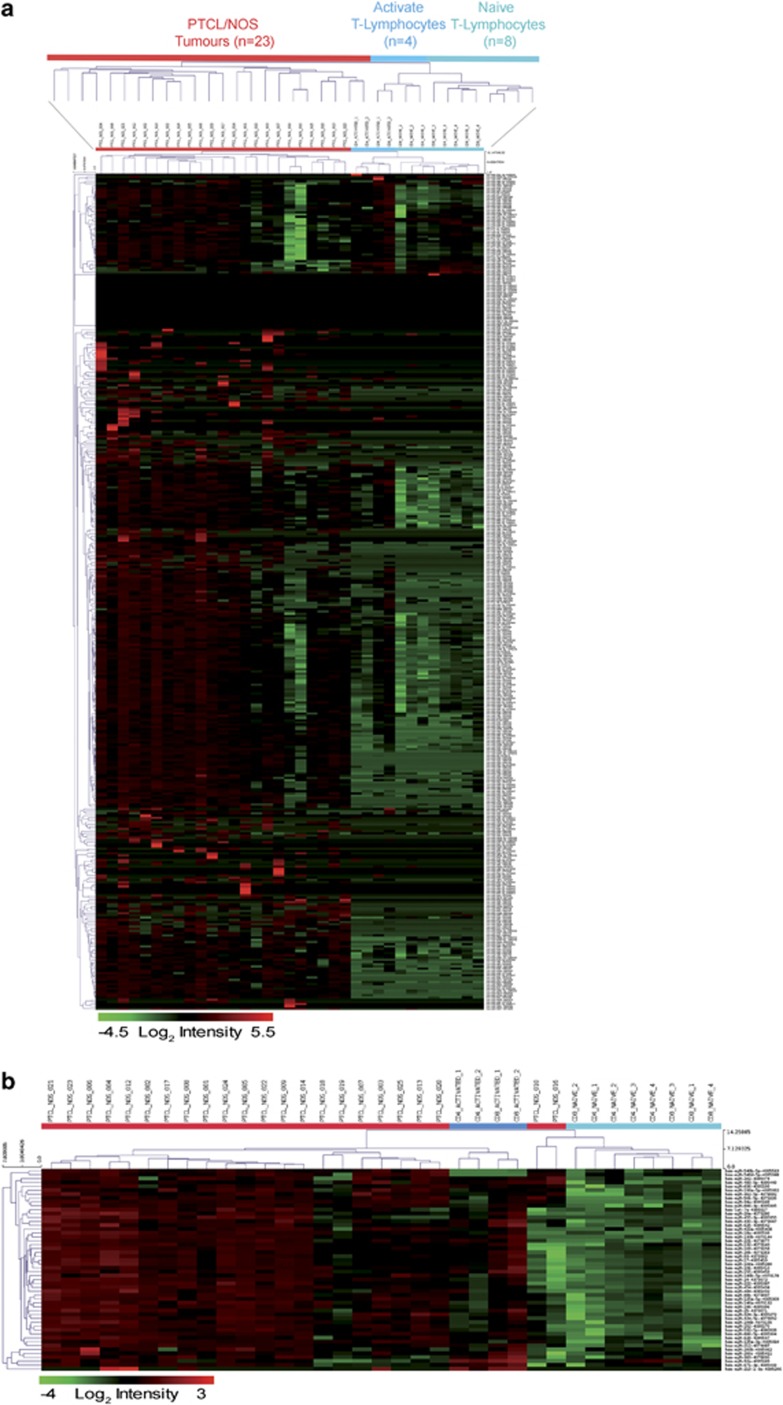
We studied the miRNA profile of 23 PTCLs/NOS and 12 normal T-lymphocyte samples (n=4 naive CD4+, n=4 naive CD8+, n=2 activated CD4+ and n=2 activated CD8+). First, unsupervised HC clearly distinguished PTCLs/NOS from naive and activated T-lymphocytes based on global miRNA expression patterns (Figure 1a). Second, supervised analysis (two-tailed Student's t-test P-value <0.05 and FC >2) allowed the identification of a signature consisting of 52 miRNAs that distinguished naive (CD4+, CD8+) from activated (CD4+, CD8+) T-lymphocytes. Third, this miRNA signature was applied to PTCL/NOS samples using a cell-type classifier based on a support vector machine algorithm: such analysis showed that activated CD4+ and CD8+ T-lymphocytes represented the closest normal counterpart of the tumour (Figure 1b). This result was consistent with previous studies based on gene expression profiling.18, 40, 41 Accordingly, for further analyses, activated CD4+ and CD8+ T-lymphocytes were regarded as the normal counterpart of PTCL/NOS.
Figure 1.
(a) Unsupervised analysis comparing PTCL/NOS vs naive and activated CD4+ and CD8+ normal T-lymphocytes. The heat map diagram shows the result of the two-way unsupervised HC of miRNAs and samples based on the expression of 379 miRNAs. HC, clearly discriminated PTCLs and CD4+ and CD8+ T-lymphocytes based on the miRNA expression patterns. In the matrix, each row represents an miRNA and each column represents a sample. The colour scale illustrates the relative expression level of an miRNA across all samples: red represents an expression level above the mean and green represents expression lower than the mean. (b) Supervised analyses identified 52 differentially expressed miRNAs comparing naive and activated CD4+ and CD8+ T-lymphocytes. HC shows miRNA expression pattern of PTCL/NOS is closest to activated CD4+ and CD8+ T-lymphocytes. In the matrix, each row represents an miRNA and each column represents a sample. The colour scale illustrates the relative expression level of an miRNA across all samples: red represents an expression level above the mean and green represents expression lower than the mean.
PTCL/NOS can be distinguished from normal activated T-lymphocytes based on global miRNA and GEPs
First, to exclude that bias was introduced by comparing FFPE samples (PTCL/NOS) and fresh normal lymphocytes, we excluded from the analysis cases with RNU6B Ct value >23; further, we compared the RNU6B Ct values in the two classes of samples and they turned out to be quite similar (P=0.33).24
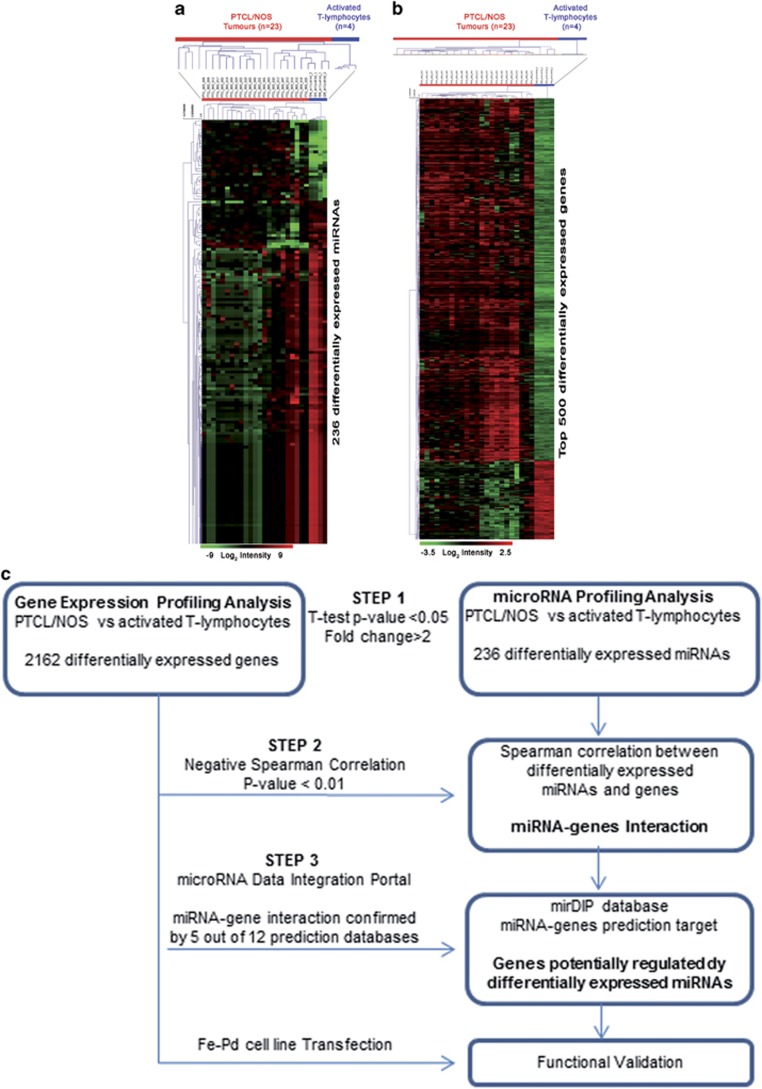
Based on the above, we performed a supervised analysis (two-tailed Student's t-test P-value <0.05 and FC >2) between PTCLs/NOS and activated CD4+ and CD8+ T-lymphocytes, which allowed the distinction of neoplastic and normal samples into two different clusters based on the differential expression of 236 miRNAs (32 miRNAs upregulated and 204 miRNAs downregulated in PTCLs/NOS) (Figure 2a and Supplementary Table 2). Furthermore, the GEP of the same 23 PTCLs/NOS and activated CD4+ and CD8+ T-lymphocytes was interrogated by supervised analysis (t-test, P-value <0.05, FC >2, false discovery rate adjusted Bonferroni correction) aiming to identify differentially expressed genes between PTCLs/NOS and activated CD4+ and CD8+ T-lymphocytes. Specifically, we found 2162 differentially expressed genes (1225 upregulated and 937 downregulated in PTCLs/NOS) (Figure 2b and Supplementary Table 3).
Figure 2.
Supervised analyses identified (a) differentially expressed miRNAs between PTCL/NOS and activated CD4+ and CD8+ T-lymphocytes. miRNAs with differences in expression were selected by the comparative cycling threshold methods (ΔΔCT) and filtered for statistical significance (two-tailed Student's t-test P-value <0.05 and FC >2). (b) Genes differentially expressed between PTCL/NOS and activated CD4+ and CD8+T-lymphocytes (t-test, P-value <0.05, FC >2, false discovery rate (FDR) adjusted Bonferroni correction). Top 500 genes in ranking are plotted. In the matrix, each row represents an miRNA/gene and each column represents a sample. The colour scale illustrates the relative expression level of an miRNA and genes across all samples: red represents an expression level above the mean and green represents expression lower than the mean. (c) Bioinformatic approach (three steps). Schematic representation of miRNA profiling and gene expression profiling data integration. The flow diagram is constituted by three steps indicated in the figure by three arrows.
A multistep bioinformatics approach: miRNA-related genes fall in cancer pathways
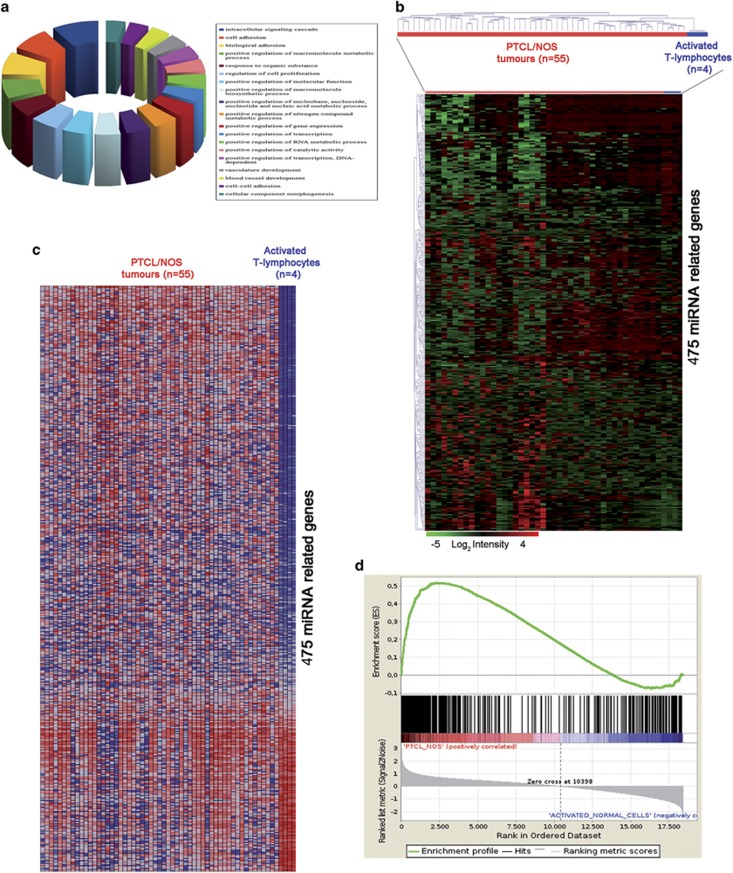
We then sought to assess whether miRNAs deregulated in PTCLs/NOS might affect the transcriptional profile of the tumours. To this aim, we adopted a multistep approach integrating miRNA and GEP data (schematic representation in Figure 2c). First, we integrated miRNA and GEP data by using Spearman's correlation (236 differentially expressed miRNA with 2162 differentially expressed genes) and selected only the inverse miRNA–gene paired combinations (i.e. miRNA up/gene down and vice versa) with high statistical significance (negative Spearman's correlation, P-value <0.01) for a total of 137 609 miRNA–genes pairs. Second, using the microRNA Data Integration Portal database we selected among the 137 609 miRNA–gene pairs only those confirmed by at least 5 out of 12 miRNA prediction algorithms and found 1184 miRNA–gene pairs corresponding to 475 inversely correlated genes that were the predicted targets of 158 miRNAs (Supplementary Table 4). To assess the potential role of such 475 miRNA-related genes, we predicted their biological functions by mapping biomolecular networks using DAVID bioinformatics resources (http://david.abcc.ncifcrf.gov/). We found that they were involved in biological processes—such as intracellular signalling cascade, cell adhesion, response to endogenous stimulus, regulation of cell proliferation and positive regulation of protein catalytic activity (Figure 3a)—that may be relevant in neoplastic transformation and progression. These results suggest that miRNAs may contribute to PTCL/NOS pathogenesis by inducing significant transcriptional changes.
Figure 3.
Four hundred and seventy-five miRNA-related genes. (a) Pie chart shows the distribution of several biological processes in which are involved the 475 ‘miRNA-driven' genes. Each colour represents a different biological process. (b) HC created on the 475 gene signatures and applied on 55 PTCLs/NOS shows a clear distinction of PTCL/NOS from activated CD4+ and CD8+ T-lymphocytes. In the matrix, each row represents a gene and each column represents a sample. The colour scale illustrates the relative expression level of genes across all samples: red represents an expression level above the mean and green represents expression lower than the mean. GSEA on the 475 miRNA-related genes: (c) heat map shows the distribution of 475 miRNA-related gene expressions in PTCL/NOS and activated CD4+ and CD8+ T-lymphocytes. (d) GSEA enrichment plot shows that the 475 genes are significantly enriched in PTCL/NOS vs activated CD4+ and CD8+ T-lymphocytes. In the enrichment plot, the x axis shows the rank order of genes from the most upregulated to the most downregulated between PTCL/NOS and activated CD4+ and CD8+ T-lymphocytes. The barcode indicates the position of 475 miRNA-related genes in the ranking list. The y axis shows the distribution of the running enrichment score generated by walking down the list of ranked genes.
miRNA expression significantly affects the transcriptome of PTCL/NOS
Subsequently, to validate the hypothesis that miRNA deregulation may affect the transcriptome of PTCL/NOS, we studied the expression of the above-mentioned 475 miRNA-related genes in 55 PTCLs/NOS in comparison with the profile of CD4+ and CD8+ T-lymphocytes (n=4). HC showed a clear distinction of PTCLs/NOS from activated CD4+ and CD8+ T-lymphocytes (Figure 3b), indicating that these 475 genes significantly characterize the molecular profile of PTCL/NOS. Further, based on the expression of these 475 genes, we developed a classifier using a support vector machine algorithm leave one-out iterative algorithm (cross-validation method). Each case among the 55 PTCLs/NOS was then re-evaluated by the molecular classifier with 100% accuracy. Moreover, this gene signature was found to be specifically enriched in PTCL/NOS vs activated CD4+ and CD8+ T-lymphocytes as demonstrated by GSEA (P-value <0.0001) (Figures 3c and d). In particular, KEGG Pathway Analysis showed a significant enrichment in several pathways as WNT signalling, mitogen-activated protein kinase signalling, T-cell receptor signalling, cell cycle and chemokine signalling. Taken together, these results indicated that the deregulated expression of the identified miRNAs is likely to contribute to the abnormal transcriptional profile of PTCL/NOS.
miRNA-132-3p downregulation has influence over PTCL/NOS transcriptome
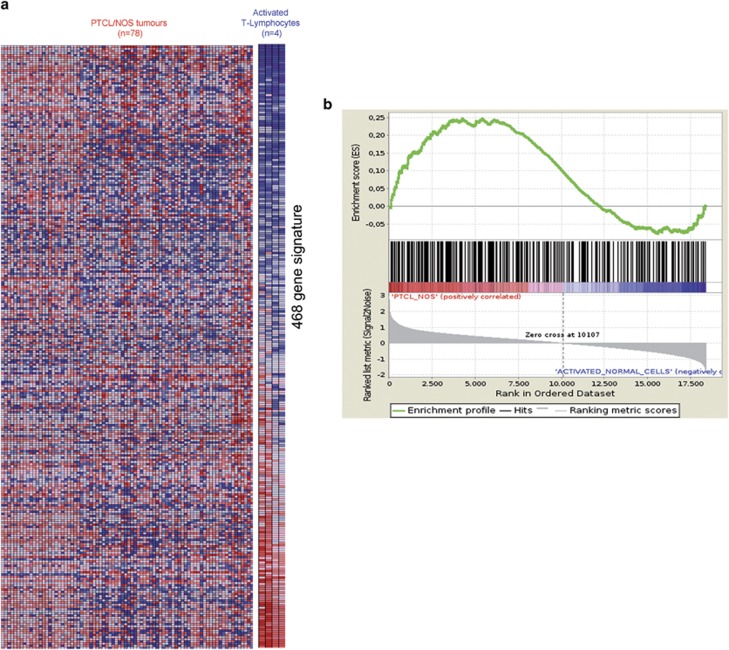
As our analyses strongly suggested that the identified miRNAs may contribute to PTCL/NOS gene expression pattern, we aimed to validate these data providing functional evidence as well. In particular, we focused our attention on the miR-132-3p that provided the highest combination number of postulated target genes (N=22) and the greatest difference in expression between PTCLs/NOS and activated CD4+ and CD8+ T-lymphocytes (log2 FC=−6.21). First, we confirmed miR-132-3p downregulation in the comparison between neoplastic and normal samples in an independent set consisting of 20 PTCLs/NOS by qRT-PCR (Supplementary Figure 1A). To investigate its biological role, we introduced miR-132-3p into Fe-Pd cells by transient transfection (100 nM mimic), transfection efficiency being confirmed by qRT-PCR (Supplementary Figure 1B). To evaluate functional effects of miR-132-3p transfection, we carried out a global GEP on transfected and untransfected Fe-Pd cells. Thereafter, we performed a supervised analysis identifying 468 genes differentiating transfected vs untransfected Fe-Pd cells (Supplementary Table 5). Then, we tested gene signature in 78 PTCL/NOS primary tumours. Indeed, HC, GSEA and a support vector machine-based classifier indicated that the genes modulated by miR-132-3p were consistently deregulated in primary cases as well (Figures 4a and b).
Figure 4.
GSEA of 468 genes differentially expressed between transfected and untransfected Fe-Pd cell line with miR-132-3p (mimic 100 nM) in 80 PTCL/NOS primary tumour and activated CD4+ and CD8+ T-lymphocytes. (a) Heat map shows the distribution of 468 genes, differentially expressed in transfected and untransfected Fe-Pd cell line, in PTCL/NOS and activated CD4+ and CD8+ T-lymphocytes. (b) GSEA enrichment plot shows that 468 genes are significantly enriched in PTCL/NOS vs activated CD4+ and CD8+ T-lymphocytes.. In the enrichment plot, the x axis shows the rank order of genes from the most upregulated to the most downregulated between PTCL/NOS and activated CD4+ and CD8+ T-lymphocytes. The barcode indicates the position of 468 miRNA-related genes in the ranking list. The y axis shows the distribution of the running enrichment score generated by walking down the list of ranked genes.
Taken together, these data indicated that miR-132-3p modulation has a significant impact on PTCL/NOS transcriptome.
miRNA expression discriminate PTCL subtypes
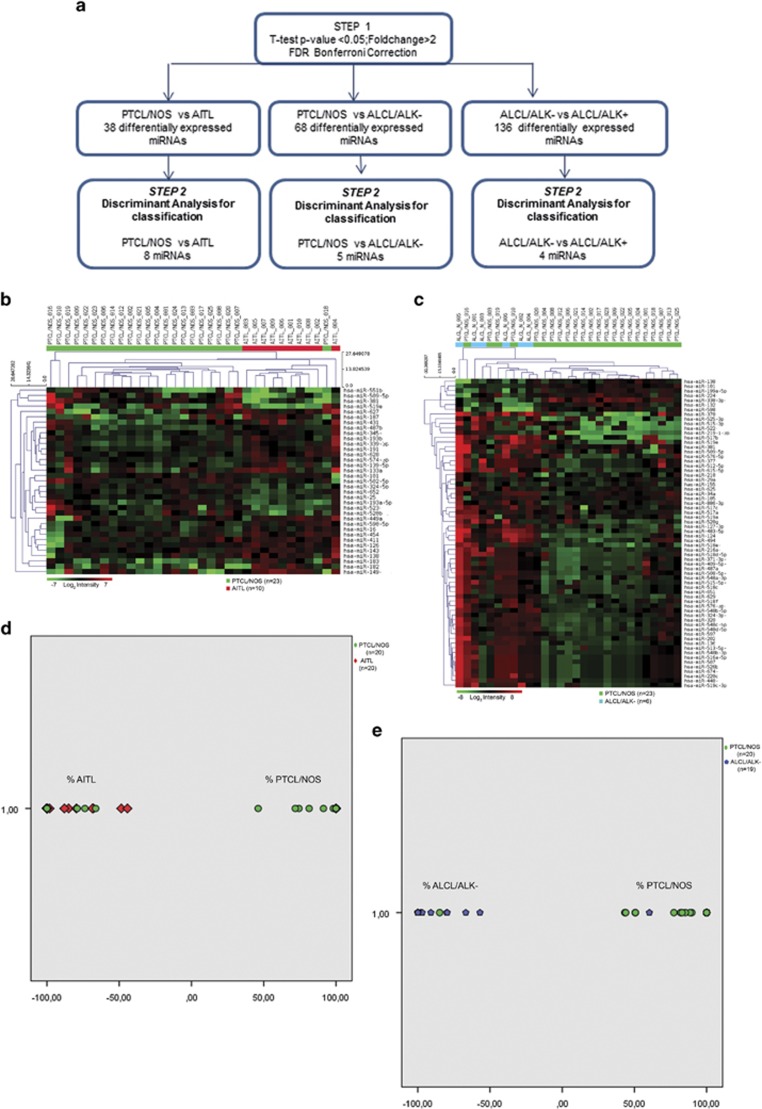
As we found that miRNAs can modulate gene expression in PTCL/NOS and GEP can be effectively used to discriminate different PTCL subtypes, we sought whether nodal PTCLs could be distinguished based on miRNA expression. To this aim, we adopted a two-step bioinformatic approach (Figure 5a). First, we performed a supervised comparison of PTCL/NOS vs AITL, PTCL/NOS vs ALCL/ALK− and ALCL/ALK− vs ALCL/ALK+ (two-tailed Student's t-test) using 23 PTCLs/NOS, 10 AITLs, 6 ALCLs/ALK− and 6 ALCLs/ALK+. Based on P-value (P<0.05) and FC (⩾2), we identified differentially expressed miRNAs among the different PTCL subtypes. In particular, we found 35 miRNAs distinguishing PTCL/NOS from AITL (Figure 5b and Supplementary Table 6), 68 miRNAs distinguishing PTCL/NOS from ALCL/ALK− (Figure 5c and Supplementary Table 7) and 136 miRNAs distinguishing ALCL/ALK− from ALCL/ALK+ (Supplementary Figure 2). Second, we focused on the possibility of developing a practical tool to be applied in the routine diagnostic workup for the differential diagnosis of nodal PTCLs. A stepwise linear discriminant analysis was then used to identify three sets of miRNAs able to efficiently distinguish PTCL/NOS vs ALCL/ALK−, PTCL/NOS vs AITL and ALCL/ALK− vs ALCL/ALK+, respectively. A discriminant function based on the linear combination of the discriminant miRNAs was created to perfectly distinguish PTCL/NOS from ALCL/ALK− using a minimal miRNA set of five miRNAs (hsa-miR-515-3p, hsa-miR-155, hsa-miR-598, hsa-miR-625 and hsa-miR-199a-5p), PTCL/NOS from AITL using eight miRNAs (hsa-miR-652, hsa-miR-627, hsa-miR-519e, hsa-miR-487b, hsa-miR-324-5p, hsa-miR-449a, hsa-miR-381 and hsa-miR-574-3p) and ALCL/ALK− from ALCL/ALK+ using four miRNAs (hsa-miR-124, hsa-miR-325, hsa-miR-181a and hsa-miR-618).
Figure 5.
Supervised analysis identify (a) ‘bioinformatic approach (two-step). Schematic representation of PTCL subtypes classification based on miRNA profiling analysis. The flow diagram is constituted by two steps indicated in the figure by three arrows'. (b) Differentially expressed miRNAs comparing PTCL/NOS vs AITL. miRNAs with differences in expression were selected by the comparative cycling threshold methods (ΔΔCT) and filtered for statistical significance (two-tailed Student's t-test P-value <0.05 and FC >2). Eight miRNAs emerging from stepwise discriminant analysis. (c) Differentially expressed miRNAs comparing PTCL/NOS vs ALK-negative anaplastic large-cell lymphoma (ALCL/ALK−). miRNAs with differences in expression were selected by the comparative cycling threshold methods (ΔΔCT) and filtered for statistical significance (two-tailed Student's t-test P-value <0.05 and FC >2). Five miRNAs emerging from stepwise discriminant analysis. For each panel in the matrix, rows represent miRNAs and columns represent samples. The colour scale illustrates the relative expression level of miRNAs across all samples: red represents an expression level above the mean and green represents expression lower than the mean. Classification of PTCL subtypes by linear discriminant analysis in an independent set of cases. Scatterplots shows the distribution of PTCL/NOS vs AITL (d) and PTCL/NOS vs ALCL/ALK− (e) according to their discriminant scores in minimum miRNA set. Each case is represented in different colour according to its molecular classification. Green circles represent PTCL/NOS cases, red rhombus represents AITL cases and blue hexagons represents ALCL/ALK− cases.
We then established the diagnostic accuracy of miRNA profiling in a phase IV study, analysing the obtained discriminant sets of miRNAs in an independent set of cases by qRT-PCR (20 PTCLs/NOS, 20 AITLs, 19 ALCLs/ALK− and 15 ALCLs/ALK+). As our aim was to develop a practical tool for the diagnostic workup of nodal PTCLs, focusing on PTCL/NOS vs AITL and PTCL/NOS vs ALCL/ALK−.
We applied the linear discriminant function to classify PTCL cases, assigning each case to either one PTCL subgroups (PTCL/NOS vs AITL and PTCL/NOS vs ALCL/ALK−). We found that 16 out of 20 AITLs and 15 out of 20 PTCLs/NOS were correctly classified, for an overall accuracy of 77.5% (P-value for Fisher's exact test=0.0012). ST and SP were, respectively, 76.2% and 78.9%, whereas PPV and NPV were 80% and 75% respectively (Figure 5d and Table 1).
Table 1. Diagnostic accuracy of PTCL/NOS vs AITL and PTCL/NOS vs ALCL/ALK− miRNA classifier evaluated in an independent set of cases.
| miRNA classification |
Histopathology |
Total | |
|---|---|---|---|
| PTCL/NOS | AITL | ||
| PTCL/NOS | 15 | 5 | 20 |
| AITL | 4 | 16 | 20 |
|
Accuracy |
|||
| Value | 95% CI | ||
| ST | 0.762 | 0.549–0.894 | |
| SP | 0.789 | 0.567–0.915 | |
| PPV | 0.8 | 0.584–0.919 | |
| NPV | 0.75 | 0.531–0.888 | |
| LR+ | 3.619 | 1.467–8.928 | |
| LR- | 0.302 | 0.136–0.671 | |
| Overall accuracy 77.5% (31 of 40) | |||
| miRNA classification |
Histopathology |
Total | |
| PTCL/NOS | ALCL/ALK− | ||
| PTCL/NOS | 17 | 3 | 20 |
| ALCL/ALK− | 1 | 18 | 19 |
|
Accuracy |
|||
| Value | 95% CI | ||
| ST | 0.857 | 0.654–0.95 | |
| SP | 0.944 | 0.742–0.999 | |
| PPV | 0.947 | 0.754–0.991 | |
| NPV | 0.85 | 0.64–0.948 | |
| LR+ | 15.429 | 2.278–104.475 | |
| LR− | 0.151 | 0.053–0.434 | |
| Overall accuracy 89.74% (35 of 39) | |||
Abbreviations: ALCL/ALK−, anaplastic large-cell lymphoma ALK− AITL, angioimmunoblastic T-cell lymphoma; LR, likelihood ratio; NPV, negative predictive value; PPV, positive predictive value; PTCL/NOS, peripheral T-cell lymphoma/not otherwise specified; SP, specificity; ST, sensitivity.
Discriminant function applied on PTCLs/NOS vs ALCLs/ALK− was able to classify correctly 18 out of 19 ALCLs/ALK− and 17 out of 20 PTCLs/NOS, for an overall accuracy of 89.74% (P-value for Fisher's exact test <0.0001). ST and SP were, respectively, 85.7% and 94.4%, whereas PPV and NPV were 94.7% and 85%, respectively (Figure 5e and Table 1).
The microRNA classifier improves categorization and prognosis of PTCL subtypes
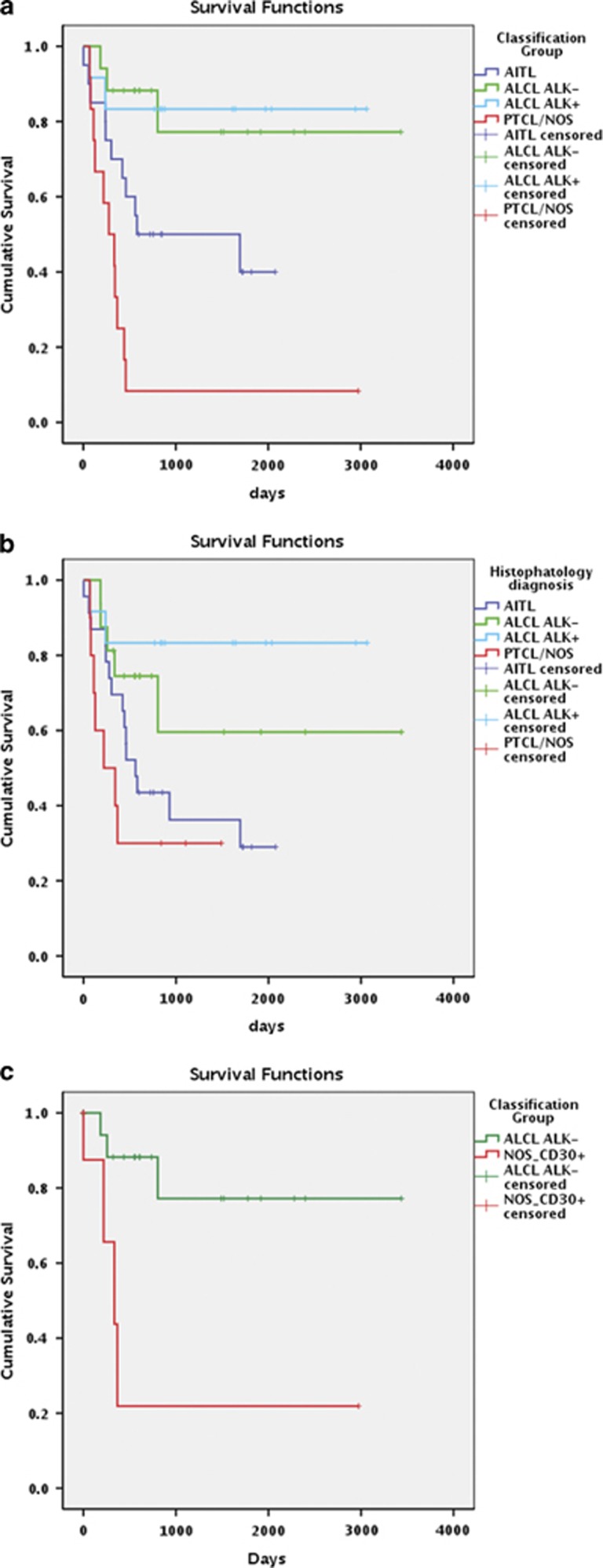
As PTCL diagnosis based on GEP is provided with relevant prognostic implications, we then assessed whether the proposed PTCLs classification based on miRNAs expression was endorsed with clinical significance as well. We could include 65/119 cases for which complete information was available; the median follow-up of alive patients was 1489 days (range 321–3435 days). The median OS for the entire population was 607 days, the 3-year OS rate being 36.2%, 68.8%, 83.3% and 27.3% for AITL, ALCL/ALK−, ALCL/ALK+ and PTCL/NOS patients, respectively. We then calculated the OS curves for cases classified according to the minimum set of miRNAs for PTCL/NOS vs AITL, PTCL/NOS vs ALCL/ALK− and ALCL/ALK− vs ALCL/ALK+. Significant survival differences were recorded when PTCL/NOS (median OS 277 days) vs AITL (median OS, 1694) subtypes were reclassified by miRNA expression (P=0.027; Figure 6a). Similarly, when PTCL/NOS (median OS 277 days) vs ALCL/ALK− (median OS not reached) subtypes were reclassified by miRNA expression, we found a statistically significant difference in terms of OS (P=0.001). The miRNA-based classifier was more effective than histopathological diagnosis in predicting prognosis (Figure 6b; P=0.001 vs P=0.05, respectively). The 3-years OS rate for reclassified samples was 50%, 77.20%, 83.3% and 8.33% for AITL, ALCL/ALK−, ALCL/ALK+ and PTCL/NOS patients, respectively.
Figure 6.
(a) Survival analyses of PTCL subtypes divided according to histopathology diagnosis. (b) Survival analyses of PTCL subtypes divided according to the minimum miRNA set classification. (c) Survival analyses of PTCL/NOS CD30+ vs ALCL/ALK− according to the minimum miRNA set classification. P-values <0.05 for log-rank Mantel–Cox test were considered statistically significant.
Moreover, we tested six examples of PTCL/NOS with CD30 expression (positivity ⩾75% of neoplastic elements), which had been sent to the Unit of Haematopathology because of the problems encountered in the distinction between PTCL/NOS and ALCL/ALK−. All patient cases were classified as PTCL/NOS in keeping with the fact that no hallmark cells were observed on the light microscopy. Most importantly, the OS of these patients turned out to be significantly worse than that of patients with ALCL/ALK− (mean OS, 851 vs 2766 days; P=0.005; Figure 6c), indicating the usefulness of the tool in this peculiar setting. Taken together, these data showed that the miRNAs tool might have clinical relevance.
Discussion
The PTCL/NOS category includes neoplasms that cannot be attributed to any of the remaining T-cell entities of the 2008 WHO Classification. It is characterized by dismal prognosis and lack of effective therapies.42, 43, 44 The pathobiology of PTCLs/NOS is still largely unknown. GEP studies have provided some hints to better understand their histogenesis, pathogenesis and prognostication, and to identify new potential therapeutic targets.3, 45, 46, 47 In particular, the molecular profile of PTCL/NOS differs from those of AITL and ALCL, and is characterized by the recurrent deregulation of genes involved in relevant functions (e.g. matrix deposition, cytoskeleton organization, cell adhesion, apoptosis, proliferation, transcription and signalling transduction).18, 19, 20, 21, 22, 23
In recent years, it has become clear that microRNAs have a pivotal role in the regulation of gene expression. Several human microRNAs identified so far show tissue or development stage-specific expression profiles, suggesting that they are integrated in the regulatory networks defining the cellular characteristics of the immune system.48, 49, 50 Our study aimed to identify the possible involvement of microRNAs in the regulation of PTCL/NOS GEP, through the analysis of the miRNomes obtained from 23 PTCL/NOS and 12 normal naive and activated CD4+ and CD8+ T-lymphocyte samples.
Only a few studies have been reported so far, dealing with miRNA expression in PTCLs, all focusing in particular on ALCLs.28, 29, 30, 31, 32, 33, 34
First, we compared naive CD4+ and CD8+ vs activated CD4+ and CD8+ T-lymphocytes to determinate an miRNA signature that could distinguish these two groups. Then, we investigated if the microRNA profile of PTCL/NOS was more similar to the one of naive or activated T-lymphocytes. As previously demonstrated by gene expression profiling,18, 40, 41 PTCL/NOS showed an miRNA expression profile closest to activated CD4+ and CD8+ T-lymphocytes.
Thus, we focused on the identification of those microRNAs differentially expressed between PTCLs/NOS and activated CD4+ and CD8+ T-lymphocytes identifying a signature consisting of 236 miRNAs.
We adopted a multistep integrated analysis of miRNA profiling data with gene expression profiling and several computational prediction tools to identify specifically miRNAs deregulated in PTCL/NOS vs normal T-lymphocytes that might affect the transcriptional profile of these tumours. Through a filtering step using microRNA Data Integration Portal database, we selected only miRNA–gene pairs inversely correlated by identifying 1184 miRNA–gene pairs, corresponding to 475 genes and 158 miRNAs. Notably, on the basis of gene ontology enrichment analysis, we found that these 475 miRNA-related genes were involved in several relevant biological processes as the intracellular signalling cascade, cell morphogenesis, cell cycle, apoptosis, regulation of cell adhesion, Ras protein signal transduction, T-cell receptor signalling, chemokine pathway and mitogen-activated protein kinase signalling. Furthermore, our findings suggest that these 158 miRNAs significantly affect the transcriptome of PTCL/NOS. To this end, we validated the 475 miRNA-related genes by GSEA in an independent set of 55 PTCLs/NOS by confirming that this gene signature was effectively enriched in PTCLs/NOS vs activated CD4+ and CD8+ T-lymphocytes.
Among the 158 miRNAs, we focused our attention on mir-132-3p that showed: (1) a higher number of mRNA targets and (2) the greatest difference in expression in PTCLs/NOS vs activated CD4+ and CD8+ T-lymphocytes. Wang et al.51 described novel miRNAs deregulated in tumours by meta-analysis of miRNA microarray, and mir-132-3p was downregulated in human tumours. Moreover, Zhang et al.52 found miR-132-3p downregulated in pancreatic cancer, and its expression was remarkably influenced by promoter methylation in pancreatic cell lines.52 Functional studies of cell transfection and subsequent GEP on transfected and untransfected cells allowed us to verify whether or not mir-132-3p had an impact on the transcriptome in PTCL/NOS. By supervised analysis, 468 differentially expressed genes were identified between transfected and untransfected cells. To evaluate the relevance of such gene signature, we applied GSEA to 78 PTCL/NOS primary tumours and found that they were enriched in these genes.
These data highlight that miR-132-3p modulation has a significant impact on PTCL/NOS transcriptome. More in general, they further stress that PTCL/NOS is a neoplasm characterized by a complex array of molecular interactions at the genome, transcriptome and proteome levels, which eventually have an important role in the process of lymphomagenesis. When the signature obtained upon miR-132-3p modulation was compared with that discriminating PTCL/NOS and normal T cells, we found nine specific genes (ADRA2A, ANKS1B, FRMPD1, FZD4, GRM3, KIRREL, NCOA7, PON1 and SPARC) that overlapped. This is reasonably consistent with the potential impact of a single miRNA, although a formal biochemical validation is warranted to confirm possible direct interactions.
Another aim of this study was to identify a specific set of miRNAs that might be used for the differential diagnosis between PTCL/NOS on the one hand and AITL and ALCL/ALK− on the other hand, which may be of remarkable clinical interest and not always straightforward based on the conventional criteria.53 In this regard, a large international study reported that up to 30% of PTCLs remain unclassified.54 Recently, our group showed that PTCL/NOS, AITL and ALCL/ALK− can be distinguish based on their GEP.7 Accordingly, we tried to develop a similar approach aiming to distinguish these PTCL subtypes on the basis of miRNA profiles. Previously, Liu et al.34 provided the basic evidence that 11 miRNAs could discriminate ALCL/ALK− from PTCL/NOS and AITL, although they did not test the potential diagnostic accuracy of their observation. Of note, the signature we identified discriminating PTCL/NOS and ALCL/ALK− (68 miRNA) only partially overlapped (3/68) with the one of Liu et al.,34 this being expected as the comparison did not include ALCL/ALK+ and AITL. We could confirm the previously reported differential expression of miR-155 between ALCL/ALK+ and ALCL/ALK−,28 although our analysis did not specifically focus on ALCL/ALK+. In our series, miRNA profiling was able to discriminate AITL and ALK− ALCL from PTCL/NOS with a specific signature of 35 miRNAs and 68 miRNAs, respectively. By linear discriminant analysis, we could restrict the number of miRNAs needed for the distinction of PTCL/NOS from ALCL/ALK− and AITL to five and eight, respectively. These sets of miRNAs were confirmed in an independent set of PTCLs by qRT-PCR, an assay that can be easily performed in most if not all molecular laboratories at the time of diagnosis. After the application of linear discriminant function, we obtained a remarkable efficiency, with an overall diagnostic accuracy of 83.54%.
Our findings propose the use of a minimum set of miRNAs as an additional tool in the diagnostic workup of nodal PTCLs, which can integrate the morphology and phenotype.
Notably, this set was also provided with prognostic relevance by distinguishing groups of tumours characterized by significantly different outcomes.
Finally, we focused on a small series of CD30+ PTCLs/NOS, which could not be easily classified according to the conventional criteria (ALCL/ALK− vs CD30+ PTCL/NOS). Based on their miRNA profile, they were all included among PTCLs/NOS.
Interestingly, although the number is relatively small, they revealed a much more aggressive clinical course than ALCLs/ALK−. These findings underline the usefulness of innovative tools, which contribute to the clearcut distinction between ALCL/ALK− and CD30+ PTCL/NOS, two conditions provided with significantly different prognostic impact. Conversely, the difference recorded between PTCL/NOS and AITL was quite surprising and probably reflected the limited number of AITL cases included.
Recently, we described a gene signature with diagnostic and prognostic potential.7 We then compared the results obtained with the miRNA-based classifier to the previous gene-based one. We found 97.5% concordance. Therefore, we considered them equivalent on the clinical/diagnostic point of view. As the miRNA classifier is technically easier to be applied, this may represent a step forward in routine diagnostics.
In conclusion, our study identified miRNA signatures capable to differentiate efficiently PTCL/NOS from activated CD4+ and CD8+ T-lymphocytes. In this setting, miR-132-3p turned out to be an important modulator of PTCL/NOS transcriptome. Finally, a set of miRNAs was developed that is applicable to FFPE tissue samples by improving the differential diagnosis between PTCL/NOS on the one hand and AITL and ALCL/ALK− on the other.
Acknowledgments
This work was supported by Centro Interdipartimentale per la Ricerca sul Cancro ‘G Prodi', BolognAIL, AIRC (IG4987; 5xMille N.10007; IG 2013 N.14355), RFO (Professor Pileri and Professor Piccaluga) and FIRB Futura 2011 RBFR12D1CB (Professor Piccaluga).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Blood Cancer Journal website (http://www.nature.com/bcj)
AIRC 5xMille consortium ‘Genetics-driven targeted management of lymphoid malignancies':
Robin Foà, Filippo Berardelli, Brunangelo Falini, Giorgio Inghirami, Gianluca Gaidano, Stefano A Pileri.
The European T-Cell Lymphoma Study Group: Italy: Giorgio Inghirami, Domenico Novero and Roberto Piva (Azienda Ospedaliera Città della Salute e della Scienza di Torino, University of Torino); Marco Chilosi and Alberto Zamó (University of Verona); Fabio Facchetti and Silvia Lonardi (University of Brescia); Anna De Chiara and Franco Fulciniti (National Cancer Institute, Napoli); Claudio Doglioni, Andrés Ferreri and Maurilio Ponzoni (San Raffaele Institute, Milano); Claudio Agostinelli, Pier Paolo Piccaluga and Stefano A Pileri (University of Bologna); Brunangelo Falini, Stefano Ascani, and Enrico Tiacci (University of Perugia); Belgium: Peter Van Loo, Thomas Tousseyn and Christiane De Wolf-Peeters (University of Leuven); Germany: Eva Geissinger and Andreas Rosenwald and Hans-Konrad Müller-Harmelink (University of Wuerzburg); Martin Leo Hansmann and Sylvia Hartmann (University of Frankfurt); Spain: Miguel Angel Piris and Maria E Rodriguez (Hospital Universitario Marqués de Valdecilla, IFIMAV, Santander and Instituto de Investigaciones Biomédicas Alberto Sols, CSIC-UAM, Madrid).
Supplementary Material
References
- 1Jaffe ES, Harris NL, Stein H, Campo E, Pileri SA, Swerdlow SH et al. Introduction and overview of the classification of the lymphoid neoplasms. In Swerdlow S, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H et al. (eds) WHO Classification of Tumors of Hematopoietic and Lymphoid Tissues, 4th edn. IARC Lyon, France, 2008, pp 158–166. [Google Scholar]
- 2Dyer MJS, Siebert R. Peripheral T-cell non-Hodgkin's lymphoma NOS: naming of parts. Leukemia 2006; 20: 208–209. [DOI] [PubMed] [Google Scholar]
- 3Pileri SA, Weisenburger DD, Sng I, Jaffe ES, Ralfkiaer E, Nakamura S et al. Peripheral T-cell lymphoma, not otherwise specified. In Swerdlow S, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H et al. (eds) WHO Classification of Tumors of Hematopoietic and Lymphoid Tissues, 4th edn. IARCLyon, France, 2008, pp 306–308. [Google Scholar]
- 4Went P, Agostinelli C, Gallamini A, Piccaluga PP, Ascani S, Sabattini E et al. Marker expression in peripheral T-cell lymphoma: a proposed clinical–pathologic prognostic score. J Clin Oncol 2006; 24: 2472–2479. [DOI] [PubMed] [Google Scholar]
- 5Mak V, Hamm J, Chhanabhai M, Shenkier T, Klasa R, Sehn LH et al. Survival of patients with peripheral T-cell lymphoma after first relapse or progression: spectrum of disease and rare long-term survivors. J Clin Oncol 2013; 16: 1970–1976. [DOI] [PubMed] [Google Scholar]
- 6Cuadros M, Dave SS, Jaffe ES, Honrado E, Milne R, Alves J et al. Identification of a proliferation signature related to survival in nodal peripheral T-cell lymphomas. J Clin Oncol. 2007; 25: 3321–3329. [DOI] [PubMed] [Google Scholar]
- 7Piccaluga PP, Fuligni F, De Leo A, Bertuzzi C, Rossi M, Bacci F et al. Molecular profiling improves classification and prognostication of nodal peripheral T-cell lymphomas: results of a phase III diagnostic accuracy study. J Clin Oncol 2013; 31: 3019–3025. [DOI] [PubMed] [Google Scholar]
- 8Roncolato F, Gazzola A, Zinzani PL, Pileri SA, Piccaluga PP. Targeted molecular therapy in peripheral T-cell lymphomas. Expert Rev Hematol 2011; 4: 551–562. [DOI] [PubMed] [Google Scholar]
- 9Corradini P, Tarella C, Zallio F, Dodero A, Zanni M, Valagussa P et al. Long-term follow-up of patients with peripheral T-cell lymphomas treated up-front with high-dose chemotherapy followed by autologous stem cell transplantation. Leukemia 2006; 20: 1533–1538. [DOI] [PubMed] [Google Scholar]
- 10Zettl A, Rudiger T, Konrad MA, Chott A, Simonitsch-Klupp I, Sonnen R et al. Genomic profiling of peripheral T-cell lymphoma, unspecified, and anaplastic large T-cell lymphoma delineates novel recurrent chromosomal alterations. Am J Pathol 2004; 164: 1837–1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11Feldman AL, Law M, Remstein ED, Macon WR, Erickson LA, Grogg KL et al. Recurrent translocations involving the IRF4 oncogene locus in peripheral T-cell lymphomas. Leukemia 2009; 23: 574–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12Thorns C, Bastian B, Pinkel D, Roydasgupta R, Fridlyand J, Merz H et al. Chromosomal aberrations in angioimmunoblastic T-cell lymphoma and peripheral T-cell lymphoma unspecified: a matrix-based CGH approach. Genes Chromosomes Cancer 2007; 46: 37–44. [DOI] [PubMed] [Google Scholar]
- 13Renedo M, Martinez-Delgado B, Arranz E, Garcia M J, Urioste M, Martinez-Ramirez A et al. Chromosomal changes pattern and gene amplification in T cell non-Hodgkin's lymphomas. Leukemia 2001; 10: 1627–1632. [DOI] [PubMed] [Google Scholar]
- 14Hartmann S, Gesk S, Scholtysik R, Kreuz M, Bug S, Vater I et al. High resolution SNP array genomic profiling of peripheral T cell lymphomas, not otherwise specified, identifies a subgroup with chromosomal aberrations affecting the REL locus. Br J Haematol 2010; 148: 402–412. [DOI] [PubMed] [Google Scholar]
- 15Streubel B, Vinatzer U, Willheim M, Raderer M, Chott A. Novel t(5;9)(q33;q22) fuses ITK to SYK in unspecified peripheral T-cell lymphoma. Leukemia 2006; 20: 313–318. [DOI] [PubMed] [Google Scholar]
- 16Fujiwara S-I, Yamashita Y, Nakamura N, Choi Y L, Ueno T, Watanabe H et al. High-resolution analysis of chromosome copy number alterations in angioimmunoblastic T-cell lymphoma and peripheral T-cell lymphoma, unspecified, with single nucleotide polymorphism-typing microarrays. Leukemia 2008; 22: 1891–1898. [DOI] [PubMed] [Google Scholar]
- 17Martinez-Delgado B, Melendez B, Cuadros M, Alvarez J, Castrillo JM, Ruiz De La Parte A et al. Expression profiling of T-cell lymphomas differentiates peripheral and lymphoblastic lymphomas and defines survival related genes. Clin Cancer Res 2004; 10: 4971–4982. [DOI] [PubMed] [Google Scholar]
- 18Piccaluga PP, Agostinelli C, Califano A, Rossi M, Basso K, Zupo S et al. Gene expression analysis of peripheral T cell lymphoma, unspecified, reveals distinct profiles and new potential therapeutic targets. J Clin Invest 2007; 117: 823–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19Piva R, Agnelli L, Pellegrino E, Todoerti K, Grosso V, Tamagno I et al. Gene expression profiling uncovers molecular classifiers for the recognition of anaplastic large-cell lymphoma within peripheral T-cell neoplasms. J Clin Oncol 2010; 28: 1583–1590. [DOI] [PubMed] [Google Scholar]
- 20Iqbal J, Weisenburger DD, Greiner TC, Vose JM, McKeithan T, Kucuk C et al. Molecular signatures to improve diagnosis in peripheral T-cell lymphoma and prognostication in angioimmunoblastic T-cell lymphoma. Blood 2010; 115: 1026–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21De Leval L, Rickman DS, Thielen C, Reynies Ad, Huang YL, Delsol G et al. The gene expression profile of nodal peripheral T-cell lymphoma demonstrates a molecular link between angioimmunoblastic T-cell lymphoma (AITL) and follicular helper T (TFH) cells. Blood 2007; 109: 4952–4963. [DOI] [PubMed] [Google Scholar]
- 22Ballester B, Ramuz O, Gisselbrecht C, Doucet G, Loï L, Loriod B et al. Gene expression profiling identifies molecular subgroups among nodal peripheral T-cell lymphomas. Oncogene 2006; 25: 1560–1570. [DOI] [PubMed] [Google Scholar]
- 23Iqbal J, Wright G, Wang C, Rosenwald A, Gascoyne RD, Weisenburger DD et al. Gene expression signatures delineate biological and prognostic subgroups in peripheral T-cell lymphoma. Blood 2014; 123: 2915–2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24Agnelli L, Mereu E, Pellegrino E, Limongi T, Kwee I, Bergaggio E et al. Identification of a 3-gene model as a powerful diagnostic tool for the recognition of ALK-negative anaplastic large-cell lymphoma. Blood 2012; 120: 1274–1281. [DOI] [PubMed] [Google Scholar]
- 25Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004; 116: 281–297. [DOI] [PubMed] [Google Scholar]
- 26Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA 2006; 103: 2257–2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D et al. MicroRNA expression profiles classify human cancers [letter]. Nature 2005; 435: 834–838. [DOI] [PubMed] [Google Scholar]
- 28Merkel O, Hamacher F, Laimer D, Sifft E, Trajanoski Z, Scheideler M et al. Identification of differential and functionally active miRNAs in both anaplastic lymphoma kinase (ALK)+ and ALK− anaplastic large-cell lymphoma. Proc Natl Acad Sci USA 2011; 107: 16228–16233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29Dejean E, Renalier MH, Foisseau M, Agirre X, Joseph N, de Paiva GR et al. Hypoxia-microRNA-16 downregulation induces VEGF expression in anaplastic lymphoma kinase (ALK)-positive anaplastic large-cell lymphomas. Leukemia 2011; 25: 1882–1890. [DOI] [PubMed] [Google Scholar]
- 30Matsuyama H, Suzuki HI, Nishimori H, Noguchi M, Yao T, Komatsu N et al. miR-135b mediates NPM-ALK-driven oncogenicity and renders IL-17-producing immunophenotype to anaplastic large cell lymphoma. Blood 2011; 118: 6881–6892. [DOI] [PubMed] [Google Scholar]
- 31Desjobert C, Renalier MH, Bergalet J, Dejean E, Joseph N, Kruczynski A et al. MiR-29a down-regulation in ALK-positive anaplastic large cell lymphomas contributes to apoptosis blockade through MCL-1 overexpression. Blood 2011; 117: 6627–6637. [DOI] [PubMed] [Google Scholar]
- 32Spaccarotella E, Pellegrino E, Ferracin M, Ferreri C, Cuccuru G, Liu C et al. STAT3-mediated activation of microRNA cluster 17~92 promotes proliferation and survival of ALK-positive anaplastic large cell lymphoma. Haematologica 2014; 99: 116–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33Yan Z-X, Wu L-L, Xue K, Zhang Q-L, Guo Y, Romero M et al. MicroRNA187 overexpression is related to tumor progression and determines sensitivity to bortezomib in peripheral T-cell lymphoma. Leukemia 2014; 28: 880–887. [DOI] [PubMed] [Google Scholar]
- 34Liu C, Iqbal J, Teruya-Feldstein J, Shen Y, Dabrowska MJ, Dybkaer K et al. MicroRNA expression profiling identifies molecular signatures associated with anaplastic large cell lymphoma. Blood 2013; 122: 2083–2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35April C, Klotzle B, Royce T, Wickham-Garcia E, Boyaniwsky T, Izzo J et al. Whole-genome gene expression profiling of formalin-fixed, paraffin-embedded tissue samples. PloS One 2009; 4: e8162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36Waddell N, Cocciardi S, Johnson J, Healey S, Marsh A, Riley J et al. Gene expression profiling of formalin-fixed, paraffin-embedded familial breast tumours using the whole genome-DASL assay. J Pathol 2010; 221: 452–461. [DOI] [PubMed] [Google Scholar]
- 37Shirdel EA, Xie W, Mak TW, Jurisica I. NAViGaTing the micronome-using multiple microRNA prediction databases to identify signalling pathway-associated microRNAs. PLoS One 2011; 6: e17429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 2005; 102: 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39Kaplan E, Meier P. Non-parametric estimation from incomplete observation. JAMA 1958; 58: 457–481. [Google Scholar]
- 40Piccaluga PP, Agostinelli C, Tripodo C, Gazzola A, Bacci F, Sabattini E et al. Peripheral T-cell lymphoma classification: the matter of cellular derivation. Expert Rev Hemat 2011; 4: 415–425. [DOI] [PubMed] [Google Scholar]
- 41Piccaluga PP, Agostinelli C, Califano A, Carbone A, Fantoni L, Ferrari S et al. Gene expression analysis of angioimmunoblastic lymphoma indicates derivation from T follicular helper cells and vascular endothelial growth factor deregulation. Cancer Res 2007; 67: 10703–10710. [DOI] [PubMed] [Google Scholar]
- 42Wilcox RA, Sun DX, Novak A, Dogan A, Ansell SM, Feldman AL. Inhibition of Syk protein tyrosine kinase induces apoptosis and blocks proliferation in T-cell non-Hodgkin's lymphoma cell lines. Leukemia 2010; 24: 229–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43Savage KJ. Aggressive peripheral T-cell lymphomas (specified and unspecified types). Hematology Am Soc Hematol Educ Program 2005, 267–277. [DOI] [PubMed]
- 44Feldman AL, Sun DX, Law ME, Novak AJ, Attygalle AD, Thorland EC et al. Overexpression of Syk tyrosine kinase in peripheral T-cell lymphomas. Leukemia 2008; 22: 1139–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45Pileri SA, Piccaluga PP. New molecular insights into peripheral T cell lymphomas. J Clin Invest 2012; 122: 3448–3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46Vose J, Armitage J, Weisenburger D. International peripheral T-cell and natural killer/T-cell lymphoma study: pathology findings and clinical outcomes. J Clin Oncol 2008; 26: 4124–4130. [DOI] [PubMed] [Google Scholar]
- 47Zhao W-L. Targeted therapy in T-cell malignancies: dysregulation of the cellular signaling pathways. Leukemia 2010; 24: 13–21. [DOI] [PubMed] [Google Scholar]
- 48Lodish HF, Zhou B, Liu G, Chen CZ. Micromanagement of the immune system by microRNAs. Nat Rev Immunol 2008; 8: 120–130. [DOI] [PubMed] [Google Scholar]
- 49O'Connell RM, Rao DS, Chaudhuri AA, Baltimore D. Physiological and pathological roles for microRNAs in the immune system. Nat Rev Immunol 2010; 10: 111–122. [DOI] [PubMed] [Google Scholar]
- 50Suzuki H I, Matsuyama H, Noguchi M, Yao T, Komatsu N, Mano H et al. Computational dissection of distinct microRNA activity signatures associated with peripheral T cell lymphoma subtypes [letter]. Leukemia 2013; 27: 2107–2111. [DOI] [PubMed] [Google Scholar]
- 51Wang W, Peng B, Wang D, Ma X, Jiang D, Zhao J et al. Human tumor microRNA signatures derived from large-scale oligonucleotide microarray datasets. Int J Cancer 2011; 129: 1624–1634. [DOI] [PubMed] [Google Scholar]
- 52Zhang S, Hao J, Xie F, Hu X, Liu C, Tong J et al. Downregulation of miR-132 by promoter methylation contributes to pancreatic cancer development. Carcinogenesis 2011; 32: 1183–1189. [DOI] [PubMed] [Google Scholar]
- 53Swerdlow S, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H et al. (eds). WHO Classification of Tumours of Hematopoietic and Lymphoid Tissues, 4th edn. IARC: Lyon, France, 2008. [Google Scholar]
- 54Federico M, Bellei M, Pesce EA, Zucca E, Pileri S, Montoto S et al.. T-cell project: an international, prospective, observational study of patients with aggressive peripheral T-cell lymphoma—analysis of first 524 patients. Ann Oncol 2011; 22(Suppl. 4)iv164–iv167(abstract 241). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.