Nuclear pore complexes (NPCs) are composed of several copies of proteins called nucleoporins (Nups). NPCs penetrate the nuclear envelope (NE) and regulate the nucleocytoplasmic trafficking of macromolecules, and NPC components influence genome functions in a transport-independent manner. Here, Ibarra and Hetzer discuss genome-related functions of Nups and their impact on essential DNA metabolism processes such as transcription, chromosome duplication, and segregation.
Keywords: nuclear genome, nuclear envelope, nuclear pore complex, nucleoporin, transcription control, mitosis, DNA damage
Abstract
Nuclear pore complexes (NPCs) are composed of several copies of ∼30 different proteins called nucleoporins (Nups). NPCs penetrate the nuclear envelope (NE) and regulate the nucleocytoplasmic trafficking of macromolecules. Beyond this vital role, NPC components influence genome functions in a transport-independent manner. Nups play an evolutionarily conserved role in gene expression regulation that, in metazoans, extends into the nuclear interior. Additionally, in proliferative cells, Nups play a crucial role in genome integrity maintenance and mitotic progression. Here we discuss genome-related functions of Nups and their impact on essential DNA metabolism processes such as transcription, chromosome duplication, and segregation.
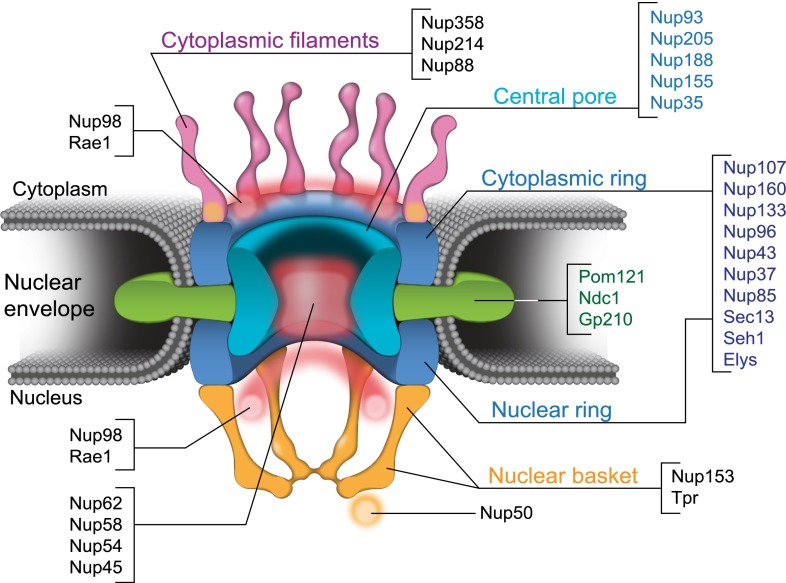
Eukaryotic chromosomes are enclosed by the double-lipid bilayer of the nuclear envelope (NE), which is perforated at multiple sites by nuclear pore complexes (NPCs). NPCs are large protein structures mediating macromolecular trafficking between the nuclear and cytoplasmic compartments. Each NPC is composed of multiple copies of ∼30 different subunits, called nucleoporins (Nups), arranged in octagonal rotational symmetry around the central transport channel. In vertebrates, this molecular assembly has a molecular weight of ∼120 MDa and can be structurally divided into a core scaffold formed by the Nup93/Nup205 complex and two rings composed of the Nup107/Nup160 complex flanking the former in the vertical plane (Fig. 1; D’Angelo and Hetzer 2008; Hoelz et al. 2011). Attached to the inner side of this structure are the so-called FG-Nups, which contain natively unfolded phenylalanine–glycine (FG) domains such as the Nup62 complex, establish the selective permeability barrier, and regulate receptor-mediated translocation through the NPC (Wente and Rout 2010; Grossman et al. 2012). Somewhat surprising for a membrane-embedded complex, there are only three known transmembrane Nups (Pom121, Ndc1, and Gp210), which link the NPC to the NE (Fig. 1). The nuclear pore is completed by peripheral filamentous protein complexes that extend into the cytoplasm and nucleoplasm, referred to as the cytoplasmic filaments and nuclear basket, respectively (D’Angelo and Hetzer 2008; Hoelz et al. 2011). Cytoplasmic filaments are mainly formed by Nup358/RanBP2, Nup214, and Nup88, while the nuclear basket is composed of Nup153 and Tpr (Fig. 1; for Nup othologs, see Rothballer and Kutay 2012).
Figure 1.
NPC structure and molecular composition. Representation and predicted molecular composition of the vertebrate NPC. For the corresponding orthologs in other model organisms, see Rothballer and Kutay (2012). The NPC core scaffold and its components are represented in dark blue and light blue, the transmembrane subunits are shown in green, the cytoplasmic structures are represented in magenta, and the basket is shown in orange. Peripheral Nups showing higher mobility by FRAP, and other approaches are depicted in black.
The selective access of regulatory factors into the nucleus and export of specific RNA molecules mediated by the NPC is required for the accurate progression of most major cellular processes. However, our perception of the NPC components is rapidly evolving, as accumulating evidence indicates that they can also directly impact DNA metabolism by genome-related functions (Liang and Hetzer 2011). Among these, one of the most remarkable and well-conserved roles of Nups is to associate with specific target genes to regulate their transcriptional activity (Casolari et al. 2004; Taddei et al. 2006; Capelson et al. 2010; Kalverda and Fornerod 2010; Vaquerizas et al. 2010; Liang et al. 2013; Pascual-Garcia and Capelson 2014). Importantly, these gene regulatory functions are not limited to NE-embedded NPCs in metazoans, as peripheral Nups can perform the same tasks in the nuclear interior (Capelson et al. 2010; Kalverda et al. 2010). Although initial reports mainly highlighted the function of Nups in transcriptional activation, recent evidence supports NPC components as general transcriptional regulators associated with both transcriptionally permissive and repressive activities.
It is still uncertain how NPC components could modulate gene expression, but it has been proposed that Nups might mediate this function by influencing the chromatin landscape. Indeed, the NE, as the largest physical scaffold of the nucleus, has been historically suggested to influence the nonrandom organization of the eukaryotic chromosomes and thus their functions (Akhtar and Gasser 2007; Mekhail and Moazed 2010; Van de Vosse et al. 2011). High-resolution images of the nuclear structure of different mammalian cell types revealed that the NE is underlain by the nuclear lamina, an intermediate filament structure that generally associates with heterochromatin, interrupted by decondensed euchromatin patches under the NPCs (Belmont et al. 1993; Schermelleh et al. 2008). These observations suggested that NPC components might influence chromosome compartmentalization, chromatin structure, or chromatin organization, generating differentially regulated regions.
NPC number and structure are coordinated with cell cycle progression, suggesting that Nups might play additional roles in genome propagation and maintenance in proliferating cells. The most dramatic structural change of the metazoan NPC takes place at the onset of mitosis, when individual Nups become heavily phosphorylated, and NPCs are disassembled (Hetzer 2010; Doye 2011; Laurell and Kutay 2011). Subsequently, NPC subunits relocate to mitotic substructures, primarily the kinetochores, and many of them display mitotic-specific functions in spindle assembly or anaphase onset, for example (Wozniak et al. 2010). During mitotic exit, NPCs reassemble into the reforming NE, and their number is roughly doubled during interphase (Doucet et al. 2010). In particular, NPC components also influence the dynamics of S phase and contribute to genome integrity maintenance, avoiding the accumulation of DNA lesions (Loeillet et al. 2005; Nagai et al. 2008; Bermejo et al. 2011). Here we review recent literature highlighting functions of Nups in transcriptional regulation and chromatin organization, genome duplication and preservation, and chromosome segregation.
Gene expression regulation by NPC components
Transcriptional regulation in yeast
The association of NPCs with decondensed, presumably active, chromatin regions was initially interpreted as a mechanism to couple transcription of active genes with mRNA export, the so called “gene gating” hypothesis (Blobel 1985). According to this idea, gene tethering to the NPCs would result in mRNA nuclear export and influence mRNA production as well. Anchoring of active genes to the NPCs has been shown for a large number of yeast genes. Some of the best-characterized examples are inducible genes, whose expression can be stimulated by environmental changes, and their activation correlates with subnuclear relocalization. For instance, temperature or nutrient shifts induce the expression of genes such as HSP104, HXK1, INO1, SUC2, or the GAL genes, among others, which are specifically activated and recruited to the NPC (Brickner and Walter 2004; Casolari et al. 2004; Cabal et al. 2006; Dieppois et al. 2006; Taddei et al. 2006; Sarma et al. 2007; Light et al. 2010). Tethering to the NPC is thought to provide optimal transcriptional activity of these genes and can be facilitated by several NPC substructures, primarily the basket components, and assisted by the transcriptional activator SAGA and the mRNA export machinery (Rodriguez-Navarro et al. 2004; Cabal et al. 2006; Taddei et al. 2006; Light et al. 2010; Dieppois and Stutz 2010; García-Oliver et al. 2012). Experiments using Nup2 fused to micrococal nuclease indicated that Nups tend to bind the promoter regions of inducible genes, suggesting that Nup–promoter interactions might represent an early event in the transcriptional activation of a subset of genes such as the GAL genes (Schmid et al. 2006). The recruitment of inducible genes to the NPC also relies on the presence of specific DNA elements within the promoter region, called gene recruitment sequences (GRSs). Genes that associate with Nup2, Mlp1, Mlp2, Nup60, or Nup116 show enrichment of GRSs at their promoters (Ahmed et al. 2010; Light et al. 2010). To date, three different classes of GRSs have been identified as necessary and sufficient to regulate association with the NPC. Mutating the GRSs present in the promoter area of the inducible INO1 or TSA2 genes blocks their interaction with the NPC, and, when artificially inserted in typically nucleoplasmic loci, GRS-containing regions can relocate them to the NPC (Ahmed and Brickner 2010; Ahmed et al. 2010; Light et al. 2010). Interestingly, this mechanism might be conserved in other species, as GRS-mediated targeting to the NPC was also observed in Schizosaccharomyces pombe (Ahmed et al. 2010), and the HSP-16.2 promoter seems sufficient to anchor chromatin at the nuclear periphery in Caenorhabditis elegans (Rohner et al. 2013), although more precise technical approaches will be necessary to determine whether particular DNA motifs could mediate Nup association with chromatin in multicellular organisms.
The role of NPCs in transcriptional regulation does not seem to be restricted to inducible genes but extends to constitutively active genes. Identification of Nup-binding sites throughout the genome by chromatin immunoprecipitation (ChIP) coupled to DNA microarray analysis (ChIP–chip) suggested that NPC basket components Nup2, Nup60, and Mlp1/2 along with scaffold components Nic96 and Nup116 occupy regions with high transcriptional activity (Casolari et al. 2004, 2005). Even if some of these NPC–chromatin interactions might require further validation by imaging, highly expressed ribosomal and glycolysis-related genes are associated to the NPC, and Nup2 was detected to bind the promoter of constitutively active genes (Casolari et al. 2004, 2005; Schmid et al. 2006; Yoshida et al. 2010; Van de Vosse et al. 2013). Altogether, these observations indicate that NPC tethering correlates with transcriptional activity (Fig. 2).
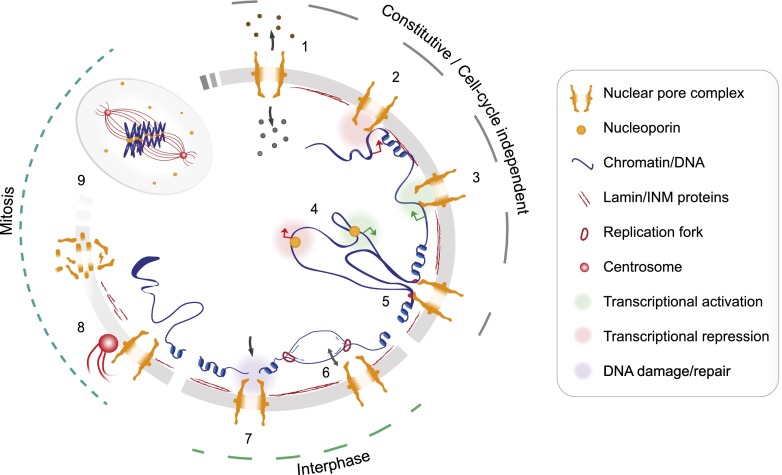
Figure 2.
How NPC components affect genome functions. Proposed roles for NPC components in eukaryotic cells regardless of the cell cycle activity (constitutive roles/cell cycle-independent) reported in cycling cells (interphase) and during mitosis (mitosis). (1) NPCs mediate nucleocytoplasmic transport in every eukaryotic nucleus. (2–4) Nups have been proposed to participate in transcriptional regulation, promoting either transcriptional activation (green circles) or repression (red circles) independently of the proliferative status of the cell. Gene expression regulation by Nups takes place mainly at the NE-embedded NPC in yeast (2,3), while in metazoans (4), a subset of mobile Nups (see also Fig. 1) can relocate to the nuclear interior to perform the same gene regulatory functions. (5) In both compartments, Nups might influence chromatin structure or organization of the tethered regions in collaboration with other factors (pink circles). (6) In proliferating cells, NPC–chromatin contacts must be regulated (arrow) during S phase to avoid replication fork collapse. (7) NPC components might facilitate the repair of a subset of persistent DNA lesions recruited to the NPC vicinity in yeast. (8) In organisms with open mitosis, NPC components assist NE breakdown and centrosome migration. (9) After NPC disassembles, some NPC components accumulate at the kinetochores, while other remain dispersed throughout the mitotic cell. By ensuring the localization and function of key mitotic components, Nups promote accurate spindle assembly, mitotic progression, and faithful chromosome segregation.
Association of NPC components with genes may result in negative regulation of transcriptional activity as well, extending the role of Nups as multifaceted players in gene expression control. In Saccharomyces cerevisiae, subtelomeric regions and silent mating type loci represent repressed regions. The ability of Nups to influence the efficient silencing and localization of these regions remains under debate and requires further clarification (Galy et al. 2000; Feuerbach et al. 2002; Hediger and Gasser 2002; Hediger et al. 2002; Gartenberg et al. 2004; Therizols et al. 2006; Ruben et al. 2011; Van de Vosse et al. 2013). However, recent reports have proposed that a subset of scaffold components, including Nup133, Nup84, and Nup170, might participate in the efficient silencing of subtelomeric regions (Therizols et al. 2006; Van de Vosse et al. 2013). Negative gene regulation has also been observed for the ribosomal protein genes (which appear to be associated with the yeast NPC core components, and down-regulation of Nup170 increases the expression of these genes [Yoshida et al. 2010; Van de Vosse et al. 2013]) as well as inducible genes (as binding of GAL1 to Nup1 down-regulates its transcriptional activity [Green et al. 2012], and Nup120 and Nup133 were reported to facilitate SUC2 repression [Sarma et al. 2011]). Therefore, NPC components can promote transcriptional repression as well and, as such, could be considered focal points for transcriptional regulation (Fig. 2).
Gene expression regulation in metazoans
While in yeast most, if not all, gene–Nup interactions occur at the NE, the situation is more complex in metazoans, where Nups are not spatially restricted to the NPC but can interact with the genome at sites that are not associated with the NE. Fluorescence recovery after photobleaching (FRAP) experiments in mammalian cells show strikingly different residence times of Nups at the NPC (Rabut et al. 2004). On the one hand, NPC core components are extremely stable, with residence times of several hours at the nuclear pore (Rabut et al. 2004) and even up to months or years in post-mitotic cells, as shown by protein turnover analyses (Fig. 1; Savas et al. 2012; Toyama et al. 2013). In contrast, most peripheral Nups show dynamic association with the NPC, with residence times ranging from seconds to minutes (Fig. 1; Rabut et al. 2004). The observed mobility of peripheral Nups is consistent with their ability to function away from the NPC as soluble regulatory modules. Interestingly, the mobility of Nups like Nup153, Nup98, or Nup50 is connected to RNA polymerase II (Pol II)-mediated transcriptional activity, indicating that off-pore Nups might actively participate in transcriptional regulation (Griffis et al. 2002; Buchwalter et al. 2014). Experimental support for this hypothesis was provided in Drosophila, where several Nups, such as Nup98, Nup50, Sec13, and Nup62, were found to bind specific chromatin regions in the nuclear interior (Capelson et al. 2010; Kalverda et al. 2010). In particular, DNA adenine methyltransferase identification (DamID), ChIP, and immunohistochemical approaches showed that these mobile Nups interact with transcriptionally active chromatin regions in the nuclear interior (Capelson et al. 2010; Kalverda et al. 2010). Depletion or overexpression of Nup98, for instance, deregulates the expression of hundreds of genes in the nuclear interior and at the NPC without altering their subnuclear localization (Capelson et al. 2010; Kalverda et al. 2010). Additionally, ChIP–chip analyses in Drosophila showed that nuclear basket components Nup153 and Mtor/Tpr associate with actively expressed regions enriched of transcriptionally permissive chromatin marks such as acetylated H3K16 and high RNA Pol II occupancy (Vaquerizas et al. 2010). Silencing of basket Nups deregulates, negatively, the expression of thousands of genes throughout the nucleus (Vaquerizas et al. 2010).
Moreover, indicative of the multidimensional relationship between Nups and gene expression regulation, metazoan Nups are also related to transcriptional silencing. For example, Nup88 associates with transcriptionally inactive regions in the nuclear interior of Drosophila salivary gland cells (Capelson et al. 2010). Based on the only study performed in human cells, ChIP–chip experiments suggested that Nup93–chromatin interactions might be enriched at repressive marks such as H3K27me3 or H3K9me3 in HeLa S3 cells (Brown et al. 2008). However, direct association with heterochromatin should be carefully interpreted, as this study used double cross-linking. This is particularly important in this case, as metazoan NPCs are usually devoid of heterochromatin (Schermelleh et al. 2008). Nevertheless, consistent with a functional role of Nups negatively regulating transcriptional activity, the Nup93 ortholog in C. elegans, NPP-13, was found in the proximity of tRNA genes, and its depletion increased the expression of these genes (Ikegami and Lieb 2013). Altogether, these observations support an evolutionarily conserved role of Nups in global gene expression regulation independently of the subnuclear localization of the transcriptional event and overcoming the restricted access of chromatin regions to the NPC in multicellular organisms (Fig. 2).
It is still unknown how Nups are recruited to intranuclear sites and whether Nup–chromatin interactions at the NPC or off-NPC might lead to differential transcriptional regulation events. However, it is possible that Nups could play specialized functions in these two different scenarios. For example, while a mutant form of Drosophila Nup98 restricted to the nucleoplasmic space shows preferential binding for actively transcribed genes, NPC-bound Nup98 does not (Kalverda et al. 2010). This suggests that, at least in some cases, Nup-mediated gene regulation may be uncoupled from the NPC in metazoans. In differentiating human cells, Nup98 participates in gene activation at the NPC, and fully active genes are then released to the nuclear interior, where intranuclear Nups maintain their transcriptional levels (Liang et al. 2013). Thus, both pools of Nup98 might be essential for optimal gene expression regulation but potentially display different activities. Interestingly, Nup98, Nup62, and Nup50 were shown to co-occupy the same subset of genes in the nuclear interior in Drosophila, suggesting potential physical and functional interactions among Nups to regulate gene expression away from the NPC environment (Kalverda et al. 2010). Although continued work will be required to obtain a detailed understanding of Nup functions on-pore and off-pore, the potential role of Nups as fundamental and versatile regulators of gene expression in metazoans is already evident.
Critical gene regulatory functions of Nups are also supported by their association with diseases and cell type-specific NPC composition observed in multicellular organisms (Olsson et al. 2004; Gomez-Cavazos and Hetzer 2012). Mutations in Nups are responsible for a number of developmental disorders and cancer, including cases of acute myeloid leukemia (AML). The latter is caused by oncogenic fusions of Nup98 to a number of different DNA-binding proteins (Franks and Hetzer 2013). Interestingly, these fusion proteins retain the Nup98 FG domain, which can act as a strong regulator of gene transcription (Kasper et al. 1999; Bai et al. 2006). In addition, cardiac dysfunction has been linked to mutations in human Nup155 (Zhang et al. 2008). As mentioned above, Nup98, Nup62, Nup50, or Sec13 target genes identified in Drosphila embryonic cells and salivary glands are enriched for developmental regulators and developmentally induced genes (Capelson et al. 2010; Kalverda et al. 2010). Interestingly, Nup98 target genes in human cells change during differentiation, likely influencing specific transcriptional programs as cell identity evolves (Liang et al. 2013). Last, down-regulation of Nup50 or Gp210 abolishes the differentiation of mouse myoblasts to myotubes (D’Angelo et al. 2012; Buchwalter et al. 2014). In conclusion, considering the plasticity of Nup-mediated transcriptional control, it might be anticipated that Nups play a pivotal role in differentiation and development.
Mechanisms underlying gene expression regulation by Nups
The molecular mechanisms underlying Nup-mediated gene regulation are not well understood. Nevertheless, several studies indicate that NPC components might impact gene expression regulation by controlling the chromatin structure and accessibility of transcription factors. It has been proposed that NPCs could serve as functional hubs formed in specific chromatin loci to locally enrich the enzymatic machinery required for the multistep process leading to epigenetic reformatting and optimal transcriptional regulation of their targets. For example, transcriptional activation at the NPC involves the conserved interaction of nuclear basket components with the histone acetyltransferase SAGA, RNA export factors, and the yeast SUMO protease Ulp1 (the yeast ortholog of SENP1 and SENP2) (Rodriguez-Navarro et al. 2004; Chow et al. 2012; García-Oliver et al. 2012; Texari et al. 2013; Umlauf et al. 2013). Disruption of Ulp1 activity at the NPC or the interactions established among the NPC, SAGA, and RNA export complexes impedes the association of GAL-inducible genes with the NPC in S. cerevisiae and delocalizes the HSP70 gene in Drosophila or the HSP-16.2 locus in C. elegans away from the NE (Rodriguez-Navarro et al. 2004; Cabal et al. 2006; Kurshakova et al. 2007; Luthra et al. 2007; Rohner et al. 2013; Texari et al. 2013). It has been proposed that desumoylation events mediated by Ulp1 would be prerequisite for tethering and activation of inducible genes such as GAL1 (Texari et al. 2013). Derepressed genes would subsequently be recognized by chromatin modifiers like SAGA or other transcriptional regulators, leading to their transcriptional activation. Indeed, NPC basket components and the histone acetyltransferase SAGA co-occupy the same regions within the target genes to stimulate transcriptional activity. Nups may use a similar mechanism to promote transcriptional silencing. In relation to inducible genes, NPC components interact with the repressor Mig1, and deletion of Nup120 or Nup133 impedes the association of Mig1 with the SUC2 promoter (Sarma et al. 2011). In addition, the interaction of the RNAi machinery with chromatin at the NPC promotes silencing of stress-inducible genes in S. pombe (Woolcock et al. 2012). Last, Nup170 has been proposed to promote gene silencing and chromatin remodeling of subtelomeric regions by facilitating the function or targeting of the chromatin remodeler RSC and the Sir4 silencer (part of the SIR complex) in yeast (Van de Vosse et al. 2013). Hypothetically, similar features could be envisioned for the intranuclear Nup-bound genes, where dynamic Nups might target specific chromatin remodelers reported to interact with them, like CBP/p300 or the MSL complex (Kasper et al. 1999; Mendjan et al. 2006).
Transcriptional regulation by NPC components through chromatin structure control is also reflected by the role of Nups in transcriptional memory. Association of Nup100 in yeast or its human ortholog, Nup98, with gene promoters like INO1 or the interferon γ-inducible HLA-DRA, respectively, keeps them poised for subsequent activation events through chromatin structure alterations, including H2A.Z deposition and H3K4me2 (Brickner et al. 2007; Ahmed et al. 2010; Light et al. 2010, 2013). Interestingly, INO1 is retained at the NPC upon activation through another specific DNA sequence called memory recruitment sequence (MRS), while Nup98 interacts with HLA-DRA in the nucleoplasm, suggesting a conserved function of Nups in the regulation of chromatin structure and transcriptional memory maintenance regardless of their subnuclear localization (Light et al. 2013).
The control exerted over chromatin by NPC components to regulate gene expression further extends to higher-order chromatin organization. One form of three-dimensional (3D) chromatin architecture is gene looping, which represents another mechanism for gene expression regulation and transcriptional memory maintenance (O’Sullivan et al. 2004; Lainé et al. 2009; Tan-Wong et al. 2009, 2012). NPC basket component Mlp1 binds the promoter and 3′ end regions of the inducible gene HXK1, suggesting the formation of a loop (Tan-Wong et al. 2009). Chromosome conformation capture (3C) analyses demonstrated that Mlp1 is required for the maintenance of the looping of this gene, which is functionally important for its rapid transcriptional reactivation (Tan-Wong et al. 2009). Interestingly, ChIP–chip experiments of yeast Nups described the association of many 3′ ends to the NPC, suggesting a potentially more extensive role of the NPC components in the 5′–3′ gene looping and the stabilization of cis-chromatin interactions (Casolari et al. 2004). In addition, yeast genes present in different chromosomes but sharing the same GRS can cluster around the same area at the NPC upon activation (Brickner et al. 2012). This association is facilitated by Nup2 and mediated by specific transcription factors such as Put3, indicative of the contribution of NPCs to trans–chromatin interactions.
It has been proposed that the impact of Nups on chromatin structure and organization could originate from their role as chromatin boundary elements, defining active and silent regions. Several NPC components, and particularly Nup2, were identified to display boundary activity in yeast, isolating reporter regions from heterochromatin spreading (Ishii et al. 2002; Dilworth et al. 2005). This function might be conserved at the vertebrate NPC, as NPC–chromatin interactions are enriched in the insulator Su(hw) along the Drosophila genome (Kalverda and Fornerod 2010). In human cells, the chromatin-binding sites of the stable Nup93 show positive correlation with CTCF insulator sites, a major organizer of the human genome (Phillips and Corces 2009), in the presence of histone deacetylase inhibitor TSA (Brown et al. 2008). In addition, a role of the NPC components as chromatin boundaries has been suggested to explain the characteristic compartmentalization of the chromosomes at the nuclear periphery observed by high-resolution microscopy, where NPCs represent “heterochromatin exclusion areas” (Belmont et al. 1993; Schermelleh et al. 2008). Consistently, silencing the basket component Tpr is sufficient to allow the spreading of heterochromatin throughout the nuclear periphery in human cells (Krull et al. 2010). Altogether, these observations support a structural role for NPC components in 3D organization of the genome (Fig. 2).
In a tentative model of Nup-mediated gene regulation, Nups might impose boundaries along the chromosomes in association with other scaffolding proteins, thus compartmentalizing chromatin regions. Isolation of specific zones would favor specific chromatin interactions, bringing regulatory elements together that might be stabilized by NPC components. The ability of Nups to interact with various chromatin regulators would expose the tethered regions to the activity of specific enzymes, depending on the desired transcriptional outcome, and thus generating “repressive” and “permissive” transcriptional environments through the nucleus (Fig. 2).
DNA replication and genome integrity maintenance
Faithful replication of the genetic material during S phase is essential to ensure the integrity of the genome. The DNA replication program follows a strict temporal and spatial organization where replication origins located throughout the nuclear interior are fired early during S phase, whereas late-firing replication origins tend to accumulate at the nuclear periphery (Natsume and Tanaka 2010). Chromatin tethering and transcriptional activity taking place at the NPC-tethered regions represent a barrier to the passage of the replication fork and would increase the topological tension during DNA replication (Bermejo et al. 2011; Helmrich et al. 2013). To relieve this stress, the S-phase checkpoint machinery of S. cerevisiae, orchestrated by Mec1 and Rad53 kinases, phosphorylates Nups such as the nuclear basket component Mlp1 (Bermejo et al. 2011). Modification of this Nup would detach tethered chromatin regions from the NPC, alleviating the topological tension preventing fork collapse (Fig. 2). Deletion of Nup1 shows similar alleviation in this study, and other NPC components such as Nup153 or Nup98 have also been identified as potential targets of the vertebrate S-phase checkpoint proteins ATR/Chk1 (Matsuoka et al. 2007; Bermejo et al. 2011; Blasius et al. 2011), suggestive of a more elaborate regulation of the NPC–chromatin contacts during DNA replication. After replication fork passage, these regions should reanchor to the NPC and resume their optimal transcriptional activity. Supporting this idea, studies following the subnuclear localization of inducible genes during the yeast cell cycle show that the peripheral localization observed for these regions during G1 is prevented during S phase, when they are released into the nuclear interior, and are again anchored to the NPC in G2 in a process regulated by CDK1 phosphorylation of Nup1 (Brickner and Brickner 2011). Therefore, dynamic post-translational modifications of the NPC structure must be coordinated with cell cycle progression for accurate genome metabolism and integrity preservation.
Recent evidence links DNA replication proteins to the NPC. In Xenopus egg extracts, Elys/Mel-28, which is part of the Nup107/Nup160 complex, interacts with the MCM2–7 complex, the main component of the eukaryotic replicative helicase (Gillespie et al. 2007). This observation led the investigators to propose a potential functional coordination between NE assembly and replication origin licensing during nuclear reorganization in this organism (Gillespie et al. 2007). This interaction appears conserved in vertebrates. A mutation of the zebrafish ELYS gene reduces Mcm2 chromatin levels and sensitizes tissue progenitors to replication stress, suggesting a potential role for this Nup in the activity of dormant replication origins (Davuluri et al. 2008; Gao et al. 2011). Although functional links between NPC components and replication origin licensing or firing are unclear, the observations reported above and the strong relationship between chromatin architecture, transcriptional activity, and DNA replication reported (Chakraborty et al. 2011; Smith and Aladjem 2014; Therizols et al. 2014) make it tempting to speculate that NPC-mediated chromatin organization might affect the licensing or activity of at least a subset of replication origins present in the nuclear periphery.
DNA damage signaling and repair machineries play a fundamental role in preventing the accumulation of DNA lesions. A series of Nup mutants in yeast and metazoan cells show accumulation of DNA damage and hypersensitivity to DNA-damaging agents, suggesting their potential involvement in DNA damage response or repair pathways (Galy et al. 2000; Loeillet et al. 2005; De Souza et al. 2006; Palancade et al. 2007; Davuluri et al. 2008; Paulsen et al. 2009; Gao et al. 2011). A role of NPC components orchestrating these responses has been inferred from the particular spatial relocalization to the NE of persistent DNA lesions and collapsed replication forks in S. cerevisiae (Therizols et al. 2006; Nagai et al. 2008; Oza et al. 2009; Horigome et al. 2014). The recruitment to the NPC in particular depends on the core Nup84 complex and the checkpoint kinases Mec1 and Tel1 (Nagai et al. 2008). Interestingly, down-regulation of several subunits of the Nup107/Nup160 complex, the vertebrate ortholog of the yeast Nup84 complex, generates the accumulation of spontaneous DNA damage in human cells (Paulsen et al. 2009). Mechanistically, concentration of the SUMO machinery at the NPC could modulate proper ubiquitination levels of intervening factors and facilitate repair (Palancade et al. 2007; Nagai et al. 2008; Dou et al. 2011). Thus, the yeast NPC can also accommodate a favorable environment for DNA repair, preventing genome instability. While a subset of persistent DNA lesions relocate to the NE in the yeast nucleus, this particular behavior has not been observed in mammalian cells (Soutoglou and Misteli 2007; Nagai et al. 2010). Therefore, it will be interesting to determine whether any mobile Nup could be recruited to intranuclear damaged regions to facilitate their repair. Overall, NPC components seem to play a fundamental role in genome integrity maintenance as targets of DNA damage response pathways and even in facilitating DNA repair (Fig. 2).
The role of Nups in chromosome segregation
Early mitotic stages
Accurate segregation of sister chromatids during mitosis is critical to prevent genomic lesions and avoid the deleterious consequences of aneuploidy formation in the daughter cells (Siegel and Amon 2012). As an early stage to successfully execute mitosis, centrosomes migrate to opposite sides of the nucleus during prophase using the NE and NPCs as a sliding platform. Assisting the centrosomes in their relocation, the cytoplasmic peripheral NPC component Nup358 helps centrosome anchoring by its association with the molecular motors dynein and kinesin through the adaptor protein Bicaudal D2, which localizes at the NPC during G2 (Fig. 2; Cai et al. 2001; Payne et al. 2003; Splinter et al. 2010). More centrally located as part of the Nup107/160 complex, Nup133 also contributes to centrosome tethering, providing anchoring to the dynein/dynactin complex independently of the Nup358–Bicaudal D2-mediated pathway (Bolhy et al. 2011). In addition, silencing of the transport channel component Nup62 results in aberrant centrosome conformation with defective maturation and mitotic malfunction (Hashizume et al. 2013b).
In metazoans, nuclear compartmentalization is lost during NE breakdown (NEBD), which occurs at the prophase–prometaphase transition (Hetzer 2010). This dramatic cell remodeling process must occur in a tightly organized way to avoid fatal errors and allow proper mitotic progression. During NEBD, NPCs are rapidly disassembled in a stepwise manner (Dultz et al. 2008; Laurell and Kutay 2011). Although little is known about how this process takes place at the molecular level, it is thought to be connected to the characteristic hyperphosphorylation of NPC components during mitosis. Supporting this idea, post-translational modification of the GLFG repeats of Nup98 (one of the first Nups released from the NPC during prophase) (Lénárt et al. 2003; Dultz et al. 2008), mainly by CDK1 and NIMA-related kinases, is required for efficient NPC disassembly and NEBD kinetics (Laurell et al. 2011). Subsequently, phosphorylation-defective Nup98 mutants display delayed NPC disassembly and loss of compartmentalization (Laurell et al. 2011). Similarly, hyperphosphorylation of the transmembrane component Gp210 in C. elegans, likely by Cyclin B/Cdc2, contributes to efficient NPC disassembly and NEBD (Galy et al. 2008). In addition, Nup358 and Nup153 directly assist NEBD progression, likely by recruiting the membrane remodeler COPI coatomer complex through their zinc finger domains (Liu et al. 2003; Prunuske et al. 2006). Even if these observations implicate NPC components in NEBD orchestration, further studies would be required to clarify the events triggered by post-translational modification of NPC components in NPC disassembly, NEBD, and further mitotic events.
Spindle assembly and anaphase onset
Upon NEBD, microtubules emanating from the centrosomes bind the kinetochores aligning the chromosomes along the equatorial plate of the cell. Completion of spindle formation is monitored by the spindle assembly checkpoint (SAC), which holds anaphase onset (Musacchio and Salmon 2007). After complete disassembly of the NPC, subcomplexes such as the Nup107/160 relocate to the kinetochore, where it has been proposed to develop new functions in chromosome congression and normal spindle assembly (Belgareh et al. 2001; Loïodice et al. 2004; Zuccolo et al. 2007). Supporting this idea, Xenopus egg extracts depleted of the Nup107/160 complex show unattached sperm chromosomes, and depletion of Seh1 or Nup107 leads to impaired chromosome alignment and mitotic delay in human cells (Orjalo et al. 2006; Zuccolo et al. 2007; Platani et al. 2009; Mishra et al. 2010). Consistently, Elys/Mel-28 was identified as a dual Nup/kinetochore protein, and its depletion leads to severe defects in spindle assembly and chromosome segregation as well (Fernandez and Piano 2006; Galy et al. 2006; Rasala et al. 2006). At the molecular level, Nup107/Nup160 components and Elys were shown to recruit the microtubule nucleator γ-tubulin ring complex (γTuRC) to unattached kinetochores to drive microtubule nucleation (Mishra et al. 2010; Yokoyama et al. 2014). All of the Nups of this complex could cooperate to promote proper spindle assembly, although it has also been proposed to be a predominant role for Elys (Yokoyama et al. 2014). In addition, the Nup107/Nup160 complex might also regulate mitotic progression by controlling the spatial positioning of the chromosome passenger complex (CPC) (Platani et al. 2009). Composed of the kinase Aurora B, INCENP, Borealin, and Survivin, the localization of the mitotic regulator CPC is tightly regulated: It accumulates at the centromeres during early mitosis and relocates to the spindle mid-zone after anaphase onset and the mid-body during cytokinesis, displaying essential functions at all stages (Carmena et al. 2012). Interestingly, silencing Seh1 or Elys leads to impaired Aurora B localization, resulting in defective chromosome segregation (Platani et al. 2009; Clever et al. 2012).
A fraction of Nup358 also relocates from NPCs to kinetochores and spindle microtubules during early mitosis to play an active role in the regulation of the stability of kinetochore–microtubule interactions. This was inferred from the loss-of-function experiments in Nup358-depleted cells resulting in the appearance of unaligned chromosomes, multipolar spindles, and mitotic catastrophe (Joseph et al. 2004; Hashizume et al. 2013a). In addition, Nup358, which is a large multidomain protein that includes SUMO E3 ligase activity (Pichler et al. 2002), is also required for progression into anaphase, as it sumoylates the topoisomerase TopoIIa, promoting its targeting to the centromeres, where it is required to release the topological links during anaphase onset (Dawlaty et al. 2008). Indicative of the crucial role of Nup358 in faithful chromosome segregation, reduced levels of Nup358 lead to the appearance of aneuploidy and tumor development in mice (Dawlaty et al. 2008).
An intriguing interplay between the mitotic machinery and NPC components is the conserved interaction established between the Mad1/Mad2 complex (a terminal transducer of the SAC) and the basket Nups Tpr and Nup153 (Campbell et al. 2001; Lee et al. 2008; Lince-Faria et al. 2009; Schweizer et al. 2013; Rodriguez-Bravo et al. 2014). Mad1 and Mad2 can be detected at the NPC during interphase, and while both Tpr and Nup153 seem important for subnuclear Mad2 localization, Mad1 localization mainly depends on Tpr (Rodriguez-Bravo et al. 2014). In particular, phosphorylation of Tpr by CDK1 might regulate this interaction (Rajanala et al. 2014). Upon NEBD, Mad1 and Mad2 are recruited to the unattached kinetochores by the upstream SAC components, while Tpr and Nup153 enrichment is not evident at any particular mitotic substructure in most cell types. Thus, whether their interaction persists during mitosis is controversial, and alternative explanations have been proposed to explain why perturbed levels of NPC basket components affect the activity of the SAC, leading to mitotic defects (Schweizer et al. 2013; Rodriguez-Bravo et al. 2014). The interaction of Mad1 and Mad2 with the basket components during interphase might influence the mitotic surveillance checkpoint in the subsequent mitosis. According to this idea, the Mad1/Tpr interaction at the NPC promotes activation of Mad2, leading to Cdc20 neutralization. Accumulation of this signal during interphase would inhibit the interaction of Cdc20 with the anaphase-promoting complex (APC/C) after NEBD and during the early stages of mitosis, preventing mitotic failure (Rodriguez-Bravo et al. 2014). Alternatively, Tpr might contribute to SAC robustness and proper spindle orientation by affecting Mad2 stability and localization at unaligned kinetochores in different organisms (De Souza et al. 2009; Lince-Faria et al. 2009; Schweizer et al. 2013). Supporting a specific role of Tpr at the kinetochores, FRAP and imaging experiments in Drosophila and Aspergillus nidulans showed a specific accumulation of Tpr orthologs (Mtor/Mlp1) surrounding the spindle microtubules rather than randomly distributed throughout the mitotic cell (De Souza et al. 2009; Lince-Faria et al. 2009). Regarding Nup153, it has been proposed to display different roles during mitotic progression (Mackay et al. 2009), and unbalanced levels of this protein influence SAC activity by affecting Mad1 localization and phosphorylation status, consequently leading to mitotic aberrancies (Lussi et al. 2010).
The Nup Rae1 and its partner at the NPC, Nup98, appear redistributed throughout the cell in mitosis. However, Rae1 is crucial for normal mitotic progression and stabilization of microtubules at the kinetochore, and perturbed levels of this Nup lead to chromosome segregation and SAC defects (Babu et al. 2003; Blower et al. 2005). This is not surprising, considering its extensive homology with SAC component Bub3 (Babu et al. 2003) and the reported interactions with the checkpoint kinase Bub1, the spindle assembly factor NuMA (nuclear mitotic apparatus), and microtubules (Wang et al. 2001; Babu et al. 2003; Wong et al. 2006). Nup98 also facilitates bipolar spindle formation in Xenopus, interacting with microtubules and antagonizing MCAK (mitotic centromere-associated kinesin) activity, a microtubule depolymerizing enzyme (Cross and Powers 2011). In addition, Nup98 and Rae1 regulate APC/Cdh1 activity during prometaphase/metaphase, ensuring timely securin degradation and preventing premature cohesin cleavage and therefore precocious sister chromatin separation (Jeganathan et al. 2005). Importantly, the role of Rae1/Nup98—ensuring the assembly of a normal spindle and timely anaphase onset—might partially contribute to the prevention of genome segregation defects and malignant transformation observed in Nup98-associated mutations (Salsi et al. 2014).
Telophase, NE reformation, and cytokinesis
By late anaphase, Nups start to be recruited to the chromosomes driving NPC and NE reassembly (Doucet et al. 2010; Mackay and Ullman 2011). Simultaneously, the central spindle is compacted into the midbody, eventually leading to abscission. The coordination of the mitotic exit network with NE reassembly is far from being fully characterized at the molecular level. However, it is noteworthy that reduced levels of NPC components such as Nup153, Elys, and Nup107/Nup160 complex members generate an accumulation of cells with unresolved midbodies (Rasala et al. 2006; Mackay et al. 2009; Platani et al. 2009). One possible explanation of this phenomenon is that Nups could influence the mitotic exit, which to a large extent involves massive dephosphorylation of mitotic kinase targets. Supporting the latter, the coordinated activity of several protein phosphatases is required to recycle the nuclear components after mitosis. Nup153 and Nup50 interact with the protein phosphatase PP1 and its targeting subunit, Repo-Man (Moorhead et al. 2008; Vagnarelli et al. 2011), suggesting that they might influence proper PP1 localization and/or activity affecting the dephosphorylation cascades.
In summary, Nups show essential transport-independent functions during mitosis, and disruption of their activities results in a collection of mitotic aberrations. NPC components impact several mitotic stages but seem to mainly concentrate their efforts around the kinetochores during prometaphase to stabilize their interactions with the microtubules by controlling the spatial localization and activity of specific components, ensuring normal spindle assembly and anaphase onset (Fig. 2).
Conclusions and perspectives
In their capacity to form macromolecular transport channels in the NE, regulation of nucleocytoplasmic transport is a critical function of Nups (Fig. 2). Molecular trafficking affects virtually every process in the cell, and, consequently, any observation made in the presence of unbalanced levels of NPC components or Nup mutant forms should be carefully interpreted. However, increasing evidence suggests additional roles of Nups affecting fundamental genome functions, some of which are likely to be independent of nuclear transport (Fig. 2). In eukaryotic cells, NPC components can associate with specific genes and regulate their expression. In yeast, this function might be restricted to the NE-embedded NPC, and chromatin regions would relocate to the nuclear periphery, while in multicellular organisms, this function has apparently extended throughout the nucleus, as mobile Nups can detach from the NPC and relocate to intranuclear regions as well (Fig. 2). While the recruitment of specific genes to the NPC is at least partially understood, the mechanisms underlying the recruitment of Nups to intranuclear regions are still uncharacterized. Theoretically, mobile Nups could recognize and bind specific DNA sequences (Ahmed and Brickner 2010), particular epigenetic marks, or specific chromatin remodelers. Comprehensive characterization of intranuclear and NE-associated NPC–chromatin interactions will be required to better understand the transport-independent functions of Nups. Additionally, it would also clarify whether mobile Nups collaborate in the nuclear interior to regulate transcriptional activity, mimicking the regulation exerted at the NPC, as inferred from the observations made in Drosophila nuclei reporting the co-occupation of several mobile Nups at specific genes (Kalverda et al. 2010).
Nup-mediated gene expression regulation might likely be caused by the role of NPC components in chromatin structure and organization (Fig. 2). However, further evidence directly linking Nups to genome architecture and chromatin structure is essential for determining their precise roles, especially in multicellular organisms. It will be particularly interesting to study the molecular connections between the NPC components and the reorganizing chromosomes during nuclear reformation after mitosis. Tethering of specific chromatin regions by NPC components during late anaphase might potentially represent an early event in the nonrandom organization of the interphase chromosomes, thus linking mitotic Nup–genome interactions to nuclear function.
In proliferating cells, Nups have been shown to play a central role in the preservation of genome integrity, influencing replication fork progression, DNA repair, and accurate chromosome segregation (Fig. 2). First, the observations regarding the post-translational modification of NPC components to allow fork passage during S phase indicate the relevance of a precise and dynamic regulation of NPC–chromatin contacts, which would likely affect different DNA metabolic processes and Nup functions (Regot et al. 2013). Second, the recruitment of persistent DNA lesions to the NPCs reported in yeast might be reflective of the ability of Nups to gather particular loci and enzymatic machineries together, as shown for transcriptional regulation, potentially facilitating DNA repair. Interestingly, NPC-associated chromatin in human cells is repaired in a different way compared with the lamin-associated regions (Lemaître et al. 2014). However, while transport-dependent roles of Nups in DNA repair are being uncovered (Lemaître et al. 2012; Moudry et al. 2012), transport-independent involvement of Nups in DNA repair in vertebrates remains undefined. Last, indicative of the potential roles of the off-pore Nups, an increasing number of NPC components has been shown to display mitotic-specific roles, ensuring accurate chromosome segregation. Again, one general interpretation of these observations is that Nups seem to form a platform to coordinate the action of specific enzymes with their targets. How Nups adapt their functions during mitosis and whether their phosphorylation by mitotic kinases is linked to that adaptation remain to be explored. Nevertheless, accurate regulation of Nup levels and functions seems crucial to avoid the accumulation of DNA lesions and aneuploidy and likely prevent malignant transformation.
Acknowledgments
We thank the Hetzer laboratory members for critical reading of the manuscript. A.I. is supported by EMBO and Human Frontier Science Program fellowships. M.W.H. is supported by National Institutes of Health grant R01GM098749, the Glenn Aging Foundation, and American Cancer Society Award number P30CA014195.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.256495.114.
References
- Ahmed S, Brickner JH. 2010. A role for DNA sequence in controlling the spatial organization of the genome. Nucleus 1: 402–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed S, Brickner DG, Light WH, Cajigas I, McDonough M, Froyshteter AB, Volpe T, Brickner JH. 2010. DNA zip codes control an ancient mechanism for gene targeting to the nuclear periphery. Nat Cell Biol 12: 111–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhtar A, Gasser SM. 2007. The nuclear envelope and transcriptional control. Nat Rev Genet 8: 507–517. [DOI] [PubMed] [Google Scholar]
- Babu JR, Jeganathan KB, Baker DJ, Wu X, Kang-Decker N, van Deursen JM. 2003. Rae1 is an essential mitotic checkpoint regulator that cooperates with Bub3 to prevent chromosome missegregation. J Cell Biol 160: 341–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai XT, Gu BW, Yin T, Niu C, Xi XD, Zhang J, Chen Z, Chen SJ. 2006. Trans-repressive effect of NUP98–PMX1 on PMX1-regulated c-FOS gene through recruitment of histone deacetylase 1 by FG repeats. Cancer Res 66: 4584–4590. [DOI] [PubMed] [Google Scholar]
- Belgareh N, Rabut G, Baï SW, van Overbeek M, Beaudouin J, Daigle N, Zatsepina OV, Pasteau F, Labas V, Fromont-Racine M, et al. 2001. An evolutionarily conserved NPC subcomplex, which redistributes in part to kinetochores in mammalian cells. J Cell Biol 154: 1147–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmont AS, Zhai Y, Thilenius A. 1993. Lamin B distribution and association with peripheral chromatin revealed by optical sectioning and electron microscopy tomography. J Cell Biol 123: 1671–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermejo R, Capra T, Jossen R, Colosio A, Frattini C, Carotenuto W, Cocito A, Doksani Y, Klein H, Gómez-González B, et al. 2011. The replication checkpoint protects fork stability by releasing transcribed genes from nuclear pores. Cell 146: 233–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasius M, Forment JV, Thakkar N, Wagner SA, Choudhary C, Jackson SP. 2011. A phospho-proteomic screen identifies substrates of the checkpoint kinase Chk1. Genome Biol 12: R78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobel G. 1985. Gene gating: a hypothesis. Proc Natl Acad Sci 82: 8527–8529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blower MD, Nachury M, Heald R, Weis K. 2005. A Rae1-containing ribonucleoprotein complex is required for mitotic spindle assembly. Cell 121: 223–234. [DOI] [PubMed] [Google Scholar]
- Bolhy S, Bouhlel I, Dultz E, Nayak T, Zuccolo M, Gatti X, Vallee R, Ellenberg J, Doye V. 2011. A Nup133-dependent NPC-anchored network tethers centrosomes to the nuclear envelope in prophase. J Cell Biol 192: 855–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickner DG, Brickner JH. 2011. Gene positioning is regulated by phosphorylation of the nuclear pore complex by Cdk1. Cell Cycle 10: 392–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickner JH, Walter P. 2004. Gene recruitment of the activated INO1 locus to the nuclear membrane. PLoS Biol 2: e342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickner DG, Cajigas I, Fondufe-Mittendorf Y, Ahmed S, Lee PC, Widom J, Brickner JH. 2007. H2A.Z-mediated localization of genes at the nuclear periphery confers epigenetic memory of previous transcriptional state. PLoS Biol 5: e81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickner DG, Ahmed S, Meldi L, Thompson A, Light W, Young M, Hickman TL, Chu F, Fabre E, Brickner JH. 2012. Transcription factor binding to a DNA zip code controls interchromosomal clustering at the nuclear periphery. Dev Cell 22: 1234–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CR, Kennedy CJ, Delmar VA, Forbes DJ, Silver PA. 2008. Global histone acetylation induces functional genomic reorganization at mammalian nuclear pore complexes. Genes Dev 22: 627–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchwalter AL, Liang Y, Hetzer MW. 2014. Nup50 is required for cell differentiation and exhibits transcription-dependent dynamics. Mol Biol Cell 25: 2472–2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabal GG, Genovesio A, Rodriguez-Navarro S, Zimmer C, Gadal O, Lesne A, Buc H, Feuerbach-Fournier F, Olivo-Marin JC, Hurt EC, et al. 2006. SAGA interacting factors confine sub-diffusion of transcribed genes to the nuclear envelope. Nature 441: 770–773. [DOI] [PubMed] [Google Scholar]
- Cai Y, Singh BB, Aslanukov A, Zhao H, Ferreira PA. 2001. The docking of kinesins, KIF5B and KIF5C, to Ran-binding protein 2 (RanBP2) is mediated via a novel RanBP2 domain. J Biol Chem 276: 41594–41602. [DOI] [PubMed] [Google Scholar]
- Campbell MS, Chan GK, Yen TJ. 2001. Mitotic checkpoint proteins HsMAD1 and HsMAD2 are associated with nuclear pore complexes in interphase. J Cell Sci 114: 953–963. [DOI] [PubMed] [Google Scholar]
- Capelson M, Liang Y, Schulte R, Mair W, Wagner U, Hetzer MW. 2010. Chromatin-bound nuclear pore components regulate gene expression in higher eukaryotes. Cell 140: 372–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmena M, Wheelock M, Funabiki H, Earnshaw WC. 2012. The chromosomal passenger complex (CPC): from easy rider to the godfather of mitosis. Nat Rev Mol Cell Biol 13: 789–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casolari JM, Brown CR, Komili S, West J, Hieronymus H, Silver PA. 2004. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell 117: 427–439. [DOI] [PubMed] [Google Scholar]
- Casolari JM, Brown CR, Drubin DA, Rando OJ, Silver PA. 2005. Developmentally induced changes in transcriptional program alter spatial organization across chromosomes. Genes Dev 19: 1188–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty A, Shen Z, Prasanth SG. 2011. ‘ORCanization’ on heterochromatin: linking DNA replication initiation to chromatin organization. Epigenetics 6: 665–670. [DOI] [PubMed] [Google Scholar]
- Chow K-H, Elgort S, Dasso M, Ullman KS. 2012. Two distinct sites in Nup153 mediate interaction with the SUMO proteases SENP1 and SENP2. Nucleus 3: 349–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clever M, Funakoshi T, Mimura Y, Takagi M, Imamoto N. 2012. The nucleoporin ELYS/Mel28 regulates nuclear envelope subdomain formation in HeLa cells. Nucleus 3: 187–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross MK, Powers MA. 2011. Nup98 regulates bipolar spindle assembly through association with microtubules and opposition of MCAK. Mol Biol Cell 22: 661–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Angelo MA, Hetzer MW. 2008. Structure, dynamics and function of nuclear pore complexes. Trends Cell Biol 18: 456–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Angelo MA, Gomez-Cavazos JS, Mei A, Lackner DH, Hetzer MW. 2012. A change in nuclear pore complex composition regulates cell differentiation. Dev Cell 22: 446–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davuluri G, Gong W, Yusuff S, Lorent K, Muthumani M, Dolan AC, Pack M. 2008. Mutation of the zebrafish nucleoporin elys sensitizes tissue progenitors to replication stress. PLoS Genet 4: e1000240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawlaty MM, Malureanu L, Jeganathan KB, Kao E, Sustmann C, Tahk S, Shuai K, Grosschedl R, van Deursen JM. 2008. Resolution of sister centromeres requires RanBP2-mediated SUMOylation of topoisomerase IIα. Cell 133: 103–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Souza CP, Hashmi SB, Horn KP, Osmani SA. 2006. A point mutation in the Aspergillus nidulans sonBNup98 nuclear pore complex gene causes conditional DNA damage sensitivity. Genetics 174: 1881–1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Souza CP, Hashmi SB, Nayak T, Oakley B, Osmani SA. 2009. Mlp1 acts as a mitotic scaffold to spatially regulate spindle assembly checkpoint proteins in Aspergillus nidulans. Mol Biol Cell 20: 2146–2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieppois G, Stutz F. 2010. Connecting the transcription site to the nuclear pore: a multi-tether process that regulates gene expression. J Cell Sci 123: 1989–1999. [DOI] [PubMed] [Google Scholar]
- Dieppois G, Iglesias N, Stutz F. 2006. Cotranscriptional recruitment to the mRNA export receptor Mex67p contributes to nuclear pore anchoring of activated genes. Mol Cell Biol 26: 7858–7870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilworth DJ, Tackett AJ, Rogers RS, Yi EC, Christmas RH, Smith JJ, Siegel AF, Chait BT, Wozniak RW, Aitchison JD. 2005. The mobile nucleoporin Nup2p and chromatin-bound Prp20p function in endogenous NPC-mediated transcriptional control. J Cell Biol 171: 955–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou H, Huang C, Van Nguyen T, Lu L-S, Yeh ETH. 2011. SUMOylation and de-SUMOylation in response to DNA damage. FEBS Lett 585: 2891–2896. [DOI] [PubMed] [Google Scholar]
- Doucet CM, Talamas JA, Hetzer MW. 2010. Cell cycle-dependent differences in nuclear pore complex assembly in metazoa. Cell 141: 1030–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doye V. 2011. Mitotic phosphorylation of nucleoporins: dismantling NPCs and beyond. Dev Cell 20: 281–282. [DOI] [PubMed] [Google Scholar]
- Dultz E, Zanin E, Wurzenberger C, Braun M, Rabut G, Sironi L, Ellenberg J. 2008. Systematic kinetic analysis of mitotic dis- and reassembly of the nuclear pore in living cells. J Cell Biol 180: 857–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez AG, Piano F. 2006. MEL-28 is downstream of the Ran cycle and is required for nuclear-envelope function and chromatin maintenance. Curr Biol 16: 1757–1763. [DOI] [PubMed] [Google Scholar]
- Feuerbach F, Galy V, Trelles-Sticken E, Fromont-Racine M, Jacquier A, Gilson E, Olivo-Marin JC, Scherthan H, Nehrbass U. 2002. Nuclear architecture and spatial positioning help establish transcriptional states of telomeres in yeast. Nat Cell Biol 4: 214–221. [DOI] [PubMed] [Google Scholar]
- Franks TM, Hetzer MW. 2013. The role of Nup98 in transcription regulation in healthy and diseased cells. Trends Cell Biol 23: 112–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galy V, Olivo-Marin JC, Scherthan H, Doye V, Rascalou N, Nehrbass U. 2000. Nuclear pore complexes in the organization of silent telomeric chromatin. Nature 403: 108–112. [DOI] [PubMed] [Google Scholar]
- Galy V, Askjaer P, Franz C, López-Iglesias C, Mattaj IW. 2006. MEL-28, a novel nuclear-envelope and kinetochore protein essential for zygotic nuclear-envelope assembly in C. elegans. Curr Biol 16: 1748–1756. [DOI] [PubMed] [Google Scholar]
- Galy V, Antonin W, Jaedicke A, Sachse M, Santarella R, Haselmann U, Mattaj I. 2008. A role for gp210 in mitotic nuclear-envelope breakdown. J Cell Sci 121: 317–328. [DOI] [PubMed] [Google Scholar]
- Gao N, Davuluri G, Gong W, Seiler C, Lorent K, Furth EE, Kaestner KH, Pack M. 2011. The nuclear pore complex protein Elys is required for genome stability in mouse intestinal epithelial progenitor cells. Gastroenterology 140: 1547–1555.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Oliver E, García-Molinero V, Rodriguez-Navarro S. 2012. mRNA export and gene expression: the SAGA–TREX-2 connection. Biochim Biophys Acta 1819: 555–565. [DOI] [PubMed] [Google Scholar]
- Gartenberg MR, Neumann FR, Laroche T, Blaszczyk M, Gasser SM. 2004. Sir-mediated repression can occur independently of chromosomal and subnuclear contexts. Cell 119: 955–967. [DOI] [PubMed] [Google Scholar]
- Gillespie PJ, Khoudoli GA, Stewart G, Swedlow JR, Blow JJ. 2007. ELYS/MEL-28 chromatin association coordinates nuclear pore complex assembly and replication licensing. Curr Biol 17: 1657–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Cavazos JS, Hetzer MW. 2012. Outfits for different occasions: tissue-specific roles of Nuclear Envelope proteins. Curr Opin Cell Biol 24: 775–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green EM, Jiang Y, Joyner R, Weis K. 2012. A negative feedback loop at the nuclear periphery regulates GAL gene expression. Mol Biol Cell 23: 1367–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffis ER, Altan N, Lippincott-Schwartz J, Powers MA. 2002. Nup98 is a mobile nucleoporin with transcription-dependent dynamics. Mol Biol Cell 13: 1282–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman E, Medalia O, Zwerger M. 2012. Functional architecture of the nuclear pore complex. Annu Rev Biophys 41: 557–584. [DOI] [PubMed] [Google Scholar]
- Hashizume C, Kobayashi A, Wong RW. 2013a. Down-modulation of nucleoporin RanBP2/Nup358 impaired chromosomal alignment and induced mitotic catastrophe. Cell Death Dis 4: e854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashizume C, Moyori A, Kobayashi A, Yamakoshi N, Endo A, Wong RW. 2013b. Nucleoporin Nup62 maintains centrosome homeostasis. Cell Cycle 12: 3804–3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hediger F, Gasser SM. 2002. Nuclear organization and silencing: putting things in their place. Nat Cell Biol 4: E53–E55. [DOI] [PubMed] [Google Scholar]
- Hediger F, Dubrana K, Gasser SM. 2002. Myosin-like proteins 1 and 2 are not required for silencing or telomere anchoring, but act in the Tel1 pathway of telomere length control. J Struct Biol 140: 79–91. [DOI] [PubMed] [Google Scholar]
- Helmrich A, Ballarino M, Nudler E, Tora L. 2013. Transcription-replication encounters, consequences and genomic instability. Nat Struct Mol Biol 20: 412–418. [DOI] [PubMed] [Google Scholar]
- Hetzer MW. 2010. The nuclear envelope. Cold Spring Harb Perspect Biol 2: a000539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoelz A, Debler EW, Blobel G. 2011. The structure of the nuclear pore complex. Annu Rev Biochem 80: 613–643. [DOI] [PubMed] [Google Scholar]
- Horigome C, Oma Y, Konishi T, Schmid R, Marcomini I, Hauer MH, Dion V, Harata M, Gasser SM. 2014. SWR1 and INO80 chromatin remodelers contribute to DNA double-strand break perinuclear anchorage site choice. Mol Cell 55: 626–639. [DOI] [PubMed] [Google Scholar]
- Ikegami K, Lieb JD. 2013. Integral nuclear pore proteins bind to Pol III-transcribed genes and are required for Pol III transcript processing in C. elegans. Mol Cell 51: 840–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii K, Arib G, Lin C, Van Houwe G, Laemmli UK. 2002. Chromatin boundaries in budding yeast: the nuclear pore connection. Cell 109: 551–562. [DOI] [PubMed] [Google Scholar]
- Jeganathan KB, Malureanu L, van Deursen JM. 2005. The Rae1–Nup98 complex prevents aneuploidy by inhibiting securin degradation. Nature 438: 1036–1039. [DOI] [PubMed] [Google Scholar]
- Joseph J, Liu S-T, Jablonski SA, Yen TJ, Dasso M. 2004. The RanGAP1–RanBP2 complex is essential for microtubule-kinetochore interactions in vivo. Curr Biol 14: 611–617. [DOI] [PubMed] [Google Scholar]
- Kalverda B, Fornerod M. 2010. Characterization of genome-nucleoporin interactions in Drosophila links chromatin insulators to the nuclear pore complex. Cell Cycle 9: 4812–4817. [DOI] [PubMed] [Google Scholar]
- Kalverda B, Pickersgill H, Shloma VV, Fornerod M. 2010. Nucleoporins directly stimulate expression of developmental and cell-cycle genes inside the nucleoplasm. Cell 140: 360–371. [DOI] [PubMed] [Google Scholar]
- Kasper LH, Brindle PK, Schnabel CA, Pritchard CE, Cleary ML, van Deursen JM. 1999. CREB binding protein interacts with nucleoporin-specific FG repeats that activate transcription and mediate NUP98–HOXA9 oncogenicity. Mol Cell Biol 19: 764–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krull S, Dörries J, Boysen B, Reidenbach S, Magnius L, Norder H, Thyberg J, Cordes VC. 2010. Protein Tpr is required for establishing nuclear pore-associated zones of heterochromatin exclusion. EMBO J 29: 1659–1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurshakova MM, Krasnov AN, Kopytova DV, Shidlovskii YV, Nikolenko JV, Nabirochkina EN, Spehner D, Schultz P, Tora L, Georgieva SG. 2007. SAGA and a novel Drosophila export complex anchor efficient transcription and mRNA export to NPC. EMBO J 26: 4956–4965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lainé JP, Singh BN, Krishnamurthy S, Hampsey M. 2009. A physiological role for gene loops in yeast. Genes Dev 23: 2604–2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurell E, Kutay U. 2011. Dismantling the NPC permeability barrier at the onset of mitosis. Cell Cycle 10: 2243–2245. [DOI] [PubMed] [Google Scholar]
- Laurell E, Beck K, Krupina K, Theerthagiri G, Bodenmiller B, Horvath P, Aebersold R, Antonin W, Kutay U. 2011. Phosphorylation of Nup98 by multiple kinases is crucial for NPC disassembly during mitotic entry. Cell 144: 539–550. [DOI] [PubMed] [Google Scholar]
- Lee SH, Sterling H, Burlingame A, McCormick F. 2008. Tpr directly binds to Mad1 and Mad2 and is important for the Mad1–Mad2-mediated mitotic spindle checkpoint. Genes Dev 22: 2926–2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaître C, Fischer B, Kalousi A, Hoffbeck A-S, Guirouilh-Barbat J, Shahar OD, Genet D, Goldberg M, Betrand P, Lopez B, et al. 2012. The nucleoporin 153, a novel factor in double-strand break repair and DNA damage response. Oncogene 31: 4803–4809. [DOI] [PubMed] [Google Scholar]
- Lemaître C, Grabarz A, Tsouroula K, Andronov L, Furst A, Pankotai T, Heyer V, Rogier M, Attwood KM, Kessler P, et al. 2014. Nuclear position dictates DNA repair pathway choice. Genes Dev 28: 2450–2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lénárt P, Rabut G, Daigle N, Hand AR, Terasaki M, Ellenberg J. 2003. Nuclear envelope breakdown in starfish oocytes proceeds by partial NPC disassembly followed by a rapidly spreading fenestration of nuclear membranes. J Cell Biol 160: 1055–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y, Hetzer MW. 2011. Functional interactions between nucleoporins and chromatin. Curr Opin Cell Biol 23: 65–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y, Franks TM, Marchetto MC, Gage FH, Hetzer MW. 2013. Dynamic association of NUP98 with the human genome. PLoS Genet 9: e1003308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Light WH, Brickner DG, Brand VR, Brickner JH. 2010. Interaction of a DNA zip code with the nuclear pore complex promotes H2A.Z incorporation and INO1 transcriptional memory. Mol Cell 40: 112–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Light WH, Freaney J, Sood V, Thompson A, D’Urso A, Horvath CM, Brickner JH. 2013. A conserved role for human Nup98 in altering chromatin structure and promoting epigenetic transcriptional memory. PLoS Biol 11: e1001524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lince-Faria M, Maffini S, Orr B, Ding Y, Florindo C, Sunkel CE, Tavares A, Johansen J, Johansen KM, Maiato H. 2009. Spatiotemporal control of mitosis by the conserved spindle matrix protein Megator. J Cell Biol 184: 647–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Prunuske AJ, Fager AM, Ullman KS. 2003. The COPI complex functions in nuclear envelope breakdown and is recruited by the nucleoporin Nup153. Dev Cell 5: 487–498. [DOI] [PubMed] [Google Scholar]
- Loeillet S, Palancade B, Cartron M, Thierry A, Richard G-F, Dujon B, Doye V, Nicolas A. 2005. Genetic network interactions among replication, repair and nuclear pore deficiencies in yeast. DNA Repair 4: 459–468. [DOI] [PubMed] [Google Scholar]
- Loïodice I, Alves A, Rabut G, Van Overbeek M, Ellenberg J, Sibarita J-B, Doye V. 2004. The entire Nup107–160 complex, including three new members, is targeted as one entity to kinetochores in mitosis. Mol Biol Cell 15: 3333–3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lussi YC, Shumaker DK, Shimi T, Fahrenkrog B. 2010. The nucleoporin Nup153 affects spindle checkpoint activity due to an association with Mad1. Nucleus 1: 71–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luthra R, Kerr SC, Harreman MT, Apponi LH, Fasken MB, Ramineni S, Chaurasia S, Valentini SR, Corbett AH. 2007. Actively transcribed GAL genes can be physically linked to the nuclear pore by the SAGA chromatin modifying complex. J Biol Chem 282: 3042–3049. [DOI] [PubMed] [Google Scholar]
- Mackay DR, Ullman KS. 2011. Coordinating postmitotic nuclear pore complex assembly with abscission timing. Nucleus 2: 283–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay DR, Elgort SW, Ullman KS. 2009. The nucleoporin Nup153 has separable roles in both early mitotic progression and the resolution of mitosis. Mol Biol Cell 20: 1652–1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, et al. 2007. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 316: 1160–1166. [DOI] [PubMed] [Google Scholar]
- Mekhail K, Moazed D. 2010. The nuclear envelope in genome organization, expression and stability. Nat Rev Mol Cell Biol 11: 317–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendjan S, Taipale M, Kind J, Holz H, Gebhardt P, Schelder M, Vermeulen M, Buscaino A, Duncan K, Mueller J, et al. 2006. Nuclear pore components are involved in the transcriptional regulation of dosage compensation in Drosophila. Mol Cell 21: 811–823. [DOI] [PubMed] [Google Scholar]
- Mishra RK, Chakraborty P, Arnaoutov A, Fontoura BMA, Dasso M. 2010. The Nup107-160 complex and γ-TuRC regulate microtubule polymerization at kinetochores. Nat Cell Biol 12: 164–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorhead GBG, Trinkle-Mulcahy L, Nimick M, De Wever V, Campbell DG, Gourlay R, Lam YW, Lamond AI. 2008. Displacement affinity chromatography of protein phosphatase one (PP1) complexes. BMC Biochem 9: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moudry P, Lukas C, Macurek L, Neumann B, Heriche J-K, Pepperkok R, Ellenberg J, Hodny Z, Lukas J, Bartek J. 2012. Nucleoporin NUP153 guards genome integrity by promoting nuclear import of 53BP1. Cell Death Differ 19: 798–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musacchio A, Salmon ED. 2007. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol 8: 379–393. [DOI] [PubMed] [Google Scholar]
- Nagai S, Dubrana K, Tsai-Pflugfelder M, Davidson MB, Roberts TM, Brown GW, Varela E, Hediger F, Gasser SM, Krogan NJ. 2008. Functional targeting of DNA damage to a nuclear pore-associated SUMO-dependent ubiquitin ligase. Science 322: 597–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai S, Heun P, Gasser SM. 2010. Roles for nuclear organization in the maintenance of genome stability. Epigenomics 2: 289–305. [DOI] [PubMed] [Google Scholar]
- Natsume T, Tanaka TU. 2010. Spatial regulation and organization of DNA replication within the nucleus. Chromosome Res 18: 7–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Sullivan JM, Tan-Wong SM, Morillon A, Lee B, Coles J, Mellor J, Proudfoot NJ. 2004. Gene loops juxtapose promoters and terminators in yeast. Nat Genet 36: 1014–1018. [DOI] [PubMed] [Google Scholar]
- Olsson M, Schéele S, Ekblom P. 2004. Limited expression of nuclear pore membrane glycoprotein 210 in cell lines and tissues suggests cell-type specific nuclear pores in metazoans. Exp Cell Res 292: 359–370. [DOI] [PubMed] [Google Scholar]
- Orjalo AV, Arnaoutov A, Shen Z, Boyarchuk Y, Zeitlin SG, Fontoura B, Briggs S, Dasso M, Forbes DJ. 2006. The Nup107–160 nucleoporin complex is required for correct bipolar spindle assembly. Mol Biol Cell 17: 3806–3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oza P, Jaspersen SL, Miele A, Dekker J, Peterson CL. 2009. Mechanisms that regulate localization of a DNA double-strand break to the nuclear periphery. Genes Dev 23: 912–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palancade B, Liu X, Garcia-Rubio M, Aguilera A, Zhao X, Doye V. 2007. Nucleoporins prevent DNA damage accumulation by modulating Ulp1-dependent sumoylation processes. Mol Biol Cell 18: 2912–2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual-Garcia P, Capelson M. 2014. Nuclear pores as versatile platforms for gene regulation. Curr Opin Genet Dev 25: 110–117. [DOI] [PubMed] [Google Scholar]
- Paulsen RD, Soni DV, Wollman R, Hahn AT, Yee MC, Guan A, Hesley JA, Miller SC, Cromwell EF, Solow-Cordero DE, et al. 2009. A genome-wide siRNA screen reveals diverse cellular processes and pathways that mediate genome stability. Mol Cell 35: 228–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne C, Rawe V, Ramalho-Santos J, Simerly C, Schatten G. 2003. Preferentially localized dynein and perinuclear dynactin associate with nuclear pore complex proteins to mediate genomic union during mammalian fertilization. J Cell Sci 116: 4727–4738. [DOI] [PubMed] [Google Scholar]
- Phillips JE, Corces VG. 2009. CTCF: master weaver of the genome. Cell 137: 1194–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichler A, Gast A, Seeler JS, Dejean A, Melchior F. 2002. The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell 108: 109–120. [DOI] [PubMed] [Google Scholar]
- Platani M, Santarella-Mellwig R, Posch M, Walczak R, Swedlow JR, Mattaj IW. 2009. The Nup107–160 nucleoporin complex promotes mitotic events via control of the localization state of the chromosome passenger complex. Mol Biol Cell 20: 5260–5275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prunuske AJ, Liu J, Elgort S, Joseph J, Dasso M, Ullman KS. 2006. Nuclear envelope breakdown is coordinated by both Nup358/RanBP2 and Nup153, two nucleoporins with zinc finger modules. Mol Biol Cell 17: 760–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabut G, Doye V, Ellenberg J. 2004. Mapping the dynamic organization of the nuclear pore complex inside single living cells. Nat Cell Biol 6: 1114–1121. [DOI] [PubMed] [Google Scholar]
- Rajanala K, Sarkar A, Jhingan GD, Priyadarshini R, Jalan M, Sengupta S, Nandicoori VK. 2014. Phosphorylation of nucleoporin Tpr governs its differential localization and is required for its mitotic function. J Cell Sci 127: 3505–3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasala BA, Orjalo AV, Shen Z, Briggs S, Forbes DJ. 2006. ELYS is a dual nucleoporin/kinetochore protein required for nuclear pore assembly and proper cell division. Proc Natl Acad Sci 103: 17801–17806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regot S, de Nadal E, Rodriguez-Navarro S, González-Novo A, Pérez-Fernandez J, Gadal O, Seisenbacher G, Ammerer G, Posas F. 2013. The Hog1 stress-activated protein kinase targets nucleoporins to control mRNA export upon stress. J Biol Chem 288: 17384–17398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Bravo V, Maciejowski J, Corona J, Buch HK, Collin P, Kanemaki MT, Shah JV, Jallepalli PV. 2014. Nuclear pores protect genome integrity by assembling a premitotic and Mad1-dependent anaphase inhibitor. Cell 156: 1017–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Navarro S, Fischer T, Luo MJ, Antúnez O, Brettschneider S, Lechner J, Pérez-Ortín JE, Reed R, Hurt E. 2004. Sus1, a functional component of the SAGA histone acetylase complex and the nuclear pore-associated mRNA export machinery. Cell 116: 75–86. [DOI] [PubMed] [Google Scholar]
- Rohner S, Kalck V, Wang X, Ikegami K, Lieb JD, Gasser SM, Meister P. 2013. Promoter- and RNA polymerase II-dependent hsp-16 gene association with nuclear pores in Caenorhabditis elegans. J Cell Biol 200: 589–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothballer A, Kutay U. 2012. SnapShot: the nuclear envelope II. Cell 150: 1084–1084.e1. [DOI] [PubMed] [Google Scholar]
- Ruben GJ, Kirkland JG, MacDonough T, Chen M, Dubey RN, Gartenberg MR, Kamakaka RT. 2011. Nucleoporin mediated nuclear positioning and silencing of HMR. PLoS ONE 6: e21923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salsi V, Ferrari S, Gorello P, Fantini S, Chiavolelli F, Mecucci C, Zappavigna V. 2014. NUP98 fusion oncoproteins promote aneuploidy by attenuating the mitotic spindle checkpoint. Cancer Res 74: 1079–1090. [DOI] [PubMed] [Google Scholar]
- Sarma NJ, Haley TM, Barbara KE, Buford TD, Willis KA, Santangelo GM. 2007. Glucose-responsive regulators of gene expression in Saccharomyces cerevisiae function at the nuclear periphery via a reverse recruitment mechanism. Genetics 175: 1127–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarma NJ, Buford TD, Haley T, Barbara-Haley K, Santangelo GM, Willis KA. 2011. The nuclear pore complex mediates binding of the Mig1 repressor to target promoters. PLoS ONE 6: e27117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savas JN, Toyama BH, Xu T, Yates JR, Hetzer MW. 2012. Extremely long-lived nuclear pore proteins in the rat brain. Science 335: 942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schermelleh L, Carlton PM, Haase S, Shao L, Winoto L, Kner P, Burke B, Cardoso MC, Agard DA, Gustafsson MGL, et al. 2008. Subdiffraction multicolor imaging of the nuclear periphery with 3D structured illumination microscopy. Science 320: 1332–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Arib G, Laemmli C, Nishikawa J, Durussel T, Laemmli UK. 2006. Nup-PI: the nucleopore-promoter interaction of genes in yeast. Mol Cell 21: 379–391. [DOI] [PubMed] [Google Scholar]
- Schweizer N, Ferrás C, Kern DM, Logarinho E, Cheeseman IM, Maiato H. 2013. Spindle assembly checkpoint robustness requires Tpr-mediated regulation of Mad1/Mad2 proteostasis. J Cell Biol 203: 883–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel JJ, Amon A. 2012. New insights into the troubles of aneuploidy. Annu Rev Cell Dev Biol 28: 189–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith OK, Aladjem MI. 2014. Chromatin structure and replication origins: determinants of chromosome replication and nuclear organization. J Mol Biol 426: 3330–3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soutoglou E, Misteli T. 2007. Mobility and immobility of chromatin in transcription and genome stability. Curr Opin Genet Dev 17: 435–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Splinter D, Tanenbaum ME, Lindqvist A, Jaarsma D, Flotho A, Yu KL, Grigoriev I, Engelsma D, Haasdijk ED, Keijzer N, et al. 2010. Bicaudal D2, dynein, and kinesin-1 associate with nuclear pore complexes and regulate centrosome and nuclear positioning during mitotic entry. PLoS Biol 8: e1000350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei A, Van Houwe G, Hediger F, Kalck V, Cubizolles F, Schober H, Gasser SM. 2006. Nuclear pore association confers optimal expression levels for an inducible yeast gene. Nature 441: 774–778. [DOI] [PubMed] [Google Scholar]
- Tan-Wong SM, Wijayatilake HD, Proudfoot NJ. 2009. Gene loops function to maintain transcriptional memory through interaction with the nuclear pore complex. Genes Dev 23: 2610–2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan-Wong SM, Zaugg JB, Camblong J, Xu Z, Zhang DW, Mischo HE, Ansari AZ, Luscombe NM, Steinmetz LM, Proudfoot NJ. 2012. Gene loops enhance transcriptional directionality. Science 338: 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Texari L, Dieppois G, Vinciguerra P, Contreras MP, Groner A, Letourneau A, Stutz F. 2013. The nuclear pore regulates GAL1 gene transcription by controlling the localization of the SUMO protease Ulp1. Mol Cell 51: 807–818. [DOI] [PubMed] [Google Scholar]
- Therizols P, Fairhead C, Cabal GG, Genovesio A, Olivo-Marin JC, Dujon B, Fabre E. 2006. Telomere tethering at the nuclear periphery is essential for efficient DNA double strand break repair in subtelomeric region. J Cell Biol 172: 189–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Therizols P, Illingworth RS, Courilleau C, Boyle S, Wood AJ, Bickmore WA. 2014. Chromatin decondensation is sufficient to alter nuclear organization in embryonic stem cells. Science 346: 1238–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyama BH, Savas JN, Park SK, Harris MS, Ingolia NT, Yates JR, Hetzer MW. 2013. Identification of long-lived proteins reveals exceptional stability of essential cellular structures. Cell 154: 971–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umlauf D, Bonnet J, Waharte F, Fournier M, Stierle M, Fischer B, Brino L, Devys D, Tora L. 2013. The human TREX-2 complex is stably associated with the nuclear pore basket. J Cell Sci 126: 2656–2667. [DOI] [PubMed] [Google Scholar]
- Vagnarelli P, Ribeiro S, Sennels L, Sanchez-Pulido L, de Lima Alves F, Verheyen T, Kelly DA, Ponting CP, Rappsilber J, Earnshaw WC. 2011. Repo-Man coordinates chromosomal reorganization with nuclear envelope reassembly during mitotic exit. Dev Cell 21: 328–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Vosse DW, Wan Y, Wozniak RW, Aitchison JD. 2011. Role of the nuclear envelope in genome organization and gene expression. Wiley Interdiscip Rev Syst Biol Med 3: 147–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Vosse DW, Wan Y, Lapetina DL, Chen WM, Chiang JH, Aitchison JD, Wozniak RW. 2013. A role for the nucleoporin Nup170p in chromatin structure and gene silencing. Cell 152: 969–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaquerizas JM, Suyama R, Kind J, Miura K, Luscombe NM, Akhtar A. 2010. Nuclear pore proteins nup153 and megator define transcriptionally active regions in the Drosophila genome. PLoS Genet 6: e1000846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Babu JR, Harden JM, Jablonski SA, Gazi MH, Lingle WL, de Groen PC, Yen TJ, van Deursen JM. 2001. The mitotic checkpoint protein hBUB3 and the mRNA export factor hRAE1 interact with GLE2p-binding sequence (GLEBS)-containing proteins. J Biol Chem 276: 26559–26567. [DOI] [PubMed] [Google Scholar]
- Wente SR, Rout MP. 2010. The nuclear pore complex and nuclear transport. Cold Spring Harb Perspect Biol 2: a000562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong RW, Blobel G, Coutavas E. 2006. Rae1 interaction with NuMA is required for bipolar spindle formation. Proc Natl Acad Sci 103: 19783–19787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolcock KJ, Stunnenberg R, Gaidatzis D, Hotz HR, Emmerth S, Barraud P, Bühler M. 2012. RNAi keeps Atf1-bound stress response genes in check at nuclear pores. Genes Dev 26: 683–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak R, Burke B, Doye V. 2010. Nuclear transport and the mitotic apparatus: an evolving relationship. Cell Mol Life Sci 67: 2215–2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama H, Koch B, Walczak R, Ciray-Duygu F, González-Sánchez JC, Devos DP, Mattaj IW, Gruss OJ. 2014. The nucleoporin MEL-28 promotes RanGTP-dependent γ-tubulin recruitment and microtubule nucleation in mitotic spindle formation. Nat Commun 5: 3270. [DOI] [PubMed] [Google Scholar]
- Yoshida T, Shimada K, Oma Y, Kalck V, Akimura K, Taddei A, Iwahashi H, Kugou K, Ohta K, Gasser SM, et al. 2010. Actin-related protein Arp6 influences H2A.Z-dependent and -independent gene expression and links ribosomal protein genes to nuclear pores. PLoS Genet 6: e1000910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Chen S, Yoo S, Chakrabarti S, Zhang T, Ke T, Oberti C, Yong SL, Fang F, Li L, et al. 2008. Mutation in nuclear pore component NUP155 leads to atrial fibrillation and early sudden cardiac death. Cell 135: 1017–1027. [DOI] [PubMed] [Google Scholar]
- Zuccolo M, Alves A, Galy V, Bolhy S, Formstecher E, Racine V, Sibarita JB, Fukagawa T, Shiekhattar R, Yen T, et al. 2007. The human Nup107–160 nuclear pore subcomplex contributes to proper kinetochore functions. EMBO J 26: 1853–1864. [DOI] [PMC free article] [PubMed] [Google Scholar]