Wei et al. found that Cas9, previously identified as the nuclease responsible for ultimate invader destruction, is also essential for adaptation in Streptococcus thermophilus. Cas9 nuclease activity is dispensable for adaptation. Wei et al. also revealed extensive, unbiased acquisition of the self-targeting host genome sequence by the CRISPR–Cas system that is masked in the presence of active target destruction.
Keywords: CRISPR, adaptation, Cas9, PAM, Streptococcus thermophilus
Abstract
To acquire the ability to recognize and destroy virus and plasmid invaders, prokaryotic CRISPR–Cas systems capture fragments of DNA within the host CRISPR locus. Our results indicate that the process of adaptation by a Type II-A CRISPR–Cas system in Streptococcus thermophilus requires Cas1, Cas2, and Csn2. Surprisingly, we found that Cas9, previously identified as the nuclease responsible for ultimate invader destruction, is also essential for adaptation. Cas9 nuclease activity is dispensable for adaptation. In addition, our studies revealed extensive, unbiased acquisition of the self-targeting host genome sequence by the CRISPR–Cas system that is masked in the presence of active target destruction.
CRISPR–Cas systems provide prokaryotes with adaptive immunity against invaders such as viruses/phages and plasmids (Terns and Terns 2011; Barrangou and Marraffini 2014; Heler et al. 2014; van der Oost et al. 2014). CRISPR–Cas systems target invaders using information stored in CRISPRs: loci that contain alternating units of an identical repeat (repeats) and short invader-derived sequences (spacers) (Fig. 1A). CRISPR transcripts are processed to a battery of CRISPR RNAs (crRNAs) that each contains a unique invader guide sequence (and common repeat sequence). The crRNAs associate with Cas proteins to form effector complexes that recognize and degrade invading nucleic acids to effect immunity (Terns and Terns 2011; Barrangou and Marraffini 2014; Heler et al. 2014; van der Oost et al. 2014). Diverse CRISPR–Cas systems are prevalent in bacteria and archaea and are categorized into three compositionally distinct groups (Types I–III), with multiple subtypes within each group (Haft et al. 2005; Makarova et al. 2011).
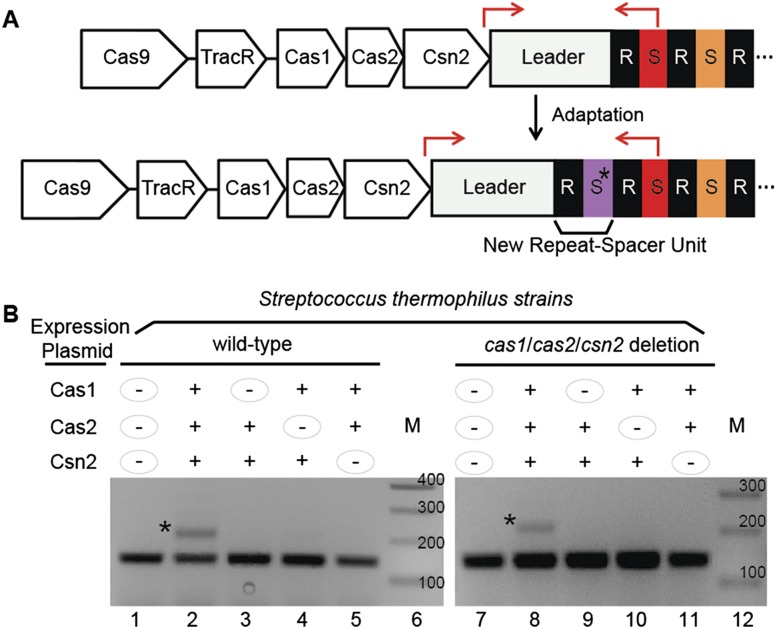
Figure 1.
Cas1, Cas2, and Csn2 are critical for adaptation. (A) Illustration of CRISPR1 module and adaptation. (Top) The S. thermophilus DGCC7710 CRISPR1 module encodes four Cas proteins: Cas1, Cas2, Csn2, and Cas9. The tracrRNA is encoded between Cas9 and Cas1. The CRISPR leader sequence (gray) is followed by 32 repeat (R, black)–spacer (S, colored) units (only a few of which are shown for simplicity). The locations of primers used in PCR for detection of new spacer acquisition are indicated by red arrows. (Bottom) The product of acquisition of a novel spacer (asterisk; purple) and an associated repeat is illustrated. (B) Analysis of spacer acquisition in wild-type (left) and cas1/cas2/csn2 deletion (right) strains expressing combinations of Cas1, Cas2, and Csn2 proteins from plasmids. The leader-proximal region of CRISPR1 was PCR-amplified with primers indicated in A. Plasmid expression of individual proteins is indicated with “+” or “−” signs. The PCR product corresponding to addition of one repeat–spacer unit is indicated with an asterisk. Sizes of DNA standards (M) are indicated. Data representative of multiple experiments are shown.
The initial step of capturing short fragments of invasive DNA into CRISPR loci (“adaptation” or “spacer acquisition”) is the least understood aspect of the CRISPR immune pathway. Adaptation appears to be a rare event but generates subpopulations of organisms that can survive infection. It has been proposed that the mechanism involves identification of “foreign” sequences for incorporation into the CRISPR (Datsenko et al. 2012; Yosef et al. 2012; Diez-Villasenor et al. 2013; Nunez et al. 2014), although host genome sequences have also been observed in CRISPRs at very low frequencies (Stern et al. 2010; Jiang et al. 2013; Paez-Espino et al. 2013). Selection of invader DNA fragments (protospacers) by the adaptation machinery requires the presence of a short (3- to 7-base-pair [bp]) neighboring motif called a protospacer-adjacent motif (PAM) (Mojica et al. 2009; Shah et al. 2013; Heler et al. 2014). Incorporation of each new spacer into a CRISPR locus is also accompanied by generation of a new repeat and occurs predominantly at the leader/repeat junction (Barrangou et al. 2007; Deveau et al. 2008; Garneau et al. 2010; Datsenko et al. 2012; Erdmann and Garrett 2012; Swarts et al. 2012; Yosef et al. 2012; Diez-Villasenor et al. 2013; Li et al. 2014).
An important goal toward understanding CRISPR adaptation is identifying the proteins (Cas and non-Cas) responsible for novel spacer acquisition in CRISPR loci in diverse CRISPR–Cas systems. Genetic studies indicate that overexpression of Cas1 and Cas2—the only Cas proteins universal to all CRISPR–Cas systems—is sufficient to induce adaptation in the absence of other Cas proteins in Type I systems such as that found in Escherichia coli (Datsenko et al. 2012; Yosef et al. 2012; Diez-Villasenor et al. 2013; Nunez et al. 2014).
Limited information is available regarding the trans-acting factors required for adaptation in Type II CRISPR–Cas systems (and no work has yet been done regarding adaptation in Type III systems). The three distinct Type II subtypes (Chylinski et al. 2014) each contain (1) three Cas proteins (Cas1, Cas2, and Cas9), (2) a trans-activating crRNA (tracrRNA) (Deltcheva et al. 2011), and (3) the CRISPR array (Fig. 1A). Interestingly, a fourth Cas protein is found in Type II-A (Csn2) and Type II-B (Cas4) systems but not Type II-C systems (Chylinski et al. 2014). Cas1 and Cas2 are presumed to be essential for adaptation in Type II systems, given their vital role in adaptation in Type I systems (Datsenko et al. 2012; Yosef et al. 2012; Diez-Villasenor et al. 2013; Nunez et al. 2014). Genetic studies (cas gene disruptions) in Streptococcus thermophilus suggest a specific requirement for Csn2 in Type II-A adaptation (Barrangou et al. 2007). Expanded CRISPR loci were not observed in a csn2 disruption strain challenged by lytic phage infection (Barrangou et al. 2007). crRNA production (Carte et al. 2014) and invader defense (Barrangou et al. 2007) were unaffected in the csn2 disruption strain. Cas9 (common to Type II systems) has been found to function in crRNA biogenesis and accumulation (Deltcheva et al. 2011; Carte et al. 2014) and invader defense (Barrangou et al. 2007; Garneau et al. 2010); however, the potential role of Cas9 in adaptation has not been examined.
As the effector nuclease of Type II CRISPR–Cas systems, Cas9 (guided by a crRNA/tracrRNA duplex) cuts opposing strands of complementary invading DNA using two nuclease domains (RuvC and HNH) (Garneau et al. 2010; Gasiunas et al. 2012; Jinek et al. 2012). The nuclease activity of the Cas9/guide RNA complex has been adapted as a powerful genome-editing tool in a variety of cell types and organisms (for review, see Terns and Terns 2014). Mutation of the nuclease domains results in a catalytically defective form of Cas9 (dCas9) that has been applied to control gene expression as an RNA-guided DNA-binding protein (Terns and Terns 2014).
In this study, we examined adaptation by a Type II-A CRISPR–Cas system in S. thermophilus. We expressed increased levels of Cas1, Cas2, and Csn2 to attain levels of adaptation that were detectable without selection in populations of cells. We tested the role of all four Type II-A Cas proteins in adaptation and unexpectedly found that Cas9 (the effector nuclease) is also required for adaptation. The nuclease activity of Cas9 is not required for adaptation, and, in the absence of nuclease destruction of the targets of acquired sequences, we found that the host genome is targeted without discrimination (i.e., equivalently to an invader) by the CRISPR–Cas system.
Results and Discussion
Cas1, Cas2, and Csn2 are required for adaptation
Adaptation (illustrated in Fig. 1A) is rarely detected within prokaryotic populations under laboratory conditions (Barrangou et al. 2007; Deveau et al. 2008; Garneau et al. 2010). We increased the frequency of adaptation by the Type II-A CRISPR–Cas system (CRISPR1) in S. thermophilus by increasing levels of Cas1, Cas2, and Csn2 (three proteins hypothesized to mediate adaptation). Adaptation events within the S. thermophilus population at CRISPR1 can be monitored by PCR amplification of the leader-proximal region (Fig. 1B, with primers at red arrows in A). Expansion of CRISPR1 (increase in the size of the PCR product by the unit length of the added spacer and repeat noted with asterisks in Fig. 1B) was observed in a detectable fraction of the population of the wild-type strain containing the pCas1/Cas2/Csn2 plasmid but not an empty plasmid (Fig. 1B, lanes 1,2), indicating that increasing expression of Cas1, Cas2, and Csn2 increases adaptation frequency. To assess whether all three proteins are required to observe adaptation, we systematically eliminated each one (Fig. 1B, lanes 3–5). CRISPR expansion was observed only when all three Cas proteins were expressed (Fig. 1B). Similar results were obtained for expression of the various combinations of Cas proteins in an S. thermophilus strain lacking endogenous cas1, cas2, and csn2 genes (Fig. 1B, right panel).
These results are the first direct genetic evidence indicating that Cas1, Cas2, and Csn2 each functions in CRISPR adaptation in Type II systems. Recent structural and biochemical studies on E. coli Type I-E system components suggest that Cas1 and Cas2 form an integrase-type complex that uses the nuclease active site of Cas1 to carry out spacer integration at CRISPRs (Datsenko et al. 2012; Yosef et al. 2012; Nunez et al. 2014). Csn2 proteins purified from diverse organisms form tetrameric ring-like structures that encircle and bind dsDNA, but no relevant enzymatic activities have yet been identified (Nam et al. 2011; van der Oost et al. 2014). Sequences at the junction of the leader and first repeat of the CRISPR direct the integration of new spacers in S. thermophilus CRISPR1 (Wei et al. 2015). The Cas1/Cas2 complex (possibly in association with Csn2) may recognize the leader/repeat junction sequence element to mediate specific integration into CRISPRs at the leader end.
Cas9 is required for adaptation
Surprisingly, we also found that Cas9 plays a role in adaptation in the S. thermophilus Type II CRISPR–Cas system. Cas9 was previously found to be the nuclease responsible for invader DNA cleavage in Type II systems and was not expected to play a role in adaptation (Barrangou et al. 2007; Gasiunas et al. 2012; Jinek et al. 2012). Previous studies examined adaptation by methods that required the invader nuclease function of Cas9 (e.g., survival of lytic phage or cleavage of exogenous plasmid) (Barrangou et al. 2007; Deveau et al. 2008; Garneau et al. 2010; Lopez-Sanchez et al. 2012; Paez-Espino et al. 2013) and therefore could not examine adaptation in the absence of Cas9. However, the assay used in this study (detection of adaptation by PCR amplification of the CRISPR locus in cell populations) does not require active defense.
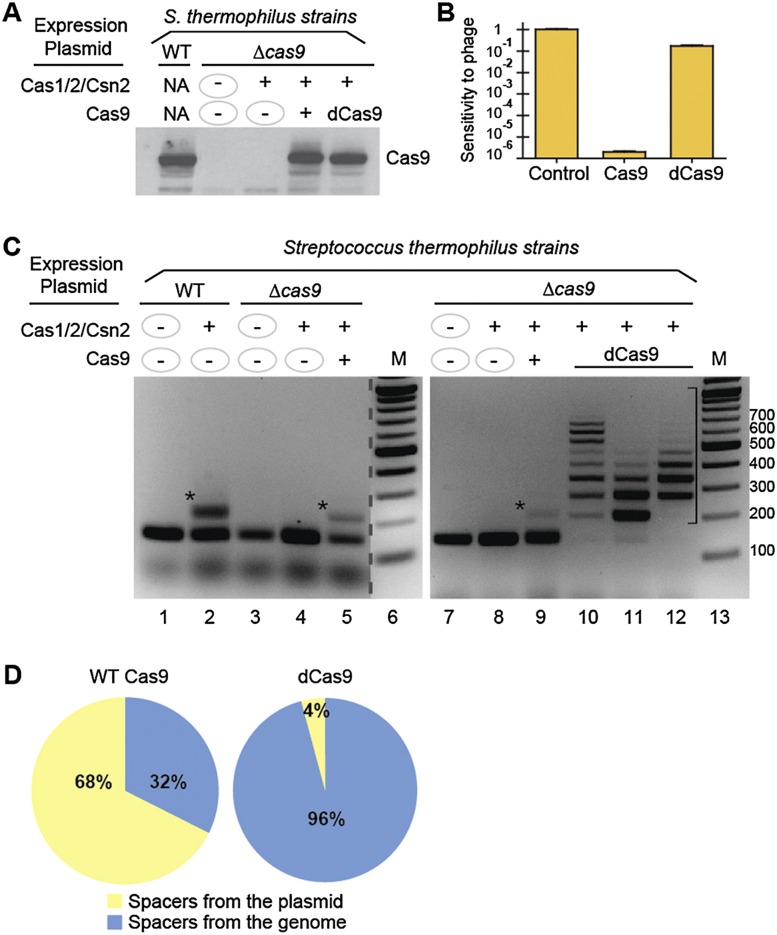
We deleted cas9 from the genome and, surprisingly, found that the adaptation observed with Cas1/Cas2/Csn2 expression (Fig. 2C, lane 2) was lost in the absence of Cas9 (Fig. 2C, lane 4). Adaptation was rescued by expression of Cas9 from a plasmid (with native promoter) (Fig. 2C, lane 5). Elimination and restoration of Cas9 protein expression in the deletion and plasmid rescue strains, respectively, was confirmed by Western analysis (Fig. 2A). These results indicate that, surprisingly, Cas9 is important for acquisition of invader sequences as well as invader destruction.
Figure 2.
Lack of adaptation in the absence of Cas9 protein and more extensive adaptation in the absence of Cas9 nuclease activity. (A) Cas9 protein levels in S. thermophilus wild-type (WT) and cas9 deletion (Δcas9) strains expressing Cas1/Cas2/Csn2 and Cas9 proteins from plasmids. Cas9 was detected via Western blot using polyclonal antibodies. Plasmid expression of proteins is indicated with “+” or “−” signs. (B) The sensitivity of strains expressing functional Cas9 (middle) or nuclease-defective Cas9 (right) to lytic phage 2972 relative to a strain lacking Cas9 (left; control) measured as efficiency of plaquing. (C) Analysis of spacer acquisition in wild-type and cas9 deletion (Δcas9) strains expressing Cas1/Cas2/Csn2 and Cas9 proteins from plasmids. The leader-proximal region of CRISPR1 was PCR-amplified with primers indicated in Figure 1A. Plasmid expression of proteins is indicated with “+” or “−” signs. PCR products corresponding to addition of one or multiple repeat–spacer units are indicated with an asterisk or bracket, respectively. Sizes of DNA standards (M) are indicated. Data representative of multiple experiments are shown. Noncontiguous lanes from the same gel are separated by a dashed line. (D) Percentages of spacers derived from the plasmid (yellow) and genome (blue) in strains expressing wild-type Cas9 (left; wild-type S. thermophilus cells expressing Cas1/Cas2/Csn2) (Fig. 1B, lane 2) or dCas9 (right; cas9 deletion strain expressing dCas9 and Cas1/Cas2/Csn2) (C, lane 10). See Supplemental Tables S1 and S4 for details. One spacer matched the cas1 gene, which is present both on the plasmid and in the genome and was arbitrarily assigned to the plasmid.
Both adaptation and invader destruction depend on PAMs in Type I and Type II CRISPR–Cas systems (Mojica et al. 2009; Shah et al. 2013; Heler et al. 2014). PAMs are important for initial protospacer selection in adaptation (in which the motifs are more specifically termed SAMs [spacer acquisition motifs]) and subsequent recognition of that protospacer sequence as the target for invader destruction (in which the motifs are more specifically termed TIMs [target interference motifs]) (Shah et al. 2013). The PAM sequence required by a CRISPR–Cas system is not found in its CRISPR repeat sequence, thereby ensuring that when a sequence is incorporated into a CRISPR, the copy in the CRISPR of the host genome is not targeted for cleavage. Evidence indicates that the C-terminal domain of Cas9 interacts with the TIM in the context of the crRNA-containing invader targeting complex for Type II systems (Anders et al. 2014; Nishimasu et al. 2014; Sternberg et al. 2014). Our findings suggest Cas9 as a likely SAM recognition factor in adaptation, providing the critical link that ensures that sequences selected for acquisition will be recognized and effectively silenced by the effector complex.
The RuvC- and HNH-based nuclease activities of Cas9 are not required for adaptation
In invader defense, Cas9 uses RuvC and HNH domains for target DNA cleavage (Gasiunas et al. 2012; Jinek et al. 2012). Adaptation conceivably also involves dsDNA cleavages (to obtain the protospacer fragment from the invader and open the CRISPR for integration of the new spacer). To assess whether the ability to cleave DNA is important for Cas9 function in adaptation, we introduced nuclease domain mutations (Gasiunas et al. 2012; Jinek et al. 2012; Nishimasu et al. 2014) into Cas9 within pCas1/Cas2/Csn2/Cas9 (and refer to the mutant Cas9D9A/H599A as dCas9 for catalytically defective Cas9) and expressed the construct in the S. thermophilus cas9 deletion strain. We confirmed that the mutant Cas9 protein is expressed (Fig. 2A) and is defective in invader defense: The strain expressing the nuclease mutant fails to resist a S. thermophilus lytic phage that is effectively targeted in the presence of wild-type Cas9 (Fig. 2B). However, we found that mutations in the RuvC and HNH domains did not disrupt adaptation (Fig. 2C, lanes 10–12), indicating that the nuclease activity of Cas9 is not required for adaptation.
Moreover, when target destruction was blocked by elimination of Cas9 nuclease activity, spacer accumulation increased significantly. PCR analysis of the cultures indicates that almost all of the cells in the population have acquired at least one spacer: Very little of the product corresponding to the original unadapted CRISPR locus is observed in these cultures (Fig. 2C, lanes 10–12 are examples of three independent cultures). Furthermore, many cells acquired two, three, or more spacers, evidenced by the PCR products of increasing repeat–spacer unit size (Fig. 2C, lanes 10–12). In the presence of functional Cas9, any individual that has acquired a spacer targeting the plasmid (or the genome) is eventually expected to be lost from the population due to chloramphenicol selection against loss of the plasmid (or lethality of genome targeting). In the presence of nuclease-defective Cas9, however, individuals that have undergone adaptation can survive and acquire additional spacers. It is also possible that in the absence of nuclease activity, Cas9 becomes more available for its role in adaptation.
CRISPR1 acquires sequences from the genome as well as invader plasmid
We examined the origin and context of the new spacers that we had initially observed in the wild-type S. thermophilus cells expressing Cas1/Cas2/Csn2 (in the presence of functional Cas9 and active defense) (Fig. 1B, lane 2). Of 37 unique spacers, 25 spacers (68%) were directed against the plasmid, and, interestingly, 12 spacers (32%) were from the S. thermophilus genome (Fig. 2D; Supplemental Table S1). (One spacer matched the cas1 gene, which is present both on the plasmid and in the genome and is arbitrarily assigned to the plasmid.) A PAM/SAM sequence is crucial for selection of an invader sequence for CRISPR incorporation (Mojica et al. 2009; Shah et al. 2013; Heler et al. 2014), and, consistently, we found the predicted NNAGAAW PAM sequence (or a previously observed variation of this consensus motif) (Deveau et al. 2008; Garneau et al. 2010; Paez-Espino et al. 2013) present adjacent to each protospacer sequence in the source, whether it was the plasmid or the genome (Supplemental Tables S1, S2).
In our assay, both the genome- and plasmid-derived spacers acquired in the presence of functional Cas9 are expected to be lost from the population over time due to the deleterious effect of targeting either source. While we observed a significant extent of acquisition of spacers from the host genome, spacers from the genome are nonetheless distinctly underrepresented relative to those from the plasmid (Fig. 1B, lane 2; Supplemental Table S1). The S. thermophilus genome (∼1.8 × 106 bp) represents ∼100 times the DNA content of the plasmid within the cell (∼1.8 × 104 bp when adjusted for plasmid copy number of approximately three) (Kok et al. 1984). However, at the time point examined, more spacers found in the population are from the plasmid (68%) than from the genome (32%), suggesting active selection of plasmid-derived spacers. Alternatively, the underrepresentation of spacers from the genome could reflect a greater penalty for targeting the host genome relative to an antibiotic-selected plasmid.
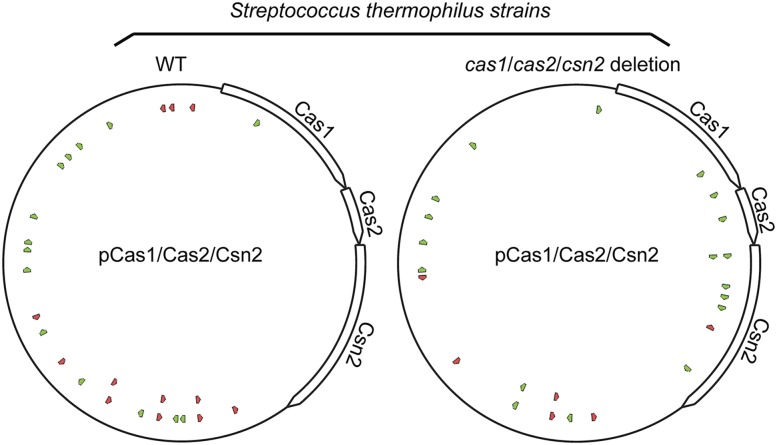
Notably, among the 25 plasmid-derived spacers, only one (4%) matched the cas1/cas2/csn2-coding region despite the fact that these genes comprise 37% of the plasmid sequence and contain 35% of the consensus (NNAGAAW) PAMs in the plasmid (Fig. 3, left panel). The underrepresentation of cas1/cas2/csn2-matching spacers could reflect specific avoidance of the cas genes in the process of adaptation or the detrimental effect of genome-matching spacers in an active CRISPR–Cas system (as spacers matching the cas1/cas2/csn2-coding region of the plasmid would also target the endogenous cas1/cas2/csn2 genes present in the genome). To determine whether the presence of genome copies impacts the abundance of spacers against the cas genes on the plasmid, we examined the sequences of spacers acquired in the strain lacking the cas1/cas2/csn2 gene targets within the genome (cas1/cas2/csn2 deletion strain) (Fig. 1B, lane 8). We found that the cas1/cas2/csn2 genes were targeted without bias in the absence of the genome copies: Ten (42%) of 24 plasmid spacers matched the cas1/cas2/csn2 genes (Fig. 3, right panel; Supplemental Table S3). The results provide further support that spacers targeting the genome (as opposed to the plasmid only) are more effectively lost from the population.
Figure 3.
Distribution of pCas1/Cas2/Csn2 plasmid protospacers in wild-type (left) and cas1/cas2/csn2 deletion strains (right). Protospacer positions and orientations (designated with green and red arrows) are marked on the pCas1/Cas2/Csn2 plasmid map. Cas1-, Cas2-, and Csn2-coding regions are labeled with open arrows.
Elimination of Cas9 nuclease activity reveals extensive, unbiased acquisition of spacers from the genome
Our findings indicate that spacers are acquired from the genome and lost from the population due to subsequent CRISPR–Cas targeting of the genome. To better determine the extent of genome spacer acquisition, we examined adaptation in the absence of target destruction. We sequenced 48 spacers from the cas9 deletion strain expressing dCas9 and Cas1/Cas2/Csn2 (Fig. 2C, lane 10). Remarkably, we found that, in the absence of defense, the overwhelming majority of spacers were derived from the genome: Forty-six of the 48 spacers (96%) were derived from the S. thermophilus genome, and two spacers (4%) originated from the plasmid (Fig. 2D, right; Supplemental Table S4). The distribution correlates well with the relative DNA content of the cell (99% genome and 1% plasmid), indicating that the host genome is targeted for spacer acquisition equivalently to the plasmid and that there is no significant bias for invader DNA in CRISPR–Cas adaptation by this system. PAM distribution in the presence of dCas9 is similar to that in the presence of wild-type Cas9 (Supplemental Table S5).
Self-targeting spacers are rarely observed at steady state in organisms with functioning CRISPR–Cas systems: Computational analysis of 23,550 spacers from 330 organisms found that 0.4% of all sequenced spacers were self-targeting (Stern et al. 2010). Moreover, in a study of long-term lytic phage infection of S. thermophilus, only 0.04% of acquired spacers observed were found to match the host genome (Paez-Espino et al. 2013). Some evidence indicates that invader sequences are actively selected and/or that genome sequences are actively avoided by CRISPR–Cas adaptation systems. In a Type I-E system in E. coli, spacers targeting plasmids were found to significantly outnumber genome-derived spacers even in the absence of CRISPR–Cas defense (Yosef et al. 2012; Diez-Villasenor et al. 2013). However, our analysis of a Type II-A system in S. thermophilus indicates that spacers are acquired from the genome more frequently than invader nucleic acid (in proportion to the relative cellular DNA content of each) and are eliminated from the population by subsequent CRISPR–Cas system targeting.
Our analysis of adaptation in S. thermophilus in the absence of defense unmasked a higher rate of ongoing adaptation (Fig. 2C, lanes 10–12) as well as extensive and unbiased genome sampling in this organism (Supplemental Table S4). Our results indicate that extensive acquisition of genome sequences occurs within populations and is normally undetected due to the lethal consequence of the event. Our findings provide strong evidence that some CRISPR–Cas systems provide prokaryotes with a bet-hedging strategy to survive nucleic acid invaders—available DNA is randomly incorporated into the CRISPR locus, and the individuals that acquire genome targeting spacers die, but those that acquire invader targeting spacers gain a fitness advantage that can ensure survival. The lack of discrimination in spacer acquisition may illuminate the need for the low rate of adaptation generally observed among prokaryotic populations: avoiding loss of great numbers of the population due to acquisition of self-targeting spacers while providing for a rate of acquisition sufficient to ensure survival upon encountering a damaging invader.
Materials and methods
Strains and plasmids
S. thermophilus DGCC7710 was kindly provided by Dr. Sylvain Moineau. The cas9 deletion and cas1/cas2/csn2 deletion strains were constructed using methods developed by Renye and Somkuti (2009) using plasmid pINTRS. Sequences of the CRISPR locus and deleted regions are shown in that Supplemental Material. S. thermophilus was maintained in M17 medium (Oxiod) supplemented with 0.5% lactose (LM17). S. thermophilus cultures were grown overnight at 37° and during the day (8 h) at 42°. E. coli Top10 (grown in Luria broth) was used for cloning and plasmid maintenance. For strains harboring pWAR228-derived plasmids (kindly provided by Dr. Michael Federle), chloramphenicol was supplemented at 10 µg/mL for E. coli, at 2 µg/mL in LM17 broth, and at 5 µg/mL in LM17 plates (1% agar) for S. thermophilus. Strains and plasmids used in this study are listed in Supplemental Table S6.
DNA manipulation
Standard protocols were used for cloning. Phusion polymerase, restriction enzymes, and T4 DNA ligase were from New England Biolabs. Taq polymerase with Crimson Taq buffer (New England Biolabs) was used for colony PCR. Zymoclean gel DNA recovery kit (Zymo Research) was used for gel extraction. QIAprep Spin miniprep kit and QIAfilter plasmid midi kit (Qiagen) were used for plasmid preparations. DNA Clean and Concentrator-5 (Zymo Research) was used for concentrating plasmids.
Construction of plasmids
Plasmid pWAR228 was used as the backbone for plasmid constructions. The Ppgm promoter was PCR-amplified from plasmid pTRK882 (kindly provided by Dr. Todd Klaenhammer) (Duong et al. 2011). The Cas1/Cas2/Csn2-coding region adjacent to CRISPR1 was PCR-amplified from the S. thermophilus genome and linked to the Ppgm promoter via overlap PCR. The final PCR product was assembled onto pWAR228 to yield plasmid pCas1/Cas2/Csn2 via restriction digestion (XhoI and NotI sites) and ligation. Inverse PCR was used to delete each of the three cas genes on pCas1/Cas2/Csn2. To construct pCas1/Cas2/Csn2/Cas9, the Cas9 promoter and coding region were PCR-amplified from the S. thermophilus genome and assembled onto pCas1/Cas2/Csn2 via restriction digestion (using a XhoI site) and ligation to yield pCas1/Cas2/Csn2/Cas9. Active sites of Cas9 were mutagenized (D9A and H599A) via QuikChange PCR using pCas1/Cas2/Csn2/Cas9 as the template. Plasmid constructions were confirmed by sequencing and transformed into S. thermophilus via electroporation (Garneau et al. 2010). Oligos used for cloning are listed in Supplemental Table S7.
Population-based adaptation assay and spacer sequence analysis
S. thermophilus cells harboring the indicated plasmids were grown in LM17 supplemented with 2 µg/mL chloramphenicol for 16 h. Cells from each strain were pelleted, resuspended in water by vortexing, and used as PCR templates. Primers matching the leader and the first spacer of CRISPR1 (Supplemental Table S7) were used for PCR amplification of CRISPR1. PCR products were run on 2% TAE-agarose gels, stained with ethidium bromide, and examined under UV light to assess CRISPR expansion. Gel bands were excised, purified, cloned into Topo vector using the Zero Blunt TOPO PCR cloning kit (Life Technology), and sequenced (Eurofins Genomics).
Western blotting
Wild-type S. thermophilus or S. thermophilus strains harboring the indicated plasmids were grown overnight. Cells were pelleted and lysed with a Mini-Beadbeater (Biospec Products). Thirty micrograms of total protein from each lysate was separated on a 10% SDS-PAGE gel and blotted onto a nitrocellulose membrane (Bio-Rad). Cas9 protein was detected using polyclonal chicken antibodies raised against Cas9 and HRP-conjugated donkey anti-chicken IgY (Gallus Immumotech).
Phage defense assay
An S. thermophilus cas9 deletion strain harboring pCas1/Cas2/Csn2/Cas9 was infected with lytic phage 2972. A surviving strain, which was confirmed to have obtained a phage-matching spacer in CRISPR1 (protospacer, AAATCAGTTTTTTGTTCAGAAACTTGTTCT; PAM, TTAGAAA), was allowed to lose the plasmid through passages on growth medium without antibiotics. The resulting strain (without the plasmid) was back-transformed with pCas1/Cas2/Csn2, pCas1/Cas2/Csn2/Cas9, or pCas1/Cas2/Csn2/dCas9 and tested for sensitivity to lytic phage 2972 as previously described (Barrangou et al. 2007).
Supplementary Material
Acknowledgments
We thank members of the Terns laboratory and Claiborne Glover for helpful discussions, and Anthony Schmalstig, Xinfu Zhang, and Erin Hollander for S. thermophilus strain construction. We also thank Rodolphe Barrangou, Sylvain Moineau, Todd Klaenhammer, Michael Federle, John Renye, and George Somkuti for generously providing strains, phage, and plasmids. This study is supported by National Institutes of Health (NIH) grant RO1 GM099876 to M.P.T. and R.M.T. Y.W. is supported by the National Institute of General Medical Sciences of the NIH under award number F32GM108263.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.257550.114.
References
- Anders C, Niewoehner O, Duerst A, Jinek M. 2014. Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature 513: 569–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrangou R, Marraffini LA. 2014. CRISPR–Cas systems: prokaryotes upgrade to adaptive immunity. Mol Cell 54: 234–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315: 1709–1712. [DOI] [PubMed] [Google Scholar]
- Carte J, Christopher RT, Smith JT, Olson S, Barrangou R, Moineau S, Glover CV 3rd, Graveley BR, Terns RM, Terns MP. 2014. The three major types of CRISPR–Cas systems function independently in CRISPR RNA biogenesis in Streptococcus thermophilus. Mol Microbiol 93: 98–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chylinski K, Makarova KS, Charpentier E, Koonin EV. 2014. Classification and evolution of type II CRISPR–Cas systems. Nucleic Acids Res 42: 6091–6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA, Pougach K, Tikhonov A, Wanner BL, Severinov K, Semenova E. 2012. Molecular memory of prior infections activates the CRISPR/Cas adaptive bacterial immunity system. Nat Commun 3: 945. [DOI] [PubMed] [Google Scholar]
- Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E. 2011. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 471: 602–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deveau H, Barrangou R, Garneau JE, Labonte J, Fremaux C, Boyaval P, Romero DA, Horvath P, Moineau S. 2008. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J Bacteriol 190: 1390–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diez-Villasenor C, Guzman NM, Almendros C, Garcia-Martinez J, Mojica FJ. 2013. CRISPR-spacer integration reporter plasmids reveal distinct genuine acquisition specificities among CRISPR–Cas I-E variants of Escherichia coli. RNA Biol 10: 792–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duong T, Miller MJ, Barrangou R, Azcarate-Peril MA, Klaenhammer TR. 2011. Construction of vectors for inducible and constitutive gene expression in Lactobacillus. Microb Biotechnol 4: 357–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdmann S, Garrett RA. 2012. Selective and hyperactive uptake of foreign DNA by adaptive immune systems of an archaeon via two distinct mechanisms. Mol Microbiol 85: 1044–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garneau JE, Dupuis ME, Villion M, Romero DA, Barrangou R, Boyaval P, Fremaux C, Horvath P, Magadan AH, Moineau S. 2010. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 468: 67–71. [DOI] [PubMed] [Google Scholar]
- Gasiunas G, Barrangou R, Horvath P, Siksnys V. 2012. Cas9–crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci 109: E2579–E2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haft DH, Selengut J, Mongodin EF, Nelson KE. 2005. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput Biol 1: e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heler R, Marraffini LA, Bikard D. 2014. Adapting to new threats: the generation of memory by CRISPR–Cas immune systems. Mol Microbiol 93: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. 2013. RNA-guided editing of bacterial genomes using CRISPR–Cas systems. Nat Biotechnol 31: 233–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. 2012. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337: 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kok J, van der Vossen JM, Venema G. 1984. Construction of plasmid cloning vectors for lactic streptococci which also replicate in Bacillus subtilis and Escherichia coli. Appl Environ Microbiol 48: 726–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Wang R, Xiang H. 2014. Haloarcula hispanica CRISPR authenticates PAM of a target sequence to prime discriminative adaptation. Nucleic Acids Res 42: 7226–7235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Sanchez MJ, Sauvage E, Da Cunha V, Clermont D, Ratsima Hariniaina E, Gonzalez-Zorn B, Poyart C, Rosinski-Chupin I, Glaser P. 2012. The highly dynamic CRISPR1 system of Streptococcus agalactiae controls the diversity of its mobilome. Mol Microbiol 85: 1057–1071. [DOI] [PubMed] [Google Scholar]
- Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, Moineau S, Mojica FJ, Wolf YI, Yakunin AF, et al. 2011. Evolution and classification of the CRISPR–Cas systems. Nat Rev Microbiol 9: 467–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, Almendros C. 2009. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology 155: 733–740. [DOI] [PubMed] [Google Scholar]
- Nam KH, Kurinov I, Ke A. 2011. Crystal structure of clustered regularly interspaced short palindromic repeats (CRISPR)-associated Csn2 protein revealed Ca2+-dependent double-stranded DNA binding activity. J Biol Chem 286: 30759–30768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimasu H, Ran FA, Hsu PD, Konermann S, Shehata SI, Dohmae N, Ishitani R, Zhang F, Nureki O. 2014. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell 156: 935–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunez JK, Kranzusch PJ, Noeske J, Wright AV, Davies CW, Doudna JA. 2014. Cas1–Cas2 complex formation mediates spacer acquisition during CRISPR–Cas adaptive immunity. Nat Struct Mol Biol 21: 528–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paez-Espino D, Morovic W, Sun CL, Thomas BC, Ueda K, Stahl B, Barrangou R, Banfield JF. 2013. Strong bias in the bacterial CRISPR elements that confer immunity to phage. Nat Commun 4: 1430. [DOI] [PubMed] [Google Scholar]
- Renye JA Jr, Somkuti GA. 2009. Insertion of a heterologous gene construct into a non-functional ORF of the Streptococcus thermophilus chromosome. Biotechnol Lett 31: 759–764. [DOI] [PubMed] [Google Scholar]
- Shah SA, Erdmann S, Mojica FJ, Garrett RA. 2013. Protospacer recognition motifs: mixed identities and functional diversity. RNA Biol 10: 891–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern A, Keren L, Wurtzel O, Amitai G, Sorek R. 2010. Self-targeting by CRISPR: gene regulation or autoimmunity? Trends Genet 26: 335–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg SH, Redding S, Jinek M, Greene EC, Doudna JA. 2014. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature 507: 62–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarts DC, Mosterd C, van Passel MW, Brouns SJ. 2012. CRISPR interference directs strand specific spacer acquisition. PLoS ONE 7: e35888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terns MP, Terns RM. 2011. CRISPR-based adaptive immune systems. Curr Opin Microbiol 14: 321–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terns RM, Terns MP. 2014. CRISPR-based technologies: prokaryotic defense weapons repurposed. Trends Gen 30: 111–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Oost J, Westra ER, Jackson RN, Wiedenheft B. 2014. Unravelling the structural and mechanistic basis of CRISPR–Cas systems. Nat Rev Microbiol 12: 479–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Chesne MT, Terns RM, Terns MP. 2015. Sequences spanning the leader-repeat junction mediate CRISPR adaptation to phage in Streptococcus thermophilus. Nucleic Acids Res doi: 10.1093/nar/gku1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yosef I, Goren MG, Qimron U. 2012. Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic Acids Res 40: 5569–5576. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.