Hainer et al. show that esBAF, a SWI/SNF family nucleosome remodeling factor, suppresses transcription of noncoding RNAs (ncRNAs) from ∼57,000 nucleosome-depleted regions (NDRs) throughout the genome of mouse embryonic stem cells. esBAF’s function to enforce nucleosome occupancy adjacent to NDRs, but not its function to maintain NDRs in a nucleosome-free state, is necessary for silencing transcription over ncDNA. Finally, the ability of a strongly positioned nucleosome to repress ncRNA depends on its translational positioning.
Keywords: esBAF, ncRNA, chromatin remodeling, nucleosome occupancy, transcription
Abstract
Approximately 75% of the human genome is transcribed, the majority of which does not encode protein. However, many noncoding RNAs (ncRNAs) are rapidly degraded after transcription, and relatively few have established functions, questioning the significance of this observation. Here we show that esBAF, a SWI/SNF family nucleosome remodeling factor, suppresses transcription of ncRNAs from ∼57,000 nucleosome-depleted regions (NDRs) throughout the genome of mouse embryonic stem cells (ESCs). We show that esBAF functions to both keep NDRs nucleosome-free and promote elevated nucleosome occupancy adjacent to NDRs. Reduction of adjacent nucleosome occupancy upon esBAF depletion is strongly correlated with ncRNA expression, suggesting that flanking nucleosomes form a barrier to pervasive transcription. Upon forcing nucleosome occupancy near two NDRs using a nucleosome-positioning sequence, we found that esBAF is no longer required to silence transcription. Therefore, esBAF’s function to enforce nucleosome occupancy adjacent to NDRs, and not its function to maintain NDRs in a nucleosome-free state, is necessary for silencing transcription over ncDNA. Finally, we show that the ability of a strongly positioned nucleosome to repress ncRNA depends on its translational positioning. These data reveal a novel role for esBAF in suppressing pervasive transcription from open chromatin regions in ESCs.
Transcription by RNA polymerase II (RNAPII) is widespread throughout the genome and not limited to coding sequences. In fact, nongenic regions account for the majority of sequences transcribed by RNAPII in higher eukaryotes (Djebali et al. 2012; Stamatoyannopoulos 2012). Noncoding RNA (ncRNA) is produced from many different regulatory regions in cells, including the 3′ ends of protein-coding genes, divergent transcription from promoters, and other distal regulatory elements, including enhancers or other gene-distal DNase I-hypersensitive sites (DHSs) (Jacquier 2009). While ncRNAs such as ribosomal RNAs (rRNAs), microRNAa (miRNAs), Piwi-interacting RNAs (piRNAs), and some long ncRNAs (lncRNAs) have well-established functions, the functions of the vast majority of ncRNAs discovered by transcriptome sequencing, if any, are unknown.
Antisense transcripts originating from promoters and aberrant transcription from intragenic regions have been well described in a diverse set of eukaryotes (Kaplan 2003; Carrozza et al. 2005; Cheung et al. 2008; Preker et al. 2008; Seila et al. 2008; Xie et al. 2011; Carvalho et al. 2013). In addition, transcription termination sites (TTSs) possess a promoter architecture sufficient to permit transcription initiation (Whitehouse et al. 2007; Murray et al. 2012; Castelnuovo et al. 2014). The mechanisms that regulate transcription start site (TSS)-associated ncRNA expression have recently been examined by several groups (Huang and Workman 2013). In mammalian cells, cotranscriptional and post-transcriptional mechanisms that mediate early termination of antisense ncRNAs and degradation by the RNA exosome complex combine to limit the abundance of TSS-associated RNAs relative to messenger RNAs (mRNAs) initiating from the same promoters (Arigo et al. 2006; Thiebaut et al. 2006; Thompson and Parker 2007; Preker et al. 2008; Flynn et al. 2011; Almada et al. 2013; Ntini et al. 2013). Furthermore, a recent study in yeast has shown that the chromatin assembly complex CAF-1 represses divergent (antisense) transcription at promoters (Marquardt et al. 2014). These findings suggest that promoters are biased toward transcription of the coding transcript in wild-type yeast cells, and this bias is enforced in part by CAF-1.
In contrast, most transcription of noncoding regions in higher eukaryotes occurs at distal regulatory regions of the genome (Djebali et al. 2012). While only ∼2% of the human genome constitutes coding sequences (International Human Genome Sequencing Consortium 2004; Elgar and Vavouri 2008), ∼75% of the human genome is transcribed, including a large fraction of gene-distal intergenic DNA (Djebali et al. 2012). Although little is known about most of these ncRNAs, short ncRNA molecules, termed eRNAs, are expressed from within enhancer regions (Kim et al. 2010; Wang et al. 2011a), suggesting that intergenic regions of open chromatin structure bound by transcription factors may be poised for RNAPII firing and ncRNA expression. However, the functions and regulation of the majority of these transcripts remain largely undefined.
Transcription by RNAPII through a chromatin template is a highly regulated process (Schwabish and Struhl 2004; Weiner et al. 2010; Adelman and Lis 2012). Nucleosomes, the basic units of chromatin, impact the regulation of transcription in several ways. Most significantly, nucleosomes interfere with binding of sequence-specific transcription factors and general transcription machinery (Almer and Hörz 1986; Boeger et al. 2003). Consequently, active enhancers and promoters generally contain nucleosome-depleted regions (NDRs) that are important for their functions (Yuan et al. 2005; Mavrich et al. 2008). Similarly, at TTSs, NDRs facilitate efficient termination of transcripts through recruitment of termination factors and promote gene looping in some contexts (Tan-Wong et al. 2012; Yadon et al. 2013). Therefore, NDRs within chromatin are critical for multiple aspects of transcriptional regulation.
One key gene regulatory mechanism shared among eukaryotes is the control of access to regulatory sequences by transcription factors through alteration of nucleosome occupancy or positioning. Nucleosome remodeling factors use the energy from ATP hydrolysis to reposition, deposit, or remove nucleosomes at regulatory regions and therefore can inhibit or promote binding of sequence-specific proteins to DNA (Racki and Narlikar 2008). The embryonic stem cell (ESC)-specific chromatin remodeling complex esBAF, a member of the SWI/SNF family of remodelers, plays a key role in regulating transcription of protein-coding genes and is required for the maintenance of pluripotency (Ho et al. 2009b). While esBAF has established roles in regulation of the expression of coding genes, the majority of esBAF binding occurs at gene-distal DHSs, many of which are regulatory regions bound by specific transcription factors (Ho et al. 2009a, 2011). Furthermore, esBAF is necessary for maintaining open chromatin structure over many DHSs in ESCs, consistent with its role as an activator of transcription (Ho et al. 2011) and raising the possibility that esBAF may activate expression of ncRNAs throughout the genome. However, the mechanisms by which esBAF regulates binding of transcription factors to these regions and how this contributes to regulation of gene expression are not well understood. Furthermore, while esBAF binds many regions of the genome where ncRNAs are expressed, whether and how it regulates ncRNA expression at these sites has not been examined.
Here, we test the hypothesis that esBAF is necessary for pervasive ncRNA expression observed throughout the genome of mammalian cells. We predicted that esBAF’s established function to maintain chromatin in an accessible state at gene-distal DHSs and over promoter-proximal regions of coding genes would facilitate ncRNA expression from these sites. Contrary to expectations, we found that esBAF functions mainly as a repressor of ncRNA expression from ∼57,000 NDRs throughout the ESC genome, the majority of which are located far from coding regions. Consistent with these findings, we observed similar alterations in chromatin structure in ESCs depleted of esBAF at gene-distal and gene-proximal NDRs from which ncRNAs are expressed. In the absence of esBAF, NDRs typically gain higher levels of nucleosome occupancy, and nucleosome occupancy is reduced over regions immediately adjacent to NDRs. Upon forcing elevated nucleosome occupancy adjacent to two such NDRs using a nucleosome-positioning sequence, we found that esBAF is no longer required to prevent transcription of the underlying ncRNAs. Therefore, we conclude that esBAF suppresses noncoding transcription from NDRs by enhancing the occupancy of flanking nucleosomes. Interestingly, depletion of the BAF catalytic subunit Brg1 has a minimal effect on ncRNA expression in mouse embryonic fibroblasts (MEFs), suggesting that either an alternative BAF complex or a BAF-independent pathway regulates ncRNA expression in differentiated cells. Together, our data show a novel role for esBAF in repressing ncRNA expression through targeted enhancement of nucleosome occupancy at the boundaries of NDRs across the ESC genome.
Results
esBAF represses transcription of ncRNAs throughout the ESC genome
esBAF localizes to regions of open chromatin throughout the murine ESC genome, with >80% of its peaks of binding occurring within DHSs, including both gene-distal DHSs and TSSs of genes (Ho et al. 2009a, 2011). Furthermore, esBAF is necessary for maintaining a DNase I-accessible chromatin configuration at many of these sites (Ho et al. 2011). These findings raise the possibility that esBAF may activate expression of ncRNAs expressed from these locations by maintaining open chromatin structure. To test this possibility, we performed two complementary RNA sequencing (RNA-seq) techniques, CapSeq and whole-transcript, strand-specific RNA-seq. CapSeq sequences capped RNA species from their 5′ end and therefore identifies the precise start sites of capped transcripts, allowing us to determine the genomic features from which ncRNAs initiate as well as measure their relative abundance genome-wide (Gu et al. 2012).
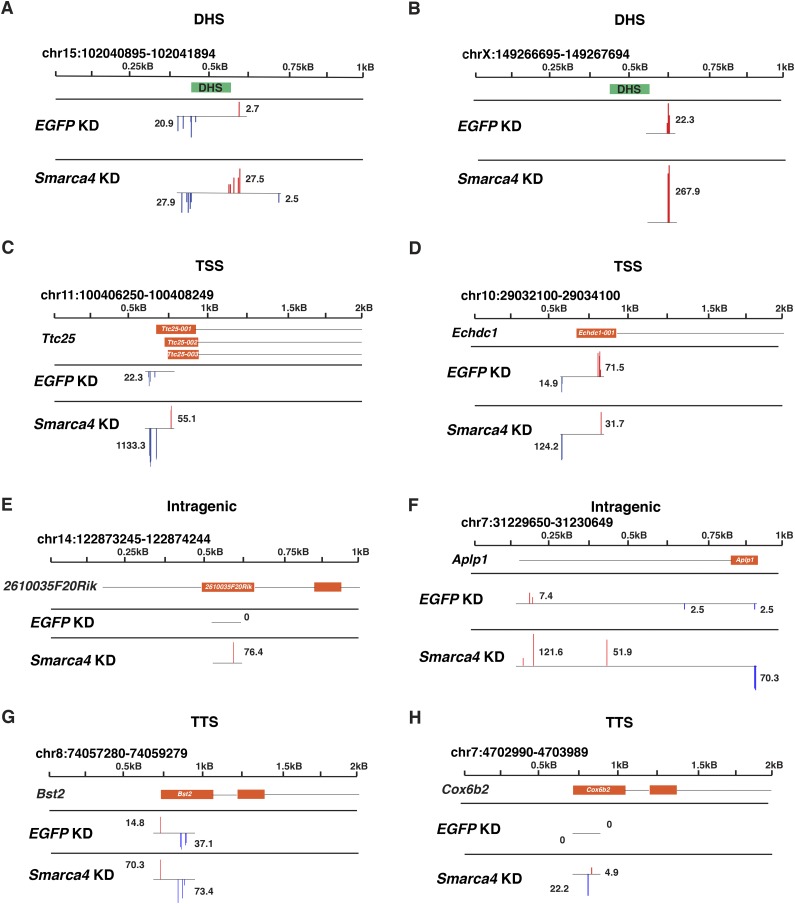
We compared the locations and abundance of capped transcripts in control (EGFP knockdown) ESCs and Smarca4 knockdown ESCs, which were depleted of the esBAF catalytic subunit Brg1 (gene name Smarca4) (Supplemental Fig. S1A,B). We performed acute knockdowns for 48 h, a time point at which we observed robust Brg1 depletion in ESCs (Supplemental Fig. S1A,B) but before self-renewal is impaired (Supplemental Fig. S1C,D). Using CapSeq, we observed clusters of ncRNAs initiating from within or near several genomic features associated with ncRNA expression, including gene-distal DHSs (Fig. 1A,B), TSSs (in the antisense orientation) (Fig. 1C,D), intragenic regions (Fig. 1E,F), and TTSs (Fig. 1G,H) in duplicate CapSeq data sets from each knockdown.
Figure 1.
The esBAF complex inhibits production of noncoding transcripts initiating from multiple genomic features. Genome browser tracks of two DHSs (A,B), two TSSs (C,D), two intragenic regions (E,F), and two TTSs (G,H). Bars corresponding to normalized CapSeq reads from EGFP knockdown (KD) and Smarca4 knockdown are shown in log2 scale at their initiation sites, and the (real space) number of normalized reads per cluster of transcripts is noted in each track. Browser tracks of normalized CapSeq reads of one replicate from EGFP knockdown and Smarca4 knockdown are shown. Isoforms are shown in an orange box below the scale, with introns indicated as black lines. DHSs are indicated in green boxes. Blue bars indicate transcription from the Crick strand, while red bars indicate transcription from the Watson strand. It should be noted that transcripts near TSSs in the same orientation as the coding sequence likely depict mRNAs not ncRNAs.
Contrary to our expectations, when we quantified capped ncRNAs originating from these genomic features, we observed a significant and global increase in noncoding transcripts at each of these regions (Supplemental Fig. S2A–D). In addition to regulation of ncRNA expression, we considered the possibility that esBAF regulates the locations of transcription initiation of both coding and noncoding transcripts. Therefore, we examined the distribution of TSS locations within the CapSeq data to test whether knockdown of Smarca4 alters TSS usage. We observed modest but significant changes (P = 1.532 × 10−14; Kolmogorov-Smirnov test) in the distribution of transcript initiation sites upon Smarca4 knockdown both upstream of and downstream from TSSs (Supplemental Fig. S3A–C,G), indicating that esBAF likely regulates alternative TSS usage at some genomic loci. In sum, CapSeq analyses demonstrate both a broader range of sequences at which transcription initiates upon esBAF loss and an increase in abundance of ncRNAs at multiple classes of genomic regions, including gene-distal DHSs, promoters, gene bodies, and transcriptional termini.
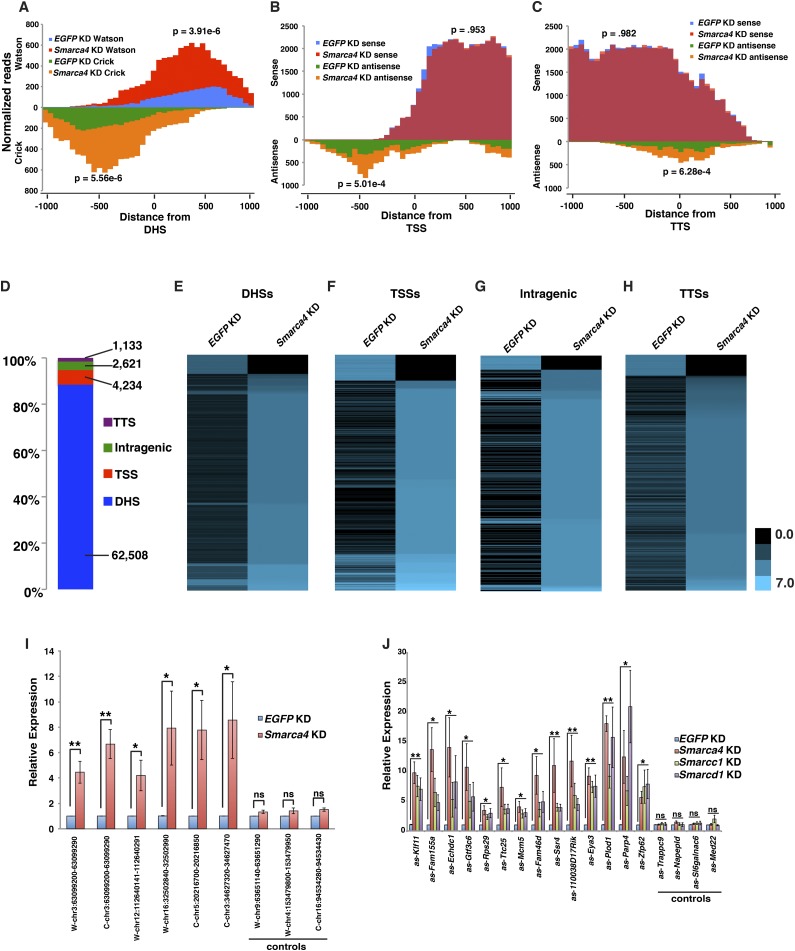
Having identified multiple genomic locations from which ncRNAs are up-regulated upon esBAF depletion, we wished to characterize these transcripts in greater detail. Although CapSeq provides insight into the start sites of ncRNAs, it provides no information about the lengths of these transcripts. Therefore, to confirm the role of esBAF in repression of pervasive ncRNA transcription by an independent assay and better understand the nature of these transcripts, we performed whole-transcript, strand-specific RNA-seq on duplicate samples of EGFP and Smarca4 knockdown ESCs. We confirmed the increase in abundance of noncoding transcripts observed by CapSeq in Smarca4 knockdown surrounding DHSs, TSSs, and TTSs (Fig. 2A–C). Importantly, ncRNAs expressed from DHSs and TSSs bound by esBAF were strongly derepressed upon Smarca4 knockdown, whereas DHSs and TSSs not bound by esBAF were not affected (Supplemental Fig. S4). These data strongly suggest that the majority of these transcripts are directly regulated by esBAF, although the possibility that some individual transcripts are indirectly affected by esBAF depletion cannot be excluded. On average, we observed ncRNA reads extending ∼0.5–1 kb from their initiation sites at each genomic feature (Fig. 2A–C), consistent with the size distribution of several classes of ncRNAs, including eRNAs and lncRNAs (Natoli and Andrau 2012; Rinn and Chang 2012). Next, we calculated the number of transcripts, both coding and noncoding, that were significantly altered upon Smarca4 knockdown using a false discovery rate of 10% as calculated by DEseq (Anders and Huber 2010). Interestingly, while acute Smarca4 knockdown resulted in significantly reduced expression of 2843 mRNAs and increased expression of 1856 mRNAs (Supplemental Fig. S5A,B), Smarca4 knockdown had a much stronger global effect on ncRNA expression, suggesting that esBAF regulates far more ncRNAs than mRNAs (Supplemental Fig. S5A). Specifically, we observed significant changes in the expression of ∼15-fold more ncRNAs than mRNAs upon Smarca4 knockdown throughout the genome, with 64,282 ncRNA transcripts significantly up-regulated and 6214 ncRNA transcripts significantly down-regulated relative to control cells (Fig. 2D–H). Importantly, the majority (87%) of ncRNAs up-regulated upon Smarca4 knockdown were expressed from gene-distal DHSs, consistent with prior data showing that the largest class of genomic regions enriched for esBAF binding are gene-distal DHSs (Ho et al. 2009a). Interestingly, at promoters, where the relationship between sense (mRNA) and antisense ncRNA transcription can be examined, we observed no correlation between mRNA and antisense ncRNA expression upon esBAF knockdown (Supplemental Fig. S5C–E), suggesting that most sense/antisense pairs of transcripts are regulated independently in ESCs. Together, these data indicate that, in ESCs, esBAF plays a much broader role in silencing expression of ncRNAs than its well-established function in regulation of protein-coding transcripts.
Figure 2.
Up-regulation of ncRNAs from gene-distal and gene-proximal regions upon esBAF depletion. (A–C) Histogram of normalized sense and antisense transcripts obtained from RNA-seq analysis surrounding DHSs (A), TSSs (B), and TTSs (C) in EGFP knockdown (KD) and Smarca4 knockdown ESCs. (D) Distribution of ncRNAs significantly misregulated upon Smarca4 knockdown in ESCs. (E–H) Heat maps quantifying noncoding transcripts surrounding DHSs (±500 bp) (E), TSSs (−500 to +100 bp) (F), intragenic regions (antisense only, >500 bp from the TSS) (G), or TTSs (antisense only, −500 to +500 bp from the TTS) (H) in averaged biological replicates of EGFP knockdown and Smarca4 knockdown ESC RNA-seq experiments. Expression is indicated as log2(normalized reads). (I) RT-qPCR validation of ncRNA transcripts initiating from gene-distal DHSs. Expression levels (mean ± SD values of three biological replicates) upon Smarca4 knockdown are shown relative to the EGFP knockdown. (J) Increased antisense transcript production from 14 coding gene promoters upon knockdown of multiple esBAF subunits. Analyses were performed as in I. (*) P < 0.05; (**) P < 0.01; (ns) not significant.
We confirmed these findings by randomly primed and strand-specific RT-qPCR at a subset of DHS-flanking regions and regions immediately upstream of promoters (Fig. 2I; Supplemental Fig. S1E,F), validating the data from both RNA-seq techniques. Control promoters and DHSs, which did not show significant changes in ncRNA expression in our genomic data sets, were also not changed in RT-qPCR analyses (Fig. 2I; Supplemental Fig. S1E–F). We performed oligo-dT RT-qPCR and observed similar results, demonstrating that at least some ncRNA transcripts are polyadenylated (Supplemental Fig. S1G).
Next, we tested the specificity of the knockdown phenotype in two ways. First, we used a nonoverlapping pool of Smarca4 endoribonuclease III-digested siRNAs (esiRNAs) and observed similar changes in ncRNA expression (Supplemental Fig. S6A–C). Second, we tested whether the increased ncRNA expression observed upon Smarca4 knockdown could also be observed upon knockdown of two additional esBAF subunits, Smarcc1 (Baf155) and Smarcd1 (Baf60a) (Supplemental Fig. S1H). We observed similar increases in ncRNA expression upon knockdown of Smarca4, Smarcc1, or Smarcd1 (Fig. 2J), confirming that Brg1 functions through esBAF. Therefore, the fact that nonoverlapping Smarca4 esiRNAs as well as individual knockdowns of multiple esBAF subunits all result in the same effect on ncRNA expression demonstrates the specificity of the knockdown phenotypes. Together, these data reveal a novel role for esBAF in repressing ncRNAs that, in the absence of esBAF, are expressed broadly throughout the ESC genome from both gene-distal and gene-proximal locations.
Brg1 is dispensable for repressing ncRNA expression in MEFs
Distinct forms of the BAF complex are expressed in different cell types, each with individual regulatory functions (Lessard et al. 2007; Wu et al. 2007; Ho et al. 2009b). To test whether the function of esBAF to suppress ncRNA expression is conserved in a somatic cell type, we performed CapSeq in EGFP knockdown and Smarca4 knockdown MEFs in duplicate (Supplemental Fig. S7A,B). In comparison with ESCs, we found far fewer changes in the locations of transcription initiation sites upon Smarca4 knockdown in MEFs (Supplemental Fig. S3D–F). Furthermore, in MEFs, we observed low levels of ncRNA expression at DHSs, promoters, intragenic regions, and TTSs irrespective of Smarca4 depletion (Supplemental Figs. S2A–D, S7C–E), indicating that ncRNA expression is maintained at low levels in MEFs, similar to those in wild-type ESCs, independently of Brg1. However, it remains possible that an alternative BAF complex in MEFs that contains the Brm ATPase in place of Brg1 may perform this repressive function. Consistent with our findings in MEFs, we found that multilineage differentiation of ESCs within embryoid bodies reduced ncRNA expression genome-wide, suggesting that most ncRNAs that are detectable in ESCs are silenced in many differentiated cell types (Supplemental Fig. S2E–H). In sum, these data suggest a cell type-specific role for BAF in repression of spurious transcripts in ESCs and raises the possibility that distinct mechanisms for silencing ncRNAs may operate in different cell types.
esBAF regulates nucleosome occupancy within and adjacent to NDRs
SWI/SNF complexes have established functions in many organisms to disrupt nucleosome architecture in order to increase accessibility of regulatory sequences and activate gene expression (Hirschhorn et al. 1992; Khavari et al. 1993; Laurent et al. 1993; Ho et al. 2009a; Yildirim et al. 2011). esBAF binds many gene-distal DHSs and promoters and functions at these sites to maintain chromatin in an accessible state (Ho et al. 2011). Consequently, given our finding that esBAF represses rather than activates ncRNAs, the mechanism of ncRNA regulation by esBAF at these regions was not obvious. Therefore, to address this question, we examined the changes in genome-wide nucleosome architecture in ESCs upon Smarca4 knockdown using micrococcal nuclease (MNase) digestion of chromatin followed by deep sequencing (MNase-seq). Smarca4 knockdown ESCs were digested with MNase side by side with previously described control (EGFP knockdown) ESCs (Supplemental Fig. S8A; Carone et al. 2014) and paired-end sequenced (see the Supplemental Material for details).
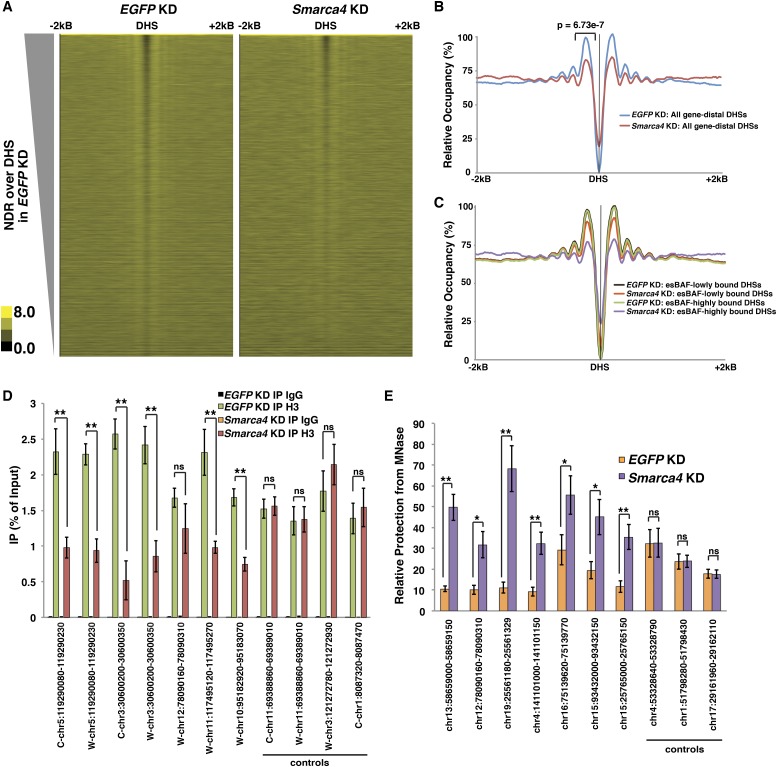
Previously, we showed that control knockdown ESCs exhibit well-established features of chromatin structure that are conserved in a wide variety of eukaryotes (Hughes and Rando 2014). Regulatory elements generally exhibit dramatic nucleosome depletion, often flanked by well-positioned nucleosomes upstream (−1 nucleosome) and downstream (+1 nucleosome) followed by less well-positioned nucleosomes (Carone et al. 2014). This is particularly notable for gene-distal DHSs, the genomic feature most strongly enriched for esBAF binding (Ho et al. 2009a), where control knockdown cells exhibit a large NDR and well-positioned flanking nucleosomes (Fig. 3A,B). Similar features are observed at promoters and at binding sites for the genomic insulator CTCF (see below), while TTSs are generally followed by a large NDR (Carone et al. 2014).
Figure 3.
esBAF promotes nucleosome depletion over DHSs and elevated nucleosome occupancy flanking DHSs in ESCs. (A) Heat maps of nucleosome occupancy obtained by MNase-seq over gene-distal DHSs ±2 kb in EGFP knockdown (KD) (left) and Smarca4 knockdown (right) ESCs. Nucleosome occupancy is indicated in yellow. DHSs were called from GSM1014154 with TSSs removed, since they are considered separately (see below). (B,C) Aggregation plot of relative nucleosome occupancy upon EGFP knockdown or Smarca4 knockdown over all gene-distal DHSs (B) or over gene-distal DHSs that are either highly or lowly bound by esBAF (C). The P-value for the change in flanking nucleosome is indicated. (D) Validation of changes in occupancy of nucleosomes immediately flanking DHSs. Histone H3 levels of nucleosomes flanking seven DHSs found to exhibit reduced occupancy in the Smarac4 knockdown MNase-seq data set were determined in EGFP knockdown and Smarca4 knockdown ESCs by ChIP-qPCR along with four control DHS-proximal nucleosomes that were not altered upon Smarca4 knockdown. Histone H3 levels are expressed as a fraction of the input. Shown are the mean ± SD values of three biological replicates. (E) Validation of changes in nuclease accessibility over DHSs. Accessibility to MNase treatment was determined over seven DHSs altered in the Smarca4 knockdown MNase-seq data set in EGFP knockdown and Smarca4 knockdown ESCs by MNase-qPCR along with three control DHSs that were not altered upon Smarca4 knockdown. Protection relative to undigested chromatin is shown as the mean ± SD values of three biological replicates. For D and E, statistical significance is indicated by an asterisk. (*) P < 0.05; (**) P < 0.01; (ns) not significant.
By comparison, Smarca4 knockdown ESCs show significant changes in nucleosome occupancy at several classes of regulatory elements. Consistent with the role of esBAF in maintaining accessibility of gene-distal DHSs (Ho et al. 2011), we observed a significant increase in nucleosome occupancy over DHS peaks upon Smarca4 knockdown (Fig. 3A,B). Interestingly, we found that Smarca4 knockdown ESCs also exhibit a significant decrease in nucleosome occupancy immediately flanking DHSs (Fig. 3A,B), which was apparent for two to three nucleosomes on each side of the NDR, on average (Fig. 3B). Upon Smarca4 knockdown, DHSs with high levels of esBAF binding exhibited much larger increases in nucleosome occupancy over NDRs and decreases in nucleosome occupancy flanking NDRs relative to DHSs lowly bound by esBAF (Fig. 3C), demonstrating that esBAF directly mediates these effects on chromatin structure. Conversely, we found that knockdown of Smarca4 in MEFs was not accompanied by a reduction in nucleosome occupancy at multiple genomic regions tested in MEFs (Supplemental Fig. S7F), consistent with the low levels of ncRNA expression with or without Smarca4 knockdown in this cell type.
We validated these findings in two additional ways. First, the effect of Smarca4 knockdown is not a result of technical variation in this protocol, as knockdown of an independent chromatin regulator, Mbd3, reveals no noticeable effects on occupancy of nucleosomes flanking DHSs and reduced occupancy over DHSs themselves (Supplemental Fig. S8B), consistent with the known role of Mbd3/NuRD as a transcriptional repressor that functions at both promoters and enhancers (Reynolds et al. 2011; Yildirim et al. 2011; Whyte et al. 2012). Second, we independently confirmed the differences that we observed in flanking nucleosome occupancy and nucleosome depletion over DHSs upon Smarca4 knockdown at multiple genomic targets using histone H3 chromatin immunoprecipitation (ChIP) and MNase protection assays, respectively (Fig. 3D,E).
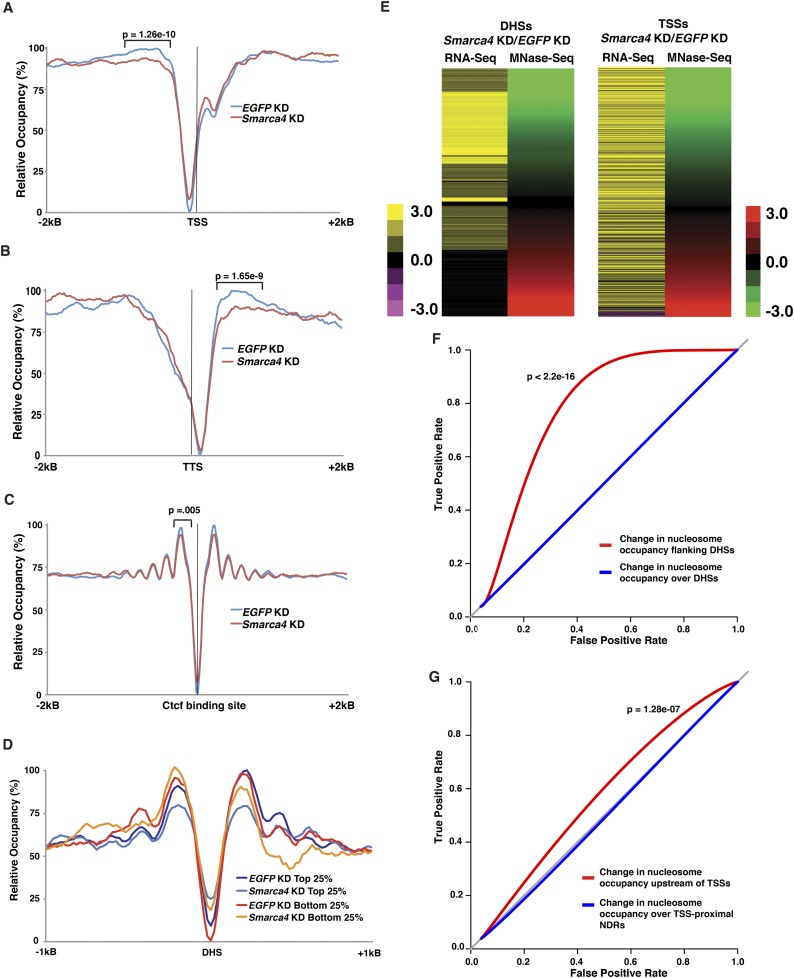
Significantly, nucleosome occupancy over other genomic regions that exhibit enhanced ncRNA expression upon esBAF depletion were similarly affected. Noncoding regions upstream of promoters and downstream from TTSs exhibited significantly reduced nucleosome occupancy upon Smarca4 depletion, consistent with the enhanced expression of ncRNAs observed at these sites (Fig. 4A,B). These sites are affected less strongly than gene-distal DHSs, consistent with the fact that esBAF binds many more DHSs (Ho et al. 2009a) and regulates many more ncRNAs at these sites than promoters or TTSs. In contrast, we observed a small but significant increase in nucleosome occupancy over the NDR and +1 nucleosome of coding genes, consistent with the combined and opposing effects of down-regulation of 11% of coding genes and up-regulation of 7% of coding genes upon Smarca4 knockdown (Fig. 4A). Unlike our findings at gene-distal DHSs, we observed only minor alterations in nucleosome occupancy over CTCF-binding sites (which represent a subset of DHSs), indicating that the changes that we observed in nucleosome occupancy were not found at all genomic locations (Fig. 4C).
Figure 4.
esBAF regulates NDRs and flanking nucleosome occupancy of promoters and TTSs. (A–C) Aggregation plot of relative nucleosome occupancy upon EGFP knockdown (KD) or Smarca4 knockdown over TSSs (A), TTSs (B), or Ctcf-binding sites (C) ±2 kb in ESCs. Ctcf-binding sites were based on published data (Chen et al. 2008). P-values indicating the statistical significance of changes in nucleosome occupancy are indicated. To calculate P-values, a standard t-test summing reads from −500 to −240 upstream of TSSs and +240 to +500 downstream from TTSs in EGFP knockdown and Smarca4 knockdown was performed. (D) Aggregation plot of relative nucleosome occupancy upon EGFP knockdown or Smarca4 knockdown over DHSs grouped by changes in ncRNA levels in Smarca4 knockdown ESCs. Displayed are the top 25% of DHSs with increased ncRNA levels upon Smarca4 knockdown and the bottom 25% of DHSs with decreased or no change in ncRNA levels upon Smarca4 knockdown. (E) Log2 fold change of Smarca4 knockdown/EGFP knockdown ncRNA expression originating from annotated DHSs (left panel) or upstream of TSSs (right panel) from averaged ESC RNA-seq experiments, sorted by the log2 fold change of Smarca4 knockdown/EGFP knockdown nucleosome occupancy adjacent to DHSs or upstream of TSSs (−1 position). (F,G) Receiver of operating characteristic (ROC) curves for changes in nucleosome occupancy relative to changes in expression for DHSs (F) and TSSs (G). The P-values indicating the significance of changes in flanking nucleosome occupancy curves relative to scrambled data are shown.
As with gene-distal DHSs, we confirmed the differences in occupancy of the −1 nucleosome, the +1 nucleosome, and NDRs upon Smarca4 knockdown at multiple promoter-proximal regions using histone H3 ChIP and MNase protection assays (Supplemental Fig S9). Over promoters, we examined the effects of Smarca4 knockdown over a broad range of gene expression (Supplemental Fig. S8C–G) and found that esBAF plays a larger role in chromatin regulation at genes regulated and/or occupied by esBAF (Supplemental Fig. S10), just as we found for gene-distal DHSs.
In conclusion, these results reveal that esBAF not only maintains DHSs in a relatively nucleosome-depleted state but also functions to maintain high levels of nucleosome occupancy immediately flanking these regions. Stabilization of nucleosomes flanking regulatory elements is an unanticipated function for esBAF in control of chromatin architecture and suggests a potential mechanism by which ATP-dependent nucleosome remodeling may regulate expression of ncRNAs throughout the genome.
esBAF-dependent nucleosome occupancy adjacent to NDRs correlates with repression of ncRNAs
The decrease in nucleosome occupancy flanking gene-distal DHSs upstream of TSSs and downstream from TTSs in Smarca4 knockdown cells suggests that esBAF may regulate the transcription of ncRNAs through a mechanism that is distinct from its well-described function to maintain open chromatin structure over transcription factor-binding sites. These findings raise the possibility that increased nucleosome occupancy (catalyzed by esBAF) adjacent to NDRs may repress transcription of ncRNAs throughout the genome. Consistent with this possibility, when we grouped DHSs into quartiles based on their levels of ncRNA expression upon esBAF depletion, we found that DHSs that express ncRNAs most strongly upon esBAF depletion lose flanking nucleosome occupancy to a much greater degree than DHSs at which ncRNAs are not derepressed upon esBAF depletion (Fig. 4D). In contrast, average nucleosome occupancy increased over DHS NDRs upon Smarca4 knockdown whether or not up-regulation of ncRNAs was observed (Fig. 4D). To further investigate the possible connection between nucleosome occupancy flanking NDRs and repression of aberrant transcription, we compared alterations in nucleosome occupancy flanking gene-distal DHSs upon Smarca4 knockdown (by MNase-seq) with alterations in ncRNA levels at these sites, expressing each as a ratio (log2[Smarca4 knockdown/EGFP knockdown]) (Fig. 4E). We found that the vast majority of DHSs with decreased flanking nucleosome occupancy exhibited an increase in ncRNA expression upon Smarca4 knockdown (Fig. 4E, left). A similar relationship was observed for ncRNA expression that occurs antisense to the promoters of protein-coding genes and loss of nucleosome occupancy upstream of promoter regions (−1 nucleosome) (Fig. 4E, right). Furthermore, the correlation between reduced nucleosome occupancy (green in Fig. 4E) and increased ncRNA expression (yellow in Fig. 4E) was highly significant (P < 2.2 × 10−16 for DHSs and P = 3.9 × 10−16 for TSSs, Spearman’s rank order correlation). We observed similar results when we compared ratios of CapSeq reads at DHSs and TSSs with alterations in nucleosome occupancy (data not shown). Finally, we assessed the performance of changes in nucleosome occupancy upon Smarca4 knockdown as a predictor of ncRNA expression, plotting the true positive rate versus the false positive rate for each. While alterations in occupancy over DHS NDRs were poor predictors of ncRNA expression (Fig. 4F, blue line), alterations in flanking occupancy were strongly predictive (Fig. 4F, red line), consistent with the correlation analyses shown above. We observed a similar, but weaker, effect on alterations in nucleosome occupancy upstream of gene promoters as a predictor of antisense ncRNA expression, whereas alterations in occupancy over TSS NDRs were uninformative (Fig. 4G). Taken together, these data suggest that esBAF directly represses expression of ncRNA by enhancing nucleosome occupancy surrounding gene regulatory regions.
Forced nucleosome occupancy adjacent to an NDR represses ncRNA expression independently of esBAF
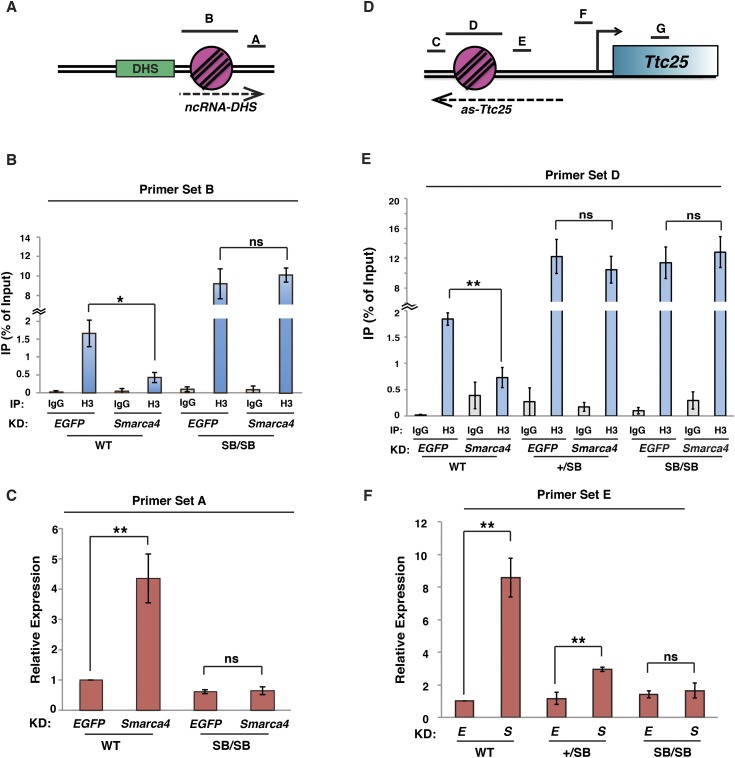
To directly test whether esBAF represses ncRNA expression by enhancing nucleosome occupancy flanking NDRs, we independently replaced the DNA sequence of two NDR-flanking nucleosomes with the previously described superbinder (SB) nucleosome-positioning sequence (Wang et al. 2011b) using CRISPR/Cas9-stimulated recombination (Cong et al. 2013; Wang et al. 2013). For these analyses, we targeted two loci that depend on esBAF for maintenance of NDR-adjacent nucleosome occupancy and silencing: a gene-distal DHS on chromosome 2 and the Ttc25 gene (Fig. 5A,D). Using this method, we obtained multiple independent ESC lines in which the nucleosome immediately adjacent to DHS-chr2 or the −1 nucleosome immediately upstream of the Ttc25 promoter was replaced with the SB sequence. First, we performed histone H3 ChIP-qPCR using primers flanking the targeted nucleosome at each site (Fig. 5B–E) within the endogenous (nontargeted, wild-type) nucleosome sequence (Supplemental Figs. S11D, S12D) or within the SB sequence (Supplemental Figs. S11E, S12E). In wild-type ESCs, these ChIPs confirm a decrease in nucleosome occupancy upon Smarca4 knockdown at both genomic loci. Remarkably, in three independent homozygous targeted (SB/SB) lines each, there is a large increase in nucleosome occupancy (Fig. 5B,E) that is specific for the SB sequence (Supplemental Figs. S11D,E, S12D,E). Furthermore, we observed no noticeable effect of Smarca4 knockdown on nucleosome occupancy over the SB sequence in these lines (Fig. 5B,E). These data show that, as in yeast (Wang et al. 2011b), the SB sequence functions as a nucleosome positioning sequence at two functionally distinct loci in mammalian cells, establishing a high-occupancy nucleosome adjacent to a DHS and upstream of the Ttc25 promoter that does not depend on esBAF.
Figure 5.
esBAF silences ncRNA expression by increasing nucleosome occupancy adjacent to NDRs. (A) Diagram of the DHS-chr2 locus with qPCR amplicons depicted. (B) Histone H3 ChIP-qPCR over the −1 nucleosome in wild-type (WT) or nucleosome SB homozygote (SB/SB) lines upon either EGFP knockdown (KD) or Smarca4 knockdown. H3 levels are expressed as a fraction of the input. Shown are the mean ± SD values of three biological replicates. (C) Randomly primed RT-qPCR of DHS transcripts in EGFP knockdown or Smarca4 knockdown cells. Expression levels were normalized to GAPDH and are shown relative to wild-type EGFP knockdown. Shown are the mean ± SD values of three biological replicates. (D) Diagram of the Ttc25 locus with qPCR amplicons depicted. (E) Histone H3 ChIP-qPCR over the −1 nucleosome in wild-type, SB heterozygote (+/SB), or SB/SB lines upon either EGFP knockdown or Smarca4 knockdown. Data are depicted as in B. (F) Randomly primed RT-qPCR of antisense transcripts in EGFP knockdown (E) or Smarca4 knockdown (S) cells, as in C. Statistical significance of alterations in expression or occupancy is indicated. (*) P < 0.05; (**) P < 0.01; (ns) not significant.
The ability of the SB sequence to enforce a high level of nucleosome occupancy independently of esBAF allowed us to test the hypothesis that high nucleosome occupancy adjacent to NDRs, and not some independent role of esBAF, directly represses ncRNAs expressed from these sites. Consistent with our hypothesis, compared with wild-type ESCs, SB/SB ESC lines exhibited low levels of antisense transcription adjacent to the targeted DHS and upstream of the Ttc25 promoter that were not increased upon Smarca4 knockdown (Fig. 5C,F). At the Ttc25 locus, we also examined the response of three independent heterozygous (+/SB) lines and found that these lines had a moderate increase in as-Ttc25 expression upon Smarca4 knockdown (Fig. 5F), indicating that the wild-type and SB allele are regulated independently. These data suggest that a strongly positioned nucleosome adjacent to an NDR blocks antisense transcription originating from NDRs at both a gene-distal DHS and the Ttc25 promoter. Therefore, in wild-type cells, we conclude that esBAF suppresses antisense transcription at diverse loci by promoting high levels of nucleosome occupancy adjacent to NDRs.
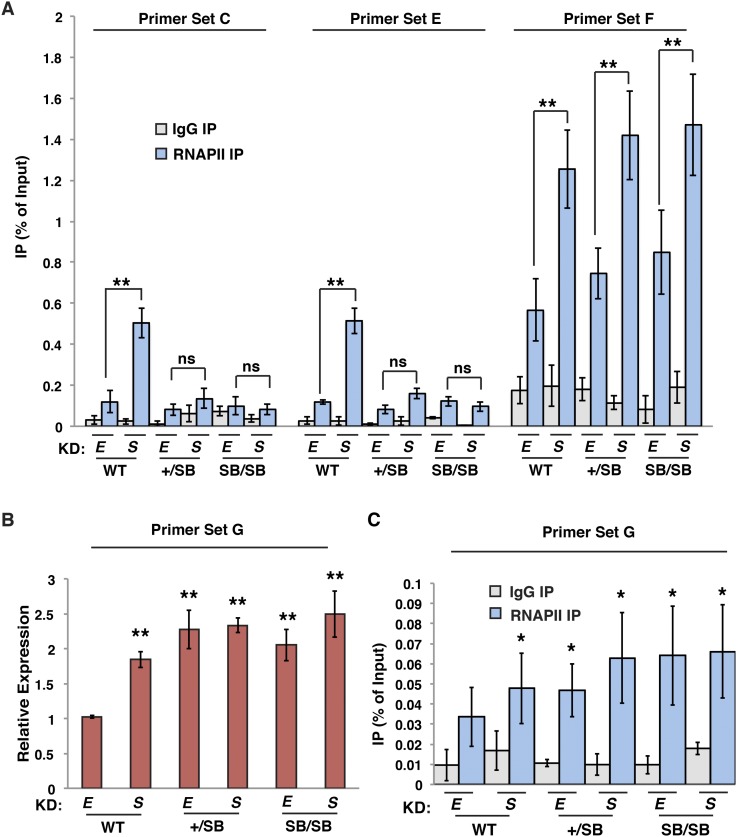
One explanation for these findings could be that by replacing the endogenous sequence with the SB sequence, we eliminated the binding sites for one or more transcription factors necessary for RNAPII recruitment to the DHS or Ttc25 promoter, thereby impairing transcription by a mechanism independent of nucleosome occupancy. To ensure that knock-in of the SB sequence did not simply disrupt the Ttc25 promoter, we examined occupancy of RNAPII over as-Ttc25 and the promoter-proximal NDR as well as expression of Ttc25 mRNA in wild-type, +/SB, and SB/SB lines and tested their dependence on esBAF. Consistent with our finding that esBAF silences ncRNA antisense to Ttc25, RNAPII levels were elevated over the ncRNA transcript upon Smarca4 knockdown in wild-type lines (Fig. 6A). In contrast, we observed low levels of RNAPII over the Ttc25 ncRNA region in SB/SB lines whether or not Smarca4 was depleted, consistent with our expression data.
Figure 6.
The nucleosome-mediated barrier to antisense ncRNA expression stimulates sense transcription at Ttc25 gene. (A) RNAPII or IgG control ChIP-qPCR upstream of the Ttc25 locus (primer sets indicated as in Fig. 5D) in wild-type (WT), +/SB, or SB/SB strains upon knockdown (KD) of either EGFP (E) or Smarca4 (S), expressed as a fraction of the input. Shown are the mean ± SD values of three biological replicates. (B) Random primed RT-qPCR of Ttc25 mRNA. Expression levels were normalized to GAPDH and are shown relative to wild-type EGFP knockdown. Shown are the mean ± SD values of three biological replicates. (C) RNAPII or IgG control ChIP-qPCR over the Ttc25 locus in wild-type, +/SB, or SB/SB strains upon either EGFP knockdown or Smarca4 knockdown for three biological replicates, displayed as in A. Statistical significance of alterations in expression or occupancy is indicated. (*) P < 0.05; (**) P < 0.01; (ns) not significant.
In wild-type cells, we observed a slight increase in RNAPII occupancy over the Ttc25 promoter and a modest increase in Ttc25 mRNA expression upon Smarca4 knockdown (Fig. 6A,B), consistent with our previous expression studies (Yildirim et al. 2011). Interestingly, we observed a modest increase in both RNAPII occupancy over the Ttc25 coding region and Ttc25 mRNA expression in the SB/SB lines that is independent of esBAF (Fig. 6A–C). This finding rules out the possibility that knock-in of the SB sequence disrupts the Ttc25 promoter. Most interestingly, these data suggest that by creating a barrier to antisense expression, RNAPII fires more frequently in the opposite direction.
Position-dependent effects of nucleosome occupancy on ncRNA regulation
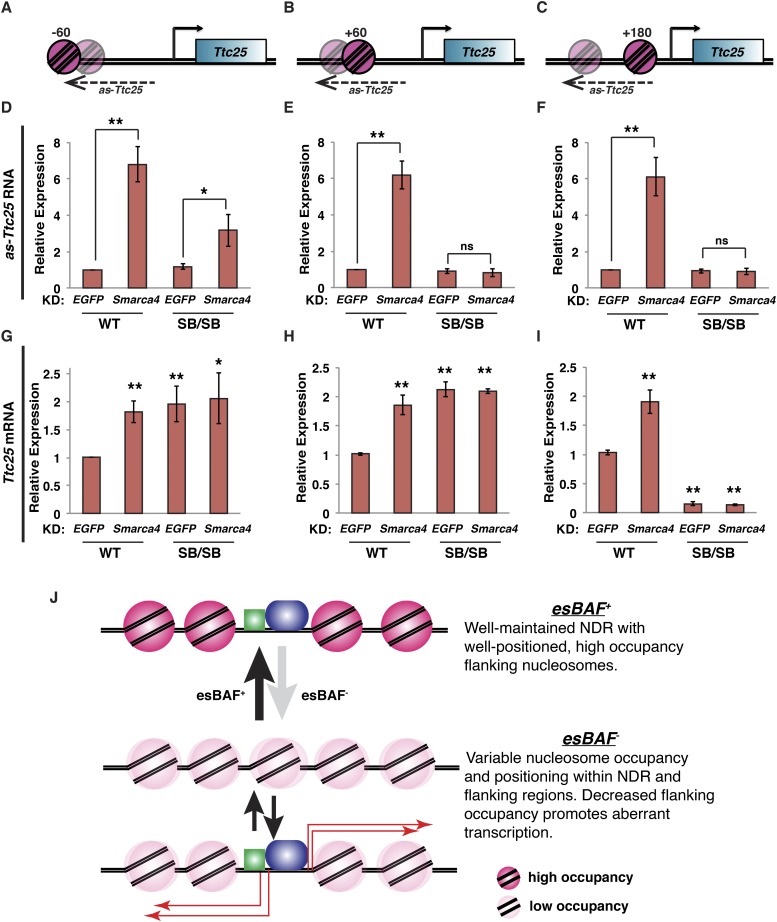
Recently, Weber et al. (2014) showed that the +1 nucleosome presents a much stronger barrier to expression of coding genes than downstream nucleosomes, raising the possibility that the positions of highly occupied nucleosomes vis-à-vis ncRNA start sites might be important for repression of these transcripts. To test this possibility, we made three additional sets of SB/SB knock-in lines upstream of the Ttc25 promoter, in which we moved the SB sequence 60 base pairs (bp) upstream (−60), 60 bp downstream (+60), or 180 bp downstream (+180) relative to the normal −1 nucleosome position (Fig. 7A–C). These alterations represent modest shifting upstream (−60) of or downstream (+60) from the normal −1 nucleosome position (hereafter the “+0” position) as well as a dramatic downstream shift (+180) that partially overlaps with the normal NDR at the Ttc25 promoter. As with the +0 location (Fig. 5), knock-in of the SB sequence at these ectopic sites promoted high levels of nucleosome occupancy that were not affected by Smarca4 knockdown (Supplemental Fig. S13A–F). Interestingly, while the two sets of downstream-shifted SB lines exhibited esBAF-independent silencing of as-Ttc25 expression similar to that of the +0 SB lines, we found that in lines harboring the upstream-shifted nucleosome (−60), as-Ttc25 was derepressed upon Smarca4 knockdown, albeit to levels lower than in wild-type cells (Fig. 7D–F). Consistent with these findings, we observed an increase in RNAPII occupancy over the as-Ttc25 sequence upon Smarca4 knockdown in the −60 lines but not in the +60 or +180 lines (Supplemental Fig. S13G–I). These data suggest that shifting the highly occupied nucleosome to a position further away from the as-Ttc25 TSS reduces its ability to serve as a barrier to transcription.
Figure 7.
Position-dependent effects of upstream nucleosomes on sense and antisense expression at the Ttc25 gene. (A–C) Diagram of the Ttc25 locus with alternative SB positions. The SB was moved 60 bp 5′ of the original −1 nucleosome (−60; A,D,G), 60 bp 3′ of the original −1 nucleosome (+60; B,E,H), or 180 bp 3′ of the original −1 nucleosome (+180; C,F,I). The location of the original −1 nucleosome is depicted as a transparent nucleosome. (D–F) Randomly primed RT-qPCR of antisense Ttc25 transcripts (as-Ttc25 RNA). Expression levels were normalized to GAPDH and are shown relative to wild-type (WT) EGFP knockdown (KD). Shown are the mean ± SD values of three biological replicates. (G–I) Randomly primed RT-qPCR of Ttc25 mRNA in alternative SB lines. (J) Model for esBAF regulation of ncRNAs. Upon loss of esBAF, nucleosome occupancy is altered, with increased occupancy over NDRs and decreased occupancy flanking NDRs. The latter change results in increased transcription of ncRNAs surrounding NDRs throughout the genome. Transcription factors and RNAPII are represented by green and blue objects.
In contrast, for the −60 and +60 SB lines (like the +0 SB lines), we observed a moderate, esBAF-independent stimulation of Ttc25 mRNA expression and RNAPII occupancy over the coding sequence, suggesting that precise positioning of the upstream nucleosome is not required for stimulation of Ttc25 mRNA expression (Fig. 7G,H; Supplemental Fig. S13J,K). However, when the SB sequence was placed far downstream (+180), we observed very low Ttc25 mRNA expression and RNAPII occupancy over the coding sequence, suggesting that increasing nucleosome occupancy too close to the TSS impairs promoter activity at this site (Fig. 7I; Supplemental Fig. S13L). Together, these data demonstrate that esBAF represses transcription of ncRNAs by promoting nucleosome occupancy flanking NDRs and that this repressive function is sensitive to the distance of the highly occupied nucleosome from the start site of the ncRNA. Furthermore, we show that this repressive function occurs throughout the ESC genome at not only gene-proximal locations but also the vastly larger number of NDRs located in intergenic regions.
Discussion
Changes in transcription of protein-coding genes are highly correlated with changes in chromatin structure; however, the extent to which chromatin regulatory factors affect production of ncRNAs throughout the mammalian genome has not been examined. Here we report the unexpected finding that esBAF functions as a global repressor of ncRNA expression at both gene-distal and gene-proximal sites—bidirectionally from gene-distal DHSs and cryptic promoters of gene bodies and antisense to protein-coding TSSs and TTSs. Furthermore, we show that esBAF accomplishes this repressive function by enhancing nucleosome occupancy adjacent to NDRs from which these ncRNAs are expressed.
While the majority of the noncoding portion of the genome has been shown to be transcribed in mammals, the mechanisms underlying regulation of ncRNA expression remain poorly understood. Previous studies examining regulation of ncRNAs have focused mainly on antisense transcription upstream of promoters (Whitehouse et al. 2007; Preker et al. 2008; Almada et al. 2013; Marquardt et al. 2014) or within coding genes (Kaplan 2003; Carrozza et al. 2005). However, the vast majority of noncoding regions transcribed in higher eukaryotes are intergenic regions found distal to promoters or gene bodies (Djebali et al. 2012), raising the question of whether cells use mechanisms to regulate transcription from gene-distal regions of the genome similar to those used in enforcing promoter directionality. In addition, the mechanisms determining whether gene-distal NDRs express ncRNA transcripts in both directions from the NDR or only one are not clear.
The SWI/SNF family of nucleosome remodeling complexes use the energy of ATP hydrolysis to perturb nucleosome structure (Bartholomew 2014). This activity can result in alterations in the translational positioning of nucleosomes, nucleosome loss, or perturbations in the underlying nucleosome architecture. In general, this activity acts locally to create open chromatin in order to promote transcription of target genes (Hirschhorn et al. 1992; Khavari et al. 1993; Laurent et al. 1993; Ho et al. 2009b, 2011; Yildirim et al. 2011). Our data support these findings, where esBAF creates an open NDR at many of its binding sites throughout the genome to permit binding of transcription factors. However, we also uncovered an unexpected role of this remodeling complex to maintain occupancy of nucleosomes flanking a large fraction of NDRs regardless of the state of nucleosome occupancy over the NDR (Fig. 4D–G). At these sites, a reduction in nucleosome occupancy flanking NDRs upon Smarca4 knockdown correlates with elevated expression of ncRNA transcripts. Remarkably, ∼15-fold more ncRNA transcripts than mRNAs are affected by esBAF depletion, suggesting that a major function of this essential nucleosome remodeling complex is to restrict RNAPII to regions of the genome where transcription is productive.
Based on these data, we propose that a major function of esBAF is to suppress transcription of ncRNAs via maintenance of defined nucleosome-free and nucleosome-bound segments within small open chromatin domains (∼200-bp NDRs plus a few flanking nucleosomes) throughout the genome (Fig. 7J). Although esBAF regulates nucleosome occupancy at many gene-distal DHSs, we found that DHSs with transcripts regulated by esBAF exhibited the most dependence on esBAF for flanking nucleosome occupancy (Fig. 4D), consistent with this model. We further propose that esBAF acts by similar mechanisms at both gene-distal and gene-proximal NDRs. To directly test our model in which esBAF represses ncRNA expression by promoting high levels of nucleosome occupancy adjacent to NDRs, we inserted an artificial nucleosome-binding sequence to strongly position a nucleosome over two of these NDR-flanking regions. At these model loci, we found that esBAF is no longer required for suppression of ncRNA expression, demonstrating that esBAF regulates these transcripts by promoting nucleosome occupancy. This mechanism is fundamentally distinct from the well-established role of esBAF as an activator of gene expression, which is thought to function by increasing chromatin accessibility.
By moving the position of the SB sequence 60 bp further upstream of its normal (−1) location upstream of the Ttc25 gene, we found that repression of as-Ttc25 by esBAF was partially restored. These data suggest that the −1 nucleosome upstream of promoters and the first nucleosome adjacent to the NDR of a DHS may be critical for repression of ncRNA transcription at these sites. In addition, we observed that different SB positions have uncorrelated effects on sense and antisense transcription. For example, while SB −60 exhibited partial derepression of as-Ttc25 upon Smarca4 knockdown, it caused esBAF-independent up-regulation of Ttc25 mRNA to the same extent as SB +0 and SB +60. In addition, SB +180 reduced expression of Ttc25 mRNA to very low levels but was equally effective as other SB positions in repression of as-Ttc25. These data argue that there is limited coordination of sense/antisense transcription at this site.
Previous work has shown that the outputs of most mammalian promoters are mainly unidirectional due to RNA cleavage and polyadenylation of TSS-proximal antisense transcripts after a few hundred bases of transcription (Almada et al. 2013). Our data demonstrate an additional layer of regulation in the form of cis-acting regulatory proteins, such as esBAF. Furthermore, we show that this regulation occurs not only upstream of bona fide promoters but at NDRs in multiple genomic contexts.
Additional work is required to understand the functional roles (if any) for pervasive transcription of most ncRNA. Furthermore, given the inability to perturb esBAF function in a manner in which only its mRNA or ncRNA targets are misregulated but not both, it is impossible to determine the relative contributions of each to the self-renewal defect observed upon esBAF loss (Fazzio et al. 2008; Gao et al. 2008; Yan et al. 2008; Ho et al. 2009a,b). However, our findings reveal that mammalian cells have evolved at least one global mechanism for suppressing ncRNA expression that can occur from thousands of NDRs across the genome. These data suggest that either pervasive ncRNA expression is deleterious to cellular proliferation or viability or ncRNAs are only beneficial in specific, as yet undetermined cell types. Our work focused on one specific nucleosome remodeling factor that functions as an important regulator of ncRNA expression throughout the ESC genome. However, we found that ncRNA expression was kept at similarly low levels in MEFs independently of Brg1, suggesting that the Brg1/BAF complex is not necessary for ncRNA repression in MEFs, although Brm/BAF could carry out this regulatory function in this cell type. Therefore, regulation of ncRNA expression by ATP-dependent nucleosome remodeling complexes may not be limited to esBAF, as alternative nucleosome remodeling factors may perform the same function in somatic cells. Furthermore, given that multiple factors have been shown to regulate promoter directionality in yeast (Whitehouse et al. 2007; Marquardt et al. 2014), it seems likely that additional factors may contribute to the repression of ncRNA transcripts in ESCs.
Materials and methods
Cell culture
E14 mouse ESCs and SV40 T-antigen immortalized MEFs were cultured as previously described (Chen et al. 2013).
RNAi-mediated knockdown
RNAi-mediated knockdown was performed with esiRNAs as previously described (Fazzio et al. 2008) using Lipofectamine 2000 (Invitrogen) for ESC knockdown and Lipofectamine RNAiMax (Invitrogen) for MEF knockdown. Knockdowns were performed for 48 h.
RT-qPCR
RNA was isolated using TRIzol reagent (Invitrogen). Total RNA was treated with DNase I (New England Biolabs) for 15 min at 37°C. One microgram of RNA was used in a cDNA synthesis reaction using either random hexamers (Promega), oligo-dT, or strand-specific primers (see Supplemental Table S1) where indicated, with reverse transcriptase or a no-RT control. cDNA was used as a template in qPCR reactions using a FAST SYBR mix (KAPA Biosystems) on an Eppendorf Realplex with specific primers (see Supplemental Table S1). GAPDH was used as a loading control for randomly primed and oligo-dT-amplified cDNA RT-qPCR reactions. For strand-specific reactions, cDNA was prepared in parallel with random hexamers, and a GAPDH loading control was performed.
Western blotting
Whole-cell lysates were extracted using WE16th buffer (25 mM Tris at pH 7.5, 125 mM NaCl, 2.5 mM EDTA, 0.05% SDS, 0.5% NP-40, 10% [w/v] glycerol). Equal amounts of lysate were separated by SDS-PAGE, transferred to nitrocellulose (Life Sciences), and assayed by immunoblotting. The antibodies used to detect Brg1 and actin were as follows: anti-Brg1 (1:1000; Bethyl Laboratories, A300-813A), anti-Oct4 (1:2000; Santa Cruz Biotechnology, sc-8628), anti-Nanog (1:500; Bethyl Laboratories, A300-398A), and anti-actin (1:50,000; Sigma, A1978).
ChIP
Formaldehyde cross-linked chromatin was immunoprecipitated with IgG (Abcam, ab37415), anti-H3 (Abcam, ab1791), or anti-RNAPII (Santa Cruz Biotechnology, sc-9001) antibodies. Immunoprecipitated DNA was used as template for qPCR reactions after reversal of cross-links. See the Supplemental Material for details.
MNase digestion
Formaldehyde-cross-linked, permeablized cells were digested with MNase (control samples were not digested). After reversal of cross-links, DNA digestion was quantified by qPCR. Undigested DNA was used as a control and to calculate a standard curve. See the Supplemental Material for details.
MNase-seq and analysis
MNase-seq was performed similar to the methods described in Henikoff et al. (2011) and Carone et al. (2014). See the Supplemental Material for details.
CapSeq and analysis
CapSeq experiments were performed essentially as in Gu et al. (2012). See the Supplemental Material for details.
Whole-transcript, strand-specific RNA-seq and analysis
RNA-seq experiments were performed as described in Levin et al. (2010) and Kumar et al. (2012) with slight modifications, and data analysis was performed similarly to CapSeq analysis. See the Supplemental Material for details.
Generation of DHS-chr2 and Ttc25 nucleosome SB lines.
The CRISPR/Cas9 system was used to stimulate recombination of the SB sequence into the genomic regions described (Cong et al. 2013) using guide RNAs designed with the available software (Hsu et al. 2013). We used gene synthesis (Invitrogen and IDT) to create homology constructs that surround the nucleosome SB sequence (Wang et al. 2011b) with short (500- to 600-bp) homology arms for recombination (see Supplemental Table S2). See the Supplemental Material for details.
Accession numbers
MNase-seq, CapSeq, and RNA-seq data sets have been assigned to Gene Expression Omnibus accession number GSE57170.
Supplementary Material
Acknowledgments
We thank Ly-Sha Ee, Kurtis McCannell, Po-shen Chen, Craig Peterson, Toshi Tsukiyama, Job Dekker, and Jennifer Benanti for insightful discussions and critical reading of this manuscript, and Scot Wolfe and Hsiuyi Chen for technical advice. This work was supported by the T32CA130807 training grant and a Leukemia and Lymphoma post-doctoral fellowship to S.J.H., National Institutes of Health (NIH) HD080224 to O.J.R., NIH GM058800 to C.C.M., and NIH HD072122 to T.G.F. C.C.M. is a Howard Hughes Medical Institute Investigator. T.G.F. is a Pew Scholar in the Biomedical Sciences and a Leukemia and Lymphoma Society Scholar. The funders had no role in the study design, data collection, analysis, decision to publish, or preparation of the manuscript. All deep sequencing was performed at the University of Massachusetts Medical School Core facility on a HiSeq2000 supported by 1S10RR027052-01. S.J.H., O.J.R., C.C.M., and T.G.F. designed all experiments. S.J.H. carried out all experiments with minor assistance from T.G.F. B.D.L. performed cloning of CRISPR donor and targeting vectors for the Ttc25 locus. S.J.H. performed analyses of genomic data with assistance from W.G. and C.C.M. for CapSeq and RNA-seq and from B.R.C. and O.J.R. for MNase-seq. S.J.H. and T.G.F. wrote the paper.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.253534.114.
References
- Adelman K, Lis JT. 2012. Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat Rev Genet 13: 720–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almada AE, Wu X, Kriz AJ, Burge CB, Sharp PA. 2013. Promoter directionality is controlled by U1 snRNP and polyadenylation signals. Nature 499: 360–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almer A, Hörz W. 1986. Nuclease hypersensitive regions with adjacent positioned nucleosomes mark the gene boundaries of the PHO5/PHO3 locus in yeast. EMBO J 5: 2681–2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Huber W. 2010. Differential expression analysis for sequence count data. Genome Biol 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arigo JT, Carroll KL, Ames JM, Corden JL. 2006. Regulation of yeast NRD1 expression by premature transcription termination. Mol Cell 21: 641–651. [DOI] [PubMed] [Google Scholar]
- Bartholomew B. 2014. Regulating the chromatin landscape: structural and mechanistic perspectives. Annu Rev Biochem 83: 671–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeger H, Griesenbeck J, Strattan JS, Kornberg RD. 2003. Nucleosomes unfold completely at a transcriptionally active promoter. Mol Cell 11: 1587–1598. [DOI] [PubMed] [Google Scholar]
- Carone BR, Hung J-H, Hainer SJ, Chou M-T, Carone DM, Weng Z, Fazzio TG, Rando OJ. 2014. High-resolution mapping of chromatin packaging in mouse embryonic stem cells and sperm. Dev Cell 30: 11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia W-J, Anderson S, Yates J, Washburn MP, et al. 2005. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 123: 581–592. [DOI] [PubMed] [Google Scholar]
- Carvalho S, Raposo AC, Martins FB, Grosso AR, Sridhara SC, Rino J, Carmo-Fonseca M, de Almeida SF. 2013. Histone methyltransferase SETD2 coordinates FACT recruitment with nucleosome dynamics during transcription. Nucleic Acids Res 41: 2881–2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castelnuovo M, Zaugg JB, Guffanti E, Maffioletti A, Camblong J, Xu Z, Clauder-Münster S, Steinmetz LM, Luscombe NM, Stutz F. 2014. Role of histone modifications and early termination in pervasive transcription and antisense-mediated gene silencing in yeast. Nucleic Acids Res 42: 4348–4362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB. 2008. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 133: 1106–1117. [DOI] [PubMed] [Google Scholar]
- Chen PB, Hung J-H, Hickman TL, Coles AH, Carey JF, Weng Z, Chu F, Fazzio TG. 2013. Hdac6 regulates Tip60-p400 function in stem cells. eLife 2: e01557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung V, Chua G, Batada NN, Landry CR, Michnick SW, Hughes TR, Winston F. 2008. Chromatin- and transcription-related factors repress transcription from within coding regions throughout the Saccharomyces cerevisiae genome. PLoS Biol 6: e277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. 2013. Multiplex genome engineering using CRISPR/Cas systems. Science 339: 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, et al. 2012. Landscape of transcription in human cells. Nature 489: 101–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgar G, Vavouri T. 2008. Tuning in to the signals: noncoding sequence conservation in vertebrate genomes. Trends Genet 24: 344–352. [DOI] [PubMed] [Google Scholar]
- Fazzio TG, Huff JT, Panning B. 2008. An RNAi screen of chromatin proteins identifies Tip60-p400 as a regulator of embryonic stem cell identity. Cell 134: 162–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn RA, Almada AE, Zamudio JR, Sharp PA. 2011. Antisense RNA polymerase II divergent transcripts are P-TEFb dependent and substrates for the RNA exosome. Proc Natl Acad Sci 108: 10460–10465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Tate P, Hu P, Tijan R, Skarnes WC, Wang Z. 2008. ES cell pluripotency and germ-layer formation require the SWI/SNF chromatin remodeling component BAF250a. Proc Natl Acad Sci 105: 6656–6661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Lee H-C, Chaves D, Youngman EM, Pazour GJ, Conte D Jr, Mello CC. 2012. CapSeq and CIP-TAP identify Pol II start sites and reveal capped small RNAs as C. elegans piRNA precursors. Cell 151: 1488–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff JG, Belsky JA, Krassovsky K, MacAlpine DM, Henikoff S. 2011. Epigenome characterization at single base-pair resolution. Proc Natl Acad Sci 108: 18318–18323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschhorn JN, Brown SA, Clark CD, Winston F. 1992. Evidence that SNF2/SWI2 and SNF5 activate transcription in yeast by altering chromatin structure. Genes Dev 6: 2288–2298. [DOI] [PubMed] [Google Scholar]
- Ho L, Jothi R, Ronan JL, Cui K, Zhao K, Crabtree GR. 2009a. An embryonic stem cell chromatin remodeling complex, esBAF, is an essential component of the core pluripotency transcriptional network. Proc Natl Acad Sci 106: 5187–5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L, Ronan JL, Wu J, Staahl BT, Chen L, Kuo A, Lessard J, Nesvizhskii AI, Ranish J, Crabtree GR. 2009b. An embryonic stem cell chromatin remodeling complex, esBAF, is essential for embryonic stem cell self-renewal and pluripotency. Proc Natl Acad Sci 106: 5181–5186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L, Miller EL, Ronan JL, Ho WQ, Jothi R, Crabtree GR. 2011. esBAF facilitates pluripotency by conditioning the genome for LIF/STAT3 signalling and by regulating polycomb function. Nat Cell Biol 13: 903–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O, et al. 2013. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol 31: 827–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F, Workman JL. 2013. Directing transcription to the right way. Cell Res 23: 1153–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL, Rando OJ. 2014. Mechanisms underlying nucleosome positioning in vivo. Annu Rev Biophys 43: 41–63. [DOI] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium 2004. Finishing the euchromatic sequence of the human genome. Nature 431: 931–945. [DOI] [PubMed] [Google Scholar]
- Jacquier A. 2009. The complex eukaryotic transcriptome: unexpected pervasive transcription and novel small RNAs. Nat Rev Genet 10: 833–844. [DOI] [PubMed] [Google Scholar]
- Kaplan CD. 2003. Transcription elongation factors repress transcription initiation from cryptic sites. Science 301: 1096–1099. [DOI] [PubMed] [Google Scholar]
- Khavari PA, Peterson CL, Tamkun JW, Mendel DB, Crabtree GR. 1993. BRG1 contains a conserved domain of the SWI2/SNF2 family necessary for normal mitotic growth and transcription. Nature 366: 170–174. [DOI] [PubMed] [Google Scholar]
- Kim T-K, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, et al. 2010. Widespread transcription at neuronal activity-regulated enhancers. Nature 465: 182–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R, Ichibhashi Y, Kimura S, Chitwood DH, Headland LR, Peng J, Maloof JN, Sinha NR. 2012. A high-throughput method for Illumina RNA-seq library preparation. Front Plant Sci. 3: 202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent BC, Treich I, Carlson M. 1993. The yeast SNF2/SWI2 protein has DNA-stimulated ATPase activity required for transcriptional activation. Genes Dev 7: 583–591. [DOI] [PubMed] [Google Scholar]
- Lessard J, Wu JI, Ranish JA, Wan M, Winslow MM, Staahl BT, Wu H, Aebersold R, Graef IA, Crabtree GR. 2007. An essential switch in subunit composition of a chromatin remodeling complex during neural development. Neuron 55: 201–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin JZ, Yassour M, Adiconis X, Nusbaum C, Thompson DA, Friedman N, Gnirke A, Regev A. 2010. Comprehensive comparative analysis of strand-specific RNA sequencing methods. Nat Methods 7: 709–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquardt S, Escalante-Chong R, Pho N, Wang J, Churchman LS, Springer M, Buratowski S. 2014. A chromatin-based mechanism for limiting divergent noncoding transcription. Cell 157: 1712–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrich TN, Ioshikhes IP, Venters BJ, Jiang C, Tomsho LP, Qi J, Schuster SC, Albert I, Pugh BF. 2008. A barrier nucleosome model for statistical positioning of nucleosomes throughout the yeast genome. Genome Res 18: 1073–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray SC, Serra Barros A, Brown DA, Dudek P, Ayling J, Mellor J. 2012. A pre-initiation complex at the 3′-end of genes drives antisense transcription independent of divergent sense transcription. Nucleic Acids Res 40: 2432–2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natoli G, Andrau JC. 2012. Noncoding transcription at enhancers: general principles and functional models. Annu Rev Genet 46: 1–19. [DOI] [PubMed] [Google Scholar]
- Ntini E, Jarvelin AI, Bornholdt J, Chen Y, Boyd M, Jorgensen M, Andersson R, Hoof I, Schein A, Andersen PR, et al. 2013. Polyadenylation site-induced decay of upstream transcripts enforces promoter directionality. Nat Struct Mol Biol 20: 923–928. [DOI] [PubMed] [Google Scholar]
- Preker P, Nielsen J, Kammler S, Lykke-Andersen S, Christensen MS, Mapendano CK, Schierup MH, Jensen TH. 2008. RNA exosome depletion reveals transcription upstream of active human promoters. Science 322: 1851–1854. [DOI] [PubMed] [Google Scholar]
- Racki LR, Narlikar GJ. 2008. ATP-dependent chromatin remodeling enzymes: two heads are not better, just different. Curr Opin Genet Dev 18: 137–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds N, Salmon-Divon M, Dvinge H, Hynes-Allen A, Balasooriya G, Leaford D, Behrens A, Bertone P, Hendrich B. 2011. NuRD-mediated deacetylation of H3K27 facilitates recruitment of Polycomb repressive complex 2 to direct gene repression. EMBO J 31: 593–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Chang HY. 2012. Genome regulation by long noncoding RNAs. Annu Rev Biochem 81: 145–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwabish MA, Struhl K. 2004. Evidence for eviction and rapid deposition of histones upon transcriptional elongation by RNA polymerase II. Mol Cell Biol 24: 10111–10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seila AC, Calabrese JM, Levine SS, Yeo GW, Rahl PB, Flynn RA, Young RA, Sharp PA. 2008. Divergent transcription from active promoters. Science 322: 1849–1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatoyannopoulos JA. 2012. What does our genome encode? Genome Res 22: 1602–1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan-Wong SM, Zaugg JB, Camblong J, Xu Z, Zhang DW, Mischo HE, Ansari AZ, Luscombe NM, Steinmetz LM, Proudfoot NJ. 2012. Gene loops enhance transcriptional directionality. Science 338: 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiebaut M, Kisseleva-Romanova E, Rougemaille M, Boulay J, Libri D. 2006. Transcription termination and nuclear degradation of cryptic unstable transcripts: a role for the nrd1–nab3 pathway in genome surveillance. Mol Cell 15: 853–864. [DOI] [PubMed] [Google Scholar]
- Thompson DM, Parker R. 2007. Cytoplasmic decay of intergenic transcripts in Saccharomyces cerevisiae. Mol Cell Biol 27: 92–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Garcia-Bassets I, Benner C, Li W, Su X, Zhou Y, Qiu J, Liu W, Kaikkonen MU, Ohgi KA, et al. 2011a. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature 474: 390–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Bryant GO, Floer M, Spagna D, Ptashne M. 2011b. An effect of DNA sequence on nucleosome occupancy and removal. Nat Struct Mol Biol 18: 507–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R. 2013. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153: 910–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber CM, Ramachandran S, Henikoff S. 2014. Nucleosomes are context specific, H2A.Z-modulated barriers to RNA polymerase. Mol Cell 53: 819–830. [DOI] [PubMed] [Google Scholar]
- Weiner A, Hughes A, Yassour M, Rando OJ, Friedman N. 2010. High-resolution nucleosome mapping reveals transcription-dependent promoter packaging. Genome Res 20: 90–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehouse I, Rando OJ, Delrow J, Tsukiyama T. 2007. Chromatin remodelling at promoters suppresses antisense transcription. Nature 450: 1031–1035. [DOI] [PubMed] [Google Scholar]
- Whyte WA, Bilodeau S, Orlando DA, Hoke HA, Frampton GM, Foster CT, Cowley SM, Young RA. 2012. Enhancer decommissioning by LSD1 during embryonic stem cell differentiation. Nature 482: 221–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JI, Lessard J, Olave IA, Qiu Z, Ghosh A, Graef IA, Crabtree GR. 2007. Regulation of dendritic development by neuron-specific chromatin remodeling complexes. Neuron 56: 94–108. [DOI] [PubMed] [Google Scholar]
- Xie L, Pelz C, Wang W, Bashar A, Varlamova O, Shadle S, Impey S. 2011. KDM5B regulates embryonic stem cell self-renewal and represses cryptic intragenic transcription. EMBO J 30: 1473–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadon AN, Singh BN, Hampsey M, Tsukiyama T. 2013. DNA looping facilitates targeting of a chromatin remodeling enzyme. Mol Cell 50: 93–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z, Wang Z, Sharova L, Sharov AA, Ling C, Piao Y, Aiba K, Matoba R, Wang W. 2008. Baf250b-associated SWI/SNF chromatin-remodeling complex is required to maintain undifferentiated mouse embryonic stem cells. Stem Cells 26: 1155–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildirim O, Li R, Hung J-H, Chen PB, Dong X, Ee L-S, Weng Z, Rando OJ, Fazzio TG. 2011. Mbd3/NURD complex regulates expression of 5-hydroxymethylcytosine marked genes in embryonic stem cells. Cell 147: 1498–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan GC, Liu Y, Dion MF, Slack MD, Wu LF, Altschuler SJ, Rando OJ. 2005. Genome-scale identification of nucleosome positions in S. cerevisiae. Science 309: 626–630. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.