GLP and G9a are major H3K9 dimethylases essential for mouse early embryonic development. Here, Liu et al. report that the histone methyltransferase activities of GLP and G9a are stimulated by neighboring nucleosomes that are premethylated at H3K9. In mouse embryonic stem cells harboring a mutant GLP that lacks H3K9me1-binding activity, critical pluripotent genes displayed inefficient establishment of H3K9me2 and delayed gene silencing during differentiation. Mice carrying the H3K9me1-binding mutant form of GLP displayed embryonic growth retardation and defects in calvaria bone formation.
Keywords: GLP, G9a, H3K9 methylation, gene repression, cell differentiation
Abstract
GLP and G9a are major H3K9 dimethylases and are essential for mouse early embryonic development. GLP and G9a both harbor ankyrin repeat domains that are capable of binding H3K9 methylation. However, the functional significance of their recognition of H3K9 methylation is unknown. Here, we report that the histone methyltransferase activities of GLP and G9a are stimulated by neighboring nucleosomes that are premethylated at H3K9. These stimulation events function in cis and are dependent on the H3K9 methylation binding activities of ankyrin repeat domains of GLP and G9a. Disruption of the H3K9 methylation-binding activity of GLP in mice causes growth retardation of embryos, ossification defects of calvaria, and postnatal lethality due to starvation of the pups. In mouse embryonic stem cells (ESCs) harboring a mutant GLP that lacks H3K9me1-binding activity, critical pluripotent genes, including Oct4 and Nanog, display inefficient establishment of H3K9me2 and delayed gene silencing during differentiation. Collectively, our study reveals a new activation mechanism for GLP and G9a that plays an important role in ESC differentiation and mouse viability.
GLP and G9a are SET domain-containing histone methyltransferases that play vital roles during early mouse development (Tachibana et al. 2001, 2002, 2005) and are also involved in many other developmental processes, such as hematopoiesis, adipogenesis, and retrovirus silencing. (Chen et al. 2012; Maksakova et al. 2013; Wang et al. 2013; Balemans et al. 2014). GLP and G9a are widely expressed and are responsible for the majority of H3K9me2 at facultative heterochromatin regions (Mermoud et al. 2002; Tachibana et al. 2002; Trojer and Reinberg 2007). Facultative heterochromatin regions often span up to millions of base pairs and, unlike the centromeric constitutive heterochromatin, vary among cell types (Wen et al. 2009; Lienert et al. 2011). During cell fate transition events such as the differentiation of the embryonic stem cells (ESCs) or X-chromosome inactivation, new blocks of facultative heterochromatin are established (Mermoud et al. 2002; Chow and Heard 2009; Wen et al. 2009). However, it remains challenging to pinpoint different mechanisms involved in efficiently establishing the facultative heterochromatin marks during cell fate transition. H3K27me3, another histone modification marking distinct facultative heterochromatic regions, can be efficiently established by two different allosteric activation mechanisms of the H3K27-specific histone methyltransferase complex PRC2, which can sense repressive histone modifications (Margueron et al. 2009) and dense chromatin (Yuan et al. 2012). H3K9me2 covers more than one-third of the genome and undergoes drastic changes during cell fate transitions (Wen et al. 2009; Lienert et al. 2011; Chen et al. 2012). However, GLP and G9a exhibit very low activities toward nucleosomal H3 in vitro (Tachibana et al. 2005). This leads to questions of whether any activation mechanisms are involved in efficiently establishing H3K9me2 during cell fate transitions and whether the enzymatic activities of GLP and G9a are also regulated by chromatin environment.
Intriguingly, in addition to the catalytic activities for methylating H3K9, GLP and G9a also carry the ankyrin repeat domains that specifically recognize H3K9me1 and H3K9me2, respectively (Brent and Marmorstein 2008; Collins et al. 2008). We reason that GLP and G9a may use their H3K9 methylation-binding activities to sense methylated H3K9 and achieve their full activity as well as maintain H3K9me2 levels during mitotic divisions or efficiently establish H3K9me2 at genomic regions that are to be silenced during cell fate transition events.
Using oligonucleosome substrates containing unmodified nucleosomes and nucleosomes premodified at the H3K9 position to various states, we observed strong stimulation of the enzymatic activities of GLP and G9a by neighboring nucleosomes containing H3K9me1 and H3K9me2, respectively. The binding abilities to H3K9me1 and H3K9me2 of GLP and G9a were required for the enzymatic stimulation. ESCs harboring the H3K9me1-binding mutant form of GLP failed to efficiently establish H3K9me2 during retinoic acid (RA)-induced differentiation across a large number of genes, and a subset of these genes displayed delayed transcription repression. Notably, several genes encoding critical pluripotent factors, including Oct4 and Nanog, were among them, suggesting that GLP’s H3K9 methylation-binding activity is required for efficient establishment of H3K9me2 and rapid shutdown of the pluripotency transcription program in ESCs undergoing differentiation. Homozygous knock-in mice carrying a H3K9me1-binding mutant form of GLP exhibited postnatal lethality. These mice displayed embryonic growth retardation and defects in calvaria bone formation. Together, these results provided evidences for the significance of GLP’s H3K9me1-binding activity in efficient H3K9me2 establishment and gene silencing during cell differentiation and mouse viability but not in mitotic inheritance of H3K9 methylation.
Results
Histone methyltransferase activities of GLP and G9a are stimulated by neighboring nucleosomes premethylated at H3K9 in an ankyrin repeat-dependent manner
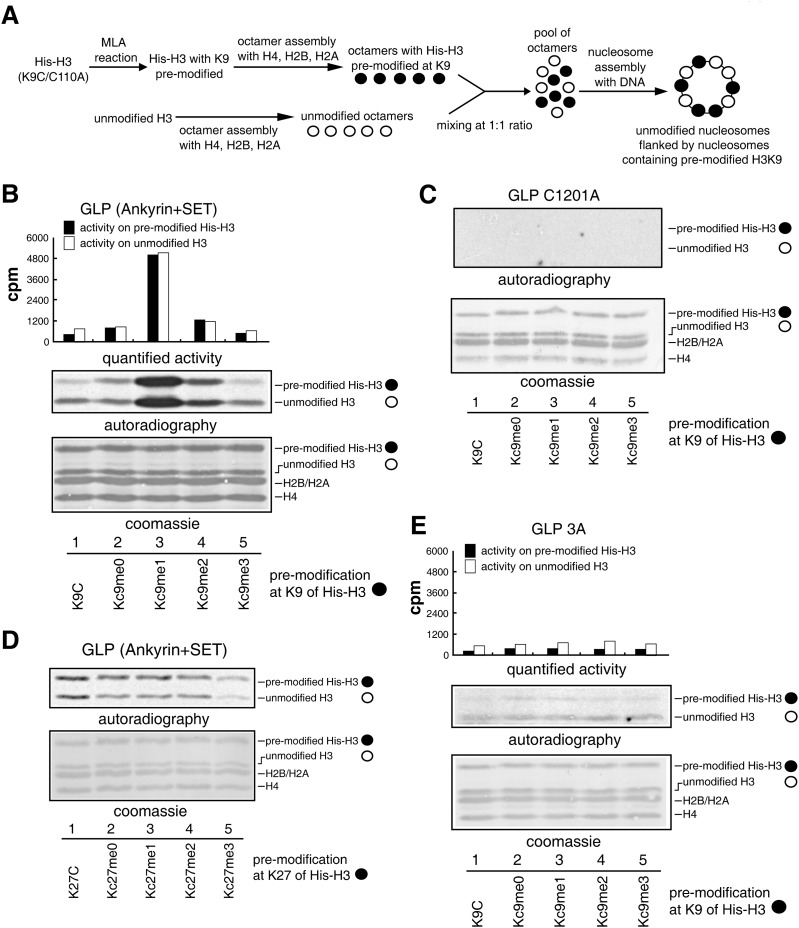
To test whether GLP and G9a can be stimulated by neighboring nucleosomes premethylated at H3K9, we developed a histone methyltransferase activity stimulation assay. We prepared premethylated histones using chemical reactions to install methyl-lysine analogs (MLAs) at the K9 position of histone H3 (Simon et al. 2007). The MLAs installed by MLA reactions preserve most of the features of the methyl-lysine, including correct recognition by specific methyl-lysine-binding proteins (Simon et al. 2007) and compatibility in serving as substrates for histone methyltransferases and demethylases (Jia et al. 2009). Histone octamers that contained H3 premodified at the K9 position with various modifications (Kc9me0/1/2/3 or unreacted K9C) and histone octamers that contained unmodified H3 were assembled separately and then pooled together at a 1:1 ratio. The octamers were assembled with DNA into oligonucleosomes. The assembled oligonucleosomes were purified by gel filtration on a Sepharose CL-4B column to remove free histone octamers. The final substrates contained unmodified nucleosomes that were flanked by nucleosomes harboring premodified H3K9 (Fig. 1A). To distinguish the two types of histone H3, the premodified H3 histones were tagged and therefore migrated slower on SDS-PAGE gels (Fig. 1B).
Figure 1.
Stimulation of GLP activity by nucleosomes that are premethylated at H3K9. (A) Substrate description. (B,C) Histone methyltransferase activity assay at nucleosomes flanked by premodified nucleosomes with the wild-type GLP (B) or GLP catalytic mutant (C). (D) Histone methyltransferase activity assay at nucleosomes flanked by nucleosomes premodified at H3K27 with GLP. (E) Histone methyltransferase activity assay at nucleosomes flanked by H3K9 premodified nucleosomes with GLP 3A. (One microgram of recombinant enzymes and 1 μg of recombinant oligonucleosomes were used in the assay.)
Recombinant GLP and G9a proteins covering ankyrin repeats and SET domains were purified from Escherichia coli and assayed for methyltransferase activity using the above-described substrates. Strikingly, despite the fact that equal amounts of enzymes and unmodified H3 histones were present in the reactions, G9a displayed 20-fold more activity on the unmodified nucleosomes that were flanked by H3Kc9me2-containing nucleosomes than on unmodified nucleosomes that were flanked by nucleosomes containing H3K9C, H3Kc9me0, or H3Kc9me3 (Supplemental Fig. 1A). These results are consistent with the fact that H3K9me2 is G9a’s favored binding partner (Collins et al. 2008). Similarly, GLP displayed the best activity on unmodified nucleosomes flanked by nucleosomes containing H3Kc9me1 (Fig. 1B), which was in agreement with its preferred binding to H3K9me1 (Collins et al. 2008). Catalytic mutants of GLP (C1201A) or G9a (C1007A) displayed no detectable activity on any of the substrates (Fig. 1C; Supplemental Fig. S1B). Notably, G9a also displayed much higher enzymatic activity at the tagged premethylated H3 histones in the case of H3Kc9me2 (Supplemental Fig. 1A, lane 4). Similar observations were also obtained for GLP at the tagged premethylated H3 histones in the case of H3Kc9me1 (Fig. 1B, lane 3). This is most likely due to G9a’s and GLP’s in vitro catalytic activities at H3K27, as previously reported (Tachibana et al. 2001; Wu et al. 2011). Indeed, when residue Lys27 of H3 was mutated to alanine, the stimulation activity was observed at only the unmodified H3 histones but not the tagged premethylated H3 histones (Supplemental Fig. 1C, lane 2). Furthermore, neither GLP nor G9a activity was stimulated by nucleosomes premethylated at H3K27 (Fig. 1D; Supplemental Fig. 1D), in agreement with their inability to recognize H3K27 methylation (Collins et al. 2008). Together, these results suggested that the activities of GLP and G9a on nucleosomal histones could be strongly stimulated when their substrates were flanked by neighboring nucleosomes harboring H3K9 premethylation.
Ankyrin repeats of GLP and G9a were responsible for their recognition toward H3K9 methylation (Collins et al. 2008). GLP W877A/W882A/E885A and G9a W791A/W796A/E799A mutants (abbreviated as GLP 3A and G9a 3A mutants, respectively) lost methylated H3K9-binding activity due to the disruption of their aromatic cages and hydrogen bonds within the ankyrin repeats that confine the methylated lysine (Collins et al. 2008), which was confirmed by isothermal titration calorimetry (ITC) (Supplemental Fig. 2). Neither GLP 3A (Fig. 1E) nor G9a 3A (Supplemental Fig. 1E) was stimulated by premethylated neighboring nucleosomes, indicating that recognition of premethylated neighboring nucleosomes by ankyrin repeats is an essential step for stimulation.
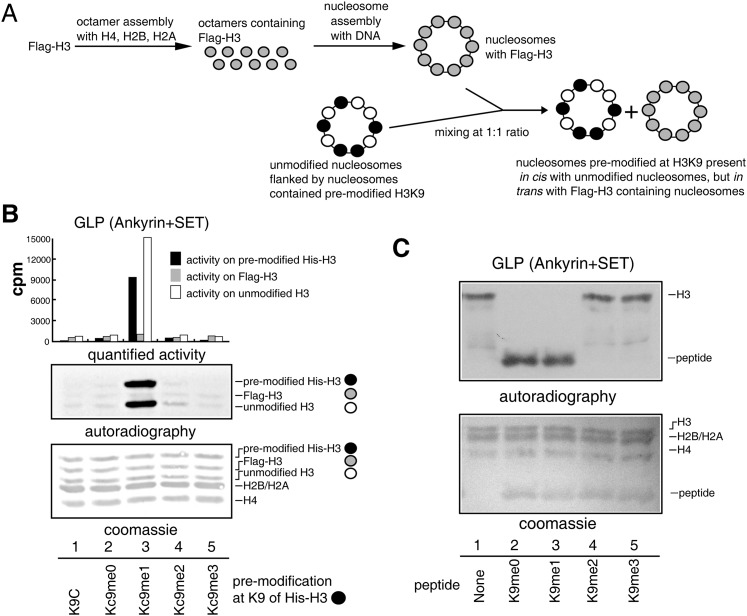
The stimulation of GLP and G9a activities by premethylated nucleosome functions in cis
A second set of substrates was assembled to allow simultaneous testing of stimulation events at nucleosomes existing in cis and in trans. Oligonucleosomes containing Flag-H3 were mixed with the substrates described in Figure 1A at a 1:1 ratio (Fig. 2A). In the final substrate mixture, the unmodified H3 histones existed in cis with the premodified His-H3 histones, while the Flag-H3 histones existed in trans with the premodified His-H3 histones (Fig. 2A). These three types of H3 histones could be distinguished in SDS-PAGE gels due to their different migration velocities (Fig. 2B). GLP (Fig. 2B) and G9a (Supplemental Fig. 3A) displayed robust activity on unmodified H3 histones existing in cis with nucleosomes containing H3Kc9me1 and H3Kc9me2, respectively, but not on Flag-H3 nucleosomes existing in trans with the premodified nucleosomes. To exclude that the Flag tag might affect the enzymatic activities, control experiments were performed using substrates in which Flag-H3 histones were placed in cis with the pre-modified His-H3 histones, and untagged H3 histones were placed in trans with the premodified His-H3 histones. GLP and G9a displayed robust activities on Flag-H3 nucleosomes existing in cis with the premethylated H3 and minimal activities on H3 nucleosomes existing in trans with the premethylated H3 (Supplemental Fig. 3B,C). These results indicated that the stimulation events were dependent on spatial position of the nucleosomes and functioned in cis.
Figure 2.
Stimulation of GLP activity by premethylated neighboring nucleosomes is a cis event. (A) Substrate description. (B) Methyltransferase activity assay of GLP at nucleosomes existing in cis and in trans with premodified nucleosomes. (C) Methylated H3K9 peptides did not stimulate the methyltransferase activity of GLP. (One microgram of recombinant enzymes, 0.5 μg of peptide, and 1 μg of recombinant oligonucleosomes were used in the assay.)
H3K27 methyltransferase (PRC2) can be allosterically activated by peptides containing H3K27me3 (Margueron et al. 2009). However, neither GLP nor G9a was activated by the addition of methylated H3K9 peptides (Fig. 2C; Supplemental Fig. 3D), which suggests that GLP and G9a adapted distinct strategies for spreading chromatin modifications different from the allosteric activation approach that PRC2 uses. Taken together, GLP and G9a were able to sense H3K9me1/2 through their ankyrin repeats and facilitate their H3K9 methylation activities in vitro.
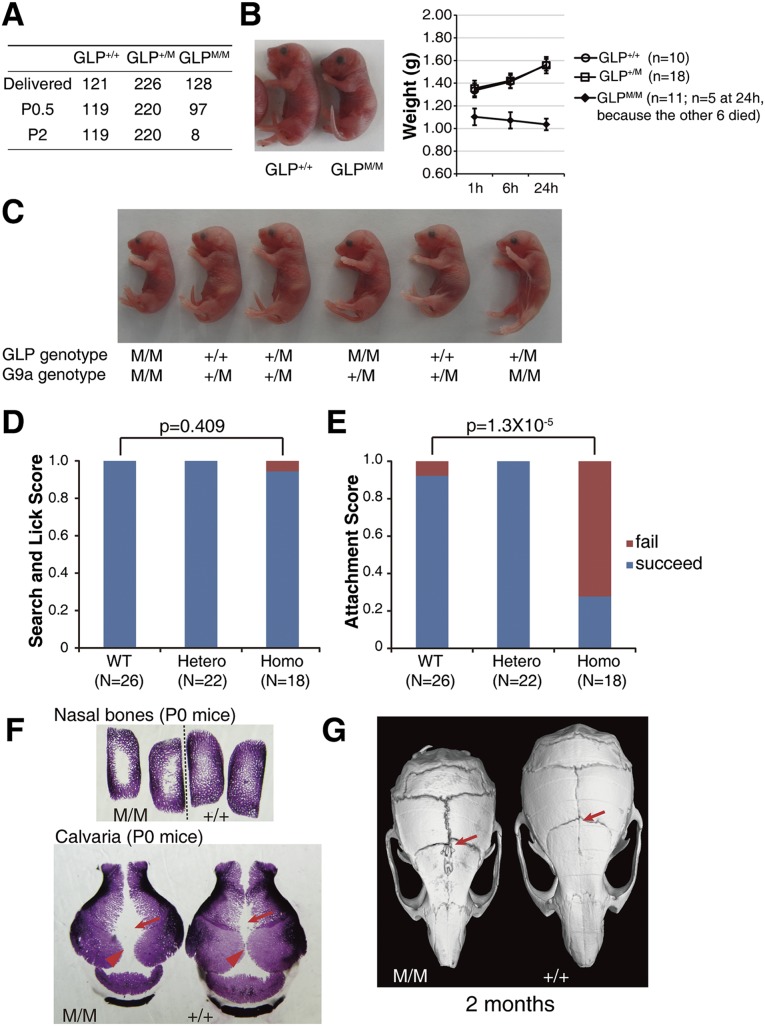
GLP 3A mutant mice were postnatal-lethal with growth retardation and craniofacial defects
To understand the in vivo role of the H3K9 methylation recognition activities of GLP and G9a, we generated knock-in mice bearing the GLP 3A or G9a 3A mutations. Mutations were introduced into R1 ESCs by homologous recombination (Supplemental Fig. 4). We confirmed that the targeting events did not affect the expression level of the targeted GLP and G9a alleles in the corresponding heterozygote ESC lines (Supplemental Fig. 4). F1 heterozygous mice of the GLP 3A and G9a 3A mutants (termed GLP+/M and G9a+/M) were healthy and fertile. Surprisingly, homozygous mice (GLPM/M and G9aM/M) displayed strikingly different phenotypes. G9aM/M mice were born at the Mendelian ratio (Supplemental Fig. 5A) and were healthy and fertile. In contrast, GLPM/M mice were mostly postnatal-lethal (Fig. 3A). All GLPM/M mice were born smaller in size (Fig. 3B), and 120 out of 128 of them died within 2 d of birth (Fig. 3A). Only eight GLPM/M mice survived to adulthood, and they suffered from severe growth retardation within the first 4 wk after birth (Supplemental Fig. 5B). GLP and G9a double-mutant mice displayed a phenotype similar to that of GLPM/M mice (Fig. 3C), suggesting that the H3K9 methylation recognition activity of GLP played a dominant role in vivo. Therefore, we focused our study on GLPM/M in all subsequent experiments.
Figure 3.
The H3K9 methylation-binding activity of GLP was required for mouse viability and calvaria ossification. (A) Mouse counts revealed the postnatal lethality of GLPM/M mice. (B) Growth retardation of GLPM/M mice at birth. Error bars represent the standard error. (C) Newborn mice of GLP and G9a single and double mutants. (D,E) Suckling assays showing the abilities of newborn mice to search and lick nipples (D) and remain stably attached to the nipples (E). (F) Alizarin red staining of neonatal mouse calvaria showing delayed ossification at the nasal bones and sagittal (arrowhead) and metopic (arrow) structures in GLPM/M mice. (G) MicroCT scanning showing bone loss in metopic sutures (arrow) of an adult GLPM/M mouse that survived.
Consistent with the smaller size of GLPM/M newborns, the GLPM/M embryos also showed growth retardation as early as embryonic day 10.5 (E10.5) (Supplemental Fig. 5C). Although no drastic abnormalities of the main organs were observed and no catastrophic events occurred in breathing and heart beating, we consistently observed that no milk was present in the stomachs of GLPM/M pups, so it was highly possible that GLPM/M mice died of starvation. To understand at which stage the GLPM/M pups failed to be fed, we adapted an assay to dissect multiple steps of suckling processes (Hongo et al. 2000). The GLPM/M mice were capable of actively searching and licking the nipples of mother mice (Fig. 3D), suggesting that their olfaction and sensory–motor systems were probably fine. However, these pups were unable to remain stably attached to the nipples; while most wild-type and heterozygote pups can be stably attached to the nipples for up to 20 min, the GLPM/M pups failed to remain attached for even just 1 min in ∼70% of the attempts (Fig. 3E), which suggested that they might suffer from craniofacial development abnormalities (Turgeon and Meloche 2009). Consistent with this hypothesis, skeletal preparations at various developmental stages showed obvious delayed calvaria and nasal bone ossification in GLPM/M mice (Fig. 3F; Supplemental Fig. 5D). The most severe delayed ossification occurred in the sagittal and metopic sutures as well as in the nasal bones (Fig. 3F). Even for the few surviving adult GLPM/M mice, bone loss was observed in their metopic sutures (Fig. 3G; Supplemental Fig. 5E), which was likely caused by ossification defects during embryonic development. In addition, several of the surviving adult GLPM/M mice (two out of eight) displayed an obviously bent nose and malocclusion (Supplemental Fig. 5E,F).
The H3K9me1-binding activity of GLP is largely dispensable for maintenance of global H3K9me2 levels in ESCs and tissues
To study the in vivo role of H3K9 methylation-binding activity of GLP at molecular level, we derived wild-type and GLP 3A mutant ESCs (termed wild-type and GLP 3A ESCs, respectively) from littermate embryos at the blastocyst stage. These ESCs displayed the correct karyotypes (Supplemental Fig. 6A) and similar expression levels of GLP (Supplemental Fig. 6B). Western analysis revealed comparable bulk H3K9me2 levels between wild-type and GLP 3A ESCs (Supplemental Fig. 6C). Next, we analyzed brain and liver samples harvested from multiple littermate pairs of wild-type and GLPM/M newborn pups (Supplemental Fig. 6D). We did not observe consistent changes of H3K9me2 between wild-type and GLPM/M livers or brains (Supplemental Fig. 6D). Considering that Western analysis is not an ideal quantitative assay, especially for subtle changes, we performed H3K9me2 ChIP-seq (chromatin immunoprecipitation [ChIP] combined with deep sequencing) experiments using ESCs and livers from newborn pups with wild-type and GLP mutant backgrounds. We first divided the mouse genome into 10-kb windows. This window size was chosen because H3K9me2 often exist in large blocks of chromatin regions termed LOCKs, and the minimal size of LOCKs is ∼20 kb (Wen et al. 2009). We plotted the H3K9me2 fold enrichment changes between mutant and wild-type samples for all of the 10-kb windows. In ESCs and livers from newborn pups, only 1% and 2.2% of the windows displayed H3K9me2 level changes >1.5-fold, respectively (Supplemental Fig. 6E). The above results collectively suggest that the H3K9me1-binding activity of GLP is largely dispensable for maintenance of global H3K9me2 levels in ESCs and tissues.
Nevertheless, among the 3940 10-kb windows that displayed a reduction of H3K9me2 in mutant newborn livers, 2430 localized to genes (62%). In contrast, among the 770 10-kb windows that displayed an increase of H3K9me2 in mutant newborn livers, 196 localized to genes (25%). In ESCs, >70% of 10-kb windows with increased or decreased H3K9me2 levels localized to genes. These results suggest that H3K9me2 levels at genic regions are preferentially affected upon the loss of H3K9me1-binding activity of GLP.
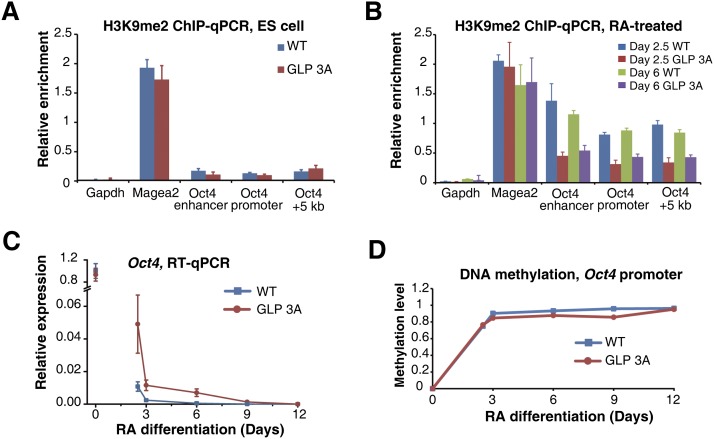
The H3K9me1-binding activity of GLP is critical for efficient establishment of H3K9me2 and gene silencing at the Oct4 gene during ESC differentiation
Considering that the H3K9me1-binding activity of GLP stimulated its catalytic activity in vitro (Fig. 1), it displayed a minimal effect in its contribution to total H3K9me2 levels in several cell types (Supplemental Fig. 6D,E). We hypothesized that the H3K9 methylation-binding activity of GLP and its enzymatic stimulation may be more important for the kinetics of H3K9me2 establishment than the maintenance of H3K9me2. To test this hypothesis, we adapted a well-studied process involving dynamic establishment of H3K9me2, which is the RA-induced differentiation of ESCs (Wen et al. 2009; Lienert et al. 2011). During RA-induced differentiation of ESCs, the expression of Oct4 gene is quickly reduced, and its final silencing requires GLP, G9a, and H3K9me2 establishment at the Oct4 locus (Feldman et al. 2006; Epsztejn-Litman et al. 2008). We first examined the H3K9me2 establishment and gene silencing at the Oct4 gene during RA-induced differentiation of ESCs with wild-type and GLP 3A backgrounds. Prior to RA treatment, H3K9me2 levels were comparably low at the Oct4 gene in wild-type and GLP 3A ESCs (Fig. 4A). After RA treatment, H3K9me2 levels were significantly increased at the Oct4 locus in wild-type cells (Fig. 4B), in agreement with previous reports (Feldman et al. 2006). However, in RA-treated GLP 3A cells, H3K9me2 levels at the Oct4 locus were much lower than that in the wild-type cells (Fig. 4B). In contrast, such a difference was not observed at the Magea2 gene (Fig. 4A,B), which is constitutively repressed by GLP and G9a (Tachibana et al. 2002).
Figure 4.
H3K9me2 establishment and transcription silencing of Oct4 during ESC differentiation were delayed in GLP 3A cells. (A,B) ChIP-qPCR results showing H3K9me2 occupancy at the indicated loci in ESCs (A) or RA-treated cells at the indicated time points (B). (C) RT–PCR results showing Oct4 expression levels during differentiation. (D) DNA methylation levels at the Oct4 promoter during differentiation. Error bars represent the standard error.
Wild-type and GLP 3A ESCs displayed comparable levels of Oct4 expression (Fig. 4C), and, in both cells, Oct4 expression was quickly reduced within the first 3 d of RA treatment, which most likely reflects changes of transcription factor association at the Oct4 gene. However, GLP 3A cells displayed an obvious delay in shutting down Oct4 expression, and complete silencing of Oct4 transcription was delayed for ∼6 d in GLP 3A cells (Fig. 4C), although establishment of DNA methylation at the Oct4 promoter was largely normal in these cells during RA-induced differentiation (Fig. 4D). To rule out a potential clonal effect of ESCs, the above experiments were repeated in ESCs derived from a separate litter of mice, and similar results were observed (Supplemental Fig. 7A,B).
We also monitored H3K9me2 establishment and Oct4 repression in a related differentiation system involving the formation of embryoid bodies (EBs) from ESCs, which to some extent mimics the in vivo post-implantation embryo development (Desbaillets et al. 2000). In this system, we observed similar defects in H3K9me2 establishment and transcription repression at the Oct4 gene (Supplemental Fig. 7C–E). These results collectively suggested that the H3K9me1-binding activity of GLP was required for efficient establishment of H3K9me2 and rapid repression of the Oct4 gene during differentiation.
The H3K9me1-binding activity of GLP is required for efficient establishment of H3K9me2, but not H3K9me1, at genic regions during ESC differentiation
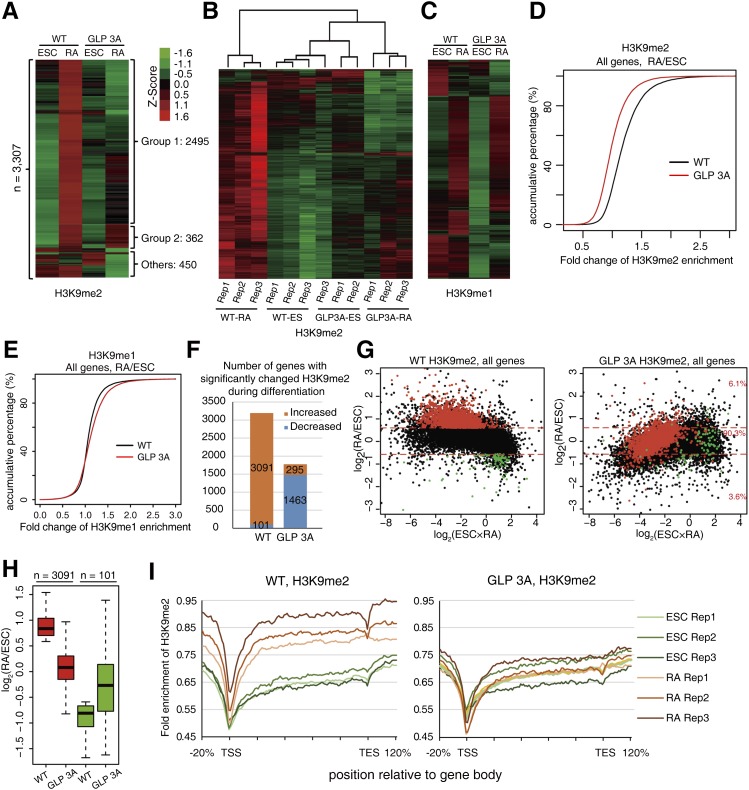
We then asked whether a similar role of GLP’s H3K9me1-binding activity occurred at the genome-wide level. Given that GLP is essential for H3K9me2 levels in vivo (Tachibana et al. 2005), we first performed ChIP-seq experiments for H3K9me2 with ESCs and RA-treated cells at 2.5 d, at which stage the GLP protein levels were comparable between wild-type and GLP 3A cells (Supplemental Fig. 8A). Three biological replicates were performed for each analysis. We divided the mouse genome into 10-kb windows. For each window, the fold enrichment of H3K9me2 was calculated as the ratio of normalized read density between ChIP and input samples, and the mean H3K9me2 fold enrichment value was calculated with data from the three replicates. Windows with at least one case of more than twofold change between any pair of samples (3307 in total) were selected for clustering analysis (Fig. 5A). The majority of these regions (86%) displayed an increase in H3K9me2 during RA-induced differentiation in wild-type cells (Fig. 5A, groups 1 and 2), suggesting that H3K9me2 was actively established at these regions in normal differentiating cells. Although wild-type and GLP 3A cells contained similar H3K9me2 levels at these regions in ESCs, ∼75% (2495 of 3307) of these regions displayed obviously lower H3K9me2 levels in RA-treated GLP 3A cells in comparison with their wild-type counterparts (Fig. 5A, group 1). On the other hand, only 11% (362 of 3307) of these regions displayed comparable or higher H3K9me2 levels in RA-treated GLP 3A cells (Fig. 5A, group 2). These results clearly indicate a global defect in establishing H3K9me2 in GLP 3A cells.
Figure 5.
The H3K9 methylation-binding activity of GLP was required for efficient H3K9me2 establishment during ESC differentiation at a genome-wide level. (A) Clustering of the 10-kb genomic windows according to their H3K9me2 levels for wild-type (WT) and GLP 3A cells with or without RA treatment. The fold enrichment values of three biological replicates were averaged, centered to the mean of rows, and normalized to standard deviation by rows (Z-score in A–C). (B) Clustering of the 10-kb genomic windows according to their H3K9me2 levels for all replicates in wild-type and GLP 3A cells with or without RA treatment. (C) Clustering of the 3307 10-kb genomic windows (shown in A) according to their H3K9me1 levels for wild-type and GLP 3A cells with or without RA treatment. The values are averages of three biological replicates. (D,E) Accumulative distribution of H3K9me2 (D) and H3K9me1 (E) fold changes between RA-treated cells and ESCs at genic regions. (F) Number of genes with significantly changed H3K9me2 in wild-type or GLP 3A cells after RA treatment. (G) MA plot of H3K9me2 fold changes against the average H3K9me2 fold enrichment. Red dots represent genes with significantly increased H3K9me2 levels after RA treatment in wild-type cells. (H) Box plot of H3K9me2 fold changes between ESCs and RA-treated cells at genes acquired by H3K9me2 (shown in G) in wild-type and GLP 3A cells. (I) H3K9me2 profile at genic regions of genes acquired by H3K9me2 (shown in G) in ESCs or RA-treated wild-type (left) and GLP 3A (right) cells. “Rep 1–3” represents the three biological repeats. Lengths of genes were normalized to 100 percentiles and extended 20% upstream of and downstream from the gene body, respectively.
To test the reproducibility of the above analysis, we performed clustering analysis for the same 3307 10-kb windows for all 12 experiments (two cell types × two treatments × three replicates). Essentially, almost all replicates clustered together, indicating good reproducibility among them (Fig. 5B). The only exception was that one experiment of the GLP 3A cells at the ESC stage was grouped together with the wild-type ESCs, which was in agreement with our observation that wild-type and GLP 3A ESCs were highly similar for their H3K9me2 patterns (Fig. 5B). These observations were also supported by quantitative results calculating the correlation coefficiency between each experiment pair (Supplemental Fig. 8B). Interestingly, RA-treated GLP 3A cells, which failed to establish H3K9me2 at most of these regions, were clustered together with ESCs but not RA-treated wild-type cells (Fig. 5B), implying that the differentiation process was impaired.
Given that GLP can also generate H3K9me1 products and used them as substrates for further methylation (Tachibana et al. 2005), we performed ChIP-seq experiments for H3K9me1 using the same samples. The same above-mentioned 3307 10-kb genomic regions were first chosen for analysis, and ∼66% of them displayed obvious gain of H3K9me1 after RA treatment in wild-type cells (Fig. 5C). Most of these regions displayed correctly established H3K9me1 in GLP 3A cells after RA treatment (Fig. 5C). However, some of the regions that are defective in establishing H3K9me2 during differentiation displayed a reduction of H3K9me1 at the ESC stage of the GLP 3A cells (Fig. 5C). To analyze whether the reduced H3K9me1 is correlated with reduced H3K9me2 in GLP 3A cells before or after RA-induced differentiation, we first selected all 10-kb regions with more than twofold increase of H3K9me2 during RA-induced differentiation in wild-type cells, categorized them into five subgroups according to their H3K9me1 ratio (GLP 3A/wild type) at the ESC stage, and calculated their H3K9me2 ratio (GLP 3A/wild type) before or after RA treatment. Regions with lower H3K9me1 levels tended to display relatively lower H3K9me2 levels in GLP 3A ESCs (Supplemental Fig. S8C). On the other hand, all of these regions displayed similar degrees of defect in their H3K9me2 levels in GLP 3A cells after RA treatment regardless of their initial levels of H3K9me1 defect at the ESC stage.
We also directly analyzed the H3K9me1 levels in the four samples above. All 10-kb windows with at least one case of more than twofold change between any pair of samples (5642 in total) were selected for clustering analysis. Clearly, the main differences among the four samples resided between ESCs and RA-treated cells but not between wild-type and GLP 3A cells (Supplemental Fig. S8D). Taken together, the above results collectively support that the primary defect of GLP 3A cells during RA treatment was the efficient establishment of H3K9me2 but not of H3K9me1.
Notably, 89% (2955 of 3307) of the regions with the highest H3K9me2 variation among the four samples were genic regions of 3079 genes (P < 1E-300, binomial test), which was consistent with previous studies reporting that GLP and G9a primarily functioned at genic regions during differentiation of ESCs (Lienert et al. 2011). Therefore, we focused on the establishment of H3K9 methylation at genic regions for further analysis. Globally, H3K9me2 levels at genic regions were comparable between wild-type and GLP 3A ESCs (Supplemental Fig. 8E). However, after RA treatment, GLP 3A cells displayed significantly lower H3K9me2 levels (P < 2.2E-16) at genic regions than wild-type cells (Supplemental Fig 8F). We also calculated the H3K9me2 ratio between RA-treated and untreated cells and plotted the accumulative percentage curve against the fold change of the H3K9me2 ratio. GLP 3A cells displayed a clear shift in comparison with wild-type cells (Fig. 5D), indicating a wide range of defects in establishing H3K9me2 at genic regions in GLP 3A cells. Such a defect was not observed for H3K9me1; instead, the GLP 3A cells displayed modestly higher H3K9me1 enrichment at genic regions after RA treatment than their wild-type counterparts (Fig. 5E), and this probably reflected that less H3K9me1 was converted to H3K9me2 in the GLP 3A cells.
To further analyze the defect of H3K9me2 establishment at genes in detail, we calculated the H3K9me2 ratio between RA-treated cells and ESCs for each gene and defined genes with significantly changed H3K9me2 levels during RA-induced differentiation in wild-type cells and GLP 3A cells (P < 0.05 in three biological replicates with at least 1.5-fold change). Clearly, far more genes gained H3K9me2 than those lost H3K9me2 (3091 vs. 101). In contrast, in GLP 3A cells, only 295 genes gained H3K9me2, while 1463 genes displayed decreased H3K9me2 levels (Fig. 5F).
We next plotted the H3K9me2 changes against the average H3K9me2 fold enrichment of ESCs and RA-treated cells. First, the overall distribution of the plot for the wild-type cells was much higher than the one for GLP 3A cells (Fig. 5G), indicating that the GLP 3A cells suffered from a wide range of defects in establishing H3K9me2 at genes during differentiation. Strikingly, 93.9% of the genes that acquired H3K9me2 in wild-type cells displayed obviously reduced H3K9me2 levels in GLP 3A cells (Fig. 5G, right panel), and the average difference in H3K9me2 changes between wild-type and GLP 3A cells at these genes was 0.8 (log2) (Fig. 5H). We next plotted the average fold enrichment of H3K9me2 at these genes for all 12 samples, and the results clearly indicated defective H3K9me2 establishment in GLP 3A cells (Fig. 5I).
We also analyzed genes with significantly changed H3K9me2 levels during differentiation in GLP 3A cells. The results showed that a large number of genes in GLP 3A cells failed to maintain the H3K9me2 levels during differentiation (Supplemental Fig. 9A). These results again implicated that the GLP 3A cells failed to establish H3K9me2 at a wide range of genes during differentiation. Notably, at the 295 genes that acquired H3K9me2 in GLP 3A cells, the average difference in H3K9me2 changes between wild-type and GLP 3A cells was only 0.2 (log2) (Supplemental Fig. 9B), much more modest than the difference observed in Figure 5H.
Together, these results indicated that GLP 3A cells had normal H3K9me2 levels at the ESC stage but failed to correctly establish H3K9me2 levels at genic regions during RA-induced cell differentiation.
The H3K9me1-binding activity of GLP is required for efficient silencing of pluripotent genes during ESC differentiation
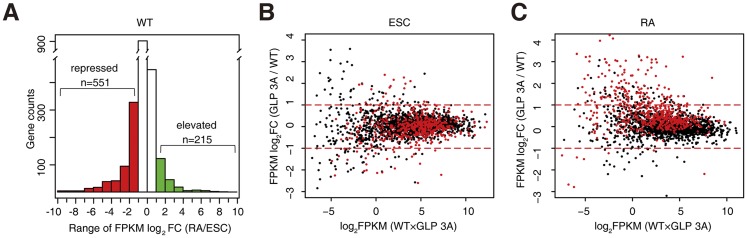
To examine whether the defective establishment of H3K9me2 in GLP 3A cells was associated with inefficient gene repression, we performed RNA sequencing (RNA-seq) experiments for the above-mentioned samples. We chose genes acquired by H3K9me2 during RA-induced differentiation in wild-type cells (Fig. 5G, red dots) and plotted their expression fold changes against their expression levels. The majority of these genes did not show more than twofold changes during RA treatment in wild-type cells, probably because H3K9me2 changes do not lead to strict linear changes in gene expression (Lienert et al. 2011), and sometimes H3K9me2 can also occur during transcription (Vakoc et al. 2005, 2006; Lee et al. 2006; Yuan et al. 2007; Chaturvedi et al. 2009). Nevertheless, 551 genes were repressed (Fig. 6A, red bars), while only 215 genes were activated (Fig. 6A, green bars) during RA treatment in wild-type cells, in line with a role of H3K9me2 establishment in silencing a subset of genes during differentiation. Remarkably, these 551 genes displayed similar expression levels in wild-type and GLP 3A ESCs (Fig. 6B), but 110 of them were expressed at much higher levels (more than twofold) in GLP 3A cells than in wild-type cells after RA treatment (Fig. 6C). Moreover, even for the rest of the 441 genes that did not cross the twofold threshold, we observed an overall trend of upshift, which suggests that inefficient establishment of H3K9me2 contributed to inefficient silencing of these genes.
Figure 6.
Inefficient gene repression in GLP 3A cells during RA-induced differentiation. (A) Histogram showing the expression change of genes acquired by H3K9me2 (Fig. 5G) during differentiation in wild-type cells. (Black bars) Unchanged; (red bars) repressed; (green bars) activated. (B,C) Expression comparison of the genes acquired by H3K9me2 in wild-type cells (Fig. 5G) between wild-type and GLP 3A ESCs (B) or RA-treated cells (C). Note: Red dots in B and C represent the same set of genes, and all of the dots shown represent the red dots in Figure 5G.
As a control, we also studied genes displaying significantly elevated H3K9me2 levels during RA-induced differentiation in GLP 3A cells (Supplemental Fig. 9A, red dots). Among these genes (295 in total; 151 have detectable expression), 70 genes were repressed in GLP 3A cells during RA-induced differentiation (Supplemental Fig. 9C, red bars), and their expression differences between GLP 3A and wild-type cells did not show a particularly biased shift before or after RA treatment (Supplemental Fig. 9D).
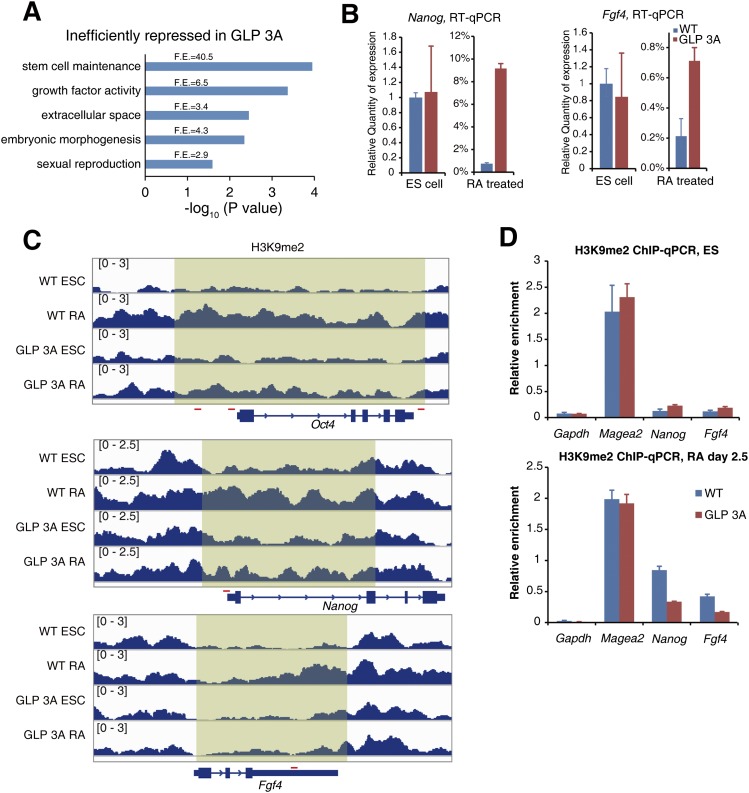
Notably, among the 110 genes that failed to be efficiently repressed in GLP 3A cells, the gene ontology (GO) term “stem cell maintenance” was enriched for >40-fold (Fig. 7A), and several critical pluripotent genes, including Oct4, Nanog, and Fgf4, were found within the list (Supplemental Table 1). The silencing defect of these genes in GLP 3A cells during RA-induced differentiation was validated by RT-qPCR results (Fig. 7B). ChIP-seq results at these gene loci are shown in Figure 7C and were verified by ChIP-qPCR (Fig. 7D).
Figure 7.
Examples of pluripotent genes with defects in H3K9me2 establishment and gene repression. (A) GO term analysis of the 110 genes that failed to be efficiently repressed in RA-treated GLP 3A cells. (F.E.) Fold enrichment. (B) RT-qPCR results showing the expression levels of the indicated genes in ESCs and RA-treated cells. (C) Normalized H3K9me2 profiles at the indicated gene loci. The red lines represent amplified regions used in ChIP-qPCR validation experiments. (D) ChIP-qPCR verified the H3K9me2 level at the indicated genes in ESCs and RA-treated cells. Error bars represent the standard error.
In conclusion, >100 genes, including several critical pluripotent genes that should have been repressed during RA-induced differentiation, were not efficiently repressed due to impaired H3K9me2 establishment when the H3K9me1-binding activity of GLP was lost.
We also found that 329 genes failed to be activated in GLP 3A cells during RA-induced differentiation, unlike in the wild-type cells. Genes with GO terms related to differentiation such as “embryonic morphogenesis” and “pattern specification process” were enriched among these genes, including Sox1, Kdr, foxa1, Bmp, and Wnt family genes, indicating a defect in differentiation (Supplemental Fig. 9E). We reason that these results are in line with the impairment in efficiently shutting down the pluripotent genes during RA-induced differentiation in GLP 3A cells.
Discussion
GLP and G9a have relatively poor activities on nucleosomes in vitro; whether they catalyze H3K9 methylation on nucleosome substrates or histone octamers prior to nucleosome assembly during replication remains an open question (Shinkai and Tachibana 2011). Our results demonstrated that GLP and G9a are capable of exerting strong catalytic activities on nucleosomal substrates when they are flanked by neighboring nucleosomes carrying H3K9 methylation (Fig. 1). This suggests that GLP and G9a can use nucleosomes as their optimal substrates.
The enzymatic activation of GLP and G9a by neighboring nucleosomes methylated at H3K9 leads to several interesting questions: Is this a directional, processive methylation-spreading activity or a dose effect? Does this contribute to the mitotic inheritance of H3K9 methylation? What are the functional roles of such H3K9 methylation-mediated activation of H3K9 methyltransferases?
The activation of G9a and GLP by neighboring methylated nucleosomes is unlikely to be a directional, processive methylation-spreading activity
Our observation that GLP and G9a can sense neighboring nucleosomes premethylated at H3K9 suggests that GLP and G9a can efficiently spread H3K9 methylation. This partially resembles the H3K9 methylation-spreading activity of Clr4 in fission yeast (Al-Sady et al. 2013). However, unlike Clr4, which guides the spreading toward one specific direction (Al-Sady et al. 2013), we believe that the spreading activity of GLP is most probably not a directional, processive event. Otherwise, it would be predicted that we could observe unaffected nucleating centers flanked by impaired H3K9me2 establishment in their surrounding regions in GLP 3A cells during RA-induced ESC differentiation, which we did not observe. Instead, we observed a general reduction of H3K9me2 within the affected regions (Fig. 7C). This is not surprising, because a mechanism that can reinforce a directional and processive spreading activity would require much more sophisticated machinery.
Instead, we favor the idea that the stimulation activities observed were due to the stabilization of GLP and G9a association with nucleosomes bearing H3K9 methylation, which leads to increased local concentration of GLP and G9a and efficient methylation of surrounding nucleosomes. Interestingly, a recent study reported that heterozygote GLP knockout mice displayed developmental delay and cranial abnormalities (Balemans et al. 2014). These phenotypes, including the bent nose phenotype observed in ∼30% of the heterozygote GLP knockout mice (Balemans et al. 2014), are highly similar to the phenotypes of the GLPM/M mice observed in our study, which suggests that both dosage and H3K9me1-mediated stimulation activity of GLP are critical for bone formation and craniofacial development. It remains unclear why bone formation appears to be one of the development processes most sensitive to the impairment of GLP-mediated H3K9 methylation. It is highly conceivable that the loss of H3K9me1 binding and the subsequent stimulation activity of GLP may affect biological processes similar to those affected by the dose reduction of GLP. This can be well explained by our understanding that the H3K9me1-binding activity of GLP helps to stabilize its association with nucleosomes bearing H3K9me1 and increase its local concentration for efficient methylation. Moreover, we note that the heterozygote GLP knockout mice display much milder defects than the GLPM/M mice in this study. The heterozygote GLP knockout mice are viable despite some cranial abnormalities (Balemans et al. 2014), which indicates that the defect in H3K9me1-binding activity of GLP has a greater impact than losing one allele of GLP.
Mitotic inheritance of H3K9 methylation does not require GLP’s and G9a’s methylation-spreading activity
H3K9me2 occupies more than 30% of the genome (Lienert et al. 2011; Voigt et al. 2012) but can be rapidly re-established during mitosis (Xu et al. 2011). Considering that G9a localizes at the replication foci (Esteve et al. 2006) and G9a and GLP are activated by premethylated neighboring nucleosomes, it is very attractive to hypothesize that GLP and G9a may travel with the replication machinery and copy–paste H3K9 methylation from the parental nucleosomes to the newly deposited nucleosomes. Knockout mice for either GLP or G9a are embryonic-lethal (Tachibana et al. 2002, 2005), indicating that proper establishment and maintenance of H3K9 methylation are critical for mouse embryonic development. However, double-mutant mice losing H3K9 methylation-binding activities of both G9a and GLP can still undergo fairly normal embryonic development and be born alive (Fig. 3C). This suggests that the H3K9-binding activity and H3K9 methylation-mediated stimulation activity of G9a and GLP are largely dispensable for mitotic inheritance of H3K9me2 because defects in mitotic inheritance of histone modifications should lead to significant reduction of the modification and more severe phenotypes. Indeed, the changes of H3K9me2 levels in GLP 3A ESCs and mouse tissues were quite modest at the global level (Supplemental Fig. S6D,E). Therefore, it is much more likely that the fast re-establishment of H3K9 methylation during replication was mediated by the association between G9a, GLP, the replication machinery, and DNMT1 via a reinforcement mechanism (Esteve et al. 2006; Shinkai and Tachibana 2011; Zhu and Reinberg 2011) rather than copying of the existing marks from the parental histones. This conclusion is consistent with our previous observations that histone lysine methylations are generally not inherited at high resolution (Xu et al. 2010, 2011; Chen et al. 2011; Huang et al. 2013).
Efficient establishment of H3K9me2 during ESC differentiation requires GLP’s H3K9me1-mediated stimulation activity
Besides mitotic inheritance of H3K9 methylation, another scenario involving rapid establishment of H3K9me2 occurs during cell differentiation. As a facultative heterochromatin marker, H3K9me2 undergoes global changes during the differentiation processes of ESCs, mesenchymal stem cells, and hematopoietic stem cells (Tan et al. 2009; Wen et al. 2009; Lienert et al. 2011; Chen et al. 2012; Wang et al. 2013). Our results demonstrated that the H3K9me1-mediated stimulation of GLP played an important role in rapid establishment of H3K9me2 during RA-induced differentiation of ESCs. Upon loss of H3K9me1 binding and subsequent enzymatic stimulation of GLP, many genes, including several critical pluripotent genes, displayed defective H3K9me2 establishment and delayed repression during differentiation of ESCs. In vivo, the GLPM/M embryos went through the entire embryonic development stage, which may argue against a role of an H3K9me2 establishment defect observed in the in vitro ESC differentiation system. There are several potential explanations. First, the GLPM/M embryos may indeed suffer from a delayed differentiation process even at the early development stage because the GLPM/M embryos were lighter than their wild-type littermates starting from E10.5, which may be a consequence of delayed differentiation processes at earlier stages. Second, although bone ossification appears to be the most vulnerable process, milder but similar impacts could well be perceived in other cell types. It would be interesting to interrogate other cell differentiation systems, especially the osteoblasts differentiation process, which is highly relevant to the observed bone ossification defects in GLPM/M mice.
Distinct roles of GLP and G9a
GLP and G9a share high sequence similarity and similar biochemical activities and are both required for H3K9me2 levels in vivo (Tachibana et al. 2005; Collins et al. 2008). However, GLP 3A mutant mice and G9a 3A mutant mice displayed dramatic phenotype differences. The H3K9 methylation-binding activity of GLP appeared to be much more important than that of G9a. It remains unclear whether this is due to their differential expression profile or their binding affinity difference toward H3K9me1 and H3K9me2. The ankyrin repeat domain of G9a preferentially associates with H3K9me2, whereas the ankyrin repeat domain of GLP preferentially associates with H3K9me1 (Collins et al. 2008). Engineering a GLP knock-in mouse line bearing an H3K9me2-binding ankyrin repeat domain may help to resolve this question in future studies. Another potential explanation is the distinct expression profile between GLP and G9a; although these two proteins can form heterodimers (Tachibana et al. 2005), they often display different tissue-specific expression profiles (Ohno et al. 2013), suggesting that they may have functions independent of each other.
Role of GLP’s H3K9me1-mediated stimulation activity in bone ossification
Bone ossification is mediated by osteocytes, which are originated from sequential differentiation of mesenchymal stem cells and osteoblasts. Chromatin modifications, including histone deacetylation and H3K9 methylation, regulate bone formation (Tan et al. 2009; Bradley et al. 2011). During mouse embryonic development, histone H3K9 methyltransferases G9a and GLP emerge on E16.5 in prehypertrophic and hypertrophic chondrocytes in the growth plate as well as mesenchyme cells in the tooth germ, resulting in increased H3K9 methylation, which may be important in regulating chondrocyte differentiation and tooth development (Ideno et al. 2013; Kamiunten et al. 2014). It is highly probable that during the differentiation process of mesenchymal stem cells and/or osteoblasts in GLPM/M mice, defective establishment of H3K9me2 at genes that should be repressed led to inefficient gene silencing, incomplete differentiation of osteocytes, and defective ossification.
Finally, we note the potential relevance between the nonhistone substrates of GLP or G9a and the phenotypes that we observed in this study. In addition to histone H3, G9a has been reported to methylate other substrates, including histone H1.4 and a number of nonhistone proteins (Sampath et al. 2007; Rathert et al. 2008; Collins and Cheng 2010). It is easily conceivable that GLP may also have additional substrates. To the best of our knowledge, no other methylated proteins have been reported to be specifically recognized by the ankyrin repeats of GLP or G9a. Considering H3K9 methylation remains the only known methyl mark that GLP recognizes via its ankyrin repeats, it is most likely that the cellular and developmental defects of GLP 3A mutants observed in this study are the consequences of impaired H3K9 methylation recognition and subsequent stimulation. However, we certainly cannot completely rule out the possibility that a fraction of the defects observed in this study may be attributed to the impaired association between GLP and an unknown methylated protein.
Materials and methods
Preparation of recombinant enzymes and substrates
For histone methyltransferase activity assays, His-tagged recombinant G9a (amino acids 601–1210, NP_006700) and recombinant GLP (amino acids 635–1296, NP_001012536) covering both the SET domain and ankyrin repeats were expressed and purified from E. coli. Briefly, His-tagged G9a or His-tagged GLP were expressed in BL21 (DE3) cells upon IPTG induction. The cells were pelleted and resuspended in lysis buffer (20 mM Tris-HCl at pH 8.0, 150 mM NaCl, 1 mM EDTA, 1 mM PMSF) and then sonicated to release cellular proteins. The cell lysates were incubated with Ni-NTA beads (Qiagen). The beads were washed with 50 column volumes of lysis buffer containing 15 mM imidazole, and recombinant proteins were then eluted with lysis buffer containing 250 mM imidazole. The purified proteins were dialyzed against lysis buffer, snap-frozen, and stored at −80°C.
All histones were purified under denatured conditions. Histone H3K9C/C110A with a C-terminal Flag-His dual tag was purified with Ni-NTA beads and then used for MLA reactions.
Recombinant oligonucleosomes were assembled via sequential salt dialysis using pG5E4 plasmid DNA and histone octamers (Yuan et al. 2012). All oligonucleosomes were purified by gel filtration with a 3-mL CL-4B gel filtration column to remove free histone octamers.
ITC
Recombinant G9a (amino acids 730–965, NP_665829) and recombinant GLP (amino acids 763–997, NP_001012536) covering the ankyrin repeats were expressed and purified from E. coli. The proteins were dialyzed against ITC buffer (10 mM Tris-HCl at pH8.0, 50 mM NaCl). The following peptides were synthesized with Scilight-Peptide, Inc. : H3K9me0, ARTKQ TARKS TGGKA P; H3K9me1, ARTKQ TARK(me1)S TGGKA P; H3K9me2, ARTKQ TARK(me2)S TGGKA P; and H3K9me3, ARTKQ TARK(me3)S TGGKA P.
ITC experiments were performed with ITC200 (MicroCal). Briefly, titration cells and syringes were washed with distilled water and ITC buffer. A total volume of 220 μL of protein (80–120 μM) was injected into the cell, 40 μL of peptides (∼1.6 mM) was loaded into the syringe, and the system was equilibrated at 25°C. The titration was performed with the following settings: 2 μL of peptide and 4-sec duration for each injection, 20 injections in total, 180-sec interval between each injection, and a stirring speed of 1000 rpm. The acquired data were processed with Origin software as instructed.
MLA reactions
The MLA reactions were performed as described previously (Simon et al. 2007). Briefly, 5 mg of histones was dissolved in 1 M HEPES (pH 8.0), 4 M urea, and 10 mM methionine and then reduced with DTT for 1 h. Next, the chemicals (N-methylethylamines with different methyl states) were added into the solutions for the alkylating reactions. Finally, β-mercaptoethanol was added to 5% to quench the reactions. All MLA reaction products were verified by MALDI-TOF mass spectrometry analysis for quality assurance.
Histone methyltransferase assays
The reactions were performed as previously described (Yuan et al. 2012). A 40-μL reaction mixture containing S-[methl-3H] adenosyl-methionine (PerkinElmer Life Sciences), recombinant oligonucleosomes, and enzymes in HKMT assay buffer (50 mM Tris at pH 8.5, 20 mM KCl, 10 mM MgCl2, 10 mM DTT, 250 mM sucrose) was incubated for 1 h at 25°C. The reaction products were separated by 13% SDS-PAGE, transferred to PVDF membranes, and then subjected to autoradiography. For quantification, the membranes were stained by Coomassie blue G250 followed by liquid scintillation counting for each stained histone bands.
Generation of knock-in mouse model
The targeting vectors were constructed according to previously published methods (Liu et al. 2003). Vectors were verified by sequencing and electrotransfected into R1 ESCs. After selection with G418 and ganciclovir, surviving clones were picked and then screened by genomic PCR. To confirm that the targeting events resulted in a functional and correctly mutated allele, a large fragment that covered the targeting region was amplified from cDNA and sequenced. The correct ESC clones were used to produce chimeras. F1 heterozygous mice were obtained by crossing chimera with C57BL/6 female mice.
ESC derivation and cell culture
ESCs were derived from littermate blastocysts obtained by intercrossing GLP+/M. The obtained clones were genotyped and karyotyped. Two pairs of ESC lines were obtained. ESCs were maintained in the presence of feeders in standard ES medium. Prior to ChIP-seq and RNA-seq experiments, ESCs were cultured in feeder-free conditions in ES medium supplemented with 2i for two passages to eliminate feeder cells. For RA induction, 1 μM all-trans RA was added into the medium, and LIF was withdrawn simultaneously. The RA induction experiments were performed in triplicate. For EB differentiation, 20-μL liquid drops containing 1000 ESCs were cultured for 2 d in ES medium without LIF. Next, the cell aggregates were transferred to ultralow attachment plates and cultured for various days in the presence of 1 μM RA.
ChIP-seq and ChIP-qPCR
ChIP was performed according to the Diagenode iDeal ChIP-seq kit with minor modifications. Briefly, 2 million cells were harvested in PBS and cross-linked with 1% formaldehyde for 2 min. After sonication, chromatin from 1 million cells was subjected to ChIP using the iDeal ChIP-seq kit (Diagenode). For ChIP-qPCR assays, the H3K9me2 enrichment was normalized to Olfr18 (a constitutively inactive gene) promoter (Magklara et al. 2011). For ChIP-seq, libraries were constructed with KAPA library preparation kits and then single-end-sequenced with HiSeq 2000. Three biological replicate experiments were performed using independently cultured cells. The antibodies used are listed in Supplemental Table 2.
RT-qPCR and RNA-seq
RNA samples were extracted using Trizol reagent, and genomic DNA contaminations were eliminated with Turbo DNase (Life Technologies). cDNA samples were synthesized using the Takara PrimeScript first strand cDNA synthesis kit. The qPCR results were normalized against a housekeeping gene, Gapdh. For high-throughput sequencing, total RNA were extracted, and the subsequent library preparation steps were carried out according to standard Illumina procedures at our in-house sequencing center and subjected to single-end sequencing with HiSeq 2000. Two biological replicates were performed.
Bisulfite sequencing
Bisulfite conversion was performed with the EpiTect bisulfite kit (Qiagen). The Oct4 promoter region was amplified with nested primers, and the PCR fragments were purified and ligated to pMD18-T vectors. At least eight clones per sample were sequenced.
Bioinformatics analysis
Sequencing depths were between 24 million and 61 million reads for the four RNA-seq experiments and between 43 million and 180 million reads for ChIP-seq experiments. The sequencing depth and mapping percentage are listed in Supplemental Table 3.
H3K9me2 ChIP-seq reads were mapped to a mouse genome (mm9), and only uniquely mapped and nonredundant reads were kept for further analysis. The mapped reads were extended to the average fragment size and binned into 25-base-pair (bp) windows for visualization in the Integrative Genomics Viewer (IGV) browser. For the analysis of genomic regions, the mouse genome was divided into 10-kb windows, and the fold enrichment of H3K9me2 for each window was calculated as the ratio of normalized read numbers between ChIP and input samples. Any window that had <100 reads in input samples was filtered because of insufficient information. For each window, the fold enrichment of H3K9me2 was calculated as the ratio of normalized read density between ChIP and input samples, and the final H3K9me2 enrichment value was an average of values from three replicate experiments. Ten-kilobase windows with at least one case of more than a twofold change between any pair of samples were selected for average linkage clustering performed with Cluster 3.0 software.
RNA-seq data were mapped to the mouse genome (mm9) with TopHat (version 1.4.1), and the transcription levels were quantified with Cufflinks (version 2.0.2) software with two replicates. The ChIP-seq and RNA-seq data were deposited in the Gene Expression Omnibus database of NCBI under accession number GSE54412.
Mouse phenotype analysis
All of our study procedures complied with the guideline of the National Institute of Biological Sciences, Beijing, for the care and use of laboratory animals.
MicroCT scanning was performed with an Aloka Latheta LCT200. Skeleton preparation and Alizarin red staining were carried out according to a previous study (Dobreva et al. 2006). Briefly, sacrificed animals were skinned and fixed in EtOH and then stained in 0.1% Alizarin red solution for at least 3 d. Next, the samples were transferred into 1% KOH to remove tissues. The calvaria of animals were carefully dissected and photographed.
Mouse suckling assays were adapted from previous studies (Hongo et al. 2000). Briefly, shortly after giving birth to newborns, female mice were given 0.05 mg/g pentobarbital sodium intraperitoneally for anesthesia and laid on a heating block (37°C). GLP+/+ and GLPM/M pups were put on their mother’s abdomen at 0.5 cm distance from nipples. We divided the suckling process into two steps: (1) searching and licking nipples (known as the rooting reflex) and (2) stably attaching to the nipples with rhythmic suckling. Successfully accomplishing each step within 1 min was scored as 1, otherwise, it was scored as 0. Each newborn mouse was given two to three trials. At least seven newborns of each genotype were tested. The final “search and lick score” and “attachment score” were calculated as summed scores divided by trials. Statistical significance was tested using Fisher’s exact test.
Supplementary Material
Acknowledgments
We thank Y. Shinkai at RIKEN for cDNA and discussion. This work was supported by China National Science Foundation grant 31425013, Chinese Ministry of Science and Technology grants 2015CB856200 and 2011CB965300, Strategic Priority Research Program of the Chinese Academy of Sciences grant XDB08010103, and the Howard Hughes Medical Institute International Early Career Scientist Program to B.Z. This work was also supported by China National Science Foundation grant 81301803 to H.W., and grant 31401114 to Z.Z.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.254425.114.
References
- Al-Sady B, Madhani Hiten D, Narlikar Geeta J. 2013. Division of labor between the chromodomains of HP1 and Suv39 methylase enables coordination of heterochromatin spread. Mol Cell 51: 80–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balemans MCM, Ansar M, Oudakker AR, van Caam APM, Bakker B, Vitters EL, van der Kraan PM, de Bruijn DRH, Janssen SM, Kuipers AJ, et al. 2014. Reduced euchromatin histone methyltransferase 1 causes developmental delay, hypotonia, and cranial abnormalities associated with increased bone gene expression in Kleefstra syndrome mice. Dev Biol 386: 395–407. [DOI] [PubMed] [Google Scholar]
- Bradley EW, McGee-Lawrence ME, Westendorf JJ. 2011. Hdac-mediated control of endochondral and intramembranous ossification. Crit Rev Eukaryot Gene Expr 21: 101–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brent MM, Marmorstein R. 2008. Ankyrin for methylated lysines. Nat Struct Mol Biol 15: 221–222. [DOI] [PubMed] [Google Scholar]
- Chaturvedi CP, Hosey AM, Palii C, Perez-Iratxeta C, Nakatani Y, Ranish JA, Dilworth FJ, Brand M. 2009. Dual role for the methyltransferase G9a in the maintenance of β-globin gene transcription in adult erythroid cells. Proc Natl Acad Sci 106: 18303–18308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Xiong J, Xu M, Chen S, Zhu B. 2011. Symmetrical modification within a nucleosome is not required globally for histone lysine methylation. EMBO Rep 12: 244–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Skutt-Kakaria K, Davison J, Ou Y-L, Choi E, Malik P, Loeb K, Wood B, Georges G, Torok-Storb B, et al. 2012. G9a/GLP-dependent histone H3K9me2 patterning during human hematopoietic stem cell lineage commitment. Genes Dev 26: 2499–2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow J, Heard E. 2009. X inactivation and the complexities of silencing a sex chromosome. Curr Opin Cell Biol 21: 359–366. [DOI] [PubMed] [Google Scholar]
- Collins R, Cheng X. 2010. A case study in cross-talk: the histone lysine methyltransferases G9a and GLP. Nucleic Acids Res 38: 3503–3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins RE, Northrop JP, Horton JR, Lee DY, Zhang X, Stallcup MR, Cheng X. 2008. The ankyrin repeats of G9a and GLP histone methyltransferases are mono- and dimethyllysine binding modules. Nat Struct Mol Biol 15: 245–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desbaillets I, Ziegler U, Groscurth P, Gassmann M. 2000. Embryoid bodies: an in vitro model of mouse embryogenesis. Exp Physiol 85: 645–651. [PubMed] [Google Scholar]
- Dobreva G, Chahrour M, Dautzenberg M, Chirivella L, Kanzler B, Fariñas I, Karsenty G, Grosschedl R. 2006. SATB2 is a multifunctional determinant of craniofacial patterning and osteoblast differentiation. Cell 125: 971–986. [DOI] [PubMed] [Google Scholar]
- Epsztejn-Litman S, Feldman N, Abu-Remaileh M, Shufaro Y, Gerson A, Ueda J, Deplus R, Fuks F, Shinkai Y, Cedar H, et al. 2008. De novo DNA methylation promoted by G9a prevents reprogramming of embryonically silenced genes. Nat Struct Mol Biol 15: 1176–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteve PO, Chin HG, Smallwood A, Feehery GR, Gangisetty O, Karpf AR, Carey MF, Pradhan S. 2006. Direct interaction between DNMT1 and G9a coordinates DNA and histone methylation during replication. Genes Dev 20: 3089–3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman N, Gerson A, Fang J, Li E, Zhang Y, Shinkai Y, Cedar H, Bergman Y. 2006. G9a-mediated irreversible epigenetic inactivation of Oct-3/4 during early embryogenesis. Nat Cell Biol 8: 188–194. [DOI] [PubMed] [Google Scholar]
- Hongo T, Hakuba A, Shiota K, Naruse I. 2000. Suckling dysfunction caused by defects in the olfactory system in genetic arhinencephaly mice. Neonatology 78: 293–299. [DOI] [PubMed] [Google Scholar]
- Huang C, Xu M, Zhu B. 2013. Epigenetic inheritance mediated by histone lysine methylation: maintaining transcriptional states without the precise restoration of marks? Philos Trans R Soc Lond B Biol Sci 368: 20110332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ideno H, Shimada A, Imaizumi K, Kimura H, Abe M, Nakashima K, Nifuji A. 2013. Predominant expression of H3K9 methyltransferases in prehypertrophic and hypertrophic chondrocytes during mouse growth plate cartilage development. Gene Expr Patterns 13: 84–90. [DOI] [PubMed] [Google Scholar]
- Jia G, Wang W, Li H, Mao Z, Cai G, Sun J, Wu H, Xu M, Yang P, Yuan W, et al. 2009. A systematic evaluation of the compatibility of histones containing methyl-lysine analogues with biochemical reactions. Cell Res 19: 1217–1220. [DOI] [PubMed] [Google Scholar]
- Kamiunten T, Ideno H, Shimada A, Nakamura Y, Kimura H, Nakashima K, Nifuji A. 2014. Coordinated expression of H3K9 histone methyltransferases during tooth development in mice. Histochem Cell Biol doi: 10.1107/s00418-014-1284-0. [DOI] [PubMed] [Google Scholar]
- Lee DY, Northrop JP, Kuo MH, Stallcup MR. 2006. Histone H3 lysine 9 methyltransferase G9a is a transcriptional coactivator for nuclear receptors. J Biol Chem 281: 8476–8485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lienert F, Mohn F, Tiwari VK, Baubec T, Roloff TC, Gaidatzis D, Stadler MB, Schübeler D. 2011. Genomic prevalence of heterochromatic H3K9me2 and transcription do not discriminate pluripotent from terminally differentiated cells. PLoS Genet 7: e1002090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Jenkins N, Copeland N. 2003. A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res 13: 476–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magklara A, Yen A, Colquitt Bradley M, Clowney EJ, Allen W, Markenscoff-Papadimitriou E, Evans Zoe A, Kheradpour P, Mountoufaris G, Carey C, et al. 2011. An epigenetic signature for monoallelic olfactory receptor expression. Cell 145: 555–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maksakova I, Thompson P, Goyal P, Jones S, Singh P, Karimi M, Lorincz M. 2013. Distinct roles of KAP1, HP1 and G9a/GLP in silencing of the two-cell-specific retrotransposon MERVL in mouse ES cells. Epigenet Chromatin 6: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Justin N, Ohno K, Sharpe ML, Son J, Drury Iii WJ, Voigt P, Martin SR, Taylor WR, De Marco V, et al. 2009. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature 461: 762–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mermoud JE, Popova B, Peters AHFM, Jenuwein T, Brockdorff N. 2002. Histone H3 Lysine 9 methylation occurs rapidly at the onset of random X chromosome inactivation. Curr Biol 12: 247–251. [DOI] [PubMed] [Google Scholar]
- Ohno H, Shinoda K, Ohyama K, Sharp LZ, Kajimura S. 2013. EHMT1 controls brown adipose cell fate and thermogenesis through the PRDM16 complex. Nature 504: 163–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathert P, Dhayalan A, Murakami M, Zhang X, Tamas R, Jurkowska R, Komatsu Y, Shinkai Y, Cheng X, Jeltsch A. 2008. Protein lysine methyltransferase G9a acts on non-histone targets. Nat Chem Biol 4: 344–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampath SC, Marazzi I, Yap KL, Krutchinsky AN, Mecklenbrauker I, Viale A, Rudensky E, Zhou MM, Chait BT, Tarakhovsky A. 2007. Methylation of a histone mimic within the histone methyltransferase G9a regulates protein complex assembly. Mol Cell 27: 596–608. [DOI] [PubMed] [Google Scholar]
- Shinkai Y, Tachibana M. 2011. H3K9 methyltransferase G9a and the related molecule GLP. Genes Dev 25: 781–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon MD, Chu F, Racki LR, de la Cruz CC, Burlingame AL, Panning B, Narlikar GJ, Shokat KM. 2007. The site-specific installation of methyl-lysine analogs into recombinant histones. Cell 128: 1003–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana M, Sugimoto K, Fukushima T, Shinkai Y. 2001. Set domain-containing protein, G9a, is a novel lysine-preferring mammalian histone methyltransferase with hyperactivity and specific selectivity to lysines 9 and 27 of histone H3. J Biol Chem 276: 25309–25317. [DOI] [PubMed] [Google Scholar]
- Tachibana M, Sugimoto K, Nozaki M, Ueda J, Ohta T, Ohki M, Fukuda M, Takeda N, Niida H, Kato H, et al. 2002. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev 16: 1779–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana M, Ueda J, Fukuda M, Takeda N, Ohta T, Iwanari H, Sakihama T, Kodama T, Hamakubo T, Shinkai Y. 2005. Histone methyltransferases G9a and GLP form heteromeric complexes and are both crucial for methylation of euchromatin at H3-K9. Genes Dev 19: 815–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J, Lu J, Huang W, Dong Z, Kong C, Li L, Gao L, Guo J, Huang B. 2009. Genome-wide analysis of histone H3 lysine9 modifications in human mesenchymal stem cell osteogenic differentiation. PLoS ONE 4: e6792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trojer P, Reinberg D. 2007. Facultative heterochromatin: is there a distinctive molecular signature? Mol Cell 28: 1–13. [DOI] [PubMed] [Google Scholar]
- Turgeon B, Meloche S. 2009. Interpreting neonatal lethal phenotypes in mouse mutants: insights into gene function and human diseases. Physiol Rev 89: 1–26. [DOI] [PubMed] [Google Scholar]
- Vakoc CR, Mandat SA, Olenchock BA, Blobel GA. 2005. Histone H3 Lysine 9 methylation and HP1γ are associated with transcription elongation through mammalian chromatin. Mol Cell 19: 381–391. [DOI] [PubMed] [Google Scholar]
- Vakoc CR, Sachdeva MM, Wang H, Blobel GA. 2006. Profile of histone lysine methylation across transcribed mammalian chromatin. Mol Cell Biol 26: 9185–9195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt P, LeRoy G, Drury Iii William J, Zee Barry M, Son J, Beck David B, Young Nicolas L, Garcia Benjamin A, Reinberg D. 2012. Asymmetrically modified nucleosomes. Cell 151: 181–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Xu S, Lee JE, Baldridge A, Grullon S, Peng W, Ge K. 2013. Histone H3K9 methyltransferase G9a represses PPARγ expression and adipogenesis. Embo J 32: 45–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen B, Wu H, Shinkai Y, Irizarry RA, Feinberg AP. 2009. Large histone H3 lysine 9 dimethylated chromatin blocks distinguish differentiated from embryonic stem cells. Nat Genet 41: 246–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Chen X, Xiong J, Li Y, Li H, Ding X, Liu S, Chen S, Gao S, Zhu B. 2011. Histone methyltransferase G9a contributes to H3K27 methylation in vivo. Cell Res 21: 365–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Long C, Chen X, Huang C, Chen S, Zhu B. 2010. Partitioning of histone H3–H4 tetramers during DNA replication-dependent chromatin assembly. Science 328: 94–98. [DOI] [PubMed] [Google Scholar]
- Xu M, Wang W, Chen S, Zhu B. 2011. A model for mitotic inheritance of histone lysine methylation. EMBO Rep 13: 60–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X, Feng W, Imhof A, Grummt I, Zhou Y. 2007. Activation of RNA polymerase I transcription by cockayne syndrome group B protein and histone methyltransferase G9a. Mol Cell 27: 585–595. [DOI] [PubMed] [Google Scholar]
- Yuan W, Wu T, Fu H, Dai C, Wu H, Liu N, Li X, Xu M, Zhang Z, Niu T, et al. 2012. Dense chromatin activates polycomb repressive complex 2 to regulate H3 Lysine 27 methylation. Science 337: 971–975. [DOI] [PubMed] [Google Scholar]
- Zhu B, Reinberg D. 2011. Epigenetic inheritance: uncontested? Cell Res 21: 435–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.