The transcription factor IRF8 controls both developmental and inflammatory stimulus-inducible genes in macrophages. Mancino et al. found that IRF8 is recruited to distinct sets of DNA consensus sequences before and after lipopolysaccharide (LPS) stimulation. While constitutively expressed IRF8-dependent genes contained only sites mediating basal IRF8/PU.1 recruitment, inducible IRF8-dependent genes contained variable combinations of constitutive and inducible sites.
Keywords: IRF8, interferon, chromatin, epigenome, inflammation, macrophages, transcription
Abstract
The transcription factor (TF) interferon regulatory factor 8 (IRF8) controls both developmental and inflammatory stimulus-inducible genes in macrophages, but the mechanisms underlying these two different functions are largely unknown. One possibility is that these different roles are linked to the ability of IRF8 to bind alternative DNA sequences. We found that IRF8 is recruited to distinct sets of DNA consensus sequences before and after lipopolysaccharide (LPS) stimulation. In resting cells, IRF8 was mainly bound to composite sites together with the master regulator of myeloid development PU.1. Basal IRF8–PU.1 binding maintained the expression of a broad panel of genes essential for macrophage functions (such as microbial recognition and response to purines) and contributed to basal expression of many LPS-inducible genes. After LPS stimulation, increased expression of IRF8, other IRFs, and AP-1 family TFs enabled IRF8 binding to thousands of additional regions containing low-affinity multimerized IRF sites and composite IRF–AP-1 sites, which were not premarked by PU.1 and did not contribute to the basal IRF8 cistrome. While constitutively expressed IRF8-dependent genes contained only sites mediating basal IRF8/PU.1 recruitment, inducible IRF8-dependent genes contained variable combinations of constitutive and inducible sites. Overall, these data show at the genome scale how the same TF can be linked to constitutive and inducible gene regulation via distinct combinations of alternative DNA-binding sites.
Developmental specification of macrophages requires the activity of a well-defined panel of transcription factors (TFs) (Rosenbauer and Tenen 2007) that act sequentially at specific stages, from the initial commitment of hematopoietic stem cells (HSCs) to the myeloid fate and then to the terminally differentiated progeny (Lichtinger et al. 2012). The ETS family TF PU.1 has a central role at all stages of myeloid development, starting from the transition from HSCs to common myeloid progenitors (CMPs) (Rosenbauer and Tenen 2007). PU.1 is broadly expressed across both the myeloid and lymphoid lineages. However, mainly due to post-translational control mechanisms, it reaches its highest intracellular concentration in macrophages (Kueh et al. 2013). In these cells, it directly promotes and maintains the accessibility of the genomic cis-regulatory information specifically available for constitutive and stimulus-inducible transcriptional regulation in this cell type (Ghisletti et al. 2010; Heinz et al. 2010; Barozzi et al. 2014). The generation of the macrophage-specific repertoire of transcriptional enhancers also requires the ability of PU.1 to interact with partner TFs that critically contribute to its recruitment to genomic binding sites (Heinz et al. 2010; Barozzi et al. 2014; Gosselin and Glass 2014) and whose expression or activity is also dictated by cues coming from the tissue microenvironment (Gosselin et al. 2014; Lavin et al. 2014).
Among the PU.1 partners, a unique role is exerted by interferon regulatory factor 8 (IRF8), a member of the IRF family (Tamura et al. 2008) whose expression is restricted to the hematopoietic system. Within the myeloid compartment, IRF8 is expressed from the GMP (granulocyte–macrophage progenitor) stage onward and is exquisitely selective for the monocyte–macrophage branch (Tamura et al. 2000). In keeping with this expression profile, Irf8 gene deletion in mice results in a myeloproliferative syndrome with a massive accumulation of granulocytes and reduced monocyte–macrophages, thus suggesting that IRF8 is required for GMP differentiation into monocytes and that, in its absence, myeloid progenitors generate granulocytes at a higher rate. Moreover, IRF8 is also required for the development of CD8α+ and plasmacytoid dendritic cells (pDCs) (Schiavoni et al. 2002; Tsujimura et al. 2003). Consistent with these phenotypes, IRF8 loss-of-function mutations in humans (Hambleton et al. 2011) are also associated with major defects of the monocyte and dendritic cell compartment.
In contrast with a more common dichotomy between TFs involved in development and TFs involved in environmental responses, IRF8 is required for not only macrophage differentiation but also the stimulus-inducible expression of some critical immune response genes such as Il12p40 (which controls T-lymphocyte polarization) and Ifnb1. The autocrine and paracrine activities of lipopolysaccharide (LPS)-induced IFNβ greatly contribute to the transcriptional response to LPS, since a large fraction of the genes activated by LPS (including many antimicrobial genes) are in fact secondary genes induced by IFNβ (Thomas et al. 2006). Such a functional duality also explains the immunodeficiency syndrome of Irf8 mutant mice and humans and specifically their high susceptibility to intracellular macrophage pathogens (Turcotte et al. 2005; Marquis et al. 2009; Hambleton et al. 2011). How these different IRF8 activities in development and inflammatory responses are hardwired into the genome remains to be understood.
Because of structural divergences in their IRF domain (Escalante et al. 2002), IRF8 and its closest paralog, IRF4, are virtually unable to bind DNA with high affinity unless associated with DNA-binding partners such as PU.1, other IRFs, and JUN/AP-1 family proteins (Escalante et al. 2002; Tamura et al. 2005; Glasmacher et al. 2012; Li et al. 2012; Tussiwand et al. 2012). A C-terminal region of IRF8 named IAD (IRF association domain) is specifically involved in making protein–protein contacts with binding partners. Consistently, a point mutation at a single amino acid in the IRF8 IAD (Arg294>Cys) in the Bxh2 mouse causes a phenotype that is almost indistinguishable from the total loss of IRF8 (Turcotte et al. 2005), the only exception being the conservation of pDCs in Bxh2 mice (Tailor et al. 2008). A direct implication of IRF4/8’s dependence on partner TFs for high-affinity DNA binding is their ability to recognize DNA sequences distinct from the canonical IRF-binding site (known as ISRE [interferon-stimulated response element]), including the ETS/IRF composite elements (EICEs) in association with PU.1 (Taniguchi et al. 2001) and the AP-1/IRF composite elements (AICEs) in association with ATF and JUN/AP-1 family proteins (Glasmacher et al. 2012; Li et al. 2012).
While some of the biological roles of IRF8 during myeloid development are well established, the molecular bases of its dual role during lineage specification and acute environmental responses are unclear but are likely linked to its unique DNA-binding properties. To specifically address this question, we investigated the genomic distribution of IRF8 and its role in controlling the chromatin landscape and the gene expression programs of mouse macrophages before and after LPS and type I IFN stimulation. Our data indicate that the genomic distribution of IRF8 in macrophages has two distinct components; namely, a constitutive component dependent on the interaction of PU.1–IRF8 complexes with EICEs and an inducible component that is restricted to multimerized IRF sites and composite IRF/AP-1 sites and that possibly depends on the LPS-induced increased expression of IRF8, IRF1, and AP-1 family TFs. Constitutive IRF8–PU.1 binding maintained the basal expression of a large panel of macrophage genes, including IFNβ-inducible genes. In turn, upon LPS stimulation, IRF8 not only was required for efficient Ifnb1 gene induction but also collaborated with IFNβ-activated STAT1 to promote chromatin and gene expression changes in response to type I IFN. Overall, these data provide the mechanistic basis underlying the functional duality of IRF8 in both macrophage development and acute antimicrobial responses. More generally, they provide detailed and mechanistic insight into how a single TF can be coupled to multiple gene expression programs.
Results
Constitutive and inducible IRF8 recruitment to distinct DNA-binding sites
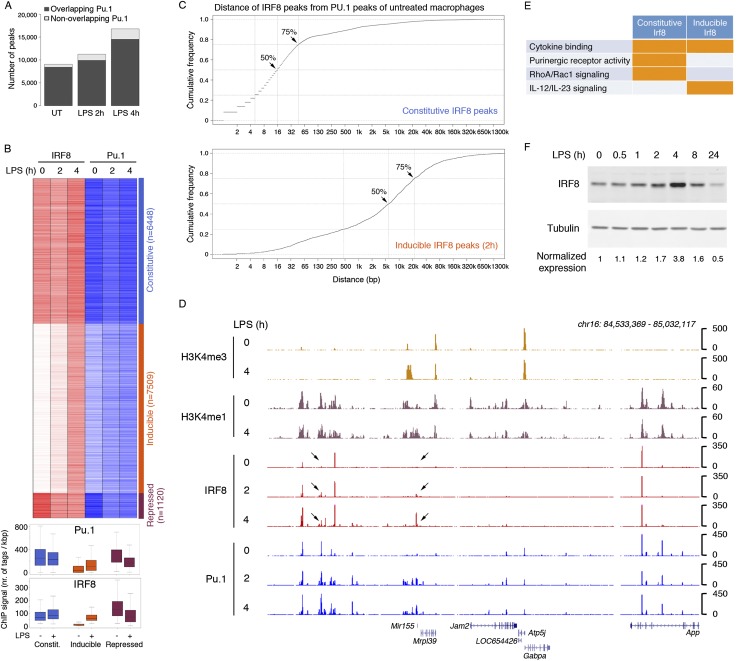
We first analyzed the genomic distribution of IRF8 in untreated and LPS-treated mouse bone marrow-derived macrophages using chromatin immunoprecipitation (ChIP) coupled to high-throughput sequencing (ChIP-seq). IRF8 was constitutively bound to thousands of sites in untreated macrophages (9056 sites using a MACS P-value of 1 × 10−10) (Fig. 1A). Although ChIP-seq data obtained with different antibodies may not be directly comparable, the genomic distribution of IRF8 appeared to be much more restricted than that of PU.1, since, using the same parameters for peak calling, we could detect 60,931 PU.1 peaks. This difference was not accounted for by a different efficiency of the PU.1 and IRF8 ChIPs, since the signal to noise ratio (measured as the ratio between the number of sequencing reads contained in peaks [signal] and those outside of peaks [noise]) was similar. LPS stimulation increased the number of IRF8 peaks and caused a doubling in their number after a 4-h treatment (Fig. 1A). These data indicate that two distinct components account for the IRF8 genomic distribution (Fig. 1B); namely, a constitutive component (n = 6448) and an inducible component (n = 7509). An additional minor component was represented by genomic regions from which IRF8 was released after LPS stimulation (repressed peaks, n = 1120) (Supplemental Table 1). Both constitutive and inducible IRF8 peaks in general overlapped with PU.1 peaks (Fig. 1A). However, most inducible IRF8 peaks occurred at genomic regions that, in unstimulated macrophages, were not prebound by PU.1, as indicated by both the heat map in Figure 1B and an analysis of the distance between the summit of PU.1 and IRF8 ChIP-seq peaks (Fig. 1C): Using the PU.1 peaks detected in unstimulated macrophages as viewpoints, 75% of constitutive IRF8 peaks (Fig. 1C, top panel) but only <20% of the inducible ones (Fig. 1C, bottom panel) were detected within a distance of 32 nucleotides (nt). Overall, these data suggest that cobinding of IRF8 and PU.1 in unstimulated macrophages is a most common event. However, LPS-induced new binding events occurred mainly in regions that were not constitutively bound by PU.1. These data suggest a fundamentally different mechanism of IRF8 recruitment to constitutively and inducibly bound sites. A representative genomic region showing both constitutive and inducible IRF8 peaks and their relationship with PU.1 is displayed in Figure 1D.
Figure 1.
Constitutive and inducible IRF8 binding in macrophages. (A) Number of IRF8 peaks identified by ChIP-seq in untreated (UT) and LPS-treated macrophages (MACS P < 1 × 10−10). (B) Heat map of constitutive, LPS-inducible, and LPS-repressed IRF8 peaks. Pu.1 ChIP-seq data at the same time points are also shown. The box plots at the bottom show the intensity of ChIP-seq signals in the different groups. (C) Cumulative distribution of the distances between constitutive IRF8 peaks in untreated macrophages and LPS-inducible (2-h) IRF8 peaks relative to PU.1 peaks in untreated macrophages. (D) Representative ChIP-seq snapshot showing constitutive and LPS-inducible (arrows) IRF8 peaks. (E) Selected ontology categories enriched in the two sets of IRF8-bound genomic regions according to GREAT. (F) IRF8 protein levels in LPS-treated macrophages. Tubulin was used as loading control. Bands were quantified using the Li-Cor system.
The two distinct types of IRF8 peaks carried functional specificities, as shown by the overrepresentation of distinct ontology terms associated with the neighboring genes. GREAT (genomic regions enrichment of annotations tool) computes the enrichment of ontology terms in a set of genomic regions extracted from sequencing data (McLean et al. 2010). When considering the genomic regions associated with the constitutive IRF8 peaks, GREAT retrieved, among the others, ontology terms related to purinergic receptor signaling, while the inducible IRF8 peaks were specifically associated with IL-12 and IL-23 signaling (Fig. 1E; Supplemental Table 2). These results are in keeping with the role of IRF8 in controlling the expression of the corresponding genes (see below).
Consistent with previous reports (Xu et al. 2012), the IRF8 protein was transiently induced with a peak (3.8× increase) at 4 h after LPS stimulation (Fig. 1F). Therefore, the overall increase in genomic IRF8 occupancy after LPS treatment may be driven by both its increased expression (see below) and LPS-mediated induction of other IRF8-binding partners (including AP-1 and ATF family TFs) (Glasmacher et al. 2012; Li et al. 2012) as well as other IRF proteins (such as IRF1).
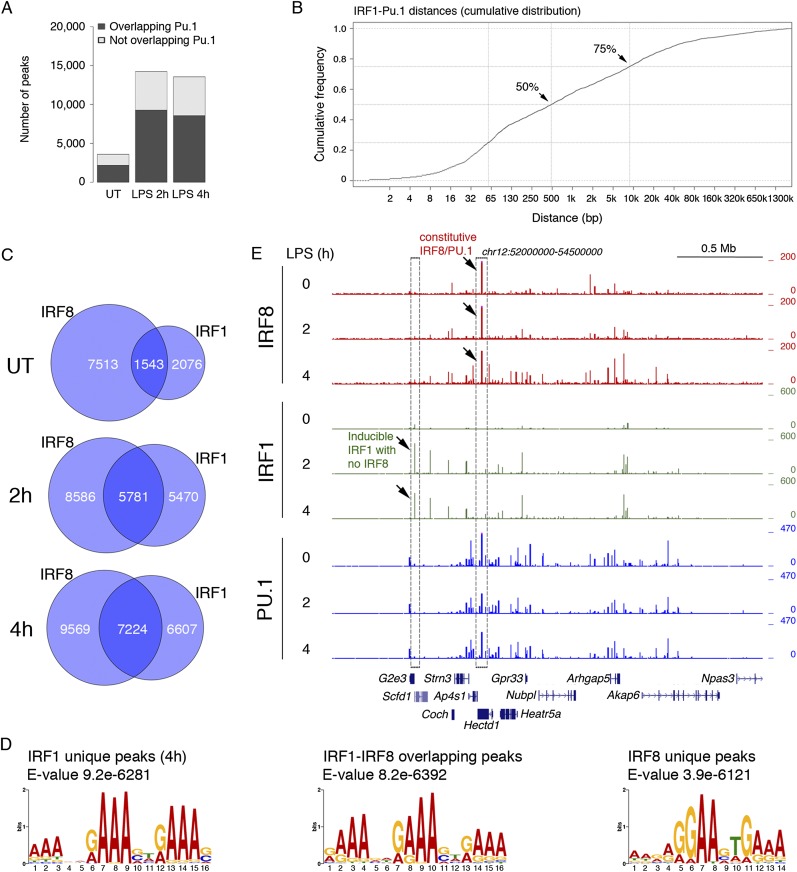
To contribute to our understanding of this issue and also determine how the different DNA-binding domains of IRF8 and IRF1 impact their genomic distribution, we analyzed the genomic occupancy of IRF1 in unstimulated macrophages and after LPS stimulation, which greatly increases IRF1 expression. IRF1 binding was limited to a few thousand sites in unstimulated macrophages but increased about three times in response to LPS stimulation (Fig. 2A; Supplemental Table 3), which is consistent with its increased expression. Compared with IRF8, the overlap with PU.1 was more limited (Fig. 2A), and the distance from PU.1 peaks was higher (Fig. 2B). Overall, the genomic distribution of IRF1 was largely distinct from that of IRF8 in unstimulated macrophages (with only 17% of IRF8 peaks overlapping IRF1 peaks) (Fig. 2C), but the overlap strongly increased in response to stimulation (40.2% at 2 h and 43% at 4 h). These data suggest that the DNA-binding sites (and the cooperating TFs) that control basal IRF8 and IRF1 recruitment were largely distinct. However, in response to stimulation, the two factors bound to a large number of shared regions, suggesting the usage of similar binding sites. A de novo motif discovery analysis showed that IRF1 unique sites and those regions in which IRF1 and IRF8 overlapped at 4 h after LPS treatment were associated with a very similar multimerized IRF-binding site, while the IRF8 unique peaks were associated with a composite ETS/IRF-binding site containing at the 5′ end the 5′-GGAA-3′ moiety characteristic of the core binding site of all ETS family TFs and at the 3′ end the 5′-GAAA-3′ that represents the core IRF-binding site (Fig. 2D). A representative snapshot with some differential peaks highlighted is shown in Figure 2E. Therefore, a specific type of ETS/IRF site defines a subgroup of unique IRF8-binding events that are not shared with other IRF family TFs.
Figure 2.
Relationship between basal and LPS-inducible IRF1 and IRF8 binding. (A) Number of IRF1 peaks before and after LPS stimulation (2 h and 4 h) (MACS P < 1 × 10−10). (B) Cumulative distribution of the distances between the summits of IRF1 and PU.1 peaks in untreated (UT) macrophages. (C) Venn diagrams show the overlap between IRF1 and IRF8 peaks in untreated and LPS-treated macrophages. (D) PWMs identified by de novo motif discovery at the indicated groups of IRF1 and IRF8 peaks (4 h after LPS stimulation). (E) A representative snapshot in which some differences between the genomic distributions of IRF1 and IRF8 were highlighted.
A limited number of DNA-binding sites predict basal vs. inducible IRF8 binding
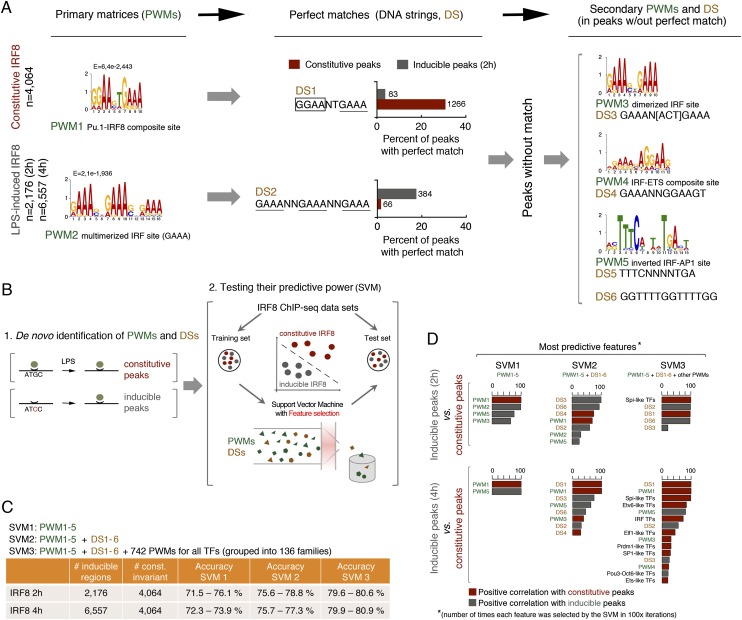
When considering the relationship between IRF8 and PU.1, a clear difference between constitutive and inducible peaks became apparent. First, a higher fraction of inducible than constitutive IRF8 peaks was not associated with PU.1 at all (Fig. 1A, light gray). Second, when considering the inducible IRF8 peaks, the median number of tags of the overlapping PU.1 peaks was much lower than that of PU.1 peaks coinciding with constitutive IRF8 (2.2-fold; P < 2.2 × 10−16 Wilcoxon rank sum test) (Fig. 1B, box plots). Third, inducible IRF8 peaks often occurred at genomic regions that were not prebound by PU.1 in unstimulated macrophages (Fig. 1C, bottom panel). These data suggest that the mode of binding of IRF8 at constitutive and inducible peaks may differ. Consistently, a de novo motif discovery analysis revealed that while constitutive peaks were mainly associated with PU.1/IRF8 composite sites (PWM1) (Fig. 3A, left panel), the inducible ones showed an overrepresentation of a multimerized 5′-GAAA-3′ motif that corresponds to a canonical IRF half-site (PWM2) (Fig. 3A, left panel; Taniguchi et al. 2001). Consistently, overexpression of IRF8 by retroviral transduction of macrophages promoted its recruitment to genomic regions that recruited IRF8 only in response to LPS stimulation and contained multimerized IRF sites (Supplemental Fig. 1A). Using an in vitro pull-down assay with an immobilized biotinylated oligonucleotide containing a multimerized IRF site, we could retrieve IRF8, albeit with a comparatively lower efficiency than observed with a composite PU.1/IRF8 probe (Supplemental Fig. 1B). LPS-inducible IRF8 recruitment via multimerized ISRE-like sites to regions of the genome that are PU.1-negative before stimulation (Fig. 1C) suggests that PU.1 recruitment in these cases may be mediated by protein–protein interactions with IRF8 rather than direct recognition of cognate DNA-binding sites, which would also explain its lower signal intensity at these regions (Fig. 1B).
Figure 3.
A small number of IRF-like DNA-binding sites predict constitutive versus LPS-inducible IRF8 recruitment. (A) Outline of the computational procedure used to retrieve sites bound by IRF8 in a constitutive or LPS-inducible manner. A de novo motif discovery approach (MEME) first retrieved PWM1 and PWM2, which were converted into DSs (DS1 and DS2, respectively) to determine the fraction of peaks in the two groups with a perfect match. Reiteration of the de novo motif discovery on the peaks without match retrieved PWM3–5 and the corresponding DSs. (B) Scheme of the SVM with feature selection used in this study. (C) The SVM was run using a combination of three sets of DNA-binding sites of increasing complexity: only PWM1–5 (SVM1), PWM1–5 and DS1–6 (SVM2), and finally, PWM1–5+DS1–6 and a set of 742 PWMs corresponding to annotated high-quality TF DNA-binding specificities (SVM3). (D) The most predictive DNA-binding sites (namely, those most frequently retrieved in 100 iterations of the three SVMs) and their association with either constitutive or LPS-inducible IRF8 peaks are shown.
To mechanistically understand the impact of different DNA-binding sites on basal versus inducible IRF8 binding, we first measured how many peaks in the two groups contained either of these two DNA-binding consensus sites. To increase the precision of this analysis, we transformed the PWMs into two DNA strings (DS1 and DS2) (Fig. 3A, middle panel) and determined the fraction of peaks with a perfect match. The two DSs showed a very skewed association with the two groups of peaks, but since they were detected in only a fraction of them, they were clearly not sufficient to mechanistically explain differential IRF8 recruitment. Therefore, we identified three additional (“secondary”) PWMs (and the corresponding DSs) in all of those peaks without a match (Fig. 3A, right panel). PWM3/DS3 correspond to a dimeric IRF site, PWM4/DS4 correspond to a composite site in which the position of the ETS and IRF sites are inverted relative to PWM1, and PWM5/DS5 correspond to an IRF–AP-1 site (Glasmacher et al. 2012; Li et al. 2012). A sixth DS (DS6) was generated from a relatively degenerated matrix that has no obvious match to annotated TF DNA-binding sites.
Next, we set out to determine to what extent these PWMs and DSs predict constitutive versus inducible IRF8 binding at a genome scale. To this aim, we used support vector machines (SVMs) (Cortes and Vapnik 1995) coupled with a feature selection procedure (Guyon and Elisseeff 2003) that would allow us to determine which DNA-binding sites have the highest predictive power (Supplemental Material) (Fig. 3B). Given a set of examples (namely, a training set), an SVM learning algorithm builds a model that can then be used to classify new data (namely, a test set) (Fig. 3B). We trained the SVM using 50% of the constitutive and inducible IRF8 sites and then used the trained SVM to predict constitutive versus inducible IRF8 binding to the remaining 50%. When the SVM was fed with only the five PWMs (SVM1), it reached a remarkable prediction accuracy between 71% and 76% (Fig. 3C); the addition of the six DSs (SVM2) increased the SVM accuracy to a minimum of 75% and a maximum of 78.8%. Finally, when a library of PWMs representative of hundreds of TF DNA-binding specificities was added (SVM3), an additional gain in accuracy was obtained, although it is clear that the five PWMs and the corresponding DSs are themselves sufficient for an accurate prediction. The most predictive features (namely, those binding sites that were more frequently retrieved in 100 independent instances of the feature selection) and their association with constitutive or inducible peaks are shown in Figure 3D. Interestingly at 4 h after LPS, only PWM1 (PU.1/IRF) and PWM5 (IRF/AP-1) were sufficient for maximal prediction accuracy. Instead, at 2 h, the multimerized IRF site had a strong impact on prediction accuracy, suggesting that cooperative interactions with different TFs control inducible IRF8 binding during the course of the LPS response. Importantly, the gain in accuracy obtained using the additional 742 PWMs was almost exclusively due to PU.1 (Spi-like)-binding sites, which were relevant to predict constitutive IRF8 binding. Therefore, constitutive IRF8 peaks were associated with composite PU.1/IRF sites and canonical PU.1 sites (since IRF8 recruitment in this case is likely due to protein–protein contacts with PU.1 in the absence of direct DNA recognition), while the inducible ones were linked to multimerized IRF sites and IRF/AP-1 composite sites.
IRF8 binding to chromatin in Bxh2 macrophages
To determine the impact of IRF8 on the macrophage epigenome and gene expression programs, we took advantage of the mutated (IRF8 R294C) Bxh2 cells. Indeed, the macrophage yield of Irf8−/− bone marrow cells is very low (approximately one-fifth of wild-type cells), which implies the occurrence of some selection process during the differentiation of progenitors into macrophages. Conversely, the macrophage generation efficiency of Bxh2 and wild-type bone marrow was indistinguishable, which likely reflects the preservation of a subset of IRF8-binding events in this mutant. The IRF8 protein amount was slightly but reproducibly lower in Bxh2 macrophages and was not increased in response to LPS stimulation (Supplemental Fig. 2A). Consistently, IRF8 constitutively bound its own locus (Supplemental Fig. 2B), mainly at an upstream enhancer whose LPS-induced acetylation was greatly reduced in Bxh2 macrophages. These data indicate that IRF8 controls its own LPS-inducible expression.
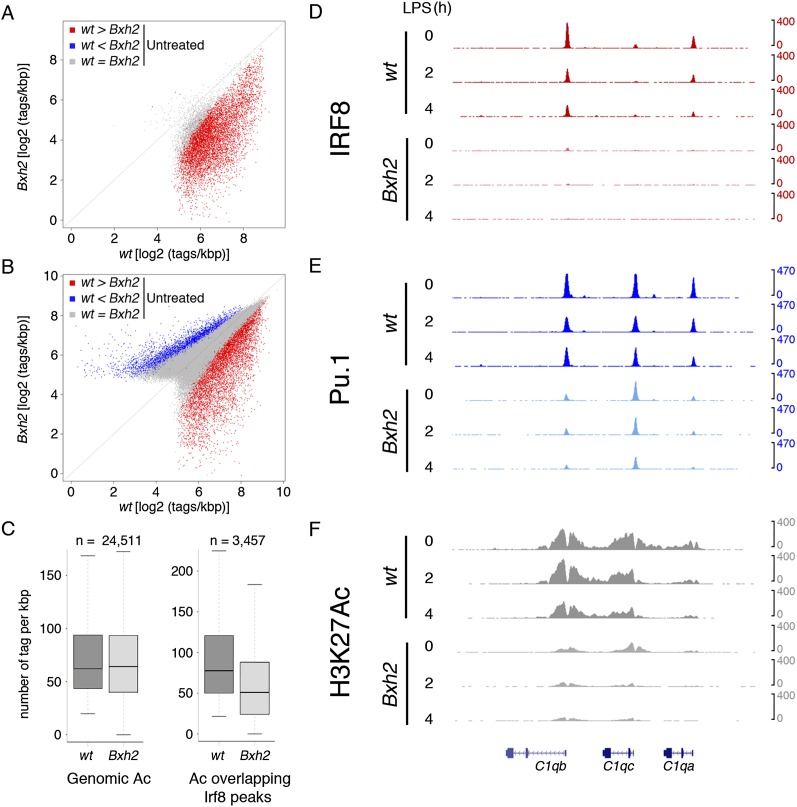
Overall, IRF8 binding was abolished or greatly reduced genome-wide in Bxh2 macrophages compared with their wild-type counterpart (Fig. 4A,D, red dots), although a relatively small population of peaks was not significantly affected by the mutation (Fig. 4A, gray peaks close to the diagonal). This residual IRF8 binding may contribute to explaining the milder phenotype of the Bxh2 mutation relative to the complete loss of IRF8 and, specifically, the more efficient generation of macrophages by Bxh2 mutant bone marrow cells.
Figure 4.
Impact of the Bxh2 mutation on IRF8, PU.1, and histone acetylation. Scatter plots indicating IRF8 (A) and PU.1 (B) levels in Bxh2 macrophages relative to wild-type cells. (C) H3K27Ac sequencing tag counts in Bxh2 macrophages relative to wild-type macrophages. The box plot on the left shows H3K27Ac data at all locations where H3K27Ac was detected in wild-type macrophages, while the one on the right shows H3K27Ac tags at IRF8 peaks reduced in the Bxh2 mutant (P = 2.9× 10−152 Wilcoxon rank sum test). The ChIP-seq snapshot on the right shows representative behaviors of IRF8 (D), PU.1 (E), and H3K27Ac (F) at a selected genomic location.
We next tested the effects of the global IRF8-binding reduction on PU.1 genomic occupancy (Fig. 4B,E). Although the genomic distribution of PU.1 was mostly unperturbed, 8.7% of PU.1 peaks (n = 6586) (Fig. 4B,E, red dots) were strongly reduced. At all time points, IRF8-dependent PU.1 peaks showed a strong overrepresentation of the composite PU.1/IRF8 site (PWM1) when compared with the PU.1 peaks that were unaffected (E-value = 1 × 10−5069 at the 4-h time point). We also detected PU.1 peaks that were increased in Bxh2 macrophages, but enhanced binding was at an overall lower magnitude and frequency than loss of binding, suggesting that it may represent an indirect effect of the mutation.
Finally, while global genomic histone acetylation (specifically, H3K27Ac) was unperturbed in Bxh2 macrophages, it was strongly (albeit not uniformly) reduced at genomic regions where IRF8 binding was abrogated or reduced in Bxh2 cells (Fig. 4C,F), which indicates that IRF8 critically contributes to the maintenance of an active chromatin state at a subset of cis-regulatory elements of unstimulated macrophages. Scatter plots obtained from LPS-stimulated macrophages showed very similar results (Supplemental Fig. 3).
Basal and inducible gene expression programs regulated by IRF8
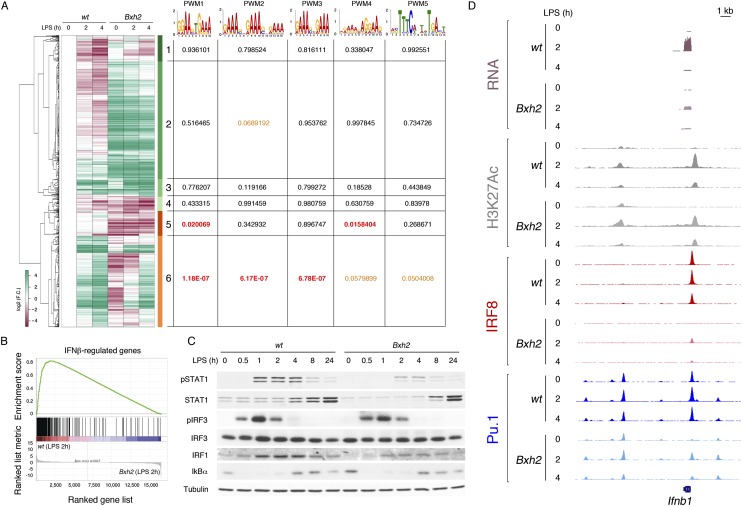
RNA sequencing (RNA-seq) experiments in untreated and LPS-treated Bxh2 macrophages showed two major trends (Fig. 5A; Supplemental Table 4). First, a large cluster (#2) of 323 genes and two smaller clusters (#1 and #3) that were down-regulated after LPS stimulation were expressed at higher levels in unstimulated Bxh2 macrophages relative to their wild-type counterpart. Clusters #1 and #2 were enriched for gene ontology (GO) terms related to cell cycle and mitosis. Increased expression of cell cycle and mitotic genes may contribute to the development of the chronic myelogenous leukemia-like syndrome caused by the IRF8 deficiency (Turcotte et al. 2005). The genomic regions surrounding (±2.5 kb) the transcription start sites (TSSs) of these genes were not enriched for IRF TF-binding sites, including the five PWMs shown above (Fig. 5A). Up-regulation of these genes was therefore likely to be an indirect effect of the loss of IRF8 activity. Second, a large group of genes was down-regulated in Bxh2 macrophages, including constitutively expressed and noninducible genes (clusters #4 and #5) (Fig. 5A) and LPS-inducible genes (cluster #6). Cluster #6 was enriched for GO terms related to IFN signaling. Consistently, gene set enrichment analysis (GSEA) (Fig. 5B) detected a strong enrichment of an IFNβ-stimulated gene (ISG) set in wild-type macrophages. In particular, of the 251 genes in cluster #6, 132 (52.5%) were ISGs (based on either their inducibility by type I IFNs or the requirement of the type I IFN receptor for their activation in response to LPS) (Raza et al. 2010; Cheng et al. 2011). Seventy-eight of these 132 ISGs were down-regulated also in untreated cells (Fig. 5A). The remaining 119 genes included a heterogeneous group of LPS-inducible genes whose promoters were enriched for EGR1–3-binding sites. Consistently, IRF8 was reported to activate the expression of EGR family TFs (Kurotaki et al. 2013), and expression of EGR1 and EGR3 was reduced in Bxh2 macrophages (Supplemental Fig. 4). When the promoters of the ISGs in this cluster were compared with those of non-ISGs, a very strong enrichment for IRF PWMs was detected (E-value = 2.01 × 10−34 for the canonical ISRE).
Figure 5.
Impairment of the LPS response in Bxh2 macrophages. (A) Differentially expressed genes in wild-type versus Bxh2 macrophages were identified by RNA-seq. Data in the heat map are expressed as log2 (fold change) and are hierarchically clustered. The numbers at the right of the heat map indicate the identified gene expression clusters. Clusters #1–3 are genes overexpressed in Bxh2 macrophages, and clusters #4–6 are genes down-regulated in Bxh2 macrophages. The E-values associated with the five PWMs described in Figure 3 (in a region including ±2.5 kb from the TSS) are shown at the right. (B) An IFNβ-regulated gene set was down-regulated in Bxh2 macrophages. (C) Western blot analysis showing the effect of the Bxh2 mutation on STAT1, IRF3, IRF1, and ΙκBα activation by LPS. (D) RNA-seq and ChIP-seq snapshot at the Ifnb1 gene locus.
We next analyzed the presence of PWM1–5 in the regions surrounding the TSSs (±2.5 kb) of the genes whose expression depends on IRF8 (clusters #4–#6). Cluster #6 (mainly LPS-inducible genes) showed a strong overrepresentation of PWM1–3 (with PWM4 and PWM5 reaching a lower statistical significance) (Fig. 5A), while cluster #5 (mainly constitutively expressed genes) showed a strong overrepresentation of only PWM1 and PWM4, corresponding to the PU.1/IRF8 PWM and an IRF/ETS composite site, respectively (Fig. 3), and both predictive of constitutive IRF8 binding in the SVM (Fig. 3D). Lack of enrichment of any of the five PWMs in cluster #4 suggests that down-regulation of these genes in Bxh2 macrophages may represent an indirect effect of IRF8 inactivation. This analysis indicates that inducible genes and, specifically, ISGs were associated with both constitutive binding sites (which enable their premarking by IRF8/PU.1) and inducible sites (which enable additional IRF8 recruitment, possibly together with other IRFs and AP-1 proteins, in response to stimulation). Loss of constitutive IRF8/PU.1 binding may contribute to reduced constitutive expression of many of the ISGs in unstimulated Bxh2 macrophages (Fig. 5A). Conversely, constitutively expressed and noninducible genes contained exclusively binding sites mediating constitutive IRF8/PU.1 recruitment.
Defective induction of ISGs in Bxh2 macrophages may be explained by the defective induction of the Ifnb1 gene in Bxh2 macrophages (Supplemental Table 4), which is in keeping with the data reported in dendritic cells (Schiavoni et al. 2002; Tsujimura et al. 2003). Reduced production of IFNβ was also indirectly confirmed by the reduced phosphorylation of STAT1 and reduced induction of IRF1 in response to LPS stimulation (Fig. 5C).
Visual inspection of the ChIP-seq data (Fig. 5D) confirmed that the Ifnb1 gene promoter was constitutively bound by IRF8 and that PU.1 association with it was largely IRF8 dependent. Binding of IRF8 and PU.1 to the IFNβ promoter in human and mouse monocytes was previously mapped to promoter sites resembling PWM1 (a composite ETS/IRF site that differs from PWM1 because of a 4-nt spacer between the 5′-GGAA-3′ ETS consensus and the 5′-GAAA-3′ IRF consensus) and PWM4 (Li et al. 2011) and is fully consistent with our ChIP-seq data. Moreover, constitutive IRF8/PU.1 binding was shown to be required for IRF3 recruitment, thus explaining reduced IFNβ induction in cells depleted of IRF8 (Li et al. 2011). Macrophages and many other cells are also known to constitutively produce extremely low levels of IFNβ (close or below the detection limit) that are, however, important to maintain the basal expression of some genes relevant for the subsequent response to both type I and type II IFNs; notably STAT1 and its binding partner, IRF9 (Ivashkiv and Donlin 2014). At least in fibroblasts, basal expression of IFNβ was attributed to JUN/AP-1-mediated regulation (Gough et al. 2010), but, in vivo, it may result from more complex regulatory circuits, such as the tonic macrophage stimulation by the microbiome (Ivashkiv and Donlin 2014). However, while IRF8 was clearly essential for maximal production of IFNβ in response to LPS, we have no clear evidence of its possible role in maintaining basal IFNβ expression, since STAT1 levels in unstimulated conditions were similar in wild-type and Bxh2 macrophages (Fig. 5C), and its genomic recruitment in response to IFNβ and IFNγ stimulation was largely unaffected (see below). Therefore, we favor the hypothesis that reduced basal expression of many ISGs in unstimulated Bxh2 cells was not due to reduced basal IFNβ secretion but mainly lack of constitutive binding of their promoters and regulatory elements by IRF8/PU.1.
In contrast to STAT1, the activation of IRF3, which is mediated by protein kinases directly activated upon LPS stimulation and is required to induce Ifnb1 gene transcription, was unaffected in the Bxh2 mutant (as assessed by normal levels of IRF3 phosphorylation) (Fig. 5C). Similarly, activation of the NF-kB pathway, as measured by degradation and NF-kB-dependent resynthesis of IkBα, was identical in wild-type and Bxh2 macrophages.
Finally, since the GREAT analysis shown in Figure 1E demonstrated a strong association of constitutive IRF8 peaks with several ontology terms related to purinergic receptor signaling, we analyzed the impact of the Bxh2 mutation on these genes. Strikingly, nine purinergic receptor genes belonging to different subgroups with distinct nucleotide recognition specificity (e.g., Adora3, P2rx4, and P2ry12) were significantly down-regulated in Bxh2 macrophages—in most cases, both before and after LPS treatment (Supplemental Table 5). These genes were usually constitutively bound by IRF8 and PU.1 at either their promoter or cis-regulatory regions nearby (Supplemental Table 5), thus suggesting that IRF8 directly controls the responsiveness of macrophages to a large number of nucleotide species. IRF8 also directly controlled the expression of Entpd1, which encodes the only ectonucleotidase (CD39) catalyzing the hydrolysis of ATP and ADP at the macrophage surface (Levesque et al. 2010), implicating IRF8 in the regulation of ATP signaling and metabolism.
IRF8 controls the IFN response downstream from Ifnb1 gene induction
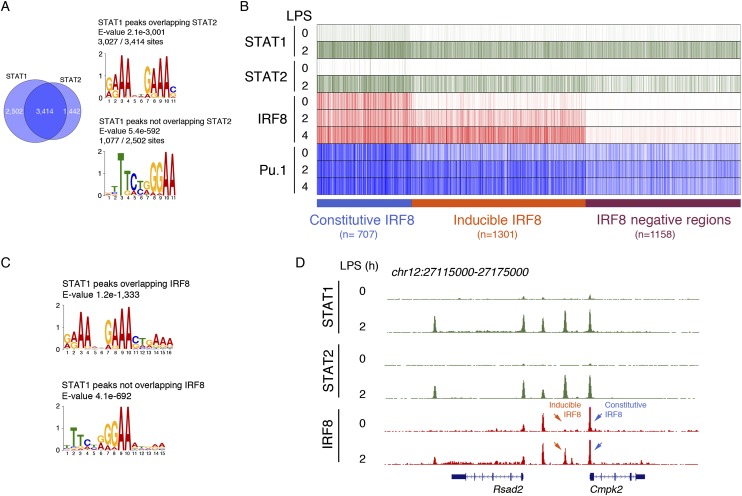
The predominant transcriptional outcomes in LPS-stimulated Bxh2 macrophages were an impairment of the interferon response and a reduced expression of non-ISGs likely regulated by EGR family TFs (Kurotaki et al. 2013). We set out to obtain additional mechanistic insight into the role of IRF8 in the control of IFNβ-regulated gene expression and epigenomic changes. Specifically, we asked whether IRF8 is exclusively required for Ifnb1 gene activation or instead collaborates with IFNβ-activated STATs in the induction of IFNβ-activated genes (Ivashkiv and Donlin 2014). According to current models, IFNα/β binding to their receptor (IFNAR) mainly promotes the release of STAT1/STAT2/IRF9 trimers that preferentially bind IRF-like sites (ISREs) because their binding specificity is determined by the IRF9 subunit. Conversely, IFNγ activates STAT1 homodimers that preferentially bind GAS (γ-activated sites) sequences (Ivashkiv and Donlin 2014). To address this question, we first generated STAT1 and STAT2 ChIP-seq data in macrophages stimulated with LPS and evaluated whether STAT1 and STAT2 landing sites in the genome were prebound by IRF8. STAT1/STAT2/IRF9 activation in this context is dependent on IRF3-mediated activation of the Ifnb1 gene (Doyle et al. 2002) and the subsequent autocrine and paracrine activities of newly synthesized IFNβ (Thomas et al. 2006). Overall, the overlap between STAT1 and STAT2 ChIP-seq data sets was very high (Fig. 6A), thus indirectly confirming that the main STAT1 species released upon LPS stimulation is the canonical STAT1/STAT2/IRF9 trimer binding IRF-like sites (Fig. 6A). Nevertheless, a substantial fraction of STAT1 peaks did not overlap STAT2 peaks and showed a clear preference toward GAS-like sites (Fig. 6A). Based on their relationship with IRF8, we identified three distinct groups of genomic regions contacted by STAT1/STAT2 in an LPS-inducible manner (Fig. 6B); namely, regions associated with constitutive (n = 707) or inducible (n = 1301) IRF8 and regions negative for IRF8 (or with low IRF8 levels that were below the threshold selected for peak calling; n = 1158). Overall, about two-thirds of the genomic regions bound by STAT1 in response to LPS were also associated with IRF8 (in either a constitutive or an inducible manner).
Figure 6.
Correlation between STAT1/STAT2 and IRF8 genomic occupancy in LPS-treated macrophages. (A) Overlap between inducible STAT1 and STAT2 ChIP-seq peaks at 2 h after LPS stimulation. The PWMs at the right were retrieved by de novo motif discovery on either STAT1/STAT2 overlapping peaks (top panel) or STAT1 peaks that did not overlap STAT2 (bottom panel). (B) STAT1 and STAT2 genomic distributions in LPS-activated macrophages. In the heat map, IRF8 ChIP-seq data for the same genomic regions where STAT1 binding was detected are shown. Data were ordered on the basis of the presence or absence of IRF8 signals and the behavior (constitutive or inducible) of IRF8 peaks. STAT2 binding to the same regions is shown. (C) PWMs overrepresented in LPS-activated STAT1 peaks that either overlap or do not overlap with IRF8. (D) ChIP-seq snapshot showing the recruitment of STAT1, STAT2, and IRF8 to a representative genomic region containing the LPS-inducible and IFNβ-dependent genes Rsad2 and Cmpk2.
The heat map in Figure 6B also shows that the behavior of PU.1 at these regions paralleled that of IRF8. In fact, regulatory elements constitutively associated with IRF8 were also constitutively bound by PU.1 (Fig. 6B, blue bar, left); regions with inducible IRF8 showed low basal PU.1 binding that was increased in response to stimulation (Fig. 6B, orange bar, middle), and STAT1/STAT2-bound regions with no IRF8 binding were, in general, characterized by comparatively low PU.1 signals that were not significantly (or only slightly) affected by stimulation.
A motif discovery analysis on STAT1 peaks that overlapped and peaks that did not overlap IRF8 identified again two distinct types of DNA-binding sites (Fig. 6C). IRF8-associated STAT1 peaks were mainly associated with ISRE-like sites, while IRF8-negative STAT1 peaks were strongly enriched for a GAS-like site (Jolma et al. 2013; Ivashkiv and Donlin 2014), thus suggesting that the STAT1 species contacting these sites may be STAT1 homodimers like those released upon IFNγ stimulation. A representative snapshot in Figure 6D shows a genomic region containing multiple STAT1 and STAT2 peaks associated with either constitutive or inducible IRF8 peaks.
Overall, the IRF8 requirement for the induction of the Ifnb1 gene and even more so the extensive overlap between the genomic distribution of IFNβ-activated STAT1/STAT2 and IRF8 indicate a close functional relationship between the main IFNβ-activated TF and IRF8.
IRF8 synergizes with STAT1 in the induction of IFNβ-activated genes
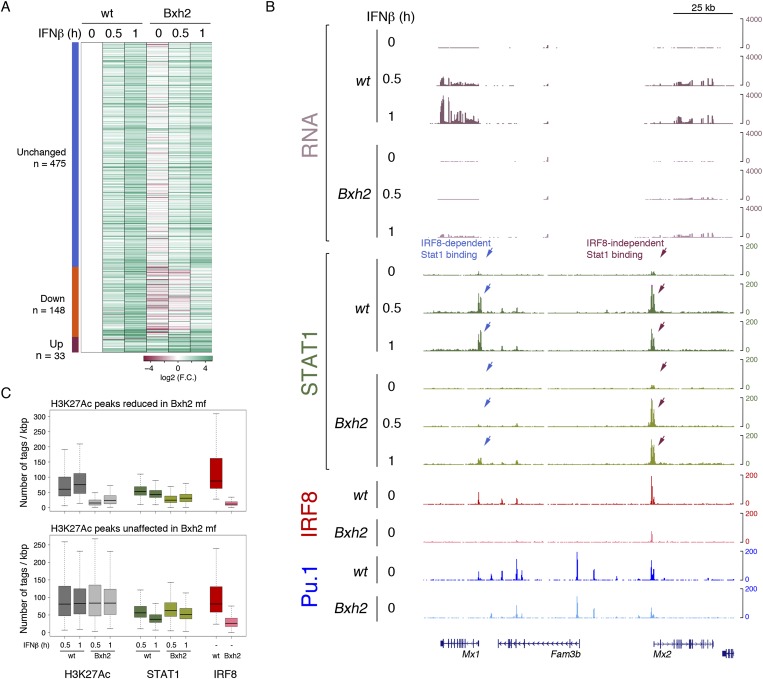
Because of the extensive overlap of LPS-induced STAT1 peaks and IRF8, we asked whether and to what extent IRF8 collaborates with STAT1 in the induction of IFNβ-activated genes. To address this question, we could not use LPS stimulation of Bxh2 macrophages, since the IFNβ response was greatly impaired in these cells (Fig. 5). Therefore we stimulated macrophages with recombinant IFNβ and tested the impact of the Bxh2 mutation on gene expression, STAT1 binding, and histone acetylation of activated macrophages.
The response of Bxh2 macrophages to IFNβ stimulation was almost indistinguishable from that of their wild-type counterpart, as indicated by STAT1 phosphorylation over a 4-h time course (Supplemental Fig. 5A). Importantly, IFNβ induced IRF8 protein expression (Supplemental Fig. 5A). This effect was correlated with a prominent binding of STAT1 to both the Irf8 gene promoter and an upstream enhancer (Supplemental Fig. 5B) and the induction of the Irf8 mRNA (approximately twofold at the time points used for RNA-seq). Neither STAT1 binding nor increased Irf8 mRNA after IFNβ stimulation was reduced in Bxh2 macrophages (in fact, they were both slightly augmented). Therefore, while the up-regulation of IRF8 in response to LPS was impaired in Bxh2 macrophages, its induction by IFNβ was unaffected.
Since the peak of STAT1 phosphorylation in response to IFNβ occurred at 30 min and strongly declined after 60 min (Supplemental Fig. 5A), we focused our initial analysis on the first hour after stimulation. Of the 656 genes induced in response to IFNβ stimulation in this time window, 148 (22.6%) showed reduced activation in Bxh2 macrophages (Fig. 7A; Supplemental Table 6). As discussed above, the transcripts of many IRF8-dependent IFNβ-inducible genes were less abundant already before stimulation (heat map in Fig. 7A), suggesting a role for IRF8 in maintaining their basal level of activity. Genes down-regulated in Bxh2 macrophages included canonical IFNβ response genes directly bound and activated by STAT1, such as Mx1 and Mx2, whose induction was almost completely abrogated in IRF8 mutant cells (Fig. 7B). Conversely, only a small group of 33 IFNβ-activated genes (5%) showed a mild increase in expression in IFNβ-stimulated Bxh2 macrophages, which may relate to the previously described activity of IRF8 as a negative regulator of interferon-stimulated genes (Rosenbauer et al. 1999).
Figure 7.
IRF8 requirement for binding of and transactivation by STAT1 in IFNβ-stimulated macrophages. (A) Heat map showing RNA-seq data in wild-type and Bxh2 macrophages stimulated with IFNβ as indicated. Genes were divided in three groups based on their behavior in Bxh2 relative to wild-type macrophages. (B) Genomic snapshot of the Mx1–Mx2 locus showing IFNβ-induced STAT1 peaks affected or not affected in Bxh2 macrophages. (C) Box plot showing H3K27Ac, STAT1, and IRF8 tag densities at genomic regions showing reduced (top box plot) or unaffected (bottom box plot) H3K27Ac in Bxh2 versus wild-type macrophages.
The complexity of the effects of the Bxh2 mutation on STAT1 binding is exemplified by its behavior at the Mx1–Mx2 locus (Fig. 7B). While STAT1 binding to the Mx2 promoter was virtually unaffected, its recruitment to the Mx1 promoter was almost completely abrogated in Bxh2 macrophages. Both promoters, however, were constitutively bound by IRF8, whose occupancy dropped in Bxh2 cells. The possibility that the residual IRF8 and PU.1 binding observed in Bxh2 macrophages at the Mx2 gene promoter may suffice to enable STAT1 recruitment should not be discounted. However, in spite of the different effects on STAT1 recruitment, the induction of both genes was strongly reduced or nearly completely abrogated in Bxh2 cells.
Genome-wide, of the 13,261 STAT1 peaks observed at 30 min after IFNβ stimulation, 38.4% (5094) overlapped with IRF8 peaks. However, only 326 peaks were significantly reduced in Bxh2 macrophages. Therefore, although the recruitment of STAT1 at some typical IFNβ-activated genes (such as Mx1) required IRF8, the overall genomic STAT1 landscape was only marginally perturbed. While the invariant STAT1 peaks were enriched (E-value = 4.1 × 10−793) for a GAS-like site similar to the one shown in Figure 6C, the peaks reduced in Bxh2 cells were enriched for the canonical composite PU.1–IRF8 site (E-value = 4.3 × 10−394). We also analyzed the impact of IRF8 on STAT1 recruitment using a more extended kinetics of IFNβ stimulation (up to 4 h). Overall, also at later time points, most STAT1-binding events were not significantly affected by the IRF8 mutation: Two-hundred-eighty-eight STAT1 peaks were significantly reduced at 2 h, and only 60 were significantly reduced at 4 h after IFNβ stimulation. A representative snapshot is shown in Supplemental Figure 6.
We next analyzed histone acetylation after IFNβ stimulation at genomic regions associated with IRF8 occupancy (Fig. 7C). While 13,128 acetylated regions at 30 min post-stimulation (15,040 at 60 min) were not significantly affected by the Bxh2 mutation, 896 regions at 30 min (980 at 60 min) were strongly down-regulated, a result that is consistent with the effects of the Irf8 mutation on IFNβ-induced gene expression. STAT1 binding at regions whose acetylation was unaffected did not display clear and significant differences in wild-type and Bxh2 macrophages. Conversely, median STAT1 binding was strongly reduced at regions whose acetylation was lower in Bxh2 macrophages (2.2-fold; P = 4.9 × 10−54, Wilcoxon rank sum test) (Fig. 7C). Therefore, STAT1 recruitment was critically reduced at those regions whose activation (measured by inducible histone acetylation) was impaired in IRF8 mutant cells even though the overall genomic STAT1-binding landscape was only minimally affected by the IRF8 mutation.
Finally, we tested the effects of the Bxh2 mutation on IFNγ-induced STAT1 binding that, as discussed above, is mainly mediated by GASs. IFNγ treatment induced 5993 STAT1 peaks at 1 h and 1293 peaks at 2 h. However, only 211 STAT1 peaks were reduced in the Bxh2 macrophages at 1 h and three peaks were reduced at 2 h post-stimulation (Supplemental Fig. 7). Overall, IRF8 had very marginal effects on recruitment of STAT1 homodimers after IFNγ stimulation.
Discussion
The objective of this study was to understand at a genome scale the mechanistic basis for the involvement of the same TF in both developmental and inducible gene expression programs. IRF8 represents a paradigmatic example, since it is required for terminal macrophage differentiation and, at the same time, is necessary for the activation of some crucial inflammatory genes, notably Ifnb1.
The analysis of the IRF8 genomic distribution before and after LPS stimulation allowed us to identify two distinct sets of binding events together with the cis-regulatory code explaining them. Rather unusually for a TF involved in developmental decisions, IRF8 displayed a genomic distribution with two clearly distinguishable components; namely, a constitutive component associated with composite PU.1/IRF8 sites or canonical PU.1 sites and therefore with high-level constitutive occupancy by PU.1 and an inducible component that was stimulus-dependent and relied on the existence of a completely different set of binding sites, including multimerized IRF sites and IRF/AP-1 composite sites. Probably because of both their low affinity and the requirement for additional binding partners (namely, other IRF or AP-1 family TFs), such sites were unable to bind IRF8 in basal conditions. When the expression and protein amount of IRF8 were increased because of IFNβ release and autocrine activity (Ivashkiv and Donlin 2014) together with increased expression of other IRFs (notably IRF1) and AP-1 proteins, IRF8 became able to contact these IRF sites, a phenomenon that closely resembles the recently described activation-inducible IRF4 recruitment in B lymphocytes (Ochiai et al. 2013). In turn, these sites appear to be relevant for the implementation of the IFNβ response by STAT1, since a sizeable fraction of the interferon-regulated genes required IRF8 for maximal induction. Since IRF8 is itself required for IFNβ induction in response to LPS, this regulatory circuit configures a feed-forward loop with a self-reinforcement component by which STAT1 controls the increased Irf8 transcription.
In addition to the Ifnb1 gene, the constitutive component of the IRF8 genomic distribution was associated with the regulation of a number of genes critical for not only basal macrophage functions but also the response to stimulation, including complement factors, pattern recognition receptors (such as TLR9, whose expression was almost completely abolished in Bxh2 macrophages), and several purinergic receptors. The association of IRF8 with genes encoding purinergic receptors was particularly striking in more than one respect. On the one hand, many of the highest-affinity IRF8 sites detected in the macrophage genome occurred at loci containing purinergic receptor genes, to the point that the most enriched GO terms identified as associated with basal IRF8-binding events were all related to purinergic receptor signaling. On the other hand, about half of the genes encoding purinergic receptors as well as the only macrophage ectonucleotidase (CD39) that degrades extracellular ATP and ADP were affected by the loss of IRF8 function. Therefore, a major critical role of IRF8 in macrophages is to regulate all of those functions (including, but not only, macrophage chemotaxis and IL1β secretion) that are directly controlled by purines (Kronlage et al. 2010; Ulmann et al. 2010; Bours et al. 2011). Previous data in Irf8−/− monocyte–dendritic cell progenitors (MDPs) suggested a role for IRF8 in controlling the expression of Klf4, an additional TF required for macrophage development, as well as some EGR family TFs, which are rapidly induced in response to many stimuli, including LPS (Kurotaki et al. 2013). In terminally differentiated macrophages such as those used in our study, the expression of Klf4 was substantially identical in Bxh2 and wild-type macrophages (data not shown). This discrepancy may be due to the residual IRF8 activity in Bxh2 cells, which may also explain their higher ability to differentiate into macrophages as compared with Irf8−/− bone marrow cells. Alternatively, the defect in Klf4 expression observed in Irf8−/− MDPs may reflect a transient impairment that is overcome when differentiation progresses. Conversely, the expression of EGR1 and EGR3 was substantially reduced in Bxh2 macrophages. Consistent with this observation, almost half of the LPS-inducible genes that were down-regulated in Bxh2 macrophages were not ISGs, and their promoters were enriched in EGR TF-binding sites.
This study also revealed an unexpected complexity in the interplay between two master regulators of macrophage development; namely, PU.1 and IRF8. PU.1 has a very broad role in controlling the macrophage cis-regulatory repertoire (Ghisletti et al. 2010; Heinz et al. 2010), since it is also pervasively required to maintain nucleosome depletion (and therefore the accessibility of the underlying regulatory information) at macrophage enhancers (Barozzi et al. 2014). Many TFs that are activated or induced in response to extracellular stimuli (including inflammatory agonists) were shown to bind regions that were premarked and made accessible by PU.1 (Ghisletti et al. 2010), and PU.1 binding was, in fact, necessary for their recruitment (Escoubet-Lozach et al. 2011; Heinz et al. 2013). The inability of TFs such as NF-kB to contact sites embedded in a nucleosomal context (Natoli 2009; Lone et al. 2013) provides a mechanistic basis for these observations. The general model suggested by these studies is that the regulatory landscape created by lineage-determining TFs determines and, in fact, limits the activity of stimulus-inducible TFs by defining the fraction of the genomic regulatory repertoire that is available for them to bind (Natoli 2010). This model is in keeping with the well-established notion that the enhancer repertoire of distinct cells types is very specific, is distinct from that of other cells, and exists before stimulation (Heintzman et al. 2009). The discovery of regulatory elements (latent or de novo enhancers) (Kaikkonen et al. 2013; Ostuni et al. 2013) that are not associated with either histone marks or constitutively bound TFs and whose emergence from latency requires activation with specific stimuli explains why the exposure to environmental changes has a direct impact on the regulatory information made available for gene regulation (Gosselin et al. 2014; Lavin et al. 2014). IRF8 directly contributes to the appearance of latent enhancers, since about one-third of the latent enhancers induced by a 4-h LPS stimulation coincided with LPS-induced IRF8 peaks associated with PWMs enabling inducible IRF8 recruitment. In general, many of the novel and LPS-inducible IRF8-binding events occurred at genomic regions that were not premarked by PU.1 in unstimulated macrophages and were at a considerable distance from constitutively PU.1-bound regions. Therefore, IRF8 may represent one of those few TFs that can directly invade chromatinized and originally inaccessible, not premarked, sites and get them involved in gene regulation.
Materials and methods
Mice
Animal experiments were performed in accordance with the Italian laws (D.L.vo 116/92 and following additions), which enforce the EU 86/609 directive. The Bxh2/TyJ mouse strain was obtained from the Jackson Laboratory in a C3H/HeJ background that is resistant to LPS stimulation due to a mutation in TLR4 (Tlr4Lps-d). BXH2/TyJ males were therefore crossed with C57BL/6 females to generate F1 mice that were then intercrossed to produce an F2 progeny. F2 littermates were crossed to generate mice not carrying Tlr4Lps-d and either homozygous for IRF8R294C (BXH2 mice) or wild type for IRF8. TaqMan probes were designed to perform SNP analysis of mouse tail DNA; in particular, 5′-CTACACCATGAATAAA-3′ for Tlr4Lps-d, 5′-CTACACCAGGAATAAA -3′ for Tlr4wt, 5′-AACACGCAGCCCTG-3′ for IRF8R294C, and 5′-AACACGCGGCCCTG-3′ for IRF8wt.
Cell culture
Macrophage cultures were carried out as described (Austenaa et al. 2012). LPS from Escherichia coli serotype EH100 (Alexis) was used at 10 ng/mL. Mouse recombinant IFNβ (Millipore, no. IF011) and IFNγ (R&D Systems, no. 485-MI) were used at a concentration of 100 U/mL.
Antibodies
An anti-IRF8 rabbit polyclonal antibody was raised in-house and affinity-purified. The anti-Pu.1 antibody has been previously described (Ostuni et al. 2013). The following antibodies were also used: phospho-STAT1 (Tyr701; Cell Signaling Technology, no. 9171), STAT1 (Cell Signaling Technology, no. 9172), IkBα (Santa Cruz Biotechnology, sc-371), phospho-IRF3 (Cell Signaling Technology, no. 4947), IRF3 (Cell Signaling Technology, no. 4302), IRF1 (sc-640), and tubulin (Sigma, T9026). For quantified images, secondary IRDye antibodies from Li-Cor were used (catalog nos. 926-68021 and 926-32210).
ChIP-seq and RNA-seq
Fixed macrophages (5 × 106 to 15 × 106 [ChIP-seq for H3K27Ac and Pu.1], 30 × 106 [STAT2 and IRF1], or 100 × 106 [IRF8 and STAT1]) were lysed with RIPA buffer and, after chromatin shearing by sonication, incubated overnight at 4°C with protein G Dynabeads (Invitrogen) that were previously coupled with 3–10 μg of antibody (Ghisletti et al. 2010; Austenaa et al. 2012). Antibodies used for ChIP-seq included STAT1 (sc-592), STAT2 (sc-950), IRF1 (sc-640), H3K27Ac (Abcam, ab4729), homemade PU.1, and IRF8 antibodies. DNA yield was ∼200 ng per 107 cells for H3K27Ac, 20 ng per 107 cells for PU1, and 1 ng per 107 cells for IRF1, IRF8, STAT1, and STAT2. Library preparation for Illumina sequencing was carried out using a previously described protocol (Garber et al. 2012) with slight modifications (Ostuni et al. 2013). Total RNA was extracted from 5 × 106 cells using RNAeasy kit (Qiagen), and libraries were prepared after oligo-dT selection using the TruSeq RNA sample preparation kit (Illumina).
Computational methods
Short reads obtained from Illumina HiSeq 2000 runs were quality-filtered according to the Illumina pipeline. Analysis of the data sets was automated using the “Fish the ChIPs” pipeline (Barozzi et al. 2011).
For RNA-seq, after quality filtering according to the Illumina pipeline, 51-base-pair (bp) paired-end reads were aligned to the mm9 reference genome and the Mus musculus transcriptome (Ensembl build 63) (Flicek et al. 2012) using TopHat (Trapnell et al. 2012). Transcript abundance was quantified, and differentially expressed genes were called using Cufflinks 1.2.1 (Trapnell et al. 2012). Detailed computational methods are described in the Supplemental Material.
The SVM approach has been recently described (Barozzi et al. 2014). In this specific case, we split the original data sets into a training set and a test set of identical size.
Accession numbers
Raw data sets are available for download at the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/gds) under the accession number GSE56123.
Supplementary Material
Acknowledgments
We thank Ido Amit, Philippe Gros, and Silvia Monticelli for critically reading the manuscript. This work was supported by the European Research Council (ERC grant NORM to G.N.). A.M. generated most data sets and the additional experimental data with contributions by S.G., R.O., and E.P.; A.T. computationally analyzed all data sets; I.B. made the SVM analysis; K.O. provided critical reagents; and G.N. designed and supervised the study and wrote the manuscript with input from all coauthors. A.M. and A.T. contributed equally to this work and are listed in alphabetical order.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.257592.114.
References
- Austenaa L, Barozzi I, Chronowska A, Termanini A, Ostuni R, Prosperini E, Stewart AF, Testa G, Natoli G. 2012. The histone methyltransferase Wbp7 controls macrophage function through GPI glycolipid anchor synthesis. Immunity 36: 572–585. [DOI] [PubMed] [Google Scholar]
- Barozzi I, Termanini A, Minucci S, Natoli G. 2011. Fish the ChIPs: a pipeline for automated genomic annotation of ChIP-Seq data. Biol Direct 6: 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barozzi I, Simonatto M, Bonifacio S, Yang L, Rohs R, Ghisletti S, Natoli G. 2014. Coregulation of transcription factor binding and nucleosome occupancy through DNA features of mammalian enhancers. Mol Cell 54: 844–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bours MJ, Dagnelie PC, Giuliani AL, Wesselius A, Di Virgilio F. 2011. P2 receptors and extracellular ATP: a novel homeostatic pathway in inflammation. Front Biosci 3: 1443–1456. [DOI] [PubMed] [Google Scholar]
- Cheng CS, Feldman KE, Lee J, Verma S, Huang DB, Huynh K, Chang M, Ponomarenko JV, Sun SC, Benedict CA, et al. 2011. The specificity of innate immune responses is enforced by repression of interferon response elements by NF-κB p50. Sci Signal 4: ra11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes C, Vapnik V. 1995. Support-vector networks. Mach Learn 20: 273–297. [Google Scholar]
- Doyle S, Vaidya S, O’Connell R, Dadgostar H, Dempsey P, Wu T, Rao G, Sun R, Haberland M, Modlin R, et al. 2002. IRF3 mediates a TLR3/TLR4-specific antiviral gene program. Immunity 17: 251–263. [DOI] [PubMed] [Google Scholar]
- Escalante CR, Brass AL, Pongubala JM, Shatova E, Shen L, Singh H, Aggarwal AK. 2002. Crystal structure of PU.1/IRF-4/DNA ternary complex. Mol Cell 10: 1097–1105. [DOI] [PubMed] [Google Scholar]
- Escoubet-Lozach L, Benner C, Kaikkonen MU, Lozach J, Heinz S, Spann NJ, Crotti A, Stender J, Ghisletti S, Reichart D, et al. 2011. Mechanisms establishing TLR4-responsive activation states of inflammatory response genes. PLoS Genet 7: e1002401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flicek P, Amode MR, Barrell D, Beal K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fairley S, Fitzgerald S, et al. 2012. Ensembl 2012. Nucleic Acids Res 40: D84–D90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garber M, Yosef N, Goren A, Raychowdhury R, Thielke A, Guttman M, Robinson J, Minie B, Chevrier N, Itzhaki Z, et al. 2012. A high-throughput chromatin immunoprecipitation approach reveals principles of dynamic gene regulation in mammals. Mol Cell 47: 810–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghisletti S, Barozzi I, Mietton F, Polletti S, De Santa F, Venturini E, Gregory L, Lonie L, Chew A, Wei CL, et al. 2010. Identification and characterization of enhancers controlling the inflammatory gene expression program in macrophages. Immunity 32: 317–328. [DOI] [PubMed] [Google Scholar]
- Glasmacher E, Agrawal S, Chang AB, Murphy TL, Zeng W, Vander Lugt B, Khan AA, Ciofani M, Spooner CJ, Rutz S, et al. 2012. A genomic regulatory element that directs assembly and function of immune-specific AP-1-IRF complexes. Science 338: 975–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosselin D, Glass CK. 2014. Epigenomics of macrophages. Immunol Rev 262: 96–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosselin D, Link VM, Romanoski CE, Fonseca GJ, Eichenfield DZ, Spann NJ, Stender JD, Chun HB, Garner H, Geissmann F, et al. 2014. Environment drives selection and function of enhancers controlling tissue-specific macrophage identities. Cell 159: 1327–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gough DJ, Messina NL, Hii L, Gould JA, Sabapathy K, Robertson AP, Trapani JA, Levy DE, Hertzog PJ, Clarke CJ, et al. 2010. Functional crosstalk between type I and II interferon through the regulated expression of STAT1. PLoS Biol 8: e1000361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyon I, Elisseeff A. 2003. An introduction to variable and feature selection. J Mach Learn Res 3: 1157–1182. [Google Scholar]
- Hambleton S, Salem S, Bustamante J, Bigley V, Boisson-Dupuis S, Azevedo J, Fortin A, Haniffa M, Ceron-Gutierrez L, Bacon CM, et al. 2011. IRF8 mutations and human dendritic-cell immunodeficiency. N Engl J Med 365: 127–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, et al. 2009. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459: 108–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38: 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Romanoski CE, Benner C, Allison KA, Kaikkonen MU, Orozco LD, Glass CK. 2013. Effect of natural genetic variation on enhancer selection and function. Nature 503: 487–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivashkiv LB, Donlin LT. 2014. Regulation of type I interferon responses. Nat Rev Immunol 14: 36–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolma A, Yan J, Whitington T, Toivonen J, Nitta KR, Rastas P, Morgunova E, Enge M, Taipale M, Wei G, et al. 2013. DNA-binding specificities of human transcription factors. Cell 152: 327–339. [DOI] [PubMed] [Google Scholar]
- Kaikkonen MU, Spann NJ, Heinz S, Romanoski CE, Allison KA, Stender JD, Chun HB, Tough DF, Prinjha RK, Benner C, et al. 2013. Remodeling of the enhancer landscape during macrophage activation is coupled to enhancer transcription. Mol Cell 51: 310–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronlage M, Song J, Sorokin L, Isfort K, Schwerdtle T, Leipziger J, Robaye B, Conley PB, Kim HC, Sargin S, et al. 2010. Autocrine purinergic receptor signaling is essential for macrophage chemotaxis. Sci Signal 3: ra55. [DOI] [PubMed] [Google Scholar]
- Kueh HY, Champhekar A, Nutt SL, Elowitz MB, Rothenberg EV. 2013. Positive feedback between PU.1 and the cell cycle controls myeloid differentiation. Science 341: 670–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurotaki D, Osato N, Nishiyama A, Yamamoto M, Ban T, Sato H, Nakabayashi J, Umehara M, Miyake N, Matsumoto N, et al. 2013. Essential role of the IRF8-KLF4 transcription factor cascade in murine monocyte differentiation. Blood 121: 1839–1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavin Y, Winter D, Blecher-Gonen R, David E, Keren-Shaul H, Merad M, Jung S, Amit I. 2014. Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell 159: 1312–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levesque SA, Kukulski F, Enjyoji K, Robson SC, Sevigny J. 2010. NTPDase1 governs P2X7-dependent functions in murine macrophages. Eur J Immunol 40: 1473–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Wong JJ, Sum C, Sin WX, Ng KQ, Koh MB, Chin KC. 2011. IRF8 and IRF3 cooperatively regulate rapid interferon-β induction in human blood monocytes. Blood 117: 2847–2854. [DOI] [PubMed] [Google Scholar]
- Li P, Spolski R, Liao W, Wang L, Murphy T, Murphy K, Leonard W. 2012. BATF–JUN is critical for IRF4-mediated transcription in T cells. Nature 490: 543–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtinger M, Ingram R, Hannah R, Muller D, Clarke D, Assi SA, Lie ALM, Noailles L, Vijayabaskar MS, Wu M, et al. 2012. RUNX1 reshapes the epigenetic landscape at the onset of haematopoiesis. EMBO J 31: 4318–4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lone IN, Shukla MS, Charles Richard JL, Peshev ZY, Dimitrov S, Angelov D. 2013. Binding of NF-κB to nucleosomes: effect of translational positioning, nucleosome remodeling and linker histone H1. PLoS Genet 9: e1003830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquis JF, LaCourse R, Ryan L, North RJ, Gros P. 2009. Disseminated and rapidly fatal tuberculosis in mice bearing a defective allele at IFN regulatory factor 8. J Immunol 182: 3008–3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G. 2010. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol 28: 495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natoli G. 2009. Control of NF-κB-dependent transcriptional responses by chromatin organization. Cold Spring Harb Perspect Biol 1: a000224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natoli G. 2010. Maintaining cell identity through global control of genomic organization. Immunity 33: 12–24. [DOI] [PubMed] [Google Scholar]
- Ochiai K, Maienschein-Cline M, Simonetti G, Chen J, Rosenthal R, Brink R, Chong A, Klein U, Dinner A, Singh H, et al. 2013. Transcriptional regulation of germinal center B and plasma cell fates by dynamical control of IRF4. Immunity 38: 918–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostuni R, Piccolo V, Barozzi I, Polletti S, Termanini A, Bonifacio S, Curina A, Prosperini E, Ghisletti S, Natoli G. 2013. Latent enhancers activated by stimulation in differentiated cells. Cell 152: 157–171. [DOI] [PubMed] [Google Scholar]
- Raza S, McDerment N, Lacaze PA, Robertson K, Watterson S, Chen Y, Chisholm M, Eleftheriadis G, Monk S, O’Sullivan M, et al. 2010. Construction of a large scale integrated map of macrophage pathogen recognition and effector systems. BMC Syst Biol 4: 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbauer F, Tenen D. 2007. Transcription factors in myeloid development: balancing differentiation with transformation. Nat Rev Immunol 7: 105–117. [DOI] [PubMed] [Google Scholar]
- Rosenbauer F, Waring JF, Foerster J, Wietstruk M, Philipp D, Horak I. 1999. Interferon consensus sequence binding protein and interferon regulatory factor-4/Pip form a complex that represses the expression of the interferon-stimulated gene-15 in macrophages. Blood 94: 4274–4281. [PubMed] [Google Scholar]
- Schiavoni G, Mattei F, Sestili P, Borghi P, Venditti M, Morse HC 3rd, Belardelli F, Gabriele L. 2002. ICSBP is essential for the development of mouse type I interferon-producing cells and for the generation and activation of CD8α+ dendritic cells. J Exp Med 196: 1415–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tailor P, Tamura T, Morse HC 3rd, Ozato K. 2008. The BXH2 mutation in IRF8 differentially impairs dendritic cell subset development in the mouse. Blood 111: 1942–1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura T, Nagamura-Inoue T, Shmeltzer Z, Kuwata T, Ozato K. 2000. ICSBP directs bipotential myeloid progenitor cells to differentiate into mature macrophages. Immunity 13: 155–165. [DOI] [PubMed] [Google Scholar]
- Tamura T, Thotakura P, Tanaka T, Ko M, Ozato K. 2005. Identification of target genes and a unique cis element regulated by IRF-8 in developing macrophages. Blood 106: 1938–1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura T, Yanai H, Savitsky D, Taniguchi T. 2008. The IRF family transcription factors in immunity and oncogenesis. Annu Rev Immunol 26: 535–584. [DOI] [PubMed] [Google Scholar]
- Taniguchi T, Ogasawara K, Takaoka A, Tanaka N. 2001. IRF family of transcription factors as regulators of host defense. Annu Rev Immunol 19: 623–655. [DOI] [PubMed] [Google Scholar]
- Thomas KE, Galligan CL, Newman RD, Fish EN, Vogel SN. 2006. Contribution of interferon-β to the murine macrophage response to the toll-like receptor 4 agonist, lipopolysaccharide. J Biol Chem 281: 31119–31130. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7: 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsujimura H, Tamura T, Ozato K. 2003. Cutting edge: IFN consensus sequence binding protein/IFN regulatory factor 8 drives the development of type I IFN-producing plasmacytoid dendritic cells. J Immunol 170: 1131–1135. [DOI] [PubMed] [Google Scholar]
- Turcotte K, Gauthier S, Tuite A, Mullick A, Malo D, Gros P. 2005. A mutation in the Icsbp1 gene causes susceptibility to infection and a chronic myeloid leukemia-like syndrome in BXH-2 mice. J Exp Med 201: 881–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tussiwand R, Lee W-L, Murphy TL, Mashayekhi M, KC W, Albring JC, Satpathy AT, Rotondo JA, Edelson BT, Kretzer NM, et al. 2012. Compensatory dendritic cell development mediated by BATF–IRF interactions. Nature 490: 502–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmann L, Hirbec H, Rassendren F. 2010. P2X4 receptors mediate PGE2 release by tissue-resident macrophages and initiate inflammatory pain. EMBO J 29: 2290–2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Zhu J, Smith S, Foldi J, Zhao B, Chung AY, Outtz H, Kitajewski J, Shi C, Weber S, et al. 2012. Notch-RBP-J signaling regulates the transcription factor IRF8 to promote inflammatory macrophage polarization. Nat Immunol 13: 642–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.