Miyazaki et al. show that Id2 and Id3 suppress the development and expansion of innate variant TFH cells by acting upstream of the Hif1a/Foxo/AKT/mTORC1 pathway as well as the c-myc/p19Arf module. Mice depleted for Id2 and Id3 expression developed colitis and αβ T-cell lymphomas, and the transcription signatures of Id2- and Id3-depleted lymphomas revealed similarities to genetic deficiencies associated with Burkitt lymphoma.
Keywords: Id proteins; T-cell quiescence; T-cell lymphoma,; tumor suppressor; FOXO/mTORC1 pathway; c-Myc/p19Arf
Abstract
It is now well established that the E and Id protein axis regulates multiple steps in lymphocyte development. However, it remains unknown how E and Id proteins mechanistically enforce and maintain the naïve T-cell fate. Here we show that Id2 and Id3 suppressed the development and expansion of innate variant follicular helper T (TFH) cells. Innate variant TFH cells required major histocompatibility complex (MHC) class I-like signaling and were associated with germinal center B cells. We found that Id2 and Id3 induced Foxo1 and Foxp1 expression to antagonize the activation of a TFH transcription signature. We show that Id2 and Id3 acted upstream of the Hif1a/Foxo/AKT/mTORC1 pathway as well as the c-myc/p19Arf module to control cellular expansion. We found that mice depleted for Id2 and Id3 expression developed colitis and αβ T-cell lymphomas. Lymphomas depleted for Id2 and Id3 expression displayed elevated levels of c-myc, whereas p19Arf abundance declined. Transcription signatures of Id2- and Id3-depleted lymphomas revealed similarities to genetic deficiencies associated with Burkitt lymphoma. We propose that, in response to antigen receptor and/or cytokine signaling, the E–Id protein axis modulates the activities of the PI3K–AKT–mTORC1–Hif1a and c-myc/p19Arf pathways to control cellular expansion and homeostatic proliferation.
The differentiation of T-lineage cells is initiated in the thymus. Soon after arriving in the thymus, T-cell progenitors initiate TCRβ locus rearrangement, undergo limited expansion, and initiate a T-lineage-specific program of gene expression. This developmental stage is characterized by the lack of expression of the coreceptors CD4 and CD8 and is commonly referred to as the double-negative (DN) stage. Once a functional TCRβ chain has been generated and the pre-TCR is assembled and expressed, DN cells undergo rapid expansion and give rise to CD4+CD8+ double-positive (DP) cells (Carpenter and Bosselut 2010). Upon reaching the DP compartment, thymocytes exit the cell cycle, initiate TCRα locus rearrangement, and undergo positive and negative selection (Singer et al. 2008; Kreslavsky et al. 2010). The lymphoid populations can be segregated into adaptive or innate immune cells. The selection process permits the developmental progression of a selected group of adaptive T-lineage cells that have acquired a TCR with moderate affinity for major histocompatibility complex (MHC) class II (CD4 single positive [CD4SP]) or class I (CD8SP) associated with self-antigens (Klein et al. 2014). On the other hand, innate T-lineage cells are selected by CD1 for invariant natural killer T (iNKT) cells and by MHC-related protein MR1 for mucosal-associated invariant T lymphocytes (MAIT) (Bendelac et al. 2007; Gold and Lewinsohn 2013)
Adaptive B and T cells express an enormously diverse antigen receptor repertoire. They maintain a naïve lymphoid cell state until they encounter invading pathogens, upon which they expand and differentiate into effector cells. Innate lymphoid cells (ILCs) carry germline-encoded receptors or express a limited antigen receptor repertoire and have the potential to rapidly induce cytokine expression (Cerutti et al. 2013; Verykokakis et al. 2014). The innate lymphoid system is comprised of multiple cell types, including natural killer (NK), lymphoid tissue inducer (LTi), type 2 ILCs, and innate-like B and T cells (Diefenbach et al. 2014). The innate-like B- and T-cell compartment consists of marginal zone B cells, iNKT cells, MAIT cells, and subsets of γδ T cells. ILCs act primarily by modulating the activities of adaptive immune cells.
It is now established that a large majority of developmental trajectories in the thymus involve regulation by members of the helix–loop–helix (HLH) family (Rothenberg 2014). These include E proteins as well as Id proteins. Four E proteins have been identified and characterized. They include E12, E47, HEB, and E2-2. E12 and E47 are encoded by the E2A locus and are generated by differential splicing. E protein DNA-binding activity is regulated by the Id gene products, named Id1–4. Id proteins contain an HLH dimerization domain but lack the basic DNA-binding region. They function predominantly by antagonizing the DNA-binding activities of E proteins (Benezra et al. 1990; Lazorchak et al. 2005; Miyazaki et al. 2014).
E protein levels are abundant in T-cell progenitors, where they activate TCRβ V(D)J locus rearrangement and induce the expression of genes encoding for members of the Notch and pre-TCR signaling cascade (Ikawa et al. 2006; Agata et al. 2007). E47 expression declines in resting DP cells and decreases further upon maturating beyond the TCR checkpoint (Engel et al. 2001; Miyazaki et al. 2011). High E2A expression prevents developmental progression, whereas decreasing E2A and HEB levels promote positive selection (Rivera et al. 2000; Jones and Zhuang 2007). HEB acts in the DP compartment to promote the development of NKT cells (D’Cruz et al. 2010). The traversal of these checkpoints in response to pre-TCR, γδ TCR, and αβ TCR signals is facilitated by the induction of Id3 expression. Beyond the pre-TCR, γδ TCR, and αβ TCR checkpoints, Id3 expression is required to maintain the naïve state (Verykokakis et al. 2010, 2014; Miyazaki et al. 2011; Li et al. 2013). The combined activities of Id2 and Id3 are required to promote efficient developmental progression of CD8SP cells (Jones-Mason et al. 2012).
Here we examined how Id2 and Id3 act mechanistically beyond the TCR checkpoint to orchestrate T-cell fate. We found that the activation of Id3 and Id2 expression in DP cells is sequential and that Id2 and Id3 suppressed the development and expansion of innate variant follicular helper T (TFH)-like cells acting in turn to promote the ectopic development of germinal center (GC) B cells. The innate TFH-like cells carried a highly restricted antigen receptor repertoire indicative of a self-renewing population. We identified a genetic network involving the Id–E protein, AKT–FOXO–mTOR, and Myc–p19Arf modules, which orchestrate a self-renewal-specific program of gene expression. Finally, mice depleted for Id2 and Id3 in T cells developed colitis as well as T-cell lymphoma. Collectively, these data point to a regulatory circuitry that underpins the mechanism by which Id2 and Id3 act to antagonize an innate variant TFH-specific program of gene expression, maintain thymocyte quiescence, and suppress the development of lymphoma.
Results
Expression patterns of Id2 and Id3 in positively selected thymocytes
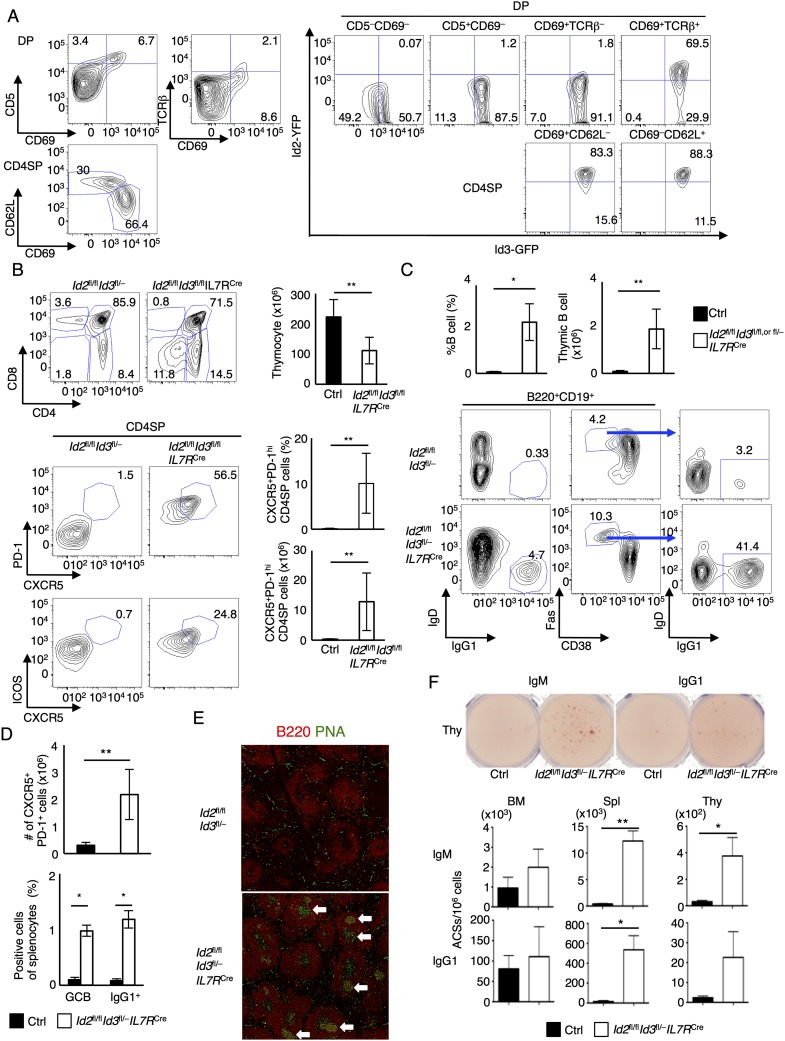
Previous studies have demonstrated that Id3 expression is induced at the pre-TCR checkpoint and further elevated during the positive selection process, whereas Id2 expression is low in positively selected DP cells but elevated in CD4SP or CD8SP cells (Bain et al. 2001; Engel et al. 2001; Miyazaki et al. 2011; Jones-Mason et al. 2012). To examine in greater detail how Id2 and Id3 expression is regulated during positive selection, we used Id2-YFP and Id3-GFP reporter mice (Yang et al. 2011a). Positively selected cells, identified as CD5+CD69− or CD69+TCRβ− DP cells, expressed higher levels of Id3 but did not display significant levels of Id2-YFP (Fig. 1A). Id2 expression was only detectable in TCRβ+ DP cells (Fig. 1A). The majority of mature CD62L+ CD4SP or CD8SP cells displayed abundant levels of Id2 and Id3 expression (Fig. 1A). Collectively, these data indicate that the induction of Id2 and Id3 expression during positive selection is sequential: Id3 expression is activated by TCR signaling in positively selected cells, whereas Id2 expression is induced at a later stage by a separate pathway, which remains to be revealed.
Figure 1.
Development of CXCR5+PD-1+ αβ T cells and IgG1 class-switched B cells in thymi derived from Id2fl/flId3fl/flIL7RCre mice. (A) Flow cytometric analysis of CD69 versus CD5 expression and CD69 versus TCRβ expression gated on CD4+CD8+ DP cells (top left); CD69 versus CD62L expression gated on CD4SP (CD4+CD8−TCRβ+) cells (bottom left); and GFP versus YFP expression gated on CD5−CD69−, CD5+CD69−, CD69+TCRβ−, CD69+TCRβ+ (DP), and CD69+CD62L− and CD69−CD62L+ (CD4SP) cells (right). Numbers in quadrants indicate percentages of cells in each compartment. Data are representative of two independent experiments. (B, top left) Flow cytometric analysis of CD4 versus CD8 expression in total thymocytes derived from 5-wk-old Id2fl/flId3fl/flIL7RCre and Id2fl/flId3fl/− control mice. (Top right) The graph shows the number of total thymocytes of 5-wk-old Id2fl/flId3fl/flIL7RCre and Id2fl/flId3fl/− control mice. The bottom panels show the expression of CXCR5 and PD-1 (middle) and CXCR5 and ICOS (bottom) gated on CD4SP (CD4+CD8−TCRβ+) cells. The absolute number and frequency of gated cells are shown in adjacent panels. Numbers in plots refer to CXCR5+PD-1+ and CXCR5+ICOS+ cells. Data represent the mean ± SD from two independent experiments analyzing four 5-wk-old mice. (C) The top graphs show the percentage and absolute number of B cells (B220+CD19+) in thymi derived from 5-wk-old Id2fl/flId3fl/flIL7RCre and littermate control mice. Flow cytometric analysis of IgG1 versus IgD expression and CD38 versus Fas expression gated on the B220+CD19+ population. The bottom right panel shows IgG1 and IgD expression gated on the CD38−Fashi cells. Data represent the mean ± SD from two independent experiments analyzing four 5-wk-old mice. (D) The absolute number of CXCR5+PD-1+ CD4T cells and frequency of GC (Fas+GL7+) B cells and IgG1 class-switched (IgG1+IgD−) B cells in spleens derived from 5-wk-old Id2fl/flId3fl/flIL7RCre mice. Data represent the mean ± SD from two independent experiments analyzing four 5-wk-old mice. (E) Representative immunofluorescence staining with anti-B220 antibody and PNA in spleens derived from 5-wk-old Id2fl/flId3fl/flIL7RCre mice. Arrows indicate PNA+ GCs. (F) Representative ELISPOT wells using thymocytes (top) and the frequency of IgM- or IgG1-secreting cells in bone marrow (BM), spleen (Spl), and thymus (Thy) derived from 6-mo-old Id2fl/flId3fl/flIL7RCre and littermate control mice (middle and bottom) are shown. (E) Data are representative of two independent experiments with three mice each. (*) P < 0.05; (**) P < 0.01 (Student’s t-test).
Development of TFH-like cells and GC formation in primary and peripheral lymphoid organs derived from Id2fl/flId3fl/flIL7RCre mice
Previous studies have demonstrated key roles of E and Id proteins in enforcing and modulating the pre-TCR and TCR checkpoints (Bain et al. 1999; Engel et al. 2001; D’Cruz et al. 2010; Miyazaki et al. 2011; Jones-Mason et al. 2012). To evaluate the roles of Id2 and Id3 throughout thymocyte development, we used Id2fl/flId3fl/flIL7RCre mice to ablate Id2 and Id3 expression in common lymphoid progenitors (CLPs) (Schlenner et al. 2010; Jones-Mason et al. 2012; Niola et al. 2012). Consistent with previous observations, the CD8SP compartment was virtually absent in Id2fl/flId3fl/flIL7RCre mice (Fig. 1B; Jones-Mason et al. 2012). Additionally, Id2fl/flId3fl/flIL7RCre mice displayed an increase in the total number of DN cells that expressed TCRβ (Supplemental Fig. 1A). Since a previous study showed aberrant development of TFH-like cells in thymi derived from Id3-null mutant mice, we examined Id2fl/flId3fl/flIL7RCre mice for CXCR5, PD-1, and ICOS expression (Miyazaki et al. 2011). We found that a large fraction of CD4SP and DN TCRβ+ as well as iNKT cells expressed CXCR5, PD-1, and ICOS in thymi derived from 5-wk-old Id2fl/flId3fl/flIL7RCre mice (Fig. 1B; Supplemental Fig. 1A,B). CXCR5+PD-1+ TFH-like cells were detectable in Id3−/−, Id3fl/−CD4Cre, and Id3fl/flIL7RCre mice but not in Id2fl/flIL7RCre mice (Supplemental Fig. 1C,D). The TFH-like populations were accompanied by increased numbers of B220+CD19+ thymic B cells, FashiCD38− GC B cells, and IgG1 class-switched cells (Fig. 1C). We found that the segregation of cortical and medullary regions was completely abolished, and B220+ cells were observed throughout the thymi derived from 5-wk-old Id2fl/flId3fl/flIL7RCre mice (Supplemental Fig. 1E). In addition, we noted spontaneous GCs in the spleens derived from Id2fl/flId3fl/flIL7RCre mice, accompanied by fewer naïve CD4 T cells and an increased number of TFH cells (Fig. 1D,E; Supplemental Fig. 2A,B). The elevated number of TFH cells was associated with the development of GC B cells, IgG1 class-switched B cells, plasma blasts, and plasma cells (Fig. 1D; Supplemental Fig. 2C). Furthermore, substantial numbers of IgM- and IgG1-secreting cells were detected in thymi and spleens, but not in the bone marrow, derived from 4- to 6-mo-old Id2fl/flId3fl/flIL7RCre mice (Fig. 1F). Taken together, these data indicate that Id2 and Id3 suppress the development and/or selection of TFH-like cells and GC B cells in primary and peripheral lymphoid organs.
Development of innate TFH-like cells in Id2fl/flId3fl/flIL7RCre mice
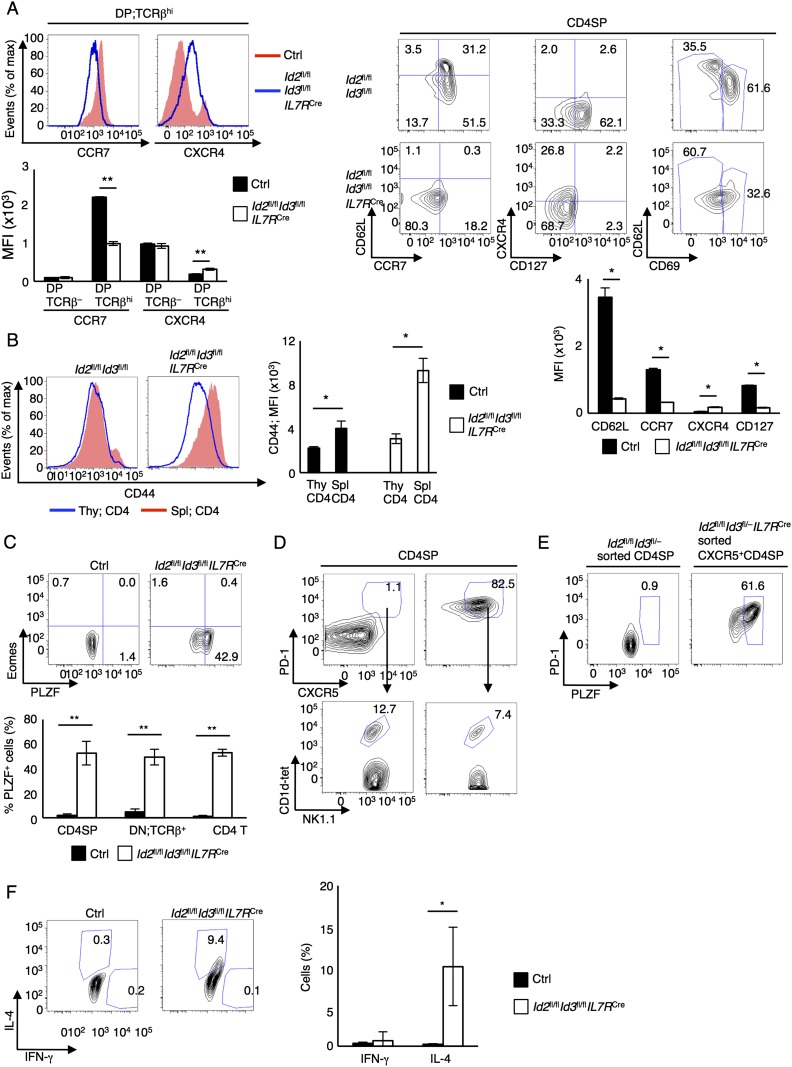
To examine in greater detail the phenotypes associated with the development of TFH-like cells, CD4SP cells were analyzed for the expression of markers associated with maturation and migration. In line with previous studies, we found that TCRβhi DP and CD4SP thymocytes displayed aberrant CCR7, CXCR4, CD62L, and CD69 expression in Id2fl/flId3fl/flIL7RCre mice (Fig. 2A; Jones-Mason et al. 2012). The level of CD44 expression displayed by Id2fl/flId3fl/flIL7RCre CD4SP thymocytes was distinct from that observed for splenic CD4+ T cells, suggesting that the CD4SP cells in the thymus were not derived from recirculating CD4+ T cells (Fig. 2B). Previous studies have demonstrated that a fraction of CD4SP cells derived from Id3−/− thymi aberrantly expressed PLZF or Eomes (Verykokakis et al. 2010, 2013; Miyazaki et al. 2011; Li et al. 2013; D’Cruz et al. 2014). The fraction of thymocytes expressing PLZF was increased in the thymi and spleens derived from Id2fl/flId3fl/flIL7RCre mice, while Eomes expressors were mostly lacking (Fig. 2C). Since PLZF is the transcription factor that is expressed in innate T cells such as iNKT cells, we examined whether the innate TFH-like population was distinct from that of iNKT cells. We found that only a small fraction of CXCR5+ CD4SP cells represented iNKT cells and that the majority of CXCR5+ CD4SP cells expressed PLZF (Fig. 2D,E). Furthermore, we found significantly increased numbers of IL-4- but not IFN-γ-producing CD4SP cells (Fig. 2F). Taken together, these data indicate that depletion of Id2 and Id3 expression at an early developmental stage results in the development of an innate TFH-like population in the thymus.
Figure 2.
Id2 and Id3 suppress the development of PLZF-expressing non-iNKT αβ T cells. (A, left) Flow cytometric analysis of CCR7 and CXCR4 gated on TCRβ+ DP cells derived from 6-wk-old Id2fl/flId3fl/flIL7RCre and littermate control mice. (Bottom left) The graph shows CCR7 and CXCR4 expression in CD69−TCRβ− and TCRβ+ DP cells presented as mean fluorescence intensity (MFI). Flow cytometric analysis of CCR7 versus CD62L, CD127 versus CXCR4, and CD69 versus CD62L gated on CD4SP cells derived from 6-wk-old Id2fl/flId3fl/flIL7RCre and littermate control mice. Numbers adjacent to the outlined areas or in quadrants indicate the percentage of cells in the population that was examined. (Bottom right) The graph shows the CD62L, CCR7, CXCR4, and CD127 expression in CD4SP cells, presented as MFI. Data are representative of one experiment with 6-wk-old mice (mean ± SD; n = 3). (B) Flow cytometric analysis of CD44 expression in CD4SP thymocytes (blue) and splenic CD4 T cells (red) derived from 6- to 8-wk-old Id2fl/flId3fl/flIL7RCre and Id2fl/flId3fl/fl control mice. The right panel shows the expression of CD44 in CD4SP thymocytes and splenic CD4 T cells, presented as MFI. Data are representative of one experiment with 6-wk-old mice (mean ± SD; n = 4). (C, top) Flow cytometric analysis of PLZF versus Eomes expression gated on CD4SP cells derived from Id2fl/flId3fl/flIL7RCre and control thymus. (Bottom) The graph shows the percentage of PLZF-expressing cells in CD4SP, DN;TCRβ+ in the thymus and CD4 T cells in the spleen. Numbers in quadrants indicate the percentages of cells. Data are representative of three independent experiments with 6-wk-old mice (mean ± SD; n = 4 biological replicates). (D) Representative flow cytometric analysis of CXCR5 versus PD-1 expression gated on CD4SP cells and CD1d-tet versus TCRβ expression gated on CXCR5+PD-1+ CD4SP cells derived from 8-wk-old thymus. (E) Flow cytometric analysis of PLZF and PD-1 expression in sorted CXCR5+ CD4SP cells (CD4+CD8−TCRβ+CD1d-tet−CXCR5+) from Id2fl/flId3fl/−IL7RCre thymus and sorted control CD4SP cells. Numbers indicate PLZF+ cells. (F, top) Cytokine expression in CD4SP cells derived from Id2fl/flId3fl/flIL7RCre and control thymi. (Bottom) The graph shows the percentage of IFN-γ+ and IL-4+ cells in CD4SP cells. Data are representative of three independent experiments with 6-wk-old mice (mean ± SD; n = 3 biological replicates). (*) P < 0.05; (**) P < 0.01 (Student’s t-test).
Rapidly expanding innate TFH-like cells in Id2fl/flId3fl/flIL7RCre mice
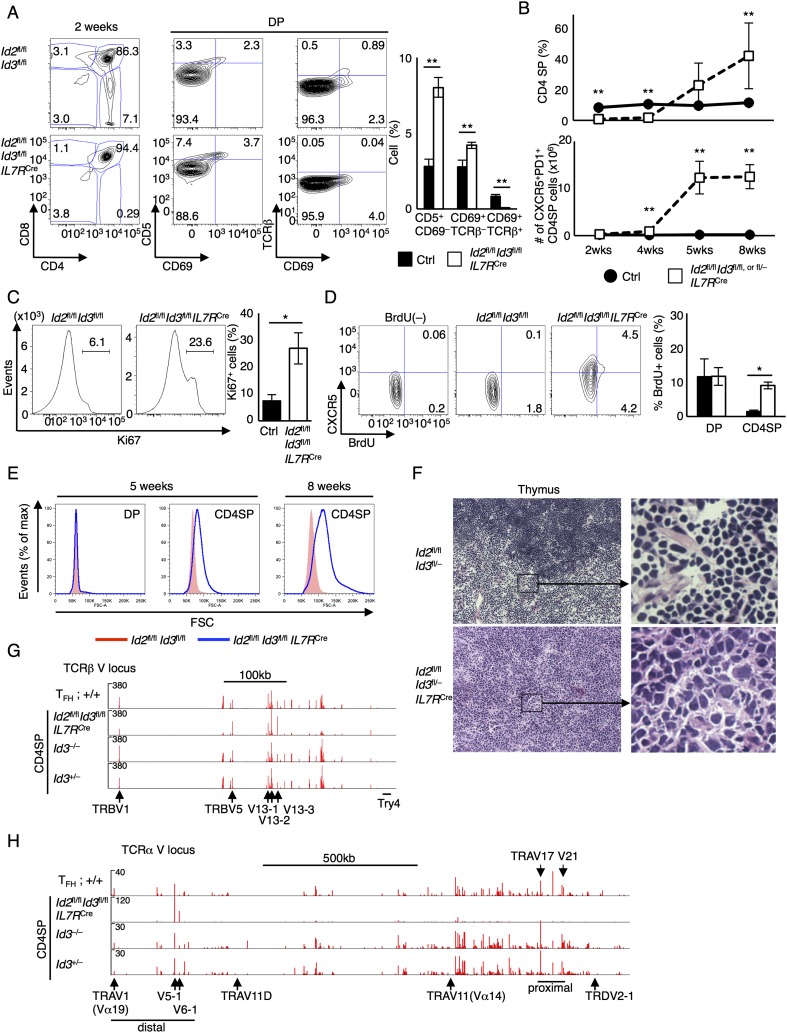
To obtain further insight into the mechanism that underpins the roles of Id2 and Id3 in thymocyte development, Id2fl/flId3fl/flIL7RCre thymi were examined for abnormalities upon aging. We found that at 2 wk after birth, thymi derived from Id2fl/flId3fl/flIL7RCre mice displayed a complete block in positive selection, with a pronounced defect during the CD69+TCRβ−-to-CD69+TCRβhi transition (Fig. 3A). For comparison, we analyzed DP cells derived from TCRα−/− mice for CD5 or CD69 expression. We found that DP cells derived from TCRα−/− thymi did not display either CD69 or CD5 expression (Supplemental Fig. 3A). These data suggest that DP thymocytes that have received a TCR signal required Id2 and Id3 expression to fully differentiate into the TCRβhi stage. However, unlike the complete block in positive selection observed in 2-wk-old mice, we found that in adult (4- to 8-wk-old) mice, CD4SP cells in Id2fl/flId3fl/flIL7RCre mice readily were detectable and continued to expand (Fig. 3B, top; Supplemental Fig. 3B). A significant fraction of the expanding CD4SP populations displayed CXCR5 and PD-1 expression (Fig. 3B, bottom; Supplemental Fig. 3B). The expanding CD4SP population showed relatively high levels of Ki67 expression and BrdU incorporation, whereas proliferating cells in the control CD4SP compartment were barely detectable (Fig. 3C,D). Likewise, DN TCRβ+ cells derived from Id2fl/flId3fl/flIL7RCre mice displayed a substantial increase in the fraction of BrdU+ cells (Supplemental Fig. 3C). Annexin V expression was not affected in these populations (Supplemental Fig. 3D). We also observed a substantial increase in cell size across the CD4SP but not in the DP compartment in Id2fl/flId3fl/flIL7RCre thymocytes (Fig. 3E). Over time, the majority of thymocytes expressed CXCR5 and PD-1 associated with large cell size (Supplemental Fig. 3E). Sections of thymi derived from 6-mo-old Id2fl/flId3fl/flIL7RCre mice showed effacement of the cortico–medullary junction due to proliferation of neoplastic cells with large, hyperchromatic, irregular nuclei and scant cytoplasm (Fig. 3F).
Figure 3.
An expanding population of CXCR5+ cells in thymi derived from Id2fl/flId3fl/flIL7RCre mice. (A) Flow cytometric analysis of CD4 versus CD8 expression in total thymocytes and CD69 versus CD5 expression and CD69 versus TCRβ expression gated on DP cells derived from 2-wk-old Id2fl/flId3fl/flIL7RCre or littermate control mice. The graph shows the frequency of CD5+CD69−, CD69+TCRβ−, and CD69+TCRβ+ cells in the DP compartment. Numbers adjacent to the outlined areas or in quadrants indicate the percentages of cells in the population. Data are representative of one experiment with 2-wk-old mice (mean ± SD; n = 5 [Ctrl] and 3 [Id2fl/flId3fl/flIL7RCre] biological replicates). (B) The frequency of CD4SP cells (top) and the number of CXCR5+PD-1+ CD4SP cells (bottom) are plotted. Data were derived from five independent experiments (mean ± SD; n = 5 or 3 [2-wk], 3 [4-wk], and 4 [5- or 8-wk] biological replicates). (C) Intracellular staining of Ki67 in CD4SP cells derived from 8-wk-old Id2fl/flId3fl/flIL7RCre or littermate control mice. Numbers above lines indicate percentages of Ki67-expressing cells. The graph shows the frequency of Ki67-expressing cells derived from 6- to 8-wk-old Id2fl/flId3fl/flIL7RCre or littermate control mice. Data were derived from two independent experiments (mean ± SD; n = 4 biological replicates). (D) Flow cytometric analysis of BrdU incorporation and CXCR5 expression gated on CD4SP (TCRβ+CD25−) cells derived from 8-wk-old Id2fl/flId3fl/flIL7RCre or littermate control mice. Mice were injected intraperitoneally with 1 mg of PBS (BrdU−) or BrdU twice (24 h and 2 h prior to harvesting). Numbers in quadrants indicate BrdU-incorporated cells. The graph shows the frequency of BrdU+ cells in DP and CD4SP cells. Data were derived from two independent experiments (mean ± SD; n = 4 biological replicates). (E) Flow cytometric analysis of cell size (FSC) in DP and CD4SP cells derived from 5- or 8-wk-old Id2fl/flId3fl/flIL7RCre or littermate control mice. Data were derived from two (5-wk) or four (8-wk) independent experiments. (F) Representative hematoxylin and eosin (H&E) staining of thymi derived from 6-mo-old Id2fl/flId3fl/flIL7RCre or littermate control mice. Original magnification, 200×. Data are representative of three independent experiments. (G,H) RNA-seq data for the TCRα V locus (H) and TCRβ V locus (G). mRNA was isolated from sorted CXCR5+CD44hi CD4 T cells from SRBC-immunized wild-type spleens (TFH) and CD4SP thymocytes derived from 8-wk-old Id2fl/flId3fl/flIL7RCre, Id3−/−, and Id3+/− thymi and analyzed using RNA-seq. Numbers of reads are indicated for each of the tracks. (G,H) Two independent experiments with three mice each. (*) P < 0.05; (**) P < 0.01 (Student’s t-test).
To determine whether the expanding population was derived from a small fraction of selected thymocytes, the TCR repertoires were examined using RNA sequencing (RNA-seq). To this end, RNA was isolated from CD4SP cells from Id3+/−, Id3−/−, and Id2fl/flId3fl/flIL7RCre thymi as well as wild-type TFH cells. We found that Vβ and Vα transcripts associated with wild-type TFH cells and CD4SP thymocytes isolated from Id3+/− and Id3−/− thymi displayed a similar polyclonal Vβ or Vα repertoire (Fig. 3G,H). On the other hand, CD4SP cells derived from Id2fl/flId3fl/flIL7RCre mice were enriched for TRBV5, TRBV13-2, and TRBV13-3 as well as distally located TRAV5-1 and TRAV6-1 transcripts (Fig. 3G,H). We also observed a substantial enrichment for transcripts encoding 3′ Jα elements (Supplemental Fig. 4A). Vα and Vβ gene usage observed in Id2fl/flId3fl/flIL7RCre CD4SP cells, however, was not invariant, as described for iNKT or MAIT cells, but rather differed between the various Id2fl/flId3fl/flIL7RCre mice that were examined (Supplemental Fig. 4B–D; Le Bourhis et al. 2011). To exclude the possibility that the oligclonal TCRα and TCRβ repertoires were caused by aberrant TCR recombination at the DP stage, we examined the CD69−TCRβ− DP cells from Id2fl/flId3fl/flIL7RCre mice for TCRβ and TCRα rearrangements. No obvious differences were detected for TCRβ and TCRα loci in Id2fl/flId3fl/flIL7RCre DP cells as compared with control cells (Supplemental Fig. 5A,B). Collectively, these observations indicate that Id2 and Id3 suppress the oligoclonal expansion of innate variant TFH-like populations.
Id2 and Id3 suppress the expansion of innate variant TFH-like cells beyond the TCR checkpoint
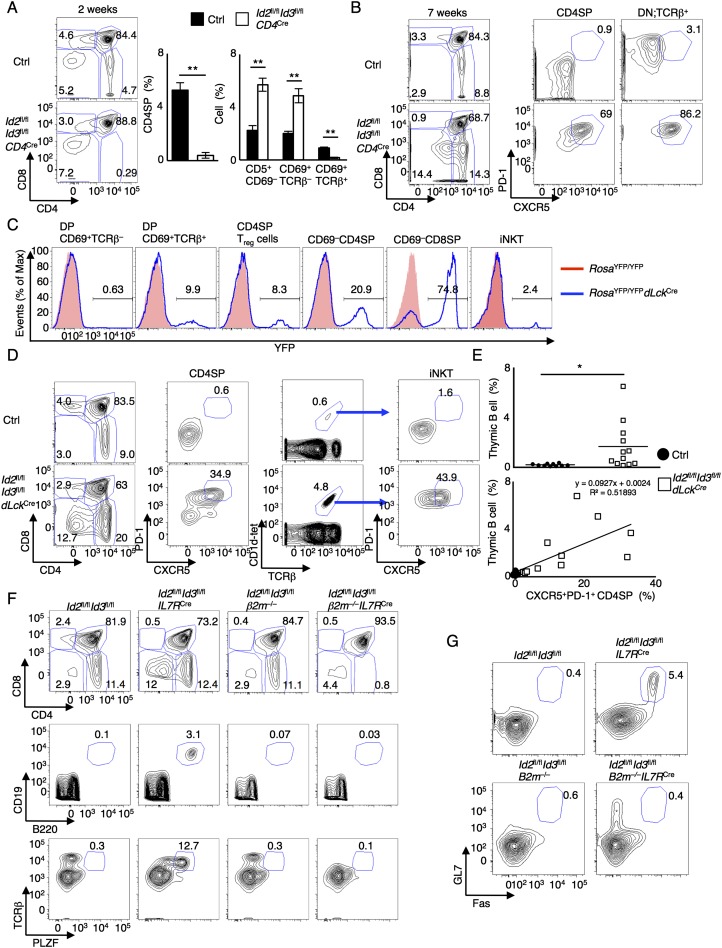
To deplete Id2 and Id3 expression in T-lineage cells, Id2fl/flId3fl/flCD4-Cre mice were generated. Similar to as described above for Id2fl/flId3fl/flIL7RCre mice, at 2 wk after birth, positive selection was severely impaired in Id2fl/flId3fl/flCD4Cre thymi (Fig. 4A). However, 7-wk-old Id2fl/flId3fl/flCD4Cre mice displayed CD4SP, DN TCRβhi, and iNKT populations that expressed high levels of CXCR5, PD-1, ICOS, and PLZF expression (Fig. 4B; Supplemental Fig. 6A,B). Again, these populations were associated with elevated numbers of thymic B cells as well as IgG1 class-switched GC B cells (Supplemental Fig. 6C).
Figure 4.
Id2 and Id3 suppress the expansion of innate αβ T cells beyond the TCR checkpoint. (A) Flow cytometric analysis of CD4 versus CD8 expression derived from 2-wk-old Id2fl/flId3fl/flCD4Cre or littermate control mice. Numbers indicate the percentage of cells in each population. Graphs show the frequency of CD4SP cells in total thymocytes and CD5+CD69−, CD69+TCRβ,− and CD69+TCRβ+ cells gated on the DP compartment. Data are representative of one experiment with 2-wk-old mice (mean ± SD; n = 5 biological replicates). (B) Flow cytometric analysis of CD4 versus CD8 expression in total thymocytes and CXCR5 versus PD-1 expression gated on CD4SP cells (CD4+CD8−TCRβ+CD1d-tet−) or DN;TCRβ+ cells (CD4−CD8−TCRβ+CD1d-tet−) derived from 7-wk-old Id2fl/flId3fl/flCD4Cre or littermate control mice. Data were obtained from two independent experiments. (C) Representative flow cytometric analysis of YFP expression gated on each compartment derived from RosaYFP/YFP (red) or RosaYFP/YFPdLckCre (blue) mice. Numbers above the lines indicate percentages of YFP-expressing cells. Data were derived from two independent experiments. (D) Flow cytometric analysis of CD4 versus CD8 expression (left) and TCRβ versus CD1d-tet expression (middle right) in total thymocytes and CXCR5 versus PD-1 expression gated on CD4SP (CD4+CD8−TCRβ+CD1d-tet−) cells (middle left) or iNKT (CD1d-tet+TCRβ+) cells (right) derived from 10-wk-old Id2fl/flId3fl/fldLckCre or littermate control mice. Data were derived from four independent experiments. (E) Graphs show the frequencies of thymic B (B220+CD19+) cells and the correlation between thymic B cells and CXCR5+PD-1+ CD4SP cells. Data were derived from seven independent experiments. Ten-week-old to 16-wk-old mice were analyzed. (mean ± SD; n = 10 [Ctrl] and 12 [Id2fl/flId3fl/fldLckCre] biological replicates). (F) Flow cytometric analysis of CD4 versus CD8 expression, B220 versus CD19 expression, and PLZF versus TCRβ expression in total thymocytes derived from Id2fl/flId3fl/fl, Id2fl/flId3fl/flIL7RCre, Id2fl/flId3fl/flβ2m−/−, and Id2fl/flId3fl/flβ2m−/−IL7RCre mice. (G) Fas and GL7 expression gated on B220+CD19+ splenic B cells derived from Id2fl/flId3fl/fl, Id2fl/flId3fl/flIL7RCre, Id2fl/flId3fl/flβ2m−/−, and Id2fl/flId3fl/flβ2m−/−IL7RCre mice. Numbers adjacent to the outlined areas indicate the percentage of cells in each population. Data are representative of three independent experiments. (*) P < 0.05; (**) P < 0.01 (Student’s t-test).
Since recent studies have demonstrated a critical role for Id2 and Id3 in regulatory T (Treg) cells, it remained possible that the innate TFH-like population developed because of systemic inflammatory conditions (Miyazaki et al. 2014). To exclude this possibility and investigate the role of Id2 and Id3 in thymocyte development beyond the TCR checkpoint, Id2fl/flId3fl/fldLckCre mice were generated (Zhang et al. 2005). We found that Cre-mediated deletion in dLckCre mice was initiated beyond the CD69+TCRβ+ DP stage (Fig. 4C; Supplemental Fig. 7A). We found that a substantial fraction of peripheral Treg cells did not exhibit Cre activity, indicative of Treg cells expressing wild-type levels of Id2 and Id3 in dLckCre mice (Supplemental Fig. 7A). As observed for Id2fl/flId3fl/flIl7RCre and Id2fl/flId3fl/flCD4Cre mice, in thymi derived from Id2fl/flId3fl/fldLckCre mice, we found increased percentages of CD4SP cells and iNKT cells that expressed CXCR5, PD-1, and PLZF and a variant but limited TCRVα and TCRVβ repertoire (Fig. 4D; Supplemental Fig. 7B–D). Again, we noted an increase in the percentages of thymic B cells and a significant relationship between the proportions of thymic B cells and CXCR5+PD-1+ CD4SP cells (R2 = 0.51893) (Fig. 4E). Notably, we detected transcripts initiated from the first exon of the Id2 and Id3 genes in sorted CXCR5−PD-1− CD4SP cells but not in sorted CXCR5+PD-1+ CD4SP cells from identical Id2fl/flId3fl/fldLckCre mice, consistent with the notion that the aberrant activation of CXCR5 and PD-1 expression was caused by depletion of Id2 and Id3 expression (Supplemental Figure 7E). Taken together, these data indicate that Id2 and Id3 expression is required to suppress the development and expansion of an innate variant TFH-like population beyond the TCR checkpoint.
To determine how Id2- and Id3-depleted innate variant TFH-like cells are selected, β2m−/−Id2fl/flId3fl/flIL7RCre mice were generated. We found that the development of the innate variant TFH cell population required β2-microglobulin expression (Fig. 4F). Specifically, CD4SP and CD8SP thymocytes were barely detectable in thymi derived from 8- to 10-wk-old β2m−/−Id2fl/flId3fl/flIL7RCre mice (Fig. 4F). Likewise, CXCR5+PD-1+ CD4SP cells, DN TCRβ+ cells, PLZF-expressing T cells, and thymic B cells were virtually absent in β2m−/−Id2fl/flId3fl/flIL7RCre thymi (Fig. 4F; Supplemental Fig. 8A). Depletion of β2m expression in Id2fl/flId3fl/flIl7RCre mice also abolished the development of GC and IgG1 class-switched B cells (Fig. 4G; Supplemental Fig. 8A). Thymi derived from Id2fl/flId3fl/flβ2m−/−IL7RCre mice showed a normal thymic cortex but abnormal medulla (Supplemental Fig. 8B). These data indicate that selection of Id2- and Id3-depleted innate variant TFH cells requires β2-microglobulin expression and that this population is closely associated with the development of GCs even in the absence of immunization.
Gene expression signature of innate variant TFH-like cells
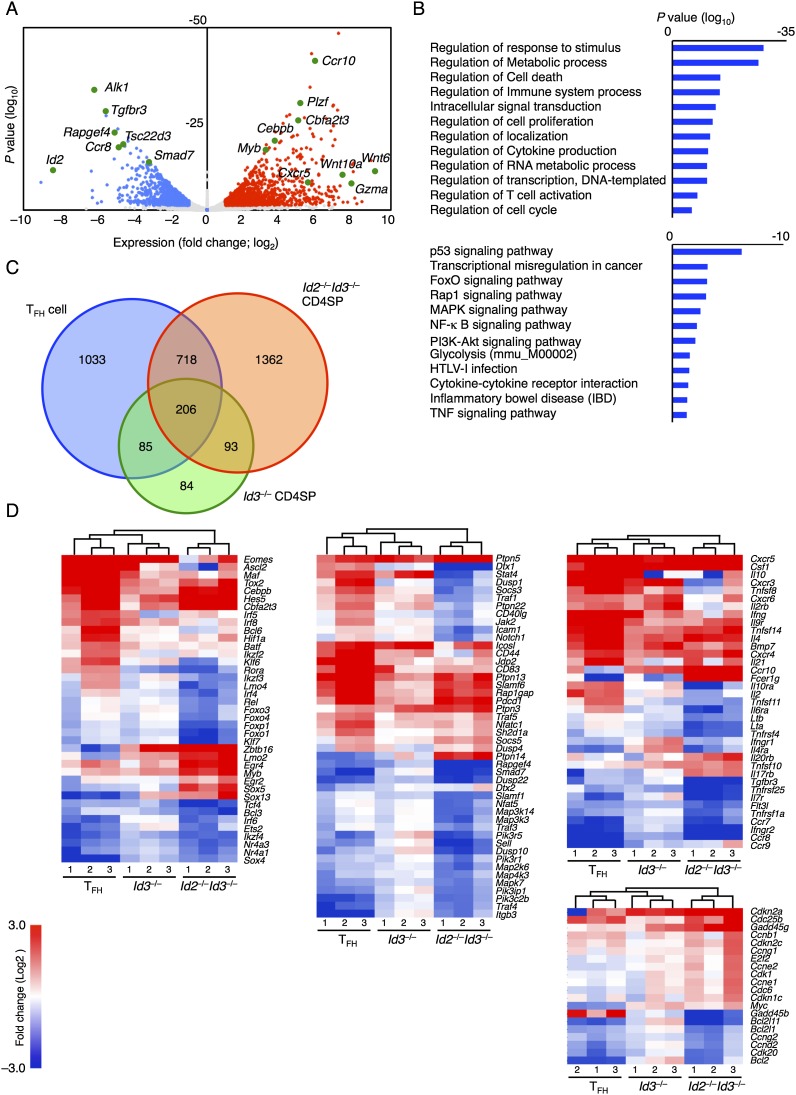
To determine how Id2 and Id3 suppressed the oligoclonal expansion of an innate variant TFH-like population, CD4SP cells derived from control and Id2fl/flId3fl/flIL7RCre thymi were isolated and analyzed by RNA-seq. Two-thousand-three-hundred-seventy-nine genes were differentially expressed (greater than twofold, P < 0.05; 1291 up-regulated; 1088 down-regulated) in Id-depleted CD4SP cells (Fig. 5A). Prominent among those genes were Cxcr5, Ccr10, Wnt10a, Gzma, Myb, Cebpb, Tgfbr3, and Smad7 (Fig. 5A). Gene ontology (GO) analysis revealed that a large fraction of differentially expressed transcripts encoded for proteins associated with metabolism, cytokine production, RNA metabolism, T-cell activation, and cell cycle progression (Fig. 5B). Furthermore, Kyoto Encyclopedia of Genes and Genomics (KEGG) pathway analysis revealed p53 and genes associated with the PI3K–AKT, MAPK, and Rap1 pathways as well as genes involved in the suppression of inflammatory bowel disease as being affected by depletion of Id2 and Id3 expression (Fig. 5B). Next, we compared the transcription signatures of Id2/Id3-depleted CD4SP thymocytes with Id3−/− CD4SP and peripheral TFH cells. We found that 924 genes showed overlap between TFH and Id2- and Id3-deficient CD4SP cells as compared with wild-type CD4SP thymocytes. Two-hundred-ninety-nine genes showed overlap between CD4SP cells isolated from Id3−/− and Id2fl/flId3fl/flIL7RCre thymi, whereas the expression of 1362 genes was modulated only in innate TFH-like cells when compared with control CD4SP cells (Fig. 5C; Supplemental Fig. 9A). Transcription signatures derived from Id3−/− CD4SP cells showed an intermediate pattern when comparing TFH and CD4SP cells derived from Id2fl/flId3fl/flIL7RCre thymi (Fig. 5C). The expression patterns of FcεR1 and IL-17rβ in control and mutant thymocytes were validated using flow cytometry (Supplemental Fig. 9B). Notably, genes associated with TFH cell function were altered across the three populations, including Maf, Batf, Foxo1, Foxp1, Sh2d1a, Cxcr5, Il4, and Il21 but not Bcl6 and Ascl2 (Fig. 5D; Ma et al. 2012; Wang et al. 2014; Xiao et al. 2014). As predicted, the expression levels of genes associated with an innate-specific program of gene expression were also elevated, including Zbtb16 (Plzf), Myb, and Egr2 (Kovalovsky et al. 2008; Hu et al. 2010; Seiler et al. 2012). Finally, consistent with an expanding population of Id2- and Id3-depleted CD4SP thymocytes, the expression patterns of genes associated with cell cycle regulation were affected (Fig. 5D). Taken together, these observations indicate that gene expression patterns of Id-depleted CD4SP thymocytes overlap, albeit partially, with that of TFH and innate T-lineage cells.
Figure 5.
Transcription signature of innate TFH-like cells. (A) Volcano plot of RNA-seq analysis of mRNA expression in sorted CD4SP thymocytes derived from 8-wk-old littermate control or Id2fl/flId3fl/llIL7Rcre mice. Red dots represent genes up-regulated in Id2fl/flId3fl/flIL7RCre CD4SP cells (more than twofold, P < 0.05, calculated by raw count value). Blue dots represent genes down-regulated in Id2fl/flId3fl/flIL7RCre CD4SP cells (more than twofold, P < 0.05). Gray dots represent genes whose expression was not significantly altered. (B) Clusters of genes whose expression was modulated in Id2- and Id3-depleted CD4SP thymocytes were identified using GO terms (top) and KEGG pathways (bottom). P-values are shown. (C) RNA-seq data of mRNA expression isolated from CXCR5+CD44hi CD4T cells from SRBC-immunized wild-type spleen (TFH) and CD4SP thymocytes derived from 8-wk-old Id2fl/flId3fl/flIL7RCre (Id2−/−Id3−/− CD4SP), Id3−/− (Id3−/− CD4SP), and control thymi. Expression was normalized to control CD4SP cells. The Venn diagram shows quantification of genes significantly changed in TFH cells, Id3−/− CD4SP cells, and Id2fl/flId3fl/llIL7Rcre CD4SP cells compared with control CD4SP cells. (D) Heat maps are presented to visualize differences in gene expression patterns for TFH cells, CD4SP cells sorted from Id3−/− thymocytes, and CD4SP cells sorted from IL7R-Cre;Id2F/F;Id3F/F mice as compared with CD4SP control thymocytes. Bars above the heat maps indicate similarities between the different samples presented in each of the lanes. Differentially expressed genes are shown only for those that displayed greater than twofold differences (P < 0.05) as compared with control CD4SP thymocytes. The heat maps represent different ontology groups, including genes encoding for factors that regulate transcription (left), signal transduction (middle), and cell cycle (bottom right). (Top right) The heat map displays the KEGG pathway for chemokines, cytokines, and associated receptors.
A genetic circuitry links Id2 and Id3 and the AKT–FOXO–mTORC1 axis
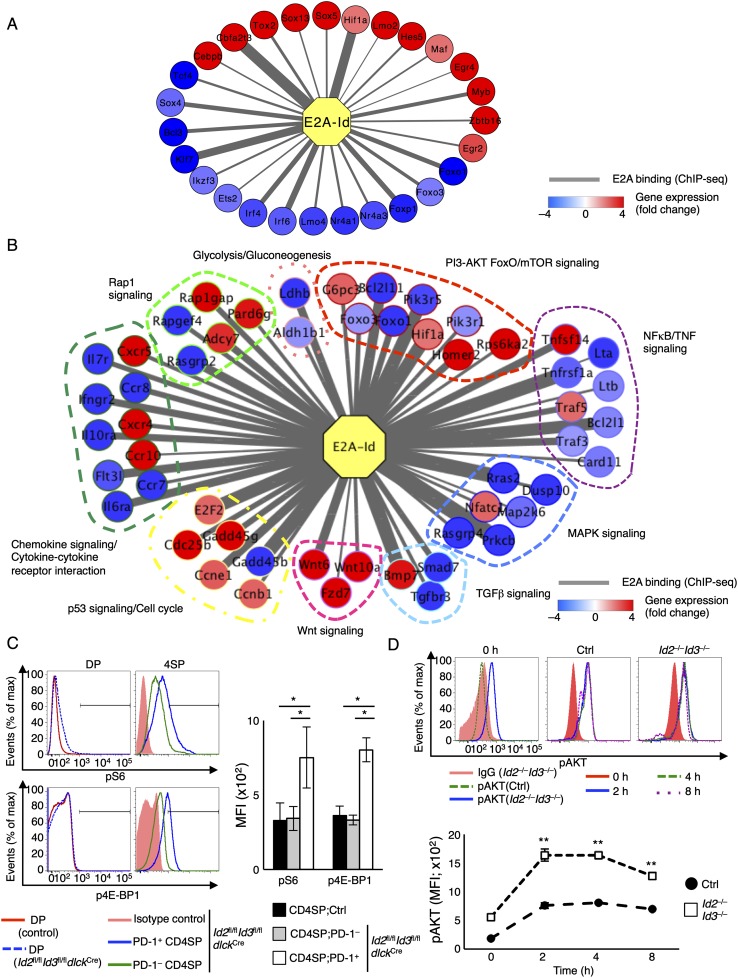
In previous studies, we had identified a spectrum of target genes regulated by the Id–E protein module in T-lineage cells, including Cxcr5, Il10, Hif1a, Ikzf3, Myb, Il10ra, E2f2, and Bmp7 (Miyazaki et al. 2014). Here we found additional target genes that are directly regulated by the E–Id protein axis, including Foxo1, Foxo3, Foxp1, Slamf6, Il7r, Il6ra, Cxcr4, Tgfbr3, Wnt10a, Cdc25b, Gadd45b, Bcl2l1, Bcl2l11, and Rps6ka2 (Supplemental Fig. 9C; data not shown). To examine for molecular pathways or modules associated with depletion of Id2 and Id3 expression, transcription signatures derived from Id-depleted CD4SP thymocytes and E protein occupancy were linked into a common framework using Cytoscape software (Fig. 6A). Using this approach, we identified an ensemble of E protein targets—including Foxo1, Foxo3 and Foxp1—associated with the maintenance of a naïve and quiescent state. Additionally, a cluster of target genes was identified closely linked with an innate-like transcription signature, including Myb, Plzf, Egr2, and Sox13 (Fig. 6A; Melichar et al. 2007; Kerdiles et al. 2009; Feng et al. 2011; Hedrick et al. 2012). Finally, we found that clusters of genes associated with the PI3K–AKT/FoxO/mTOR, NFκB/TNF, MAPK, Rap1, cytokine/chemokine, and p53/cell cycle pathways were targeted by the E and Id protein module (Fig. 6B).
Figure 6.
Id2, Id3, and the FOXO/mTOR axis. (A) Regulatory network that connects the activity of E proteins and an ensemble of transcriptional regulators in Id2- and Id3-depleted CD4SP thymocytes into a common framework. E2A ChIP-seq (chromatin immunoprecipitation [ChIP] combined with deep sequencing) data were derived from E2A-deficeint T-cell lymphoma cells transduced with E47 (Lin et al. 2010). The widths of the connectors reflect relative peak scores of E2A occupancy. Colors reflect higher (red) or lower (blue) gene expression levels in Id2fl/flId3fl/flIL7RCre CD4SP cells as compared with control CD4SP thymocytes. (B) Networks that link distinct pathways, as defined by KEGG analysis, to E2A occupancy. (C) Flow cytometric analysis of phosphorylation of S6 (pS6) and 4E-BP1 (p4E-BP1) in DP and CD4SP cells for the indicated populations derived from control and Id2fl/flId3fl/fldLckCre mice. The graph shows the level of phosphorylated S6 and 4E-BP1, presented as MFI. Data are representative of three independent experiments (mean ± SD; n = 3 biological replicates). (D) Flow cytometric analysis of phosphorylation of AKT. Sorted CD4SP cells (CD4+CD8−TCRβ+CD1d-tet−) derived from the Id2fl/flId3fl/flIL7RCre thymus were stimulated with plate-coated anti-CD3ε (2 μg/mL) and anti-CD28 (5 μg/mL) antibodies and analyzed at the indicated time points for pAKT expression. (*) P < 0.05; (**) P < 0.01 (Student’s t-test).
To validate these findings, we examined activation of the mTOR and AKT pathways in developing thymocytes using flow cytometry. We found significant increases in phospho-S6 and phospho-4E-BP1 levels in Id2- and Id3-depleted PD-1+ CD4SP cells derived from Id2fl/flId3fl/fldLckCre thymi as compared with control thymi (Fig. 6C). As predicted, phospho-S6 and phospho-4E-BP1 levels were not altered in PD-1− CD4SP or DP cells derived from Id2fl/flId3fl/fldLckCre versus control mice (Fig. 6C). To determine whether the E–Id protein axis also regulates the activity of the mTORC pathway to control the expansion of innate γδ T cells, Id3−/− Vγ1.1+ cells were examined for the expression of phospho-S6 and phospho-4E-BP1. Indeed, Id3−/− Vγ1.1+ cells displayed significantly increased levels of phopho-S6 and phospho-4E-BP1 (Supplemental Fig. 10). In contrast, Id3−/− Vγ2+ cells were not associated with elevated levels of phopho-S6 and phospho-4E-BP1 (Supplemental Fig. 10). Next, we examined the phosphorylation status of AKT (pAKT) in sorted control and Id-depleted CD4SP thymocytes. We found that resting control CD4SP cells lacked detectable levels of pAKT (Fig. 6D, left panel). However, activating TCR signaling readily elevated levels of pAKT (Fig. 6D, middle and right panels). Notably, Id2- and Id3-depleted CD4SP cells exhibited pAKT expression in the absence of TCR stimulation, and TCR-mediated signaling resulted in even higher levels of pAKT (Fig. 6D, bottom). Taken together, these data indicate that the E–Id protein axis modulates the PI3K–AKT–FOXO–mTORC1 pathway at multiple steps.
Id2 and Id3 suppress the development of αβ T-cell lymphomas
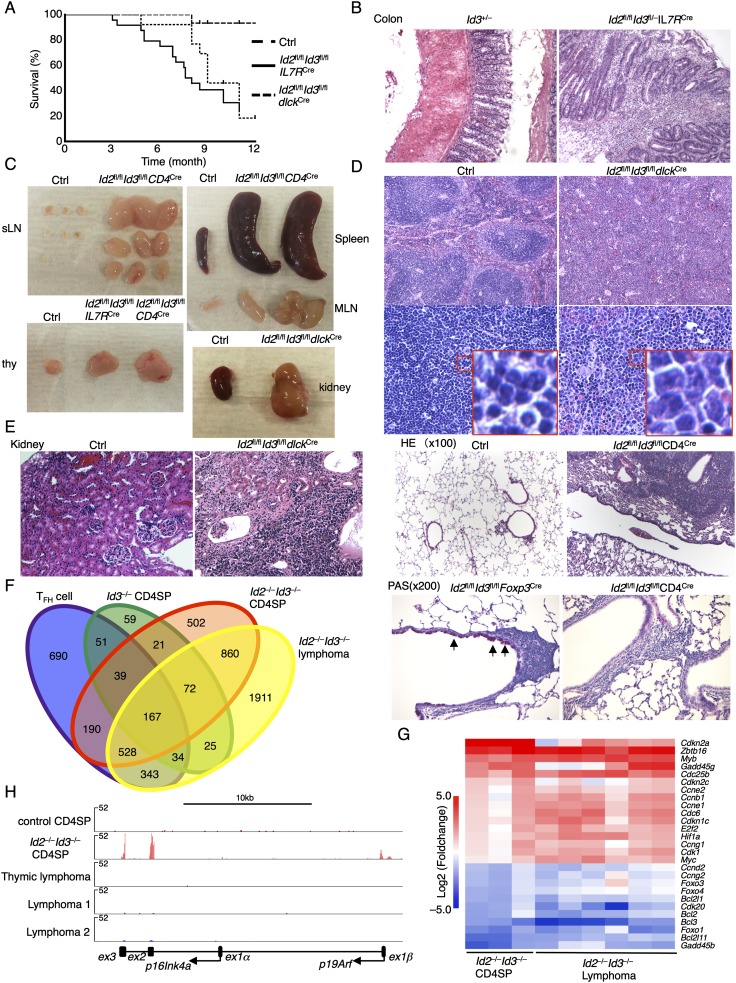
Previous studies have demonstrated that the development of Burkitt lymphoma is closely associated with mutations across the Id3 HLH region, that forced expression of Id2 in a murine model of BCR–ABL interferes with the development of chronic myeloid leukemia, and that Id3-deficient mice develop γδ T-cell lymphomas (Ko et al. 2008; Li et al. 2010; Schmitz et al. 2012). To determine whether the combined loss of Id2 and Id3 expression in T-lineage cells leads to the development of lymphoma, aged Id2fl/flId3fl/flIL7RCre mice were monitored for signs of distress. We found that a substantial fraction of 6- to 8-mo-old Id2fl/flId3fl/flIL7RCre mice exhibited ruffled fur, hunched posture, and rectal prolapse (data not shown). The majority of Id2fl/flId3fl/flIL7RCre or Id2fl/flId3fl/fldLckCre mice died within 1 yr (Fig. 7A). Histological analysis showed moderate to severe colitis in 6- to 8-mo-old Id2fl/flId3fl/flIL7RCre, Id2fl/flId3fl/flCD4Cre, and Id2fl/flId3fl/fldLckCre mice (Fig. 7B; Supplemental Fig. 11A). In addition to colitis, we noted splenomegaly and large subcutaneous and/or mesenteric lymph nodes in 21 out of 41 8- to 14-mo-old Id2fl/flId3fl/flIL7RCre, Id2fl/flId3fl/flCD4Cre, and Id2fl/flId3fl/fldLckCre mice (Fig. 7C). Four out of 21 Id2fl/flId3fl/flIL7RCre, Id2fl/flId3fl/flCD4Cre, and Id2fl/flId3fl/fldLckCre mice also developed thymic lymphoma (Fig. 7C; Supplemental Fig. 11B). Histopathological analysis of the peripheral lymphoid organs displayed a disorganized architecture, such as blurring of the demarcation between red and white pulp in the spleen, with infiltrating monomorphic lymphoid cells containing large polymorphic nuclei and scant cytoplasm (Fig. 7D; Supplemental Fig. 11C). The infiltrate in the lungs derived from Id2fl/flId3fl/flCD4Cre mice was composed of neoplastic nucleated cells (Fig. 7E). We note that lymphocyte infiltration was not caused by inflammatory disease, since there was no evidence of elevated mucin levels in the bronchial epithelium (lack of periodic acid Schiff [PAS]-positive goblet cells) (Fig. 7E). The lymphomas that developed in lymph nodes and spleens were mainly composed of TCRβ+CD3+ T-lineage cells associated with a CD8lowCD4−, CD8−CD4− or CD8−CD4+ phenotype (Supplemental Fig. 11E). As expected, a large fraction of these cells showed CXCR5 and PD-1 expression (Supplemental Fig. 11E). Finally, we found that lymphoma cells were highly malignant, as evidenced by metastasis across the liver (17 out of 21), kidney (three out of 10), and lung (10 out of 21) tissues (Fig. 7C,E; Supplemental Fig. 11B,D). Collectively, these observations indicate that depletion of Id2 and Id3 in T-lineage cells leads to increased levels of morbidity caused by the development of lymphoma and/or inflammatory disease.
Figure 7.
Id2 and Id3 suppress the development of T-cell lymphoma. (A) Kaplan-Meier curve of a survival plot of Id2fl/flId3fl/flIL7RCre, Id2fl/flId3fl/fldLckCre, and littermate control mice. (Id2fl/flId3fl/flIL7RCre, n = 24; Id2fl/flId3fl/fldLckCre, n = 14; control, n = 16). (B) Representative H&E staining of colons derived from 6-mo-old Id3+/− or Id2fl/flId3fl/−IL7RCre mice. Original magnification, 100×. (C) Representative images of subcutaneous lymph nodes (sLN), thymi, spleens, mesenteric LNs (MLN), and kidneys derived from 8- to 12-mo-old control, Id2fl/flId3fl/flIL7RCre, Id2fl/flId3fl/flCD4Cre, and Id2fl/flId3fl/fldLckCre mice are shown. (D) Representative H&E stainings of spleens derived from control and Id2fl/flId3fl/fldLckCre mice are displayed. Original magnifications: top, 100×; bottom, 400×. (E) Representative H&E stainings of kidneys (left) and lungs (top right) derived from control, Id2fl/flId3fl/fldLckCre, and Id2fl/flId3fl/flCD4Cre mice are shown. The kidney section was derived from metastasis of lymphoma cells as shown in C. (Bottom right) PAS stainings of the lungs from Id2fl/flId3fl/flFoxp3Cre and Id2fl/flId3fl/flCD4Cre mice are shown. Arrows indicate mucus-producing goblet cells. Original magnifications: kidney, 200×; lung (H&E), 100×; lung (PAS), 200×. (F) The Venn diagram shows overlaps among TFH cells, Id3−/− CD4SP cells, Id2−/−Id3−/− CD4SP cells, and Id2−/−Id3−/− lymphoma cells as compared with control CD4SP cells, as shown in Figure 5C. (G) Heat map for selected significantly differentially expressed genes that are differentially expressed in Id2- and Id3-depleted CD4SP thymocytes as well as lymphomas. Clusters of genes associated with cell cycle and tumor suppression are indicated. (H) Visualized RNA-seq data across the Cdkn2a locus, presented in reads per million reads aligned (RPM). Arrows indicate transcription start site of p19Arf and p16Ink4a gene and direction of transcription.
The c-Myc and p19Arf axis and the development of Id2- and Id3-deficient lymphomas
To gain insight into the mechanism that underpins the development of lymphoma in Id2- and Id3-deficient mice, transcription signatures were analyzed from a set of lymphomas using RNA-seq. We found that Id2- and Id3-deficient lymphomas revealed transcription profiles that overlapped with those derived from Id2- and Id3-deficient CD4SP cells (Fig. 7F). Specifically, ∼2100 genes were differentially expressed in Id2- and Id3-deficient lymphomas versus Id2- and Id3-depleted CD4SP cells (Supplemental Fig. 12A,B). The majority of isolated lymphomas expressed a limited TCR repertoire, arguing against polyclonal expansion (Supplemental Fig. 12C,D). We found that the differences in transcription profiles between lymphomas and CD4SP cells were closely associated with genes involved in metabolism, proliferation, and the immune response (Supplemental Fig. 12E). As predicted, we found substantial decreased levels of Foxo1 and Foxo3 as well as an elevated abundance of c-Myc and Hif1a (Fig. 7G). Conspicuous among the spectrum of aberrantly expressed genes was the Cdkn2a locus. Notably, Cdkn2a transcript abundance was high in Id2fl/flId3fl/flIL7RCre CD4SP cells but low in lymphoma cells (Fig. 7G). The Cdkn2a locus encodes for two tumor suppressors: p16Ink4a and p19Arf. p19Arf is activated by c-Myc and has been demonstrated to suppress lymphomagenesis (Kamijo et al. 1997; Zindy et al. 2003). We found that p19Arf but not p16Ink4a transcript abundance was elevated in the self-renewing innate variant TFH-like population (Fig. 7H). In contrast, p19Arf transcript levels were virtually undetectable in Id-deficient lymphomas (Fig. 7H). Taken together, these data point to a regulatory circuitry that maintains thymocyte quiescence and provide a mechanism involving c-myc and p19Arf that underpins the development of T-cell lymphoma in Id2- and Id3-deficient T cells.
Discussion
Previous studies have demonstrated that positive selection is enforced by the E and Id protein module (Bain et al. 1999; Jones and Zhuang 2007; Jones-Mason et al. 2012). Here we examined how the Id proteins orchestrate thymocyte selection. Based on this analysis, we propose two key steps that involve Id2 and Id3 during the positive selection process. The first step involves the induction of Id3 expression by TCR-mediated signaling. The second step involves the induction of Id2 expression in cells that have already received a TCR signal. We demonstrate here that Id2- and Id3-depleted thymocytes were not able to pass this second step of selection, with the exception of a slowly expanding population of cells that was characterized by an innate-like transcription signature. Innate T cells, including iNKT, T-CD4T, and MAIT cells, are selected by a mechanism that is distinct from that of conventional CD4SP thymocytes (Li et al. 2005; Lee et al. 2010; Alonzo and Sant’Angelo 2011; Gold and Lewinsohn 2013). Specifically, iNKT and MAIT cells are MHC class I-like-restricted, involving innate TCR signaling. Additionally, iNKT and T-CD4 T cells are selected by DP thymocytes rather than by thymic epithelial cells. Hence, we propose that differences in TCR-mediated signaling—conventional or innate-mediated TCR signaling—affect the duration and/or periodicity of Id3 and/or Id2 transcription and that CD4 T cells selected by MHC class II in thymic epithelial cells but not innate CD4 T cells require Id2 and Id3 expression. Differences in the strength, periodicity, and timing of Id2 and Id3 expression would then instruct progenitors to commit to either the adaptive or innate T-cell lineage.
We found that Id2- and Id3-depleted positively selected thymocytes express a TFH-like program of gene expression. Particularly intriguing was the decline of Foxo1 and Foxp1 abundance in Id-depleted CD4SP thymocytes. Foxo1 and Foxp1 are well-characterized suppressors of the TFH cell fate (Hedrick et al. 2012; Wang et al. 2014; Xiao et al. 2014). These data bring into question how Id proteins and Foxo1 and Foxp1 are linked. We found E2A-bound sites across regulatory regions associated with the Foxo1 and Foxp1 loci. Hence, we suggest that in CD4SP thymocytes, the E proteins act as transcriptional repressors that interfere with Foxo1 and Foxp1 transcription. High levels of Id2 and Id3 antagonize the DNA-binding activity of E proteins, relieving Foxo1 and Foxp1 from E2A/HEB-mediated repression. Elevated levels of Foxo1 and Foxp1 in turn would prevent the premature activation of a TFH lineage-specific program of gene expression (Supplemental Fig. 13A).
A notable feature of our findings is the development of an innate variant TFH cell population in mice depleted for Id2 and Id3 expression. Are these cells genuine TFH cells? We noted overlap but also significant differences in transcription signatures upon comparing TFH and Id-depleted innate TFH cells. Notably, Bcl6 and Ascl2 expression, closely associated with a TFH-specific program of gene expression (Crotty 2014), were not modulated, at least transcriptionally, in Id-depleted CD4SP thymocytes. Despite these differences, we found that Id-depleted innate variant TFH cells secreted high levels of IL-4, required interaction with MHC class I-like molecules, and their presence was closely associated with the spontaneous development of GCs. Hence, we suggest that these cells represent a distinct subset of TFH cells (Supplemental Fig. 13B).
A key aspect of the findings reported here involves a slowly expanding population of innate TFH-like cells. How do Id2 and Id3 regulate thymocyte quiescence? We found that multiple pathways known to maintain lymphocyte quiescence were affected by depletion of Id2 and Id3 expression. Prominent among these was the FOXO–mTOR module. The roles for the FoxO proteins in maintaining thymocyte quiescence and acting as tumor suppressors are well documented (Paik et al. 2007; Kerdiles et al. 2009; Hedrick et al. 2012). Similarly, spontaneous activation of the mTORC1 pathway in Tsc1-deficient T cells leads to an increase in cell size and loss of quiescence, phenotypes that are equivalent to that described here for Id2- and Id3-depleted CD4SP thymocytes (Yang et al. 2011b). As mentioned previously, the entire ensemble of Foxo loci, including Foxo1, Foxo3, and Foxo4, appears to be controlled by the E–Id protein module. Likewise, the AKT–FOXO–mTOR pathway was activated in Id2- and Id3-depleted CD4SP thymocytes as well as in Id3-deficient innate γδ T cells. Multiple levels of regulation by the E–Id axis appear to be involved here: (1) modulation of Foxo transcript levels, (2) activation of the AKT pathway plausibly mediated by changes in the expression of an ensemble of PTPN phosphatases that are modulated upon depletion of Id2 and Id3, and (3) activation of the mTORC pathway in part by induction of Rps6ka2 expression. We suggest that this form of regulation is not restricted to innate variant TFH and a subset of Vγ1.1 γδ T cells. Rather, we propose that the regulation of the PI3K–FOXO–mTOR pathway by the E–Id axis is a general mechanism that underpins the homeostasis of lymphoid progenitors and self-renewing committed B and T lymphoid cells.
Finally, we found that c-Myc and p19Arf levels were elevated in Id-depleted CD4SP thymocytes. p19Arf is a well-known tumor suppressor that is regulated by c-Myc (Lowe and Sherr 2003). Previous studies have demonstrated that c-myc expression is directly regulated by E proteins (Schwartz et al. 2006). Hence, we suggest that loss of Id expression leads to elevated c-Myc abundance, which in turn leads to the induction of p19Arf expression. We speculate that cellular expansion upon depletion of Id2 and Id3 expression is attenuated by the induction of p19Arf expression. This then would lead to a population that slowly self-renews. How do lymphomas develop in Id2- and Id3-depleted thymocytes? We speculate that, through mechanisms yet to be determined, p19Arf expression is inactivated in a single progenitor, releasing the brakes and ultimately leading to the development of a monoclonal αβ T-cell lymphoma (Supplemental Fig. 13C; von Boehmer 2004).
The mechanisms that underpin the development of αβ T-cell lymphomas in Id2- and Id3-depleted mice overlap with those observed in human Burkitt lymphoma and a murine model of Burkitt lymphoma (Sander et al. 2012; Schmitz et a. 2012). It is now established that the development of Burkitt lymphoma is closely associated with high levels of c-Myc expression and mutations localized across the HLH region of Id3 (Love et al. 2012; Schmitz et al. 2012). Likewise, we found that Id2- and Id3-depleted murine T-cell lymphomas expressed relatively high levels of c-Myc expression. There are also similarities between the two sets of lymphomas as they relate to the PI3K pathway. Burkitt lymphomas are associated with increased PI3K signaling and display an activated AKT pathway (Sander et al. 2012; Schmitz et al. 2012). We found that Id-deficient CD4 T cells displayed decreased abundance of Foxo1/3 expression as well as activated AKT and mTORC1 pathway. Upon examining transcription signatures in human T-cell lymphomas, we found that changes in Id2, Foxo1, and Foxo3 abundance were significantly associated with the development of human T-cell lymphoma, (Supplemental Fig. 14; Piccaluga et al. 2007). Finally, since a very high fraction of aged mice display symptoms of inflammatory disease, it is conceivable that chronic inflammation contributes to the development of T-cell lymphoma in Id2- and Id3-deficient mice similarly to as described for viral infections associated with the development of human lymphomas.
Materials and methods
Mice
C57BL/6, Id3f/f, Id2f/f, Id3−/−, Id2YFP/+, Id3GFP/+, IL7RαCre, CD4Cre, dLckCre, and ROSAYFP/YFP mice were bred and housed in specific pathogen-free conditions in accordance with the Institutional Animal Care and Use Guidelines of the University of California at San Diego.
Flow cytometry and cell sorting
Single-cell suspensions from the bone marrow, thymus, and spleen were stained with the following: FITC-labeled, PE-labeled, APC-labeled, APC-Cy7-labeled, Pacific Blue-labeled, Alexa Fluor 700-labeled, Alexa Fluor 780-labeled, PerCP-Cy5.5-labeled, PE-Cy7-labeled, or biotin-labeled monoclonal antibodies, which were purchased from BD PharMingen, including CD11c (HL3), CD44 (IM7), CXCR5 (2G8), IgG1 (A85-1), CD95 (Jo-2), CD138 (281-2), GL-7, Ki67 (B56), and phospho-AKT (M89-61). CD8 (53-6.7), CD19 (ID3), CD38 (90), CD62L (MEL-14), CD44 (IM7), CD69 (H1.2F3), CD127 (A7R34), B220 (RA3-6B2), Mac1 (M1/70), Gr1 (RB6-8C5), Nk1.1 (PK136), Ter119 (TER119), TCRβ (H57), TCRγδ (GL3), PD-1 (J43), ICOS (7E.17G9), IgD (11-26), CCR7 (4B12), CXCR4 (2B11), TBR2 (Dan11mag), phosph-S6 (supk43k), phosph-4E-BP1 (V3NTY24), and AnnexinV were purchased from eBioscience. FceR1 (MAR-1), CD3ε (2C11), CD4 (GK1.5), CD8 (53-6.7), CD11b (M1/70), CD24 (M169), CD25 (PC61), PD-1 (RMP1-30), and F4/80 (BM8) were obtained from Biolegend. CCR9 was purchased from R&D Systems. PLZF (clone D9) was obtained from Santa Cruz Biotechnology. PE-conjugated CD1d tetramer was generously provided by the National Institutes of Health Tetramer Core Facility (mCD1d/PBS57). Biotinylated antibodies were labeled with streptavidin-conjugated Qdot-665, or Qdot-605 (Invitrogen). Clone 2.4 G2 anti-CD32:CD16 (eBioscience) was used to block FcRs. Dead cells were removed from analysis and sorting by staining with propidium iodide (PI) (Sigma-Aldrich). BrdU incorporation was performed using a BD BrdU flow kit. Annexin V staining was performed using an eBioscience Annexin V apoptosis detection kit APC. Treg cell staining kit (eBioscience) was used for intracellular staining of PLZF and Foxp3 and Eomes detection. Intracellular staining kit (eBioscience) was used for phosph-S6/4E-BP1 staining. BD Cytofix fixation buffer and BD Phosflow perm buffer III were used for phospho-AKT staining. Samples were collected on a LSRII (BD Biosciences) and were analyzed with FlowJo software (Tree Star). Cells were sorted on a FACSAria. For intracellular staining of IFN-γ and IL-4, MACS-purified CD4SP and DN TCRβ+ cells from thymi were cultured in the presence of phorbol 12-myristate 13-acetate (PMA) plus ionomycin (4 h) and Golgi stop (2 h). After culture, cells were stained with anti-CD4, CD8, TCRβ, and CD3ε antibodies. Intracellular staining was performed with the BD Biosciences Cytofix/Cytopermkit.
RNA-seq analysis
Total RNA and the library preparations were described previously (Miyazaki et al. 2014). The strand-specific RNA-seq libraries were sequenced with a HiSeq 2500 sequencer (Illumina). Alignment and trimming of reads were performed using the OSA algorithm against the mm10 murine genome reference in Arraystudio (Omicsoft) (Hu et al. 2012). RNA transcripts were quantified using RSEM methods (http://deweylab.biostat.wisc.edu/rsem) as implemented in Arraystudio (Omicsoft). Abundance values (counts) were normalized and compared with calculated P-values using DESeq (http://www-huber.embl.de/users/anders/DESeq). Genes whose abundance values were <10 in all samples were removed. MultiExperiment Viewer software was used to generate heat maps and for hierarchical clustering. GO analyses and visualization files were generated using HOMER (http://biowhat.ucsd.edu/homer), and read pile-ups were visualized using the University of California at Santa Cruz Genome Browser.
Histology
Tissues were fixed in 4% paraformaldehyde (Electron Microscopy Sciences). Fixed tissues were embedded in paraffin and sliced, followed by haematoxylin and eosin (H&E) staining.
ELISPOT
Cells were cultured overnight at 37°C on 96-well MultiScreen-HA filter plates (Millipore) precoated with goat anti-murine Ig(H+L) capture antibodies (Southern Biotechnology Associates [SBA]). Spots were visualized with goat anti-murine IgM or IgG1 antibodies conjugated to HRP, and color was developed by 3-amino-9-ethyl carbazole (Sigma-Aldrich).
Statistical analyses
P-values were calculated with the two-tailed Student’s test for two-group comparison, as applicable, with Microsoft Excel software.
Data access
RNA-seq data have been deposited at Gene Expression Omnibus under accession number GSE64779.
Supplementary Material
Acknowledgments
We thank the National Institutes of Health (NIH) Tetramer Facility for providing us with CD1d tetramers. We thank L. Deng and the University of California at San Diego Histology Core for performing the histology. We thank D. Broide, M. Suzukawa, C.Y. Yang, and C. Katayama for technical advice, and Rodrigo Hess for TCRα−/− thymi. We thank A. Goldrath (University of California at San Diego) for Id2-YFP mice, Yuan Zhuang (Duke University) for Id3fl/fl mice, A. Lasorella (Columbia University) for Id2fl/fl mice, and N. Kileen for dLCKCre mice. This work was partly funded by the National Resource for Network Biology (P41 GM103504) and the San Diego Center for Systems Biology (P50 GM085764). The studies were supported by grants awarded to H.-R.R. (European Research Council advanced grant no. 233074 and DFG-SFB 938-project L) and C.M. (NIH RO1CA078384 and 1PO1AI102853 and the Genomics Core associated with 1PO1AI102853).
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.255331.114.
Freely available online through the Genes & Development Open Access option.
References
- Agata Y, Tamaki N, Sakamoto S, Ikawa T, Masuda K, Kawamoto H, Murre C. 2007. Regulation of T cell receptor β gene rearrangements and allelic exclusion by the helix–loop–helix protein, E47. Immunity 27: 871–884. [DOI] [PubMed] [Google Scholar]
- Alonzo ES, Sant’Angelo DB. 2011. Development of PLZF-expressing innate T cells. Curr Opin Immunol 23: 220–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bain G, Quong MW, Soloff RS, Hedrick SM, Murre C. 1999. Thymocyte maturation is regulated by the activity of the helix–loop–helix protein, E47. J Exp Med 190: 1605–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bain G, Cravatt CB, Loomans C, Alberola-Ila J, Hedrick SM, Murre C. 2001. Regulation of the helix–loop–helix proteins, E2A and Id3, by the Ras–ERK MAPK cascade. Nat Immunol 2: 165–171. [DOI] [PubMed] [Google Scholar]
- Bendelac A, Savage PB, Teyton L. 2007. The biology of NKT cells. Annu Rev Immunol 25: 297–336. [DOI] [PubMed] [Google Scholar]
- Benezra R, Davis RL, Lockshon D, Turner DL, Weintaub H. 1990. The protein Id: a negative regulator of helix–loop–helix DNA binding proteins. Cell 61: 49–59. [DOI] [PubMed] [Google Scholar]
- Carpenter AC, Bosselut R. 2010. Decision checkpoints in the thymus. Nat Immunol 11: 666–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerutti A, Cols M, Puga I. 2013. Marginal zone B cells: virtues of innate-like antibody-producing lymphocytes. Nat Rev Immunol 13: 118–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotty S. 2014. T follicular helper cell differentiation, function and roles in disease. Immunity 41: 529–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Cruz LM, Knell J, Fujimoto JK, Goldrath AW. 2010. An essential role for the transcription factor HEB in thymocyte survival, Tcra rearrangement and the development of natural killer T cells. Nat Immunol 11: 240–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Cruz LM, Stradner MH, Yang CY, Goldrath AW. 2014. E and Id proteins influence invariant NKT cell sublineae differentiation and proliferation. J Immunol 192: 2227–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diefenbach A, Colonna M, Koyasu S. 2014. Development, differentiation, and diversity of innate lymphoid cells. Immunity 41: 354–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel I, Johns C, Bain G, Rivera RR, Murre C. 2001. Early thymocyte development is regulated by modulation of E2A protein activity. J Exp Med 194: 733–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng X, Wang H, Takata H, Day TJ, Willen J, Hu H. 2011. Transcription factor Foxp1 exerts essential cell-intrinsic regulation of the quiescence of naive T cells. Nat Immunol 12: 544–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold MC, Lewinsohn DM. 2013. Co-dependents: MR1-restricted MAIT cells and their antimicrobial function. Nat Rev Microbiol 11: 14–19. [DOI] [PubMed] [Google Scholar]
- Hedrick SM, Hess Michelini R, Doedens AL, Goldrath AW, Stone EL. 2012. FOXO transcription factors throughout T cell biology. Nat Rev Immunol 12: 649–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu T, Simmons A, Yuan J, Bender TP, Alberola-Ila J. 2010. The transcription factor c-Myb primes CD4+CD8+ immature thymocytes for selection into the iNKT lineage. Nat Immunol 11: 435–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Ge H, Newman M, Liu K. 2012. OSA: a fast and accurate alignment tool for RNA-seq. Bioinformatics 28: 1933–1934. [DOI] [PubMed] [Google Scholar]
- Ikawa T, Kawamoto H, Goldrath AW, Murre C. 2006. E proteins and Notch signaling cooperate to promote T cell lineage specification and commitment. J Exp Med 203: 1329–1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones ME, Zhuang Y. 2007. Acquisition of a functional T cell receptor during T lymphocyte development is enforced by HEB and E2A transcription factors. Immunity 27: 860–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones-Mason ME, Zhao X, Kappes D, Lasorella A, Iavarone A, Zhuang Y. 2012. E protein transcription factors are required for the development of CD4+ lineage T cells. Immunity 36: 348–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamijo T, Zindy F, Roussel MF, Quelle DE, Downing JR, Ashmun RA, Grosveld G, Sherr CJ. 1997. Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell 91: 649–659. [DOI] [PubMed] [Google Scholar]
- Kerdiles YM, Beisner DR, Tinoco R, Dejean AS, Castrillon DH, DePinho RA, Hedrick SM. 2009. Foxo1 links homing and survival of naive T cells by regulating L-selectin, CCR7 and interleukin 7 receptor. Nat Immunol 10: 176–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein L, Kyewski B, Allen PM, Hogquist KA. 2014. Positive and negative selection of the T cell repertoire: what thymocytes see (and don’t see). Nat Rev Immunol 14: 377–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko J, Patel N, Kawamoto H, Frank O, Rivera RR, Van Etten RA, Murre C. 2008. Suppression of E-protein activity interferes with the developmen of BCR-ABL-mediated myeloproliferative disease. Proc Natl Acad Sci 105: 12967–12972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovalovsky D, Uche OU, Eladad S, Hobbs RM, Yi W, Alonzo E, Chua K, Eidson M, Kim HJ, Im JS, et al. 2008. The BTB-zinc finger transcriptional regulator PLZF controls the development of invariant natural killer T cell effector functions. Nat Immunol 9: 1055–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreslavsky T, Gleimer M, von Boehmer H. 2010. αβ versus γδ lineage choice at the first TCR-controlled checkpoint. Curr Opin Immunol 22: 185–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazorchak A, Jones ME, Zhuang Y. 2005. New insights into E-protein function in lymphocyte development. Trends Immunol 26: 334–338. [DOI] [PubMed] [Google Scholar]
- Le Bourhis L, Guerri L, Dusseaux M, Martin E, Soudais C, Lantz O. 2011. Mucosal-associated invariant T cells: unconventional development and function. Trends Immunol 32: 212–218. [DOI] [PubMed] [Google Scholar]
- Lee YJ, Jeon YK, Kang BH, Chung DH, Park CG, Shin HY, Jung KC, Park SH. 2010. Generation of PLZF+ CD4+ T cells via MHC class II-dependent thymocyte–thymocyte interaction is a physiological process in humans. J Exp Med 207: 237–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Kim MG, Gourley TS, McCarthy BP, Sant’Angelo DB, Chang CH. 2005. An altermate pathway for CD4 T cell development: thymocyte-expressed MHC class II selects a distinct T cell population. Immunity 23: 375–386. [DOI] [PubMed] [Google Scholar]
- Li J, Maruyama T, Zhang P, Konkel JE, Hoffman V, Zamarron B, Chen W. 2010. Mutation of inhibitory helix–loop–helix protein Id3 causes γδ T cell lymphoma. Blood 116: 5615–5621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Wu D, Jiang N, Zhuang Y. 2013. Combined deletion of Id2 and Id3 genes reveals multiple roles for E proteins in invariant NKT cell development and expansion. J Immunol 191: 5052–5064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YC, Jhunjhunwala S, Benner C, Heinz S, Welinder E, Mannson R, Sigvardsson M, Hagman J, Espinoza CA, Dutkowski J, et al. 2010. A global network of transcription factors, involving E2A, EBF1 and Foxo1, that orchestrates B cell fate. Nat Immunol 11: 635–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love C, Sun Z, Jima D, Li G, Zhang J, Miles R, Richards KL, Dunphy CH, Choi WW, Srivastava G, et al. 2012. The genetic landscape of mutations in Burkitt lymphoma. Nat Genet 44: 1321–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe SW, Sherr CJ. 2003. Tumor suppression by Ink4a–Arf: progress and puzzles. Curr Opin Genet Dev 13: 77–83. [DOI] [PubMed] [Google Scholar]
- Ma CS, Deenick EK, Batten M, Tangye SG. 2012. The origins, function, and regulation of T follicular helper cells. J Exp Med 209: 1241–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melichar HJ, Narayan K, Der SD, Hiraoka Y, Gardiol N, Jeannet G, Held W, Chambers CA, Kang J. 2007. Regulation of γδ versus αβ T lymphocyte differentiation by the transcription factor SOX13. Science 315: 230–233. [DOI] [PubMed] [Google Scholar]
- Miyazaki M, Rivera RR, Miyazaki K, Lin YC, Agata Y, Murre C. 2011. The opposing roles of the transcription factor E2A and its antagonist Id3 that orchestrate and enforce the naive fate of T cells. Nat Immunol 12: 992–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki K, Miyazaki M, Murre C. 2014. The establishment of B versus T cell identity. Trends Immunol 35: 205–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niola F, Zhao X, Singh D, Castano A, Sullivan R, Lauria M, Nam HS, Zhuang Y, Benezra R, Di Bernardo D, et al. 2012. Id proteins synchronize stemness and anchorage to the niche of neural stem cells. Nat Cell Biol 14: 477–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paik JH, Kollipara R, Chu G, Ji H, Xiao Y, Ding Z, Miao L, Tothova Z, Horner JW, Carrasco DR, et al. 2007. FoxOs are lineage-restricted redundant tumor suppressors and regulate endothelial cell homeostasis. Cell 128: 309–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccaluga PP, Agostinelli C, Califano A, Rossi M, Basso K, Zupo S, Went P, Klein U, Zinzani PL, Baccarani M, et al. 2007. Gene expression analysis of peripheral T cell lymphoma, unspecified, reveals distinct profiles and new potential therapeutic targets. J Clin Invest 117: 823–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera RR, Johns CP, Quan J, Johnson RS, Murre C. 2000. Thymocyte selection is regulated by the helix-loop-helix inhibitor protein, Id3. Immunity 12: 17–26. [DOI] [PubMed] [Google Scholar]
- Schlenner SM, Madan V, Busch K, Tietz A, Laufle C, Costa C, Blum C, Fehling HJ, Rodewald HR. 2010. Fate mapping reveals separate origins of T cells and myeloid lineages in the thymus. Immunity 32: 426–436. [DOI] [PubMed] [Google Scholar]
- Schwartz R, Engel I, Fallahi-Sichani M, Petrie HT, Murre C. 2006. Gene expression patterns define novel roles for E47 in cell cycle progression, cytokine-meidaed signaling and T lineage development. Proc Natl Acad Sci 103: 9976–9981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenberg EV. 2014. Transcriptional control of early T and B cell developmental choices. Annu Rev Immunol 32: 283–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander S, Calado DP, Srinivasan L, Kochert K, Zang B, Rosolowski M, Rodig SJ, Holzman K, Stilgenbauer S, Siebert R, et al. 2012. Synergy between PI3K signaling and MYC in Burkitt lymphomagenesis. Cancer Cell 22: 167–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz R, Young RM, Ceribelli M, Jhavar S, Xiao W, Zhang M, Wright G, Shaffer AL, Hodson DJ, Buras E, et al. 2012. Burkitt lymphoma pathogenesis and therapeutic targets from structural and functional genomics. Nature 490: 116–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiler MP, Mathew R, Liszewski MK, Spooner CJ, Barr K, Meng F, Singh H, Bendelac A. 2012. Elevated and sustained expression of the transcription factors Egr1 and Egr2 controls NKT lineage differentiation in response to TCR signaling. Nat Immunol 13: 264–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer A, Adoro S, Park JH. 2008. Lineage fate and intense debate: myths, models and mechanisms of CD4− versus CD8-lineage choice. Nat Rev Immunol 8: 788–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verykokakis M, Boos MD, Bendelac A, Kee BL. 2010. SAP protein-dependent natural killer T-like cells regulate the development of CD8+ T cells with innate lymphocyte characteristics. Immunity 33: 203–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verykokakis M, Krishnamoorth V, Iavarone A, Lasorella A, Sigvardsson M, Kee BL. 2013. Essential functions for ID proteins at multiple checkpoints in invariant NKT cell development. J Immunol 191: 5973–5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verykokakis M, Zook EC, Kee BL. 2014. ID’ing innate and innate-like lymphoid cells. Immunol Rev 261: 177–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Boehmer H. 2004. Selection of the T-cell repertoire: receptor-controlled checkpoints in T-cell development. Adv Immunol 84: 201–238. [DOI] [PubMed] [Google Scholar]
- Wang H, Geng J, Wen X, Bi E, Kossenkov AV, Wolf AI, Tas J, Choi YS, Takata H, Day TJ, et al. 2014. The transcription factor Foxp1 is a critical negative regulator of the differentiation of follicular helper T cells. Nat Immunol 15: 667–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao N, Eto D, Elly C, Peng G, Crotty S, Liu YC. 2014. The E3 ubiquitin ligase Itch is required for the differentiation of follicular helper T cells. Nat Immunol 15: 657–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CY, Best JA, Knell J, Yang E, Sheridan AD, Jesionek AK, Li HS, Rivera RR, Lind KC, D’Cruz LM, et al. 2011a. The transcriptional regulators Id2 and Id3 control the formation of distinct memory CD8+ T cell subsets. Nat Immunol 12: 1221–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang K, Neale G, Green DR, He W, Chi H. 2011b. The tumor suppressor Tsc1 enforces quiescence of naive T cells to promote immune homeostasis and function. Nat Immunol 12: 888–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang DJ, Wang Q, Wei J, Baimukanova G, Buchholz F, Stewart AF, Mao X, Killeen N. 2005. Selective expression of the cre recombinase in late-stage thymocytes using the distal promoter of the Lck gene. J Immunol 174: 6725–6731. [DOI] [PubMed] [Google Scholar]
- Zindy F, Williams RT, Baudino TA, Rehg JE, Skapek SX, Cleveland JL, Roussel MF, Sherr CJ. 2003. Arf tumor suppressor promoter monitors latent oncogenic signals in vivo. Proc Natl Acad Sci 100: 15930–15935. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.