The orphan nuclear receptor TLX regulates neural stem cell self-renewal in the adult brain and functions primarily as a transcription repressor through recruitment of Atrophin corepressors, which bind to TLX via a conserved peptide motif termed the Atro box. Zhi et al. report crystal structures of the human and insect TLX ligand-binding domain in complex with Atro box peptides. Mutations that weaken the TLX–Atrophin interaction compromise the repressive activity of TLX. In addition, mutations of corresponding residues in other members of this class of receptors diminish their repressor activities.
Keywords: COUP, PNR, SHP, TLX
Abstract
The orphan nuclear receptor TLX regulates neural stem cell self-renewal in the adult brain and functions primarily as a transcription repressor through recruitment of Atrophin corepressors, which bind to TLX via a conserved peptide motif termed the Atro box. Here we report crystal structures of the human and insect TLX ligand-binding domain in complex with Atro box peptides. In these structures, TLX adopts an autorepressed conformation in which its helix H12 occupies the coactivator-binding groove. Unexpectedly, H12 in this autorepressed conformation forms a novel binding pocket with residues from helix H3 that accommodates a short helix formed by the conserved ALXXLXXY motif of the Atro box. Mutations that weaken the TLX–Atrophin interaction compromise the repressive activity of TLX, demonstrating that this interaction is required for Atrophin to confer repressor activity to TLX. Moreover, the autorepressed conformation is conserved in the repressor class of orphan nuclear receptors, and mutations of corresponding residues in other members of this class of receptors diminish their repressor activities. Together, our results establish the functional conservation of the autorepressed conformation and define a key sequence motif in the Atro box that is essential for TLX-mediated repression.
Nuclear receptors form a family of ligand-inducible transcription factors that regulate a wide array of events in human physiology and pathology (Mangelsdorf et al. 1995; Evans and Mangelsdorf 2014). A subset of nuclear receptors does not have known cognate ligands and, as such, is termed orphan nuclear receptors (Mangelsdorf et al. 1995; Evans and Mangelsdorf 2014). When orphan nuclear receptors execute their physiological roles, several members mainly function as transcriptional activators (activator class of orphan nuclear receptors), while several others primarily serve as transcriptional repressors (repressor class of orphan nuclear receptors) (Zhang and Dufau 2004; Mullican et al. 2013). The last two decades have witnessed the determination of most nuclear receptor structures, which revealed the typical three-dimensional (3D) architecture of nuclear receptors and the mechanistic basis for their ligand-regulated action (Brelivet et al. 2012; Gallastegui et al. 2015). However, the knowledge remains limited regarding the ligand-free action of orphan nuclear receptors, which unusually adopt diverse strategies in assembling coregulators and regulating target gene expression (Mullican et al. 2013).
Previous studies have shown that ligand-activated receptors use the last helix (H12, also known as activation function-2 helix or AF-2 helix) in the ligand-binding domain (LBD) to recruit coactivators that contain LXXLL (L = leucine, X = any amino acid) motifs (Gampe et al. 2000; Li et al. 2005; Zhi et al. 2012) to open up repressive chromatin structures and induce transcriptional activation of target genes (Xu et al. 1999b). Dislodgement of helix H12 from the active conformation by either deletion or antagonist binding opens up an extended groove for interactions with the longer LXXXIXXX (L/I) motifs present in nuclear receptor corepressors such as NCoR and SMRT (Xu et al. 2002; Phelan et al. 2010). However, these studies could not provide insight into the molecular basis for ligand-independent repression by the repressor class of orphan nuclear receptors because they do not rely on the traditional nuclear receptor corepressors NCoR and SMRT to drive gene inhibition. Rather, they recruit other types of corepressors to execute their repressive activity (Bavner et al. 2002; Boulias and Talianidis 2004; Wang et al. 2006; Zhang et al. 2006; Sanyal et al. 2007; Takezawa et al. 2007; Yokoyama et al. 2008). Crystal structures of several repressive orphan nuclear receptors determined to date reveal a common structural feature, termed the autorepressed conformation (Wang et al. 2003; Flaig et al. 2005; Kruse et al. 2008; Sablin et al. 2008; Zhou et al. 2011; Tan et al. 2013; Zhi et al. 2014), in which helix H12 is packed into the canonical coactivator-binding groove, thus preventing both coactivators and corepressors from binding to this site. While the physiological relevance of the autorepressed conformation for repressor orphan nuclear receptors remains an open question, disruption of the autorepressed conformation by deletions or mutations of H12 impaired their abilities to repress gene transcription (Lalli et al. 1997; Bavner et al. 2002; Sun et al. 2007; Tan et al. 2013). Taken together, these earlier results suggest that the formation of an autorepressed conformation may be involved in corepressor recruitment.
TLX, a homolog of Drosophila tailless (Yu et al. 1994), is a prominent member of the repressor class of orphan nuclear receptors that also includes the photoreceptor-specific nuclear receptor (PNR), chicken ovalbumin upstream promoter transcription factors (COUP-TFs), and small heterodimer partner (SHP) (Zhang and Dufau 2004; Mullican et al. 2013). TLX is highly conserved in mammals, birds, fish, and insects (Supplemental Fig. 1). Human TLX (hTLX, hereafter) shares almost 100% sequence identity with other mammalian TLXs, 90% with fish TLXs, and >40% with insect TLXs. The Drosophila homolog of TLX, tailless (hereafter referred to as dTLX), controls the segmentation process during early embryogenesis (Pignoni et al. 1990) by directly binding to specific response elements in the promoters of several gap genes, including knirps (kni), to inhibit their expression in the embryo termini (Pankratz et al. 1989). As a result, dTLX restricts gap gene expression within the trunk region, thus allowing the terminal cell fates to be specified (Pignoni et al. 1990; Steingrimsson et al. 1991; Younossi-Hartenstein et al. 1997). In mice, TLX governs eye development and neurogenesis (Shi et al. 2004; Zhang et al. 2006, 2008; Islam and Zhang 2014). TLX also promotes neural stem cell self-renewal by down-regulating the expression of cell cycle check genes such as PTEN and p21 (Zhang et al. 2006; Sun et al. 2007). Elevated expression of TLX has also been implicated in gliomagenesis, making it a promising therapeutic target for the treatment of human brain tumors (Liu et al. 2010; Park et al. 2010; Zou et al. 2012; Xie et al. 2014). A number of corepressors of TLX have been identified, which include Atrophin proteins, LSD1 (lysine-specific demethylase), and histone deacetylases (HDACs) (Wang et al. 2006; Sun et al. 2007, 2010; Yokoyama et al. 2008), among which Atrophin proteins are directly recruited by TLX.
Atrophin proteins form a new class of nuclear receptor corepressors whose members include mammalian Atrophin-1, Atrophin-2 (also known as RERE), and Drosophila Atrophin (hereafter referred to as dAtrophin) (Wang and Tsai 2008). In contrast to Atrophin-1, both Atrophin-2 and dAtrophin have longer N termini that contain an ELM2 (EGL-27 and MTA1 homology 2) domain and a SANT (SWI3/ADA2/NCoR/TFIII-B) domain. These modules coordinately recruit histone-modifying enzymes such as HDACs and the histone methyltransferase G9a to drive gene inhibition (Wang et al. 2006, 2008). Atrophin proteins selectively bind to a number of repressor orphan nuclear receptors and influence their physiological activities (Wang et al. 2006, 2008; Zhang et al. 2006; Haecker et al. 2007; Escher et al. 2009; Vilhais-Neto et al. 2010; Davis et al. 2014). To date, the functional interaction between dTLX and dAtrophin has been best characterized in Drosophila using genetic and biochemical analysis (Wang et al. 2006; Haecker et al. 2007; Davis et al. 2014). Atrophin proteins bind to TLX via their C-terminal region, which contains the highly conserved Atro box motif (Fig. 1A; Supplemental Fig. 2; Wang et al. 2006). The Atro box sequence in mammalian Atrophin proteins is 100% identical. Its flanking sequences are also conserved across species (e.g., humans/mice/rats/cattle) but vary between Atrophin-1 and Atrophin-2. The mammalian Atro box sequence differs from its Drosophila counterpart in only one residue (T in mammal and Q in Drosophila). Mutational replacement of leucine residues in the Atro box impaired Atrophin–TLX interactions (Wang et al. 2006). The Atro box, whose sequence differs from the classical LXXLL and LXXXIXXX (L/I) motifs, binds TLX in a ligand-independent manner. To understand how TLX recruits Atrophin, we determined the crystal structure of TLX in complex with the Atro box peptide.
Figure 1.
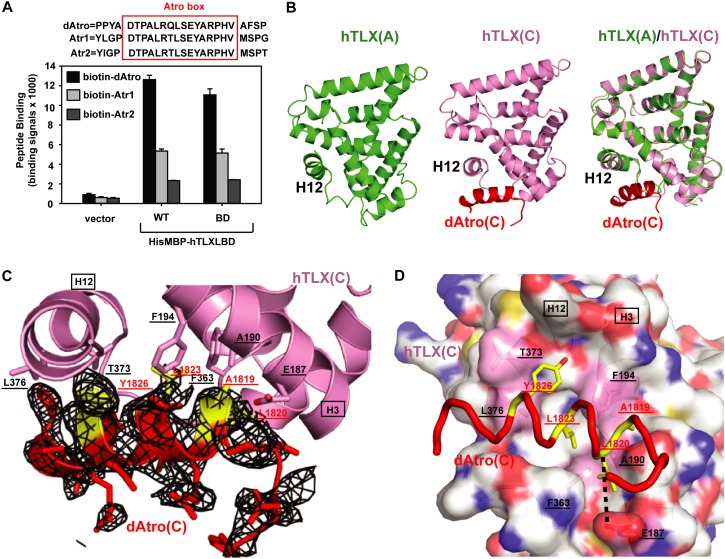
Structural analysis of the hTLX–Atro box complex. (A) Binding of hTLX wild type (WT) and BD (the mutant used in crystallization) to Atrophin peptides. His6MBP-fused hTLX LBD-WT (residues 182–385) and hTLX LBD-BD (residues 182–385, K257R/N259T/K260L/C338V) were used in AlphaScreen assays against three biotinylated peptides derived from Atrophin proteins. Error bars indicate SD (n = 3). The sequences of peptides are listed, with the Atro box sequence boxed. (B) A ribbon model of hTLX (molecule A) (apo; green), hTLX (molecule C) (dAtro complex; pink), and their superposition. (C) Representative Fo − Fc electron density omit map contoured at 1.0 σ for peptide dAtro (complex C) (red). dAtrophin amino acids involved in hTLX binding are highlighted in yellow, labeled in red, and underlined. dAtrophin-binding amino acids in hTLX are labeled in black and underlined. They are contributed to by helices H3 and H12, which are labeled in black and boxed, and also by the loop between H11 and H12. (D) Close-up presentation of the hTLX/Atrophin interface. hTLX amino acids involved in the interaction are marked by a pink transparent surface, labeled in black, and underlined. dAtrophin amino acids involved in the interaction are marked in yellow, labeled in red, and underlined. The black dashed line represents the H bond interaction between hTLX and Atrophin.
Results
Crystallization of the hTLX LBD in complex with the Atro box peptide
Among the 48 human nuclear receptors, only a very few LBD structures remain to be determined, and those including TLX are notorious for being refractory to crystallization. To overcome the technical difficulty of TLX crystallization, we developed the following strategies.
We expressed the LBD of hTLX (NP_003260) in bacteria because the TLX LBD is sufficient for binding to Atrophin proteins (Wang et al. 2006; Zhang et al. 2006). Supplemental Figure 1 shows that the amino acids of TLX corresponding to N-terminal helices H1 and H2 in the canonical nuclear receptor LBD fold are not conserved across species and contain many prolines that disfavor the formation of helices. Similarly, PNR, which is the closest homolog of TLX (sequence identity = 44% in humans; Protein Data Bank [PDB] code 4LOG) (Tan et al. 2013), as well as other ligand-free nuclear receptors, including DAX-1, TR4, and SHP (Sablin et al. 2008; Zhou et al. 2011; Zhi et al. 2014), lack H1 and H2. We therefore reasoned that the hTLX LBD likely does not have traditional helices H1 and H2. Consistently, among dozens of TLX LBD constructs, only one lacking the H1–H2 region (residues 182–385) formed crystals (see details in Supplemental Fig. 1). However, these crystals were tiny and did not diffract X-rays.
Short peptides containing coactivator or corepressor motifs have been successfully used to stabilize nuclear receptor LBDs and improve their crystallization (Xu et al. 1999a, 2001; Li et al. 2005). Therefore we turned our attention to the TLX corepressor Atrophin. Three Atro box-containing peptides (dAtro from dAtrophin, Atr1 from mammalian Atrophin-1, and Atr2 from mammalian Atrophin-2) (see Supplemental Fig. 2; Wang et al. 2006) were synthesized, and their ability to bind the hTLX LBD was tested in AlphaScreen assays (Fig. 1A). All three peptides bound to TLX and were used in cocrystallization screens. The IC50 values of the interactions determined by homologous competition demonstrated that the interactions with dAtro and Atr1 have similar binding strengths, with IC50 values of ∼30 µM, while Atr2 has an IC50 value of ∼70 µM (Supplemental Fig. 3).
We further fused a maltose-binding protein (MBP) tag at the N terminus of TLX, as this MBP fusion strategy has previously been successfully used in crystallizing the repressive orphan nuclear receptors PNR and SHP (Tan et al. 2013; Zhi et al. 2014). Only the MBP-fused hTLX LBD (residues 182–385) in complex with the dAtro peptide yielded initial crystals that diffracted poorly. This issue was later improved by the use of the homologous surface mutation strategy (Supplemental Fig. 4A; Zhi et al. 2012, 2014), in which flexible surface residues of TLX were replaced with less flexible residues derived from the corresponding region in PNR (see the Materials and Methods). Among the tested TLX LBD proteins, only one mutant protein with amino acid changes at positions B and D improved crystals in the presence of dAtro (named MBP-hTLX LBD-BD in Supplemental Fig. 4B). These crystals diffracted to 3.55 Å and allowed us to determine the hTLX–Atrophin complex crystal structure. Importantly, the BD mutant protein showed activities similar to wild-type hTLX in Atrophin-binding (Fig. 1A; Supplemental Fig. 4C) as well as transcriptional repression assays (Supplemental Fig. 4D), indicating that these mutations do not interfere with the ability of TLX to physically and functionally interact with the Atro box.
Crystal structure of the hTLX–Atrophin complex
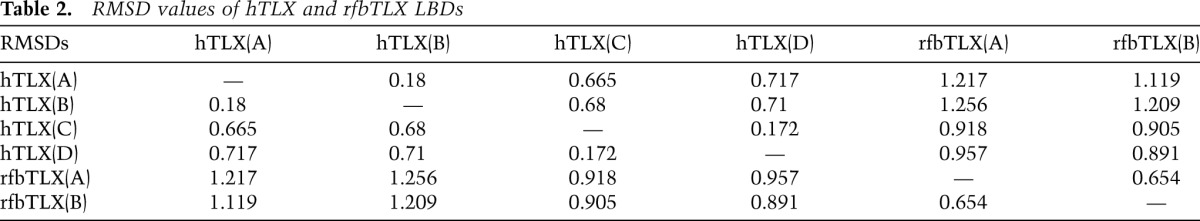
The hTLX–Atrophin complex structure was solved by molecular replacement using MBP as a model (data and structure statistics in Table 1). The noncrystallographic symmetric (NCS) unit (Supplemental Fig. 5) contained four MBP-hTLX LBDs. Interestingly, two LBDs (chains C and D) were bound with peptide dAtro and arranged in a nonsymmetric manner, while the other two LBDs (chains A and B), which were not bound with the dAtro box peptide, were arranged in a symmetric manner. All four LBDs showed an architecture similar to one another (root mean square deviation [RMSD] in Table 2), with helix H12 fitting into the canonical coactivator-binding groove, indicating an autorepressed conformation (Fig. 1B). Most interestingly, the position of helix H12 created an unexpected Atro box-binding pocket in both complex structures, in which the dAtro peptide lies beneath helix H12 and extends to helix H3 (Fig. 1B). We termed the Atrophin-binding pocket in TLX as the autorepressed pocket to distinguish it from the classic AF-2/H12 pocket. Figure 1, C and D, shows that the dAtro-binding site in TLX consists of residues E187, A190, and F194 from helix H3, F363 from the loop between H11 and H12 (the H11–H12 loop), and T373 and L376 from H12. These TLX residues mainly make hydrophobic contacts with dAtrophin residues A1819, L1820, L1823, and Y1826. E187 of TLX also forms hydrogen bonds with the main chain of A1819 and L1820 and serves as the N-cap of the Atro box helix. The TLX and Atrophin residues that mediate the TLX–Atrophin interaction are identical in the two complexes within crystals and are conserved across species (Fig. 1A; Supplemental Figs. 1, 2), in support of the biological relevance of the observed interactions.
Table 1.
Data collection and refinement statistics
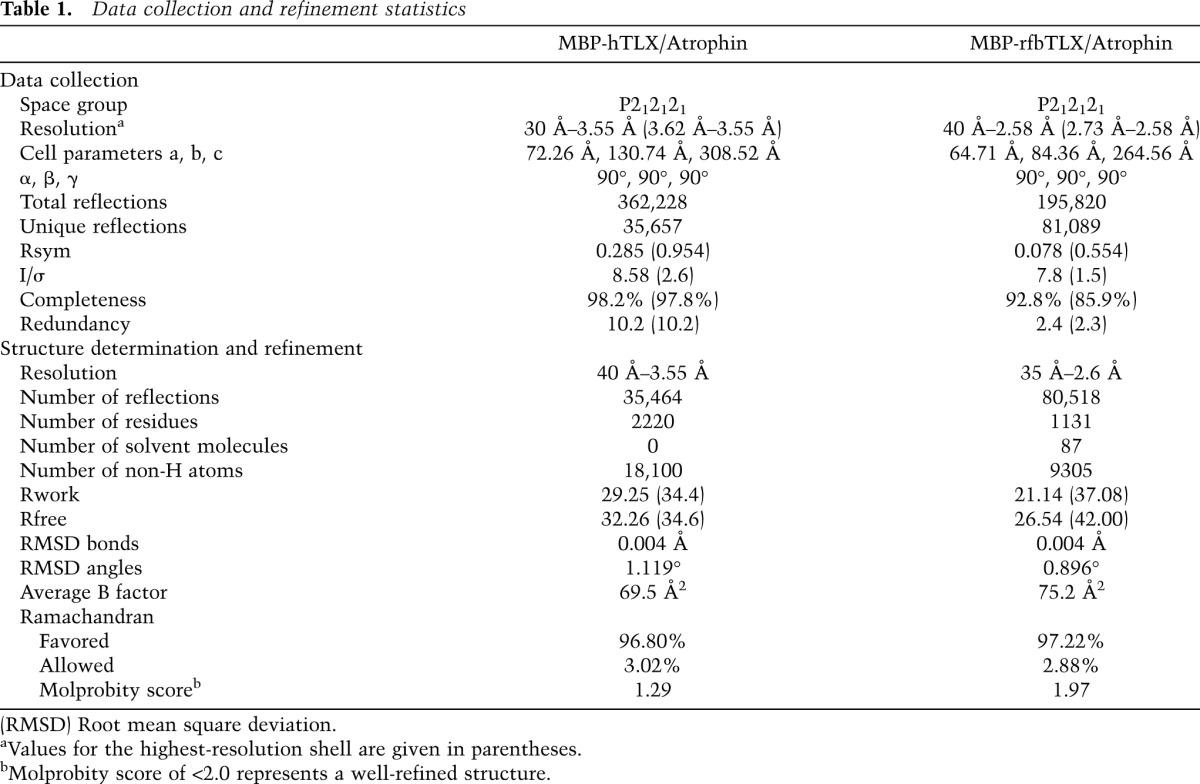
Table 2.
RMSD values of hTLX and rfbTLX LBDs
Another interesting feature of the hTLX structure is that a clear kink is observed between helices H10 and H11, which results in the collapse of helix H11 into the space that corresponds to the ligand-binding pocket of ligand-regulated nuclear receptors (Supplemental Fig. 6, cf. hTLX and ligand-bound RXR), leaving no room for ligand binding. Thus, the hTLX structure is in an autorepressed and ligand-free conformation, resembling those of several other orphan nuclear receptors, including DAX-1, COUP-TF2, TR4, PNR, and SHP (Kruse et al. 2008; Sablin et al. 2008; Zhou et al. 2011; Tan et al. 2013; Zhi et al. 2014).
Crystal structure of TLX from red flour beetles in complex with the Atro box peptide
Repeated attempts to achieve a higher-resolution hTLX–Atrophin structure were not successful. We noticed that inclusion of Atrophin peptides is required for TLX crystallization and that the TLX-binding strength of Atrophin peptides correlated with the ease of TLX crystallization. Since the LBDs of vertebrate TLX proteins are nearly 100% conserved (Supplemental Fig. 1), we screened a number of insect TLX LBDs in order to identify a species whose TLX binds to Atrophin peptides with an affinity that is higher than that of hTLX. As shown in Supplemental Figure 7A, the LBD from red flour beetle TLX (rfbTLX, NP_001034502) yielded a threefold higher dAtro-binding signal than hTLX in an in vitro binding assay. The rfbTLX LBD shares 43% and 39% sequence identity with the hTLX and dTLX LBDs (Fig. 2A). Red flour beetles also contain a homolog of Atrophin with a conserved Atro box (Supplemental Fig. 2). Importantly, coincubation of both the MBP-fused rfbTLX LBD and dAtro peptide readily yielded crystals that diffracted up to 2.6 Å, which allowed us to solve the TLX/Atrophin structure at a higher resolution than that of the hTLX–Atrophin complex (Supplemental Fig. 7B).
Figure 2.
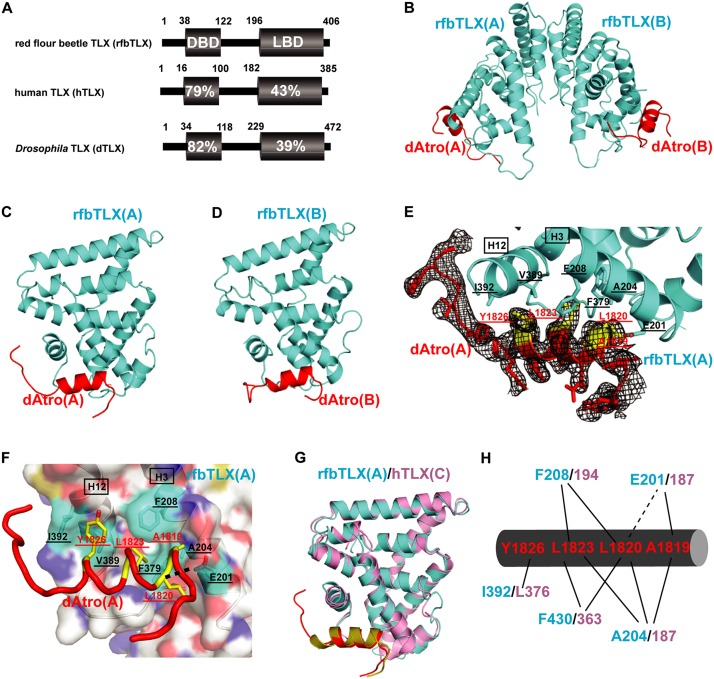
Structural analysis of the rfbTLX–Atro box complex. (A) Sequence identities of rfbTLX (NP_001034502), dTLX (NP_524596), and hTLX (NP_003260). Numbers refer to the amino acid position in the DBD or LBD of each TLX protein. (B) A ribbon model of rfbTLX (cyan) in complex with peptide dAtro (red). rfbTLX forms a dimer via the helix H10–H10 interaction. MBP has been omitted from the structure. (C) A ribbon model of rfbTLX(A) in complex with peptide dAtro. (D) A ribbon model of rfbTLX(B) in complex with peptide dAtro. (E) Representative Fo − Fc electron density omit map contoured at 1.0 σ for peptide dAtro(A) (red). The dAtrophin amino acids involved in hTLX binding are highlighted in yellow, labeled in red, and underlined. dAtrophin-binding amino acids in hTLX are labeled in black and underlined. They are contributed to by helices H3 and H12, which are labeled in black and boxed, and also by the loop between H11 and H12. (F) Close-up presentation of the rfbTLX/Atrophin interface. rfbTLX amino acids involved in the interaction are marked by a cyan transparent surface, labeled in black, and underlined. dAtrophin amino acids involved in the interaction are marked in yellow, labeled in red, and underlined. The black dashed line represents the H bond interaction between rfbTLX and Atrophin. (G) A ribbon model of rfbTLX (complex A; cyan) superposed to hTLX (complex C; pink). Peptide dAtro (complex A) bound to rfbTLX (complex A) is shown in red, and peptide dAtro (complex C) bound to hTLX (complex C) is shown in yellow. (H) Schematic presentation of the interactions between rfbTLX/hTLX and peptide dAtro. dAtro residues are labeled in red, rfbTLX residues are shown in cyan, and human residues are labeled in pink. Hydrophobic interactions are illustrated by solid lines, and H bonds are indicated by dashed lines.
The NCS unit of the hTLX–Atro structure contained two MBP-rfbTLX LBDs, and both were in complex with the dAtro peptide (Fig. 2B; Supplemental Fig. 8A). The rfbTLX LBD has an architecture similar to that of the hTLX LBD and also forms the Atrophin-bound autorepressed pocket, indicating that the novel corepressor-binding mode is structurally conserved in TLX proteins (Fig. 2C,D). The residues that constitute the Atrophin-binding site in two NCS complexes were mostly the same except for those from helix H12 (cf. Fig. 2E,F and Supplemental Fig. 8B), indicating that insect TLX helix H12 is flexible and can use different residues to accommodate the binding peptide. Superposition of the rfbTLX–dAtro complex to the hTLX–dAtro complex confirms that TLX uses conserved structural elements for Atrophin binding (Fig. 2G,H).
Analysis of the TLX–Atrophin interaction
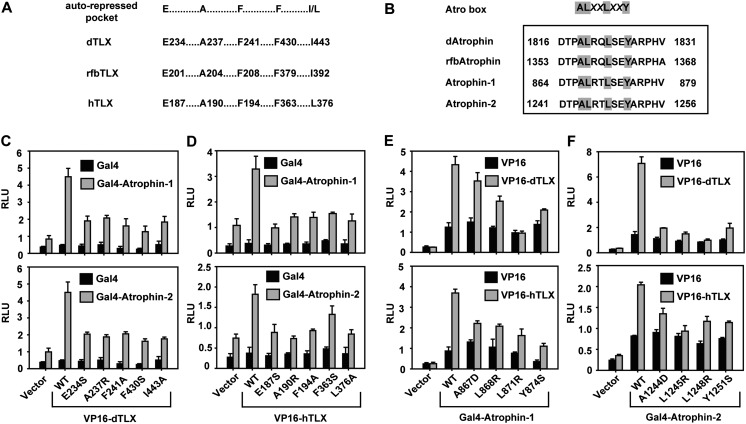
Based on structural comparison of hTLX and rfbTLX complexes and protein sequence alignment of hTLX, dTLX, and rfbTLX (see Fig. 2G,H; Supplemental Fig. 1), we identified the dTLX residues that correspond to the hTLX and rfbTLX Atrophin-interacting residues as E234, A237, and F241 from helix H3; F430 from the H11–H12 loop; and I443 from helix H12 (Fig. 3A). These conserved amino acids define an autorepressed pocket, which can accommodate the Atro box of corepressors. The Atrophin-1 residues that interact with TLX are A867, L868, L871, and Y874, and the Atrophin-2 residues are A1244, L1245, L1248, and Y1251 (Fig. 3B). These amino acids are highly conserved and define a new ALXXLXXY motif, which mediates Atrophin binding to the autorepressed pocket of TLX.
Figure 3.
Analysis of the TLX–Atrophin interaction. (A) Atrophin contact residues are conserved in dTLX, rfbTLX, and hTLX, which define an autorepressed pocket. (B) TLX contact residues are conserved in dAtrophin (NP_996025), rfbAtrophin (XP_008194914), Atrophin-1 (NP_031907), and Atrophin-2 (NP_001078961), which define an ALXXLXXY motif (Atro motif; highlighted in gray) in the Atro box sequence. (C) Mutations in the dTLX autorepressed pocket affected its interactions with Atrophin-1 and Atrophin-2 in mammalian two-hybrid assays. (D) Mutations in the hTLX autorepressed pocket compromised its interactions with Atrophin-1 and Atrophin-2 in mammalian two-hybrid assays. (E) Mutations in the Atro motif of Atrophin-1 reduced its interactions with dTLX and hTLX in mammalian two-hybrid assays. (F) Mutations in the Atro motif of Atrophin-2 reduced its interactions with dTLX and hTLX in mammalian two-hybrid assays. All error bars indicate SD (n = 3). All experiments were repeated at least twice.
To validate these structural observations, we mutated individual Atrophin contact residues in dTLX /hTLX or individual TLX contact residues in Atrophin-1/Atrophin-2 and measured their effects on TLX–Atrophin interactions in mammalian two-hybrid assays (Fig. 3C–F). Consistent with our structural findings, mutations of these key residues in TLX and Atrophin greatly compromised TLX–Atrophin interactions in cells.
Functional analysis of the TLX–Atrophin interface
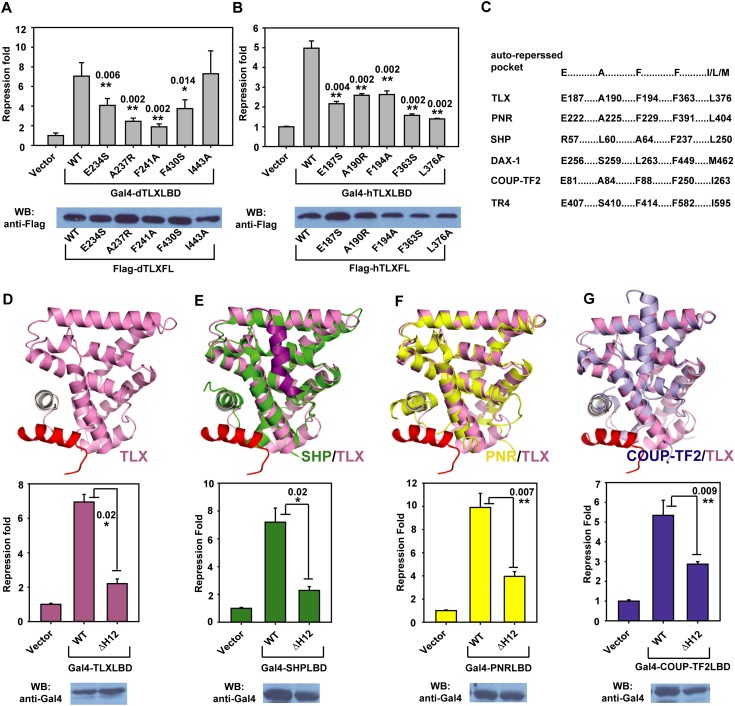
TLX functions primarily as a transcriptional repressor, and we therefore further investigated the importance of the integrity of the TLX–Atrophin interface in TLX-mediated repression. The LBD of TLX is sufficient for repressing Gal4 transcriptional activity (Zhang et al. 2006; Sun et al. 2007). Hence, we used Gal4-fused dTLX or hTLX LBDs to study repressor activity of the TLX wild type and mutants. We used repression fold to represent the ability of TLX to repress basal transcriptional activity of Gal4, which is equal to relative light unit (RLU) with Gal4 divided by RLU with Gal4-TLX. Figure 4, A and B, shows that mutations of Atrophin-contact residues in dTLX or hTLX significantly decreased their repressive activities, except for I443 in dTLX (see the Discussion). Interestingly, these TLX residues are conserved in other repressive orphan nuclear receptors, including COUP-TFII, SHP, PNR, and TR4, suggesting that these receptors also contain a similar autorepressed pocket for corepressor recruitment (Fig. 4C). In support of this notion, disruption of their potential autorepressed pockets by deletion of helix H12 greatly compromised their repressor activities (Fig. 4D–G), indicating a general mechanism of repression for this class of orphan nuclear receptors.
Figure 4.
Functional analysis of the TLX–Atrophin interface. (A) Gal4-dTLXLBD (residues 229–472) was used in reporter assays to examine the repressor activity of dTLX mutants. Western blot was performed to check the expression level of full-length dTLX mutants in cells. Repression fold was calculated by comparing RLU with Gal4-dTLX to RLU with Gal4. (B) Gal4-hTLXLBD (residues 182–385) was used in reporter assays to examine the repressor activity of hTLX mutants. Western blot was performed to check the expression level of full-length hTLX mutants in cells. Repression fold was calculated by comparing RLU with Gal4-hTLX to RLU with Gal4. (C) Atrophin contact residues are also conserved in other repressive orphan nuclear receptors, including PNR, SHP, DAX-1, COUP-TF2, and TR4. (D) Deletion of hTLX helix H12 affected its repressor activity. hTLX H12 is shown in gray. Gal4-hTLXWT (NP_003260, residues 182–385) and Gal4-hTLX∆H12 (residues 182–371) were used in reporter assays, and their expression levels were checked by Western blot using an anti-Gal4 antibody. (E) Deletion of SHP helix H12 compromised its repressor activity. Superposition of SHP (green) (PDB code 4NUF) onto hTLX suggests a potential autorepressed pocket in SHP. Mouse SHP wild type (WT) (NP_035980, residues 55–260) and SHP∆H12 (residues 55–248) fused to Gal4 were used in reporter assays, and their expression levels were checked by Western blot using an anti-Gal4 antibody. (F) Deletion of PNR helix H12 compromised its repressor activity. Superposition of PNR (yellow) (PDB code 4LOG) onto hTLX suggests a potential autorepressed pocket in PNR. hPNR wild type (NP_055064, residues 217–410) and PNR∆H12 (residues 217–399) fused to Gal4 were used in reporter assays, and their expression levels were checked by Western blot using an anti-Gal4 antibody. (G) Deletion of COUP-TF2 helix H12 compromised its repressor activity. Superposition of COUP-TF2 (light blue) (PDB code 3CJW) onto hTLX suggests a potential autorepressed pocket in COUP-TF2. Human COUP-TF2 wild type (NP_001138627, residues 76–281) and COUP-TF2∆H12 (residues 76–258) were used in reporter assays, and their expression levels were checked by Western blot using an anti-Gal4 antibody.
Discussion
Both TLX and Atrophin are conserved proteins that play important physiological roles in mammals and insects (Gui et al. 2011; Islam and Zhang 2014). In adult mouse brains, TLX functions as a transcriptional repressor that is critical for the regulation of neurogenesis and the progression of neural stem cell-dependent gliomagenesis (Sun et al. 2007; Liu et al. 2008, 2010; Zou et al. 2012). Thus, TLX has been suggested as a therapeutic target for the treatment of human neurological disorders and brain tumors (Islam and Zhang 2014), and a recent study showed that TLX activity can be regulated by small molecules (Benod et al. 2014). These exciting findings suggest that our crystal structures of TLX may also provide a rational template to develop drugs for the treatment of TLX-related diseases. The methods that we used to obtain hTLX crystals, including MBP fusion, N-terminal truncations, homologous surface mutations, and inclusion of short binding peptides, have also been successfully used by us to obtain the previous structure of the orphan nuclear receptor SHP and could serve as future tools to tackle other difficult protein structures (Zhi et al. 2014).
The rfbTLX structure reveals a dimerization mode for TLX that is mediated by hydrophobic residues from helices H10 of both monomers (Supplemental Fig. 9A). This H10–H10 interaction mode represents a canonical mode of dimerization in nuclear receptors and is also observed in the PNR structure (Supplemental Fig. 9B). The residues that form the dimer interface are conserved in rfbTLX, hPNR, and hTLX (Supplemental Fig. 9C,D). This H10–H10 dimerization mode is not observed in the hTLX structure due to crystal packing interference of MBP fusion. Because hTLX formed a dimer in solution, as shown by size exclusion chromatography (Supplemental Fig. 9E), we reasoned that hTLX also dimerizes via helix H10. Consistently, mutation of corresponding H10 residues compromised hTLX-repressive activity (Supplemental Fig. 9F), suggesting that the TLX dimer is active and conserved across species.
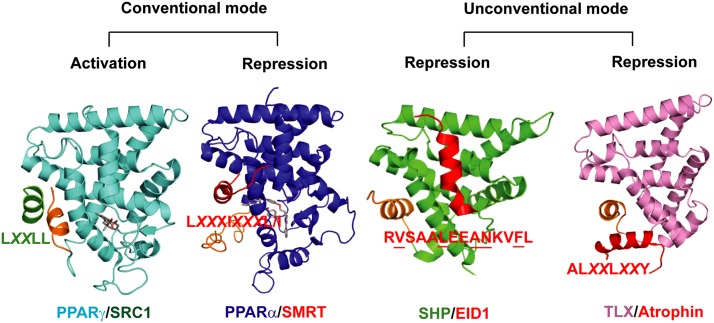
The TLX corepressor-binding mode revealed in this study is distinct from the binding of nuclear receptors to the common corepressors NCoR and SMRT, whose corepressor motifs fit into the extended coactivator-binding groove (Fig. 5; Xu et al. 2002). Instead, the Atro box is bound to the pocket underneath helix H12 and forces H12 into the “autorepressed” state (Fig. 5). The binding mode of TLX to corepressors is also different from that of the orphan nuclear receptor SHP to its corepressor, EID1, where EID1 binds to SHP by mimicking nuclear receptor helix H1 (Fig. 5; Zhi et al. 2014). Although TLX appears to lack helix H1 and therefore could have an open pocket to bind corepressors, Atrophin proteins do not bind to TLX in the SHP–EID1 manner. Instead, they bind to a new pocket underneath H12 that is at the boundary to H3. Because this pocket is closely related to the autorepressed conformation of TLX, we named it the autorepressed pocket. Together, the SHP–EID1 structure and the TLX–Atrophin structure represent two unconventional corepressor-binding modes used by ligand-free orphan nuclear receptors that are distinct from the conventional binding modes of ligand-bound receptors. The two unusual modes indicate a diversity of strategies used by orphan nuclear receptors to repress gene transcription and achieve their biological functions. It is also possible that the two modes can be present in the same receptor, given that their binding sites are not overlapping on the surface of the LBD (Fig. 5).
Figure 5.
Conventional and unconventional modes of nuclear receptor action. Comparison of the modes of action used by nuclear receptors for transcriptional activation and repression. In the conventional mode, agonist binding positions the helix H12 (shown in orange) to an active site that favors the coactivator binding. Antagonist binding dislodges H12 from the active site and opens up an extended groove to promote the corepressor binding. In the unconventional mode, the orphan nuclear receptor SHP lacks the traditional helix H1, which leads to an H1 pocket to recruit the corepressor EID1 that is otherwise covered by H1. The orphan nuclear receptor TLX uses another pocket underneath helix H12 to recruit the corepressor Atrophin, which is consistent with its autorepressed conformation. It is possible that both repressive modes are present in the same one of several ligand-free orphan nuclear receptors, considering they share similar structural features, including lack of H1 and an autorepressed conformation. Representative structures from left to right show PPARγ/SRC1 (PDB code 1FM6), PPARα/SMRT (PDB code 1KKQ), SHP/EID1 (PDB code 4NUF), and TLX/Atrophin (PDB code in this study). X indicates any amino acid. The EID1 residues that mediate the SHP interaction are underlined.
Consistent with this notion, mutation of the residues that correspond to the SHP/EID1 interface in TLX impaired TLX-repressive activity (Supplemental Fig. 10A,B). On the other hand, SHP was able to interact with Atrophin proteins such as Atrophin-2 in a coimmunoprecipitation assay (Supplemental Fig. 10C,D). Interestingly, deletion of the Atro box in Atrophin-2 greatly compromised the Atrophin-2–SHP interaction, while deletion of the Atro box in Atrophin-1 had a marginal effect on the Atrophin-1–SHP interaction, suggesting that elements other than the Atro box play a role in regulating the interaction between Atrophin-1 and SHP (Supplemental Fig. 10E). Importantly, deletion of helix H12 in SHP decreased the interaction between SHP and Atrophin-2 (Supplemental Fig. 10F). Together, our results support the notion that two unconventional corepressor-binding modes can be present in the same receptor, such as TLX and SHP. Additionally, Atrophin proteins might represent general corepressors to a group of orphan nuclear receptors, including TLX, COUP-TF2, and SHP (Wang et al. 2006; Zhang et al. 2006; Vilhais-Neto et al. 2010).
The functionality of the autorepressed pocket has been validated by our biochemical and cellular studies, as mutations of key Atrophin-contacting residues in dTLX or hTLX affected the TLX–Atrophin interactions and repressive activity. Of note, helix H12 in insect TLX might be more flexible than in hTLX because we found different sets of Atro box-contacting H12 residues in the rfbTLX complex structure. This likely explains why mutation of dTLX H12 I443 barely affected the receptor repressor activity (Fig. 4A) because other H12 residues could substitute for its loss. Nevertheless, mutation of I443 also decreased the interactions of dTLX with Atrophin-1 and Atrophin-2 (Fig. 3C), which could be a result of reduced protein expression (Fig. 4A).
The autorepressed conformation is a general feature of a number of repressor orphan nuclear receptors, as revealed by their crystal structures (Kruse et al. 2008; Sablin et al. 2008; Zhou et al. 2011; Tan et al. 2013; Zhi et al. 2014). While the physiological relevance of this conformation had not been clear previously, studies consistently showed that deletions or mutations of key structural elements of the autorepressed conformation compromised the receptor repressor function (Lalli et al. 1997; Bavner et al. 2002; Sun et al. 2007; Tan et al. 2013). Our studies here provide key insights into the functional properties associated with the autorepressed conformation. As revealed in our structures, the autorepressed conformation creates a pocket to recruit nuclear receptor corepressors, such as Atrophin proteins, which might serve as general corepressors for the repressive group of orphan nuclear receptors. Deletions or mutations of key structural elements disrupted the autorepressed pocket and consequently affected corepressor recruitment and function. The residues that form the autorepressed pocket are also conserved in repressor orphan nuclear receptors, such as PNR, SHP, DAX-1, COUP-TF2, and TR4. Therefore, our results reveal a general structural mechanism for corepressor recruitment and gene repression by this important class of orphan nuclear receptors.
Materials and methods
Plasmids and reagents
The mutants used for crystallization, AlphaScreen assays, or cotransfection assays were created by site-directed mutagenesis using the QuikChange method (Stratagene) and verified by sequencing.
Protein preparation and crystallization
For biochemical assays, the hTLX LBD (residues 182–385) and rfbTLX LBD (residues 196–406) were cloned into an engineered pETDuet-1 (Novagen) plasmid in which HisMBP was introduced in front of its original multiple cloning sites. Biotinylated MBP-hTLX LBD (residues 182–385) was constructed as described elsewhere (Zhi et al. 2014). Homologous surface mutations of TLX were based on the sequence alignment of hTLX and hPNR (NP_055064) (Supplemental Fig. 4A). Six TLX residues with flexible side chains that were predicted to be solvent-accessible were mutated either singly or in combination to the corresponding PNR residues that have less-flexible side chains.
For crystallization trials, hTLX (residues 182–385) and rfbTLX (residues 196–406) were cloned into the MBP vector described previously (Zhi et al. 2014). The proteins were expressed in BL21 (DE3) cells, first purified by amylose resin chromatography (Biolabs) and then followed by size exclusion chromatography.
The MBP-hTLX-Atrophin crystals were grown at 20°C in sitting drops containing 1.0 μL of the protein solution (10 mg/mL) and 1.0 μL of the well solution containing 0.2 M sodium/potassium phosphate and 20% (w/v) PEG3350. The MBP-rfbTLX-Atrophin crystals were grown at 20°C in sitting drops containing 1.0 μL of the protein solution (14 mg/mL) and 1.0 μL of the well solution containing 0.04 M citric acid, 0.06 M BIS-TRIS propane (pH 6.4), and 20% (w/v) PEG3350. The molecular ratio of MBP-hTLX or MBP-rfbTLX to peptide dAtro (PPYADTPALRQLSEYARPHVAFSP) was 1:1.5. In general, crystals appeared within 2 d and grew to the final size in ∼1 wk, at which time they were flash-frozen and stored in liquid nitrogen.
Data collection and structure determination
The diffraction data were collected with a MAR225 CCD detector at the 21-ID beamline (LS-CAT) at the Advanced Photon Source at Argonne National Laboratory. The observed reflections of the hTLX and rfbTLX crystals were reduced, merged, and scaled with DENZO and SCALEPACK in the HKL2000 package and XDS package, respectively (Otwinowski and Minor 1997; Kabsch 2010). The rfbTLX structure was determined with the PHASER program (Bailey 1994) by molecular replacement using the crystal structures of MBP and PNR as the models. The hTLX structure was determined with the PHASER program by molecular replacement using the crystal structure of MBP and rfbTLX as the models. Manual model building was carried out with the programs O (Jones et al. 1991) and COOT (Emsley and Cowtan 2004), and the structure was refined with crystallography NMR software (Brunger et al. 1998) and the Phenix program refmac5 (Adams et al. 2010). The model of hTLX was further improved using DEN-assisted refinement from the CNS package (Schroder et al. 2010) and Rosetta integrated in the Phenix package (DiMaio et al. 2013). RMSD values of hTLX/rfbTLX LBDs were calculated using PyMOL.
AlphaScreen binding assays
The binding of Atrophin peptides to hTLX/rfbTLX was determined by AlphaScreen luminescence proximity assays as described (Zhi et al. 2012, 2014). Reaction mixtures consisted of 50 nM His-MBP fusion proteins, 200 nM biotinylated peptides/proteins, 10 μg/mL nickel chelate-coated acceptor beads (PerkinElmer Life Sciences), and 10 μg/mL streptavidin-coated donor beads (PerkinElmer Life Sciences) in a buffer containing 50 mM 3-(N-morpholino) propane sulfonic acid (MOPS; pH 7.4), 50 mMNaF, 50 mM 3-[(3-cholamidopropyl) dimethylammonio]-1-propanesulfonate (CHAPS), and 0.1 mg/mL BSA.
Cell reporter assays, statistics analysis, and coimmunoprecipitation assays
AD293 cells were cultured and cotransfected in 24-well plates as reported (Zhi et al. 2014). hTLX (residues 182–385) wild type and mutants were cloned into the Gal4 or VP16 vector. dTLX (residues 229–472) wild type and mutants were cloned into the Gal4 or VP16 vector. Gal4-Atrophin-1 and Gal4-Atrophin-2 were constructed by genetically fusing mouse Atrophin-1 (residues 823–950) or mouse Atrophin-2 (residues 1197–1334) to the Gal4-DBD. Statistical analysis was performed using Excel (Zhi et al. 2014). Comparisons were performed using a Student’s independent-sample t-test (two-tailed distribution and two-sample equal variance). The statistical significance level was set at P < 0.05 or P < 0.01.
For coimmunoprecipitation assays, Flag- or HA-fused hTLX full-length, mouse SHP full-length (NP_035980), Atrophin-1 (NP_001007027, residues 720–1191), and Atrophin-2 (NP_001036046, residues 1087–1566) constructs were generated as described elsewhere (Wang et al. 2008; Zhi et al. 2014).
PDB codes
The PDB code is 4XAI for the structure of rfbTLX and 4XAJ for the structure of hTLX.
Supplementary Material
Acknowledgments
We thank S. Kliewer for discussions, and Z. Wawrzak and J.S. Brunzelle for assistance in data collection at the beamlines of sector 21 (Life Sciences Collaborative Access Team), which is funded in part by Michigan Economic Development Corporation and Michigan Technology Tri-Corridor grant 085P1000817. Use of the Advanced Photon Source was supported by the Office of Science of the US Department of Energy. This work was supported in part by Jay and Betty Van Andel Foundation, Ministry of Science and Technology (China), grants 2012ZX09301001, 2012CB910403, 2013CB910600, XDB08020303, and 2013ZX09507001, and National Institute of Health grants DK071662 (to H.E.X.), NS070981 and NS088095 (to C.-L.Z.), and GM102545 and GM104212 (to K.M.).
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.254904.114.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66: 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey S. 1994. The Ccp4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr 50: 760–763. [DOI] [PubMed] [Google Scholar]
- Bavner A, Johansson L, Toresson G, Gustafsson JA, Treuter E. 2002. A transcriptional inhibitor targeted by the atypical orphan nuclear receptor SHP. EMBO Rep 3: 478–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benod C, Villagomez R, Filgueira CS, Hwang PK, Leonard PG, Poncet-Montange G, Rajagopalan S, Fletterick RJ, Gustafsson JA, Webb P. 2014. The human orphan nuclear receptor tailless (TLX, NR2E1) is druggable. PLoS ONE 9: e99440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulias K, Talianidis I. 2004. Functional role of G9a-induced histone methylation in small heterodimer partner-mediated transcriptional repression. Nucleic Acids Res 32: 6096–6103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brelivet Y, Rochel N, Moras D. 2012. Structural analysis of nuclear receptors: from isolated domains to integral proteins. Mol Cell Endocrinol 348: 466–473. [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. 1998. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr 54: 905–921. [DOI] [PubMed] [Google Scholar]
- Davis SM, Thomas AL, Nomie KJ, Huang L, Dierick HA. 2014. Tailless and Atrophin control Drosophila aggression by regulating neuropeptide signalling in the pars intercerebralis. Nat Commun 5: 3177. [DOI] [PubMed] [Google Scholar]
- DiMaio F, Echols N, Headd JJ, Terwilliger TC, Adams PD, Baker D. 2013. Improved low-resolution crystallographic refinement with Phenix and Rosetta. Nat Methods 10: 1102–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132. [DOI] [PubMed] [Google Scholar]
- Escher P, Gouras P, Roduit R, Tiab L, Bolay S, Delarive T, Chen SM, Tsai CC, Hayashi M, Zernant J, et al. 2009. Mutations in NR2E3 can cause dominant or recessive retinal degenerations in the same family. Hum Mutat 30: 342–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans RM, Mangelsdorf DJ. 2014. Nuclear receptors, RXR, and the big bang. Cell 157: 255–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaig R, Greschik H, Peluso-Iltis C, Moras D. 2005. Structural basis for the cell-specific activities of the NGFI-B and the Nurr1 ligand-binding domain. J Biol Chem 280: 19250–19258. [DOI] [PubMed] [Google Scholar]
- Gallastegui N, Mackinnon JA, Fletterick RJ, Estebanez-Perpina E. 2015. Advances in our structural understanding of orphan nuclear receptors. Trends Biochem Sci 40: 25–35. [DOI] [PubMed] [Google Scholar]
- Gampe RT Jr, Montana VG, Lambert MH, Miller AB, Bledsoe RK, Milburn MV, Kliewer SA, Willson TM, Xu HE. 2000. Asymmetry in the PPARγ/RXRα crystal structure reveals the molecular basis of heterodimerization among nuclear receptors. Mol Cell 5: 545–555. [DOI] [PubMed] [Google Scholar]
- Gui H, Li ML, Tsai CC. 2011. A tale of tailless. Dev Neurosci 33: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haecker A, Qi D, Lilja T, Moussian B, Andrioli LP, Luschnig S, Mannervik M. 2007. Drosophila brakeless interacts with atrophin and is required for tailless-mediated transcriptional repression in early embryos. PLoS Biol 5: e145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam MM, Zhang CL. 2014. TLX: a master regulator for neural stem cell maintenance and neurogenesis. Biochim Biophys Acta 1849: 210–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M. 1991. Improved methods for building protein models in electron-density maps and the location of errors in these models. Acta Crystallogr A 47: 110–119. [DOI] [PubMed] [Google Scholar]
- Kabsch W. 2010. XDS. Acta Crystallogr D Biol Crystallogr 66: 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse SW, Suino-Powell K, Zhou XE, Kretschman JE, Reynolds R, Vonrhein C, Xu Y, Wang L, Tsai SY, Tsai MJ, et al. 2008. Identification of COUP-TFII orphan nuclear receptor as a retinoic acid-activated receptor. PLoS Biol 6: e227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalli E, Bardoni B, Zazopoulos E, Wurtz JM, Strom TM, Moras D, Sassone-Corsi P. 1997. A transcriptional silencing domain in DAX-1 whose mutation causes adrenal hypoplasia congenita. Mol Endocrinol 11: 1950–1960. [DOI] [PubMed] [Google Scholar]
- Li Y, Choi M, Suino K, Kovach A, Daugherty J, Kliewer SA, Xu HE. 2005. Structural and biochemical basis for selective repression of the orphan nuclear receptor liver receptor homolog 1 by small heterodimer partner. Proc Natl Acad Sci 102: 9505–9510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HK, Belz T, Bock D, Takacs A, Wu H, Lichter P, Chai M, Schutz G. 2008. The nuclear receptor tailless is required for neurogenesis in the adult subventricular zone. Genes Dev 22: 2473–2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HK, Wang Y, Belz T, Bock D, Takacs A, Radlwimmer B, Barbus S, Reifenberger G, Lichter P, Schutz G. 2010. The nuclear receptor tailless induces long-term neural stem cell expansion and brain tumor initiation. Genes Dev 24: 683–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, et al. 1995. The nuclear receptor superfamily: the second decade. Cell 83: 835–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullican SE, DiSpirito JR, Lazar MA. 2013. The orphan nuclear receptors at their 25-year reunion. J Mol Endocrinol 51: T115–T140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Macromol Crystallogr Pt A 276: 307–326. [DOI] [PubMed] [Google Scholar]
- Pankratz MJ, Hoch M, Seifert E, Jackle H. 1989. Kruppel requirement for knirps enhancement reflects overlapping gap gene activities in the Drosophila embryo. Nature 341: 337–340. [DOI] [PubMed] [Google Scholar]
- Park HJ, Kim JK, Jeon HM, Oh SY, Kim SH, Park MJ, Soeda A, Nam DH, Kim H. 2010. The neural stem cell fate determinant TLX promotes tumorigenesis and genesis of cells resembling glioma stem cells. Mol Cells 30: 403–408. [DOI] [PubMed] [Google Scholar]
- Phelan CA, Gampe RT Jr, Lambert MH, Parks DJ, Montana V, Bynum J, Broderick TM, Hu X, Williams SP, Nolte RT, et al. 2010. Structure of Rev-erbα bound to N-CoR reveals a unique mechanism of nuclear receptor-co-repressor interaction. Nat Struct Mol Biol 17: 808–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pignoni F, Baldarelli RM, Steingrimsson E, Diaz RJ, Patapoutian A, Merriam JR, Lengyel JA. 1990. The Drosophila gene tailless is expressed at the embryonic termini and is a member of the steroid receptor superfamily. Cell 62: 151–163. [DOI] [PubMed] [Google Scholar]
- Sablin EP, Woods A, Krylova IN, Hwang P, Ingraham HA, Fletterick RJ. 2008. The structure of corepressor Dax-1 bound to its target nuclear receptor LRH-1. Proc Natl Acad Sci 105: 18390–18395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyal S, Bavner A, Haroniti A, Nilsson LM, Lundasen T, Rehnmark S, Witt MR, Einarsson C, Talianidis I, Gustafsson JA, et al. 2007. Involvement of corepressor complex subunit GPS2 in transcriptional pathways governing human bile acid biosynthesis. Proc Natl Acad Sci 104: 15665–15670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder GF, Levitt M, Brunger AT. 2010. Super-resolution biomolecular crystallography with low-resolution data. Nature 464: 1218–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Chichung Lie D, Taupin P, Nakashima K, Ray J, Yu RT, Gage FH, Evans RM. 2004. Expression and function of orphan nuclear receptor TLX in adult neural stem cells. Nature 427: 78–83. [DOI] [PubMed] [Google Scholar]
- Steingrimsson E, Pignoni F, Liaw GJ, Lengyel JA. 1991. Dual role of the Drosophila pattern gene tailless in embryonic termini. Science 254: 418–421. [DOI] [PubMed] [Google Scholar]
- Sun G, Yu RT, Evans RM, Shi Y. 2007. Orphan nuclear receptor TLX recruits histone deacetylases to repress transcription and regulate neural stem cell proliferation. Proc Natl Acad Sci 104: 15282–15287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun G, Alzayady K, Stewart R, Ye P, Yang S, Li W, Shi Y. 2010. Histone demethylase LSD1 regulates neural stem cell proliferation. Mol Cell Biol 30: 1997–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takezawa S, Yokoyama A, Okada M, Fujiki R, Iriyama A, Yasuo Y, Ito H, Takada I, Kishimoto M, Miyajima A, et al. 2007. A cell cycle-dependent co-repressor mediates photoreceptor cell-specific nuclear receptor function. EMBO J 26: 764–774. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Tan MH, Zhou XE, Soon FF, Li X, Li J, Yong EL, Melcher K, Xu HE. 2013. The crystal structure of the orphan nuclear receptor NR2E3/PNR ligand binding domain reveals a dimeric auto-repressed conformation. PLoS ONE 8: e74359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilhais-Neto GC, Maruhashi M, Smith KT, Vasseur-Cognet M, Peterson AS, Workman JL, Pourquie O. 2010. Rere controls retinoic acid signalling and somite bilateral symmetry. Nature 463: 953–957. [DOI] [PubMed] [Google Scholar]
- Wang L, Tsai CC. 2008. Atrophin proteins: an overview of a new class of nuclear receptor corepressors. Nucl Recept Signal 6: e009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZL, Benoit G, Liu JS, Prasad S, Aarnisalo P, Liu XH, Xu HD, Walker NPC, Perlmann T. 2003. Structure and function of Nurr1 identifies a class of ligand-independent nuclear receptors. Nature 423: 555–560. [DOI] [PubMed] [Google Scholar]
- Wang L, Rajan H, Pitman JL, McKeown M, Tsai CC. 2006. Histone deacetylase-associating Atrophin proteins are nuclear receptor corepressors. Genes Dev 20: 525–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Charroux B, Kerridge S, Tsai CC. 2008. Atrophin recruits HDAC1/2 and G9a to modify histone H3K9 and to determine cell fates. EMBO Rep 9: 555–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Q, Flavahan WA, Bao SD, Rich J. 2014. The tailless root of glioma: cancer stem cells. Cell Stem Cell 15: 114–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu HE, Lambert MH, Montana VG, Parks DJ, Blanchard SG, Brown PJ, Sternbach DD, Lehmann JM, Wisely GB, Willson TM, et al. 1999a. Molecular recognition of fatty acids by peroxisome proliferator-activated receptors. Mol Cell 3: 397–403. [DOI] [PubMed] [Google Scholar]
- Xu L, Glass CK, Rosenfeld MG. 1999b. Coactivator and corepressor complexes in nuclear receptor function. Curr Opin Genet Dev 9: 140–147. [DOI] [PubMed] [Google Scholar]
- Xu HE, Lambert MH, Montana VG, Plunket KD, Moore LB, Collins JB, Oplinger JA, Kliewer SA, Gampe RT, McKee DD, et al. 2001. Structural determinants of ligand binding selectivity between the peroxisome proliferator-activated receptors. Proc Natl Acad Sci 98: 13919–13924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu HE, Stanley TB, Montana VG, Lambert MH, Shearer BG, Cobb JE, McKee DD, Galardi CM, Plunket KD, Nolte RT, et al. 2002. Structural basis for antagonist-mediated recruitment of nuclear co-repressors by PPARα. Nature 415: 813–817. [DOI] [PubMed] [Google Scholar]
- Yokoyama A, Takezawa S, Schule R, Kitagawa H, Kato S. 2008. Transrepressive function of TLX requires the histone demethylase LSD1. Mol Cell Biol 28: 3995–4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younossi-Hartenstein A, Green P, Liaw GJ, Rudolph K, Lengyel J, Hartenstein V. 1997. Control of early neurogenesis of the Drosophila brain by the head gap genes tll, otd, ems, and btd. Dev Biol 182: 270–283. [DOI] [PubMed] [Google Scholar]
- Yu RT, Mckeown M, Evans RM, Umesono K. 1994. Relationship between Drosophila gap gene tailless and a vertebrate nuclear receptor Tlx. Nature 370: 375–379. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Dufau ML. 2004. Gene silencing by nuclear orphan receptors. Vitam Horm 68: 1–48. [DOI] [PubMed] [Google Scholar]
- Zhang CL, Zou Y, Yu RT, Gage FH, Evans RM. 2006. Nuclear receptor TLX prevents retinal dystrophy and recruits the corepressor atrophin1. Genes Dev 20: 1308–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang CL, Zou Y, He W, Gage FH, Evans RM. 2008. A role for adult TLX-positive neural stem cells in learning and behaviour. Nature 451: 1004–1007. [DOI] [PubMed] [Google Scholar]
- Zhi X, Zhou XE, Melcher K, Motola DL, Gelmedin V, Hawdon J, Kliewer SA, Mangelsdorf DJ, Xu HE. 2012. Structural conservation of ligand binding reveals a bile acid-like signaling pathway in nematodes. J Biol Chem 287: 4894–4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhi X, Zhou XE, He Y, Zechner C, Suino-Powell KM, Kliewer SA, Melcher K, Mangelsdorf DJ, Xu HE. 2014. Structural insights into gene repression by the orphan nuclear receptor SHP. Proc Natl Acad Sci 111: 839–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou XE, Suino-Powell KM, Xu Y, Chan CW, Tanabe O, Kruse SW, Reynolds R, Engel JD, Xu HE. 2011. The orphan nuclear receptor TR4 is a vitamin A-activated nuclear receptor. J Biol Chem 286: 2877–2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Y, Niu W, Qin S, Downes M, Burns DK, Zhang CL. 2012. The nuclear receptor TLX is required for gliomagenesis within the adult neurogenic niche. Mol Cell Biol 32: 4811–4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.