In Bacillus subtilis, nitrogen homeostasis is controlled by a unique circuitry composed of the regulator TnrA and the repressor GlnR. Here, Schumacher et al. describe a comprehensive molecular dissection of this network that reveals novel mechanisms, including oligomeric transformations, by which their inducible signal transduction domains are employed to provide a readout of nitrogen levels.
Keywords: glutamine synthetase, dodecamer, tetradecamer, TnrA/GlnR family, MerR family
Abstract
All cells must sense and adapt to changing nutrient availability. However, detailed molecular mechanisms coordinating such regulatory pathways remain poorly understood. In Bacillus subtilis, nitrogen homeostasis is controlled by a unique circuitry composed of the regulator TnrA, which is deactivated by feedback-inhibited glutamine synthetase (GS) during nitrogen excess and stabilized by GlnK upon nitrogen depletion, and the repressor GlnR. Here we describe a complete molecular dissection of this network. TnrA and GlnR, the global nitrogen homeostatic transcription regulators, are revealed as founders of a new structural family of dimeric DNA-binding proteins with C-terminal, flexible, effector-binding sensors that modulate their dimerization. Remarkably, the TnrA sensor domains insert into GS intersubunit catalytic pores, destabilizing the TnrA dimer and causing an unprecedented GS dodecamer-to-tetradecamer conversion, which concomitantly deactivates GS. In contrast, each subunit of the GlnK trimer “templates” active TnrA dimers. Unlike TnrA, GlnR sensors mediate an autoinhibitory dimer-destabilizing interaction alleviated by GS, which acts as a GlnR chaperone. Thus, these studies unveil heretofore unseen mechanisms by which inducible sensor domains drive metabolic reprograming in the model Gram-positive bacterium B. subtilis.
Nitrogen is an essential macronutrient for all life. However, its bioavailability is scarce and often a growth-limiting factor. As a result, organisms have evolved effective systems for nitrogen sensing, acquisition, and utilization that are highly regulated (Fisher 1999; Reitzer 2003). When nitrogen levels are low, genes encoding nitrogen transport and metabolic factors need to be up-regulated, and when nitrogen levels rise, these genes must be repressed to conserve cellular resources. In the model Gram-positive bacterium Bacillus subtilis, nitrogen homeostasis is regulated by a unique protein network distinct from the two-component systems employed by enteric bacteria, which is comprised of the central enzyme of nitrogen metabolism, glutamine synthetase (GS); the global transcription regulator TnrA; and the repressor GlnR. Recent data suggest that GlnK, which is a component of the ammonium transport complex, also plays a role in this network (Gunka and Commichau 2012). These proteins form a highly integrated system that permits B. subtilis to detect nitrogen levels and transmit this signal to effect intracellular enzyme activity and gene regulation. However, the molecular mechanisms that control this complex circuitry are unknown.
GS sits at the center of the B. subtilis nitrogen regulatory hub. In B. subtilis, GS, which produces glutamine (Gln) from ammonium, ATP, and glutamate, is the only enzyme capable of assimilating nitrogen into usable cellular metabolites (Deuel et al. 1970; Eisenberg et al. 2000). The B. subtilis enzyme is also unusual for a bacterial GS in that it is regulated by feedback inhibition by its product, Gln. Recent structural data revealed the basis for B. subtilis catalysis and product feedback inhibition. These studies showed that while the B. subtilis enzyme has an overall structure and dodecameric arrangement similar to those of Gram-negative GS, it is distinct from these proteins in that it undergoes large conformational changes during catalysis that are required to construct a functional active site, and binding of its product, Gln, locks in a conformation that is unable to transit to the active state (Murray et al. 2013). Feedback-inhibited GS also interacts with and regulates the activities of the transcription factors TnrA and GlnR, thus acting as a transcription coregulator (Wray et al. 2001; Fisher and Wray 2008). During nitrogen limitation, TnrA is in its DNA-binding active state and turns on the transcription of genes required for nitrogen assimilation. Under conditions of nitrogen excess, feedback-inhibited GS forms a stable complex with TnrA, which inhibits its DNA-binding activity. In contrast, feedback-inhibited GS acts as a chaperone to stabilize the DNA-binding activity of GlnR, which represses the transcription of nitrogen assimilation genes (Wray et al. 2001; Fisher and Wray 2008). Thus, B. subtilis GS is an unusual multitasking protein that functions as an enzyme, transcription coregulator, and chaperone. GlnK appears to play an ancillary role in this pathway during nitrogen depletion by binding and stabilizing TnrA (Heinrich et al. 2006; Kayumov et al. 2008, 2011).
The predicted N-terminal DNA-binding domains of TnrA and GlnR share 62% sequence identity, which is congruent with their ability to bind the same DNA sites with the consensus TGTNAN7TNACA (Wray et al. 1996, 2000; Wray and Fisher 2008). Because the two proteins are active under different conditions as dictated by effector-binding interactions, they produce distinct transcriptional outcomes. TnrA functions primarily as an activator by binding operator DNA sites and recruiting RNA polymerase (RNAP) (Wray et al. 1996, 2000; Yoshida et al. 2003). GlnR does not bind RNAP and hence functions as a repressor. The amino acid sequences of the predicted DNA-binding domains of TnrA and GlnR have resulted in their placement in the MerR family of winged–HTH (helix–turn–helix)–coiled-coil DNA-binding proteins. However, unlike MerR proteins, which activate transcription by distorting and realigning DNA promoters with nonoptimal spacing between the −10 and −35 boxes, the promoters bound by TnrA and GlnR are optimally arranged (Heldwein and Brennan 2001). These data suggest that TnrA and GlnR may regulate transcription using molecular mechanisms distinct from MerR proteins. Indeed, the C-terminal domains of TnrA and GlnR are not predicted to form coiled coils and instead bind target proteins. The C-terminal 15 residues of the 110-residue TnrA protein bind GS, while a region within TnrA residues 75–90 interacts with GlnK (Wray et al. 2001; Kayumov et al. 2011). The C-terminal domain of the 135-residue GlnR protein is sequentially distinct from TnrA and contains an extra 15 residues. In some unknown manner, this region prevents GlnR from binding DNA. The interaction of GlnR with feedback-inhibited GS shifts the equilibrium from this inhibited state to the DNA-bound state (Wray and Fisher 2008). Thus, the TnrA and GlnR C-terminal domains function as nitrogen-sensing, signal transduction modules.
Despite the fact that the proteins and interactions involved in regulating B. subtilis nitrogen homeostasis have been extensively investigated from genetic and cellular standpoints, the molecular mechanisms controlling this circuitry have remained elusive. Here we describe a comprehensive molecular dissection of this network that unveils TnrA and GlnR as founding members of a new family of transcription regulators and reveals novel mechanisms, including heretofore unseen oligomeric transformations, by which their inducible signal transduction domains are employed to provide a readout of nitrogen levels in the model Gram-positive bacterium B. subtilis.
Results
Structures of the global nitrogen transcription regulator TnrA bound to DNA
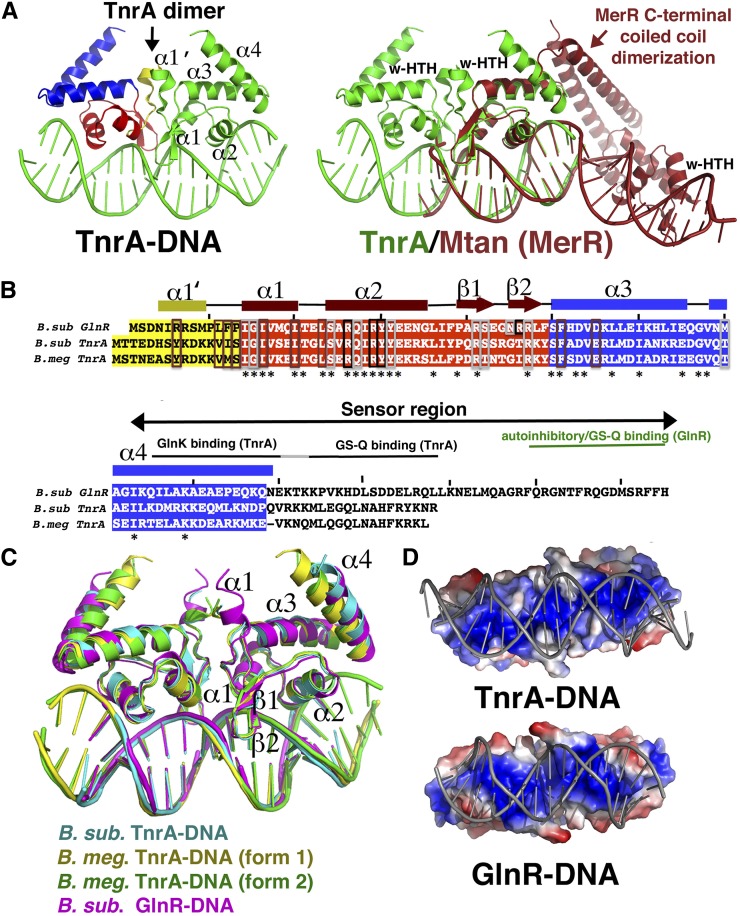
Comparative genomic studies have shown that TnrA is highly conserved in Bacilli, indicating its ubiquitous role as a master regulator of nitrogen metabolism (Gunka and Commichau 2012). To deduce the DNA-binding mechanism of TnrA, we determined structures of the B. subtilis and Bacillus megaterium TnrA proteins (60% identical) bound to cognate DNA sites. The structure of the full-length (FL) B. subtilis TnrA bound to the 17mer TGTAAGATTTCCTGACA was solved to 3.50 Å by multiple wavelength anomalous diffraction (MAD) (Supplemental Fig. S1A). This structure was then used to solve two structures of FL B. megaterium TnrA in complex with the 21mer DNA CGTGTAAGATTTCCTGACACG to 2.25 Å and 2.70 Å resolution (Supplemental Material; Supplemental Table S1). The B. subtilis and B. megaterium TnrA–DNA structures are essentially identical (root mean squared deviation [RMSD] = 0.6 Å) and reveal that TnrA binds its palindromic DNA site as a symmetric dimer (Fig. 1A–C). Each TnrA subunit can be divided into three regions: a short N-terminal helical region, a DNA-binding winged–HTH motif, and a C-terminal domain comprised of two α helices and a disordered tail (Fig. 1A). The overall topology is [α1′–(6–11)–α1(16–23)–α2(26–38)–β1(40–44)–β2(47–50)–α3(52–67)–α4(72–90)].
Figure 1.
Structures of TnrA–DNA and GlnR–DNA complexes. (A) The TnrA–DNA complex. TnrA is composed of three structural regions, colored yellow (α1′), red (winged–HTH), and blue (C-terminal region). (Right) Superimposition of one TnrA winged–HTH (green) onto that of the MerR protein MtaN (dark red) underscoring that, although the winged–HTH regions are highly similar, the proteins employ dramatically different dimerization and DNA-binding modes. This figure and all ribbon diagrams were made using PyMOL. (B) Sequence alignment of the TnrA and GlnR proteins solved in this study. Structural elements are indicated above the sequences and are colored according to the regional division in A. Residues boxed in brown, black, and silver are involved in dimerization, DNA base, and DNA phosphate contacts, respectively. (C) Overlay of TnrA–DNA and GlnR–DNA structures. (D) Electrostatic surface representation of GlnR and TnrA highlighting their strong electropositive (blue) DNA-binding surfaces.
Database searches show that the overall structure of TnrA is unique. Indeed, while its winged–HTH domain is very similar to MerR proteins (RMSD = 1.1 Å in a comparison with MerR protein MtaN) (Newberry and Brennan 2004), the remainder of the TnrA protein, its mode of dimerization, and DNA binding are strikingly different (Fig. 1A). In particular, TnrA does not harbor a C-terminal coiled-coil dimerization motif, which is a hallmark of MerR proteins. In fact, TnrA C-terminal residues 91–110 are disordered. The lack of a C-terminal coiled coil in TnrA is consistent with data showing that this region functions as a sensor domain and hence likely folds only upon effector protein binding. TnrA dimer formation is instead mediated by hydrophobic residues located on its winged–HTH (α3) and residues in its N-terminal helix (α1′), the latter of which is notably absent in MerR proteins. However, only ∼490 Å2 of total surface area is buried (BSA) upon TnrA dimer formation, which is significantly less than the >2000 Å2 of BSA shielded by the MerR coiled coil and other biologically relevant dimers (Krissinel and Hendrick 2007). The small TnrA oligomer BSA is suggestive of a weak dimer. Because dimerization by TnrA is essential for binding its palindromic cognate site, its regulation could provide an attractive mechanism to control TnrA function.
In the TnrA–DNA structure, the winged–HTH regions, which actually participate in dimerization, are closely juxtaposed, and the two wings from the dimer bind the same minor groove. In contrast, in MerR proteins, the winged–HTHs are located ∼50 Å apart, at opposite ends of the bound DNA, and function as separate entities (Fig. 1A). As a result, although TnrA causes some DNA distortion, unlike MerR proteins, which dramatically reshape their DNA sites outward, TnrA bends the DNA inward at an overall angle of 23°. The TnrA DNA is slightly underwound, with an average twist of 33.8° (compared with 36° for B-DNA). The snug fit of the TnrA recognition helices into the major grooves and the dual insertion of both wings of the dimer into the same minor groove result in a slight expansion of the major groove (12–13 Å compared with 11 Å for B-DNA) and compression of the minor grooves at the ends of the DNA site (3.8 Å compared with 5.9 Å for B-DNA). This tight fit is augmented by electrostatic complementarity between the protein and the DNA (Fig. 1D).
Structure of the B. subtilis nitrogen repressor protein GlnR bound to DNA
Fluorescence polarization (FP) studies revealed that FL GlnR bound a DNA consensus site with >10-fold reduced affinity compared with GlnRΔ18, consistent with previous studies suggesting that the C-terminal region inhibits DNA binding (Supplemental Fig. S2). Thus, GlnRΔ18 was used to obtain a structure of a GlnR–DNA complex to 2.55 Å (Supplemental Fig. S1B; Supplemental Table S1). With the exception of a slightly extended N-terminal α1′ helix, the GlnR–DNA structure is very similar to the TnrA–DNA complexes; the proteins form the same dimers and have identical topologies (Fig. 1C). GlnR induces bend and conformational changes in the DNA similar to those in TnrA and mirrors TnrA in harboring a strong electrostatic complementarity with DNA (Fig. 1D). Similar to TnrA, GlnR dimerization also buries a meager ∼460 Å2 of surface from solvent, suggesting that both proteins may be regulated by modulation of their dimerization.
TnrA and GlnR: founders of a new family of DNA-binding proteins
The TnrA–DNA and GlnR–DNA structures reveal a heretofore unseen DNA-binding mode, marking them as founding members of a new family of DNA-binding proteins, here termed the TnrA/GlnR family. The structures indicate that a 17mer constitutes the minimal binding site for these proteins, which also distinguishes them from MerR members, which require longer DNA sites for high-affinity binding (>22 base pairs [bp]) (Heldwein and Brennan 2001; Newberry and Brennan 2004). The B. megaterium TnrA–21mer structures revealed only two additional phosphate contacts to the DNA ends compared with the complexes with the 17mer. Consistent with this, FP studies showed only a small enhancement in TnrA and GlnR binding to the 21mer compared with the 17mer. TnrA bound the 17mer and 21mer with Kds of 15.7 nM ± 0.9 nM and 11.6 nM ± 1.8 nM, respectively, while the Kds of GlnRΔ18 for the 17mer and 21mer were 8.1 nM ± 0.3 nM and 6.0 nM ± 0.9 nM (Supplemental Fig. S3A,B).
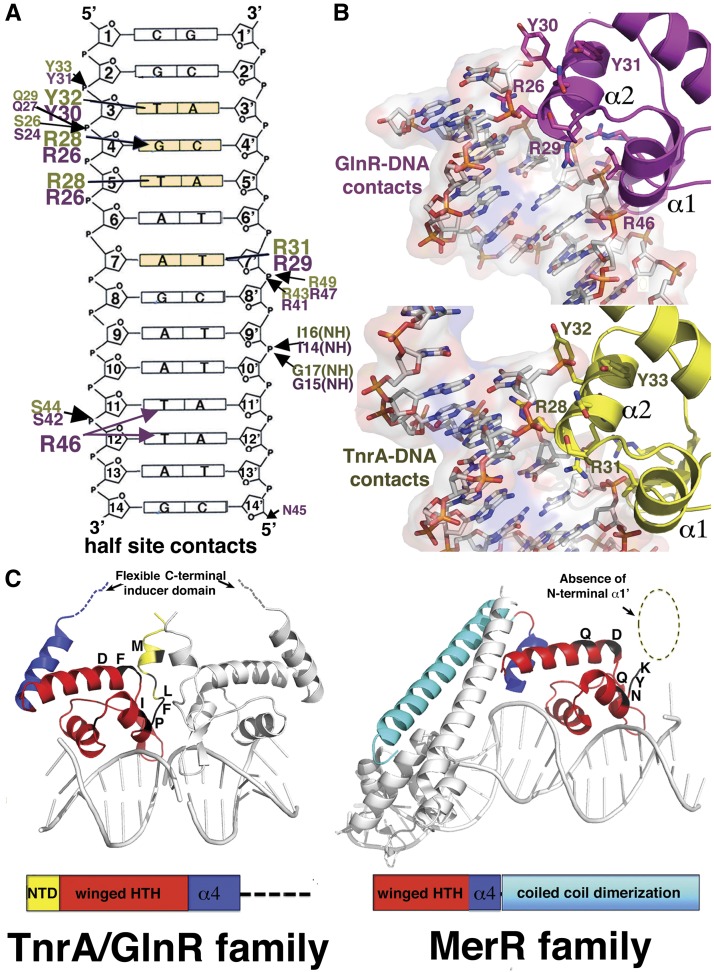
Combined in vivo and in vitro studies on the DNA-binding and nucleotide preferences of TnrA and GlnR revealed that only 4 nucleotides (nt) in each operator half-site (indicated in bold for the 21mer N1N2T3G4T5N6A7N(7)T7′N6′A5′C4′A3′N2′N1′, where N indicates any nucleotide) are required for their specific DNA binding (Gutowski and Schreier 1992; Brown and Sonenshein 1996; Wray and Fisher 2008; Wray et al. 1996, 2000; Yoshida et al. 2003). These findings are explained by the structures, which reveal that TnrA and GlnR contact bases in the DNA half-sites in identical manners using conserved residues from their recognition helices. The Thy3 methyl group is read by hydrophobic contacts from the aromatic ring of the conserved tyrosine residue Tyr32 and Tyr30 in TnrA and GlnR, respectively. TnrA/GlnR residue Arg28/Arg26 is central to their DNA-binding functions, as it contacts two of the conserved bases; it makes specific hydrogen bonds to Gua4, and its side chain, Cδ, makes hydrophobic interactions with the methyl moiety of Thy5 (Fig. 2A,B). This dual DNA-binding contact by TnrA/GlnR Arg28/Arg26 is somewhat reminiscent of the so-called 5′-pyrimidine–guanine-3′ (5′-YpG-3′) interaction identified in multiple protein–DNA structures by Glover and coworkers (Lamoureux et al. 2004). However, in a 5′-YpG-3′ interaction, the arginine makes specific hydrogen bonds to a guanine while simultaneously contacting the preceding pyrimidine, which becomes unstacked (Lamoureux et al. 2004). In TnrA/GlnR, the Arg28/Arg26 interacts with a thymine that is 3′ to the guanine that it contacts, and the bases are not significantly unstacked. This additional type of dual contact from Arg28/Arg26 expands the repertoire of DNA interactions made by this flexible side chain, including the ability to specify multiple bases in a cognate site (Supplemental Fig. S4). Finally, the side chain methylene carbons of TnrA/GlnR residue Arg31/Arg29 contact the methyl group of Thy7′ (Fig. 2A,B).
Figure 2.
TnrA and GlnR: founders of a new DNA-binding family. (A) Schematic showing residues from GlnR (magenta) and TnrA (yellow) that contact DNA (conserved bases in TnrA/GlnR sites are colored light yellow). (B) Close-up of these interactions. (C) The elements in TnrA/GlnR proteins that delineate them from MerR proteins are indicated and include the presence of an extra α1′ helix (not found in MerR proteins, as indicated by the dashed circle), a C-terminal flexible sensor region in TnrA/GlnR proteins in place of the characteristic MerR coiled-coil dimerization motif, and hydrophobic residues that mediate TnrA/GlnR dimerization and are primarily polar or charged in MerR proteins.
The finding that Gua4 is critical for the TnrA/GlnR interaction is consistent with previous studies showing that its substitution to any other nucleotide abrogated TnrA binding, while mutations of the other conserved nucleotides—3, 5, and 7—led to 16-fold, 17-fold, and 84-fold reductions in DNA binding (Wray et al. 2000). Also consonant with the structures, mutation of TnrA residues Arg28 and Arg31 to alanine essentially abolishes DNA binding (>1000-fold reduction in binding) (Wray et al. 2000). The structures also show that a 7-bp spacing between half-sites is essential for proper DNA docking by TnrA/GlnR dimers, consistent with previous data (Wray et al. 2000). Notably, even the TnrA/GlnR phosphate interactions are provided by residues conserved between the proteins (Fig. 2A,B). The only differences in DNA contacts between the complexes arise from the less conserved wing residues. In the GlnR structure, wing residue Arg46 makes nonspecific contacts to bases in the minor groove, while the corresponding residues in the B. subtilis and B. megaterium TnrA proteins threonine and isoleucine do not interact with DNA.
The unique structural features revealed in the TnrA–DNA and GlnR–DNA complexes led us to perform database searches of putative MerR proteins to query whether TnrA/GlnR may represent a larger family of DNA-binding proteins distinct from the established MerR grouping. Strikingly, we found that there are more than a hundred proteins categorized as “MerR” proteins that likely represent TnrA/GlnR members, as they contain ∼11–20 extra N-terminal residues, and their C-terminal regions are not predicted to form coiled coils. Modeling shows that the absence of α1′ alone would prevent proper TnrA/GlnR dimerization. However, perhaps the strongest evidence for the distinct TnrA/GlnR and MerR family designations is the finding that residues involved in TnrA/GlnR dimerization are conserved as hydrophobic amino acids in these proteins, while they are primarily polar or charged in canonical MerR proteins (Fig. 2C; Supplemental Fig. S5A,B).
The TnrA–DNA and GlnR–DNA structures reveal the mechanisms by which these proteins bind their operators to effect transcription. However, studies have demonstrated that the C-terminal domains of these proteins, which the structures show are largely disordered in the DNA-bound forms, function as protein–protein interaction modules to sense nitrogen levels and do so by somehow modulating the function of the distant (>40 Å) DNA-binding domains. Inducible activation domains, also called intrinsically disordered regions, play key roles in multiple processes in all domains of life, including transcription, yet little structural information is available for such domains (Dunker et al. 2009). Thus, we next performed studies to deduce the nitrogen-sensing mechanisms of the inducible C-terminal domains of TnrA and GlnR.
Regulation of GlnR function by its nitrogen sensor domain
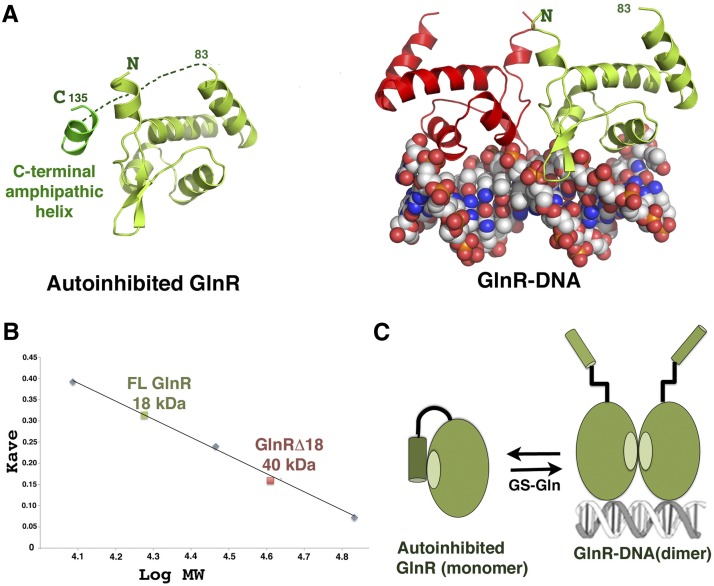
The GlnR C terminus contains an extension not found in TnrA that inhibits its DNA-binding function (Supplemental Fig. S2; Wray and Fisher 2008). The GlnRΔ18–DNA crystal structure provided insight into the GlnR autoinhibition mechanism, as, in addition to a protein–DNA complex, the structure also contained an apo (non-DNA-bound) GlnR subunit that is monomeric. In the crystal, the hydrophobic dimerization region of the apo GlnR is shielded from solvent by interactions with a short amphipathic helix from an adjacent GlnR molecule in the crystal (Fig. 3A). The finding that a short helix can stabilize apo GlnR and compete with the DNA-bound form of the GlnR dimer supports the notion that this dimer is weak. Notably, the C-terminal autoinhibitory region of GlnR is also predicted to form a short amphipathic helix, suggesting that the interaction observed in the structure may mimic the autoinhibited state of GlnR and function by preventing dimerization. To test this hypothesis, we carried out size exclusion chromatography (SEC) studies. These analyses showed that GlnRΔ18 forms dimers at 3 mg/mL, indicating that the weak GlnRΔ18 dimer can form at sufficiently high concentrations (Wray and Fisher 2008). In contrast, FL GlnR was monomeric (Fig. 3B; Supplemental Fig. S6).
Figure 3.
Molecular mechanism of GlnR autoinhibition. (A, left) Structure of apo GlnR with proposed autoinhibitory contacts present in the crystal structure. (Right) The DNA-bound form showing the subunit in the same orientation. (B) SEC of FL GlnR and GlnRΔ18 at 3 mg/mL showing that FL GlnR is a monomer and that GlnRΔ18 is a dimer. (C) Schematic model for GlnR autoinhibition and activation by GS. The C-terminal region of GlnR folds back and forms an autoinhibitory helix that inhibits dimer formation. Binding of this C-terminal tail by GS–Gln relieves autoinhibition, shifting the equilibrium to the DNA-binding active form.
Our data indicate a model for GlnR function in which the GlnR C-tail interacts with the dimer domain to prevent formation of the DNA-binding active dimer during low-nitrogen conditions (Fig. 3C). During nitrogen excess, the GlnR equilibrium shifts to the DNA-binding active form. This switch is expedited by a transient interaction between the GlnR autoinhibitory domain and feedback-inhibited GS (Wray and Fisher 2008). To try to gain insight into this unique GS chaperoning function, we carried out structural studies. Attempts to obtain a FL GlnR–GS–Gln structure, however, were not successful, likely due to the unstable nature of the complex. Hence, as only the GlnR autoinhibitory region is required for complex formation, we crystallized GS–Gln in the presence of a large excess of a GlnR C-terminal peptide, RFQRGNTFRQGDMSRFFH, and solved the structure by molecular replacement (MR) to 3.8 Å.
The GS in the GS–Gln–GlnR structure is essentially identical to previous B. subtilis GS structures (Murray et al. 2013), revealing a dodecamer with two stacked hexameric rings (Supplemental Fig. S7A). Each B. subtilis GS subunit contains 19 β strands and 15 α helices. All GS structures solved to date are dodecamers or decamers that are comprised of two stacked rings (either hexamers or pentamers) (Liaw and Eisenberg 1994; Eisenberg et al. 2000; Unno et al. 2006; van Rooyen et al. 2011). However, because GS active sites are formed at the interfaces between two subunits within a single ring, hexamer, or pentamer, it has been unclear what role the double-ring structure may play in GS function. Strikingly, the GS–GlnR peptide structure provides a function for this specific oligomeric arrangement, as it shows that the GlnR peptide binds in a surface-exposed region between stacked hexamers (Supplemental Fig. S7B,C). Upon GS binding, the GlnR peptide folds into a helix. These findings suggest that GS functions as a chaperone by sequestering the GlnR C-terminal tail within the exposed cavity and preventing the intramolecular GlnR autoinhibitory interaction. Therefore, these data indicate that a GS unable to form the stacked ring structure would not function as a GlnR chaperone. To test this hypothesis, we generated a GS truncation mutant lacking C-terminal residues 432–444 that mediates the so-called “helical thong” interaction that glues the two rings together (Supplemental Fig. S8A). Circular dichroism showed that the truncation mutant was folded (Supplemental Fig. S8B). Hence, the truncated and wild-type GS proteins were used in FP studies examining their ability to stimulate DNA binding by FL GlnR. In these studies, FL GlnR was titrated into the reaction cell with fluoresceinated DNA, and then either wild-type GS or the truncation mutant was added. As noted, FL GlnR shows only weak binding to DNA. This binding was stimulated significantly by the addition of FL GS. However, as predicted from the structure, titration of the GS truncation did not enable DNA binding by FL GlnR (Supplemental Fig. S8C).
Structural studies on the GlnK–TnrA–DNA complex
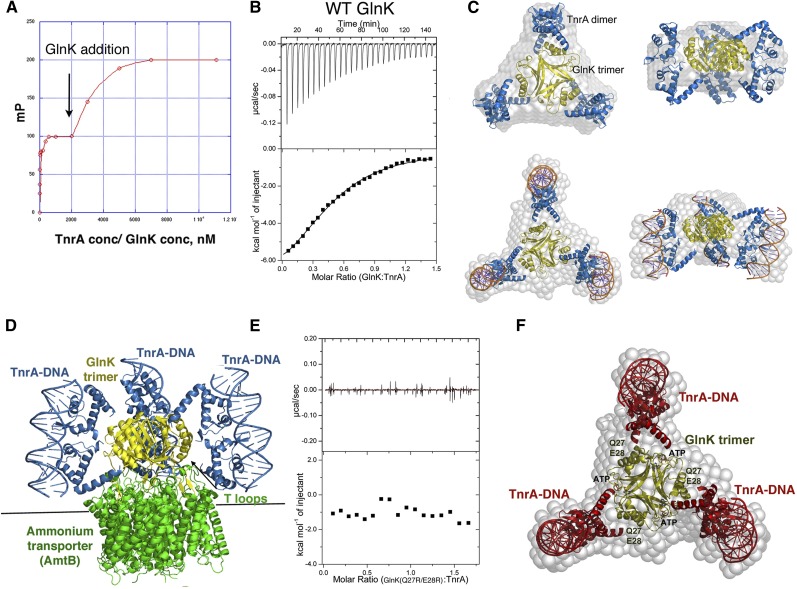
Like GlnR, the main regulator of TnrA activity is GS. However, when nitrogen is depleted, TnrA is released from GS, activating it for DNA binding. Interestingly, under these conditions, TnrA (but not GlnR) is further stabilized by an interaction with GlnK (Heinrich et al. 2006; Kayumov et al. 2008, 2011). The GlnK- and GS-interacting regions in TnrA (residues 75–90 and 95–110, respectively) are highly conserved, while the short region that connects them is not, suggesting that the mechanisms involved in TnrA regulation would be applicable in Bacilli and other Gram-positive bacteria. To gain insight into the role of GlnK in TnrA function, we first assessed the ability of TnrA and GlnK to form a stable complex using FP. In these experiments, TnrA was first bound to fluoresceinated cognate DNA. When GlnK was titrated into the mixture, a clear and saturable second binding event was revealed (Fig. 4A).
Figure 4.
Characterization of the TnrA–GlnK interaction. (A) FP analysis of GlnK binding to a preformed TnrA–DNA complex. TnrA–DNA complex formation is represented by the first binding event. Subsequent addition of GlnK leads to a second, saturable binding curve. (B) ITC analysis of the TnrA–GlnK interaction. The binding isotherm reveals a binding stoichiometry of two TnrA subunits (TnrA dimer):one GlnK subunit. (C) SAXS analyses of GlnK–TnrA and GlnK–TnrA–DNA complexes. Shown are the SAXS envelopes (gray) with docked GlnK (yellow) and TnrA (blue). The DNA is shown as sticks for the GlnK–TnrA–DNA complex. (D) Model of the GlnK–AmtB–TnrA–DNA quaternary complex constructed using the structure of GlnK–AmtB and the GlnK–TnrA–DNA SAXS model. (E) ITC analysis of the GlnK(Q27R/E28R) mutant showing no binding to TnrA. (F) SAXS GlnK–TnrA model, with the locations of the GlnK(Q27R/E28R) mutations and ATP, which disrupt TnrA binding by overlapping with its binding site, labeled.
To determine the GlnK:TnrA-binding stoichiometry, we next employed isothermal titration calorimetry (ITC) (Materials and Methods). These experiments revealed a binding stoichiometry of one TnrA dimer to one GlnK subunit and a Kd of 2.5 μM (Fig. 4B). How GlnK may interact with TnrA is unclear, as all PII/GlnK proteins that have been structurally characterized to date are trimers, and the active form of TnrA is a dimer (Arcondeguy et al. 2001). However, the B. subtilis GlnK shows limited sequence homology (<25%) with these proteins, suggesting that it could harbor a different fold and oligomeric arrangement. To address this possibility, we determined structures of the B. subtilis GlnK protein. Two crystal structures were obtained to 2.52 Å and 2.65 Å resolution (Supplemental Material; Supplemental Fig. S9A,B). Both revealed the characteristic trimeric arrangement observed in other GlnK/PII structures. Each GlnK subunit consists of five β strands and three helices. The long, central β sheets, composed of β2–β3, intertwine to form a trimeric β-sheet scaffold. The ends of this central sheet and its connecting loop form the so-called T-loop region, which has been shown to be flexible and mediate interactions with PII effector proteins (Arcondeguy et al. 2001).
The best-characterized PII-binding partner is the ammonium transporter (AmtB). In enteric bacteria, GlnK binds AmtB by inserting its T loops into the AmtB channel, blocking ammonium import during nitrogen excess (Supplemental Fig. S9C; Conroy et al. 2007; Gruzwitz et al. 2007). When nitrogen levels drop, a tyrosine in the T loops is uridylylated, causing it to be released from AmtB, allowing ammonium transport. The B. subtilis GlnK protein is not uridylylated and is also unique in that it interacts with TnrA (Heinrich et al. 2006). Interestingly, under certain conditions, B. subtilis GlnK appears to be able to bind both AmtB and TnrA, perhaps fine-tuning nitrogen homeostasis (Heinrich et al. 2006). To determine the molecular mechanism behind the GlnK–TnrA interaction, we next carried out small-angle X-ray scattering (SAXS) experiments on GlnK–TnrA and GlnK–TnrA–17mer DNA complexes.
The SAXS data for the GlnK–TnrA and GlnK–TnrA–DNA complexes permitted the construction of de novo envelopes of high quality (Schneidman-Duhovny et al. 2010; Tuukkanen and Svergun 2014). The GlnK trimer could be readily docked into the center of both envelopes, with extra density extending from the trimer (Fig. 4C; Supplemental Material; Supplemental Fig. S10). The DNA-bound dimeric conformation of TnrA provided an excellent fit to the remaining density at the sides of the GlnK–TnrA envelope, and, strikingly, compared with the GlnK–TnrA complex, the GlnK–TnrA–17mer envelope had additional density at each end of the knobs, which could be well accommodated by a 17mer DNA duplex (Fig. 4C). The resulting models show that each GlnK subunit is asymmetrically gripped between the ends of two 4α helices from a TnrA dimer. Unlike previous PII–effector complexes, TnrA does not interact with the GlnK T loops but rather its α helices. Notably, this model is consistent with data showing that residues in TnrA α4 interact with GlnK (Kayumov et al. 2011). Moreover, it is also consonant with the 1:2 GlnK subunit:TnrA subunit stoichiometry obtained from our ITC analyses and data indicating simultaneous binding of TnrA and AmtB by GlnK (Fig. 4D).
To test the SAXS model, we mutated GlnK residues Gln27 and Glu28, which the model predicts are critical for TnrA binding, and performed ITC analyses. As shown in Figure 4E, the GlnK (Q27R/E28R) mutant was defective in TnrA binding, supporting the structural model (Fig. 4F). Studies have shown that, like other PII proteins, B. subtilis GlnK binds ATP, serving as a sensor and integrator of cellular energetic and metabolic processes (Arcondeguy et al. 2001). Interestingly, these studies also demonstrated that, when in complex with ATP, the B. subtilis GlnK protein is unable to bind TnrA (Kayumov et al. 2011). Thus, to gain additional structural constraints on the GlnK–TnrA interaction and further test the SAXS model, we determined the B. subtilis GlnK–ATP structure (Supplemental Table S2). Strikingly, the ATP molecules, which bind at the interface between GlnK subunits (Supplemental Fig. S11A,B), overlap the TnrA-binding region predicted from the SAXS model, indicating why the GlnK–ATP complex cannot bind TnrA (Fig. 4F). Thus, our combined data, including crystallographic, mutagenesis, fluorescence, and ITC, support the GlnK–TnrA SAXS model. Importantly, these data suggest that GlnK stabilizes TnrA by acting as a templater of its active dimeric conformation.
Structure of the GS–Gln–TnrA complex: mechanism of TnrA deactivation and unprecedented GS oligomeric transformation
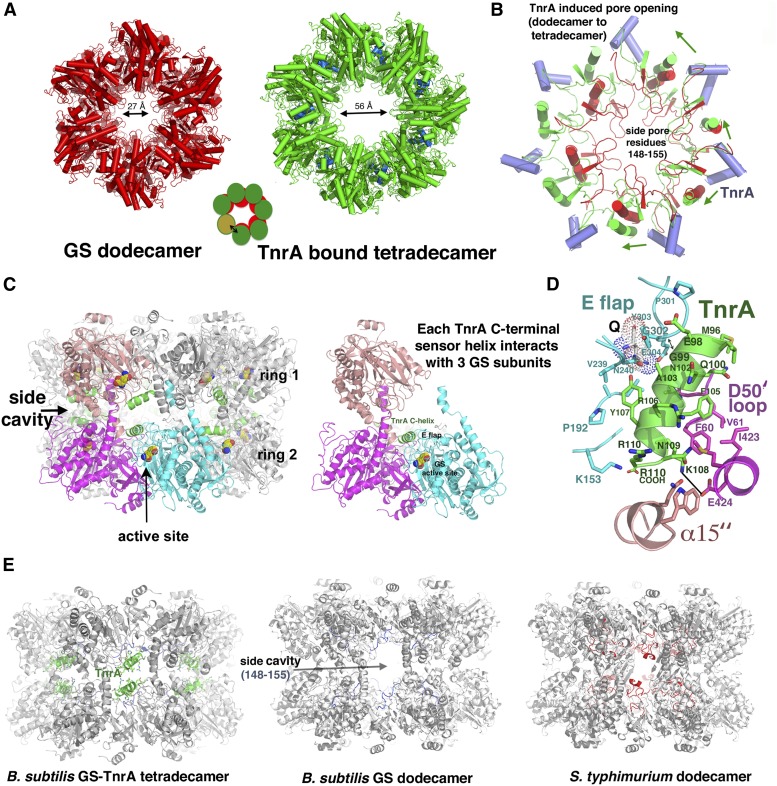
Under nitrogen-rich conditions, GS–Gln forms a tight complex with TnrA, disabling its DNA-binding activity. The C-terminal 15 residues of TnrA have been shown to be necessary and sufficient to mediate this interaction (Wray et al. 2001; Kayumov et al. 2011). To deduce how GS deactivates TnrA’s ability to bind DNA, we performed structural studies. Well-ordered crystals were obtained of TnrA(74–110) in complex with GS–Gln, and data from these crystals were used to determine the structure to 3.5 Å (Supplemental Material). Unexpectedly, the structure could not be solved using the B. subtilis GS dodecamer and instead was determined using a single GS subunit in a multicopy search (Murray et al. 2013; Bunkoczi et al. 2013). Remarkably, the combined solution of individual subunits resulted in the generation of a GS tetradecamer (Fig. 5A). Initial refinement of this model resulted in Rwork/Rfree = 31.9%/36.0% compared with the best dodecamer solution, Rwork/Rfree = 48.9%/59.8%, supporting the tetradecamer as the correct solution (Fig. 5A,B; Supplemental Table S2). A tetradecameric GS structure is unprecedented, as all bacterial GS structures solved to date, including B. subtilis GS structures in the absence of TnrA, are dodecamers (Liaw and Eisenberg 1994; Eisenberg et al. 2000; Murray et al. 2013). The TnrA-bound tetradecameric GS subunits are similar to those in the dodecameric GS structures (RMSD = 0.6–0.9 Å); however, conversion to the tetradecamer results in shifts in the subunit interfaces, which, notably, form the GS active sites. Consistent with studies showing that the 15-residue C-terminal tail of TnrA mediates GS binding, helical electron density is visible for ∼15 residues for 14 TnrA molecules near the intersubunit GS cavities, here termed “side cavities,” indicating that these TnrA residues fold into a helix upon binding GS (Fig. 5C; Supplemental Fig. S12; Wray et al. 2001; Wray and Fisher 2007; Kayumov et al. 2011). Each GS side cavity binds two noninteracting TnrA molecules, and each TnrA-binding pocket is constructed from three different GS subunits: two from one heptamer and the third from a subunit in the adjacent stacked heptamer (Fig. 5C).
Figure 5.
The GS–Gln–TnrA complex; unprecedented GS oligomeric conversion and basis for TnrA inhibition by Gram-positive GS. (A) Comparison of apo B. subtilis GS (red) and GS (green) bound to TnrA (blue). (B) One subunit of the TnrA-bound GS (green) was overlaid onto that of the dodecamer GS (red) to underscore how TnrA (blue helices) binding causes a shift in the α15″ helices and pore loops. This cascading effect leads to pore opening and the ultimate insertion of a seventh subunit in each GS layer. (C) The TnrA binding pocket is near the GS active site and is composed of residues from three different GS subunits: two from one heptamer and the third from the adjacently stacked heptamer. (D) Close-up view of the TnrA–GS–Gln interaction. TnrA is green, and the three interacting GS subunits are colored (as in C) magenta, salmon, and cyan. The Gln is labeled Q and is shown as dots. (E) Molecular explanation for why TnrA binds Gram-positive but not Gram-negative GS. GS molecules are shown rotated 90° compared with A to show the stacked ring structures and side cavity locations where TnrA binds. The Gram-negative GS side regions contain a large insertion, which overlaps the TnrA-binding site.
The finding that GS can undergo a massive interconversion from a dodecamer to a tetradecamer upon TnrA binding suggests that the enzyme must be able to transit between oligomeric states in solution. In fact, previous SEC and electron microscopy studies demonstrated that the GSs from Escherichia coli, Ruminococcus albus 8, Neurospora crassa, and the plant Phaseolus vulgaris are present in solution as both monomers and their requisite higher-order oligomers (Dávila et al. 1980; Denman and Wedler 1984; Amaya et al. 2005; Llorca et al. 2006). However, to assess the solution oligomeric status of the B. subtilis enzyme, we performed SEC studies. Our analyses showed that the apo B. subtilis GS (or GS–Gln complex) is present in solution as both monomers and dodecamers (Supplemental Figs. S13, S14). Specifically, SEC on apo GS and the GS–Gln complex revealed peaks corresponding to molecular weights (MWs) of ∼40 kDa/44 kDa and 490 kDa/480 kDa, respectively (compared with the calculated MW for monomer/dodecamer of 50 kDa/600 kDa). These data provide an explanation for where the extra monomers come from to build the GS tetradecamer when bound to TnrA, as they indicate that the GS enzyme exists in an equilibrium between monomer and higher-order oligomeric states. Strikingly, SEC analyses on the GS–Gln–TnrA complex showed a peak corresponding to a MW of ∼1.1 MDa, which is consistent with the tetradecamer crystal structure (the tetradecameric GS–Gln–TnrA complex corresponds to ∼990 kDa, while a GS[dodecamer]–TnrA complex is ∼830 kDa) (Materials and Methods; Supplemental Figs. S13, S14). Thus, GS ring opening caused by TnrA binding (schematized in Fig. 5A) would allow the insertion of two GS monomers that are free in solution.
The binding site for TnrA directly abuts the GS active site, located at the interface between GS subunits. In fact, this finding is consonant with previous genetic screens and alanine scanning studies, which indicated that substitution of GS residues in the catalytic E flap, Gly59 from the D50′ loop, and Glu424 inhibited TnrA binding (Wray et al. 2001; Fisher et al. 2002; Fisher and Wray 2006; Wray and Fisher 2007; Kayumov et al. 2011; Murray et al. 2013). However, because most of these mutations were in the active site, it has been unclear whether they directly affect TnrA binding or have indirect effects (i.e., alter Gln binding). The structure shows that the combined residues implicated in these studies, including Glu424, form a continuous binding pocket for TnrA. Notably, most of the GS contacts to TnrA are mediated by the E flap (residues 301–306) and D50′ loop (residues 52–66), which are arguably the most important enzymatic loops in GS; the D50′ loop provides the aspartic acid that abstracts a proton from ammonium, while the E flap harbors the catalytic base Glu304 (Fig. 5C,D; Supplemental Fig. S15). Residues in α14′ (residues 400–430) and the C-terminal region of α15″ (where ′ refers to the neighboring subunit and ″ indicates the subunit from the heptamer from the adjacent layer) (Fig. 5D) also contact TnrA. The tight apposition of TnrA and the E flap necessitates that TnrA residue 99 and GS E flap residue 302 are both glycines, which explains why mutation of either residue abolishes binding (Fig. 5D; Wray et al. 2001; Wray and Fisher 2007). The helical face of TnrA opposite to that interacting with the E flap contacts the D50′ loop; Gln100 interacts with the backbone atoms of residues 50–52, and Phe105 packs against Phe60 and Val61, while TnrA residue Met96 interacts with Ile63 (Fig. 5D). Finally, Glu424 from α14′ makes an electrostatic interaction with TnrA residue Lys108, providing an explanation for why a GS E424K mutation, which would lead to charge–charge repulsion, inhibits TnrA binding (Fisher et al. 2002).
The insertion of the TnrA sensor helix into the GS side cavity requires shifts in both the adjacent C-terminal α15″ helix and the pore loop (residues 148–155). These structural changes allow contacts between the TnrA Arg110 side chain and residues from α15″ as well as electrostatic interactions between the Arg110 C-terminal carboxyl group and the pore-lining residue Lys153 (Fig. 5D). The cumulative shifts triggered by these interactions appear to drive the dramatic oligomeric GS reconfiguration, which is underscored by the expansion of the intrapore size from 27 Å in the dodecamer to ∼56 Å in the tetradecamer (Fig. 5A). The structure also explains the long-standing conundrum of why TnrA cannot bind Gram-negative GS enzymes. Comparison of the B. subtilis and Gram-negative GS shows that the primary difference between their overall structures is that the Gram-negative GS contains an extra ∼30-residue insertion in its side cavities compared with the B. subtilis GS. This addition overlaps the TnrA-binding site. Hence, TnrA would be unable to bind the Gram-negative GS side cavities (Fig. 5E).
Biochemical analyses of the TnrA–GS interaction
Our SEC analyses support the structural data showing that the GS–Gln–TnrA complex is tetradecameric. However, to provide additional tests of the model, we generated mutant TnrA and GS proteins and performed biochemical studies. As the GS–TnrA structure revealed an electrostatic interaction between TnrA residue Lys108 and GS residue Glu424, we generated TnrA(K108E) and GS(E424K) mutants and used the resultant proteins in FP experiments. In these studies, TnrA proteins were first bound to fluoresceinated DNA (both wild-type and K108E mutant TnrA bound DNA with essentially the same affinity), and then increasing concentrations of wild-type GS or GS(E424K) were added. The results (Supplemental Fig. S16) showed that, as expected, wild-type GS was able to bind and remove wild-type TnrA from the DNA, while GS(E424K) was not. In contrast, TnrA(K108E) showed little to no interaction with wild-type GS, as it remained bound to the DNA. Addition of GS(E424K) was able to partially remove bound TnrA(K108E), suggesting that the charge-swapped proteins interacted. The binding of TnrA(K108E) to GS(E424K) might be expected to be weaker than the wild-type interaction due to the presence of additional negatively charged residues in the binding pocket that also provide electrostatic contributions to Lys108 binding, including Glu27 and Glu158, the latter of which makes direct contacts to the Lys108 side chain in some pockets. Thus, the combined results are consistent with the structure.
A key finding from the GS–TnrA structure is that TnrA binds in the central cavity between heptamers. Hence, as another assessment of the model, we next used the GS truncation mutant in combination with FP. In these assays, TnrA was first bound to a fluoresceinated 21mer cognate DNA, and then increasing concentrations of wild-type or mutant GS proteins were added. Addition of FL GS led to binding and hence removal of TnrA from the DNA; however, the GS truncation mutant showed no interaction with TnrA, as ascertained by its inability to dissociate TnrA from cognate DNA even at millimolar concentrations, supporting the structural data (Fig. 6A).
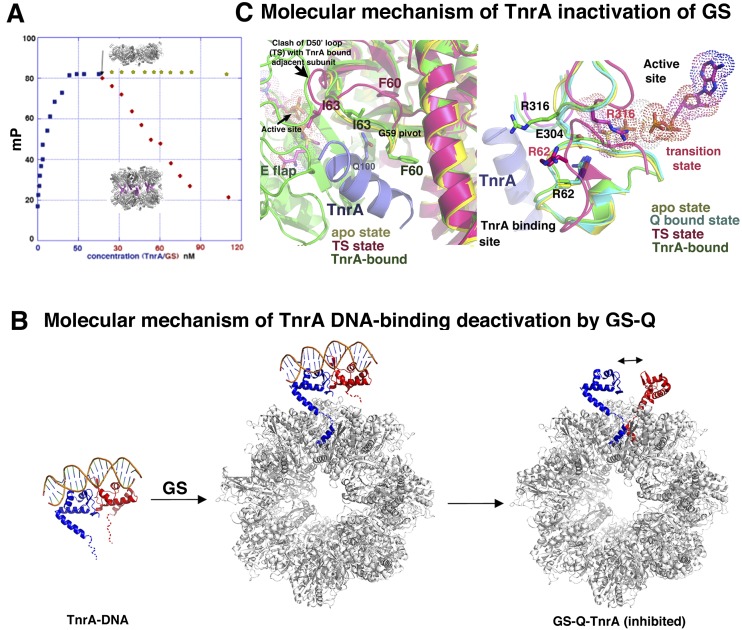
Figure 6.
Dual deactivation of TnrA and GS by formation of the GS–TnrA complex. (A) FP analysis in which TnrA was first bound to fluoresceinated DNA, and then increasing concentrations of FL GS or the GS truncation were added. FL GS dissociated TnrA from the DNA, but the truncation GS mutant could not. (B) Molecular mechanism for GS inhibition of TnrA DNA binding. For clarity, only one TnrA dimer (subunits colored red and blue) is shown, although each GS can bind seven TnrA dimers. GS binding of one TnrA subunit places the second TnrA sensor of the dimer in proximity to another binding pocket. The TnrA dimer would be disrupted to permit the second TnrA sensor to bind. (C) TnrA inhibition of GS. (Left) Superposition of one subunit of the transition state (TS) GS (magenta), apo GS (yellow), and GS–Gln–TnrA (green) structures. The bound TnrA (blue) makes multiple contacts to the D50′ loop that would prevent TS formation and hence catalysis. Tetradecamer formation, which creates different intersubunit interfaces at the active site, would also prevent TS formation. (Right) The same overlay as at left but highlighting that the GS apo and Gln-bound state harbor the same structure. TS changes, including Arg316 relocation, are prevented by TnrA binding.
Molecular mechanism of GS inactivation of TnrA
The GS–Gln–TnrA structure shows that the TnrA sensor, residues 95–110, folds into a helix upon GS binding. Therefore, when bound to GS, the FL TnrA would consist of two ordered regions—the N-terminal winged–HTH domain (residues 1–90) and the GS-interacting helix (residues 95–110)—that are connected by a four-residue linker. Even when fully extended, such a linker would reach only ∼12 Å. Modeling shows that binding the one TnrA subunit to GS would juxtapose the C-terminal tail of the second TnrA in the dimer near a GS-binding site, hence facilitating its interaction with GS (Fig. 6B). However, when the TnrA–DNA dimer structure is docked onto the GS–TnrA complex with one bound TnrA subunit, the closest achievable distance between residues 90 and 95 in the second TnrA subunit is ∼26 Å (Fig. 6B). As a consequence, the TnrA dimer would be disfavored when both of its sensors simultaneously bind GS. Dimer disruption would be further enabled by the relatively weak TnrA dimer (Fig. 6B).
TnrA functions as a novel allosteric regulator of GS
Interestingly, TnrA has been shown to bind apo GS, albeit with reduced affinity. Moreover, these studies revealed that formation of this complex inhibits GS enzyme activity (Wray and Fisher 2002; Fedorova et al. 2013). This reciprocal GS/TnrA deactivation could provide tight control of the generation of nitrogen metabolic factors during nitrogen excess. However, the basis for TnrA-mediated inactivation of GS has been unclear. Our structural data provide an explanation for this unusual catalytic inhibition, as the data show that TnrA interacts with the GS active site loops, in particular the D50′ loop, and freezes them in the inactive state (Fig. 6C). Indeed, the D50′ loop, which is in the same conformation in the apo as the Gln-bound states, undergoes a large conformational change during catalysis, which is required to create an optimal architecture for the GS reaction (Murray et al. 2013). Gly59 serves as a pivot for the D50′ loop switch, but, in the TnrA bound structure, this residue cannot move, as it forms a tight interaction with TnrA. This finding explains why a GS G59R mutation abrogates TnrA binding (Fig. 6C; Fisher et al. 2002). In addition to TnrA freezing the GS D50′ loop in its inactive state, the large GS oligomeric conversion caused by TnrA binding results in altered subunit interfaces, which form the active site. Thus, these data reveal how TnrA functions as a novel allosteric regulator of GS.
Discussion
Cell-biological and genetic studies have demonstrated that, in sharp contrast to the NtrBC/σ54 system employed by enteric bacteria, B. subtilis uses an unrelated set of transcription regulators (TnrA and GlnR) to detect and respond to intracellular nitrogen levels. The subsequent finding that the activities of both proteins are regulated by feedback-inhibited GS revealed a novel regulatory paradigm in which a central enzyme in metabolism (GS) directly transduces nutrient availability to global transcription regulators (Wray et al. 2001; Fisher and Wray 2008). Recent work indicated a secondary role for GlnK in this circuitry as a TnrA-interacting protein (Kayumov et al. 2011). Thus, these combined studies defined the B. subtilis nitrogen regulatory network. However, there are no related signaling systems that can be used to glean molecular insight into this circuitry. Hence, the mechanisms that control it have remained unknown. In this study, we provide a molecular dissection of this network, as schematized in Figure 7.
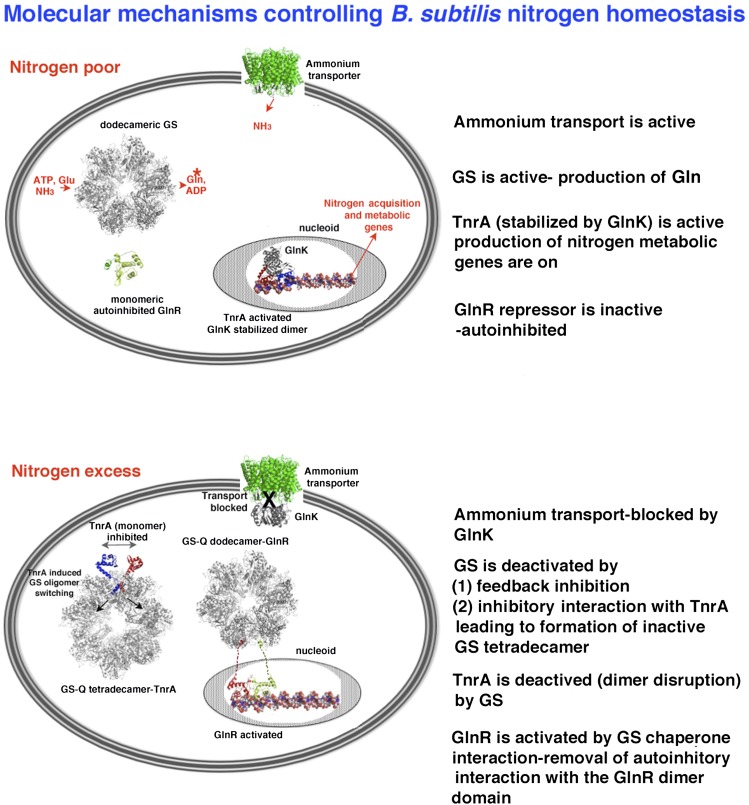
Figure 7.
Schematic summarizing the molecular details that control B. subtilis nitrogen homeostasis. (Top) B. subtilis regulatory proteins and machinery during low-nitrogen conditions. (Bottom) Circuitry and their interactions during nitrogen excess.
To understand the regulatory mechanisms used by TnrA and GlnR, we obtained multiple structures of both proteins in complex with DNA. These structures revealed TnrA and GlnR as founding members of a new family of DNA-binding proteins that appear to have evolved from or represent a separate branch of the MerR family of transcription regulators. Indeed, although TnrA/GlnR and MerR proteins have highly related winged–HTH, their mode of DNA-binding and dimerization are completely unrelated. Whereas MerR proteins form tight dimers via their extended C-terminal coiled coils, TnrA/GlnR form weak dimers on cognate DNA by contacts between their N-terminal DNA-binding domains. Instead of dimerization, the C-terminal regions of TnrA and GlnR function as nitrogen sensors that perform multiple functions, including autoinhibition and effector binding, the ultimate outcome of which determines their DNA-binding activity. These functions were unveiled in molecular detail by structures of TnrA and GlnR in various states and in complex with effector proteins. These structures revealed that the functions of the inducer domains are all intimately tied to the weak dimerization of the GlnR and TnrA proteins by which the sensors themselves or their interaction with effectors act as switches to shift the equilibrium of the dimer forms of the proteins.
GS–Gln formation signals the presence of excess nitrogen and transmits that signal by interacting with and affecting the DNA-binding and transcription programs of both GlnR and TnrA. GS–Gln activates GlnR through a chaperoning interaction. In contrast, GS–Gln forms a stable complex with TnrA, inhibiting its DNA-binding function. How the large GS oligomer mediates these functions has been a mystery. Indeed, all bacterial GSs, including the B. subtilis enzyme, form dodecamers composed of two stacked hexamers. However, there has been no known function for this specific structure. Here we show that B. subtilis GS uses the interring cavity as a protein–protein interaction site, with multiple functional consequences. Specifically, this cavity acts as a chaperone for GlnR, folding its C-terminal tail into a helix and preventing its autoinhibition interaction. Binding of the TnrA sensor to this region also results in a helical transition; however, the complex that is formed causes TnrA dimer disengagement. Intriguingly, the altered TnrA “dimer” that would be bound to GS bears some resemblance to a canonical MerR dimer, except that the C-terminal helices in TnrA are not as tightly intertwined as the MerR coiled coils, suggesting again an evolutionary link between these structural families (Supplemental Fig. S17).
In the GS–TnrA structure, the TnrA helix interacts with three subunits of GS: two subunits that combine to form an active site and an adjacent subunit from the stacked heptamer layer. An unexpected result of this deep insertion is the concomitant movement of GS pore elements and dramatic expansion of the GS dodecamer. This expansion opens the GS oligomer, leaving an unoccupied site within each stacked layer that can be precisely fit with an extra GS subunit. The remarkable result is the generation of a GS tetradecamer, an oligomeric state that has never been observed for any GS but can be explained by the presence, in solution, of GS monomers in equilibrium with higher-order oligomers. These structural changes have multiple functional outcomes. First, they convert GS into a catalytically inactive state. Second, the folding of the C-terminal 15 residues of two TnrA sensor domains deep within each GS side cavity provides the switch that turns off TnrA DNA-binding activity, as the region connecting TnrA α4 to the newly folded α5 helices is too short to permit the dimer configuration (Fig. 5D). Thus, the binding of the GlnR and TnrA sensor domains to GS–Gln affects their transcription activity by stabilizing the formation of the dimer of the GlnR repressor and inhibiting TnrA dimerization (Fig. 7).
Upon shifting to nitrogen-limiting conditions, Gln dissociates from GS, allowing its C-terminal tail to form the intramolecular autoinhibitory interaction that prevents GlnR dimer formation and DNA binding. This shuts off GlnR repression. Under these conditions, TnrA is released from GS–Gln, allowing TnrA dimerization and activation of its transcription program. GlnK appears to play an ancillary role in this process by acting as a templating agent for TnrA. This stabilization function is also consistent with data showing that GlnK can protect TnrA against proteolysis (Kayumov et al. 2008). In conclusion, our combined data provide detailed molecular snapshots of all of the nitrogen regulatory components and their germane complexes from the model Gram-positive bacteria B. subtilis. These structures reveal a novel paradigm for metabolic regulation in which inducible sensor domains are harnessed via multiple disparate mechanisms under different environmental queues to directly impact distant DNA-binding domains of master transcription regulators and hence drive the reprograming of an entire metabolic pathway.
Materials and methods
Protein purification, crystallization, and structure determination
The genes encoding B. subtilis and B. megaterium TnrA, B. subtilis GlnR, B. subtilis GS, and B. subtilis GlnK were purchased from Genscript Corporation and subcloned into pET15b such that a His tag was expressed for purification. For detailed descriptions of the purification, crystallization, structure determination, and refinement protocols, see the Supplemental Material.
ITC analyses on GlnK–TnrA interactions
ITC experiments were performed using a VP-ITC system. For these experiments, TnrA and GlnK were dialyzed into the same buffer, consisting of 100 mM NaCl and 20 mM Tris HCl (pH 7.5). The GlnK protein, either wild-type GlnK or the GlnK(Q27R–E28R) mutant, at a concentration of 74 μM was titrated into the sample cell containing 11 μM TnrA. The data were fit using Origin7 software.
SAXS analyses of the GlnK–TnrA and GlnK–TnrA–DNA complexes
SAXS data were collected at Advance Light Source (ALS) beamline 12.3.1 at a wavelength of 1 Å. The details of the experiments are provided in the Supplemental Material. Briefly, GlnK–TnrA and GlnK–TnrA–DNA complexes were formed with stoichiometric ratios of 1:2 and 1:2:1, respectively. SAXS data were collected over a concentration range of 1–4 mg/mL, and the profiles were evaluated for aggregation using Guinier analyses (Schneidman-Duhovny et al. 2010). Kratky plots indicated that the samples were homogeneous and well-folded. The resultant molecular envelopes displayed threefold volumes. Hence, threefold symmetry was enforced for the final envelopes used for final molecular docking (Tuukkanen and Svergun 2014). BUNCH was used to produce and evaluate three-dimensional models in which the GlnK trimer and TnrA crystal structures were docked into envelopes, and fits were assessed via χ2 values.
Accession numbers
Coordinates and structure factor amplitudes have been deposited with the Protein Data Bank under accession codes 4R22, 4R24, 4R25, 4R4E, and 4S0R.
Supplementary Material
Acknowledgments
We thank Dr. Sue Fisher and Dr. Lewis Wray for past collaborative interactions. SAXS and X-ray crystallographic data were collected at the Advanced Light Source (ALS). ALS is a national user facility operated by Lawrence Berkeley National Laboratory on behalf of the Department of Energy (DOE), Office of Basic Energy Sciences, through the Integrated Diffraction Analysis Technologies program supported by the DOE Office of Biological and Environmental Research. Additional support comes from project MINOS (Macromolecular Insights on Nucleic Acids Optimized by Scattering) grant R01GM105404. This work was supported by an M.D. Anderson Trust Fellowship and Duke start-up funds (to M.A.S.).
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.254714.114.
References
- Amaya KR, Kocherginskaya SA, Mackier RI, Cann IKO. 2005. Biochemical and mutational analysis of glutamine synthetase type III from the rumen anaerobe Ruminococcus albus 8. J Bacteriol 187: 7481–7491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arcondeguy T, Jack R, Merrick M. 2001. PII signal transduction proteins, pivotal players in microbial nitrogen control. Microbiol Mol Biol Rev 65: 80–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SW, Sonenshein AL. 1996. Autogeneous regulation of the Bacillus subtilis glnRA operon. J Bacteriol 178: 2450–2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunkoczi G, Echols N, McCoy AJ, Oeffner RD, Adams PD, Read RJ. 2013. Phaser MRage: automated molecular replacement. Acta Crystallogr D Biol Crystallogr 69: 2276–2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conroy MJ, Durand A, Lupo D, Li XD, Bullough PA, Winkler FK, Merrick M. 2007. The crystal structure of the Escherichia coli AmtB–GlnK complex reveals how GlnK regulates the ammonia channel. Proc Natl Acad Sci 104: 1213–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dávila G, Lara M, Guzmán J, Mora J. 1980. Relation between structure and function of Neurospora crassa glutamine synthetase. Biochem Biophys Res Commun 92: 134–140. [DOI] [PubMed] [Google Scholar]
- Denman RB, Wedler FC. 1984. Association-dissociation of mammalian brain glutamine synthetase: effects of metal ions and other ligands. J Biol Chem 232: 427–440. [DOI] [PubMed] [Google Scholar]
- Deuel TF, Ginsburg A, Yeh J, Shelton E, Stadtman ER. 1970. Bacillus subtilis glutamine synthetase. Purification and physical characterization. J Biol Chem 245: 5195–5205. [PubMed] [Google Scholar]
- Dunker AK, Silman I, Uversky VN, Sussman JL. 2009. Function and structure of inherently disordered proteins. Curr Opin Struct Biol 18: 756–764. [DOI] [PubMed] [Google Scholar]
- Eisenberg D, Gill HS, Pfluegl GM, Rotstein SH. 2000. Structure function relationships of glutamine synthetases. Biochim Biophys Acta 1477: 122–145. [DOI] [PubMed] [Google Scholar]
- Fedorova K, Kayumov A, Woyda K, Ilinskaja O, Forchhammer K. 2013. Transcription factor TnrA inhibits the biosynthetic activity of glutamine synthetase in Bacillus subtilis. FEBS Lett 587: 1293–1298. [DOI] [PubMed] [Google Scholar]
- Fisher SH. 1999. Regulation of nitrogen metabolism in Bacillus subtilis: vive la difference! Mol Microbiol 32: 223–232. [DOI] [PubMed] [Google Scholar]
- Fisher SH, Wray LV Jr. 2006. Feedback-resistant mutations in Bacillus subtilis glutamine synthetase are clustered in the active site. J Bacteriol 188: 5966–5974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher SH, Wray LV Jr. 2008. Bacillus subtilis glutamine synthetase regulates its own synthesis by acting as a chaperone to stabilize GlnR–DNA complexes. Proc Natl Acad Sci 105: 1014–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher SH, Brandenburg JL, Wray LV Jr. 2002. Mutations in Bacillus subtilis glutamine synthetase that block its interaction with transcription factor TnrA. Mol Microbiol 45: 627–635. [DOI] [PubMed] [Google Scholar]
- Gruzwitz F, O’Connell J 3rd, Stroud RM. 2007. Inhibitory complex of the transmembrane ammonia channel, AmtB, and the cytosolic regulatory protein, GlnK, at 1.96 Å. Proc Natl Acad Sci 104: 42–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunka K, Commichau FM. 2012. Control of glutamate homeostasis in Bacillus subtilis: a complex interplay between ammonium assimilation, glutamate biosynthesis and degradation. Mol Microbiol 86: 213–224. [DOI] [PubMed] [Google Scholar]
- Gutowski JC, Schreier HJ. 1992. Interaction of the Bacillus subtilis glnRA repressor with operator and promoter sequences in vivo. J Bacteriol 174: 671–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich A, Woyda K, Brauburger K, Meiss G, Detsch C, Stülke J, Forchhammer K. 2006. Interaction of the membrane-bound GlnK–AmtB complex with the master regulator of nitrogen metabolism TnrA in Bacillus subtilis. J Biol Chem 281: 34909–34917. [DOI] [PubMed] [Google Scholar]
- Heldwein EE, Brennan RG. 2001. Crystal structure of the transcription activator BmrR bound to DNA and a drug. Nature 409: 378–382. [DOI] [PubMed] [Google Scholar]
- Kayumov A, Heinrich A, Sharipova M, Iljinskaya O, Forchhammer K. 2008. Inactivation of the general transcription factor TnrA in Bacillus subtilis by proteolysis. Microbiology 154: 2348–2355. [DOI] [PubMed] [Google Scholar]
- Kayumov A, Heinrich A, Fedorova K, Ilinskaya O, Forchhammer K. 2011. Interaction of the general transcription factor TnrA with the PII-like protein GlnK and glutamine synthetase in Bacillus subtilis. FEBS J 278: 1742–1789. [DOI] [PubMed] [Google Scholar]
- Krissinel E, Hendrick K. 2007. Inference of macromolecular assemblies from crystalline state. J Mol Biol 372: 774–797. [DOI] [PubMed] [Google Scholar]
- Lamoureux JS, Maynes JT, Glover JNM. 2004. Recognition of 5′-YpG-3′ sequences by coupled stacking/hydrogen bonding interactions with amino acid residues. J Mol Biol 335: 399–408. [DOI] [PubMed] [Google Scholar]
- Liaw SH, Eisenberg D. 1994. Structural model for the reaction mechanism of glutamine synthetase, based on five crystal structures of enzyme–substrate complexes. Biochemistry 33: 675–681. [DOI] [PubMed] [Google Scholar]
- Llorca O, Betti M, González JM, Valencia A, Márquez AJ, Valpuesta JM. 2006. The three-dimensional structure of an eukaryotic glutamine synthetase: functional implications of its oligomeric state. J Struct Biol 156: 469–479. [DOI] [PubMed] [Google Scholar]
- Murray DS, Chinnam N, Tonthat NK, Whitfill T, Wray LV Jr, Fisher SH, Schumacher MA. 2013. Structures of the Bacillus subtilis glutamine synthetase dodecamer reveal large intersubunit catalytic conformational changes linked to a unique feedback inhibition mechanism. J Biol Chem 288: 35801–35811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newberry KJ, Brennan RG. 2004. The structural mechanism for transcription activation by MerR family member multidrug transporter activation, N terminus. J Biol Chem 279: 20356–20362. [DOI] [PubMed] [Google Scholar]
- Reitzer L. 2003. Nitrogen assimilation and global regulation in Escherichia coli. Annu Rev Microbiol 57: 155–176. [DOI] [PubMed] [Google Scholar]
- Schneidman-Duhovny D, Hammel M, Sali A. 2010. FoXS: a Web server for rapid computation and fitting of SAXS profiles. Nucleic Acids Res 38: W540–W544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuukkanen AT, Svergun DI. 2014. Weak protein-ligand interactions studied by small-angle X-ray scattering. FEBS J 281: 1974–1987. [DOI] [PubMed] [Google Scholar]
- Unno H, Uchida T, Sugawara H, Kurisu G, Sugiyama T, Yamaya T, Sakakibara H, Hase T, Kusunoki M. 2006. Atomic structure of plant glutamine synthetase. A key enzyme for plant productivity. J Biol Chem 281: 29287–29296. [DOI] [PubMed] [Google Scholar]
- van Rooyen JM, Abratt VR, Belrhali H, Sewell T. 2011. Crystal structure of the type III glutamine synthetase: surprising reversal of the inter-ring interface. Structure 19: 471–483. [DOI] [PubMed] [Google Scholar]
- Wray LV Jr, Fisher SH. 2002. A feedback-resistant mutant of Bacillus subtilis glutamine synthetase with pleiotropic defects in nitrogen-regulated gene expression. J Biol Chem 280: 33298–33304. [DOI] [PubMed] [Google Scholar]
- Wray LV Jr, Fisher SH. 2007. Functional analysis of the carboxy-terminal region of Bacillus subtilis TnrA, a MerR family protein. J Bacteriol 189: 20–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray LV Jr, Fisher SH. 2008. Bacillus subtilis GlnR contains an autoinhibitory C-terminal domain required for the interaction with glutamine synthetase. Mol Microbiol 68: 277–285. [DOI] [PubMed] [Google Scholar]
- Wray LV Jr, Ferson AE, Rohrer K, Fisher SH. 1996. TnrA, a transcription factor required for global nitrogen regulation in Bacillus subtilis. Proc Natl Acad Sci 93: 8841–8845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray LV Jr, Zalieckas JM, Fisher SH. 2000. Purification and in vitro activities of the Bacillus subtilis TnrA transcription factor. J Mol Biol 200: 29–40. [DOI] [PubMed] [Google Scholar]
- Wray LV Jr, Zalieckas JM, Fisher SH. 2001. Bacillus subtilis glutamine synthetase controls gene expression through a protein–protein interaction with transcription factor TnrA. Cell 107: 427–435. [DOI] [PubMed] [Google Scholar]
- Yoshida K, Yamaguchi H, Kinehara M, Ohki YH, Nakaura Y, Fujita Y. 2003. Identification of additional TnrA-regulated genes of Bacillus subtilis associated with a TnrA box. Mol Microbiol 49: 157–165. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.