Summary
Psychiatric disorders autism and schizophrenia have a strong genetic component, and copy number variants (CNVs) are firmly implicated. Recurrent deletions and duplications of chromosome 16p11.2 confer high risk for both diseases, but the pathways disrupted by this CNV are poorly defined. Here we investigate the dynamics of 16p11.2 network by integrating physical interactions of 16p11.2 proteins with spatio-temporal gene expression from developing human brain. We observe profound changes in protein interaction networks throughout different stages of brain development and/or in different brain regions. We identify late mid-fetal period of cortical development as most critical for establishing connectivity of 16p11.2 proteins with their co-expressed partners. Furthermore, our results suggest that the regulation of KCTD13-Cul3-RhoA pathway in layer four of inner cortical plate is crucial for controlling brain size and connectivity, and its dysregulation by the de novo mutations may be a potential determinant of 16p11.2 CNV deletion and duplication phenotypes.
Introduction
Accumulating evidence suggests that rare copy number variants (CNVs) are an important risk factor to multiple psychiatric disorders (Malhotra and Sebat, 2012) including autism spectrum disorders (ASD) (Levy et al., 2011; Marshall et al., 2008; Pinto et al., 2010; Sanders et al., 2011; Sebat et al., 2007), schizophrenia (SCZ) (Consortium, 2008; Kirov et al., 2009; Stefansson et al., 2008; Walsh et al., 2008), bipolar disorder (BD) (Malhotra et al., 2011), developmental delay (DD) (Cooper et al., 2011), attention deficit hyperactivity disorder (ADHD) (Lionel et al., 2011), and intellectual disability (ID) (Girirajan et al., 2012; Merikangas et al., 2009). One of the most frequent CNVs involved in neurodevelopmental diseases is the 16p11.2 CNV locus encompassing ~600 kb (chr16:29.5–30.2 Mb). The 16p11.2 CNV was implicated in multiple psychiatric phenotypes with the deletions associated with ASD and ID, whereas the duplications have been associated with ASD, SCZ, BD and ID (Bijlsma et al., 2009; Malhotra and Sebat, 2012; Marshall et al., 2008; McCarthy et al., 2009; Weiss et al., 2008). Moreover, a reciprocal dosage effect of 16p11.2 on the head size has been reported, with macrocephaly observed in the deletion carriers and microcephaly observed in the duplication carriers (McCarthy et al., 2009). These human phenotypes were recapitulated in zebrafish by either increasing or suppressing the expression of KCTD13, respectively (Golzio et al., 2012). The mouse models of 16p11.2 CNVs have dosage-dependent changes in gene expression, brain architecture, behavior and viability (Horev et al., 2011; Portmann et al., 2014). In humans, transcriptome profiling from lymphoblasts of 16p11.2 CNV carriers identified expression dysregulation of many genes located outside of the 16p11.2 locus, in addition to the changes of genes’ dosage within the locus (Luo et al., 2012).
Despite the progress in linking 16p11.2 genetic changes with the phenotypic abnormalities in the patients and model organisms, the specific brain regions, developmental periods, networks and pathways impacted by this CNV remain unknown. To address these questions we have constructed dynamic spatio-temporal networks of 16p11.2 genes by integrating data from brain developmental transcriptome (Kang et al., 2011; Miller et al., 2014) with physical interactions of 16p11.2 proteins (Chatr-Aryamontri et al., 2013; Corominas et al., 2014; Rolland et al., 2014).
Until now, most protein-protein interaction (PPI) studies of CNVs in psychiatric disorders have been focused on analyzing static topological network properties, such as connectivity, modules and clusters (Gilman et al., 2011; Noh et al., 2013; Pinto et al., 2010). However, cells are highly dynamic entities, and protein interactions could be profoundly influenced by spatial and temporal availability of the interacting gene products, as has been previously demonstrated for yeast grown under varying experimental conditions (de Lichtenberg et al., 2005; Luscombe et al., 2004). Recent studies that analyzed genes with de novo mutations in ASD (Parikshak et al., 2013; Willsey et al., 2013) and SCZ (Gulsuner et al., 2013) have integrated transcriptome data to capture dynamic information at different brain spatio-temporal intervals. Here, we incorporate physical protein-protein interactions into spatio-temporal transcriptome analysis of 16p11.2 genes. This novel approach identifies profound changes in co-expressed and physically interacting protein pairs that are not observable from the static PPI networks. We demonstrate that 16p11.2 proteins interact with their corresponding partners primarily in four specific spatio-temporal intervals, and that the interaction patterns change across these intervals. In particular, we identify the late mid-fetal period of cortical development as crucial for establishing connectivity of 16p11.2 proteins with their partners. Our results implicate physical KCTD13-Cul3 interaction within inner cortical plate layer four in regulating RhoA levels, and possibly in influencing the brain size. Finally, we experimentally confirm that nonsense mutations in CUL3 identified in ASD patients weaken or even disrupt physical interaction between KCTD13 and Cul3 proteins. Our study places 16p11.2 interactions into a spatio-temporal context and identifies dynamic subnetworks of interacting proteins during human brain development.
Results
High-risk CNVs have distinct spatio-temporal signatures
The ability of two proteins to interact greatly depends on their spatial and temporal availability. Generally, an interacting protein pair could form only if two proteins are present in the same cellular compartment at the same time in sufficient quantities. Indeed, strong correlation between co-expression and protein interactions have been observed (Ge et al., 2001; Grigoriev, 2001), especially for the subunits of permanent protein complexes that are maintained across various cellular conditions (Jansen et al., 2002). Integration of gene expression with protein interactions could therefore identify the most plausible spatio-temporal intervals at which a biologically relevant interaction between two proteins may occur. Data integration from heterogeneous sources has been previously used to gain biological insights into various cellular processes and human diseases (Pujana et al., 2007; Segal et al., 2003).
To understand how genes from different CNVs conferring high risk for psychiatric disorders (Table S1) interact in the context of brain development, we constructed dynamic spatio-temporal networks by integrating physical protein-protein interactions with gene co-expression (Figure 1, Tables S2–3). We investigated whether these networks are enriched in co-expressed and physically interacting protein pairs across four brain regions and eight developmental periods resulting in 32 spatio-temporal intervals (Experimental Procedures, Figure 1). We observed no significant differences between the fractions of co-expressed interacting protein pairs in the combined CNV network (1,918 pairs involving 104 CNV genes from seven high-risk CNVs) vs. background control of all human brain-expressed interacting proteins (HIBE) (Experimental Procedures, Figure 2). Similarly, we do not observe a common signature in a simulated CNV dataset of 10,000 randomly selected genomic regions with the same number of genes and interactions as the high risk CNVs (Figure 2).
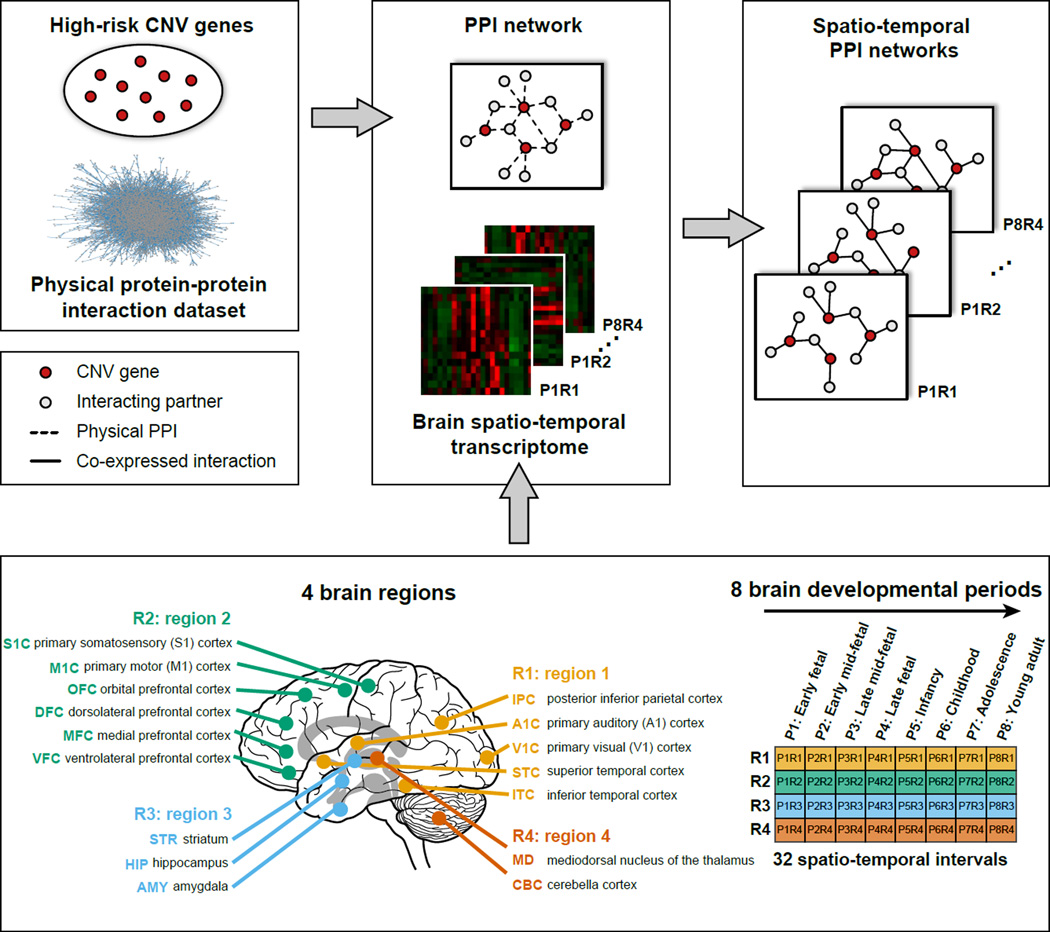
Figure 1. Spatio-temporal protein-protein interaction networks construction.
Spatio-temporal PPI networks were constructed by integrating physical protein-protein interaction data with brain spatio-temporal transcriptome. The connections (solid black lines) between CNV proteins (red circles) and their interacting partners (grey circles) within the spatio-temporal PPI networks were drawn only when two proteins were co-expressed and physically interacting (dashed black lines). Four brain regions (R1: yellow, R2: green, R3: blue and R4: orange) and eight brain developmental periods (P1-P8) were integrated to build 32 spatio-temporal PPI networks for each CNV region. See also Table S2–3.
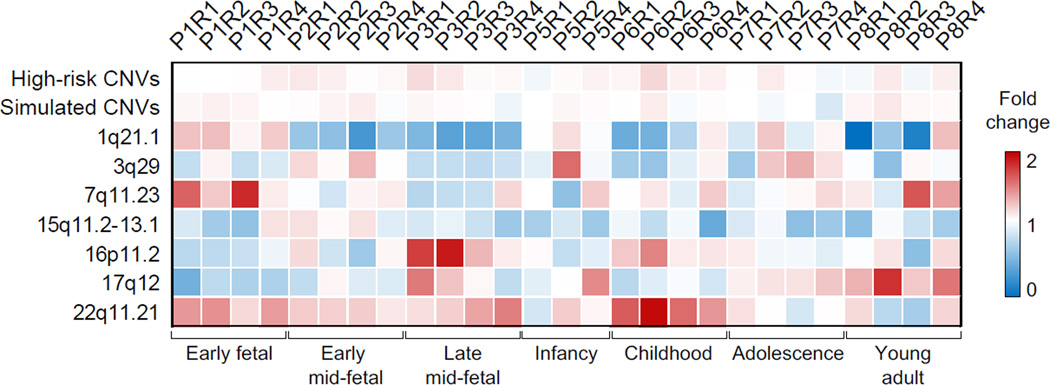
Figure 2. High-risk CNV regions have distinct spatio-temporal signatures.
Seven CNVs conferring high-risk to multiple psychiatric disorders were analyzed in combination (high-risk CNVs line) and independently to calculate the fractions of co-expressed interacting protein pairs for each spatio-temporal interval. Each cell represents fold change of the fraction of co-expressed interacting pairs of CNV network compared with the background control of co-expressed interacting pairs from HIBE network. Color scale indicates the fold change level, ranging from 0 (blue, depletion) to 2 (red, enrichment). The heatmap shows no difference between the fractions of co-expressed interacting protein pairs in the combined CNV network and in the network of simulated CNVs with the same number of genes and PPIs (±10%) as in real CNVs, compared with the background control. The median frequency of 10,000 simulated networks for each spatio-temporal interval is shown. CNVs show distinct spatio-temporal signatures when analyzed separately.
The analysis of each CNV separately demonstrated that some CNVs are characterized by distinct signatures of enriched co-expressed interacting protein pairs that are non-randomly distributed across spatio-temporal intervals (Figure 2). For example, 7q11.23 CNV is enriched in such pairs primarily during early fetal and young adult periods in the R3 brain region composed of amygdala, hippocampus and striatum, whereas 22q11.21 CNV has the strongest signal during childhood in all brain regions. Likewise, the 16p11.2 co-expressed interacting protein pairs are strongly and moderately enriched during late mid-fetal and childhood periods of brain development, respectively. This suggests that different CNVs may impact different brain regions during different periods of brain development.
16p11.2 co-expressed interacting protein pairs are enriched in late mid-fetal and childhood periods
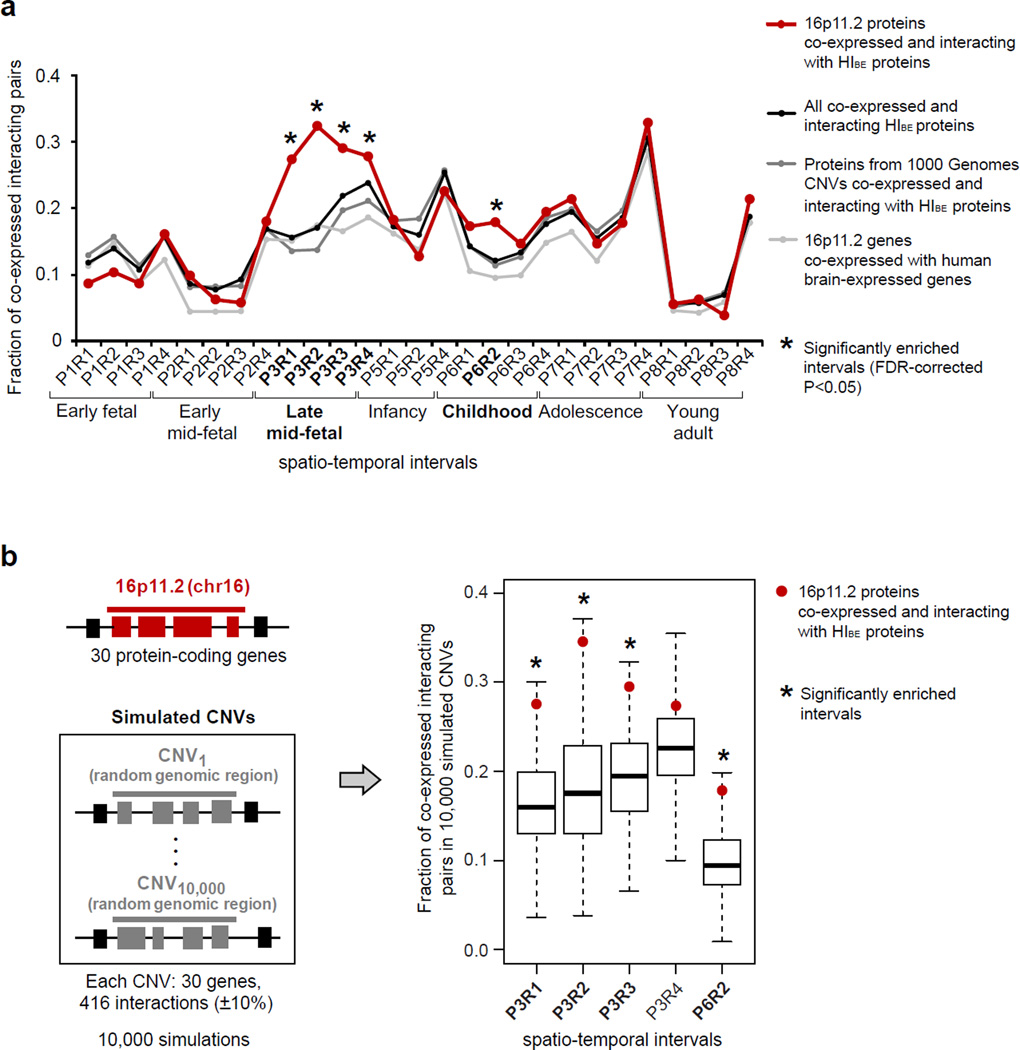
We focused our subsequent analysis on the 16p11.2 CNV as it represents the most interesting example of a region with a broad phenotypic expressivity (Weiss et al., 2008). To assess the statistical significance of the enrichment observed for 16p11.2 CNV, we calculated the fractions of co-expressed interacting protein pairs across all spatio-temporal intervals in three control datasets: all HIBE pairs; proteins from common CNVs identified in the 1000 Genomes Project (Mills et al., 2011) connected by interactions from HIBE; and all possible pairs between 16p11.2 genes and human brain-expressed genes (Experimental Procedures). These analyses consistently identified late mid-fetal and childhood periods as being significantly enriched in co-expressed interacting pairs independently of the control dataset (Figure 3a). The sequential removal of each of the 16p11.2 proteins together with their corresponding partners from the network did not influence this unique spatio-temporal signature or the enriched spatio-temporal intervals (Figure S1). This indicates that the observed enrichment is not due to random effects from PPIs, CNV or co-expression because different types of controls should have addressed these biases. After false discovery rate (FDR) correction for the multiple testing, we identified significant enrichment in five intervals: P3R1 (Fisher’s exact p=8.7×10−9); P3R2, (p=5.0×10−13); P3R3 (p=0.003); P3R4 (p=0.042) and P6R2 (p=0.013) (Figure 3a). To control for the biases from network topology, we used randomly permuted genomic regions with the same number of genes and interactions as in 16p11.2 CNV as an additional control. This analysis confirmed four out of five previously identified networks as being significantly enriched in co-expressed interacting pairs (Figure 3b). Furthermore, using a more stringent co-expression coefficient (Figure S2), restricting network to PPIs only detected by the systematic high-throughput screens, or to only co-expressed gene pairs produced similar results (Figure S3).
Figure 3. 16p11.2 co-expressed interacting protein pairs are significantly enriched in four spatio-temporal intervals.
(a) The fractions of protein pairs from the 16p11.2 CNV co-expressed and interacting with HIBE proteins (red line), all co-expressed and interacting HIBE proteins (black line), proteins from 1000 Genome project CNVs co-expressed and interacting with HIBE proteins (dark grey line), and 16p11.2 CNV genes co-expressed with all brain-expressed human genes (light grey line) are shown. Twenty-seven spatio-temporal intervals of brain development are shown on the X-axis. 16p11.2 co-expressed interacting protein pairs are significantly enriched in five spatio-temporal intervals (star symbols) compared to three control networks. The statistical enrichment was calculated using Fisher’s exact test and P-values were FDR-corrected for multiple comparisons. (b) The spatio-temporal intervals with significant enrichment were further evaluated against a background control of 10,000 simulated CNV networks with the same number of genes and interactions (±10%) as in the 16p11.2 network. Four out of five spatio-temporal intervals remained significantly enriched (P3R1, P=0.014; P3R2, P=0.009; P3R3, P=0.038; and P6R2, P=0.041; p-values are FDR-corrected). See also Figures S1–3.
16p11.2 networks change across spatio-temporal intervals
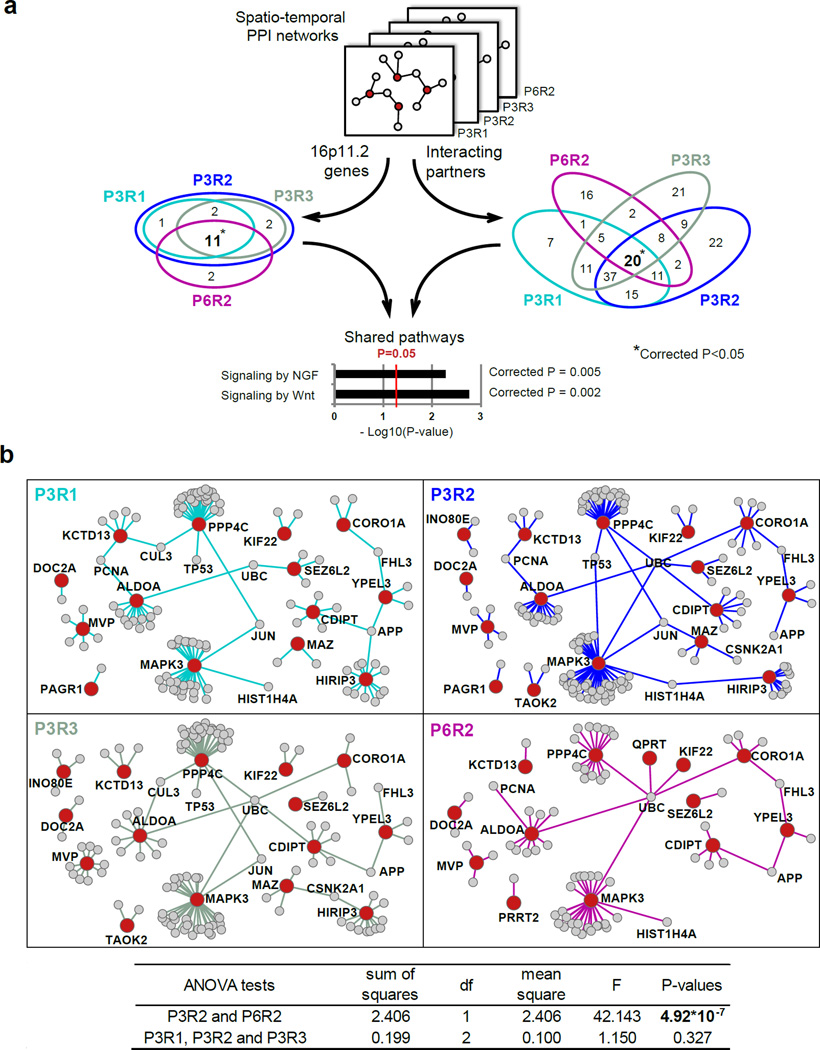
To identify commonalities between the spatio-temporal 16p11.2 networks, we investigated their convergence by calculating the fraction of shared proteins in these networks. We observed that 11/18 (61%) of the 16p11.2 CNV proteins and 20/187 (10.7%) of their co-expressed interacting partners are shared by all four networks (Figure 4a). These numbers are significantly higher than expected by chance from 10,000 randomly simulated spatio-temporal networks with the same properties (corrected empirical p=0.01 for 16p11.2 proteins and corrected empirical p=0.02 for the partners). Furthermore, co-expressed interacting protein pairs shared by all four networks are significantly, and perhaps unsurprisingly, enriched in the pathways relevant to neuronal development, signaling by nerve growth factor (NGF) (FDR-corrected p=0.005) and signaling by Wnt (FDR-corrected p=0.002) (Figure 4a). In agreement with recent findings (Willsey et al., 2013), spatio-temporal 16p11.2 networks are also enriched in cortical glutamatergic neuron markers in layers 5 and 6 (empirical p=0.0098) (Table S4) suggesting that shared neuronal circuits may be involved in autism subtypes caused by mutations affecting different genes.
Figure 4. Functional convergence and divergence of the 16p11.2 spatio-temporal networks.
(a) The overlap of 16p11.2 genes (left Venn diagram) and their co-expressed interacting partners (right Venn diagram) across four significant spatio-temporal intervals. Statistical significance of observing higher than expected number of overlapping genes (star symbol) was assessed by permutation test using simulated CNVs. The enrichment analyses of 20 interacting pairs shared by four intervals were performed using DAVID. “Signaling by NGF” and “Signaling by Wnt” pathways are significantly enriched. (b) Comparison of spatio-temporal networks across different brain regions within the same developmental period (P3R1, P3R2 and P3R3) and across different developmental periods within the same brain region (P3R2 and P6R2). 16p11 genes are shown as red nodes; their co-expressed interacting partners as grey nodes; and the PPIs between co-expressed genes at a particular developmental period are shown as colored edges: P3R1 (turquoise); P3R2 (blue); P3R3 (greenish gray); P6R2 (purple). The nodes that lost all edges were removed from the corresponding networks. Significant differences are observed across developmental periods but not across brain regions (ANOVA Table below). See also Tables S5–6.
Our subsequent analyses addressed the question of the spatio-temporal 16p11.2 network differences. The co-expressed interacting protein pairs within four spatio-temporal 16p11.2 networks are enriched across different brain regions (R1, R2 and R3) within the same developmental period (late mid-fetal P3), and also across different developmental periods (late mid-fetal P3 and childhood P6) within the same brain region (R2). We next compared network changes within the same period (P3) and within the same region (R2) by calculating fractions of co-expressed interacting pairs that are shared by different networks (Figure 4b). We found no significant difference between three regions within the same developmental period (P3R1, P3R2 and P3R3, ANOVA P=0.33, n=14) (Experimental Procedures, Table S5). However, statistically significant differences were observed when P3R2 was compared with P6R2 (ANOVA P=4.9×10−7, n=15) (Table S6). This suggests that 16p11.2 network changes are more pronounced across developmental periods than across brain regions. Our data further suggest that spatio-temporal interaction networks may undergo substantial changes in the developing brain.
De novo ASD mutations are significantly enriched in spatio-temporal networks
Recent exome sequencing studies have identified a large number of de novo mutations in ASD, SCZ and ID patients. The analysis of the interacting partners of 16p11.2 proteins using the combined set of 1,975 de novo mutations from three disorders indicates that both, the entire 16p11.2 network and four spatio-temporal intervals, are significantly enriched in genes carrying likely gene damaging (LGD) and multiple-hit de novo mutations, even after correction for gene size and GC content (Table S7, Experimental Procedures). At the same time, none of the networks is enriched in genes with mutations detected in controls; however, the number of mutations in controls is limited. Importantly, the observed effect is largely driven by the ASD mutations as no significant enrichment is observed for SCZ and ID mutations when the analysis is performed separately for each disorder. This result agrees with the study by Fromer et al. (Fromer et al., 2014) that also observed the enrichment of ASD but not SCZ LGD mutations. Given that schizophrenia is associated with 16p11.2 duplications but not with deletions, this lack of association in SCZ is not surprising.
The spatio-temporal networks are also significantly enriched in post-synaptic density genes and Fragile X Mental Retardation Protein (FMRP) target genes (Table S7), in agreement with previous studies (Fromer et al., 2014; Iossifov et al., 2012). These enrichment results provide independent lines of evidence for disease risk association and suggest that functional impact of the de novo mutations on networks needs further investigation.
Spatio-temporal KCTD13 networks identify DNA replication and RhoA pathways
One of the strongest candidates for a gene that is a major contributor to neuropsychiatric phenotypes within the 16p11.2 locus is KCTD13. A recent study in a zebrafish model has convincingly demonstrated that KCTD13 is the only gene within the 16p11.2 region capable of inducing the microcephalic phenotype associated with the 16p11.2 duplication and the macrocephalic phenotype associated with the 16p11.2 deletion (Golzio et al., 2012). Importantly, these phenotypes in the fish are capturing the mirror phenotypes of humans (McCarthy et al., 2009). Given this strong functional evidence, we focused on investigating interaction pattern of KCTD13 across four spatio-temporal networks.
The analysis of KCTD13 networks indicates that seven proteins physically interact and are co-expressed with KCTD13 across four spatio-temporal intervals (Figure 5a). Furthermore, some of these proteins also physically interact and are co-expressed with each other, thereby forming two functionally distinct modules, predominantly at P3R1 and P3R2 intervals.
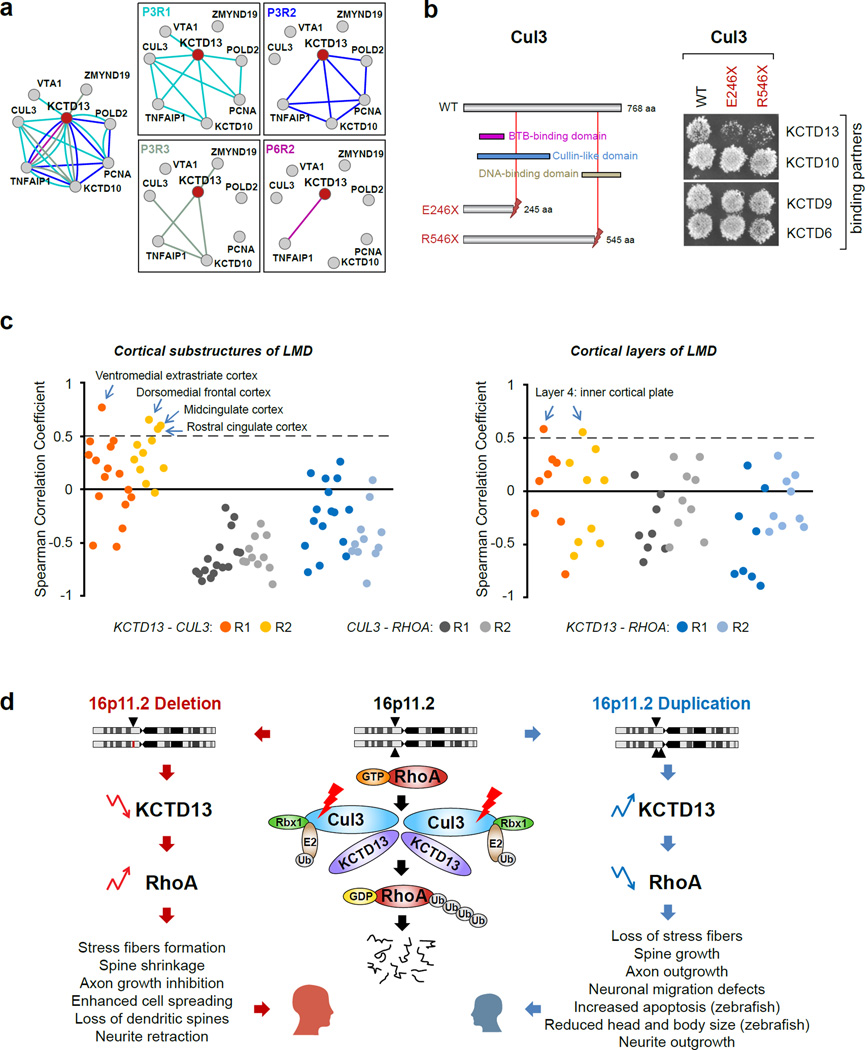
Figure 5. The 16p11.2 spatio-temporal networks implicate KCTD13-Cul3-RhoA pathway in pathogenesis of psychiatric diseases.
(a) Static (left) and dynamic (four right panels) spatio-temporal networks of the KCTD13 protein together with its co-expressed interacting partners. Edge color and spatio-temporal intervals are as in Figure 4. The P3R1 network connects ubiquitin ligase Cul3 to other network members. The interconnected members of the P3R2 network are primarily involved in the DNA replication and repair. (b) The results of Y2H experiments indicate that two truncating mutations of Cul3 detected in autism patients disrupt KCTD13-Cul3, but not KCTD10-Cul3, KCTD9-Cul3 or KCTD6-Cul3 interactions. The interaction of the wild-type Cul3 with KCTD13 remains intact. (c) Pair-wise co-expression profiles of KCTD13 CUL3 and RHOA in the laser micro-dissected prenatal human brain substructures (left) and cortical layers (right). KCTD13 and CUL3 are positively correlated in the majority of brain substructures. RHOA is negatively correlated with both, KCTD13 and CUL3 in the majority of LMD substructures. KCTD13 and CUL3 are co-expressed in layer 4 (inner cortical plate). (d) The KCTD13-Cul3-RhoA pathway may be dysregulated in both, 16p11.2 CNV carriers and in the patients with the truncating CUL3 mutations, through altered cell migration in the brain during development. When 16p11.2 CNV deletion (left side of the figure) or duplication (right side of the figure) event occurs, it may lead to a decrease (downward arrow) or an increase (upward arrow) of KCTD13 levels. Center part of the figure shows KCTD13-Cul3-RhoA pathway, where KCTD13-Cul3 interaction regulates RhoA ubiquitination and degradation. The altered RhoA levels may have opposing functional consequences during brain development, which may in turn lead to a disrupted cell migration and influence the brain size during prenatal development. See also Table S8.
The first functionally related group of proteins that interact with KCTD13 consists of PCNA-POLD2-TNFAIP1-KCTD10 (Figure 5a). KCTD13 is also known as ‘polymerase delta interacting protein 1’ (POLDIP1), because it was initially identified as a binding partner of the small subunit of polymerase delta, POLD2 (He et al., 2001). KCTD13 also directly interacts with PCNA, an auxiliary cofactor of polymerase delta, and nuclear localization of these proteins in the replication foci suggests their role in DNA replication. Furthermore, KCTD13, TNFAIP1 and KCTD10 have high sequence similarity and share the PCNA binding motif at the C-terminus, suggesting their important roles in DNA synthesis and repair (Wang et al., 2009; Yang et al., 2010).
The second functionally related group of proteins interacting with KCTD13 consists of Cul3-TNFAIP1-KCTD10 (Figure 5a). CUL3 encodes a scaffold protein cullin, a core component of E3 ubiquitin-protein ligase complexes that mediate the ubiquitination and subsequent proteasomal degradation of target proteins. These multimeric complexes play key roles in the regulation and control of the cell cycle (Genschik et al., 2013) along with other biological functions. The complex of Cul3 with adapter proteins KCTD13, TNFAIP1 and KCTD10 regulates the ubiquitination and degradation of small GTPase RhoA, which in turn is a major regulator of actin cytoskeleton and cell migration. The RNAi knockdown of either CUL3 or adapter proteins leads to abnormal RhoA accumulation and activation, resulting in excessive actin stress fiber formation and impaired cell migration (Chen et al., 2009b), whereas downregulation of RhoA activity promotes cell migration (Govek et al., 2011). Since RhoA levels are likely regulated through the formation of KCTD13-Cul3-RhoA complex, maintaining sufficient and balanced levels of its components may be crucial for proper functioning of the RhoA pathway in neuronal development, including neurite outgrowth, axon pathfinding, neuronal migration, dendritic spine formation and maintenance. It is possible that 16p11.2 deletions and duplications that lead to dosage changes of 16p11.2 genes including KCTD13 could impair this important neuronal pathway, especially during P3R1 developmental interval when this protein complex is most likely to form. In conclusion, our analyses identified two KCTD13-centered interconnected modules with different functions: one involved in DNA replication, synthesis and repair, which is primarily observed in the prefrontal and motor-sensory cortex during late mid-fetal development (P3R2); and the other involved in formation of E3 ubiquitin ligase complexes, which is primarily observed in the parietal, temporal and occipital cortex during late mid-fetal development (P3R1).
De novo truncating mutations in CUL3 disrupt its interaction with KCTD13
One of the interacting partners of KCTD13, Cul3, carries two de novo protein-truncating mutations, p.Glu246Stop (E246X) (O’Roak et al., 2012) and p.Arg546Stop (R546X) (Kong et al., 2012), that have been detected in two unrelated ASD patients. To evaluate the impact of these mutations on the physical interaction between KCTD13 and Cul3, we carried out yeast-two-hybrid (Y2H) experiments with the wild-type and two mutants of CUL3 (Experimental Procedures). We observed that both mutations significantly weaken or even abolish the KCTD13-Cul3 interaction, whereas the interaction of KCTD13 with the wild-type Cul3 is preserved (Figure 5b). Interestingly, the same mutations do not disrupt interactions of Cul3 with three other BTB/POZ domain-containing proteins, KCTD10, KCTD9 and KCTD6 (Figure 5b). Since the interaction of KCTD13 with Cul3 is required for RhoA ubiquitination and subsequent degradation, our results suggests that the disruption of KCTD13-Cul3 interaction by gene damaging mutations may impact RhoA protein levels and dysregulate RhoA pathway.
KCTD13-CUL3-RHOA co-expression patterns in the developing cortex
Deficits in cortical patterning have been recently observed in the ASD (Parikshak et al., 2013; Stoner et al., 2014; Willsey et al., 2013) and SCZ (Gulsuner et al., 2013) patients. Given variability in gene expression across different cortical layers of the brain, we further investigated pair-wise co-expression patterns of three genes, KCTD13, CUL3 and RHOA, using more detailed layer-specific expression data from laser-microdissected (LMD) prenatal human brain (Miller et al., 2014) (Experimental Procedures, Table S8). We observed that in the majority of LMD substructures from P3R1 and P3R2 networks, expression levels of KCTD13 and CUL3 are positively correlated (Figure 5c). The highest co-expression values are observed in ventromedial extrastriate cortex (P3R1), and in dorsomedial frontal, rostral cingulate and midcingulate cortex (P3R2). In addition, KCTD13 and CUL3 are co-expressed in the layer 4 corresponding to the inner cortical plate (Figure 5c). On the contrary, the KCTD13-RHOA and CUL3-RHOA pairs are negatively correlated in the majority of the substructures (Figure 5c).
These observations are in agreement with experimental results suggesting that KCTD13 and Cul3 negatively regulate RhoA levels (Chen et al., 2009b). For example, KCTD13 or CUL3 knockdowns in the cell culture systems lead to RhoA/RhoA-GTP accumulation and actin stress fibers formation (Chen et al., 2009b). The decrease of the RhoA level has been previously linked to extensive apoptosis during embryogenesis, resulting in dramatic reduction of head and body size in zebrafish model (Zhu et al., 2008). Alternatively, the upregulation of RhoA through its constitutive expression has been linked to suppression of dendritic spine morphogenesis and to dramatic loss of spines (Govek et al., 2011; Zhang and Macara, 2008). The important role of RhoA protein levels in the local regulation of axon growth has been recently demonstrated (Walker et al., 2012).
It is tempting to speculate that CUL3 mutations as well as 16p11.2 deletions and duplications that alter the dosage of KCTD13 may act via similar mechanism by influencing RhoA protein levels (Figure 5d). The changes in the RhoA levels may in turn regulate cellular processes that influence head and body size during development. However, it remains to be determined whether these two types of mutations in the genes from the same pathway may have related molecular consequences in the brains of the patients.
Discussion
We report the construction of dynamic spatio-temporal interaction networks connecting genes from the 16p11.2 CNV, a strong genetic risk factor for multiple psychiatric disorders. In contrast to traditional approaches that use static representation of protein interaction networks to establish functional connections between genes, we integrate physical PPIs with genomescale transcriptome data from developing human brain to gain new insights into pathways that may be dysregulated by CNV mutations. This novel approach enables investigation of molecular mechanisms of psychiatric disorders in the context of brain development.
One of the most intriguing observations from our study is that some CNVs have distinct spatio-temporal signatures, with enrichment of the co-expressed interacting protein pairs at different developmental stages in different brain regions. For example, a high fraction of interactions derived from the Williams-Beuren/7q11.23 microduplication syndromes CNV region implicated in both autism (Sanders et al., 2011) and schizophrenia (Mulle et al., 2014), are “turned on” (i.e. co-expressed) during early fetal development in hippocampus, amygdala and striatum (P1R3, Figure 2). This signature is quite different from the one for 3q29 CNV, also conferring high risk to several psychiatric disorders including schizophrenia (Mulle et al., 2010), when the interactions are “turned on” primarily during cortical development in infancy (P5R2, Figure 2). The 3q29 signature is, again, quite distinct from the 16p11.2 signatures (P3R1 and P3R2), or the 17q12 signature (P8R2), or the 22q11.21 signature (P6R2) (Figure 2). Although no signature has been observed for one CNV, 15q11, and some of the intervals did not withstand the stringent permutation correction, we found statistically robust spatio-temporal signatures for multiple intervals in three high-risk CNVs in addition to the 16p11.2 locus (Figure S4). These observations suggest that CNVs may have critical periods in brain development when their functional impact is most apparent, and that specific molecular pathways may be disrupted primarily during these critical developmental windows.
In the case of 16p11.2, we identified late mid-fetal developmental period as most critical for establishing connections of CNV proteins with their partners. During this period, corresponding to 19–24 post-conceptual weeks (PCW) of fetal development (Kang et al., 2011), a higher than expected number of pairs were found to be co-expressed and interacting in the majority of cortical regions (Figure 3). Our results are in agreement with a recent study by Willsey et al. (Willsey et al., 2013) that also identified the same developmental period as crucial for autism pathogenesis, albeit using a different set of genes and a different approach that did not incorporate protein interaction networks into the analyses. The observed convergence is encouraging and suggests that brain alterations during late mid-fetal period may be common for different types of autisms, even for those with diverse genetic backgrounds (i.e. 16p11.2 CNV and the de novo mutations).
Another important observation that emerges from our study is the profound interaction network changes during brain development. By comparing 16p11.2 network connectivity during different developmental periods (P3 and P6) and among different brain regions (R1, R2 and R3) we found that these networks vary to a greater extent temporarily than spatially (Figure 4b). This suggests that post-translational dysregulation during brain development may play a central role in the pathogenesis of neuropsychiatric diseases.
One of the pathways that our study proposes as being most likely impacted by the 16p11.2 CNV is KCTD13-Cul3-RhoA. The transcriptional profiles of these three genes are interdependent, with KCTD13-CUL3 being positively correlated and both KCTD13-RHOA and CUL3-RHOA being negatively correlated (Figure 5c). Furthermore, it was previously suggested that KCTD13 and Cul3 regulate RhoA protein levels through ubiquitination by the KCTD13-Cul3 ligase complex based on the observation that RhoA accumulates upon KCTD13 or CUL3 knockdowns (Chen et al., 2009b). Our study further demonstrates that KCTD13 and Cul3 physically interact, and that they are co-expressed during late mid-fetal period of cortical development (Figure 5a–c), thereby placing KCTD13-Cul3-RhoA complex into spatio-temporal context of the brain development.
Recent exome sequencing studies have identified two de novo CUL3 LGD mutations in unrelated patients with ASD (Kong et al., 2012; O’Roak et al., 2012). These loss-of-function mutations may lead to production of shorter proteins with missing C-terminus, likely disrupting normal functioning of Cul3. Alternatively, the truncations may activate nonsense-mediated decay mechanisms leading to elimination of the mutant transcripts. We hypothesized that in both cases, either through protein disruption or through alteration of the expression levels, the interaction of Cul3 with several partners may be perturbed. We tested this hypothesis and demonstrated that truncating mutations of Cul3 indeed disrupt the KCTD13-Cul3 physical interaction, possibly impacting RhoA levels (Figure 5).
Using prenatal human brain LMD transcriptome (Miller et al., 2014) we next show that KCTD13 and CUL3 are highly co-expressed in layer 4, inner cortical plate (Figure 5c). Disruption of the KCTD13-Cul3 interaction, either through dosage imbalance of KCTD13 as a result of 16p11.2 CNV or by CUL3 mutations, may potentially affect this brain layer. Interestingly, a recent study pointed to the same layer 4 as having pathological focal patches of abnormal laminar cytoarchitecture and cortical disorganization of neurons in prefrontal and temporal cortical tissues in a majority of young children with autism (Stoner et al., 2014). Further anatomical, cytological and gene expression studies of the brains from individuals with 16p11.2 CNVs would reveal whether such patches are more pronounced amongst the 16p11.2 CNVs carriers. Presently, the convergence of neuroanatomical data from Stoner et al. with genetic, co-expression and protein interaction data suggests that RhoA pathways may be involved in dysregulation of layer formation and layer-specific neuronal differentiation at prenatal developmental stages. However, further studies are needed to directly connect the 16p11.2 phenotype with the dysregulation of the RhoA pathway.
One of the limitations of our study is the lack of cellular-level resolution for the networks that we have constructed. Although spatio-temporal networks are an advancement compared to the static network models, they still lack the power to implicate specific neuronal cell types or neuronal populations into disease pathogenesis. This lack of resolution is largely due to the absence of the single-cell developmental transcriptome data that will ultimately be required to fully exploit novel approaches developed here. Further advancement of the single-cell genomic, transcriptomic and proteomic technologies (Shapiro et al., 2013), especially in the context of brain development (Kitchen et al., 2014), will open new avenues for improving the resolution of the dynamic spatio-temporal neuronal networks.
Another limitation of our study is that brain developmental transcriptome data used for the analyses was not derived from the patients with psychiatric disorders. However, postmortem brain tissues from 16p11.2 CNV carriers are scarce, and no transcriptome data is currently available for early developmental stages of these individuals. To address this shortcoming, we used lymphoblast transcriptome data from the 16p11.2 deletion and duplication carriers (Luo et al., 2012) to identify the changes in the expression patterns of interacting pairs from our spatio-temporal networks (Supplemental Experimental Procedures, Figure S5). We observed that some highly co-expressed interacting pairs, including KCTD-Cul3, have a significantly reduced expression in the lymphoblasts of deletion carriers, whereas other pairs have a significantly increased expression in the duplication carriers (Table S9).
The RhoA pathway has been intensively studied in the past decade using different cellular models and model organisms. Rho GTPases play critical roles in neuronal migration and are key regulators of actin and microtubule cytoskeleton, cell polarity, and adhesion. The data from the literature strongly suggests that the Rho GTPase signaling pathway has important functions in brain morphogenesis at early stages of brain development (Chen et al., 2009a; Govek et al., 2011; Zhu et al., 2008). Although results may vary depending on the study, general findings are that the increase in RhoA levels leads to stress fibers formation, axon growth inhibition, enhanced cell spreading, loss of dendritic spines and neurite retraction, whereas opposite effects are observed when RhoA levels are decreased (Figure 5d). In addition, knockdown of RHOA was shown to lead to reduced head and body size and increased apoptosis in a zebrafish model (Zhu et al., 2008). Integrating the results of our study into this model allows us to suggest that the functional impact of the 16p11.2 CNV may be manifested through dysregulation of the RhoA pathway. Specifically, we propose that KCTD13 dosage changes in the deletion and duplication carriers may influence RhoA levels and lead to impaired brain morphogenesis and cell migration during fetal stages of brain development. Furthermore, based on our experimental results, we suggest that the functional impact of CUL3 truncating mutations may also be manifested through the RhoA pathway. Intriguingly, activation of RhoA signaling, albeit through a different mechanism, has recently been implicated in a rare monogenic form of autism, Timothy syndrome (Krey et al., 2013). If confirmed by further studies, this convergence of different types of mutations on a common pathway will provide basis for future exploration of the role of RhoA in autism and in other neuropsychiatric diseases.
Experimental Procedures
Human high-risk CNVs for psychiatric disorders
Previous studies have demonstrated evidence of a strong association of ASD, SCZ, BD and ID with the following 11 CNV loci (deletions and/or duplications): 1q21.1, 2p16.3 (NRXN1), 3q29, 7q11.23, 15q11.2, 15q11.2–13.1, 15q13.3, 16p13.11, 16p11.2, 17q12 and 22q11.21 (Malhotra and Sebat, 2012). All these loci are implicated as highly significant risk factors for two or more psychiatric disorders. Four of the CNVs from this list (2p16.3, 15q11.2, 15q13.3, 16p13.11) contain less than 10 protein-coding genes each and were not included in current study due to limited sizes of the resulting PPI networks. The remaining CNVs containing a total of 145 genes were investigated in the current study (Table S1). The CNV boundaries were defined as previously described (Malhotra and Sebat, 2012).
Human brain transcriptome data
To build dynamic spatio-temporal PPI networks in the context of human brain development, BrainSpan transcriptome exon microarray data (Kang et al., 2011) (http://www.brainspan.org) summarized to Gencode 10 (Harrow et al., 2012) genes was used. This dataset consists of 1,340 brain samples and was generated by dissecting brain regions from 57 clinically unremarkable postmortem brain donors ranging in age from 4 PCW to 82 years. The expression levels of 17,181 protein-coding genes within each sample were assayed using the Affymetrix GeneChip Human Exon 1.1 ST Array platform as described in Kang et al. (Kang et al., 2011). To reduce noise, we included only genes with log2-intensity values greater than 6 in at least one sample, and with a coefficient of variance at least 0.07. As a result, a total of 14,619 genes were considered as brain-expressed.
The datasets of physical protein-protein interactions restricted to brain-expressed genes
A comprehensive map of physically interacting human proteins was assembled using our experimentally identified physical binary interactions (HIC) expanded with physical interactions from the BioGRID 3.2.106 (Stark et al., 2006) downloaded in October 2013. HIC was assembled to include the interactions from two datasets: (1) the Human Interactome II-14 (HI-II-14) (Rolland et al., 2014) containing ~14,000 novel physical binary interactions between ~4,300 human proteins; and (2) the gene-level binary interactions from the Autism Spliceform Interaction Network (ASIN) (Corominas et al., 2014) (ASIN later became a part of the newer version of BioGRID 3.2.116). After redundancy and self-interactions removal, the PPI network was integrated with human brain transcriptome to assemble brain-expressed Human Interactome (HIBE) consisting of 116,147 pairs of brain-expressed interacting proteins.
Construction of spatio-temporal PPI networks
The brain transcriptome data was generated across 13 dissection stages, varying from eight to 16 brain structures for each stage (Kang et al., 2011). Since well-defined anatomical structures are limited during early embryonic development, the first period (4–8 PCW) was removed from further analysis. After merging dissection stages, we defined eight non-overlapping developmental periods ranging from 8 PCW to 40 years of age (Table S3). Brain regions were grouped into four clusters using hierarchical clustering based on brain transcriptional similarity to reflect actual topological proximity and functional segregation as described in Willsey et al. (Willsey et al., 2013) (Figure 1). As a result, 27 spatio-temporal regions were defined after eliminating late fetal (P4) developmental period and one region from P5 (P5R3) due to lack of transcriptome data for analyses.
The CNV genes were mapped to HIBE network to construct a static brain-expressed PPI network for each CNV region. Subsequently, spatio-temporal co-expression data were integrated with the static PPI network. An interaction between two proteins was defined as positive if the pair-wise Spearman Correlation Coefficient (SCC) value was >0.5 (using a more stringent SCC >0.7 lead to similar results, Figure S2). Using this approach, 27 different spatio-temporal CNV networks were generated and used for further analyses for each CNV region (Figure 1). The combined 16p11.2 spatio-temporal network consisted of 416 brain-expressed and interacting protein pairs involving 21 16p11.2 proteins and 367 interacting partners from HIBE (Table S2).
Enrichment analyses in four spatio-temporal networks
To test for the enrichment of shared co-expressed interacting partners between four spatio-temporal networks in Figure 4a, 10,000 simulated CNVs with the same number of genes and interactions as in 16p11.2 were generated. For each simulated CNV, the number of shared co-expressed interacting partners was determined and compared with 16p11.2 networks. Finally, the empirical P-values were calculated based on the fraction of 10,000 simulated CNVs with equal or higher number of shared co-expressed interacting partners than in 16p11.2 networks. P-values were FDR-corrected for multiple testing.
To evaluate the differences between four spatio-temporal networks in Figure 4b, the one-way ANOVA tests were performed. To test the variance among the networks from three brain regions of the same developmental period (P3R1, P3R2 and P3R3; Table S5), three topological properties were defined for each 16p11.2 CNV gene: (1) fraction of co-expressed interacting partners unique to one network; (2) fraction of co-expressed interacting partners shared by two out of three networks; and (3) fraction of co-expressed interacting partners shared by all three networks. Similarly, to test the variance between the networks from two developmental periods of the same region (P3R2 and P6R2; Table S6), two topological properties were defined for each 16p11.2 CNV gene: (1) fraction of co-expressed interacting partners unique to one network; (2) faction of co-expressed interacting partners shared by both networks. ANOVA was used to calculate statistically significant differences between the networks.
The enrichment analyses of interacting protein partners from 16p11.2 spatio-temporal networks were performed using HIBE as a background (Table S7). The de novo mutations (DNMs) were extracted from 19 publications and network genes were classified as “likely gene damaging” if they carried nonsense, frameshift or splice site de novo mutations; and “multiple-hit” if they carried two or more LGD and/or missense mutations. The post-synaptic density genes and FMRP target genes were extracted as previously described (Corominas et al., 2014). The p-values were corrected for gene size and GC content; the empirical p-values were calculated by selecting from the HIBE 10,000 datasets with the same gene lengths (±10%) or the same GC content (±10%) as in the 16p11.2 networks; the reported P-values were FDR-corrected (Table S7).
The analysis of physical interactions of wild type and mutant Cul3
To compare interactions patterns of the protein products of the wild-type (wt) and mutant (E246X and R546X) CUL3 gene, site-directed mutagenesis and binary interaction mapping were carried out as described (Zhong et al., 2009). Each de novo mutation was introduced into a wt ORF clone by a two-step procedure using specific primers (Supplemental Experimental Procedures). Each of the corresponding CUL3 clones (wt, E246X and R546X) was introduced into pDEST-DB via Gateway LR reaction. The interacting partners in the pDEST-AD configuration were obtained from the human ORFeome collection (Yang et al., 2011). The Y2H mating was performed as follows: DBs and ADs were spotted on YEPD agar plates, replicaplated onto SC-Leu-Trp plates for diploid selection, and then replica-plated onto the phenotyping plates with 3-AT (control for interaction) and with 3-AT plus cycloheximide (CHX) (control for autoactivation). Growth intensity on 3-AT plates was compared between the wild-type and the mutants to determine the presence or the absence of interaction perturbations (Supplemental Experimental Procedures). PCR products of bait and prey ORFs of all positive colonies were Sanger-sequenced to confirm the identities of the interacting partners.
The analysis of KCTD13-CUL3-RHOA co-expression in the laser-microdissected prenatal human brain
To investigate KCTD13-CUL3-RHOA co-expression patterns in the prenatal human brain, layer-specific gene expression data were obtained from Miller et al. (Miller et al., 2014) and downloaded from the BrainSpan (http://www.brainspan.org). This dataset profiles gene expression in two brains spanning periods 2 to 3 of development (15–21 PCW). Gene expression profiles were assessed for 347 finely laser-microdissected tissues from subdivisions distributed across cortical and noncortical regions (Miller et al., 2014). Gene expression of highly discrete laser-microdissected brain regions from two 21 PCW brains were extracted for our analyses (Table S8). To reduce the noise, we only used probes with evidence of robust expression (detection p value ≤ 0.01 in at least 50% of all samples). After filtering, 36,956 probes (corresponding to 16,470 genes) were used for the analyses. Since multiple probes can cover each gene, the expressions of these probes within the same sample were averaged, resulting in a vector of expression values to represent each gene.
The neocortical substructures (a total of 27) and the layers of cortical regions (a total of 9) were defined as in Miller et al. (Miller et al., 2014) (Table S8). To investigate KCTD13-CUL3-RHOA co-expression in each layer, the pair-wise SCCs were calculated, and genes with SCC>0.5 were considered co-expressed.
Supplementary Material
Acknowledgements
We thank Katya Tsimring and Keith Happawana for technical assistance. We also thank Shuli Kang for help with protein interaction dataset processing, and Nidhi Sahni for advice during the experiments. J.T. is the recipient of an ERC Advanced Grant (# 340941). This work was supported by the NIH grants R01MH091350 (L.M.I.), R01HD065288 (L.M.I), R21MH104766 (L.M.I.) and R01MH105524 (L.M.I.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
L.M.I., G.N.L. and R.C. conceived the study and designed the experiments and analyses. G.N.L. and R.C. performed the experiments and analyses. I.L. and J.T. contributed to experiments. X.Y., D.E.H., M.V. and J.S. contributed to the analyses and discussion of the project. L.M.I. directed the project. All authors discussed the results. L.M.I., G.N.L. and R.C. wrote the manuscript.
Conflict of Interest
The authors have no conflict of interest.
References
- Bijlsma EK, Gijsbers AC, Schuurs-Hoeijmakers JH, van Haeringen A, Fransen van de Putte DE, Anderlid BM, Lundin J, Lapunzina P, Perez Jurado LA, Delle Chiaie B, et al. Extending the phenotype of recurrent rearrangements of 16p11.2: deletions in mentally retarded patients without autism and in normal individuals. Eur J Med Genet. 2009;52:77–87. doi: 10.1016/j.ejmg.2009.03.006. [DOI] [PubMed] [Google Scholar]
- Chatr-Aryamontri A, Breitkreutz BJ, Heinicke S, Boucher L, Winter A, Stark C, Nixon J, Ramage L, Kolas N, O’Donnell L, et al. The BioGRID interaction database: 2013 update. Nucleic Acids Res. 2013;41:D816–D823. doi: 10.1093/nar/gks1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Melendez J, Campbell K, Kuan CY, Zheng Y. Rac1 deficiency in the forebrain results in neural progenitor reduction and microcephaly. Dev Biol. 2009a;325:162–170. doi: 10.1016/j.ydbio.2008.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Yang Z, Meng M, Zhao Y, Dong N, Yan H, Liu L, Ding M, Peng HB, Shao F. Cullin mediates degradation of RhoA through evolutionarily conserved BTB adaptors to control actin cytoskeleton structure and cell movement. Mol Cell. 2009b;35:841–855. doi: 10.1016/j.molcel.2009.09.004. [DOI] [PubMed] [Google Scholar]
- Consortium IS. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455:237–241. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper GM, Coe BP, Girirajan S, Rosenfeld JA, Vu TH, Baker C, Williams C, Stalker H, Hamid R, Hannig V, et al. A copy number variation morbidity map of developmental delay. Nat Genet. 2011 doi: 10.1038/ng.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corominas R, Yang X, Lin GN, Kang S, Shen Y, Ghamsari L, Broly M, Rodriguez M, Tam S, Trigg SA, et al. Protein interaction network of alternatively spliced isoforms from brain links genetic risk factors for autism. Nat Commun. 2014;5:3650. doi: 10.1038/ncomms4650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lichtenberg U, Jensen LJ, Brunak S, Bork P. Dynamic complex formation during the yeast cell cycle. Science. 2005;307:724–727. doi: 10.1126/science.1105103. [DOI] [PubMed] [Google Scholar]
- Fromer M, Pocklington AJ, Kavanagh DH, Williams HJ, Dwyer S, Gormley P, Georgieva L, Rees E, Palta P, Ruderfer DM, et al. De novo mutations in schizophrenia implicate synaptic networks. Nature. 2014 doi: 10.1038/nature12929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge H, Liu Z, Church GM, Vidal M. Correlation between transcriptome and interactome mapping data from Saccharomyces cerevisiae. Nat Genet. 2001;29:482–486. doi: 10.1038/ng776. [DOI] [PubMed] [Google Scholar]
- Genschik P, Sumara I, Lechner E. The emerging family of CULLIN3-RING ubiquitin ligases (CRL3s): cellular functions and disease implications. EMBO J. 2013;32:2307–2320. doi: 10.1038/emboj.2013.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilman SR, Iossifov I, Levy D, Ronemus M, Wigler M, Vitkup D. Rare de novo variants associated with autism implicate a large functional network of genes involved in formation and function of synapses. Neuron. 2011;70:898–907. doi: 10.1016/j.neuron.2011.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girirajan S, Rosenfeld JA, Coe BP, Parikh S, Friedman N, Goldstein A, Filipink RA, McConnell JS, Angle B, Meschino WS, et al. Phenotypic heterogeneity of genomic disorders and rare copy-number variants. N Engl J Med. 2012;367:1321–1331. doi: 10.1056/NEJMoa1200395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golzio C, Willer J, Talkowski ME, Oh EC, Taniguchi Y, Jacquemont S, Reymond A, Sun M, Sawa A, Gusella JF, et al. KCTD13 is a major driver of mirrored neuroanatomical phenotypes of the 16p11.2 copy number variant. Nature. 2012;485:363–367. doi: 10.1038/nature11091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govek EE, Hatten ME, Van Aelst L. The role of Rho GTPase proteins in CNS neuronal migration. Dev Neurobiol. 2011;71:528–553. doi: 10.1002/dneu.20850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoriev A. A relationship between gene expression and protein interactions on the proteome scale: analysis of the bacteriophage T7 and the yeast Saccharomyces cerevisiae. Nucleic Acids Res. 2001;29:3513–3519. doi: 10.1093/nar/29.17.3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulsuner S, Walsh T, Watts AC, Lee MK, Thornton AM, Casadei S, Rippey C, Shahin H, Nimgaonkar VL, Go RC, et al. Spatial and temporal mapping of de novo mutations in schizophrenia to a fetal prefrontal cortical network. Cell. 2013;154:518–529. doi: 10.1016/j.cell.2013.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa A, Searle S, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H, Tan CK, Downey KM, So AG. A tumor necrosis factor alpha- and interleukin 6-inducible protein that interacts with the small subunit of DNA polymerase delta and proliferating cell nuclear antigen. Proc Natl Acad Sci U S A. 2001;98:11979–11984. doi: 10.1073/pnas.221452098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horev G, Ellegood J, Lerch JP, Son YE, Muthuswamy L, Vogel H, Krieger AM, Buja A, Henkelman RM, Wigler M, Mills AA. Dosage-dependent phenotypes in models of 16p11.2 lesions found in autism. Proc Natl Acad Sci U S A. 2011;108:17076–17081. doi: 10.1073/pnas.1114042108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iossifov I, Ronemus M, Levy D, Wang Z, Hakker I, Rosenbaum J, Yamrom B, Lee YH, Narzisi G, Leotta A, et al. De novo gene disruptions in children on the autistic spectrum. Neuron. 2012;74:285–299. doi: 10.1016/j.neuron.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen R, Greenbaum D, Gerstein M. Relating whole-genome expression data with protein-protein interactions. Genome Res. 2002;12:37–46. doi: 10.1101/gr.205602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HJ, Kawasawa YI, Cheng F, Zhu Y, Xu X, Li M, Sousa AM, Pletikos M, Meyer KA, Sedmak G, et al. Spatio-temporal transcriptome of the human brain. Nature. 2011;478:483–489. doi: 10.1038/nature10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirov G, Grozeva D, Norton N, Ivanov D, Mantripragada KK, Holmans P, Craddock N, Owen MJ, O’Donovan MC. Support for the involvement of large copy number variants in the pathogenesis of schizophrenia. Hum Mol Genet. 2009;18:1497–1503. doi: 10.1093/hmg/ddp043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitchen RR, Rozowsky JS, Gerstein MB, Nairn AC. Decoding neuroproteomics: integrating the genome, translatome and functional anatomy. Nat Neurosci. 2014;17:1491–1499. doi: 10.1038/nn.3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong A, Frigge ML, Masson G, Besenbacher S, Sulem P, Magnusson G, Gudjonsson SA, Sigurdsson A, Jonasdottir A, Wong WS, et al. Rate of de novo mutations and the importance of father’s age to disease risk. Nature. 2012;488:471–475. doi: 10.1038/nature11396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krey JF, Pasca SP, Shcheglovitov A, Yazawa M, Schwemberger R, Rasmusson R, Dolmetsch RE. Timothy syndrome is associated with activity-dependent dendritic retraction in rodent and human neurons. Nat Neurosci. 2013;16:201–209. doi: 10.1038/nn.3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy D, Ronemus M, Yamrom B, Lee YH, Leotta A, Kendall J, Marks S, Lakshmi B, Pai D, Ye K, et al. Rare de novo and transmitted copy-number variation in autistic spectrum disorders. Neuron. 2011;70:886–897. doi: 10.1016/j.neuron.2011.05.015. [DOI] [PubMed] [Google Scholar]
- Lionel AC, Crosbie J, Barbosa N, Goodale T, Thiruvahindrapuram B, Rickaby J, Gazzellone M, Carson AR, Howe JL, Wang Z, et al. Rare copy number variation discovery and cross-disorder comparisons identify risk genes for ADHD. Sci Transl Med. 2011;3:95ra75. doi: 10.1126/scitranslmed.3002464. [DOI] [PubMed] [Google Scholar]
- Luo R, Sanders SJ, Tian Y, Voineagu I, Huang N, Chu SH, Klei L, Cai C, Ou J, Lowe JK, et al. Genome-wide transcriptome profiling reveals the functional impact of rare de novo and recurrent CNVs in autism spectrum disorders. Am J Hum Genet. 2012;91:38–55. doi: 10.1016/j.ajhg.2012.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luscombe NM, Babu MM, Yu H, Snyder M, Teichmann SA, Gerstein M. Genomic analysis of regulatory network dynamics reveals large topological changes. Nature. 2004;431:308–312. doi: 10.1038/nature02782. [DOI] [PubMed] [Google Scholar]
- Malhotra D, McCarthy S, Michaelson JJ, Vacic V, Burdick KE, Yoon S, Cichon S, Corvin A, Gary S, Gershon ES, et al. High frequencies of de novo CNVs in bipolar disorder and schizophrenia. Neuron. 2011;72:951–963. doi: 10.1016/j.neuron.2011.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra D, Sebat J. CNVs: harbingers of a rare variant revolution in psychiatric genetics. Cell. 2012;148:1223–1241. doi: 10.1016/j.cell.2012.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall CR, Noor A, Vincent JB, Lionel AC, Feuk L, Skaug J, Shago M, Moessner R, Pinto D, Ren Y, et al. Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet. 2008;82:477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy SE, Makarov V, Kirov G, Addington AM, McClellan J, Yoon S, Perkins DO, Dickel DE, Kusenda M, Krastoshevsky O, et al. Microduplications of 16p11.2 are associated with schizophrenia. Nat Genet. 2009;41:1223–1227. doi: 10.1038/ng.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merikangas AK, Corvin AP, Gallagher L. Copy-number variants in neurodevelopmental disorders: promises and challenges. Trends Genet. 2009;25:536–544. doi: 10.1016/j.tig.2009.10.006. [DOI] [PubMed] [Google Scholar]
- Miller JA, Ding SL, Sunkin SM, Smith KA, Ng L, Szafer A, Ebbert A, Riley ZL, Royall JJ, Aiona K, et al. Transcriptional landscape of the prenatal human brain. Nature. 2014;508:199–206. doi: 10.1038/nature13185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills RE, Walter K, Stewart C, Handsaker RE, Chen K, Alkan C, Abyzov A, Yoon SC, Ye K, Cheetham RK, et al. Mapping copy number variation by population-scale genome sequencing. Nature. 2011;470:59–65. doi: 10.1038/nature09708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulle JG, Dodd AF, McGrath JA, Wolyniec PS, Mitchell AA, Shetty AC, Sobreira NL, Valle D, Rudd MK, Satten G, et al. Microdeletions of 3q29 confer high risk for schizophrenia. Am J Hum Genet. 2010;87:229–236. doi: 10.1016/j.ajhg.2010.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulle JG, Pulver AE, McGrath JA, Wolyniec PS, Dodd AF, Cutler DJ, Sebat J, Malhotra D, Nestadt G, Conrad DF, et al. Reciprocal duplication of the Williams-Beuren syndrome deletion on chromosome 7q11.23 is associated with schizophrenia. Biol Psychiatry. 2014;75:371–377. doi: 10.1016/j.biopsych.2013.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh HJ, Ponting CP, Boulding HC, Meader S, Betancur C, Buxbaum JD, Pinto D, Marshall CR, Lionel AC, Scherer SW, Webber C. Network topologies and convergent aetiologies arising from deletions and duplications observed in individuals with autism. PLoS Genet. 2013;9:e1003523. doi: 10.1371/journal.pgen.1003523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Roak BJ, Vives L, Girirajan S, Karakoc E, Krumm N, Coe BP, Levy R, Ko A, Lee C, Smith JD, et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012;485:246–250. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikshak NN, Luo R, Zhang A, Won H, Lowe JK, Chandran V, Horvath S, Geschwind DH. Integrative functional genomic analyses implicate specific molecular pathways and circuits in autism. Cell. 2013;155:1008–1021. doi: 10.1016/j.cell.2013.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D, Pagnamenta AT, Klei L, Anney R, Merico D, Regan R, Conroy J, Magalhaes TR, Correia C, Abrahams BS, et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010;466:368–372. doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portmann T, Yang M, Mao R, Panagiotakos G, Ellegood J, Dolen G, Bader PL, Grueter BA, Goold C, Fisher E, et al. Behavioral abnormalities and circuit defects in the Basal Ganglia of a mouse model of 16p11.2 deletion syndrome. Cell Rep. 2014;7:1077–1092. doi: 10.1016/j.celrep.2014.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujana MA, Han JD, Starita LM, Stevens KN, Tewari M, Ahn JS, Rennert G, Moreno V, Kirchhoff T, Gold B, et al. Network modeling links breast cancer susceptibility and centrosome dysfunction. Nat Genet. 2007;39:1338–1349. doi: 10.1038/ng.2007.2. [DOI] [PubMed] [Google Scholar]
- Rolland T, Tasan M, Charloteaux B, Pevzner SJ, Zhong Q, Sahni N, Yi S, Lemmens I, Fontanillo C, Mosca R, et al. A proteome-scale map of the human interactome network. Cell. 2014;159:1212–1226. doi: 10.1016/j.cell.2014.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SJ, Ercan-Sencicek AG, Hus V, Luo R, Murtha MT, Moreno-De-Luca D, Chu SH, Moreau MP, Gupta AR, Thomson SA, et al. Multiple recurrent de novo CNVs, including duplications of the 7q11.23 Williams syndrome region, are strongly associated with autism. Neuron. 2011;70:863–885. doi: 10.1016/j.neuron.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, Walsh T, Yamrom B, Yoon S, Krasnitz A, Kendall J, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal E, Wang H, Koller D. Discovering molecular pathways from protein interaction and gene expression data. Bioinformatics. 2003;19(Suppl 1):i264–i271. doi: 10.1093/bioinformatics/btg1037. [DOI] [PubMed] [Google Scholar]
- Shapiro E, Biezuner T, Linnarsson S. Single-cell sequencing-based technologies will revolutionize whole-organism science. Nat Rev Genet. 2013;14:618–630. doi: 10.1038/nrg3542. [DOI] [PubMed] [Google Scholar]
- Stark C, Breitkreutz BJ, Reguly T, Boucher L, Breitkreutz A, Tyers M. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 2006;34:D535–D539. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefansson H, Rujescu D, Cichon S, Pietilainen OP, Ingason A, Steinberg S, Fossdal R, Sigurdsson E, Sigmundsson T, Buizer-Voskamp JE, et al. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455:232–236. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoner R, Chow ML, Boyle MP, Sunkin SM, Mouton PR, Roy S, Wynshaw-Boris A, Colamarino SA, Lein ES, Courchesne E. Patches of disorganization in the neocortex of children with autism. N Engl J Med. 2014;370:1209–1219. doi: 10.1056/NEJMoa1307491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker BA, Ji SJ, Jaffrey SR. Intra-axonal translation of RhoA promotes axon growth inhibition by CSPG. J Neurosci. 2012;32:14442–14447. doi: 10.1523/JNEUROSCI.0176-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh T, McClellan JM, McCarthy SE, Addington AM, Pierce SB, Cooper GM, Nord AS, Kusenda M, Malhotra D, Bhandari A, et al. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008;320:539–543. doi: 10.1126/science.1155174. [DOI] [PubMed] [Google Scholar]
- Wang Y, Zheng Y, Luo F, Fan X, Chen J, Zhang C, Hui R. KCTD10 interacts with proliferating cell nuclear antigen and its down-regulation could inhibit cell proliferation. J Cell Biochem. 2009;106:409–413. doi: 10.1002/jcb.22026. [DOI] [PubMed] [Google Scholar]
- Weiss LA, Shen Y, Korn JM, Arking DE, Miller DT, Fossdal R, Saemundsen E, Stefansson H, Ferreira MA, Green T, et al. Association between microdeletion and microduplication at 16p11.2 and autism. N Engl J Med. 2008;358:667–675. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- Willsey AJ, Sanders SJ, Li M, Dong S, Tebbenkamp AT, Muhle RA, Reilly SK, Lin L, Fertuzinhos S, Miller JA, et al. Coexpression networks implicate human midfetal deep cortical projection neurons in the pathogenesis of autism. Cell. 2013;155:997–1007. doi: 10.1016/j.cell.2013.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Liu N, Hu X, Zhang W, Wang T, Li H, Zhang B, Xiang S, Zhou J, Zhang J. CK2 phosphorylates TNFAIP1 to affect its subcellular localization and interaction with PCNA. Mol Biol Rep. 2010;37:2967–2973. doi: 10.1007/s11033-009-9863-1. [DOI] [PubMed] [Google Scholar]
- Yang X, Boehm JS, Salehi-Ashtiani K, Hao T, Shen Y, Lubonja R, Thomas SR, Alkan O, Bhimdi T, Green TM, et al. A public genome-scale lentiviral expression library of human ORFs. Nat Methods. 2011;8:659–661. doi: 10.1038/nmeth.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Macara IG. The PAR-6 polarity protein regulates dendritic spine morphogenesis through p190 RhoGAP and the Rho GTPase. Dev Cell. 2008;14:216–226. doi: 10.1016/j.devcel.2007.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Q, Simonis N, Li QR, Charloteaux B, Heuze F, Klitgord N, Tam S, Yu H, Venkatesan K, Mou D, et al. Edgetic perturbation models of human inherited disorders. Mol Syst Biol. 2009;5:321. doi: 10.1038/msb.2009.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Korzh V, Gong Z, Low BC. RhoA prevents apoptosis during zebrafish embryogenesis through activation of Mek/Erk pathway. Oncogene. 2008;27:1580–1589. doi: 10.1038/sj.onc.1210790. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.