Abstract
This manuscript illustrates a protocol for efficiently creating integration-free human induced pluripotent stem cells (iPSCs) from peripheral blood using episomal plasmids and histone deacetylase (HDAC) inhibitors. The advantages of this approach include: (1) the use of a minimal amount of peripheral blood as a source material; (2) nonintegrating reprogramming vectors; (3) a cost effective method for generating vector free iPSCs; (4) a single transfection; and (5) the use of small molecules to facilitate epigenetic reprogramming. Briefly, peripheral blood mononuclear cells (PBMCs) are isolated from routine phlebotomy samples and then cultured in defined growth factors to yield a highly proliferative erythrocyte progenitor cell population that is remarkably amenable to reprogramming. Nonintegrating, nontransmissible episomal plasmids expressing OCT4, SOX2, KLF4, MYCL, LIN28A, and a p53 short hairpin (sh)RNA are introduced into the derived erythroblasts via a single nucleofection. Cotransfection of an episome that expresses enhanced green fluorescent protein (eGFP) allows for easy identification of transfected cells. A separate replication-deficient plasmid expressing Epstein-Barr nuclear antigen 1 (EBNA1) is also added to the reaction mixture for increased expression of episomal proteins. Transfected cells are then plated onto a layer of irradiated mouse embryonic fibroblasts (iMEFs) for continued reprogramming. As soon as iPSC-like colonies appear at about twelve days after nucleofection, HDAC inhibitors are added to the medium to facilitate epigenetic remodeling. We have found that the inclusion of HDAC inhibitors routinely increases the generation of fully reprogrammed iPSC colonies by 2 fold. Once iPSC colonies exhibit typical human embryonic stem cell (hESC) morphology, they are gently transferred to individual iMEF-coated tissue culture plates for continued growth and expansion.
Keywords: Cellular Biology, Issue 92, Induced pluripotent stem cells, iPSC, iPSC generation, human, HDAC inhibitors, histone deacetylase inhibitors, reprogramming, episomes, integration-free
Introduction
iPSCs are derived from somatic tissues via ectopic expression of a minimal set of pluripotency genes. This technique was initially demonstrated by retroviral transduction of human fibroblasts with OCT4, SOX2, KLF4, and cMYC, which are highly expressed in the pluripotent state1. These transiently expressed “reprogramming factors” alter the target cell’s epigenetic landscape and gene expression profile analogous to human embryonic stem cells2. Once created, iPSCs can potentially be differentiated into any tissue type for further investigation. Thus, they hold promise for use in regenerative medicine, disease modeling, and gene therapy applications. However, disrupting the genome with integrating viruses has the potential to alter endogenous gene expression, influence cellular phenotype, and ultimately bias scientific results. Furthermore, random viral integrations can lead to deleterious cellular effects, including the possibility of malignant transformation3 or re-expression of the oncogenic transgenes4. Future clinical applications will require non-integrating iPSC generation.
Episomes, which are extra chromosomal circular DNA molecules, offer a strategy to generate cost effective, integration-free iPSCs5. The combination of episomal vectors shown in Table 1 express the reprogramming factors OCT4, SOX2, KLF4, MYCL, and LIN28A. The pCXLE_hOCT3/4-shp53-F plasmid also contains a p53 shRNA for temporary suppression of TP53 to enhance cellular reprogramming6. The replication deficient pCXWB-EBNA1 vector promotes amplification of reprogramming factors and increased reprogramming efficiency by providing a transient increase in EBNA1 expression7. The pCXLE_EGFP plasmid can be added to the nucleofection mixture for the purpose of determining the transfection efficiency or for cell sorting applications. With the exception of pCXWB-EBNA1, the episomal plasmids used in this protocol contain the Epstein-Barr virus origin of viral replication and EBNA1 gene, which mediate replication and partition of the episome during division of the host cell8. The episomes are spontaneously lost with successive iPSC expansion7. Subcloning and characterization of iPSCs with episomal vector loss, which can be inferred from loss of eGFP expression, can lead to completely integration free iPSCs for future clinical applications.
Inherent to the process of iPSC generation is suppression of lineage specific genes and reactivation of pluripotency-associated genes. Regulation of gene expression occurs at multiple levels within the nucleus, including modifications to DNA and chromatin to allow transcription factors, regulatory DNA elements, and RNA polymerase access to target genes. Remodeling of the epigenetic landscape via global chromatin modifications is a key component to re-expression of the pluripotency genetic program. A specific chromatin modification that is important in regulation of gene expression is acetylation of histones at particular lysine residues, which allows access to target genes through decreased tension of the histone-DNA coil. HDAC inhibitors are small molecules that have been shown to enhance iPSC reprogramming and hESC self-renewal9,10, likely due to supporting the acetylated state11. The protocol described below, adapted from a prior publication using integrating lentiviruses12, provides a step-by-step method for optimized iPSC generation from peripheral blood using episomes and HDAC inhibitors. The HDAC inhibitor concentrations used here are half of those described by Ware, et al.9, and have routinely led to a 2 fold increase in fully reprogrammed iPSC colonies over standard episomal reprogramming protocols without HDAC inhibitors. This level of reprogramming is on par with the efficiency we observe with lentiviral methods. Using this protocol, we have efficiently generated iPSC from individuals as old as 87 years of age.
Protocol
Written informed consent, as approved by the Institutional Review Boards of the Fred Hutchinson Cancer Research Center and the Children"s Hospital of Philadelphia, was obtained from patients before collecting peripheral blood samples. All institutional guidelines were observed. All animal experiments including MEF generation and teratoma formation were approved by the Institutional Animal Care and Use Committee.
1. Ficoll Separation of PBMCS and Expansion of Erythroblasts – Day 0
Dilute peripheral blood 1:1 with Dulbecco’s Phosphate Buffered Saline (DPBS). In a 15 ml round-bottom polystyrene tube, carefully layer 7 ml of diluted blood onto 3 ml of room temperature Ficoll-Hypaque.
Centrifuge the sample at 400 x g for 30 min.
The PBMCs will have settled to the interface between the plasma and the Ficoll-Hypaque, seen as a cloudy white layer. Using a sterile transfer pipette, collect the PBMCs into one 15 ml conical tube. Bring the volume to 10 ml using DPBS.
Using a hemacytometer, count the PBMCs.
Pipette 2 × 106 PBMCs into a separate 15 ml conical tube and centrifuge at 300 x g for 5 min. Spin down the remaining cells and freeze at a concentration of 2 × 106 PBMCs/ml in 90% fetal bovine serum (FBS)/10% dimethyl sulfoxide (DMSO).
Prepare Expansion Medium: QBSF-60 serum free medium (protect from light) + penicillin/streptomycin (1%) + ascorbic acid (50 ng/ml) + stem cell factor (SCF, 50 ng/ml) + interleukin 3 (IL-3, 10 ng/ml) + erythropoietin (EPO, 2 U/ml) + insulin-like growth factor 1 (IGF-1, 40 ng/ml) + dexamethasone (1 μM; protect from light; thaw a fresh aliquot each media change).
Resuspend 2 × 106 PBMCs in 2 ml of freshly made Expansion Medium and place into one well of a 12 well tissue culture plate and incubate in a 37 °C humidified incubator with an atmosphere of 5% O2, 5% CO2, and 90% N2.
- On Day 3 and Day 6, replace media.
- Collect the cells and wash the well with 2 ml of QBSF-60 to collect any remaining cells.
- Centrifuge the cell suspension at 300 x g for 5 min and aspirate the supernatant.
- Resuspend the cells in 2 ml of new Expansion Medium and return them to the same 12 well plate.
2. Reprogramming Cells by Nucleofection – Day 9
Pipette the reprogramming plasmids into a sterile 1.5 ml Eppendorf tube (Table 1).
Mix the nucleofection solution with Supplement (supplied by manufacturer) and pipette into a sterile 1.5 ml Eppendorf tube.
Pipette 2 ml of Expansion Medium into one well of a 12 well plate and place in a humidified 37 °C incubator (5% O2, 5% CO2, and 90% N2) for equilibration prior to adding cells.
Collect the cultured cells and wash the well with 2 ml of QBSF-60 to collect remaining cells.
Centrifuge the cells at 300 x g for 5 min and aspirate the supernatant.
Wash the cells by resuspending in 5 ml of DPBS and centrifuging at 300 x g for 5 min.
Aspirate the supernatant.
Resuspend the cell pellet in 100 μl of nucleofection solution + Supplement, taking care to avoid generating bubbles.
Pipette the cell suspension into the tube containing the reprogramming plasmids, pipette up and down once, and pipette the sample into the bottom of the nucleofection cuvette.
Place the cuvette into the nucleofection machine and run Program T-019.
Transfer 100 μl of the prewarmed Expansion Medium (from the equilibrating plate in Step 2.3) into the nucleofection cuvette.
Immediately collect the entire sample and deposit into the well of pre-warmed Expansion Medium. Avoid collecting the white, floating dead cell debris.
Place the plate in a humidified 37 °C incubator (5% O2, 5% CO2, and 90% N2) for growth.
3. Plating Feeder Cells – Day 10 or 11
- On Day 10, plate feeder cells.
- Coat one 6 well tissue culture plate with gelatin.
- Pipette 1 m of 0.1% gelatin in Hank’s Buffered Saline Solution (HBSS) into each well of the plate.
- Place the plate in a humidified 37 °C incubator for at least 5 min.
- Immediately before use, aspirate the gelatin solution from the plate.
- Thaw one vial of 2 × 106 mouse embryonic fibroblasts irradiated at 3,000 cGy into 10 ml of prewarmed MEF Media (Dulbecco’s Modified Eagle Medium (DMEM) + FBS (10%) + penicillin/streptomycin (1%)).
- Centrifuge the cells at 300 x g for 5 min and aspirate the supernatant. Resuspend the cells in 12 ml of MEF Media and pipette 2 ml into each well of the gelatin-coated 6 well plate. Place the plate in a humidified 37 °C incubator (5% O2, 5% CO2, and 90% N2) until Day 12.
4. Plating Cells on Feeder Cells and Changing Medium – Day 12
Collect the nucleofected cells into one 15 ml conical tube and collect any remaining cells by washing the well with 2 ml of QBSF-60. Centrifuge the sample at 300 x g for 5 min.
Prepare Reprogramming Medium: Iscove’s Modified Dulbecco’s Medium (IMDM) + FBS (10%) + non-animal L-glutamine (1 mM) + non-essential amino acids (NEAA, 1%) + penicillin/streptomycin (1%) + β-mercaptoethanol (0.1 mM) + basic fibroblast growth factor (bFGF, added fresh at feeding, 10 ng/ml) + ascorbic acid (added fresh at feeding, 50 μg/ml).
Aspirate the supernatant and resuspend the cells in 12 ml of Reprogramming Medium. Pipette 2 ml per well of the cell suspension into the 6 well plate of feeder cells (as prepared in Section 3). Centrifuge the plate at 50 x g for 30 min at room temperature.
Place the plate in a humidified 37 °C incubator (5% O2, 5% CO2, and 90% N2) for growth. Change the Reprogramming Medium every other day until iPSC colonies begin appearing (1-2 weeks).
Prepare hESC Medium: DMEM/F12 1:1 + Knockout Serum Replacement (20%) + penicillin/streptomycin (1%) + NEAA (1%) + non-animal L-glutamine (1 mM) + sodium pyruvate (1%) + sodium bicarbonate (0.12%) + β-mercaptoethanol (0.1 mM) + bFGF (added fresh at feeding, 10 ng/ml)
Prepare HDAC inhibitors: sodium butyrate (added fresh at feeding, 100 μM), suberanilohydroxamic acid (SAHA, added fresh at feeding, 200 nM). Once iPSC colonies appear, begin feeding the cells with hESC Medium + HDAC inhibitors, changing medium every day. NOTE: Sodium butyrate is a small molecule that is adsorbed to plastic tubes, therefore store it in a borosilicate glass tube at 4 °C.
5. Isolating iPSC Clones
Once iPSC colonies are large enough to be isolated (20-25 cells), pick them with a P20 pipette.
Prepare HDAC inhibitors: sodium butyrate (added fresh at feeding, 100 μM), SAHA (added fresh at feeding, 200 nM)
Place the isolated colonies onto iMEFs in individual 35 mm dishes containing hESC Medium + HDAC inhibitors + Cloning & Recovery Supplement.
The day after picking the colonies, change the medium to hESC Medium + HDAC inhibitors.
Change the medium every day until iPSC colonies are ready for passaging.
Remove HDAC inhibitors from the medium once colonies stabilize at passage 2-3.
Representative Results
Three days after nucleofection and before plating the nucleofected cells onto iMEFs, the efficiency of successful nucleofection should be estimated by fluorescence microscopy for eGFP. Figure 1 shows a typical nucleofection experiment with approximately 5-10% of the total cell population expressing eGFP.
Reprogrammed iPSC colonies will begin to appear approximately two weeks after nucleofection. The colonies are generally circular with well-defined borders and may be identified by characteristic hESC morphology including (1) small, tightly packed cells with a high nuclear-cytoplasmic ratio and (2) visible nucleoli (Figure 2, Bright field). Incompletely reprogrammed cells will either deteriorate or form heterogeneous cell masses that are easily distinguished from true iPSC colonies.
Of the representative lines that we have chosen to characterize completely, all express the standard pluripotency markers (Figure 2), reactivate endogenous pluripotency genes (Figure 2), and form teratomas when injected into NOD-SCID mice.
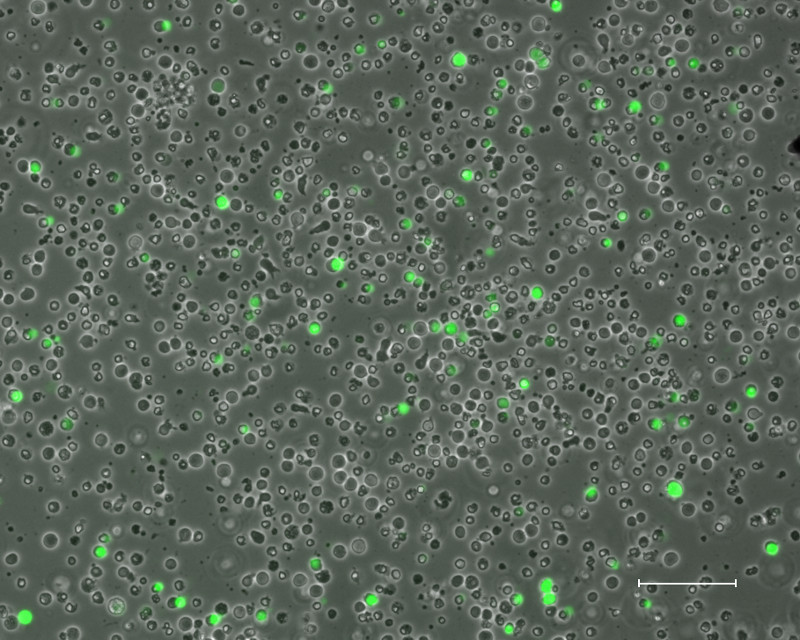
Figure 1: Nucleofected Cells. Stimulated PBMCs three days after nucleofection with plasmids containing OCT4, SOX2, KLF4, MYCL, EBNA1, p53 shRNA, and eGFP (Scale bar is 100 μm).
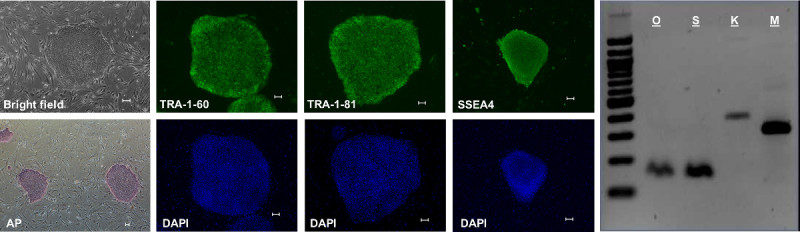 Figure 2: IPSC Characterization. IPSC generated using this method exhibit hESC-like morphology when grown on iMEF (Bright field), test positive for alkaline phosphatase activity (AP), and are positive by immunohistochemistry for pluripotency markers TRA-1-60, TRA-1-81, and SSEA4. Additionally, endogenous expression of the pluripotency genes Oct4 (O), Sox2 (S), Klf4 (K), and cMyc (M) is detectable by RT-PCR. (All scale bars are 100 μm). Please click here to view a larger version of this figure.
Figure 2: IPSC Characterization. IPSC generated using this method exhibit hESC-like morphology when grown on iMEF (Bright field), test positive for alkaline phosphatase activity (AP), and are positive by immunohistochemistry for pluripotency markers TRA-1-60, TRA-1-81, and SSEA4. Additionally, endogenous expression of the pluripotency genes Oct4 (O), Sox2 (S), Klf4 (K), and cMyc (M) is detectable by RT-PCR. (All scale bars are 100 μm). Please click here to view a larger version of this figure.
| Episome | Mass per reaction (ng) |
| pCXLE-hOct3/4-shp53-F | 830 |
| pCXLE-hSK | 830 |
| pCXLE-hUL | 830 |
| pCXLE-EGFP | 500 |
| pCXWB-EBNA1 | 500 |
Table 1: Optimal Ratio of Reprogramming Plasmids6. These plasmids are available to order through Addgene.
Discussion
For successful iPSC generation when using this protocol, there are several important caveats that should be considered. During the erythroblast expansion stage, the media change schedule should be strictly followed, as deviations may lead to inefficient stimulation of the target progenitor cell population and a lower efficiency of iPSC generation. It is important to make new expansion medium with fresh dexamethasone with each media change; and the QBSF-60 base medium and dexamethasone should be protected from light during storage. During nucleofection, the DPBS wash is critical to remove excess solutes from the cell suspension that may cause the electric current from the nucleofector to arc, resulting in widespread cell death. Approximately 10 days after nucleofection, the plate should be examined daily for iPSC colony appearance. Once several small colonies are present, the Reprogramming Medium should be changed to hESC Medium (with HDAC inhibitors) as prolonged exposure to serum may induce spontaneous differentiation.
Depending on laboratory set-up and available resources, modifications to the protocol may be considered. We prefer a low O2 incubator because hypoxic conditions have been shown to increase the efficiency of iPSC generation13; however, we have had success generating iPSCs from peripheral blood using normoxic conditions with 5% CO2. Therefore, a standard humidified CO2 incubator can also be used. The MEFs used in this protocol may be purchased from a life science vendor. However, due to commercial batch variability, we prefer to generate iMEFs from CF-1 mice following a published protocol for MEF isolation, culture, and irradiation14. The inclusion of HDAC inhibitors is a favorable aspect of this protocol as epigenetic remodeling is a crucial aspect of cellular reprogramming9,15. HDAC inhibitors may be omitted, however the number of fully reprogrammed iPSC colonies will be reduced. Lastly, we have generated iPSCs using this method with a bone marrow sample with no additional modifications needed.
In summary, this protocol uses a plasmid based reprogramming system that avoids some of the drawbacks of iPSC generation with integrating vectors, including random insertions into the host genome and safety concerns regarding the handling of recombinant virus. Additionally, small scale virus production and characterization is relatively labor intensive when compared to plasmid generation. Reprogramming efficiency can also vary significantly due to batch variability inherent to virus production, while one large scale production of quality plasmids can lead to consistent iPSC generation over time. Combined with HDAC inhibitors, this protocol offers an efficient method to generate integration free and footprint free iPSCs for stem cell research.
Disclosures
The authors declare that they have no competing financial interests.
Acknowledgments
The authors wish to acknowledge the following grants from the NIH for supporting this research: K08DK082783 (AR), P30DK56465 (BTS), U01HL099993 (BTS), T32HL00715036 (SKS), and K12HL0806406 (SKS); and the JP McCarthy Foundation (AR).
References
- Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131(5):861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Koche RP, et al. Reprogramming factor expression initiates widespread targeted chromatin remodeling. Cell Stem Cell. 2011;8(1):96–105. doi: 10.1016/j.stem.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum C, Kustikova O, Modlich U, Li Z, Fehse B. Mutagenesis and oncogenesis by chromosomal insertion of gene transfer vectors. Human Gene Therapy. 2006;17(3):253–263. doi: 10.1089/hum.2006.17.253. [DOI] [PubMed] [Google Scholar]
- Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448(7151):313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- Yu J, et al. Human induced pluripotent stem cells free of vector and transgene sequences. Science. 2009;324(5928):797–801. doi: 10.1126/science.1172482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong H, et al. Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature. 2009;460(7259):1132–1135. doi: 10.1038/nature08235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okita K, et al. A more efficient method to generate integration-free human iPS cells. Nature Methods. 2011;8(5):409–412. doi: 10.1038/nmeth.1591. [DOI] [PubMed] [Google Scholar]
- Yates JL, Warren N, Sugden B. Stable replication of plasmids derived from Epstein-Barr virus in various mammalian cells. Nature. 1985;313(6005):812–815. doi: 10.1038/313812a0. [DOI] [PubMed] [Google Scholar]
- Ware CB, et al. Histone deacetylase inhibition elicits an evolutionarily conserved self-renewal program in embryonic stem cells. Cell Stem Cell. 2009;4(4):359–369. doi: 10.1016/j.stem.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, et al. Butyrate greatly enhances derivation of human induced pluripotent stem cells by promoting epigenetic remodeling and the expression of pluripotency-associated genes. Stem Cells. 2010;28(4):713–720. doi: 10.1002/stem.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuara V, et al. Chromatin signatures of pluripotent cell lines. Nature Cell Biology. 2006;8(5):532–538. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- Sommer AG, et al. Generation of human induced pluripotent stem cells from peripheral blood using the STEMCCA lentiviral vector. J. Vis. Exp. 2012. p. e4327. [DOI] [PMC free article] [PubMed]
- Yoshida Y, Takahashi K, Okita K, Ichisaka T, Yamanaka S. Hypoxia enhances the generation of induced pluripotent stem cells. Cell Stem Cell. 2009;5:237–241. doi: 10.1016/j.stem.2009.08.001. [DOI] [PubMed] [Google Scholar]
- Abbondanzo SJ, Gadi I, Stewart CL. Derivation of Embryonic Stem Cell Lines. Methods in Enzymology. 1993;225:803–823. doi: 10.1016/0076-6879(93)25052-4. [DOI] [PubMed] [Google Scholar]
- Kim HJ, Bae SC. Histone deacetylase inhibitors: molecular mechanisms of action and clinical trials as anti-cancer drugs. American Journal of Translational Research. 2011;3(2):166–179. [PMC free article] [PubMed] [Google Scholar]


