Version Changes
Revised. Amendments from Version 1
We have included the reviewer’s suggestions in our revised version. Specifically, we have modified figure 2, summary figure 5, and supplementary figure 3 according to Monica Folgueira’s suggestions, and added clarifications in the text according to the reviewer’s suggestions.
Abstract
Background: The telencephalon shows a remarkable structural diversity among vertebrates. In particular, the everted telencephalon of ray-finned fishes has a markedly different morphology compared to the evaginated telencephalon of all other vertebrates. This difference in development has hampered the comparison between different areas of the pallium of ray-finned fishes and the pallial nuclei of all other vertebrates. Various models of homology between pallial subdivisions in ray-finned fishes and the pallial nuclei in tetrapods have been proposed based on connectional, neurochemical, gene expression and functional data. However, no consensus has been reached so far. In recent years, the analysis of conserved developmental marker genes has assisted the identification of homologies for different parts of the telencephalon among several tetrapod species.
Results: We have investigated the gene expression pattern of conserved marker genes in the adult zebrafish ( Danio rerio) pallium to identify pallial subdivisions and their homology to pallial nuclei in tetrapods. Combinatorial expression analysis of ascl1a, eomesa, emx1, emx2, emx3, and Prox1 identifies four main divisions in the adult zebrafish pallium. Within these subdivisions, we propose that Dm is homologous to the pallial amygdala in tetrapods and that the dorsal subdivision of Dl is homologous to part of the hippocampal formation in mouse. We have complemented this analysis be examining the gene expression of emx1, emx2 and emx3 in the zebrafish larval brain.
Conclusions: Based on our gene expression data, we propose a new model of subdivisions in the adult zebrafish pallium and their putative homologies to pallial nuclei in tetrapods. Pallial nuclei control sensory, motor, and cognitive functions, like memory, learning and emotion. The identification of pallial subdivisions in the adult zebrafish and their homologies to pallial nuclei in tetrapods will contribute to the use of the zebrafish system as a model for neurobiological research and human neurodegenerative diseases.
Keywords: telencephalon, teleost, Actinopterygii, amygdala, hippocampus, neuroanatomy, vertebrate brain, homology, evolution, neurogenesis
Background
The functions of the different parts of the telencephalon encompass control of sensory and motor, autonomic and endocrine functions, as well as cognitive tasks like memory, learning and emotion. The structures in the telencephalon can be assigned either to its dorsal part, the pallium, or its ventral part, the subpallium 1– 3. The telencephalon of most vertebrates forms by an evagination of the neural tube, where the central lumen of the neural tube expands to form the two paired telencephalic vesicles 1, 3, 4. However, in ray-finned fishes the rostral neural tube is thought to bend outward resulting in two telencephalic hemispheres separated by an unpaired ventricle and covered by a thin roof plate, thus referred to as “everted” 1, 3, 4. In the rayfin teleost zebrafish, it has been shown that the morphogenetic movement that creates this different layout of the telencephalon does not result from a simple lateral outward bending of the telencephalic walls 5. Instead, telencephalon morphogenesis comprises first the generation of a ventricular outfolding between telencephalon and diencephalon, followed by an enlargement of the pallial territory rostrally 5. The different development of the telencephalon results in an unpaired ventricle and a different arrangement of the parts in the pallium compared to all other vertebrates 1, 4, 6. Hence, due to its everted nature, a comparison between the parts of the pallium of rayfin fishes and all other vertebrates has been difficult. The correct determination of homologous pallial areas between teleosts and tetrapods is critical for the usage of teleost fish as neurobiological models as well as models for human neurological diseases. In recent years, a variety of different studies have demonstrated that using the expression pattern of conserved developmental regulatory genes as landmarks is a useful approach to identify homologous subdivisions of brain regions between divergent vertebrate species. The advantage of using the expression of conserved developmental genes lies in the uncoupling of anatomical and developmental differences of the brain part of interest between divergent species. This approach has been especially valuable for the telencephalon with its great variability in morphology between vertebrate species and has led to the clarification of the homology within subdivisions of the telencephalon between different vertebrate species, such as the domestic mouse, the chicken and the African clawed frog (e.g. 7– 22). For example, the gene expression of Tbr1 and Eomes ( Tbr2) has been successfully used to identify the extent of the pallium in tetrapod embryos 9, 21, 23. In addition, absence of Emx1 expression and presence of Tbr1 expression delineate the ventral pallium in tetrapod embryos 9, 10, 12, 13, 21.
The pallium in teleosts has generally been subdivided in a medial (Dm), dorsal (Dd), central (Dc), lateral (Dl) and posterior (Dp) part 4. The pallium shows a notable structural variety and different subdivisions of these broad divisions have been described in different teleost species 3, 24– 29. The subdivisions of the pallium and their homologies to nuclei in other vertebrate species have not been resolved, but different models based on neurochemical and connectional data have been suggested 3, 29– 38. Ablation experiments combined with behavioral experiments suggest that the lateral nucleus of the pallium shows a similarity in function to the hippocampus and the medial nucleus of the pallium to the amygdala of amniotes 39– 43. Based on the expression of nicotine adenine dinucleotide phosphate diphorase (NADPHd) and Parvalbumin, a new model of four subdivisions (Dm, Dc, Dl, and Dp) has been proposed for the adult zebrafish pallium 32. However, a comprehensive study of pallial subdivisions based on different conserved molecular markers is still missing in the adult zebrafish.
The object of this study was to analyze expression of conserved marker genes to identify subdivisions within the adult zebrafish pallium. Here, we investigated the expression patterns of the molecular marker genes emx1, emx2, emx3 to identify a ventral pallial subdivision both in the larval and adult zebrafish pallium. The expression of Prox1 in a dorsal subdivision of Dl caudally suggests that it is homologous to the dentate gyrus in mouse. Combinatorial expression of ascl1a, emx1, emx2, emx3, and eomesa shows four main divisions in the pallium, Dm, Dc, Dl, and Dp. The combinatorial expression pattern also suggests a subdivision of Dl in a dorsal and ventral subdivision (which we have named Dld and Dlv, respectively).
Material and methods
Fish maintenance
Fish were kept under standard conditions at a 14 hours light/10 hours dark cycle as previously described 44, 45. All procedures were in accordance with the live animal handling and research regulations of the local Animal Care and Use Committee, the Regierungspräsidium Dresden (permit AZ 24D-9168.11-1/2008-1 and -4). Wildtype experimental animals (Biotechnology Center Dresden) were adult fish from the gol-b1 line in the AB genetic background 46. Adult fish were 6–8 months old and had a 24mm–32mm body length, zebrafish larvae were 7dpf old.
Tissue preparation
Brains (either dissected or within the skull) were fixed at 4°C overnight in 2–4% paraformaldehyde/0.1M phosphate buffer (PB), pH 7.5. They were washed 1 × 10 minutes and then up to 1h in 0.1M PB and subsequently transferred for decalcification and cryoprotection to 20% sucrose/20% EDTA in 0.1M PB, pH7.5. Brains were frozen in 7.5% gelatine/20% sucrose and sectioned into 14–16 µm cryosections. Sections were stored at -20°C.
Orthology analysis between tetrapod and teleost genes
Compared to tetrapods, teleost fish have undergone an additional whole genome duplication: the teleost genome duplication (TGD) (reviewed in 47). Thus, there is the possibility of two co-orthologous genes in zebrafish compared to the single tetrapod gene.
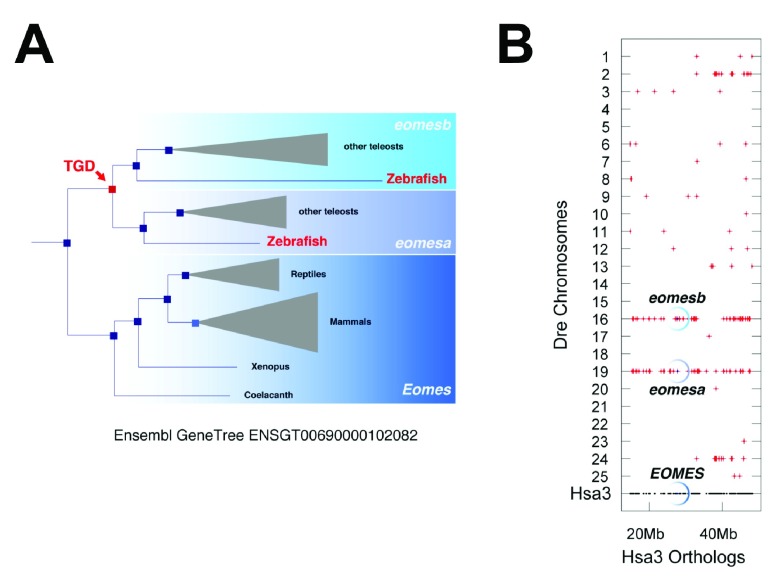
We analyzed if there are two co-orthologous genes compared to the tetrapod gene using Ensembl73 gene trees ( http://www.ensembl.org) and synteny analysis with the Synteny Database ( http://syntenydb.uoregon.edu; 48). This is clearly the case, e.g., for human EOMES with two TGD co-orthologs in zebrafish, eomesa and eomesb ( Figure S1A,B).
Figure S1. Teleost eomes genes.
A. The topology of the Ensembl Gene Tree suggest that the teleost eomes co-orthologs, eomesa and eomesb, were duplicated at the base of the teleost lineage and thus most likely during the teleost genome duplication (TGD). B. Dotplot from the Synteny Database shows that the human EOMES gene region on chromosome Hsa3 shows extensive double conserved synteny to zebrafish chromosomes Dre19 and Dre16 which contain eomesa and eomesb, respectively, providing strong evidence for the TGD origin of teleost eomes co-orthologs.
Two genes are currently termed ascl1 in zebrafish. Zebrafish ascl1a is clearly orthologous by phylogeny and conserved synteny to Ascl1 in lobefins (tetrapods and coelacanth). Zebrafish ascl1b not only has a separate ortholog in coelacanth but also shows conserved synteny to tetrapod Ascl2, suggesting that ascl1b is in fact the missing teleost ascl2 gene.
There are two prox1 genes described in zebrafish, yet while prox1a is clearly orthologous to tetrapod Prox1, teleost prox1b shows no conserved synteny to teleost prox1a or tetrapod Prox1. This suggests that teleost prox1b represent a more distant prox paralog and is not a TGD paralog of prox1a. The phylogeny of the emx genes in zebrafish has previously been determined in 49.
RNA in situ hybridization
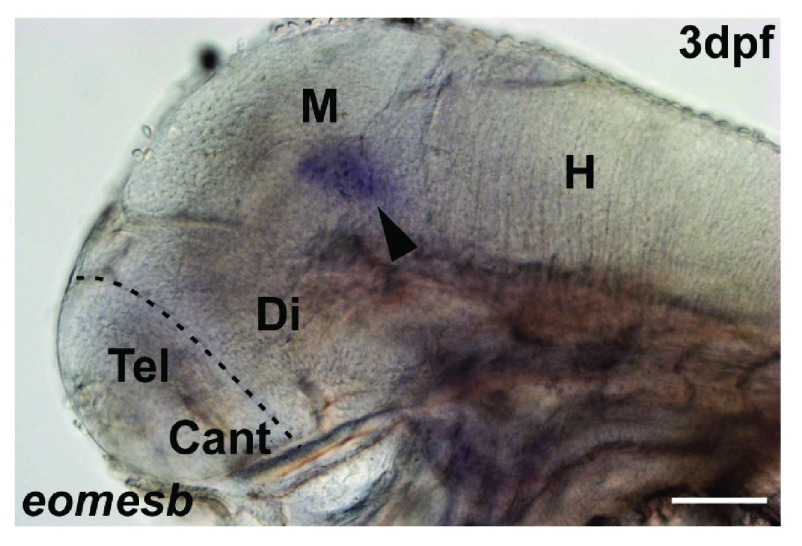
RNA in situ hybridization on sections and on whole-mount brains and RNA probe generation was essentially performed as previously described 7, 50. Briefly, after defrosting at room temperature (RT), sections were rehydrated for 15 minutes in PBS with 0.3% TritonX (PBSTx) and incubated with the probe overnight at 62–65°C. Information on the antisense in situ riboprobes can be found in 7 for ascl1a (NM_131219), emx1 (NM_198144), emx2 (NM_131280), emx3 (NM_131279), eomesa (NM_131679). The in situ probe eomesb (NM_001083575) was cloned from zebrafish embryonic cDNA with the following primers ( eomesb-F, TTTCCAAAACGAAAAGCGTA, eomesb-R, GAGCCAGAACTGGATCCTTCT). The eomesb probe was tested on 3dpf embryos and showed specific staining only in the midbrain at 3dpf ( Figure S2, Dataset 1). A sense probe of eomesb did not show any signal. The sections were washed at 60–65°C in washing solution (1 × SSC, 50% deionized formamide) for 1 × 15 minutes and 2 × 30 minutes followed by 2 × 30 minutes MAB with 0.1% Tween-20 (MABT) washes. Sections were incubated for 1h at RT in 2% DIG-blocking reagent (Roche) and incubated with anti-DIG antibody (Roche Diagnostics, sheep, polyclonal, Fab fragments conjugated to alkaline phosphatase, #11093274910) diluted 1:4000 in 2% DIG-blocking reagent overnight at 4°C. Subsequently, sections were washed 4 × 20 minutes in MABT, equilibrated with staining buffer and stained with the substrate NBT/BCIP. The staining was controlled using a stereomicroscope. Finally, sections were washed 2 × 5 minutes in PBS, postfixed with 4% PFA for 20–30 minutes, washed again 2 × 10 minutes in PBS and mounted with 70% glycerol in PBS. All washing steps were performed on a shaker, all incubation steps in a humid chamber. To test for nonspecific binding of the antibodies that detect digoxigenin, which are not endogenous to vertebrate tissue, we performed control experiments in which the labeled RNA was omitted from the hybridization mix. No signal was detected in the absence of the riboprobe, demonstrating that the antibody reacts specifically with the synthetic RNA ( Dataset 2).
Figure S2. Expression of eomesb in the embryonic zebrafish brain.
A. eomesb expression is not found in the telencephalon (T), but present in the midbrain (M), arrowhead, shown is a lateral view. Di Diencephalon, H Hindbrain.
Immunohistochemistry
Immunohistochemistry on cryosections was performed as previously described 51. Briefly, to retrieve the antigens of Prox1, sections were pre-incubated in 50 mM Tris-buffer (pH 8.0) at 99°C for 5 minutes, cooled down to RT over 15 minutes and washed for 5 minutes in PBS and twice for 10 minutes in PBSTx. The sections were then incubated in primary and secondary antibodies in PBSTx. The primary antibody Prox1 (AB5475, 1:2000, Millipore) was incubated overnight at 4°C and secondary antibodies for 1h at room temperature. The slides were washed in PBSTx and mounted. The secondary antibody (dilution 1:750) was Alexa 488-Fluor conjugated (A-11034, Invitrogen, Karlsruhe).
Image acquisition and processing
Confocal images were acquired with Leica TCS-SP5 confocal microscope. Brightfield images were acquired with Zeiss Axio Imager Z1. The images were processed using ImageJ v. 1.4.3.67 and Adobe Photoshop CS2. Composites were assembled using Adobe Photoshop CS2 and Adobe Illustrator CS2.
Nomenclature
We primarily followed the nomenclature proposed in 32 with modifications based on our combinatorial expression pattern analysis, which suggest a subdivision of Dl in a dorsal and ventral subdivision (which we have named Dld and Dlv, respectively). In the figures we employ a two-color code to separate subdivisions that can be made based on the current marker (red dashed line) from those that we make based on other markers (white dashed line). The black dashed line indicates the boundary between D and V. The distinction of ventricular zone versus neuronal layer was based on cellular morphology. The ventricular zone is the region where cells are directly facing the ventricle. The neuronal layer is characterized by cells with a round shape.
Results
Expression of eomesa in the adult zebrafish pallium
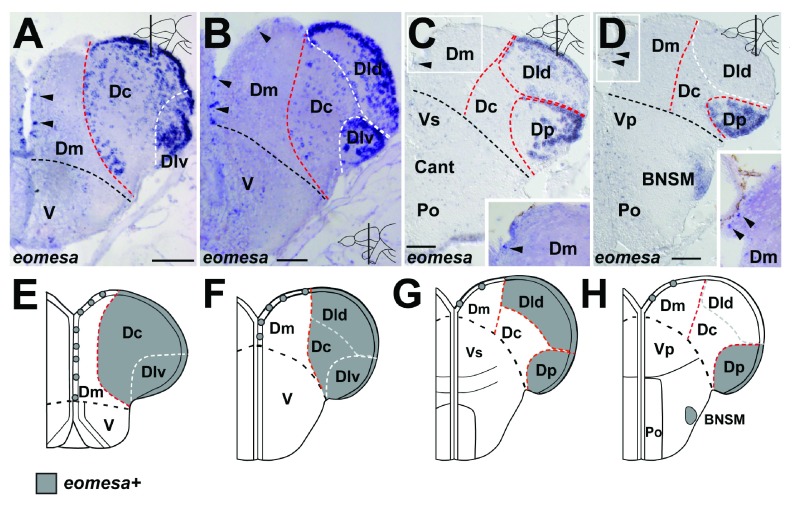
In tetrapod embryos, Eomes ( Tbr2) expression is found in the ventricular zone and mantle layer in all parts of the pallium at embryonic stages 9, 21, 23. In the zebrafish embryo, eomesa expression is present throughout the dorsal telencephalon 52, 53. Its TGD paralog, eomesb is not present in the embryonic or adult telencephalon ( Figure S2, Dataset 1) and thus was not further taken into account. In the adult zebrafish, eomesa positive cells are scattered in a salt-and-pepper pattern along the ventricular zone of Dm along the rostro-caudal axis ( Figure 1A–D, arrowheads). Further, eomesa expression is present in the ventricular zone and neuronal layer of Dc, Dlv and Dld in the rostral telencephalon and at mid-telencephalic levels and in the ventricular zone and neuronal layer of Dld and Dp at the anterior commissure (Cant; Figure 1A–C). Caudal to Cant, eomesa is expressed in the ventricular zone and neuronal layer of Dp, sporadically in Dld and in bed nucleus of the stria medullaris (BNSM, Figure 1D). In summary, eomesa is differentially expressed in the pallium along the rostro-caudal axis ( Figure 1E–H, Dataset 3; Table 1). The most abundant parenchymal eomesa expression is detected in parts of the central and lateral dorsal telencephalic nuclei.
Figure 1. Expression of eomesa in the pallium.
A. In the rostral telencephalic, eomesa expression is found in the ventricular zone (vz) of Dm (arrowheads) and in the vz and neuronal layer (nl) of Dc and Dlv. B. At mid-telencephalic levels, eomesa expression is present in the vz of Dm (arrowheads), in the nl of Dc and in the vz and nl of Dld and Dlv. C. At the anterior commissure (Cant), eomesa expression is present in the vz of Dm (arrowhead) and in the vz and nl of Dld and Dp. Inset shows close-up of vz of Dm D. Caudal to Cant, eomesa expression is present in the vz of Dm (arrowhead), in the vz and nl of Dp and in BNSM. Inset shows close-up of vz of Dm E.– H. Summary of the expression pattern of eomesa at rostral ( E.), mid-telencephalic ( F.), commissural ( G.) and postcommissural levels ( H.). A.– D. Brightfield images of cross-sections at the levels indicated through the telencephalon. Red dashed line indicates subdivisions based on the current marker. White dashed line indicates subdivisions based on other markers. The black dashed line indicates the boundary between D and V. Scale bars = 50µm in A– D.
Table 1. Summary of gene expression pattern in the adult zebrafish pallium.
| ascl1a | eomesa | emx1 | emx2 | emx3 | Prox1 | |
|---|---|---|---|---|---|---|
| Dm | 3 vz a | 3 vz a | 0 | 0 | 3vz nl | 0 |
| Dc | 0 | 3 vz nl/0 b | 0 | 0/3 nl c | 1–2 nl d | 0 |
| Dld | 0 | 3 vz nl e | 0 | 0 | 0 | 0/3 nl f |
| Dlv | 2 vz a | 3 vz nl | 0 | 0 | 2 vz nl g | 0 |
| Dp | 2 vz a | 3 vz nl | 3 vz nl | 0 | 2 vz nl | 0 |
| EN | 0 | 0 | 3 nl | 0 | 0 | 0 |
| BNSM | 0 | 3 nl | 0 | 0 | 0 | 0 |
0 expression not detected, 1 weak expression, 2 moderate expression, 3 strong expression, vz ventricular zone, nl neuronal layer,
a scattered cells in the ventricular zone; b no expression from mid-telencephalic levels; c part of Dc posterior to Cant; d scattered cells in the neuronal layer; e no expression posterior to Cant; f expression at mid-telencephalic level shortly anterior to Cant, shortly caudal to Cant, only scattered Prox1+ cells are present; g expression present in the rostralmost telencephalon in the vz, moving caudally expression in vz and nl.
Expression of emx1, emx2 and emx3 in the adult zebrafish pallium
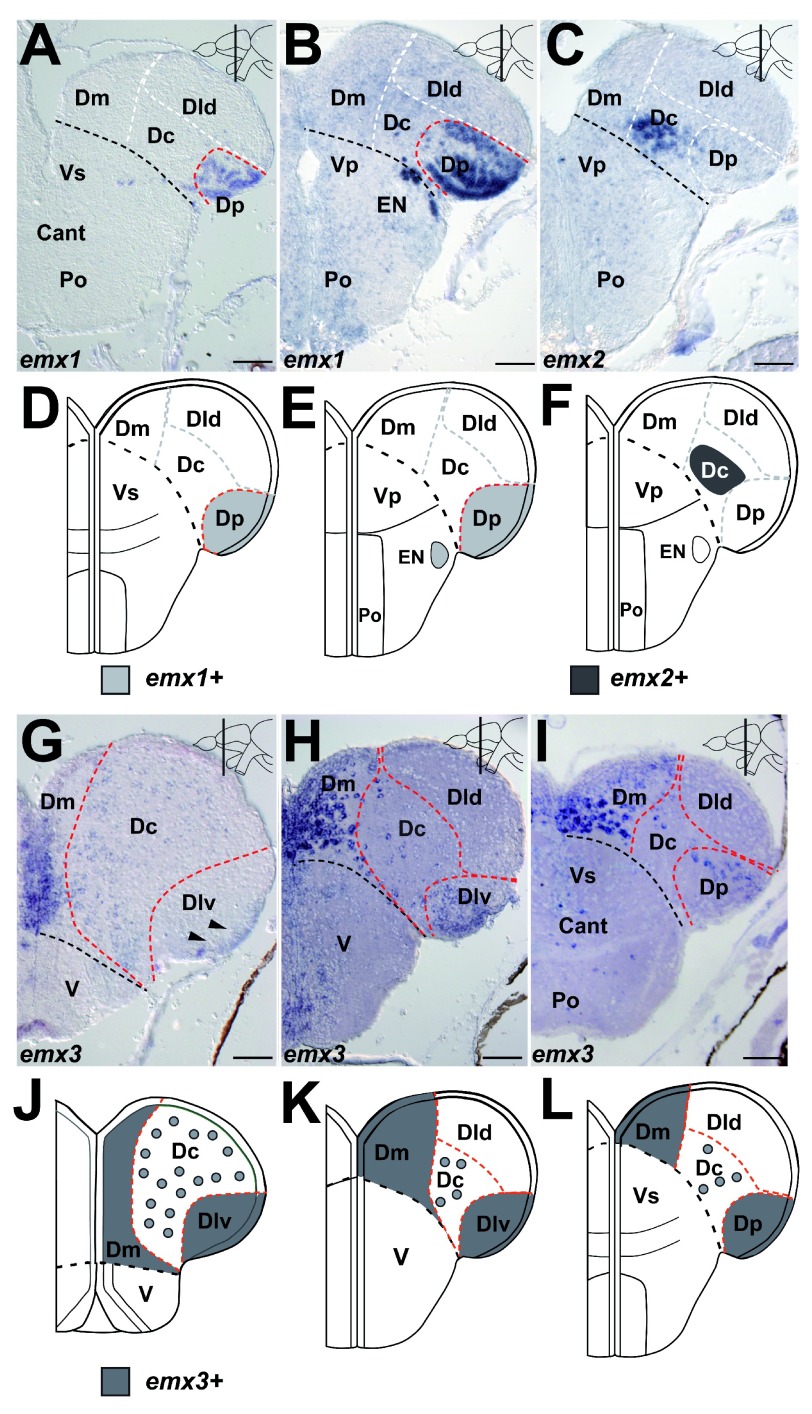
In tetrapod embryos, the ventral pallial subdivision is characterized by absence of Emx1 and presence of Tbr1 9, 10, 12, 13, 21. In embryonic and adult zebrafish, tbr1 is expressed throughout the pallium 7, 53, 54. Thus, we investigated the expression of emx1, emx2 and emx3 to identify a ventral pallial subdivision in the zebrafish pallium. In zebrafish, the three emx genes are expressed at 1 day post fertilization (dpf) in the embryo throughout the dorsal telencephalon 49, 55, 56. In 7 day-old larva, the expression of emx1 and emx2 is restricted to a small caudo-lateral area in the dorsal telencephalon ( Figure S3A,B,D, Dataset 4). The expression of emx3 is found throughout the dorsal telencephalon ( Figure S3C,E, Dataset 4). In the adult zebrafish pallium, emx1 and emx2 are expressed in a very restricted fashion. The expression of emx1 only starts in the mid-telencephalon shortly rostral to Cant, where it is restricted to the ventricular zone and neuronal layer of Dp. This expression pattern continues to Cant ( Figure 2A). Furthermore, emx1 expression is present in Dp and in EN caudal to Cant ( Figure 2B). The expression of emx2 is only found caudal to Cant in an area in Dc lateral to Vp ( Figure 2C). Similar to the larvae, emx3 shows the broadest expression of the emx genes. In the rostral telencephalon, the expression of emx3 is found in the ventricular zone and neuronal layer of Dm and weakly in scattered cells in the neuronal layer of Dc ( Figure 2G). Additionally, the expression of emx3 is weakly present rostrally in the ventricular zone of Dlv ( Figure 2G, arrowheads). At mid-telencephalic levels, emx3 expression is found in the ventricular zone and neuronal layer of Dm, in scattered cells in Dc and in the ventricular zone and neuronal layer of Dlv ( Figure 2H). At Cant, expression of emx3 is found in the ventricular zone and neuronal layer of Dm, in scattered cells in Dc and in the ventricular zone and neuronal layer of Dp ( Figure 2I). Caudal to Cant, the expression gets weaker in Dm, Dc and Dp (data not shown). In summary, emx1 and emx2 show a very restricted expression pattern in the adult zebrafish pallium ( Figure 2D–F, Dataset 5; Table 1). Expression of emx3 is present in Dm, Dc and Dlv/Dp ( Figure 2J–L, Dataset 5; Table 1).
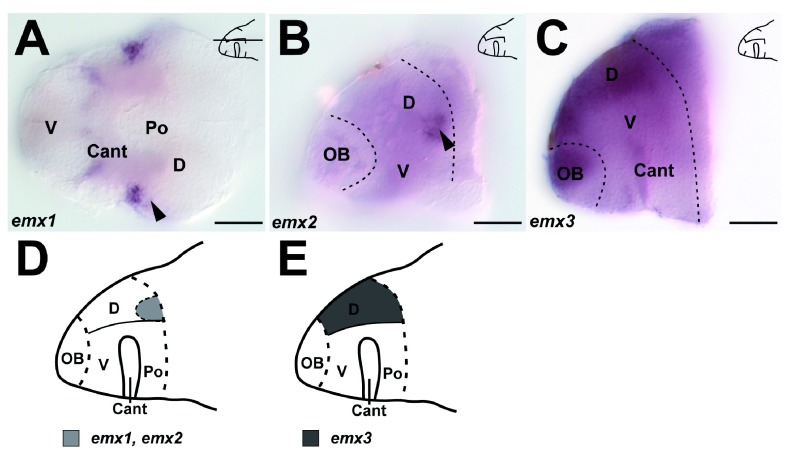
Figure S3. Expression of emx1, emx2 and emx3 in the zebrafish larval brain at 7dpf.
A. emx1 expression is present in the lateral part of the dorsal telencephalon ( D, arrowhead), shown is a ventral view. B. emx2 expression is present in the lateral part of D (arrowhead), shown is a parasaggital view. C. emx3 expression is present throughout D, shown is a parasaggital view. D. Summary of the distribution of emx1 and emx2 expression in D in a sagittal view. E. Summary of the distribution of emx3 expression in D in a sagittal view. A.– C. Brightfield whole-mount images, A. ventral, B.– C. sagittal views. Scale bars = 50µm A– C.
Figure 2. Expression of emx1, emx2 and emx3 in the pallium.
A. At the anterior commissure (Cant), emx1 expression is found in neuronal layer (nl) and ventricular zone (vz) of Dp. B. Caudal to Cant, emx1 expression is present in the vz and nl of Dp and in EN. C. Caudal to Cant, emx2 expression is present in part of Dc. D.– F. Summary of the expression pattern of emx1 and emx2 at commissural ( D.) and postcommissural levels ( E., F.). G. In the rostral telencephalon, emx3 expression is present in the vz and nl of Dm, weakly in scattered cells of the nl od Dc and in the vz of Dlv (arrowheads). H. At mid-telencephalic levels, emx3 expression is found in the vz and nl of Dm, in scattered cells in Dc, and in the vz and nl of Dlv. I. At Cant, emx3 expression is present in the vz and nl of Dm, in scattered cells in Dc and in the vz and nl of Dp. J.– L. Summary of the expression pattern of emx3 at rostral ( J.), mid-telencephalic ( K.) and commissural levels ( L.). A.– F. Brightfield images of cross-sections at the levels indicated through the telencephalon. Red dashed line indicates subdivisions based on the current marker. White dashed line indicates subdivisions based on other markers. The black dashed line indicates the boundary between D and V. Scale bars = 50µm in A– D.
Expression of Prox1 in the adult zebrafish pallium
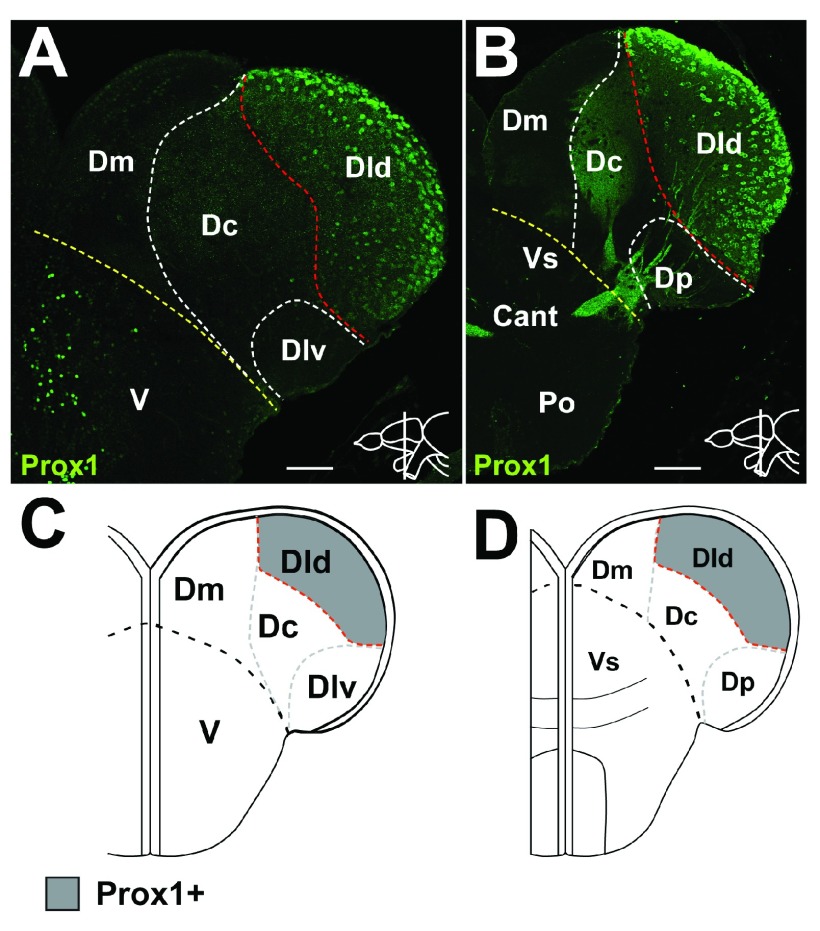
During mouse development, Prox1 expression is found in the amygdala, dentate gyrus and in the neocortex 57. At adult stages, however, strong Prox1 expression is restricted to the dentate gyrus of the hippocampus and is commonly used as a specific marker for granule cells of the hippocampus 57– 60. In the adult zebrafish pallium, Prox1+ cells only are present in the midtelencephalon shortly rostral to Cant in the neuronal layer of Dld ( Figure 3A). This expression pattern continues to the anterior commissure ( Figure 3B). Shortly caudal to Cant, only scattered Prox1+ cells are present in Dld, more caudally no Prox1+ cells can be found (data not shown). In summary, Prox1 staining is present in Dld starting at mid-telencephalic levels until shortly caudal to Cant ( Figure 3A–D, Dataset 6; Table 1).
Figure 3. Expression of Prox1 in the pallium.
A. At mid-telencephalic levels, Prox1 positive cells are found in the neuronal layer (nl) of Dld shortly before the anterior commissure (Cant). B. At Cant, Prox1 positive cells are found in the nl of Dld. C.– D. Summary of expression pattern of Prox1. A.– B. Confocal images of cross-sections at the levels indicated through the telencephalon. Red dashed line indicates subdivisions based on the current marker. White dashed line indicates subdivisions based on other markers. The yellow dashed line indicates the boundary between D and V. Scale bars = 50µm in A– B.
Expression of ascl1a in the adult zebrafish pallium
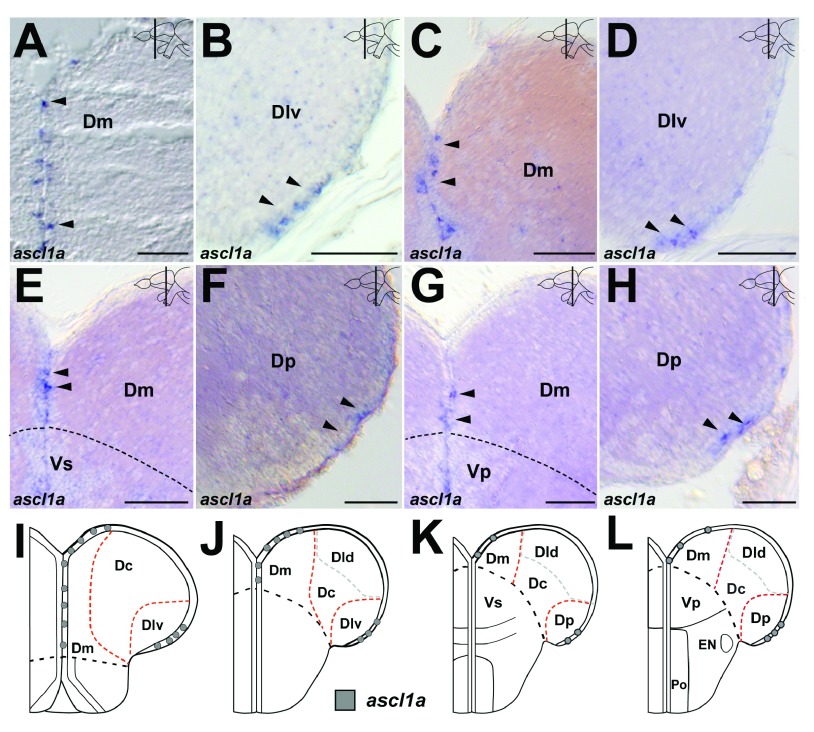
In tetrapod embryos, Ascl1 is expressed a subpopulation of progenitors in the dorsal telencephalon 61, 62. In the zebrafish embryo, ascl1a expression is present in the caudomedial ventricular zone of the dorsal telencephalon 63. In the adult zebrafish pallium, ascl1a is expressed in scattered cells in the ventricular zone of Dm and in scattered cells in the ventricular zone of Dlv ( Figure 4A–D, arrowheads, Figure 4I,J, Dataset 7; Table 1). At Cant and posterior to Cant ascl1a is present in scattered cells in Dm and Dp ( Figure 4E–H, arrowheads, Figure 4K,L, Dataset 7; Table 1).
Figure 4. Expression of ascl1a in the pallium.
A.– B. In the rostral telencephalon, ascl1a expression is present in scattered cells in the ventricular zone (vz) of Dm ( A, arrowheads) and Dlv ( B, arrowheads). C.– D. At mid-telencephalic levels, ascl1a expression is present in scattered cells in the vz of Dm ( C, arrowheads) and Dlv ( D, arrowheads). E.– F. At Cant, ascl1a expression is present in scattered cells in the vz of Dm ( E, arrowheads) and Dp ( F, arrowheads). G.– H. Posterior to Cant, ascl1a expression is present in scattered cells in the vz of Dm ( G, arrowheads) and Dp ( H, arrowheads). I.– L. Summary of the expression pattern of ascl1a rostral ( I.), mid-telencephalic ( J.), commissural ( K.) and postcommissural levels ( L.). A.- H. Brightfield images of cross-sections at the levels indicated through the telencephalon. Red dashed line indicates subdivisions based on the current marker. White dashed line indicates subdivisions based on other markers. The black dashed line indicates the boundary between D and V. Scale bars = 50µm in A– C.
Dataset 1 Expression of eomesb in the embryonic brain and the adult pallium in zebrafish. Raw data of Figure S2 and additional image files of eomesb expression in the embryo and the adult pallium.
Dataset 2 Images of negative control. No signal was detected in the absence of the riboprobe, demonstrating that the antibody reacts specifically with the synthetic RNA.
Dataset 3 Expression of eomesa in the zebrafish pallium. Raw data of Figure 1 and additional image files of eomesa expression in the adult pallium.
Dataset 4 Expression of emx1, emx2 and emx3 in the zebrafish larval brain. Raw data of Figure S3 and additional image files of emx gene expression in the zebrafish larvae.
Dataset 5 Expression of emx1, emx2 and emx3 in the zebrafish pallium. Raw data of Figure 2 and additional image files of emx gene expression in the adult pallium.
Dataset 6 Expression of Prox1 in the zebrafish pallium. Raw data of Figure 3 and additional image files of Prox1 expression in the adult pallium.
Dataset 7 Expression of ascl1a in the zebrafish pallium. Raw data of Figure 4 and additional image files of ascl1a expression in the adult pallium.
Copyright: © 2015 Ganz J et al.
Data associated with the article are available under the terms of the Creative Commons Zero "No rights reserved" data waiver (CC0 1.0 Public domain dedication).
Discussion
Subdivisions of the zebrafish pallium and their homology to other vertebrates
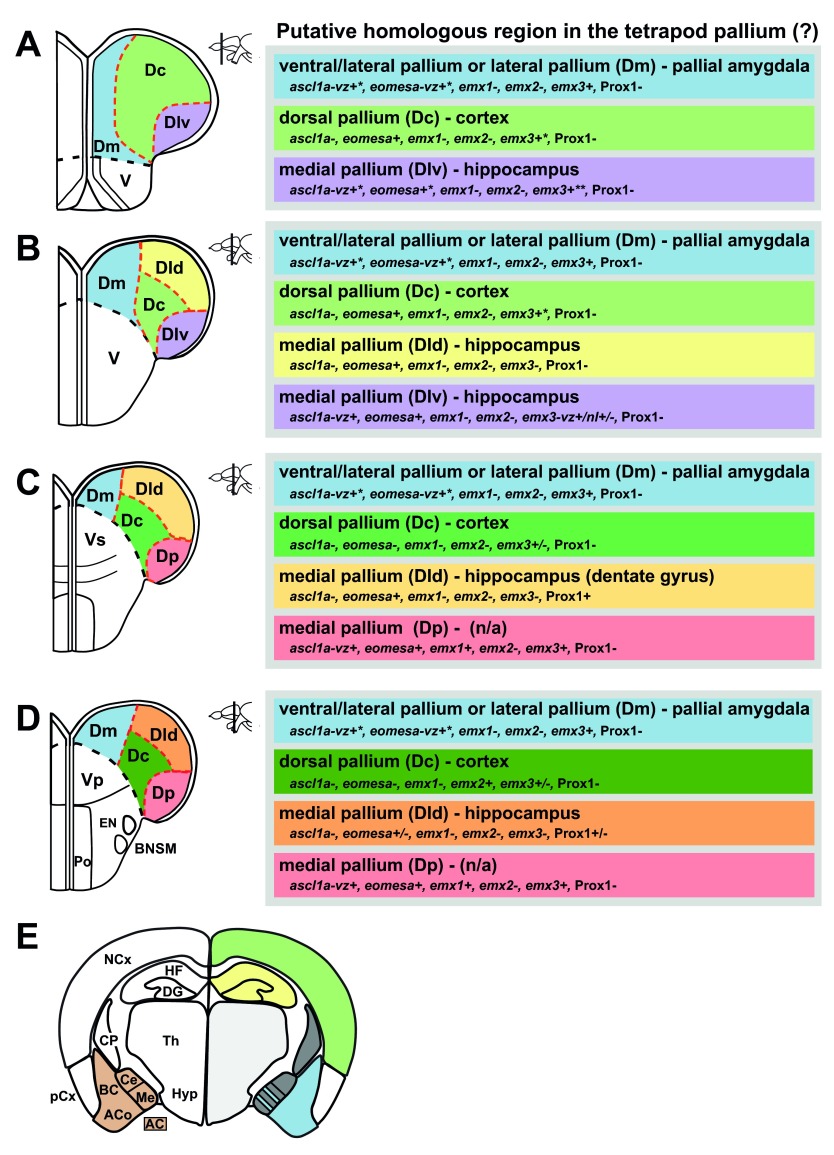
Due to the different development, the pallium of ray-finned fishes has a markedly different morphology compared to all other vertebrates, which makes the comparison between the areas of the pallium of ray-finned fishes to pallial nuclei of other vertebrates particularly challenging. Yet, the correct assignment of homologous pallial areas between teleosts and tetrapods is essential for usage of the teleost fish model in neurobiological research. We have analyzed several conserved molecular marker genes that are found in specific areas of the pallium in the domestic mouse, chicken and the African clawed frog. Based on the expression analysis we identify four main subdivisions of the pallium (Dm, Dl, Dc and Dp) and propose that Dl is subdivided in a dorsal (Dld) and ventral part (Dlv). Based on our data we also suggest putative homologies to pallial nuclei in tetrapods. We suggest that Dm is homologous to the ventral or ventral/lateral pallium, Dc to the dorsal pallium, Dl to the medial pallium, and suggest that Dp comprises a specialized part of Dl ( Figure 5). Additional marker analysis, lineage tracing experiments and functional analyses will be necessary to substantiate the proposed pallial subdivisions and their homology to pallial nuclei in tetrapods. As our study is based solely on comparative gene expression data, we have discussed our results in the framework of gene expression data of other organisms and subsequently compared them to connectional, neurochemical, and functional data in teleosts. For clarity, we discuss our results separately for each pallial subdivision.
Figure 5. Summary of the gene expression patterns in the adult zebrafish pallium.
A.– D. Cross-sections at the levels indicated through the telencephalon. E. Schematic diagram of a cross section through the mouse telencephalon for comparison (modified after 14), note that the amygdaloid complex (AC, brown) is derived both from subpallium (grey) and pallium. Light grey areas (Th/Hyp) are part of the diencephalon. Indicated is also a model of the putative homology of the subdivisions in the adult zebrafish pallium to regions in the tetrapod pallium taking the data presented in this paper into account. Additional marker analysis, lineage tracing experiments and functional analyses are necessary to substantiate the proposed homology to pallial nuclei in tetrapods. The different shades of green (Dc) and yellow (Dld) indicate gene expression changes along the rostro-caudal axis. * in scattered cells, ** in Dlv in the ventricular zone, moving caudally expression in the neuronal layer and ventricular zone, note that Prox1 positive cells are present in Dld shortly before the anterior commissure, n/a = not applicable. The black dashed line indicates the boundary between D and V. The red dashed lines indicate the boundaries between different nuclei in D. AC amygdaloid complex, ACo anterior cortical amygdalar area, BC basal amygdalar complex, BNSM bed nucleus of the stria medullaris, Ce central amygdala, CP Caudateputamen, DG dentate gyrus, EN entopeduncular nucleus, HF hippocampal formation, Hyp hypothalamus, Me medial amygdala, NCx neocortex, pCx piriform cortex, Po preoptic region, Th thalamus, V area ventralis telencephali, Vp postcommissural nucleus of the area ventralis telencephali, Vs supracommisural nucleus of the area ventralis telencephali.
Medial part of the area dorsalis telencephali (Dm)
In the African clawed frog, the chicken, and the domestic mouse, four pallial divisions have been identified in the embryo, the ventral pallium (VP), the lateral pallium (LP), the dorsal pallium (DP) and the medial pallium (MP) 9, 10, 13, 21. In the African clawed frog, chicken, and domestic mouse, the ventral pallial subdivision is characterized in the embryo by the absence of Emx1 expression and presence of the pallial marker Tbr1 9, 10, 13, 21. In embryonic and adult zebrafish, tbr1 is expressed throughout the pallium 7, 53, 54. In adult ray-finned fishes, Dm has been proposed to be homologous to the ventral pallium (pallial amygdala) based on topological, connectional and functional data 29– 31, 64– 66. In contrast, Nieuwenhuys (2009) proposed that Dm is homologous to the lateral pallium based on topology and Yamamoto et al. (2007) suggest that Dm together with Dd and Dld is homologous to the dorsal pallium. Thus, we analyzed the expression of the emx genes in the larval and adult zebrafish to determine if the absence of these markers identifies a ventral pallial subdivision in the zebrafish pallium. In zebrafish, the emx genes show a dynamic expression pattern both in the embryo and the adult. At 1dpf, the three emx genes are expressed at throughout the dorsal telencephalon 49, 55, 56. We found that expression of emx1 is restricted to a caudolateral expression domain in the zebrafish larvae at 7dpf and to Dp in the adult ( Figure 2D,E, Figure S3). Similarly, emx2 also shows a restricted expression pattern to a caudolateral domain both in the zebrafish larvae at 7dpf and in the adult zebrafish pallium ( Figure 2F, Figure S3). The expression of emx3 is present in the entire pallium in the zebrafish embryo and larvae ( 49, Figure S3C). In the adult, emx3 is present in the rostral telencephalon in Dm, Dc and in the ventricular zone of Dlv ( Figure 2J). Moving caudally, emx3 is present in Dm, in scattered cells in Dc and in the ventricular zone and neuronal layer of Dlv and Dp ( Figure 2K,L). As it has been previously suggested that zebrafish emx3 supplies the functions provided by Emx1 and Emx2 in mouse 49, we took the combined expression pattern of the three emx genes into account for our analysis. The lack of an area in Dm that is tbr1 positive and emx gene negative suggests that zebrafish might not have a distinct ventral pallial subdivision and that Dm is either homologous to the lateral pallium, or that Dm comprises a combined ventral and lateral pallium ( Figure 5A–D). Emx1 alone might not be an adequate marker for the ventral pallium in the domestic mouse and chicken, as they have lost the Emx3 gene 49.
Both the ventral pallium and the lateral pallium contribute together with subpallial derivatives to the amygdaloid complex in tetrapods 10, 67, 68. In rodents, the Cannabinoid Receptor (CB1) is expressed in a subset of neurons in the basolateral amygdala 69, 70. In the weakly electric fish Apteronotus leptorhynchus and zebrafish, cb1 is expressed in Dm 71– 73, thus supporting the model that Dm is homologous to the pallial amygdala. Based on gene expression analysis, we have suggested previously that the supracommissural nucleus of the area ventralis telencephali (Vs) is homologous to the dorsal and ventral part of the central amygdala and the bed nucleus of the stria terminalis (BST) and that the postcommissural nucleus of the area ventralis telencephali (Vp) is homologous to the dorsal part of the central amygdala and the BST 7. Thus, it is plausible that Vs and Vp (subpallial amygdala) and Dm (pallial amygdala) form the amygdaloid complex in the adult zebrafish ( Figure 5A–E).
Lateral part of the area dorsalis telencephali (Dl)
Based on topological and gene expression data it has been proposed that the entire Dl 4 or Dl excluding Dlv or Dp is homologous to the medial pallium in other vertebrates 24, 30, 32, 64. In contrast, Wullimann and Mueller (2004) considered only Dlv to be homologous to the medial pallium and Yamamoto et al. (2007) proposed that only the dorsal part of Dl is homologous to the medial pallium. Functional ablation experiments suggested that Dl is equivalent to the hippocampus of tetrapods 65. Our combinatorial expression data suggests that Dl is subdivided in Dld and Dlv rostrally and at Cant and posterior to Cant in Dld and Dp ( Figure 5A–D). Dld is different from Dc based on absence of emx3 positive cells in Dld. During mouse development, Prox1 expression is found in the amygdala, dentate gyrus and in the neocortex 57. At adult stages, however, strong Prox1 expression is restricted to the dentate gyrus of the hippocampus and is commonly used as a specific marker for granule cells of the hippocampus 57– 59. In the adult zebrafish, Prox1 positive cells are exclusively present in the caudal part of Dld. In summary, our data suggests that Dl (excluding Dp, see below) may be homologous to the medial pallium and hippocampus and the caudal part of Dld homologous to the dentate gyrus in the domestic mouse ( Figure 5A–E). We will discuss the possible homology of Dp in the subsequent part.
Posterior part of D (Dp)
It has previously been shown that Dp receives olfactory input and has been on this basis homologized to the lateral pallium in amphibians and all other gnathostomes with evaginated forebrains 4. Being homologous to the lateral pallium, it should be located next to Dm in the embryo, the presumptive ventral pallium. The discrepancy between the position of Dp in the adult and an expected location next to Dm has led to different models to explain the different position of Dp in the adult pallium: “the partial pallial eversion model” by Wullimann and Mueller 31, 32, 52, 74, the “eversion-rearrangement theory” by Northcutt and Braford 33, 64, 75, and the “new eversion model” by Yamamoto and colleagues 37. In the “partial pallial eversion model” 31, 32, 52, 74, based on connectional and gene expression data, it has been proposed that the homolog of the lateral pallium does not participate in the eversion. Wullimann and Mueller put forward that neuroblasts generated by the ventricular zone of the uneverted, medially located lateral pallium homolog migrate laterally to give rise to the submeningeally located nucleus Dp 31, 74. Our gene expression analysis does not support this part of their modified partial eversion model. Based on differential gene expression we can identify Dp, which is not located submeningeally, but has its own ventricular zone even in the adult 4, 64, 76. New neurons are still generated in the zebrafish pallium in the adult 77, 78. Thus, a small area of ventricular zone should be still present next to Dm to generate neurons for Dp. However, we have not observed neuroblasts migrating laterally across the pallium to reach Dp in the adult by BrdU pulse chase studies or genetic lineage tracing 51, 76, 77. The “eversion-rearrangement theory” by Northcutt and Braford suggests that differential expansion of the ventricular surface of some pallial zones and differential proliferation and migration of neuroblasts from the different ventricular zones might result in displacement or shifting of the different pallial subdivisions 33, 64, 75. Similar to the “partial pallial eversion model”, they propose that a small stretch of Dp is still located between Dm and Dl. Our gene expression data does not support this aspect of both models, as we do not identify a small area of ventricular zone and neuronal layer sandwiched between Dm and Dc rostrally and Dm and Dld more caudally that matches the gene expression profile found in Dp.
In the “new eversion model”, the eversion was suggested to occur in a caudolateral direction leading to a shift of the arrangement of the different pallial subdivisions 37. Yamamoto and colleagues propose that ventral pallial and lateral pallial homologs are not present in the rostral but only in the caudal pallium. In their model they suggest that Dp is homologous to the lateral pallium 37. In contrast, we find the putative ventral or ventral/lateral pallial homolog Dm contiguously from rostral to caudal. However, we also identify a separate Dp nucleus only in the posterior part of the pallium.
In contrast to the models discussed above, Nieuwenhuys has put forward that the lateral olfactory tract in vertebrates with everted telencephala is not homologous to the olfactory tract in vertebrates with evaginated telencephala 4. He bases this on data that showed considerable variation in the pattern of secondary olfactory projections among different groups of ray-finned fishes and showed secondary olfactory projections both to Dm and Dp 4, 29, 75. He suggests that the olfactory input has increased overall in Dl, with a subsequent confinement to Dp and has decreased in Dm during ray-fin fish evolution. He considers the lateral olfactory tract as an apomorphy of actinopterygian pallia 4. Even though our gene expression data does not support the models that suggest a migration or displacement of Dp from a position equivalent to the lateral pallium to its caudolateral position, we can identify Dp as a subdivision of Dl separate from Dld. Furthermore, Neuropeptide Y and Parvalbumin immunohistochemistry clearly distinguishes Dl from Dp 79. Furthermore, as discussed above, our gene expression data suggest that Dm might be homologous to the lateral pallium or comprise a combined ventral and lateral pallium, which might suggest that Dp is not homologous to the lateral pallium. Our gene expression data is consistent with Nieuwenhuys’ model that Dp comprises a specialized part of Dl that receives olfactory input, even though the other parts of Dl might be homologous to the medial pallium and hippocampus in tetrapods ( Figure 5A–D).
Central part of the area dorsalis telencephali (Dc)
In the “modified partial pallial eversion model” 32, based on expression of Parvalbumin and nicotine adenine dinucleotide phosphate diphorase (NADPHd) in Dl, it was suggested that Dc is a true histogenetic unit that has its own germinative zone rostrally, but is caudally displaced to a more central location by differential growth of Dm and Dl. Mueller and colleagues suggest that Dc is homologous to the dorsal pallium in other vertebrates and that Dd is absent in zebrafish 32. This model is consistent with connectional and gene expression data in Apteronotus leptorhynchus 24. In addition, it has been noted that Dc shows no significant immunoreactivity to Calretinin, Neuropeptide Y or Thyrosine hydroxylase separating it from the surrounding areas of the pallium 79. Other models based mainly on topology suggest that Dm, Dd and Dld are homologous to the dorsal pallium 37, whereas others have homologized Dd with the dorsal pallium 4, 29, 30, 33, 64 and have suggested that Dc is not a separate unit, but represents the deeper zone of the periventricular areas of Dm, Dd and Dl. Our gene expression data is consistent with the “modified partial pallial eversion model” of Mueller and colleagues with regard to Dc. We have adapted the nomenclature of Mueller et al. (2011) to call the displaced nucleus Dc, even though our data do not rule out the possibility that Dc is rather a displaced Dd nucleus. Accordingly, Dc does not have its own germinative zone caudally, but only in the rostral part of the pallium. However, the morphogenesis of the telencephalon has been analyzed between 1dpf and 5dpf and no such displacement of Dc has been described 5, so if it is the case, then it has to happen after 5dpf. To conclusively evaluate the displacement of Dc, detailed lineage analysis using Cre/loxP technology should be performed from embryonic stages till adulthood.
Gene expression in pallial progenitors
In the adult zebrafish, proliferating cells are found scattered in the ventricular zone of the pallium 76– 78. In the adult zebrafish pallium, ascl1a is found in scattered cells in the ventricular zone of Dm and in Dlv/Dp. In addition, eomesa is present in scattered cells in the ventricular zone of Dm and in the ventricular zone of Dld and Dlv/Dp. Emx1 is also present in the ventricular zone of Dp and emx3 is found in the ventricular zone of Dm, Dlv and Dp ( Figure 5A–D).
In mouse, Ascl1 is expressed in a subpopulation of progenitors in the dorsal telencephalon 61, 62. Eomes (Tbr2) is present in intermediate progenitors in the cortex in the embryo and in the dentate gyrus both in the embryo and the adult and is part of the glutamatergic differentiation cascade that also contains NeuroD and Tbr1 80. In the adult zebrafish, neurod and tbr1 as well as the vesicular glutamate transporters vglut1/2.1/2.2 marking glutamatergic neurons are expressed throughout the pallium, suggesting that the glutamatergic differentiation cascade may still be present in the adult zebrafish telencephalon 7, 72. The expression in scattered cells in the ventricular zone suggests that eomesa and ascl1a may label progenitor subpopulations in the pallium and eomesa could play a role in differentiation of glutamatergic neurons in the adult zebrafish brain. In addition, emx1 and emx3 might be involved in regulating neuronal differentiation processes in the pallium. It will be interesting to perform comprehensive lineage analysis using Cre/loxP technology to follow the progeny of these different markers in the adult zebrafish.
Conclusions
In this study, we present a new model of the subdivisions in the adult zebrafish pallium based on conserved marker gene expression and propose putative homologies to pallial nuclei in tetrapods. Additional marker analysis, lineage tracing experiments and functional analyses are necessary to substantiate the proposed pallial subdivisions and their homology to pallial nuclei in tetrapods. It is important to identify pallial areas in adult zebrafish and their homologies to pallial nuclei in tetrapods to improve our knowledge about the zebrafish brain, in order to implement the zebrafish system as an ideal model for neurobiological research and as a model for human neurodegenerative diseases.
List of abbreviations
- BNSM
bed nucleus of the stria medullaris
- BST
bed nucleus of the stria terminalis
- Cant
anterior commissure
- D
area dorsalis telencephali
- Dc
central part of the area dorsalis telencephali
- Dd
dorsal part of the area dorsalis telencephali
- Dl
lateral part of the area dorsalis telencephali
- Dld
dorsal part of Dl
- Dlv
ventral part of Dl
- Dm
medial part of the area dorsalis telencephali
- Dp
posterior part of the area dorsalis telencephali
- dpf
days post fertilization
- EN
entopeduncular nucleus
- nl
neuronal layer
- OB
olfactory bulb
- Po
preoptic region
- V
area ventralis telencephali
- Vp
postcommissural nucleus of the area ventralis telencephali
- Vs
supracommisural nucleus of the area ventralis telencephali
- vz
ventricular zone
Data availability
The data referenced by this article are under copyright with the following copyright statement: Copyright: © 2015 Ganz J et al.
Data associated with the article are available under the terms of the Creative Commons Zero "No rights reserved" data waiver (CC0 1.0 Public domain dedication). http://creativecommons.org/publicdomain/zero/1.0/
Figshare: Gene expression analysis in the adult zebrafish pallium. Doi: http://dx.doi.org/10.6084/m9.figshare.1266194 81
Acknowledgements
We thank the Didier Stainier lab, the Monte Westerfield lab and the Steve Wilson lab for sharing plasmids. We are thankful to Anja Menge, Sabine Hübner, Julia Ludwig and Paul Morath for technical support. We thank Salvador Martinez and Stefano Suzzi for comments on the manuscript.
Funding Statement
This work was supported by grants to MB from the Deutsche Forschungsgemeinschaft (SFB 655 A3), European Union (ZF-Health) and the Center for Regenerative Therapies Dresden (CRTD).
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
[version 2; referees: 2 approved
Supplementary figures
References
- 1. Northcutt RG: The forebrain of gnathostomes: in search of a morphotype. Brain Behav Evol. 1995;46(4–5):275–318. 10.1159/000113279 [DOI] [PubMed] [Google Scholar]
- 2. Northcutt RG: Evolution of the telencephalon in nonmammals. Annu Rev Neurosci. 1981;4:301–350. 10.1146/annurev.ne.04.030181.001505 [DOI] [PubMed] [Google Scholar]
- 3. Nieuwenhuys R, Meek J: The Telencephalon of Actinopterygian Fishes. Cereb Cortex. 1990;8A:31–73. 10.1007/978-1-4757-9622-3_2 [DOI] [Google Scholar]
- 4. Nieuwenhuys R: The forebrain of actinopterygians revisited. Brain Behav Evol. 2009;73(4):229–252. 10.1159/000225622 [DOI] [PubMed] [Google Scholar]
- 5. Folgueira M, Bayley P, Navratilova P, et al. : Morphogenesis underlying the development of the everted teleost telencephalon. Neural Dev. 2012;7:32. 10.1186/1749-8104-7-32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Butler AB, Hodos W: Comparative vertebrate neuroanatomy: evolution and adaptation. 2nd edn. Hoboken, N.J.: Wiley-Interscience;2005. 10.1002/0471733849 [DOI] [Google Scholar]
- 7. Ganz J, Kaslin J, Freudenreich D, et al. : Subdivisions of the adult zebrafish subpallium by molecular marker analysis. J Comp Neurol. 2012;520(3):633–655. 10.1002/cne.22757 [DOI] [PubMed] [Google Scholar]
- 8. Medina L, Brox A, Legaz I, et al. : Expression patterns of developmental regulatory genes show comparable divisions in the telencephalon of Xenopus and mouse: insights into the evolution of the forebrain. Brain Res Bull. 2005;66(4–6):297–302. 10.1016/j.brainresbull.2005.02.003 [DOI] [PubMed] [Google Scholar]
- 9. Brox A, Puelles L, Ferreiro B, et al. : Expression of the genes Emx1, Tbr1, and Eomes ( Tbr2) in the telencephalon of Xenopus laevis confirms the existence of a ventral pallial division in all tetrapods. J Comp Neurol. 2004;474(4):562–577. 10.1002/cne.20152 [DOI] [PubMed] [Google Scholar]
- 10. Puelles L, Kuwana E, Puelles E, et al. : Pallial and subpallial derivatives in the embryonic chick and mouse telencephalon, traced by the expression of the genes Dlx-2, Emx-1, Nkx-2.1, Pax-6, and Tbr-1. J Comp Neurol. 2000;424(3):409–438. [DOI] [PubMed] [Google Scholar]
- 11. Flames N, Pla R, Gelman DM, et al. : Delineation of multiple subpallial progenitor domains by the combinatorial expression of transcriptional codes. J Neurosci. 2007;27(36):9682–9695. 10.1523/JNEUROSCI.2750-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Puelles L, Kuwana E, Puelles E, et al. : Comparison of the mammalian and avian telencephalon from the perspective of gene expression data. Eur J Morphol. 1999;37(2–3):139–150. 10.1076/ejom.37.2.139.4756 [DOI] [PubMed] [Google Scholar]
- 13. Medina L, Legaz I, Gonzalez G, et al. : Expression of Dbx1, Neurogenin 2, Semaphorin 5A, Cadherin 8, and Emx1 distinguish ventral and lateral pallial histogenetic divisions in the developing mouse claustroamygdaloid complex. J Comp Neurol. 2004;474(4):504–523. 10.1002/cne.20141 [DOI] [PubMed] [Google Scholar]
- 14. Medina L, Abellan A: Development and evolution of the pallium. Semin Cell Dev Biol. 2009;20(6):698–711. 10.1016/j.semcdb.2009.04.008 [DOI] [PubMed] [Google Scholar]
- 15. Abellan A, Vernier B, Retaux S, et al. : Similarities and differences in the forebrain expression of Lhx1 and Lhx5 between chicken and mouse: Insights for understanding telencephalic development and evolution. J Comp Neurol. 2010;518(17):3512–3528. 10.1002/cne.22410 [DOI] [PubMed] [Google Scholar]
- 16. Abellan A, Legaz I, Vernier B, et al. : Olfactory and amygdalar structures of the chicken ventral pallium based on the combinatorial expression patterns of LIM and other developmental regulatory genes. J Comp Neurol. 2009;516(3):166–186. 10.1002/cne.22102 [DOI] [PubMed] [Google Scholar]
- 17. Moreno N, Morona R, Lopez JM, et al. : Subdivisions of the turtle Pseudemys scripta subpallium based on the expression of regulatory genes and neuronal markers. J Comp Neurol. 2010;518(24):4877–4902. 10.1002/cne.22493 [DOI] [PubMed] [Google Scholar]
- 18. Moreno N, Gonzalez A, Retaux S: Evidences for tangential migrations in Xenopus telencephalon: developmental patterns and cell tracking experiments. Dev Neurobiol. 2008;68(4):504–520. 10.1002/dneu.20603 [DOI] [PubMed] [Google Scholar]
- 19. Moreno N, Dominguez L, Retaux S, et al. : Islet1 as a marker of subdivisions and cell types in the developing forebrain of Xenopus. Neuroscience. 2008;154(4):1423–1439. 10.1016/j.neuroscience.2008.04.029 [DOI] [PubMed] [Google Scholar]
- 20. Moreno N, Bachy I, Retaux S, et al. : LIM-homeodomain genes as developmental and adult genetic markers of Xenopus forebrain functional subdivisions. J Comp Neurol. 2004;472(1):52–72. 10.1002/cne.20046 [DOI] [PubMed] [Google Scholar]
- 21. Bachy I, Berthon J, Retaux S: Defining pallial and subpallial divisions in the developing Xenopus forebrain. Mech Dev. 2002;117(1–2):163–172. 10.1016/S0925-4773(02)00199-5 [DOI] [PubMed] [Google Scholar]
- 22. Gonzalez A, Morona R, Moreno N, et al. : Identification of striatal and pallidal regions in the subpallium of anamniotes. Brain Behav Evol. 2014;83(2):93–103. [DOI] [PubMed] [Google Scholar]
- 23. Bulfone A, Martinez S, Marigo V, et al. : Expression pattern of the Tbr2 (Eomesodermin) gene during mouse and chick brain development. Mech Dev. 1999;84(1–2):133–138. 10.1016/S0925-4773(99)00053-2 [DOI] [PubMed] [Google Scholar]
- 24. Harvey-Girard E, Giassi AC, Ellis W, et al. : Organization of the gymnotiform fish pallium in relation to learning and memory: IV. Expression of conserved transcription factors and implications for the evolution of dorsal telencephalon. J Comp Neurol. 2012;520(15):3395–3413. 10.1002/cne.23107 [DOI] [PubMed] [Google Scholar]
- 25. Giassi AC, Harvey-Girard E, Valsamis B, et al. : Organization of the gymnotiform fish pallium in relation to learning and memory: I. Cytoarchitectonics and cellular morphology. J Comp Neurol. 2012;520(12):3314–3337. 10.1002/cne.23097 [DOI] [PubMed] [Google Scholar]
- 26. Giassi AC, Ellis W, Maler L: Organization of the gymnotiform fish pallium in relation to learning and memory: III. Intrinsic connections. J Comp Neurol. 2012;520(15):3369–3394. 10.1002/cne.23108 [DOI] [PubMed] [Google Scholar]
- 27. Giassi AC, Duarte TT, Ellis W, et al. : Organization of the gymnotiform fish pallium in relation to learning and memory: II. Extrinsic connections. J Comp Neurol. 2012;520(15):3338–3368. 10.1002/cne.23109 [DOI] [PubMed] [Google Scholar]
- 28. Burmeister SS, Munshi RG, Fernald RD: Cytoarchitecture of a cichlid fish telencephalon. Brain Behav Evol. 2009;74(2):110–120. 10.1159/000235613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Northcutt RG: Connections of the lateral and medial divisions of the goldfish telencephalic pallium. J Comp Neurol. 2006;494(6):903–943. 10.1002/cne.20853 [DOI] [PubMed] [Google Scholar]
- 30. Northcutt RG: Forebrain evolution in bony fishes. Brain Res Bull. 2008;75(2–4):191–205. 10.1016/j.brainresbull.2007.10.058 [DOI] [PubMed] [Google Scholar]
- 31. Wullimann MF, Mueller T: Teleostean and mammalian forebrains contrasted: Evidence from genes to behavior. J Comp Neurol. 2004;475(2):143–162. 10.1002/cne.20183 [DOI] [PubMed] [Google Scholar]
- 32. Mueller T, Dong Z, Berberoglu MA, et al. : The dorsal pallium in zebrafish, Danio rerio (Cyprinidae, Teleostei). Brain Res. 2011;1381:95–105. 10.1016/j.brainres.2010.12.089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Braford MR, Jr: Comparative aspects of forebrain organization in the ray-finned fishes: touchstones or not? Brain Behav Evol. 1995;46(4–5):259–274. 10.1159/000113278 [DOI] [PubMed] [Google Scholar]
- 34. Kaslin J, Panula P: Comparative anatomy of the histaminergic and other aminergic systems in zebrafish ( Danio rerio). J Comp Neurol. 2001;440(4):342–377. 10.1002/cne.1390 [DOI] [PubMed] [Google Scholar]
- 35. Kaslin J: The aminergic and cholinergic neurotransmitter systems in the zebrafish brain. Abo Akademi University, Department of Biology;2004. Reference Source [Google Scholar]
- 36. Folgueira M, Anadon R, Yanez J: Experimental study of the connections of the telencephalon in the rainbow trout ( Oncorhynchus mykiss). II: Dorsal area and preoptic region. J Comp Neurol. 2004;480(2):204–233. 10.1002/cne.20341 [DOI] [PubMed] [Google Scholar]
- 37. Yamamoto N, Ishikawa Y, Yoshimoto M, et al. : A new interpretation on the homology of the teleostean telencephalon based on hodology and a new eversion model. Brain Behav Evol. 2007;69(2):96–104. 10.1159/000095198 [DOI] [PubMed] [Google Scholar]
- 38. O’Connell LA, Hofmann HA: The vertebrate mesolimbic reward system and social behavior network: a comparative synthesis. J Comp Neurol. 2011;519(18):3599–3639. 10.1002/cne.22735 [DOI] [PubMed] [Google Scholar]
- 39. Broglio C, Rodriguez F, Gomez A, et al. : Selective involvement of the goldfish lateral pallium in spatial memory. Behav Brain Res. 2010;210(2):191–201. 10.1016/j.bbr.2010.02.031 [DOI] [PubMed] [Google Scholar]
- 40. Broglio C, Gomez A, Duran E, et al. : Hallmarks of a common forebrain vertebrate plan: specialized pallial areas for spatial, temporal and emotional memory in actinopterygian fish. Brain Res Bull. 2005;66(4–6):277–281. 10.1016/j.brainresbull.2005.03.021 [DOI] [PubMed] [Google Scholar]
- 41. Rodriguez F, Lopez JC, Vargas JP, et al. : Conservation of spatial memory function in the pallial forebrain of reptiles and ray-finned fishes. J Neurosci. 2002;22(7):2894–2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rodriguez F, Lopez JC, Vargas JP, et al. : Spatial memory and hippocampal pallium through vertebrate evolution: insights from reptiles and teleost fish. Brain Res Bull. 2002;57(3–4):499–503. 10.1016/S0361-9230(01)00682-7 [DOI] [PubMed] [Google Scholar]
- 43. Duran E, Ocana FM, Broglio C, et al. : Lateral but not medial telencephalic pallium ablation impairs the use of goldfish spatial allocentric strategies in a “hole-board” task. Behav Brain Res. 2010;214(2):480–487. 10.1016/j.bbr.2010.06.010 [DOI] [PubMed] [Google Scholar]
- 44. Brand M, Granato M, Nuesslein-Volhard C: Keeping and raising zebrafish. In Zebrafish: A Practical Approach Edited by Nuesslein-Volhard C, Dahm, R. Oxford: Oxford University Press;1999. Reference Source [Google Scholar]
- 45. Westerfield M: The zebrafish book. A guide for the laboratory use of zebrafish ( Danio rerio). 4 edn. Eugene: University of Oregon Press;1995. Reference Source [Google Scholar]
- 46. Streisinger G, Walker C, Dower N, et al. : Production of clones of homozygous diploid zebra fish ( Brachydanio rerio). Nature. 1981;291(5813):293–296. 10.1038/291293a0 [DOI] [PubMed] [Google Scholar]
- 47. Braasch I, Postlethwait J: Polyploidy in Fish and the Teleost Genome Duplication. In Polyploidy and Genome Evolution Edited by Soltis PS, Soltis DE: Springer Berlin Heidelberg;2012:341–383. 10.1007/978-3-642-31442-1_17 [DOI] [Google Scholar]
- 48. Catchen JM, Conery JS, Postlethwait JH: Automated identification of conserved synteny after whole-genome duplication. Genome Res. 2009;19(8):1497–1505. 10.1101/gr.090480.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Viktorin G, Chiuchitu C, Rissler M, et al. : Emx3 is required for the differentiation of dorsal telencephalic neurons. Dev Dyn. 2009;238(8):1984–1998. 10.1002/dvdy.22031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Reifers F, Bohli H, Walsh EC, et al. : Fgf8 is mutated in zebrafish acerebellar (ace) mutants and is required for maintenance of midbrain-hindbrain boundary development and somitogenesis. Development. 1998;125(13):2381–2395. [DOI] [PubMed] [Google Scholar]
- 51. Kroehne V, Freudenreich D, Hans S, et al. : Regeneration of the adult zebrafish brain from neurogenic radial glia-type progenitors. Development. 2011;138(22):4831–4841. 10.1242/dev.072587 [DOI] [PubMed] [Google Scholar]
- 52. Mueller T, Wullimann MF, Guo S: Early teleostean basal ganglia development visualized by zebrafish Dlx2a, Lhx6, Lhx7, Tbr2 (eomesa), and GAD67 gene expression. J Comp Neurol. 2008;507(2):1245–1257. 10.1002/cne.21604 [DOI] [PubMed] [Google Scholar]
- 53. Mione M, Shanmugalingam S, Kimelman D, et al. : Overlapping expression of zebrafish T-brain-1 and eomesodermin during forebrain development. Mech Dev. 2001;100(1):93–97. 10.1016/S0925-4773(00)00501-3 [DOI] [PubMed] [Google Scholar]
- 54. Costagli A, Kapsimali M, Wilson SW, et al. : Conserved and divergent patterns of Reelin expression in the zebrafish central nervous system. J Comp Neurol. 2002;450(1):73–93. 10.1002/cne.10292 [DOI] [PubMed] [Google Scholar]
- 55. Kawahara A, Dawid IB: Developmental expression of zebrafish emx1 during early embryogenesis. Gene Expr Patterns. 2002;2(3–4):201–206. 10.1016/S1567-133X(02)00062-5 [DOI] [PubMed] [Google Scholar]
- 56. Morita T, Nitta H, Kiyama Y, et al. : Differential expression of two zebrafish emx homeoprotein mRNAs in the developing brain. Neurosci Lett. 1995;198(2):131–134. 10.1016/0304-3940(95)11988-9 [DOI] [PubMed] [Google Scholar]
- 57. Lavado A, Oliver G: Prox1 expression patterns in the developing and adult murine brain. Dev Dyn. 2007;236(2):518–524. 10.1002/dvdy.21024 [DOI] [PubMed] [Google Scholar]
- 58. Lavado A, Lagutin OV, Chow LM, et al. : Prox1 is required for granule cell maturation and intermediate progenitor maintenance during brain neurogenesis. PLoS Biol. 2010;8(8):pii: e1000460. 10.1371/journal.pbio.1000460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Galeeva A, Treuter E, Tomarev S, et al. : A prospero-related homeobox gene Prox-1 is expressed during postnatal brain development as well as in the adult rodent brain. Neuroscience. 2007;146(2):604–616. 10.1016/j.neuroscience.2007.02.002 [DOI] [PubMed] [Google Scholar]
- 60. Iwano T, Masuda A, Kiyonari H, et al. : Prox1 postmitotically defines dentate gyrus cells by specifying granule cell identity over CA3 pyramidal cell fate in the hippocampus. Development. 2012;139(16):3051–3062. 10.1242/dev.080002 [DOI] [PubMed] [Google Scholar]
- 61. Britz O, Mattar P, Nguyen L, et al. : A role for proneural genes in the maturation of cortical progenitor cells. Cereb Cortex. 2006;16(Suppl 1):i138–151. 10.1093/cercor/bhj168 [DOI] [PubMed] [Google Scholar]
- 62. Fode C, Ma Q, Casarosa S, et al. : A role for neural determination genes in specifying the dorsoventral identity of telencephalic neurons. Genes Dev. 2000;14(1):67–80. [PMC free article] [PubMed] [Google Scholar]
- 63. Wullimann MF, Mueller T: Expression of Zash-1a in the postembryonic zebrafish brain allows comparison to mouse Mash1 domains. Brain Res Gene Expr Patterns. 2002;1(3–4):187–192. 10.1016/S1567-133X(02)00016-9 [DOI] [PubMed] [Google Scholar]
- 64. Braford MR, Jr: Stalking the everted telencephalon: comparisons of forebrain organization in basal ray-finned fishes and teleosts. Brain Behav Evol. 2009;74(1):56–76. 10.1159/000229013 [DOI] [PubMed] [Google Scholar]
- 65. Portavella M, Vargas JP, Torres B, et al. : The effects of telencephalic pallial lesions on spatial, temporal, and emotional learning in goldfish. Brain Res Bull. 2002;57(3–4):397–399. 10.1016/S0361-9230(01)00699-2 [DOI] [PubMed] [Google Scholar]
- 66. Portavella M, Torres B, Salas C, et al. : Lesions of the medial pallium, but not of the lateral pallium, disrupt spaced-trial avoidance learning in goldfish ( Carassius auratus). Neurosci Lett. 2004;362(2):75–78. 10.1016/j.neulet.2004.01.083 [DOI] [PubMed] [Google Scholar]
- 67. Moreno N, Gonzalez A: Evolution of the amygdaloid complex in vertebrates, with special reference to the anamnio-amniotic transition. J Anat. 2007;211(2):151–163. 10.1111/j.1469-7580.2007.00780.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Maximino C, Lima MG, Oliveira KR, et al. : “Limbic associative” and “autonomic” amygdala in teleosts: a review of the evidence. J Chem Neuroanat. 2013;48–49:1–13. 10.1016/j.jchemneu.2012.10.001 [DOI] [PubMed] [Google Scholar]
- 69. Mailleux P, Vanderhaeghen JJ: Distribution of neuronal cannabinoid receptor in the adult rat brain: a comparative receptor binding radioautography and in situ hybridization histochemistry. Neuroscience. 1992;48(3):655–668. 10.1016/0306-4522(92)90409-U [DOI] [PubMed] [Google Scholar]
- 70. Matsuda LA, Bonner TI, Lolait SJ: Localization of cannabinoid receptor mRNA in rat brain. J Comp Neurol. 1993;327(4):535–550. 10.1002/cne.903270406 [DOI] [PubMed] [Google Scholar]
- 71. Lam CS, Rastegar S, Strahle U: Distribution of cannabinoid receptor 1 in the CNS of zebrafish. Neuroscience. 2006;138(1):83–95. 10.1016/j.neuroscience.2005.10.069 [DOI] [PubMed] [Google Scholar]
- 72. Aoki T, Kinoshita M, Aoki R, et al. : Imaging of neural ensemble for the retrieval of a learned behavioral program. Neuron. 2013;78(5):881–894. 10.1016/j.neuron.2013.04.009 [DOI] [PubMed] [Google Scholar]
- 73. Harvey-Girard E, Giassi AC, Ellis W, et al. : Expression of the cannabinoid CB1 receptor in the gymnotiform fish brain and its implications for the organization of the teleost pallium. J Comp Neurol. 2013;521(4):949–75. 10.1002/cne.23212 [DOI] [PubMed] [Google Scholar]
- 74. Mueller T, Wullimann MF: An evolutionary interpretation of teleostean forebrain anatomy. Brain Behav Evol. 2009;74(1):30–42. 10.1159/000229011 [DOI] [PubMed] [Google Scholar]
- 75. Northcutt R, Braford MR, Jr: New observations on the organization and evolution of the telencephalon of the actinopterygian fishes. In Comparative Neurology of the Telencephalon Edited by SOE E. New York: Plenum Press;1980:41–98. 10.1007/978-1-4613-2988-6_3 [DOI] [Google Scholar]
- 76. Ganz J, Kaslin J, Hochmann S, et al. : Heterogeneity and Fgf dependence of adult neural progenitors in the zebrafish telencephalon. Glia. 2010;58(11):1345–1363. 10.1002/glia.21012 [DOI] [PubMed] [Google Scholar]
- 77. Grandel H, Kaslin J, Ganz J, et al. : Neural stem cells and neurogenesis in the adult zebrafish brain: origin, proliferation dynamics, migration and cell fate. Dev Biol. 2006;295(1):263–277. 10.1016/j.ydbio.2006.03.040 [DOI] [PubMed] [Google Scholar]
- 78. Adolf B, Chapouton P, Lam CS, et al. : Conserved and acquired features of adult neurogenesis in the zebrafish telencephalon. Dev Biol. 2006;295(1):278–293. 10.1016/j.ydbio.2006.03.023 [DOI] [PubMed] [Google Scholar]
- 79. Castro A, Becerra M, Manso MJ, et al. : Calretinin immunoreactivity in the brain of the zebrafish, Danio rerio: distribution and comparison with some neuropeptides and neurotransmitter-synthesizing enzymes. II. Midbrain, hindbrain, and rostral spinal cord. J Comp Neurol. 2006;494(5):792–814. 10.1002/cne.20843 [DOI] [PubMed] [Google Scholar]
- 80. Hevner RF, Hodge RD, Daza RA, et al. : Transcription factors in glutamatergic neurogenesis: conserved programs in neocortex, cerebellum, and adult hippocampus. Neurosci Res. 2006;55(3):223–233. 10.1016/j.neures.2006.03.004 [DOI] [PubMed] [Google Scholar]
- 81. Ganz J, Kroehne V, Freundenreich D, et al. : Gene expression analysis in the adult zebrafish pallium. Figshare. 2014. Data Source