Abstract
Background
Previous studies suggested genetic variations in PSCA (prostate stem cell antigen) may confer the susceptibility of cancer. Many case–control studies have reported the relationship between PSCA rs2294008 C > T polymorphism and cancer, especially gastric cancer and bladder cancer. However, the results are inconsistent. This meta-analysis is aimed at evaluating the association of rs2294008 polymorphism with cancer risk.
Methods
The databases of PubMed, ISI Web of Knowledge, EMBASE, and Chinese National Knowledge Infrastructure (CNKI) were searched for related publications. Odds ratios (ORs) and 95% confidence intervals (CIs) were used to estimate the strength of the associations. Fixed models were used when heterogeneity among studies was not detected, otherwise the random model was used.
Results
Twenty-six studies from 24 articles with 30,050 multiple cancer cases and 51,670 controls were pooled into this meta-analysis. The results showed that the rs2294008 polymorphism was associated with increased cancer risk in any genetic model (T vs C, OR: 1.18, 95% CI: 1.08–1.28; TT vs CC, OR: 1.36, 95% CI: 1.14–1.62; TC vs CC, OR: 1.29, 95% CI: 1.17–1.44; TT + TC vs CC, OR: 1.32, 95% CI: 1.18–1.49; TT vs TC + CC, OR: 1.15, 95% CI: 1.02–1.30). In stratified analysis by cancer type, we found that the T allele had a significant high risk of gastric and bladder cancer, but not in other cancers. In subgroup analysis by ethnicity, increased cancer risk was found in both Asians and Caucasians.
Conclusion
Our study suggested that the PSCA rs2294008 C > T polymorphism is a risk factor for cancer, especially in gastric and bladder cancer.
Keywords: risk, meta-analysis, prostate stem cell antigen, single nucleotide polymorphisms, SNPs
Introduction
Cancer has become one of the most serious diseases threatening human health. According data from GLOBOCAN 2008, about 12.7 million new cancer cases and 7.6 million cancer deaths have occurred.1 Unfortunately, the mechanisms of cancer still need intensive study. Interactions between genetic and environmental factors were proved to play a dominant role in the occurrence and progression of cancer.2,3 It seems that discovering the effects of genes and the environment on humans would result in a big step being taken toward healing cancer.
Prostate stem cell antigen (PSCA) is a prostate-specific gene, which was initially discovered by Reiter et al by representational difference analysis.4 PSCA encodes a 123 amino acid cell surface protein with 30% homology to stem cell antigen 2 (SCA-2) that belongs to the Thy-1/Ly-6 family and is located on chromosome 8q24.2. PSCA is not only overexpressed in prostate cancer, but was also demonstrated to be highly expressed in other malignancies such as bladder, renal, pancreatic, and ovarian cancers.4–8 The most extensively studied single nucleotide polymorphism (SNP) in PSCA is rs2294008 C > T. Although in vitro experiments have revealed that the variant rs2294008T could reduce the transcriptional activity of an upstream fragment of PSCA, its mechanism and physiological function are still unknown.5,9 Previously, genome-wide association studies (GWAS) have found that the rs2294008 polymorphism in PSCA is significantly associated with gastric and bladder cancers. Although several studies have described the relationship between the rs2294008 polymorphism and other cancers, the sample sizes of these studies were small. More recently, five meta-analyses have assessed the relationship between the polymorphism of PSCA rs2294008 C > T and the susceptibility to gastric cancer.10–14 However, the results of these meta-analyses were not entirely consistent. Especially the results of subgroup analysis by ethnicity are controversial. Furthermore, there is a lack of evidence of a relationship between the rs2294008 polymorphism and cancer overall. Additionally, the genetic variant of rs2294008 may be correlated with cancer risk in different cancer type and/or ethnicity. Herein, we carried out a meta-analysis to derive a more precise evaluation on the relationship between the rs2294008 polymorphism and cancer risk.
Materials and methods
Publication search strategy
We searched the databases of PubMed, ISI Web of Knowledge, EMBASE, and Chinese National Knowledge Infrastructure (CNKI) for related articles with the keywords: “PSCA/Prostate stem cell antigen”, “rs2294008”, “single nucleotide polymorphism/SNP/variation/genotype”, and “cancer/carcinoma/tumor/neoplasm”. The last search was updated on September 30, 2014. All published papers with available full text matching the eligible criteria were retrieved. Furthermore, the references of relevant reviews and eligible articles that our search retrieved were checked.
Inclusion and exclusion criteria
The following criteria were used to select the eligible literature for this meta-analysis: 1) original papers investigating the associations between PSCA (rs2294008) polymorphisms and cancer risk; 2) case–control studies; and 3) full-text published articles and included detailed genotyping data.
Accordingly, the exclusion criteria were: 1) no control group; 2) no available or detailed genotype frequency; and 3) reviews and duplicated studies.
Data extraction and synthesis
The literature were carefully extracted from all eligible studies independently by two reviewers according to the inclusion and exclusion criteria mentioned above. If these two reviewers could not reach an agreement, another reviewer was consulted to resolve the controversy. The following information were extracted from all included studies: first author, published year, country of origin, cancer type, ethnicity, genotyping methods, source of control, sample size of cases and controls, and the distribution of each genotype in case and control groups. The subgroups grouped according to cancer types included gastric cancer, bladder cancer, and others. With regard to ethnicity, the studies included Caucasian, Asian, and mixed ethnicity (ie, the ethnicity was unclear).
Statistical analysis
The associations between the PSCA (rs2294008) polymorphism and cancer risk were measured by the odds ratio (OR) with 95% confidence intervals (CIs) according to allele contrast (T vs C), homozygote (TT vs CC), heterozygote (TC vs CC), recessive (TT vs CC + TC), and dominant (CC vs TC + TT) models. The significance of the overall OR was determined by the Z test. Heterogeneity among studies was evaluated with the Q and I2 statistic tests. If the P-value of the heterogeneity test was greater than 0.05 (P>0.05), the pooled OR estimate of each study was calculated by the fixed effects model. Otherwise, the random effects model was used.15 Furthermore, sources of heterogeneity were investigated by stratified analysis based on cancer type and ethnicity as mentioned above. A funnel plot was used to evaluate publication bias. All statistical analyses were carried out with Review Manager (Revman; v5.2; The Cochrane Collaboration, Oxford, UK).
Results
Characteristics of studies
As shown in Figure 1, a total of 38 potential articles were extracted after the initial search. Fourteen articles were excluded after further screening. Six studies were excluded because they were systematic review articles. One article was excluded because it was not a case–control study. Seven articles were excluded because of a lack of detailed genotyping data. Finally, 26 studies from 24 publications9,16–38 including 30,050 multiple cancer cases and 51,670 controls were included in this meta-analysis.
Figure 1.
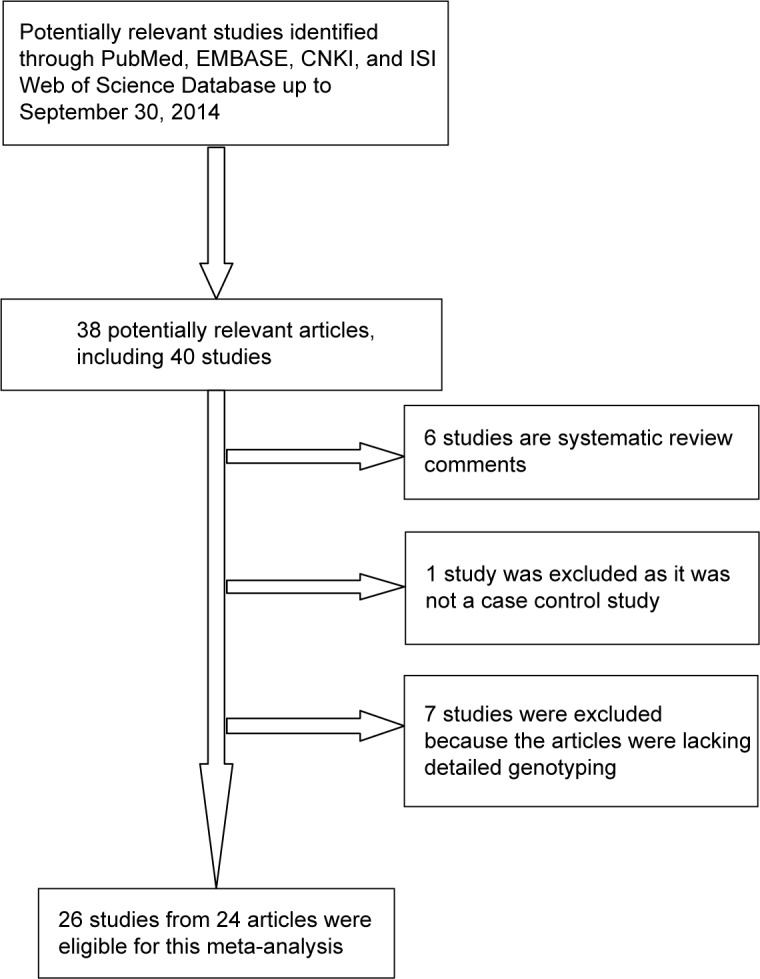
Flow chart of study selection.
Abbreviation: CNKI, Chinese National Knowledge Infrastructure.
The characteristics of the included studies are summarized in Table 1. Among the 26 studies, there were 17 studies of gastric cancer, five studies of bladder cancer, two studies of esophageal cancer, two studies of gallbladder cancer, and one study of colorectal cancer. As for subjects, 19 studies had an Asian background, seven studies had a Caucasian background, and only one had a ‘mixed’ background. Furthermore, there were eleven hospital-based studies and 15 population-based studies.
Table 1.
Characteristics of studies included in the meta-analysis
| Study | Year | Country | Ethnicity | Study design | Genotyping method | Source of control | Cancer type | Sample size (case/control) |
|---|---|---|---|---|---|---|---|---|
| Dai et al16 | 2014 | People’s Republic of China | Asian | CC | TaqMan | Population | EC | 2,083/2,220 |
| Wang et al17 | 2014 | People’s Republic of China | Asian | CC | TaqMan | Population | BC | 1,210/1,008 |
| Ma et al18 | 2013 | People’s Republic of China | Asian | CC | iPLEX | Population | BC | 184/962 |
| Zhao et al19 | 2013 | People’s Republic of China | Asian | CC | DHPLC | Population | GC | 717/951 |
| Rizzato et al20 | 2013 | Venezuela | Mixed | CC | TaqMan | Hospital | GC | 180/1,061 |
| Rai et al21 | 2013 | India | Asian | CC | TaqMan | Population | GBC | 405/247 |
| Ono et al22 | 2013 | Japan | Asian | CC | TaqMan | Hospital | GBC | 44/173 |
| Fu et al23 | 2012 | Europe | Caucasian | CC | TaqMan | Population | BC | 5,393/7,324 |
| Li et al24 | 2012 | People’s Republic of China | Asian | CC | iPLEX | Hospital | GC | 300/300 |
| Smith et al25 | 2012 | Scotland | Caucasian | CC | TaqMan | Hospital | CRC | 77/804 |
| Sala et al26 | 2012 | Europe | Caucasian | CC | SNP array | Population | GC | 411/1,530 |
| Zhao et al27 | 2012 | People’s Republic of China | Asian | CC | DHPLC | Population | GC | 185/200 |
| Tanikawa et al28 | 2012 | Japan | Asian | CC | SNP array | Hospital | GC | 2,300/16,567 |
| Song et al29 | 2011 | Korea | Asian | CC | PCR-RFLP | Hospital | GC | 3,245/1,700 |
| Zeng et al30 | 2011 | People’s Republic of China | Asian | CC | PCR-RFLP | Hospital | GC | 460/549 |
| Lochhead et al31a | 2011 | Poland | Caucasian | CC | TaqMan | Population | GC | 312/383 |
| Lochhead et al31b | 2011 | USA | Caucasian | CC | TaqMan | Population | Upper GIC | 468*/211 |
| Wang et al32 | 2010 | People’s Republic of China | Asian | CC | PCR-RFLP | Hospital | BC | 581/580 |
| Ou et al33 | 2010 | People’s Republic of China | Asian | CC | PCR/LDR | Hospital | GC | 196/246 |
| Lu et al34 | 2010 | People’s Republic of China | Asian | CC | PCR-RFLP | Population | GC | 1,053/1,100 |
| Chen et al35 | 2010 | People’s Republic of China | Asian | CC | PCR-RFLP | Hospital | GC | 460/549 |
| Wu et al36 | 2009 | Europe | Caucasian | CC | TaqMan | Population | BC | 5,038/9,363 |
| Matsuo et al37 | 2009 | Japan | Asian | CC | TaqMan | Hospital | GC | 708/708 |
| Wu et al38 | 2009 | People’s Republic of China | Asian | CC | PCR-RFLP | Population | GC | 1,736/1,020 |
| Sakamoto et al9a | 2008 | Japan | Asian | CC | TaqMan | Population | GC | 1,531/1,399 |
| Sakamoto et al9b | 2008 | Korea | Asian | CC | TaqMan | Population | GC | 871/390 |
Notes:
Including 309 gastric cancer cases and 159 esophageal cancer cases. Lochhead et al31a refers the study evaluated association of the rs2294008 polymorphism with gastric cancer risk and Lochhead et al31b refers to the study evaluated association of the rs2294008 polymorphism with upper GI cancer risk. Sakamoto et al9a refers to the study evaluated association of the rs2294008 with gastric cancer types in Japan and Sakamoto et al9b refers to the study evaluated association of the rs2294008 with gastric cancer types in Korea.
Abbreviations: BC, bladder cancer; CC, case–control; CRC, colorectal cancer; DHPLC, denaturing high-performance liquid chromatography; EC, esophageal cancer; GBC, gallbladder cancer; GIC, gastrointestinal cancers; GC, gastric cancer; LDR, ligation detection reaction; PCR, polymerase chain reaction; RFLP, restriction fragment length polymorphism; SNP, single nucleotide polymorphism.
Quantitative data synthesis
As shown in Table 2, the frequency of the minor allele varied widely across the 26 eligible studies, ranging from 0.23 to 0.71. The average frequency of the minor allele in gastric cancer, bladder cancer, and other cancers were 0.45, 0.47, and 0.27, respectively. The average frequency of the T allele was 0.40, 0.49, and 0.63 in Asians, Caucasians, and the mixed group, respectively.
Table 2.
PSCA rs2294008 polymorphism genotype distribution and allele frequency in cases and controls
| Study | Genotype (N)
|
Allele frequency (N)
|
MAF | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Case
|
Control
|
Case
|
Control
|
||||||||||
| Total | CC | CT | TT | Total | CC | CT | TT | C | T | C | T | ||
| Gastric cancer | |||||||||||||
| Zhao et al19 | 717 | 275 | 342 | 100 | 951 | 465 | 401 | 85 | 892 | 542 | 1,331 | 571 | 0.38 |
| Rizzato et al20 | 180 | 23 | 86 | 69 | 1,061 | 231 | 507 | 319 | 132 | 224 | 969 | 1,145 | 0.63 |
| Li et al24 | 300 | 124 | 141 | 35 | 300 | 168 | 111 | 21 | 389 | 211 | 447 | 153 | 0.35 |
| Sala et al26 | 411 | 93 | 198 | 118 | 1,530 | 491 | 714 | 310 | 384 | 434 | 1,696 | 1,334 | 0.53 |
| Zhao et al27 | 185 | 74 | 90 | 21 | 200 | 108 | 79 | 13 | 238 | 132 | 295 | 105 | 0.37 |
| Tanikawa et al28 | 2,300 | 1,030 | 1,073 | 197 | 16,567 | 6,620 | 7,587 | 2,360 | 3,133 | 1,467 | 20,827 | 12,307 | 0.32 |
| Song et al29 | 3,245 | 576 | 1,620 | 1,049 | 1,700 | 414 | 818 | 468 | 2,772 | 3,718 | 1,646 | 1,754 | 0.57 |
| Zeng et al30 | 460 | 202 | 216 | 42 | 549 | 289 | 223 | 37 | 620 | 300 | 801 | 297 | 0.33 |
| Lochhead et al I31 | 312 | 47 | 143 | 102 | 383 | 101 | 166 | 115 | 237 | 347 | 368 | 396 | 0.59 |
| Lochhead et al II31 | 309 | 85 | 129 | 94 | 211 | 49 | 110 | 49 | 299 | 317 | 208 | 208 | 0.51 |
| Ou et al33 | 196 | 85 | 93 | 18 | 246 | 132 | 96 | 18 | 263 | 129 | 360 | 132 | 0.33 |
| Lu et al34 | 1,053 | 547 | 404 | 72 | 1,110 | 605 | 387 | 77 | 1,498 | 548 | 1,597 | 541 | 0.27 |
| Chen et al35 | 460 | 202 | 216 | 42 | 549 | 289 | 223 | 37 | 620 | 300 | 801 | 297 | 0.33 |
| Matsuo et al37 | 708 | 330 | 329 | 49 | 708 | 273 | 338 | 97 | 989 | 427 | 884 | 532 | 0.30 |
| Wu et al38 | 1,736 | 759 | 819 | 132 | 1,020 | 506 | 412 | 77 | 2,337 | 1,083 | 1,424 | 566 | 0.32 |
| Sakamoto et al I9 | 1,531 | 96 | 700 | 728 | 1,399 | 210 | 650 | 536 | 892 | 2,156 | 1,070 | 1,722 | 0.71 |
| Sakamoto et al II9 | 871 | 133 | 461 | 277 | 390 | 122 | 176 | 92 | 727 | 1,015 | 420 | 360 | 0.58 |
| Bladder cancer | |||||||||||||
| Wang et al17 | 1,210 | 604 | 509 | 97 | 1,008 | 566 | 376 | 66 | 1,717 | 703 | 1,508 | 508 | 0.29 |
| Ma et al18 | 184 | 84 | 80 | 11 | 962 | 543 | 355 | 64 | 248 | 102 | 1,441 | 483 | 0.29 |
| Fu et al23 | 5,393 | 1,363 | 2,804 | 1,226 | 7,324 | 2,107 | 3,645 | 1,572 | 5,530 | 5,256 | 7,859 | 6,789 | 0.49 |
| Wang et al32 | 581 | 272 | 259 | 50 | 580 | 316 | 220 | 44 | 803 | 359 | 852 | 308 | 0.31 |
| Wu et al36 | 5,038 | 1,288 | 2,613 | 1,137 | 9,363 | 2,842 | 4,668 | 1,853 | 5,189 | 4,887 | 10,352 | 8,374 | 0.49 |
| Other cancers | |||||||||||||
| Esophageal cancer | |||||||||||||
| Dai et al16 | 2,083 | 1,232 | 724 | 127 | 2,220 | 1,222 | 851 | 147 | 3,188 | 978 | 3,295 | 1,145 | 0.23 |
| Lochhead et al II31 | 159 | 61 | 63 | 34 | 211 | 49 | 110 | 49 | 185 | 131 | 208 | 208 | 0.41 |
| Gallbladder cancer | |||||||||||||
| Rai et al21 | 405 | 104 | 233 | 68 | 247 | 79 | 126 | 42 | 441 | 369 | 284 | 210 | 0.46 |
| Ono et al22 | 44 | 12 | 23 | 9 | 173 | 68 | 75 | 30 | 47 | 41 | 211 | 135 | 0.47 |
| Colorectal cancer | |||||||||||||
| Smith et al25 | 77 | 25 | 39 | 13 | 804 | 287 | 387 | 130 | 89 | 65 | 961 | 647 | 0.42 |
Notes: C represents the major allele, T represents the minor allele.
Abbreviation: MAF, minor allele frequencies; M–H, Mantel–Haenszel.
In terms of overall analysis, a significant association was found in all genetic models (T vs C, OR: 1.18, 95% CI: 1.08–1.28, P=0.0001; TT vs CC, OR: 1.36, 95% CI: 1.14–1.62, P=0.0008; TC vs CC, OR: 1.29, 95% CI: 1.17–1.44, P<0.00001; dominant model TT + TC vs CC, OR: 1.32, 95% CI: 1.18–1.49, P<0.00001; recessive model TT vs TC + CC, OR: 1.15, 95% CI: 1.02–1.30, P=0.02).
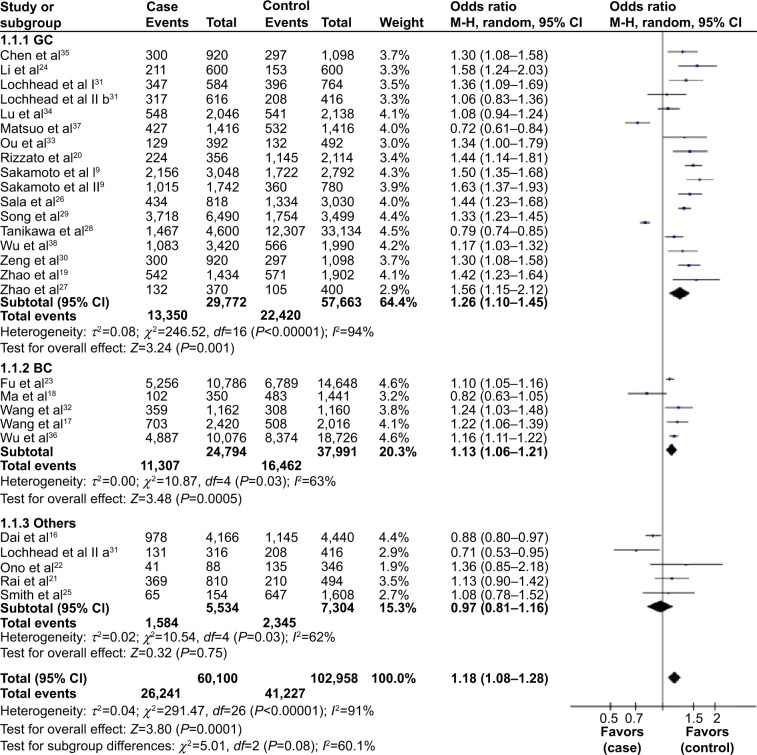
In subgroup analysis by cancer type, we found that the T allele had a significantly high risk of gastric cancer and bladder cancer, but null association in other cancers (Table 3). For gastric cancer, our meta-analysis contained 17 studies with 14,886 cases and 28,782 controls. The rs2294008 polymorphism was associated with gastric cancer risk in four genetic models (T vs C, OR: 1.26, 95% CI: 1.10–1.45, P=0.001; TT vs CC, OR: 1.51, 95% CI: 1.10–2.08, P=0.01; TC vs CC, OR: 1.39, 95% CI: 1.19–1.63, P<0.0001; TT + TC vs CC, OR: 1.44, 95% CI: 1.19–1.74, P=0.0002). However, the recessive model showed that there was no association between rs2294008 and gastric cancer (OR: 1.22, 95% CI: 0.99–1.49, P=0.06).
Table 3.
Meta-analysis results
| Comparisons | OR | 95% CI | P-value | Heterogeneity
|
Effects model | |
|---|---|---|---|---|---|---|
| I2 | P-value | |||||
| T vs C | 1.18 | 1.08–1.28 | 0.0001 | 91% | <0.00001 | R |
| Gastric cancer | 1.26 | 1.10–1.45 | 0.001 | 94% | <0.00001 | R |
| Bladder cancer | 1.13 | 1.06–1.21 | 0.0005 | 63% | 0.03 | R |
| Others | 0.97 | 0.81–1.16 | 0.75 | 62% | 0.03 | R |
| Asian | 1.19 | 1.05–1.35 | 0.007 | 93% | <0.00001 | R |
| Caucasian | 1.15 | 1.04–1.26 | 0.004 | 75% | 0.0005 | R |
| Mixed | 1.44 | 1.14–1.81 | 0.002 | NA | NA | |
| TT vs CC | 1.36 | 1.14–1.62 | 0.0008 | 90% | <0.00001 | R |
| Gastric cancer | 1.51 | 1.10–2.08 | 0.01 | 94% | <0.00001 | R |
| Bladder cancer | 1.28 | 1.20–1.37 | <0.00001 | 0% | 0.57 | F |
| Others | 0.91 | 0.75–1.10 | 0.33 | 39% | 0.16 | F |
| Asian | 1.37 | 1.03–1.82 | 0.03 | 92% | <0.00001 | R |
| Caucasian | 1.32 | 1.10–1.59 | 0.003 | 73% | 0.001 | R |
| Mixed | 2.17 | 1.32–3.59 | 0.002 | NA | NA | |
| TC vs CC | 1.29 | 1.17–1.44 | <0.00001 | 85% | <0.00001 | R |
| Gastric cancer | 1.39 | 1.19–1.63 | <0.0001 | 87% | <0.00001 | R |
| Bladder cancer | 1.23 | 1.17–1.30 | <0.00001 | 0% | 0.65 | F |
| Others | 0.98 | 0.68–1.41 | 0.90 | 77% | 0.001 | R |
| Asian | 1.36 | 1.18–1.57 | <0.0001 | 88% | <0.00001 | R |
| Caucasian | 1.13 | 0.95–1.34 | 0.16 | 79% | <0.0001 | R |
| Mixed | 1.70 | 1.05–2.77 | 0.03 | NA | NA | |
| TT + TC vs CC | 1.32 | 1.18–1.49 | <0.00001 | 90% | <0.00001 | R |
| Gastric cancer | 1.44 | 1.19–1.74 | 0.0002 | 92% | <0.00001 | R |
| Bladder cancer | 1.25 | 1.19–1.31 | <0.00001 | 0% | 0.64 | F |
| Others | 0.98 | 0.69–1.38 | 0.90 | 77% | 0.002 | R |
| Asian | 1.38 | 1.17–1.62 | 0.0001 | 92% | <0.00001 | R |
| Caucasian | 1.18 | 1.00–1.40 | 0.05 | 80% | <0.0001 | R |
| Mixed | 1.88 | 1.19–2.99 | 0.007 | NA | NA | |
| TT vs CC + TC | 1.15 | 1.02–1.30 | 0.02 | 83% | <0.00001 | R |
| Gastric cancer | 1.22 | 0.99–1.49 | 0.06 | 89% | <0.00001 | R |
| Bladder cancer | 1.13 | 1.07–1.20 | <0.0001 | 0% | 0.56 | F |
| Others | 0.95 | 0.79–1.14 | 0.57 | 0% | 0.96 | F |
| Asian | 1.13 | 0.93–1.36 | 0.87 | 87% | <0.00001 | R |
| Caucasian | 1.15 | 1.09–1.22 | <0.00001 | 47% | 0.08 | F |
| Mixed | 1.46 | 1.05–2.03 | 0.02 | NA | NA | |
Note: P-values in bold indicate nonsignificance.
Abbreviations: F, fixed; NA, not applicable; R, random; CI, confidence interval; OR, odds ratio; M–H, Mantel–Haenszel.
Five studies with 12,397 cases and 19,237 controls were used to evaluate the relationship between the rs2294008 polymorphism and bladder cancer risk. As shown in Table 3 and Figure 2, there was a significant association between rs2294008 and bladder cancer risk in all genetic models (T vs C, OR: 1.13, 95% CI: 1.06–1.21, P=0.0005; TT vs CC, OR: 1.28, 95% CI: 1.20–1.37, P<0.00001; TC vs CC, OR: 1.23, 95% CI: 1.17–1.30, P<0.00001; TT +TC vs CC, OR: 1.25, 95% CI: 1.19–1.31, P<0.00001; TT vs TC + CC, OR: 1.13, 95% CI: 1.07–1.20, P<0.0001).
Figure 2.
Forest plots of the PSCA rs2294008 polymorphism and cancer risk in the overall population and each subgroup stratified by cancer type (T vs C).
Notes: The squares and horizontal lines correspond to the study-specific OR and 95% CI. The area of the squares reflects the weight (inverse of the variance). The diamonds represent the summary OR and 95% CI.
Abbreviations: CI, confidence interval; OR, odds ratio; GC, gastric cancer; BC, bladder cancer; M–H, Mantel–Haenszel.
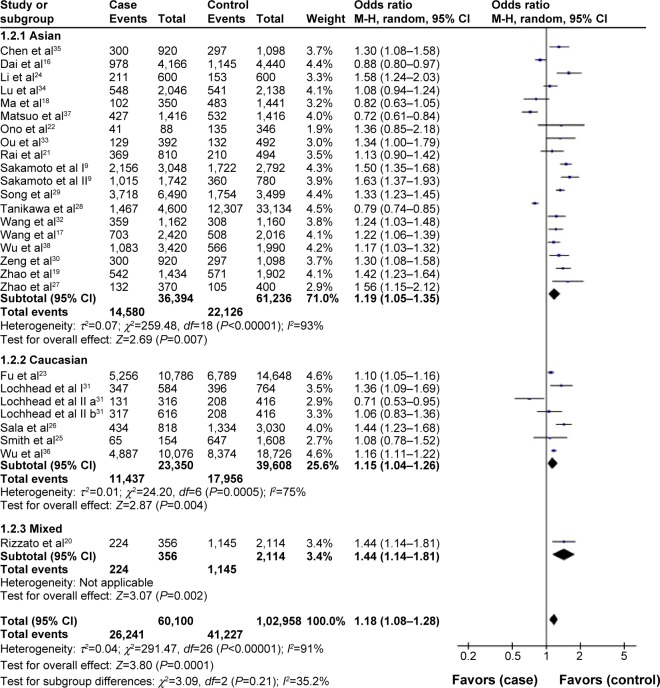
In the stratified analysis by ethnicity, pooled analysis of Asians showed that the T allele of the PSCA rs2294008 polymorphism was associated with increased cancer risk under all genetic models except for the recessive model (T vs C, OR: 1.19, 95% CI: 1.05–1.35, P=0.007, Figure 3; TT vs CC, OR: 1.37, 95% CI: 1.03–1.82, P=0.03; TC vs CC, OR: 1.36, 95% CI: 1.18–1.57, P<0.0001; TT + TC vs CC, OR: 1.38, 95% CI: 1.17–1.62, P=0.0001). PSCA rs2294008 showed a significant association among Caucasians based on three models (T vs C, OR: 1.15, 95% CI: 1.04–1.26, P=0.004, Figure 3; TT vs CC, OR: 1.32, 95% CI: 1.10–1.59, P=0.003; TT vs TC + CC, OR: 1.15, 95% CI: 1.09–1.22, P<0.00001). However, there was no significant association in Caucasians in the other two genetic models (TC vs CC, OR: 1.13, 95% CI: 0.95–1.34; TT + TC vs CC, OR: 1.18, 95% CI: 1.00–1.40).
Figure 3.
Forest plots of PSCA rs2294008 polymorphism and cancer risk in the overall population and each subgroup stratified by ethnicity (T vs C).
Notes: The squares and horizontal lines correspond to the study specific OR and 95% CI. The area of the squares reflects the weight (inverse of the variance). The diamonds represent the summary OR and 95% CI.
Abbreviations: CI, confidence interval; OR, odds ratio; M–H, Mantel–Haenszel.
Tests of heterogeneity
The Q and I2 statistic tests were used to evaluate heterogeneity. For the overall studies, there was statistically significant heterogeneity in this meta-analysis (T vs C, P<0.00001, I2=91%; TT vs CC, P<0.00001, I2=90%; TC vs CC, P<0.00001, I2=85%; dominant model TT + TC vs CC, P<0.00001, I2=90%; recessive model TT vs CC + TC, P<0.00001, I2=83%). Therefore, we carried on further subgroup analyses by cancer type and ethnicity, respectively. When the P-value of the heterogeneity test was more than 0.05 (P>0.05) in the following analyses, a fixed effects model was performed. Otherwise, the random effects model was used (shown in Table 3).
Publication bias
A funnel plot was used performed to evaluate publication bias in this meta-analysis. According to the funnel plot generated for the genetic models, there was an absence of obvious asymmetries for the distributions of ORs from every study (Figure 4). Therefore, the results indicated that publication bias had little effect in this meta-analysis.
Figure 4.
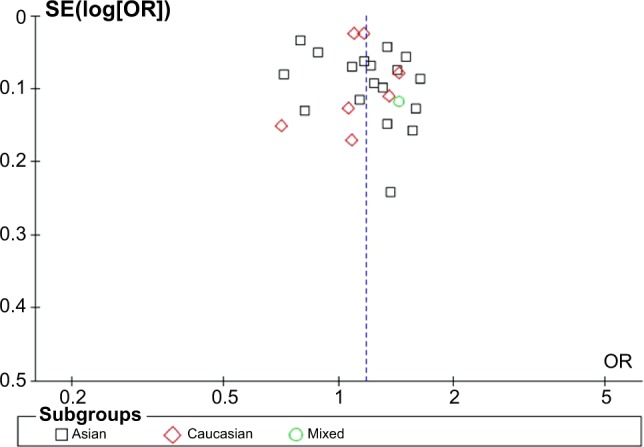
Funnel plot assessing evidence of publication bias from 26 studies (T vs C).
Abbreviations: OR, odds ratio; SE, standard error.
Discussion
PSCA is a member of the Thy-1/Ly-6 family of glycosylphosphatidylinositol (GPI)-anchored surface proteins, comprising three exons and two introns. Proteins in the Thy-1 family and GPI-anchored proteins have been proven to play a role in T cell activation.39 It has been demonstrated to be upregulated in prostate cancer and several other tumors and plays an important role in cell adhesion, proliferation, and survival.11 Recently, many cancer GWAS and replication studies have revealed the relationship between PSCA and risk of different cancers. A study performed by Sakamoto et al indicated that the mechanism of PSCA-suppressing cancers may be involved in cell proliferation inhibition and/or cell death induction.9 The findings of our meta-analysis may demonstrate that rs2294008 acts as an effect modifier in the development of different cancers. Of course, further biologically functional studies are warranted to verify the molecular mechanisms.
This comprehensive meta-analysis, involving 26 studies from 24 articles with 30,050 multiple cancer cases and 51,671 controls, showed that the rs2294008 polymorphism is significantly associated with overall cancer risk based on all genetic models. Further stratified analyses by cancer type revealed that the polymorphism was associated with an increased risk for gastric and bladder cancer; no association was found with other cancers in all genetic models. Studies from Dai et al16 and Lochhead et al31 indicated that the variant rs2294008C may have a protective role in esophageal cancer, but large well-designed studies are warranted to confirm this conclusion. The stratified analysis by ethnicity showed that the rs2294008 polymorphism was associated with an increased risk of cancer in both Asians and Caucasians. However, there was no study based on patients with an African background. Larger scale multicenter studies based on Africans are warranted to further validate the association between the rs2294008 polymorphism and cancer risk.
So far, there have been five meta-analyses that have investigated the role of the PSCA rs2294008 polymorphism in gastric cancer risk.10–14 All of them had the same finding that the rs2294008 polymorphism is associated with increased risk of gastric cancer. The stratified analyses by ethnicity were performed in four meta-analyses.10–12,14 Significantly increased risks were found for rs2294008 both among Asians and Caucasians in three articles,10–12 which is congruous with our results. However, one study showed no significant associations with the rs2294008 polymorphism and Caucasians.14 The latest research in these five published meta-analyses was performed by Gu et al.14 Their last search update was on August 2013, and in total, they identified 16 studies, including 18,820 gastric cases and 35,756 controls for the rs2294008 polymorphism. Compared to these five meta-analyses, we added another nine studies on other cancers in addition to gastric cancer. Our updated meta-analysis extracted data from all the published studies including 26 studies from 24 articles with 30,050 cancer cases and 51,671 controls. This meta-analysis provided evidence on the overall cancer risk of rs2294008 and contained the newest data and largest sample size on the relationship between rs2294008 and cancer. Thus, our results are more comprehensive and persuasive.
Although the pooled analysis was performed to show the association between rs2294008 and cancer risk, some limitations still inevitably exist in this meta-analysis. Firstly, we excluded some studies because of the limits of raw data. Secondly, we could not obtain all articles. Some unpublished literature and relevant published reports in other languages except English and Chinese that may be eligible for this meta-analysis were missed. Thirdly, there were only two studies on esophageal cancer, two on gallbladder cancer, and one on colorectal cancer. Additionally, there was a lack of studies on other types of cancers. Hence, the final OR was largely contributed by the ORs of gastric or bladder cancer; more evidence is needed to prove whether the rs2294008 polymorphism is association with cancer overall. Further large-scale multicenter studies based on a variety of cancer types are needed. Fourthly, the sources of the controls were not consistent. Both population-based healthy individuals and hospital patients without cancer were included in the control groups. Controls enrolled from hospitals may not always truly represent the underlying source populations, especially when the polymorphism was also expected to affect the risk of other diseases. Fifthly, the genotyping methods of the eligible studies were not identical, which may influence the results. Finally, because of the limit of individual data, the ORs of this meta-analysis were not strictly adjusted by the same potential confounders, such as age, sex, and stage of tumor. Additionally, a more precise analysis could be performed to eliminate the confounding bias.
Conclusion
In conclusion, the present meta-analysis demonstrated that the PSCA rs2294008 C > T polymorphism is a risk factor for cancer in both Asians and Caucasians. Furthermore, rs2294008 is associated with an increased risk of gastric and bladder cancer. Further large case–control studies are needed to assess the relationship between rs2294008 and other cancer types.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (no 81471670, 81472822); the International Cooperative Project of Shaanxi province, China (no 2013KW-32-01); the China Postdoctoral Science Foundation (no 2014M560791); the Fundamental Research Funds for the Central Universities, China (no 2014qngz-04), and the Specialized Research Fund of The Second Affiliated Hospital of Xi’an Jiaotong University, China (RC [GG] 201203).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Pharoah PD, Dunning AM, Ponder BA, Easton DF. Association studies for finding cancer-susceptibility genetic variants. Nat Rev Cancer. 2004;4(11):850–860. doi: 10.1038/nrc1476. [DOI] [PubMed] [Google Scholar]
- 3.Tang W, Qiu H, Ding H, et al. Association between the STK15F31I polymorphism and cancer susceptibility: a meta-analysis involving 43,626 subjects. PLoS One. 2013;8(12):e82790. doi: 10.1371/journal.pone.0082790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reiter RE, Gu Z, Watabe T, et al. Prostate stem cell antigen: a cell surface marker overexpressed in prostate cancer. Proc Natl Acad Sci U S A. 1998;95(4):1735–1740. doi: 10.1073/pnas.95.4.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saeki N, Gu J, Yoshida T, Wu X. Prostate stem cell antigen: a Jekyll and Hyde molecule? Clin Cancer Res. 2010;16(4):3533–3538. doi: 10.1158/1078-0432.CCR-09-3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elsamman EM, Fukumori T, Tanimoto S, et al. The expression of prostate stem cell antigen in human clear cell renal cell carcinoma: a quantitative reverse transcriptase polymerase chain reaction analysis. BJU Int. 2006;98(3):668–673. doi: 10.1111/j.1464-410X.2006.06350.x. [DOI] [PubMed] [Google Scholar]
- 7.Elsamman E, Fukumori T, Kasai T, et al. Prostate stem cell antigen predicts tumour recurrence in superficial transitional cell carcinoma of the urinary bladder. BJU Int. 2006;97(6):1202–1207. doi: 10.1111/j.1464-410X.2006.06153.x. [DOI] [PubMed] [Google Scholar]
- 8.Grubbs EG, Abdel-Wahab Z, Tyler DS, Pruitt SK. Utilizing quantitative polymerase chain reaction to evaluate prostate stem cell antigen as a tumor marker in pancreatic cancer. Ann Surg Oncol. 2006;13(12):1645–1654. doi: 10.1245/s10434-006-9029-5. [DOI] [PubMed] [Google Scholar]
- 9.Study Group of Millennium Genome Project for Cancer. Sakamoto H, Yoshimura K, Saeki N, et al. Genetic variation in PSCA is associated with susceptibility to diffuse-type gastric cancer. Nat Genet. 2008;40(6):730–740. doi: 10.1038/ng.152. [DOI] [PubMed] [Google Scholar]
- 10.Zhang QH, Yao YL, Gu T, Gu JH, Chen L, Liu Y. Association of the PSCA rs2294008 C > T polymorphism with gastric cancer risk: evidence from a meta-analysis. Asian Pac J Cancer Prev. 2012;13(6):2867–2871. doi: 10.7314/apjcp.2012.13.6.2867. [DOI] [PubMed] [Google Scholar]
- 11.Wang T, Zhang L, Li H, Wang B, Chen K. Prostate stem cell antigen polymorphisms and susceptibility to gastric cancer: a systematic review and meta-analysis. Cancer Epidemiol Biomarkers Prev. 2012;21(5):843–850. doi: 10.1158/1055-9965.EPI-11-1176. [DOI] [PubMed] [Google Scholar]
- 12.Shi D, Wang S, Gu D, et al. The PSCA polymorphisms derived from genome-wide association study are associated with risk of gastric cancer: a meta-analysis. J Cancer Res Clin Oncol. 2012;138(8):1339–1345. doi: 10.1007/s00432-012-1210-6. [DOI] [PubMed] [Google Scholar]
- 13.Zhang T, Chen YN, Wang Z, Chen JQ, Huang S. Effect of PSCA gene polymorphisms on gastric cancer risk and survival prediction: a meta-analysis. Exp Ther Med. 2012;4(1):158–164. doi: 10.3892/etm.2012.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gu X, Zhang W, Xu L, Cai D. Quantitative assessment of the influence of prostate stem cell antigen polymorphisms on gastric cancer risk. Tumour Biol. 2014;35(3):2167–2174. doi: 10.1007/s13277-013-1287-9. [DOI] [PubMed] [Google Scholar]
- 15.Dai ZJ, Kang HF, Wang XJ, et al. Association between genetic polymorphisms in AURKA (rs2273535 and rs1047972) and breast cancer risk: a meta-analysis involving 37,221 subjects. Cancer cell Int. 2014;14(1):91. doi: 10.1186/s12935-014-0091-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dai N, Zheng M, Wang C, et al. Genetic variants at 8q24 are associated with risk of esophageal squamous cell carcinoma in a Chinese population. Cancer Sci. 2014;105(6):731–735. doi: 10.1111/cas.12399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang P, Ye D, Guo J, et al. Genetic score of multiple risk-associated single nucleotide polymorphisms is a marker for genetic susceptibility to bladder cancer. Genes Chromosomes Cancer. 2014;53(1):98–105. doi: 10.1002/gcc.22121. [DOI] [PubMed] [Google Scholar]
- 18.Ma Z, Hu Q, Chen Z, et al. Systematic evaluation of bladder cancer risk-associated single-nucleotide polymorphisms in a Chinese population. Mol Carcinog. 2013;52(11):916–921. doi: 10.1002/mc.21932. [DOI] [PubMed] [Google Scholar]
- 19.Zhao J, Geng P, Li Z, et al. Prostate stem cell antigen rs2294008 polymorphism differentially contributes to Helicobacter pylori-negative gastric cancer among various populations in China. Mol Clin Oncol. 2013;1(3):493–498. doi: 10.3892/mco.2013.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rizzato C, Kato I, Plummer M, Muñoz N, Canzian F. Genetic variation in PSCA and risk of gastric advanced preneoplastic lesions and cancer in relation to Helicobacter pylori infection. PLoS One. 2013;8(9):e73100. doi: 10.1371/journal.pone.0073100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rai R, Sharma KL, Misra S, Kumar A, Mittal B. PSCA gene variants (rs2294008 and rs2978974) confer increased susceptibility of gallbladder carcinoma in females. Gene. 2013;530(2):172–177. doi: 10.1016/j.gene.2013.08.058. [DOI] [PubMed] [Google Scholar]
- 22.Ono H, Chihara D, Chiwaki F, et al. Missense allele of a single nucleotide polymorphism rs2294008 attenuated antitumor effects of prostate stem cell antigen in gallbladder cancer cells. J Carcinog. 2013;12:4. doi: 10.4103/1477-3163.109030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fu YP, Kohaar I, Rothman N, et al. Common genetic variants in the PSCA gene influence gene expression and bladder cancer risk. Proc Natl Acad Sci U S A. 2012;109(13):4974–4979. doi: 10.1073/pnas.1202189109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li F, Zhong MZ, Li JH, Liu W, Li B. Case-control study of single nucleotide polymorphisms of PSCA and MUC1 genes with gastric cancer in a Chinese. Asian Pac J Cancer Prev. 2012;13(6):2593–2596. doi: 10.7314/apjcp.2012.13.6.2593. [DOI] [PubMed] [Google Scholar]
- 25.Smith C, Lochhead P, Basavaraju U, et al. Lack of association between the rs2294008 polymorphism in the prostate stem cell antigen gene and colorectal neoplasia: a case-control and immunohistochemical study. BMC Res Notes. 2012;5:371. doi: 10.1186/1756-0500-5-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sala N, Muñoz X, Travier N, et al. Prostate stem-cell antigen gene is associated with diffuse and intestinal gastric cancer in Caucasians: results from the EPIC-EURGAST study. Int J Cancer. 2012;130(10):2417–2427. doi: 10.1002/ijc.26243. [DOI] [PubMed] [Google Scholar]
- 27.Zhao JD, Geng PL, Zhao JH, et al. Relationship between the rs2294008 polymorphism of the PSCA gene and susceptibility to gastric cancer in Tibetans. World Chin J Digestol. 2012;20:418–421. [Google Scholar]
- 28.Tanikawa C, Urabe Y, Matsuo K, et al. A genome-wide association study identifies two susceptibility loci for duodenal ulcer in the Japanese population. Nat Genet. 2012;44(4):430–434. doi: 10.1038/ng.1109. , S1–S2. [DOI] [PubMed] [Google Scholar]
- 29.Song HR, Kim HN, Piao JM, et al. Association of a common genetic variant in prostate stem-cell antigen with gastric cancer susceptibility in a Korean population. Mol Carcinog. 2011;50(11):871–875. doi: 10.1002/mc.20796. [DOI] [PubMed] [Google Scholar]
- 30.Zeng Z, Wu X, Chen F, et al. Polymorphisms in prostate stem cell antigen gene rs2294008 increase gastric cancer risk in Chinese. Mol Carcinog. 2011;50(5):353–358. doi: 10.1002/mc.20718. [DOI] [PubMed] [Google Scholar]
- 31.Lochhead P, Frank B, Hold GL, et al. Genetic variation in the prostate stem cell antigen gene and upper gastrointestinal cancer in white individuals. Gastroenterology. 2011;140(2):435–441. doi: 10.1053/j.gastro.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang S, Tang J, Wang M, Yuan L, Zhang Z. Genetic variation in PSCA and bladder cancer susceptibility in a Chinese population. Carcinogenesis. 2010;31(4):621–624. doi: 10.1093/carcin/bgp323. [DOI] [PubMed] [Google Scholar]
- 33.Ou J, Li K, Ren H, Bai H, Zeng D, Zhang C. Association and haplotype analysis of prostate stem cell antigen with gastric cancer in Tibetans. DNA Cell Biol. 2010;29(6):319–323. doi: 10.1089/dna.2009.0960. [DOI] [PubMed] [Google Scholar]
- 34.Lu Y, Chen J, Ding Y, et al. Genetic variation of PSCA gene is associated with the risk of both diffuse- and intestinal-type gastric cancer in a Chinese population. Int J Cancer. 2010;127(9):2183–2189. doi: 10.1002/ijc.25228. [DOI] [PubMed] [Google Scholar]
- 35.Chen FG, Zeng ZR, Wu XQ, Chen B, Chen MH. Association of prostate stem cell antigen gene rs2294008 polymorphism with gastric cancer in Chinese Han. Chin J Gastroenterol. 2010;15(1):17–20. [Google Scholar]
- 36.Wu X, Ye Y, Kiemeney LA, et al. Genetic variation in the prostate stem cell antigen gene PSCA confers susceptibility to urinary bladder cancer. Nat Genet. 2009;41(9):991–995. doi: 10.1038/ng.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matsuo K, Tajima K, Suzuki T, et al. Association of prostate stem cell antigen gene polymorphisms with the risk of stomach cancer in Japanese. Int J Cancer. 2009;125(8):1961–1964. doi: 10.1002/ijc.24519. [DOI] [PubMed] [Google Scholar]
- 38.Wu C, Wang G, Yang M, et al. Two genetic variants in prostate stem cell antigen and gastric cancer susceptibility in a Chinese population. Mol Carcinog. 2009;48(12):1131–1138. doi: 10.1002/mc.20565. [DOI] [PubMed] [Google Scholar]
- 39.Raff AB, Gray A, Kast WM. Prostate stem cell antigen: a prospective therapeutic and diagnostic target. Cancer Lett. 2009;277(2):126–132. doi: 10.1016/j.canlet.2008.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]