Abstract
Background
Doxorubicin (DOX), an anthracycline anticancer antibiotic, is used for treating various types of cancers. However, its use is associated with toxicity to normal cells and development of resistance due to overexpression of drug efflux pumps. Poloxamer 407 (P407) and vitamin E TPGS (D-α-tocopheryl polyethylene glycol succinate, TPGS) are widely used polymers as drug delivery carriers and excipients for enhancing the drug retention times and stability. TPGS reduces multidrug resistance, induces apoptosis, and shows selective anticancer activity against tumor cells. Keeping in view the problems, we designed a mixed micelle system encapsulating DOX comprising TPGS for its selective anticancer activity and P407 conjugated with folic acid (FA) for folate-mediated receptor targeting to cancer cells.
Methods
FA-functionalized P407 was prepared by carbodiimide crosslinker chemistry. P407-TPGS/FA-P407-TPGS-mixed micelles were prepared by thin-film hydration method. Cytotoxicity of blank micelles, DOX, and DOX-loaded micelles was determined by alamarBlue® assay.
Results
The size of micelles was less than 200 nm with encapsulation efficiency of 85% and 73% for P407-TPGS and FA-P407-TPGS micelles, respectively. Intracellular trafficking study using nile red-loaded micelles indicated improved drug uptake and perinuclear drug localization. The micelles show minimal toxicity to normal human cell line WRL-68, enhanced cellular uptake of DOX, reduced drug efflux, increased DOX–DNA binding in SKOV3 and DOX-resistant SKOV3 human ovarian carcinoma cell lines, and enhanced in vitro cytotoxicity as compared to free DOX.
Conclusion
FA-P407-TPGS-DOX micelles show potential as a targeted nano-drug delivery system for DOX due to their multiple synergistic factors of selective anticancer activity, inhibition of multidrug resistance, and folate-mediated selective uptake.
Keywords: doxorubicin nanocarriers, folate targeting, doxorubicin cytotoxicity, synergistic drug delivery, Pgp-inhibiting micelles
Introduction
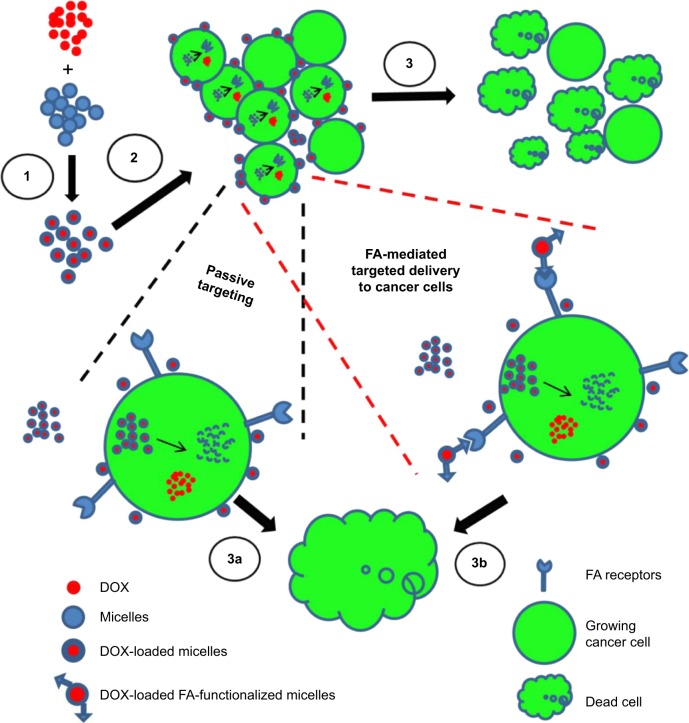
Drug delivery to a specific target at organ or cellular level has been widely explored. Although Paul Ehrlich’s magic bullet concept has not been fulfilled to the fullest, there have been substantial improvements. Delivering anticancer drug, intact, to the target site, that is, tumor, has been one such area where magic bullet approach has been substantially tried.1 Nontargeted drug delivery systems release the drug payload directly into plasma, which could result in adverse effects associated with anticancer drugs.2,3 A valuable approach to overcome this issue is the concept of ligand-mediated targeting using antibodies, peptides, carbohydrates, aptamers, and small molecules like folic acid (FA) (Figure 1). FA as an active targeting ligand has been extensively investigated4 and is a helpful approach for active targeting as the polymers conjugated with it could be preferentially taken up into cells overexpressing FA receptors.5 This selective uptake approach could limit the off-targeted adverse effects of anticancer drugs.6
Figure 1.
Schematic representation of active and passive targeting of DOX-loaded P407/FA-P407-TPGS micelles in cancer cells.
Notes: 1: DOX was loaded into micelles by solvent evaporation and thin-film hydration methods. 2: DOX-loaded micelles enter the cells by either passive transport or active transport mediated by FA ligand. Both active and passive transports across the cells are illustrated in enlarged version of cells. 3: Once inside the cells, DOX is released and acts on the target site to cause cell death. TPGS and P407 reduce drug efflux and enhance DOX–DNA binding. In both cases, that is, 3a and 3b, the ultimate goal is the cell death in which the targeted micelles result in enhanced cell death as compared to nontargeted DOX micelles.
Abbreviations: DOX, doxorubicin; P407, poloxamer 407; FA, folic acid; TPGS, D-α-tocopheryl polyethylene glycol succinate.
In recent years, the use of polymers for drug delivery, specifically to the tumor cells, has significantly increased owing to the ability of carriers to achieve desirable effects like enhanced permeability and retention and long circulation times.7 These polymers have been used to fabricate various drug delivery systems including drug–polymer conjugates,8 dendrimers,9 nanoparticles,10 and polymeric micelles11 for various purposes ranging from solubility and permeation enhancement to as an adjuvant and as an active ingredient to act synergistically with the drugs they are administered with. Block copolymers are one such class of polymers which have been used for such purposes.12 Poloxamers, commercially known as Pluronic, are a family of tri-block copolymers of polyethylene oxide (PEO) and polypropylene oxide (PPO) which have been widely used in pharmaceutical research for their desirable properties such as increased blood retention times of drugs.13 Poloxamers can arrange themselves to form micelles where the core is made up of hydrophobic PPO segment, while the outer parts termed shell are made up of PEO, which is hydrophilic. The hydrophobic compartment can be used to load hydrophobic drugs inside the micelles, whereas the presence of PEO on the surface of micelles acts to protect the micelles from recognition by the immune system and unwanted interactions with serum proteins; thus, the micelles could go undetected enhancing the drug retention times in plasma.14 Similarly, D-α-tocopheryl polyethylene glycol succinate 1000 (TPGS), which is a modified form of vitamin E or α-tocopherol, can form micelles and have been shown to enhance the stability of colloidal systems.15 In addition, TPGS has also been shown to inhibit the growth of tumor cells in vitro and in vivo. Furthermore, it also reduces the multidrug resistance by P-glycoprotein (Pgp) inhibition16 as well as induces apoptosis selectively in cancer cells.17 It could thus act synergistically with anticancer drugs to enhance their antitumor efficacy.18 Doxorubicin (DOX) is an anticancer drug used for the treatment of a variety of cancers; however, it encounters problems of cytotoxicity to normal cells as well as development of resistance to chemotherapeutic agents.19
Keeping in view the aforementioned characteristics, poloxamer 407 (P407) was functionalized by conjugation with FA adding value to selective cancer cell targeting of TPGS for a targeted and specific delivery. The mixed micelles of P407-FA and TPGS were produced by thin-film hydration method and characterized for size, zeta potential, encapsulation efficiency (EE), and in vitro drug release. Nile red-loaded micelles were used for study of intracellular trafficking. Cytotoxicity of blank micelles, DOX, and DOX-loaded micelles was determined by alamarBlue® assay. Studies on the effect of micelles on rhodamine 123 (R123) efflux, cellular uptake of DOX, and DOX–DNA binding were undertaken to investigate the enhanced cytotoxicity of DOX-loaded micelles. Findings from these studies indicated that FA-P407-TPGS-mixed micelles show enhanced drug release at acidic pH, increased DOX–DNA binding, reduced drug efflux, and improved DOX uptake and could act synergistically with DOX to enhance its cytotoxicity.
Materials and methods
Materials
P407 (Pluronic F127), TPGS, 1,1′-carbonyldiimidazole (CDI), FA, nile red, R123, and dimethyl sulfoxide (DMSO) were purchased from Sigma-Aldrich (St Louis, MO, USA). DOX hydrochloride (DOX·HCl) was purchased from EMD Biosciences (Calbiochem, San Diego, CA, USA). Triethylamine, dichloromethane (DCM), chloroform, Hoechst 33342, and 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) were purchased from Merck Schuchardt OHG (Hohenbrunn, Germany). AlamarBlue®, trypsin–ethylenediaminetetraacetic acid (EDTA), Dulbecco’s Modified Eagle’s Medium (DMEM), and fetal bovine serum (FBS) were purchased from Life Technologies (Gibco, Carlsbad, CA, USA).
Cell lines
Human hepatic cell line WRL-68 and human ovarian adenocarcinoma cell line SKOV3 were obtained from ATCC (Manassas, VA, USA). SKOV3 cells overexpress folate receptors at intermediate levels which are comparable to levels of folate receptors in malignant ovarian tissues.20 DOX-resistant SKOV3 (SKOV3-DOX) cell line was generated by continuous selection with increasing concentrations of DOX. WRL-68 and SKOV3 cells were maintained in DMEM containing 10% FBS and 1% penicillin–streptomycin at 37°C in a 95% air–5% CO2 atmosphere. SKOV3-DOX cell line was maintained in culture medium containing 800 ng/mL of DOX. SKOV3 or SKOV3-DOX cells were cultured in folate-free DMEM in experiments involving folate receptor-mediated cell uptake. All cells were regularly passaged at approximately 80% confluence using 0.25% trypsin–EDTA.
Synthesis and characterization of FA-conjugated P407
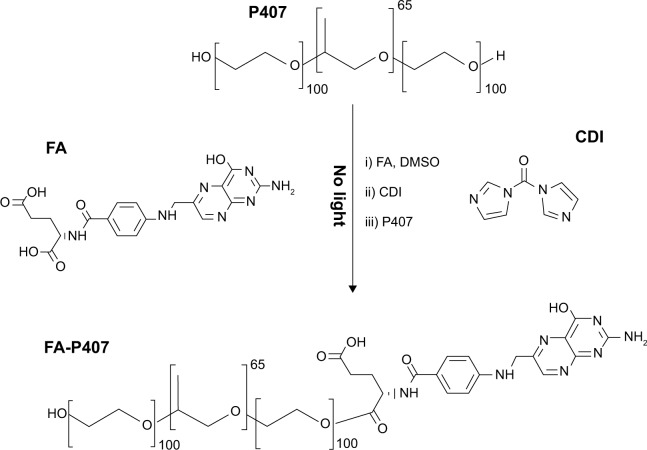
The synthesis of FA-conjugated P407 (FA-P407) is shown in Figure 2. Firstly, FA was dissolved in DMSO for overnight in a two-neck flask. CDI was then added into the solution, and the reaction mixture was stirred overnight under nitrogen atmosphere and protected from light. P407 was then added, and the reaction was stirred continuously for 24 hours under similar conditions. After the reaction completed, the mixture was dialyzed against deionized water for up to 5 days in order to remove any un-reacted FA. To recover the FA-P407 conjugate, the dialyzed solution was then freeze dried.
Figure 2.
Synthesis of FA-P407 conjugate.
Notes: FA and P407 were conjugated by CDI-mediated coupling, purified by dialysis, and recovered by freeze drying.
Abbreviations: FA, folic acid; P407, poloxamer 407; CDI, 1,1′-carbonyldiimidazole; DMSO, dimethyl sulfoxide.
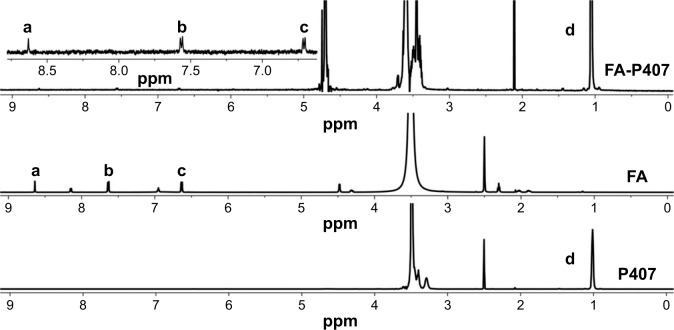
The conjugation of FA onto P407 was confirmed by recording and analyzing 1H NMR spectra of pure P407, FA, and FA-P407 conjugate. 1H NMR spectra were recorded on Bruker AVANCE 600 MHz spectrometer at 25°C. FA was dissolved in DMSO, whereas P407 and FA-P407 were dissolved in deuterated water (D2O) for acquisition of NMR spectra. Degree of substitution on a molar basis was determined by dividing the total integral values of five FA aromatic protons (Figure 3, peaks a–c) in region 6.5–9.0 ppm by methyl group protons of PPO at 1.05 ppm (Figure 3, peak d).
Figure 3.
1H NMR spectra of FA, P407, and FA-P407.
Notes: Peaks a–c represent aromatic protons in FA, whereas peak d represents methyl protons of PPO confirming the conjugation of FA to P407.
Abbreviations: FA, folic acid; P407, poloxamer 407; PPO, polypropylene oxide.
UV–vis spectra were acquired using UV-1601 spectrophotometer (Shimadzu Corp., Kyoto, Japan) as a qualitative proof of FA conjugation with P407. The degree of conjugation of FA was determined by UV–vis spectroscopy. FA solutions of known concentrations were prepared in DMSO, and their respective UV absorbance was recorded. A calibration curve was obtained by plotting UV absorbance against FA concentrations. To determine the concentration of conjugated FA, 10 mg of the sample was dissolved in DMSO, and UV absorbance at 360 nm was recorded. FA present in conjugate was then calculated from the calibration curve. Degree of conjugation of FA on a weight basis was calculated by using the following equation:
Preparation of micelles
Micelles were prepared by the thin-film hydration method. P407/FA-P407 (100 mg) and TPGS (5 mg) were dissolved in 10 mL of chloroform. These binary solutions were put in vials, and chloroform was evaporated overnight in a fume hood to get thin films. These thin films were then freeze dried for 16 hours and stored in tightly sealed containers. Thin films were then hydrated with deionized water, incubated at 37°C for 30 minutes, and filtered through 0.45 μm nylon filter to obtain micelle suspension.17
In order to prepare drug-loaded micelles, the thin films were dissolved in 3 mL DCM, and 1 mg DOX·HCl was added in the presence of TEA. This mixture was then added dropwise to 50 mL of deionized water ultrasonicated using a probe sonifier (Branson Ultrasonics Co., Danbury, CT, USA) over 2 minutes. Organic solvents were then removed using rotary evaporator, and the remaining suspension was passed through vivacell 250-ultrafiltration assembly (MWCO 10 kDa) to remove any unloaded DOX. The obtained DOX-loaded micelle suspension was freeze dried and stored in tightly sealed containers.
For the preparation of micelles incorporating nile red, P407/FA-P407 (100 mg), TPGS (5 mg), and nile red (1 mg) were dissolved in 3 mL of DCM and added dropwise to 20 mL of deionized water with ultrasonication using a probe sonifier to produce an emulsion. It was stirred overnight in darkness, and organic solvent was removed under vacuum. The suspension was then filtered through a 0.45 μm nylon filter to remove any unincorporated nile red.21 The filtered solution was freeze dried and stored under light protection.
Characterization of micelles
The particle size and surface charge of FA-P407-TPGS-mixed micelles were determined by dynamic light scattering (DLS) method using Zetasizer Nano ZS (Malvern Instruments, Malvern, UK). For the analysis of micelle morphology, transmission electron microscopy (TEM) and field emission scanning electron microscopy (FESEM) were used. For TEM, a drop of micelle suspension was placed and air dried onto a carbon-coated formvar film on a 400 mesh copper grid. TEM images were then acquired at 220 kV under different magnifications (TEM, Tecnai Spirit, FEI, People’s Republic of China). For FESEM, a drop of micelles was placed and air dried onto aluminum stubs, and images were acquired at 3 kV and different magnifications (FESEM, GEMINI II, Zeiss, Germany).
Critical micelle concentration of micelles was determined by DLS method17 using Zetasizer. Dilutions of samples from 1 μM to 5 mM were prepared in deionized water, and changes in the light intensity for each sample were recorded at a scattering angle of 90°. A graph was plotted between the molar concentration of sample and light intensity. Critical micelle concentration is the concentration at or above which unimers self-assemble to form micelles, and it is associated with a sharp increase in light scattering intensity.
To determine the EE of DOX in micelles, drug-loaded micelles were dissolved in 5 mL of DMSO to disrupt the micelles and release encapsulated DOX. UV absorbance of DOX was measured at 482 nm using UV–vis spectrophotometer (UV-1800, Shimadzu Corp.), and the amount of encapsulated DOX was calculated using calibration curve. EE and % mass of the DOX in drug-loaded micelles were calculated using the following equations:
In vitro drug release
The release studies of DOX were carried out by dialysis bag method using phosphate-buffered saline (PBS) as the release medium at pH 7 (corresponding to physiological pH) and pH 5 (to mimic the slightly acidic environment in the tumors). A known amount of DOX-loaded micelles in PBS was placed into a dialysis bag (MWCO 10,000, Sigma-Aldrich), and both ends were sealed. End-sealed dialysis bag was immersed in PBS (pH 5 or 7) shaken at 100 rpm, and temperature was maintained at 37°C. At designated time points, 2 mL of samples was taken from the release medium, and an equal volume of fresh medium was replaced. DOX concentration in the withdrawn sample was determined from the calibration curve using UV absorbance at 482 nm. Drug release curves were obtained by plotting the cumulative % drug release against time.
Drug release kinetics
The following mathematical models were applied on the data obtained in the drug release studies from DOX-loaded micelles to predict the release process involved.
Zero-order model
Zero-order model assumes that the release of drug from the formulation follows a steady pattern but is independent of the concentration or time22 described by the following equation:
Here, Mt, M0, t, and K0 represent amount of drug released at time t, initial amount of the drug, time, and the zero-order rate constant of drug release, respectively. Data from drug release study were fitted into this model by plotting a cumulative drug release (%) against time.
First-order model
According to first-order model, the drug release from a formulation depends upon time and initial concentration of the drug present.23 As a result, an exponential reduction in activity within the system is also expected as described by the following equation:
Here, k1t represents the first-order rate constant. A plot of the log of cumulative remaining drug (%) and time was used for fitting data into the first-order model.
Hixson–Crowell model
The Hixson–Crowell model24 describes the drug release from particles keeping in view the area and volume of the given particles and is represented by the equation given below:
Here, Mt/M∞, t, and kHCt represent the fractional drug release, time, and Hixson–Crowell rate constant, respectively. Data from the drug release study were fitted into this model by plotting cube root of % drug remaining against time.
Korsmeyer–Peppas model
Korsmeyer–Peppas equation could be used to predict the mechanism of drug release from a polymeric system. For evaluation of DOX release mechanism from mixed micelles system, Korsmeyer equation25 was applied by plotting log of the cumulative drug release (%) against log of time described by the following equation:
Here, Mt/M∞, t, kKP, and n represent the fractional drug release, time, Korsmeyer–Peppas rate constant, and release exponent that characterizes the mechanism of the release, respectively. The first 60% of drug release data was fitted to predict the release mechanism.
Diffusion coefficients
Diffusion coefficients of the DOX release from P407/FA-P407-TPGS-mixed micelles were calculated by using the following equation:
where D is the diffusion coefficient, dt/d∞ is the fractional drug release, L is the diameter, and t represents respective times.26
Cell viability assay for blank micelles
To study the effect or possible toxicity of P407/FA-P407-TPGS-mixed micelles in normal cells, alamarBlue® cell viability assay was used. Briefly, WRL-68 cells were seeded in 96-well plates at 2×104 cells per well in DMEM. Twenty-four hours after seeding, cells were washed with PBS, and fresh medium containing micelles at a final concentration in the range of 10–1,000 μg mL−1 was added. Medium without any treatment was used as a negative control. After 24-hour incubation, the medium was removed, and the cells were washed twice with PBS and medium containing 10% alamarBlue® was added to the wells followed by another incubation of 4 hours. The plates were then read at 570 nm (A570) with a microplate reader (Varioskan Flash; Thermo Scientific, Waltham, MA, USA), and cell viability was calculated using the following equation:
Cytotoxicity of DOX-loaded micelles
Cytotoxicity of DOX-loaded micelles was evaluated in SKOV3 and SKOV3-DOX cell lines. Cells were seeded in 96-well plates at 5×104 cells per well. The cells were treated with varying concentrations of DOX and DOX-loaded micelles. alamarBlue® assay was carried out as described earlier.
Effect of micelles on drug efflux
R123 is a substrate for drug efflux pumps or Pgps and is actively pumped out of the cells.27 The effect of micelles on drug efflux was monitored in SKOV3/SKOV3-DOX cells (106) seeded in 35 mm culture dishes. The cells were allowed to attach overnight, and then, the medium was replaced with media containing 50 μM R123 pre-incubated for 2 hours with TPGS, P407, P407/FA-P407-TPGS-mixed micelles or blank media. After 2 hours of incubation, R123-containing medium was removed, and the cells were washed twice with ice-cold PBS followed by the addition of 1% Triton X-100 to lyse the cells. The fluorescence intensity of R123 was measured using a microplate reader (Varioskan Flash; Thermo Scientific) at excitation and emission wavelengths of 485 nm and 530 nm, respectively. Bradford assay28 was used to normalize the intracellular R123 concentration to total protein content.
DNA binding of DOX
SKOV3/SKOV3-DOX cells were seeded in 12-well plates at 5×105 cells per well and incubated for 48 hours. After incubation, the cells were treated with free DOX or DOX-loaded P407/FA-P407-TPGS-mixed micelles for 4 hours. Posttreatment, the cells were washed with ice-cold PBS twice and detached using cell scraper. DOX taken up by the cells was measured by taking fluorescence reading at excitation and emission wavelengths of 471 nm and 556 nm, respectively. The recorded fluorescence corresponds to DOX fluorescence in cytoplasm (DOXcp) as at this point, DOX fluorescence is quenched due to binding with DNA. To remove DOX from its complex with DNA, the cells were then treated with 0.1% Triton X-100 for 15 minutes at 25°C, and the fluorescence readings were taken again. This reading was considered as DOXt (total DOX fluorescence) as the exposure to Triton X-100 removes DOX from its complex with DNA, and DOX fluorescence which was quenched earlier can be recorded.29 The difference between these two measurements was calculated in order to get the DNA-bound DOX (DOXDNA) by using the following equation:
DOX uptake and localization
The localization and uptake of DOX in SKOV3/SKOV3-DOX cells were investigated by treating the cells with free DOX solutions or DOX-loaded micelles. The cells were seeded onto coverslips in 35 mm cell culture dishes and then were treated with either DOX solutions or DOX-loaded micelles for 4 hours. After incubation, the cells were washed with ice-cold PBS three times and stained with Hoechst 33342 and immediately observed under fluorescence microscope to visualize the uptake of DOX.
To determine the effect of folate functionalization on DOX uptake, cells were treated with FA-P407-TPGS-mixed micelles in FA-free medium or medium containing 2 mM FA.
Statistical analysis
The data are mean values of a minimum of three repetitions with standard deviations. Statistical analysis was performed by GraphPad Prism 5.0 (GraphPad Software, Inc., La Jolla, CA, USA) using one-way analysis of variance followed by post hoc Dunnett’s multiple-comparison test.
Results and discussion
Synthesis and characterization of FA-P407
Synthesis of folate-conjugated P407 was confirmed by using 1H NMR. As shown in Figure 3, 1H NMR spectrum of FA-P407 showed the characteristic FA peaks at 6.71 ppm and 7.56 ppm (aromatic protons of FA) and 8.63 ppm corresponding to C7–H of FA.30 In addition, the peaks at 1.05 ppm correspond to –CH3 of PPO, and the peaks in the region 3.38–3.72 ppm correspond to –CH2CHO and –CH2CH2O– segments of PPO and PEO. The degree of substitution on molar-to-molar basis was 5:1 moles of FA to P407. The concentration of FA in conjugate was 5.9%, whereas the degree of substitution of FA onto P407 was ~2% as determined from UV. Figure 4 shows the UV spectra of FA, P407, and FA-P407. P407 itself does not show any absorbance peaks from 250 nm to 400 nm; however, when it was conjugated with FA, the characteristic UV absorbance peaks around 360 nm and 280 nm appeared in the spectra indicating the presence of FA in the conjugate.31
Figure 4.
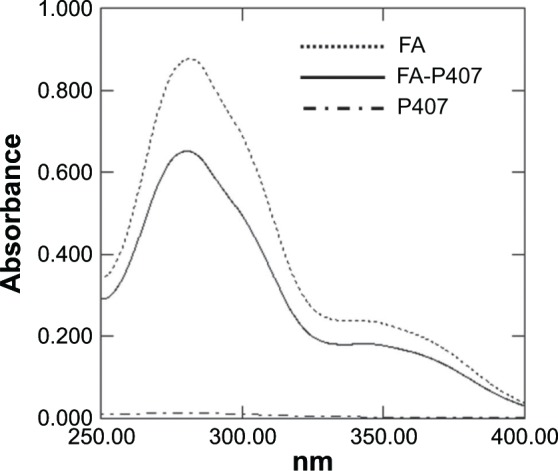
UV–vis spectra of FA, P407, and FA-P407.
Notes: FA and FA-P407 show characteristic absorption peaks at around 280 nm and 360 nm, whereas P407 has no peaks in this region.
Abbreviations: FA, folic acid; P407, poloxamer 407.
Physical characteristics of micelles
The morphology of the micelles was observed by FESEM and TEM. As shown in Figure 5, the micelles exhibited a spherical shape and smooth surface. The size of FA-functionalized micelles was found to be 173±31 nm (Table 1). The size calculated from FESEM and TEM was comparably smaller than the size observed by DLS. The difference in the mean size could be due to the fact that DLS size corresponds to the size of particles in solution, whereas the electron micrographs show the size of particles in a dried state. The zeta potential showed insignificant variations between both FA-P407-TPGS micelles and P407-TPGS micelles. Similar results were observed for critical micelle concentration, and the micelles exhibited relatively high EE as shown in Table 1. The high EE seems to have resulted from the interactions and π–π stacking between hydrophobic PPO and tocopherol segments of TPGS and the aromatic structure of DOX.32
Figure 5.
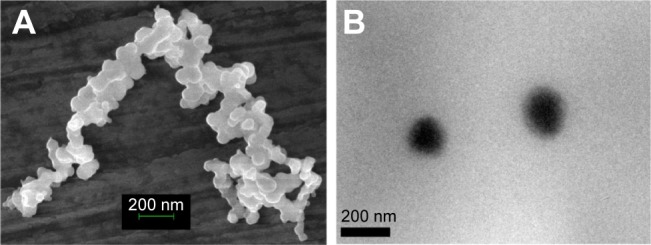
Morphology of micelles.
Notes: (A) FESEM image of FA-P407-TPGS micelles. (B) TEM image of FA-P407-TPGS micelles.
Abbreviations: FESEM, field emission scanning electron microscopy; FA, folic acid; P407, poloxamer 407; TPGS, D-α-tocopheryl polyethylene glycol succinate; TEM, transmission electron microscopy.
Table 1.
Characterization of P407/FA-P407-TPGS micelles
| Sample | CMC (M) | Hydrodynamic size (nm) | PDI | Zeta potential (mV) | EE (%) |
|---|---|---|---|---|---|
| P407-TPGS | 5×10−5 | 29±7 | 0.392±0.05 | −15±4 | 85±12 |
| FA-P407-TPGS | 5×10−5 | 173±31 | 0.211±0.07 | −18±5 | 73±9 |
Abbreviations: P407, poloxamer 407; FA, folic acid; TPGS, D-α-tocopheryl polyethylene glycol succinate; CMC, critical micelle concentration; PDI, polydispersity index; EE, encapsulation efficiency.
pH-dependent DOX release in vitro
The result of the drug release of DOX from micelles is shown in Figure 6. DOX-loaded P407/FA-P407-TPGS-mixed micelles showed similar release patterns differing only slightly in terms of cumulative drug release. The release of free DOX was found to be rapid and reached 100% cumulative almost in less than 2 hours. In case of micelles, an initial fast release was observed for first 6 hours (~30%), after which the release of DOX from micelles increased steadily up to 120 hours. It was observed that at pH 5, about 80% of DOX was released over a 5-day period, whereas only about 50% DOX was released in the same period at pH 7 which could be due to higher partitioning of DOX at acidic pH.33 This could be helpful and add to the advantage of selective and targeted uptake in cancer cells where the pH is slightly acidic compared to normal physiological pH. Analysis of the data obtained from the in vitro drug release experiments was done by data fitting in drug release models and further by Korsmeyer–Peppas power law to elucidate the release mechanism involved. Accuracy of data fitting was determined by correlation coefficient (R2) values. As shown in Table 2, the data correlated well with the Korsmeyer–Peppas power law with R2 values of 0.92 and 0.97 for P407-TPGS micelles and 0.85 and 0.97 for FA-P407-TPGS micelles at pH 7 and pH 5, respectively. Furthermore, it was observed that when the pH of release medium was changed from pH 7 to pH 5, the result was an increased release exponent (n) value which could be due to the enhanced solubility of DOX at acidic pH resulting in a higher drug release. The diffusion coefficients also increased in correlation with release exponents indicating that the rate of drug release became faster at acidic pH. It could be suggested that the pH-sensitive behavior of micelles was a result of the combined effects of DOX partitioning in acidic media and relaxation or swelling of micelles in solution over time.34
Figure 6.
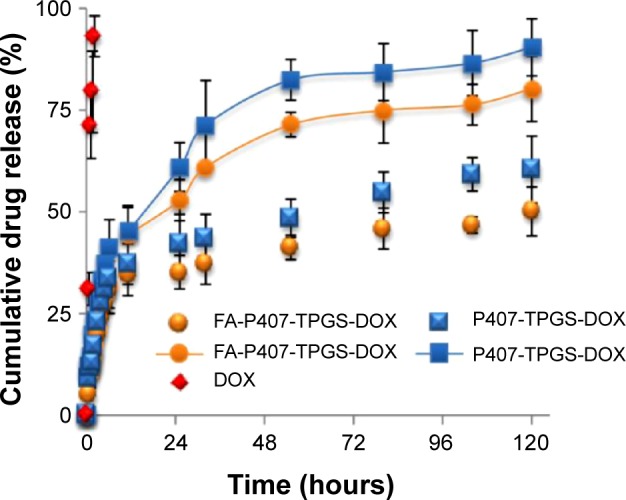
pH-dependent DOX release from P407/FA-P407-TPGS micelles.
Notes: Red symbols: free DOX; symbols with lines: DOX release at pH 5; symbols without lines: DOX release at pH 7.
Abbreviations: DOX, doxorubicin; P407, poloxamer 407; FA, folic acid; TPGS, D-α-tocopheryl polyethylene glycol succinate.
Table 2.
Kinetic parameters of DOX release from micelles
| Kinetic parameters | P407-TPGS-DOX
|
FA-P407-TPGS-DOX
|
|||
|---|---|---|---|---|---|
| pH 7 | pH 5 | pH 7 | pH 5 | ||
| Zero order | k0 | 20.75 | 26.993 | 18.261 | 22.354 |
| R2 | 0.7234 | 0.7701 | 0.6624 | 0.7835 | |
| First order | k1 | 1.89 | 1.86 | 1.909 | 1.89 |
| R2 | 0.8293 | 0.9293 | 0.7444 | 0.9035 | |
| Hixson–Crowell | kHC | 4.29 | 4.18 | 4.3341 | 4.26 |
| R2 | 0.7958 | 0.8841 | 0.7176 | 0.8674 | |
| Korsmeyer–Peppas | kKP | 1.14 | 1.21 | 1.0643 | 1.1519 |
| R2 | 0.9204 | 0.9737 | 0.8594 | 0.9766 | |
| n | 0.33 | 0.44 | 0.34 | 0.39 | |
| Diffusion coefficients | D (cm2 min−1) | 1.75E–15 | 5.08E–15 | 4.53E–14 | 1.36E–13 |
Abbreviations: DOX, doxorubicin; P407, poloxamer 407; TPGS, D-α-tocopheryl polyethylene glycol succinate; FA, folic acid.
Intracellular localization of P407/FA-P407-TPGS micelles
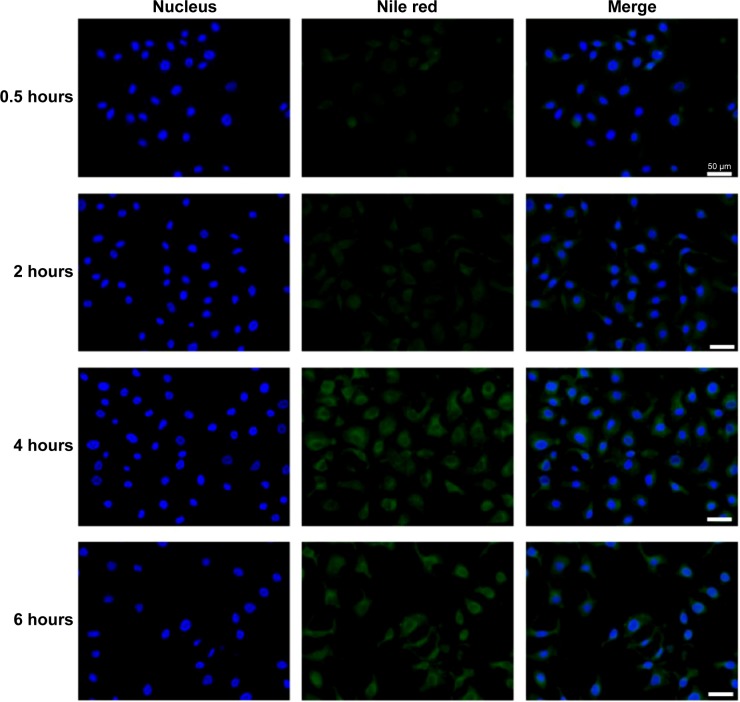
Intracellular trafficking of P407/FA-P407-TPGS-mixed micelles was studied in SKOV3 cells incubated with nile red-loaded micelles at different time points by fluorescence microscopy imaging (Figure 7). It was observed that as early as 30 minutes, the nile red fluorescence could be seen inside the cell and around nucleus. With increasing incubation times, the fluorescence intensity outside the cells gradually decreased, and the intensity of the nile red inside the cells increased with more localized fluorescence observed in the area surrounding nucleus and cytoplasm. It could be suggested that micelles enter and release the encapsulated drugs inside and maintain the drug concentration inside cells by reducing drug efflux.
Figure 7.
Nile red uptake.
Notes: SKOV3 cells were seeded on glass coverslips in 35 mm culture dishes. After 24 hours, cells were washed and treated with medium containing 0.5 μM nile red or equivalent concentration of nile red-loaded micelles. Nile red images were pseudo-colored green and merged with Hoechst 33342 images. Scale bars =50 μm.
Drug efflux in SKOV3 and SKOV3-DOX cells
Drug efflux pumps rapidly pump out the drug entering the cells rendering it ineffective which could result in failure of treatments. It was expected that TPGS reduces the drug resistance by altering drug efflux,35 and this could result in enhanced DOX effects. To assess this, SKOV3-DOX cell line was used as the cells overexpress Pgp, resulting in enhanced drug efflux and subsequently reduced DOX retention inside the cells. The control study, in which no treatment was given, showed maximum drug efflux and the least intracellular R123 levels of 1,773 ng/mg and 418 ng/mg protein, respectively. On the other hand, the intracellular R123 levels were increased by 1.43% and 24% in SKOV3 and SKOV3-DOX cells, respectively, when the cells were treated with P407 micelles alone. The small increase in the retention of R123 could have resulted from the enhanced retention of micelles inside the cells, as P407 did not show significant inhibition of drug efflux as compared to some other poloxamers. The presence of TPGS, however, resulted in a significant increase in R123 intracellular levels in both SKOV3 and SKOV3-DOX cells (14% and 264%). For both type of micelles, with or without the presence of FA, the study resulted in similar high retention of R123 and reduced efflux (Figure 8A). A significant reduction in drug efflux resulting in higher intracellular R123 levels was clearly observed in SKOV3-DOX cells, which was due to the inhibition of high drug efflux in the resistant cells. SKOV3 cell line, which would have the least expression of Pgp, showed an almost similar level of R123 efflux that further strengthened our hypothesis of reduced drug efflux by the mixed micelles resulting in enhanced drug retention inside the cells. The higher intracellular R123 levels observed in the case of P407/FA-P407-TPGS could be the result of the combined effects of inhibition of Pgp by TPGS and the enhanced drug uptake of the micelles. This synergistic effect could allow more drugs to be available inside the cells reducing the need for higher doses and in turn increasing the efficacy of the encapsulated drugs.
Figure 8.
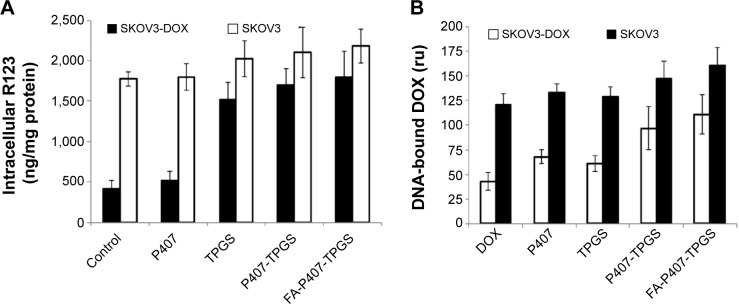
Drug efflux and DOX–DNA binding.
Notes: (A) R123 intracellular levels in both DOX-sensitive SKOV3 cells and SKOV3-DOX cells are shown after 2-hour treatment with TPGS, P407 alone, or P407-TPGS or FA-P407-TPGS micelles. (B) DOX binding with DNA in SKOV3 and SKOV3-DOX cells is shown as determined by fluorescence measurements.
Abbreviations: DOX, doxorubicin; R123, rhodamine 123; SKOV3-DOX, DOX-resistant SKOV3; TPGS, D-α-tocopheryl polyethylene glycol succinate; P407, poloxamer 407; FA, folic acid.
Enhancement of DOX binding with DNA
DOX has been shown to exert its cytotoxic effect as an intercalating agent. For this to occur, it has to enter the cells and subsequently into the nucleus to act on the topoisomerase enzyme.36 During this process, most of the drug is effluxed out of the cells. As the present micelles were able to reduce the drug efflux and increase retention of Pgp substrate R123 as described earlier, it would provide the drug (DOX) extended time to be localized close to the nucleus and greater chances to interact with topoisomerase and exert its cytotoxic action. This was confirmed by evaluating the subcellular localization of DOX inside the nucleus. The binding of DOX with DNA results in fluorescence quenching; however, this could be reversed by the addition of 0.1% Triton X-100 which displaces the drug from the DNA complex.29 SKOV3-DOX cells showed less DNA binding as compared to the SKOV3 cells which could be due to the increase in Pgp activity. Treatment of cells with P407 or TPGS resulted in slight increase in DOX–DNA binding compared to free DOX. Both P407-TPGS and FA-P407-TPGS micelles increased the DNA binding of DOX in SKOV3 cells by 21% and 33%, respectively, as shown in Figure 8B. In SKOV3-DOX cell line, the DOX– DNA binding increase was much more significant (125% and 158% by P407-TPGS and FA-P407-TPGS, respectively). This again was due to the increased time of exposure for DOX to interact with the DNA as well as enhanced drug uptake and retention inside the cells. This could perhaps also be due to the presence of multiple effects of reduced drug efflux as well as reduced entrapment inside the acidic vesicles which breakdown DOX.12,37
Enhancement of in vitro DOX cellular uptake
In order for DOX to exert its cytotoxic action as explained earlier, it is important to ensure the drug to be taken up inside the cells. Hence, cellular uptake studies of free DOX and DOX-loaded micelles were carried out in SKOV3 cells. It was observed that the uptake of DOX-loaded P407-TPGS-mixed micelles was comparatively higher compared to free DOX as shown in Figure 9. This enhanced uptake was perhaps because of the reduced efflux as well as the selective uptake due to the presence of TPGS.
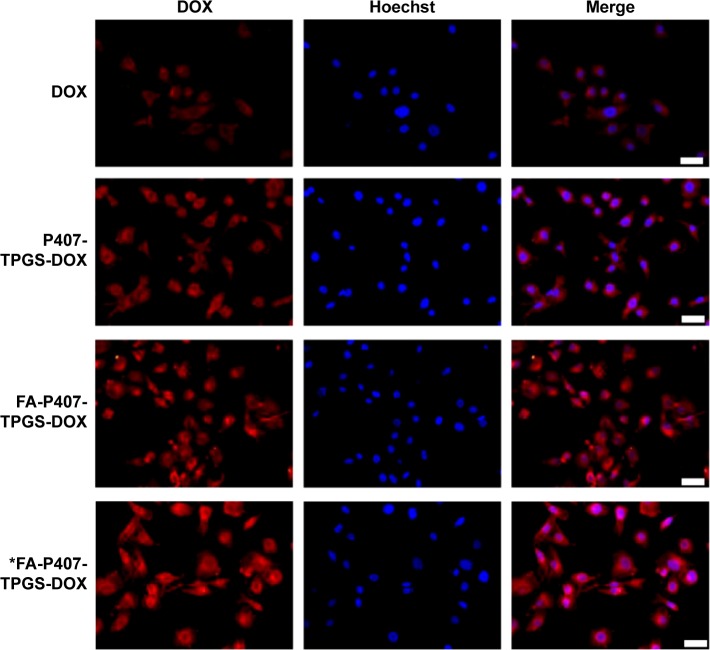
Figure 9.
DOX uptake.
Notes: SKOV3 cells cultured on glass coverslips were treated with DOX for 4 hours. Images were acquired and merged with Hoechst 33342 images. Asterisk in the last row refers to cells cultured in the FA-free medium. Scale bars =50 μm.
Abbreviations: DOX, doxorubicin; FA, folic acid; P407, poloxamer 407; TPGS, D-α-tocopheryl polyethylene glycol succinate.
The FA-mediated cellular uptake of DOX was also investigated in this study. FA has a high binding affinity (Kd ~10−10 m) for folate receptors.5 It has been shown that the FA-mediated uptake takes place by endocytosis,38 and it could be inhibited by the addition of FA into the in vitro cell culture. In order to investigate the effect of FA-mediated cellular uptake, SKOV3 cells were seeded in media containing 2 mM FA or FA-free medium. The results indicated that there was no significant difference in cellular uptake when P407-TPGS or FA-P407-TPGS-mixed micelles were used in the presence of FA in media. However, when there was no competing FA in the medium, it resulted in enhanced uptake of FA-conjugated micelles (Figure 9). This further confirmed that FA-conjugated micelles could be better taken up in the cells compared to micelles without FA conjugation. It was also observed that the uptake of free DOX and DOX-loaded P407-TPGS-mixed micelles was not influenced by the presence or absence of FA in the medium. A graphical representation of folate-mediated targeting of DOX-loaded FA-P407-TPGS-mixed micelles and passive transport across cancer cells is provided in Figure 1. The figure shows that free DOX enters the cells by diffusion process or passive transport, whereas DOX-loaded FA-P407-TPGS-mixed micelles enter the cells by FA ligand-mediated uptake. Once inside the cells, DOX is released and acts on the target site to cause cell death, whereas TPGS and P407 reduce drug efflux and enhance DOX–DNA binding. This selective and targeted uptake of DOX-loaded FA-P407-TPGS-mixed micelles results in enhanced cell death as compared to free DOX or nontargeted P407-TPGS micelles as described in the next section.
In vitro cytotoxicity of DOX-loaded P407/FA-P407-TPGS micelles
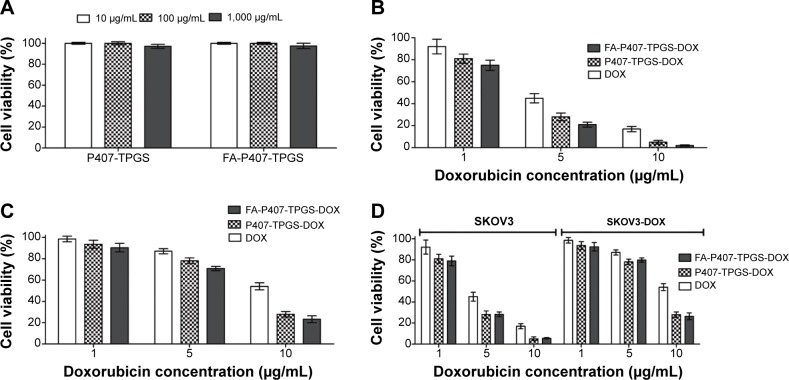
Both P407-TPGS and FA-P407-TPGS-mixed micelles exhibited minimal cytotoxicity for all the tested concentrations. More than 95% cells were viable at concentrations as high as 1 mg/mL after 24 hours of incubation in WRL-68 cells (Figure 10A). Cytotoxicity studies indicated that DOX-loaded micelles exhibited enhanced cytotoxicity compared to free DOX at similar concentrations (Figure 10B and C). It was also observed that cytotoxicity increased in the order of FA-P407-TPGS-DOX > P407-TPGS-DOX > DOX in both SKOV3 and SKOV3-DOX cell lines as shown by the decreasing IC50 values (Table 3). However, when FA-containing medium was used, no significant difference was observed among the DOX-loaded FA-P407-TPGS and P407-TPGS-mixed micelles due to the competition with free folate in the medium as shown in Figure 10D. It can be suggested that this enhanced cytotoxicity probably resulted from the combination of enhanced cellular uptake due to the activity of the micelles, selective cytotoxicity by TPGS, folate-mediated endocytosis, and reduced drug efflux by inhibition of Pgp.
Figure 10.
Cell viability of blank micelles and cytotoxicity of DOX and DOX-loaded micelles.
Notes: (A) Cell viability of WRL-68 cells after 24-hour treatment with P407-TPGS or FA-P407-TPGS micelles was determined by alamarBlue® cell viability assay. Cytotoxicity assessment of free DOX and DOX-loaded P407-TPGS or FA-P407-TPGS micelles by alamarBlue® assay is shown for (B) DOX-sensitive SKOV3 cells and (C) SKOV3-DOX cells. (D) Cytotoxicity of DOX or DOX-loaded micelles in SKOV3 and SKOV3-DOX cells in medium supplemented with 2 mM FA.
Abbreviations: DOX, doxorubicin; P407, poloxamer 407; TPGS, D-α-tocopheryl polyethylene glycol succinate; FA, folic acid; SKOV3-DOX, DOX-resistant SKOV3.
Table 3.
IC50 values of DOX in free or micelle form
| Cells | DOX (μM) | P407-TPGS-DOX (μM) | FA-P407-TPGS-DOX (μM) |
|---|---|---|---|
| SKOV3 | 7.7±0.4 | 4.8±0.5 | 3.6±0.4 |
| SKOV3-DOX | 17.5±1.3 | 6.1±0.7 | 5.7±0.8 |
Abbreviations: DOX, doxorubicin; P407, poloxamer 407; TPGS, D-α-tocopheryl polyethylene glycol succinate; FA, folic acid; SKOV3-DOX, DOX-resistant SKOV3.
Conclusion
In this study, FA-functionalized P407-TPGS-mixed micelles encapsulating DOX were successfully prepared. The micelles exhibited particle size of less than 200 nm and released DOX in a pH-dependent manner. The intracellular trafficking study of nile red-encapsulated P407/FA-P407-TPGS showed that the micelles entered and released the encapsulated drug inside the cells and resulted in enhanced concentration around the nucleus. It was also shown that P407/FA-P407-TPGS-mixed micelles reduced drug efflux activity, enhanced cellular uptake, and increased DNA binding of DOX. Furthermore, blank micelles showed minimal toxicity to normal cells, while DOX-loaded micelles had improved cytotoxicity effect against the SKOV3 and SKOV3-DOX cells as compared to free DOX. FA-P407-TPGS-DOX micelles offer a high potential as a targeted drug delivery system for DOX as a result of their multiple synergistic factors.
Acknowledgments
This work was carried out by support from research grant (GUP-SK-07-23-045) from Universiti Kebangsaan Malaysia and Science Fund (02-01-02-SF0738) from Ministry of Science, Technology and Innovation, Malaysia.
Footnotes
Disclosure
The authors declare that they have no competing interests.
References
- 1.Strebhardt K, Ullrich A. Paul Ehrlich’s magic bullet concept: 100 years of progress. Nat Rev Cancer. 2008;8(6):473–480. doi: 10.1038/nrc2394. [DOI] [PubMed] [Google Scholar]
- 2.Ashley CE, Carnes EC, Phillips GK, et al. The targeted delivery of multicomponent cargos to cancer cells by nanoporous particle-supported lipid bilayers. Nat Mater. 2011;10(5):389–397. doi: 10.1038/nmat2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nel AE, Mädler L, Velegol D, et al. Understanding biophysicochemical interactions at the nano-bio interface. Nat Mater. 2009;8(7):543–557. doi: 10.1038/nmat2442. [DOI] [PubMed] [Google Scholar]
- 4.Leamon CP, Reddy JA. Folate-targeted chemotherapy. Adv Drug Deliv Rev. 2004;56(8):1127–1141. doi: 10.1016/j.addr.2004.01.008. [DOI] [PubMed] [Google Scholar]
- 5.Yu MK, Park J, Jon S. Targeting strategies for multifunctional nanoparticles in cancer imaging and therapy. Theranostics. 2012;2(1):3–44. doi: 10.7150/thno.3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bae YH, Park K. Targeted drug delivery to tumors: myths, reality and possibility. J Control Release. 2011;153(3):198–205. doi: 10.1016/j.jconrel.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liechty WB, Kryscio DR, Slaughter BV, Peppas NA. Polymers for drug delivery systems. Annu Rev Chem Biomol Eng. 2010;1:149–173. doi: 10.1146/annurev-chembioeng-073009-100847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anbharasi V, Cao N, Feng S-S. Doxorubicin conjugated to D-α-tocopheryl polyethylene glycol succinate and folic acid as a prodrug for targeted chemotherapy. J Biomed Mater Res A. 2010;94A(3):730–743. doi: 10.1002/jbm.a.32734. [DOI] [PubMed] [Google Scholar]
- 9.Zhang C, Pan D, Luo K, et al. Dendrimer-doxorubicin conjugate as enzyme-sensitive and polymeric nanoscale drug delivery vehicle for ovarian cancer therapy. Polym Chem. 2014;5(18):5227–5235. [Google Scholar]
- 10.Shi Y, Su C, Cui W, et al. Gefitinib loaded folate decorated bovine serum albumin conjugated carboxymethyl-beta-cyclodextrin nanoparticles enhance drug delivery and attenuate autophagy in folate receptor-positive cancer cells. J Nanobiotechnology. 2014;12(1):43. doi: 10.1186/s12951-014-0043-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Amjad MW, Mohd Amin MCI, Mahali SM, et al. Nanoscale diblock copolymer micelles: characterizations and estimation of the effective diffusion coefficients of biomolecules release through cylindrical diffusion model. PLoS One. 2014;9(8):e105234. doi: 10.1371/journal.pone.0105234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alakhov V, Klinski E, Li S, et al. Block copolymer-based formulation of doxorubicin. From cell screen to clinical trials. Colloids Surf B. 1999;16:113–134. [Google Scholar]
- 13.Kabanov AV, Batrakova EV, Alakhov VY. Pluronic® block copolymers for overcoming drug resistance in cancer. Adv Drug Deliv Rev. 2002;54(5):759–779. doi: 10.1016/s0169-409x(02)00047-9. [DOI] [PubMed] [Google Scholar]
- 14.Gombotz WR, Guanghui W, Horbett TA, Hoffman AS. Protein adsorption to poly(ethylene oxide) surfaces. J Biomed Mater Res. 1991;25(12):1547–1562. doi: 10.1002/jbm.820251211. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Z, Huey Lee S, Feng S-S. Folate-decorated poly(lactide-co-glycolide)-vitamin E TPGS nanoparticles for targeted drug delivery. Biomaterials. 2007;28(10):1889–1899. doi: 10.1016/j.biomaterials.2006.12.018. [DOI] [PubMed] [Google Scholar]
- 16.Li P-Y, Lai P-S, Hung W-C, Syu W-J. Poly(L-lactide)-vitamin E TPGS nanoparticles enhanced the cytotoxicity of doxorubicin in drug-resistant MCF-7 breast cancer cells. Biomacromolecules. 2010;11(10):2576–2582. doi: 10.1021/bm1005195. [DOI] [PubMed] [Google Scholar]
- 17.Butt AM, Mohd Amin MCI, Katas H, Sarisuta N, Witoonsaridsilp W, Benjakul R. In vitro characterization of pluronic F127 and D-α-tocopheryl polyethylene glycol 1000 succinate mixed micelles as nanocarriers for targeted anticancer-drug delivery. J Nanomater. 2012;2012:11. [Google Scholar]
- 18.Xu P, Yin Q, Shen J, et al. Synergistic inhibition of breast cancer metastasis by silibinin-loaded lipid nanoparticles containing TPGS. Int J Pharm. 2013;454(1):21–30. doi: 10.1016/j.ijpharm.2013.06.053. [DOI] [PubMed] [Google Scholar]
- 19.Amjad MW, Mohd Amin MCI, Katas H, Butt AM. Doxorubicin-loaded cholic acid-polyethyleneimine micelles for targeted delivery of antitumor drugs: synthesis, characterization, and evaluation of their in vitro cytotoxicity. Nanoscale Res Lett. 2012;7(1):687. doi: 10.1186/1556-276X-7-687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Corona G, Giannini F, Fabris M, Toffoli G, Boiocchi M. Role of folate receptor and reduced folate carrier in the transport of 5-methyltetrahydrofolic acid in human ovarian carcinoma cells. Int J Cancer. 1998;75(1):125–133. doi: 10.1002/(sici)1097-0215(19980105)75:1<125::aid-ijc19>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 21.Pal S, Ghosh Roy S, De P. Synthesis via RAFT polymerization of thermo- and pH-responsive random copolymers containing cholic acid moieties and their self-assembly in water. Polym Chem. 2014;5(4):1275–1284. [Google Scholar]
- 22.Möckel J, Lippold B. Zero-order drug release from hydrocolloid matrices. Pharm Res. 1993;10(7):1066–1070. doi: 10.1023/a:1018931210396. [DOI] [PubMed] [Google Scholar]
- 23.Gibaldi M, Feldman S. Establishment of sink conditions in dissolution rate determinations. Theoretical considerations and application to nondisintegrating dosage forms. J Pharm Sci. 1967;56(10):1238–1242. doi: 10.1002/jps.2600561005. [DOI] [PubMed] [Google Scholar]
- 24.Hixson AW, Crowell JH. Dependence of reaction velocity upon surface and agitation. Ind Eng Chem Res. 1931;23(8):923–931. [Google Scholar]
- 25.Korsmeyer RW, Gurny R, Doelker E, Buri P, Peppas NA. Mechanisms of solute release from porous hydrophilic polymers. Int J Pharm. 1983;15(1):25–35. doi: 10.1002/jps.2600721021. [DOI] [PubMed] [Google Scholar]
- 26.Chouhan R, Bajpai A. Real time in vitro studies of doxorubicin release from PHEMA nanoparticles. J Nanobiotechnology. 2009;7(1):5. doi: 10.1186/1477-3155-7-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fontaine M, Elmquist WF, Miller DW. Use of rhodamine 123 to examine the functional activity of P-glycoprotein in primary cultured brain microvessel endothelial cell monolayers. Life Sci. 1996;59(18):1521–1531. doi: 10.1016/0024-3205(96)00483-3. [DOI] [PubMed] [Google Scholar]
- 28.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72(1–2):248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 29.Goto S, Ihara Y, Urata Y, et al. Doxorubicin-induced DNA intercalation and scavenging by nuclear glutathione S-transferase π. FASEB J. 2001;15(14):2702–2714. doi: 10.1096/fj.01-0376com. [DOI] [PubMed] [Google Scholar]
- 30.Wan A, Sun Y, Li H. Characterization of folate-graft-chitosan as a scaffold for nitric oxide release. Int J Biol Macromol. 2008;43(5):415–421. doi: 10.1016/j.ijbiomac.2008.07.016. [DOI] [PubMed] [Google Scholar]
- 31.Matias R, Ribeiro PRS, Sarraguca MC, Lopes JA. A UV spectrophotometric method for the determination of folic acid in pharmaceutical tablets and dissolution tests. Anal Methods. 2014;6(9):3065–3071. [Google Scholar]
- 32.Sun Y, Zou W, Bian S, et al. Bioreducible PAA-g-PEG graft micelles with high doxorubicin loading for targeted antitumor effect against mouse breast carcinoma. Biomaterials. 2013;34:6818–6828. doi: 10.1016/j.biomaterials.2013.05.032. [DOI] [PubMed] [Google Scholar]
- 33.Dalmark M, Storm HH. A Fickian diffusion transport process with features of transport catalysis. J Gen Physiol. 1981;78:349–364. doi: 10.1085/jgp.78.4.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao X, Peng Liu P. Reduction-responsive core-shell-corona micelles based on triblock copolymer: novel synthetic strategy, characterization, and application as tumor microenvironment-responsive drug delivery system. ACS Appl Mater Interfaces. 2014;7(1):166–174. doi: 10.1021/am505531e. [DOI] [PubMed] [Google Scholar]
- 35.Zhao S, Tan S, Guo Y, et al. pH-sensitive docetaxel-loaded D-α-tocopheryl polyethylene glycol succinate-poly(β-amino ester) copolymer nanoparticles for overcoming multidrug resistance. Biomacromolecules. 2013;14(8):2636–2646. doi: 10.1021/bm4005113. [DOI] [PubMed] [Google Scholar]
- 36.Liu H, Qiao C, Yang J, Weng J, Zhang X. Self-assembling doxorubicin-prodrug nanoparticles as siRNA drug delivery system for cancer treatment: in vitro and in vivo. J Mater Chem B. 2014;2:5910. doi: 10.1039/c4tb00814f. [DOI] [PubMed] [Google Scholar]
- 37.Benderra Z, Morjani H, Trussardi A, Manfait M. Characterization of H+-ATPase-dependent activity of multidrug resistance-associated protein in homoharringtonine-resistant human leukemic K562 cells. Leukemia. 1998;12:1539–1544. doi: 10.1038/sj.leu.2401166. [DOI] [PubMed] [Google Scholar]
- 38.Kelemen LE. The role of folate receptor a in cancer development, progression and treatment: cause, consequence or innocent bystander? Int J Cancer. 2006;119:243–250. doi: 10.1002/ijc.21712. [DOI] [PubMed] [Google Scholar]