Abstract
Background
Mitochondrial Transcription Factor A (TFAM) is regarded as a histone-like protein of mitochondrial DNA (mtDNA), performing multiple functions for this genome. Aging affects mitochondria in a tissue-specific manner and only calorie restriction (CR) is able to delay or prevent the onset of several age-related changes also in mitochondria.
Methods
Samples of the frontal cortex and soleus skeletal muscle from 6- and 26-month-old ad libitum-fed and 26-month-old calorie-restricted rats and of the livers from 18- and 28-month-old ad libitum-fed and 28-month-old calorie-restricted rats were used to detect TFAM amount, TFAM-binding to mtDNA and mtDNA content.
Results
We found an age-related increase in TFAM amount in the frontal cortex, not affected by CR, versus an age-related decrease in the soleus and liver, fully prevented by CR. The semi-quantitative analysis of in vivo binding of TFAM to specific mtDNA regions, by mtDNA immunoprecipitation assay and following PCR, showed a marked age-dependent decrease in TFAM-binding activity in the frontal cortex, partially prevented by CR. An age-related increase in TFAM-binding to mtDNA, fully prevented by CR, was found in the soleus and liver. MtDNA content presented a common age-related decrease, completely prevented by CR in the soleus and liver, but not in the frontal cortex.
Conclusions
The modulation of TFAM expression, TFAM-binding to mtDNA and mtDNA content with aging and CR showed a trend shared by the skeletal muscle and liver, but not by the frontal cortex counterpart.
General significance: Aging and CR appear to induce similar mitochondrial molecular mechanisms in the skeletal muscle and liver, different from those elicited in the frontal cortex.
Keywords: Mitochondrial Transcription Factor A, Mitochondrial Transcription Factor, A–mitochondrial deoxyribonucleic acid binding, Tissue-specificity, Aging rat, Calorie restriction
1. Introduction
Mitochondrial Transcription Factor A (TFAM) is a High Mobility Group protein, encoded by nuclear DNA and mainly directed to mitochondria [1], which was discovered and characterized initially for its function in mitochondrial transcription [2]. Several subsequent studies revealed a number of different roles performed by TFAM in the organ-elle [3,4]. Because of the close connection between mitochondrial DNA (mtDNA) transcription and replication [5], TFAM has become an attractive candidate for the regulation of mtDNA copy number [6]. Furthermore, TFAM is involved in the constitution of mtDNA nucleoids [7] and appears to be a likely member of a system responsible for sensing and repair of oxidative damage to mtDNA [8,9], so that it exerts histone-like functions for mtDNA. Due to the relevant involvement in the regulation of mitochondrial biogenesis and transcription, TFAM expression has been investigated in several rat tissues [10] in different physiological conditions, including aging (liver, heart, cerebellum, kidney [11,12], hind-limb muscles [13] and frontal cortex [14]). Aging is a very complex phenomenon that implies the progressive structural and functional decline of tissues also through the age-dependent dysfunction of the mitochondrial respiratory complexes, reducing the amount of synthesized adenosine triphosphate (ATP) [15]. The involvement of mitochondria in aging is tissue-specific [16] and is particularly relevant within tissues such as the brain, heart and skeletal muscle that have a high dependence on oxidative metabolism [17]. However, other metabolically very active tissues, such as the liver, are also affected by age-related mitochondrial dysfunction [15]. Another relevant and common feature of aging is an increased presence of reactive oxygen species (ROS), mainly by-products of the mitochondrial respiratory complexes, resulting in the onset of oxidative stress. This originates oxidative damage to mitochondrial proteins, lipids and DNA and contributes to the aging phenotype [18]. In particular, mtDNA can undergo several kinds of qualitative and quantitative alterations that affect its structure and function. Quantitative alterations, namely changes in mtDNA content, were reported in various tissues from humans and animals [11–14, 19–23]. To date, the only established experimental approach shown to delay or prevent the onset of several age-related alterations, also in mitochondria, in organisms ranging from yeast to man, is calorie restriction (CR) [24,25]. Such treatment involves the administration of a nutrient dense diet that reduces calorie intake by 40% [26] and exhibits a very marked tissue-specificity [16]. Among the likely multiple cellular mechanisms at the basis of CR efficacy, the reduction of age-related oxidative stress has obtained large credit [25,27]. Recently, however, the regulation of mitochondrial metabolism through activation of the PGC-1α-dependent cascade of mitochondrial biogenesis has also gained a wide consensus as one of the means by which CR counteracts the age-dependent dysfunction [28]. Little is known about the effects of CR on mitochondrial age-related phenotypic and genotypic alterations [29–33]. The tissue-specific effects of CR on mtDNA content [12,14,21] and TFAM amount [12,14] in aged rats have been reported. TFAM is a key factor in mtDNA replication and maintenance and the study of its possible role in mtDNA protection and repair from oxidative damage appears very interesting. Therefore, given the importance of TFAM-binding to mtDNA for various mitochondrial functions, it was very appealing for us to determine and compare TFAM amount and its binding to functionally relevant regions of mtDNA as well as mtDNA content during aging and CR in various rat tissues. We analyzed frontal cortex, soleus skeletal muscle and liver, featuring a distinct dependence on oxidative metabolism, with the aim of unveiling eventual tissue-specific differences in a situation of physiological oxidative stress, such as aging, and in CR. Our results demonstrated, as for the regulation of TFAM amount and mtDNA content and the modulation of TFAM-binding to mtDNA, a trend that was shared by the skeletal muscle and the liver but not by the frontal cortex counterpart.
2. Materials and methods
2.1. Samples
The study was approved by the Institutional Animal Care and Use Committee at the University of Florida. All procedures were performed in accordance with the National Institutes of Health (NIH) guidelines for the care and use of laboratory animals. The frontal cortex, soleus skeletal muscle and liver samples used in this study were, respectively, from Fischer 344 male rats obtained from the NIH barrier-raised rodent colony (frontal cortex and soleus muscle) or from Fischer 344 × Brown Norway (F344BNF1) male rats obtained from the National Institute of Aging colony (Indianapolis, IN) (liver) and housed at the Department of Aging and Geriatric Research, Division of Biology of Aging, College of Medicine, University of Florida, Gainesville, FL. The animals consisted of the following groups: 6-month-old ad libitum-fed (AL-6, n = 8 for frontal cortex; n = 5 for soleus muscle), 26-month-old ad libitum-fed (AL-26, n = 7 for frontal cortex; n = 6 for soleus muscle), and 26-month-old calorie-restricted (CR-26, n = 7 for frontal cortex; n = 5 for soleus muscle); 18-month-old ad libitum-fed (AL-18, n = 6 for liver), 28-month-old ad libitum-fed (AL-28, n = 8 for liver) and 28-month-old calorie-restricted (CR-28, n = 7 for liver) rats. CR had been initiated at 3.5 months of age (10% restriction), raised to 25% restriction at 3.75 months, and kept at 40% restriction from 4 months until the end of each animal's life. The animals were anesthetized before being sacrificed and samples from the frontal cortex, soleus muscle, and liver were immediately removed, snap-frozen in isopentane cooled by liquid nitrogen, and stored in liquid nitrogen until further analysis.
2.2. Western blotting
Total proteins were extracted from all tissue samples obtained from AL-6, AL-18, AL-26, AL-28, CR-26, and CR-28 animals. Approximately 100 mg of each frozen sample was grounded and suspended in 600 μl of lysis buffer (220 mM mannitol, 70 mM sucrose, 20 mM Tris–HCl pH 7.4, 5 mM MgCl2, 5 mM EGTA, and 1 mM EDTA). Cell lysates were pre-cleared by centrifugation in an Eppendorf microfuge at 12,000 rpm for 10 min and the supernatant fraction containing proteins was recovered. Proteins were quantified with the Bradford method (Bio-Rad Laboratories Inc., Hercules, CA, USA) according to the supplier's instructions. Total proteins (10 μg) were separated by gel electrophoresis on 4–12% Bis–Tris Criterion XT precast gels (Bio-Rad Laboratories Inc., Hercules, CA, USA) and electroblotted onto PVDF membranes (Amersham-Pharmacia Biotech Inc., Piscataway, NJ, USA). The membranes were blocked for 1 h in 5% milk in 1×-PBS/Tween 20 (0.15 M NaCl, 0.1 mM KH2PO4, 3 mM Na2HPO4, 0.1% Tween 20) and probed with TFAM (1:25,000) and β-actin (1:10,000; Sigma-Aldrich Corp., St. Louis, MO, USA). The antibody against TFAM was custom-made using as antigen in rabbit the protein expressed from the clone containing the peptide fraction corresponding to aa 35 to 201 of the rat protein and donated by Dr. H. Hinagaki (Department of Chemistry, National Industrial Research Institute of Nagoya, Japan). Membranes were then incubated with anti-rabbit secondary antibody conjugated with horseradish peroxidase (HRP) at a dilution of 1:10,000 (Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA). Membranes were washed in PBS (3 × 15 min) and proteins were subsequently visualized with an enhanced chemiluminescence kit (ECL-Plus; Amersham-Pharmacia Biotech Inc., Piscataway, NJ, USA). Autoradiographs were analyzed by laser densitometry with the Chemi Doc System and Quantity One software (Bio-Rad Laboratories Inc., Hercules, CA, USA). The densitometric value of OD units of each TFAM band was then related to the OD unit number of the respective band of the β-actin (in the corresponding lane).
2.3. Mitochondrial DNA immunoprecipitation (mIP)
The binding in vivo of TFAM to specific regions of mtDNA was analyzed using mitochondrial DNA immunoprecipitation (mIP) following the procedure described by Picca et al. [14]. 250 μl of a cold formalde-hyde cross-linking solution 1× (1% formaldehyde, 5 mM HEPES pH 8.0, 10 mM NaCl, 0.1 mM EDTA pH 8.0, 0.05 mM EGTA) was added to 100 mg of frozen tissue. Cross-linking was terminated by adding 0.125 M glycine and incubating at room temperature for 10 min. Tissues were briefly homogenized and centrifuged twice at 3000 rpm for 5 min. Each pellet was washed with 1 ml of RB (Resuspending Buffer, 135 mM NaCl, 10 mM Na2HPO4, 2.7 mM KCl, 1.8 mM KH2PO4 pH 7.4), suspended with 1 ml of homogenization buffer (100 mM Tris–HCl pH 8.0, 100 mM NaCl, 30 mM MgCl2, 0.1% NP-40, 0.1 mM PMSF), and manually homogenized. The sample was centrifuged at 3000 rpm for 5 min and the pellet was incubated in lysis buffer (0.5% Triton X-100, 300 mM NaCl, 50 mM Tris–HCl pH 7.4, containing 100 μg/ml leupeptin and 200 μM PMSF), for 10 min at RT. Cellular DNA was sheared by sonication and the size of DNA fragments, ranging between 500 and 900 base pairs, was checked by electrophoresis on an ethidium bromide-stained 1.2% agarose gel in 1× TAE buffer (20 mM Tris acetate, 50 mM EDTA pH 8.3). Each sample was diluted with FSB buffer (5 mM EDTA, 20 mM Tris–HCl pH 7.5, 50 mM NaCl) and pre-cleared with 75 μl of protein A-agarose/Salmon sperm 50% DNA (Millipore Corporate Headquarters, Billerica, MA, USA) for 2 ml of sample on a rotator at 4 °C for 30 min. After a centrifugation at 1000 rpm for 1 min, each sample was divided into four aliquots: the first one (100 μl) was not immunoprecipitated (input, stored at −80 °C until further use); the other three aliquots (100 μl each) were incubated overnight at 4 °C, respectively, with a rabbit anti-TFAM antibody (1:50 dilution), a non-specific rabbit anti-β-actin antibody (1:100 dilution) and without antibody (−Ab). On the following day, 15 μl of protein A-agarose was added to each sample for 2 h at 4 °C to isolate protein–DNA complexes. The samples were centrifuged at 1000 rpm for 1 min, and the pellets were washed: three times with 1 ml of 140 mM NaCl RIPA buffer (10 mM Tris–HCl pH 8.0, 1% Triton X-100, 0.1% SDS, 0.1% deoxycholate, 1 mM EDTA, 140 mM NaCl); three times with 1 ml of 500 mM NaCl RIPA buffer (as above but with 500 mM NaCl); three times with 1 ml of LiCl buffer (10 mM Tris–HCl pH 8.0, 0.25 M LiCl, 0.5% NP-40, 0.5% deoxycholate, 1 mM EDTA) and twice with 1 ml of TE (10 mM Tris–HCl pH 8.0, 1 mM EDTA). Each wash lasted 5 min with rotation at 4 °C, followed by centrifugation at 1000 rpm for 1 min. The last pellets, suspended with TE containing 0.5% SDS (200 μl), together with the previously stored input, were incubated at 65 °C for 6 h for the thermal reversal of the cross-links. The DNA samples treated with specific antibodies, the −Ab samples, and the input DNAs were all precipitated by adding: 20 μl of 3 M Na-acetate pH 5.2, 1 μl of 20 μg/μl glycogen, and 500 μl of cold absolute ethanol. After precipitation at −80 °C overnight, the samples were centrifuged at 13,000 rpm for 15 min at RT, washed in 300 μl of cold 70% ethanol, centrifuged at 13,000 rpm for 5 min at RT, dried and suspended in 200 μl of sterile ultrapure H2O. Ten μg of RNase A (50 μg/μl) was added to each sample and incubated for 1 h at 37 °C. Twenty μg of proteinase K (100 μg/μl) and SDS (0.25% final concentration) was added and incubated at 37 °C overnight. An extraction with phenol/chloroform/isoamyl alcohol (25:24:1) and one with chloroform/isoamyl alcohol (24:1) were performed. All DNAs were precipitated overnight at −80 °C with 20 μg of glycogen, Na-acetate pH 5.2 (0.3 M final concentration) and 2 volumes of cold absolute ethanol. On the next day, all the samples were centrifuged at 12,000 rpm for 20 min at 4 °C, washed in 300 μl of cold 70% ethanol, centrifuged at 12,000 rpm for 10 min at 4 °C, dried, and suspended in 60 μl of 10 mM Tris–HCl pH 8.0. Input and mIP mtDNAs were subjected to PCR analysis using three or five primer pairs listed in Table 1 that were designed to include, respectively, a part of the D-loop with the OriH origin of replication; the Ori-L origin together with a portion of the CO I gene; a part of the cytochrome b gene; a portion of the CO II gene upstream of the direct repeat 1 of the 4.8 kb deletion and a part of the ATPase 6 gene containing the direct repeat 1 of the same 4.8-kb deletion. PCR reactions contained 20 pmol of each primer in a 25 μl mixture (200 μM dNTPs, 1.5 mM MgCl2, 1× Taq Buffer and 1 U Taq Polymerase by Roche, F. Hoffmann-La Roche, Basel, Switzerland) using a Mastercycler PCR (Eppendorf Scientific, Hinz GmbH, Hamburg, Germany). Amplification conditions were: one cycle at 94 °C for 5 min followed by 30 cycles at 94 °C for 1 min, 58 °C for 1 min, 72 °C for 1 min, and then finally one cycle at 72 °C for 5 min. Reactions were analyzed on an ethidium bromide-stained 1.3% agarose gel in 1× TAE buffer. The intensity of each DNA band visualized on agarose gels was analyzed by densitometry and used for detection of specific TFAM-binding by subtracting the value of the intensity of the aliquot precipitated without primary antibody from that of the TFAM-immunoprecipitated aliquot, both normalized to the value of the respective input aliquot made equal to 1. Such calculated value was compared with the intensity of the non-specific β-actinimmunoprecipitated aliquot that resulted always below 1% of that with anti-TFAM antibody.
Table 1.
Oligonucleotide primer sequences.
| Primer set | Forward primer | Reverse primer | (nps) | (nps) |
|---|---|---|---|---|
| Primers for PCR of mIP | ||||
| 7.1 For/7.3 Rev | 5′-CACTAATACTAACAACAAAACrAAC-3′ | 5′-GGGATTATGTAGGAGTCAAAGC-3′ | (7133–7157) | (7376–7355) |
| 8.0 For/8.2 Rev | 5′-CAACCGACTACACTCATTTCAAC-3′ | 5′-CTCATAGGGGGATGGCTATGC-3′ | (8035–8057) | (8246–8226) |
| 15.0 For/15.2 Rev | 5′-GAGTCGTAGCCCTAATCTTATC-3′ | 5′-GAATGAGGATAATTGAAAAGTAGC-3′ | (15,004–15,025) | (15,228–15,205) |
| 16.0 For/16.2 Rev | 5′-GCTCGAAAGACTATTTTATTCATG-3′ | 5′-GCTAAGATTTAAGTTAAAATTTTGTG-3′ | (16,015–16,038) | (16,226–16,201) |
| 5.0 For/5.3 Rev | 5′-GGATTCAAACCTACGAAAATTTAG-3′ | 5′-GTGGTTAGTTGAAAAGAGTCAAC-3′ | (5092–5115) | (5358–5336) |
| Primers for RT-PCR | ||||
| MtDNA For/mtDNA Rev | 5′-GGTTCTTACTTCAGGGCCATCA-3′ | 5′-TGATTAGACCCGTTACCATCGA-3′ | (15,785–15,806) | (15,868–15,847) |
| Beta-actin For/beta-actin Rev | 5′-CCCAGCCATGTACGTAGCCA-3′ | 5′-CGTCTCCGGAGTCCATCAC-3′ | (2181–2200) | (2266–2248) |
2.4. Determination of mtDNA content
MtDNA content was measured using quantitative real time polymer-ase chain reaction (qRT-PCR). RT-PCR reactions were performed via SYBR Green chemistry on an ABI PRISM 7000 Sequence Detection System (Applied Biosystems, Foster City, CA, USA). The primers were specific, respectively, for the rat mitochondrial D-loop region (numbering is according to GenBank™ accession number AY172581) and for the rat nuclear β-actin gene (numbering is according to GenBank™ accession number VO1217.1) and are listed in Table 1. The method has been validated by primer-limiting experiments and by evaluating the equal reaction efficiency of the two amplicons. Amplification specificity was controlled by melting curve analysis and gel electrophoresis. Each sample was analyzed in triplicate in 25 μl of final volume containing: iTaq SYBR Green Supermix PCR 1× Master Mix (Bio-Rad Laboratories Inc., Hercules, CA, USA), 0.2 μM forward and reverse primers, and DNA template (2.5 μl of diluted 1:10). After 10 min of denaturation at 95 °C, amplification proceeded for 40 cycles, each consisting of denaturation at 95 °C for 15 s, annealing, and extension at 60 °C for 1 min. The quantification of the relative mtDNA content in aged AL and CR rats, compared to young or middle-age rats, all normalized to β-actin, was performed according to the Pfaffl mathematical model [34].
2.5. Statistical analysis
The statistical significance of differences between groups of animals was assessed by analysis of variance (ANOVA) using Tukey's Honestly Significant Difference (HSD) post-hoc test with the SPSS Base 11.5 software (SPSS Inc., Chicago, IL, USA). Statistical significance for all tests was set at p < 0.05.
3. Results
3.1. TFAM in the frontal cortex, soleus and liver
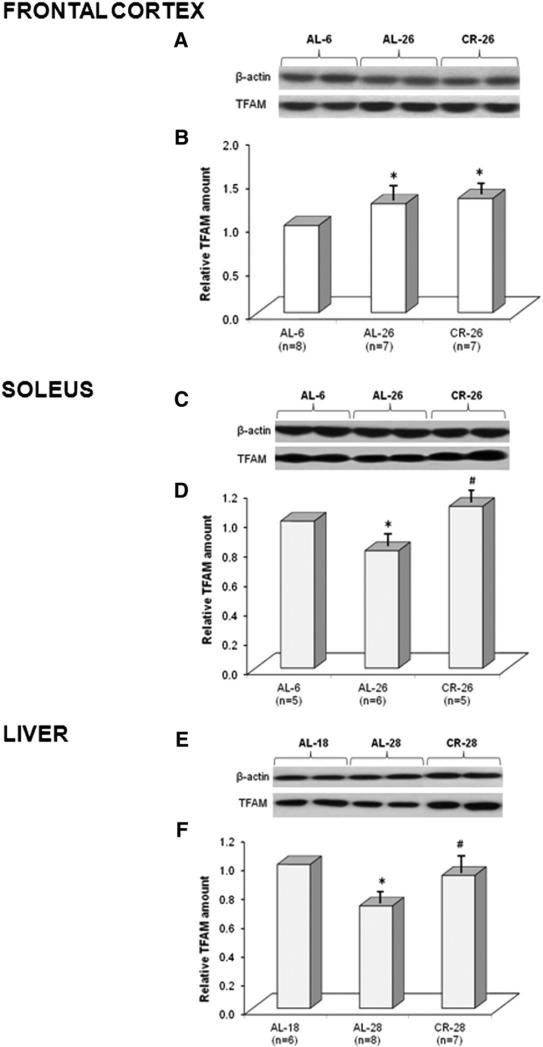
The amount of TFAM protein in the frontal cortex of ad libitum-fed (AL), 6- and 26-month-old (AL-6, AL-26), and calorie-restricted (CR), 26-month-old (CR-26), rats was measured by Western blot experiments, using β-actin as the internal standard reference for loading equal amounts of total proteins in each sample lane. Therefore, the densitometric value of OD units of the TFAM band was related to the number of OD units of the respective band of the β-actin for each analyzed sample. As reported in Fig. 1(A, B) we found a statistically significant, age-related increase (+25%) in the TFAM amount in the AL-26 animals with respect to the young AL-6 counterparts. CR-26 rats exhibited a statistically significant increase (+30%), compared to the AL-6 value, but not different from that of the AL-26 animals. We also determined, by Western blot experiments, the amount of TFAM in the soleus skeletal muscle samples from AL-6, AL-26, and CR-26 animals. An age-related, statistically significant decrease (−20%) in TFAM was found in the AL-26 rats (Fig. 1C, D). Such a decrease was completely prevented by CR because the amount of TFAM in the CR-26 rats was not statistically different from that of the young AL-6 ones, supporting the preventive effect of the CR. The amount of TFAM in the livers from three groups of animals, respectively, 18-month-old ad libitum-fed (AL-18), 28-month-old ad libitum-fed (AL-28), and 28-month-old calorie-restricted (CR-28) rats, was also determined by Western blot experiments (Fig. 1E, F). An age-related, statistically significant decrease (−29%) in the TFAM amount was found in the aged AL-28 group with respect to the younger age AL-18 counterpart. Such a decrease was completely prevented by CR because the amount of TFAM in the CR-28 rats was higher and statistically different from that of the AL-28 group, whereas it did not statistically differ from that of the AL-18 animals. Considering the tissue-specific changes of TFAM amount in relationship to aging and CR, we decided to broaden our study by analyzing the in vivo TFAM-binding activity to specific mtDNA regions (Fig. 2) by using the mtDNA immunoprecipitation (mIP) technique.
Fig. 1.
Age- and CR-related changes of TFAM amount in three rat tissues. Representative Western blot carried out in (A) the frontal cortex, (C) the soleus skeletal muscle, and (E) the liver. The bands from top to bottom show, respectively, the signals from β-actin and TFAM. (B) The histogram shows the relative amount of TFAM in AL-26 and CR-26 rats, compared to AL-6 rats, all normalized with respect to β-actin in frontal cortex samples. Bars represent the mean (±SD) of the values obtained, respectively, from analysis in triplicate of the protein extract, from each young and aged AL and CR-treated rat. *p < 0.05 versus the value of the AL-6 rats (fixed as 1); n, number of analyzed animals. (D) The histogram shows the results in the soleus samples. The details of the legend are as in (B). (F) The histogram shows the relative amount of TFAM in AL-28 and CR-28 rats, compared to that in AL-18 rats, all normalized with respect to β-actin in liver samples. *p < 0.05 versus the value of the AL-18 rats (fixed as 1); n, number of analyzed animals.
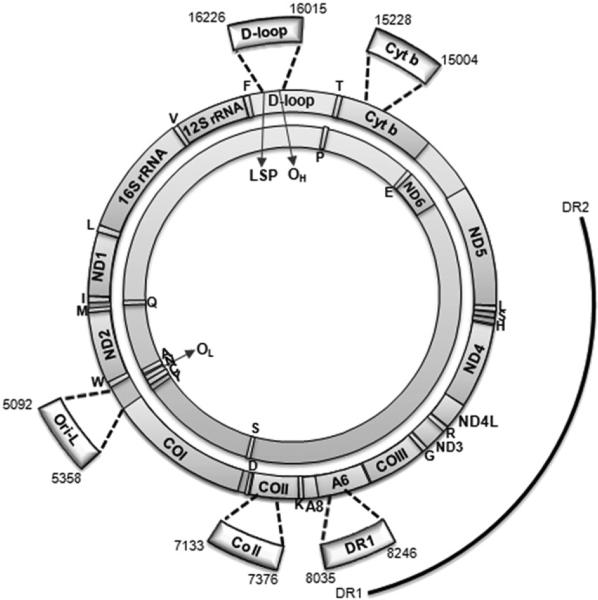
Fig. 2.
Genetic map of rat mtDNA. Rat mtDNA consists of a heavy-strand (outer circle) and a light-strand (inner circle) that code for the 12S and 16S rRNA and 22 tRNA genes (single-letter codes indicated under the corresponding tRNA genes) and subunits of the oxidative phosphorylation complexes: NADH dehydrogenase subunits (ND1–6 and ND4L), cyto-chrome b (Cyt b), cytochrome c oxidase subunits (CO I–III), and ATP synthase subunits (A6, A8). Also represented are: the origin of replication of the H strand (OH), the origin of replication of the L strand (OL), and the promoter of the L strand transcription (LSP). The thick arches extending out of the circles represent the five mIP amplified regions delimited, respectively, by the primer pairs: 16.0 For/16.2 Rev (D-loop), 5.0 For/5.3 Rev (Ori-L), 7.1 For/7.3 Rev (CO II), 8.0 For/8.2 Rev (DR1), and 15.0 For/15.2 Rev (Cyt b), listed in Table 1, and corresponding to the indicated nucleotides. Numbering is according to GenBank™ accession number AY172581. The outmost arch represents the 4.8-kb deletion, delimited by direct repeat 1 and 2 (DR1, DR2).
3.2. Frontal cortex mtDNA immunoprecipitation (mIP)
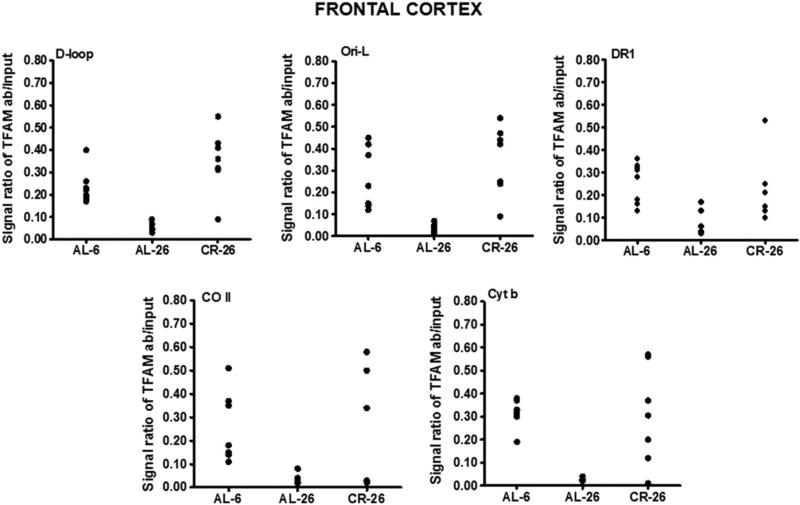
To determine whether aging and CR may influence the in vivo binding of TFAM to specific regions of mtDNA in the frontal cortex, we carried out mIP experiments. We analyzed TFAM binding to five regions: 1) a part of the D-loop with the OriH origin of replication (np 16,015–16,226); 2) the Ori-L origin together with a portion of the CO I gene (np 5092–5358); 3) a part of the cytochrome b gene (np 15,004– 15,228); 4) a portion of the CO II gene (np 7133–7376); and 5) a sequence containing the direct repeat 1 (DR1) (np 8035–8246) of the 4.8-kb deletion (Fig. 2). The D-loop and the Ori-L-containing regions were selected because of their role in mtDNA replication which, together with protection from molecular damages to mtDNA, participates to the overall maintenance of the mitochondrial genome also through the involvement of TFAM-binding. The Cyt b gene section was used as a control region, as it is not involved in particular functions requiring the binding of TFAM. The region encompassing the CO II gene section, upstream of DR1, and that including DR1 were chosen because of their possible different involvements in the origin of mtDNA deletions [35, 36]. TFAM-binding was analyzed by means of a semi-quantitative PCR assay, able to detect the presence or the absence of TFAM-binding at the assayed mtDNA region in the examined animal. In fact, the DNA template for such PCR was the aliquot obtained from immunoprecipitation for TFAM, resulting “enriched” in mtDNA bound by TFAM. The densitometric signal of this PCR product was later adequately purged from the component due to non-immunoprecipitated mtDNA template and compared with the signal derived from amplification of mtDNA aspecifically bound by proteins. Therefore, the signal for TFAM-binding was ascertained because its value was calculated by subtracting the value of the product obtained from the aliquot precipitated without primary antibody from that of the TFAM-immunoprecipitated aliquot. Such calculated value was compared with that of the signal due to the aspecific binding of the β-actin protein to mtDNA and followed by a very limited immunoprecipitation, which resulted always below 1% of that with anti-TFAM antibody. Fig. 3 reports the data about such binding of TFAM in frontal cortex at the five regions of D-loop, Ori-L, DR1, CO II, and Cyt b from all tested rats. AL-6 rats showed TFAM-binding at all analyzed regions. TFAM-binding was reduced in the aged AL-26 rats at all regions, whereas the results from the CR-26 samples were quite dispersed, suggesting a partial preservation of TFAM-binding activity by the CR.
Fig. 3.
Age- and CR-related changes of TFAM-binding to rat mtDNA regions (D-loop, Ori-L, DR1, CO II, and Cyt b) by mIP assay and following semi-quantitative PCR of mIP templates from the frontal cortex samples. Evaluation of TFAM-binding results in the three groups of animals after the mIP assay performed at five mtDNA regions. The groups of assayed animals included, respectively, eight AL-6, seven AL-26, and seven CR-26 rats, and their values were represented in the scatter-plot format. The intensity of each DNA band, produced by the specific PCR and visualized on agarose gels (not shown), was analyzed by densitometry and used for detection of TFAM-binding by subtracting the value of the intensity of the aliquot precipitated without primary antibody from that of the TFAM-immunoprecipitated aliquot, both normalized to the value of the respective input aliquot made equal to 1.
3.3. Soleus mIP
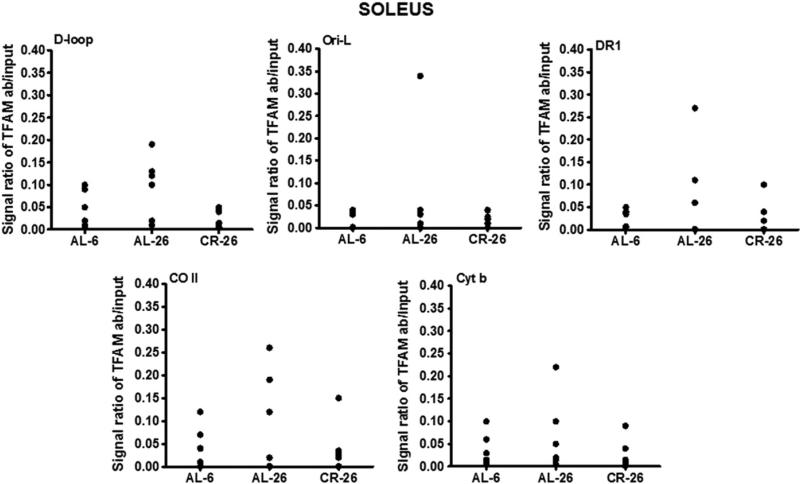
Analyzing, by mIP technique, the same five regions above indicated in the three groups of soleus skeletal muscle samples, we found an overall tendency toward increased TFAM-binding to mtDNA at all regions in the AL-26 rats, although the data from the individual animals were dispersed (Fig. 4). Such a trend of TFAM-binding activity in this post-mitotic tissue was quite different from the frontal cortex counterpart. The tissue-specificity was further evidenced by CR, completely preventing the age-related increase in TFAM-binding and maintaining values in the CR-26 animals that were very close to those from the AL-6 rats.
Fig. 4.
Age- and CR-related changes of TFAM-binding to rat mtDNA regions (D-loop, Ori-L, DR1, CO II, and Cyt b) by mIP assay and following semi-quantitative PCR of mIP templates from the soleus skeletal muscle samples. Evaluation of TFAM-binding results in the three groups of animals after the mIP assay was performed at five mtDNA regions. The groups of assayed animals included, respectively, five AL-6, six AL-26, and five CR-26 rats, and their values were represented in the scatter-plot format. The intensity of each DNA band, produced by the specific PCR and visualized on agarose gels (not shown), was analyzed by densitometry and used for detection of TFAM-binding by subtracting the value of the intensity of the aliquot precipitated without primary antibody from that of the TFAM-immunoprecipitated aliquot, both normalized to the value of the respective input aliquot made equal to 1.
3.4. Liver mIP
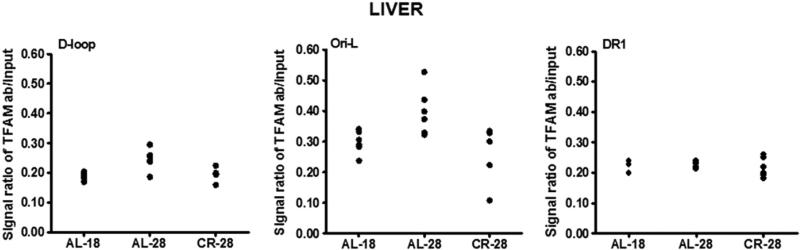
We analyzed in liver samples the eventual age- or CR-related changes in the in vivo mtDNA-binding activity of TFAM to three selected regions (D-loop, Ori-L, and DR1), with the exclusion of the two including, respectively, part of the cytochrome b gene (np 15,004–15,228) and a portion of the CO II gene (np 7133–7376). With respect to the D-loop and Ori-L regions, presumably involved in mtDNA replication, the DR1-containing region likely represented all other mtDNA regions not implicated in such process. In Fig. 5 the densito-metric evaluation of the semi-quantitative PCR assay showed the presence of TFAM-binding in all assayed samples at the different regions. An age-related increase in TFAM-binding at both D-loop and Ori-L regions was noted in the AL-28 rats. The age-matched CR-28 animals, on the contrary, did not show relevant differences with respect to the younger AL-18 group at the two regions, supporting a preventive action by the CR. The absence of age- or diet-related changes in the binding of TFAM at the DR1-containing region was also evident.
Fig. 5.
Age- and CR-related changes of TFAM-binding to rat mtDNA regions (D-loop, Ori-L, and DR1) by mIP assay and following semi-quantitative PCR of mIP templates from the liver samples. Evaluation of TFAM-binding results in the three groups of animals after the mIP assay performed at three mtDNA regions. The groups of assayed animals included, respectively, six AL-18, six AL-28, and six CR-28 rats, and their values were represented in the scatter-plot format. The intensity of each DNA band, produced by the specific PCR and visualized on agarose gels (not shown), was analyzed by densitometry and used for detection of TFAM-binding by subtracting the value of the intensity of the aliquot precipitated without primary antibody from that of the TFAM-immunoprecipitated aliquot, both normalized to the value of the respective input aliquot made equal to 1.
3.5. MtDNA content in the frontal cortex, soleus and liver
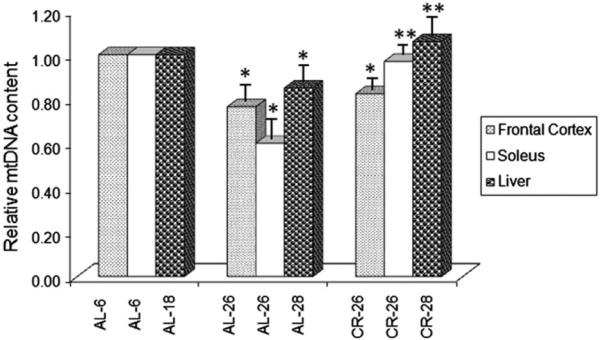
Due to the presumed great relevance of TFAM-binding for the maintenance of mtDNA, we determined, by quantitative real time-PCR, the mtDNA content in samples from the three assayed tissues. In the frontal cortex we found (Fig. 6) a statistically significant decrease (−25%) in the AL-26 animals compared to the young ones. In the CR-26 rats a smaller decrease (−17%) was reported, which was statistically signifi-cant with respect to the value of the young counterpart, but not different compared to that of the AL-26 rats. As for the soleus muscle (Fig. 6), mtDNA content presented a relevant, statistically significant decrease (−40%) with aging in the AL-26 animals, that was completely prevented in the CR-26 ones, supporting a positive effect of the diet. In the liver (Fig. 6) we showed the same kind of changes described in the soleus with aging and CR, namely a statistically significant age- related decrease (−15%) in the AL-28 rats with respect to the AL-18 younger counterpart, that was fully prevented in the CR-28 animals by treatment with CR.
Fig. 6.
Age- and CR-related changes of the relative mtDNA content. The histogram shows the mean value of the ratio mtDNA/nuclear DNA in AL and CR rats. Bars represent the mean (±SE) obtained, respectively, from analysis in triplicate of total nucleic acids from each AL-6, AL-18, AL-26, AL-28, CR-26 and CR-28 rat. The groups of assayed animals included, respectively, five AL-6, five AL-18, six AL-26, six AL-28, five CR-26 and five CR-28 rats. *p < 0.05 versus the value of the younger rats (AL-6 or AL-18 fixed as 1); **p < 0.05 versus the value of the old AL rats (AL-26 or AL-28).
4. Discussion
In this study, we analyzed TFAM amount and binding to some regions of mtDNA, as well as the mtDNA content in three different rat tissues (frontal cortex, soleus skeletal muscle and liver) to unveil tissue-specific effects induced by aging and CR. The tissues' different post-mitotic (frontal cortex and soleus) or mitotic (liver) types allowed for a very intriguing comparison. The liver samples were derived from animals sacrificed at dissimilar ages and belonging to a strain (hybrid rat Fischer 344 × Brown Norway) specifically used for aging studies due to its very long living. This allowed us to use animals (AL-18) older than the Fischer 344 counterparts (AL-6) as controls, being the former at the start of the aging process, and to make comparisons among the three tissues. Our study, as many others dealing with aging animals or humans, suggests the very broad inter-individual variability among aged rats for all assayed parameters, leading to quite dispersed values in the same group of samples or to mean values slightly different from those previously reported. As for TFAM amount, we found an age-related increase in the frontal cortex versus a decrease in the soleus and liver, confirming data previously published by our group [12,14,23]. An important and novel finding of this study is the age-related decrease in TFAM observed in the post-mitotic tissue of the soleus skeletal muscle. The CR effect in the frontal cortex, consistent with what previously reported [14], appeared very mild, leading to a TFAM increase not statistically different from that in the aged AL rats. The findings suggest that, in this cortex area and likely also in other regions of the brain, metabolic sensitivity to nutrient availability and/or diet type is very limited. This might be due to the need to maintain a life-long, constant basal metabolic activity in the cells, to guarantee the normal performance of crucial functions of the central nervous system, independent of changing external conditions. On the contrary, CR completely prevented the age-related decrease in TFAM in the other post-mitotic tissue, the soleus, suggesting a relevant sensitivity to nutrient availability. A response to CR for TFAM amount, similar to that of soleus, was shown in the liver. The latter mitotic tissue is known to be affected by long-term CR [37] as well as other diets because it can adapt its metabolism to nutrient availability and, in general, to changing external conditions. The semi-quantitative analysis of the binding activity of TFAM to mtDNA by mIP further highlighted the tissue-specific differences. In fact, we confirmed here in the frontal cortex the previously described, age-related decrease in TFAM-binding at all five assayed regions, partially prevented by CR [14]. Effectively, the dispersion of the CR-26 data is suggestive of a mild preventive effect by the treatment. On the contrary, we found here in the soleus an age-related, overall tendency for increased TFAM-binding at all assayed regions, completely prevented by CR. An age-related increase in TFAM-binding, specific for the regions including the mtDNA origins of replication and completely prevented by CR, was here shown in the liver, consistent with previous results [12]. Considering a possible link between changes in TFAM-binding to mtDNA and maintenance of the mitochondrial genome, we analyzed the mtDNA content in the same groups of samples. We found a shared age-related decrease in mtDNA content and, again, a differential response in sensitivity to CR between the frontal cortex by one side and the soleus and liver by the other. The common age-related loss of mtDNA, however, might be explained by two different tissue-specific mechanisms. In fact, the age-related decrease in TFAM-binding in the frontal cortex might imply decreased mtDNA replication and/or increased mtDNA damage, not counteracted by the usual repair mechanisms [14]. On the contrary, the age-related increased TFAM-binding at both origins of replication in the soleus and liver might explain the mtDNA loss [12,13,19,21] through a hindered mtDNA replication. A similar occur-rence was postulated by Bestwick and Shadel whereby excessive binding of TFAM at the HSP2 promoter was thought to competitively inhibit the transcription machinery [38]. As for the CR effect, the partial preservation of TFAM-binding in the frontal cortex might be consistent with the reduced mtDNA loss in the CR-26 rats. The full efficacy of CR in preventing the age-related mtDNA loss in both the soleus and liver might be linked to the preservation of TFAM-binding activities, especially at the origins of replication, similar to those of the younger animals. These data further strengthen the concept of differential responses to aging and CR between the frontal cortex by one side and the soleus and liver by the other. Furthermore, such tissue-specific difference can be supported also considering the effect of aging on mitochondrial transcription where TFAM-binding is also required, whatever model for the composition of the initiation complex might be favored [38,39]. The age-related decrease in TFAM-binding at the D-loop region, containing the light strand promoter (LSP), in the frontal cortex might explain the previously reported, brain-specific reduction in mitochondrial transcription with aging [40]. Conversely, the age-related increase in TFAM-binding at the same region, here shown in the soleus and liver, might justify the absence of effect on mitochondrial transcription in both tissues with aging [19,40]. We have thus unveiled a novel similarity between the soleus muscle and liver regarding the effects of aging and CR, leading to mitochondrial alterations completely unlike those found in the frontal cortex. The present data suggest that a fine modulation of TFAM-binding to mtDNA may be one of the mechanisms contributing to several tissue-specific mitochondrial changes involved in the aging process, as well as in the preventive effects of CR. This could have relevant consequences for future applications of CR or other nutritional regimens as a strategy to delay the onset of pathologies such as sarcopenia and neurodegeneration that are related to the age-dependent mitochondrial dysfunction. Of course, more studies are needed to explore the molecular causes of the modulation of TFAM-binding to mtDNA, which appears to be a very promising avenue for research on mitochondria.
Acknowledgements
This research was supported by grants to CL (National Institute on Aging (NIA) R01 AG17994), the University of Florida Institute on Aging and the Claude D. Pepper Older Americans Independence Center (1 P30 AG028740). We thank Isabella Maiellaro, MS, for assisting with the experiments.
Footnotes
Conflict of interest
The authors have no actual or potential conflict of interest associated with this research.
References
- 1.Larsson NG, Garman JD, Oldfors A, Barsh GS, Clayton DA. A single mouse gene encodes the mitochondrial transcription factor A and a testis-specific nuclear HMG-box protein. Nat. Genet. 1996;13:296–302. doi: 10.1038/ng0796-296. [DOI] [PubMed] [Google Scholar]
- 2.Fisher RP, Clayton DA. Purification and characterization of human mitochondrial transcription factor 1. Mol. Cell. Biol. 1988;8:3496–3509. doi: 10.1128/mcb.8.8.3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kanki T, Ohgaki K, Gaspari M, Gustafsson CM, Fukuoh A, Sasaki N, Hamasaki N, Kang D. Architectural role of mitochondrial transcription factor A in maintenance of human mitochondrial DNA. Mol. Cell. Biol. 2004;24:9823–9834. doi: 10.1128/MCB.24.22.9823-9834.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lezza AMS. Mitochondrial transcription factor A (TFAM): one actor for different roles. Front. Biol. 2012;7:30–39. [Google Scholar]
- 5.Parisi MA, Clayton DA. Similarity of human mitochondrial transcription factor 1 to high mobility group proteins. Science. 1991;252:965–969. doi: 10.1126/science.2035027. [DOI] [PubMed] [Google Scholar]
- 6.Larsson NG, Wang J, Wilhelmsson H, Oldfors A, Rustin P, Lewandaski M, Barsh GS, Clayton DA. Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nat. Genet. 1998;18:231–236. doi: 10.1038/ng0398-231. [DOI] [PubMed] [Google Scholar]
- 7.Kaufman BA, Durisic N, Mativetsky JM, Costantino S, Hancock MA, Grutter P, Shoubridge EA. The mitochondrial transcription factor TFAM coordinates the assembly of multiple DNA molecules into nucleoid-like structures. Mol. Biol. Cell. 2007;18:3225–3236. doi: 10.1091/mbc.E07-05-0404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yoshida Y, Izumi H, Ise T, Uramoto H, Torigoe T, Ishiguchi H, Murakami T, Tanabe M, Nakayama Y, Itoh H, Kasai H, Kohno K. Human mitochondrial transcription factor A binds preferentially to oxidatively damaged DNA. Biochem. Biophys. Res. Commun. 2002;295:945–951. doi: 10.1016/s0006-291x(02)00757-x. [DOI] [PubMed] [Google Scholar]
- 9.Canugovi C, Maynard S, Bayne AC, Sykora P, Tian J, de Souza-Pinto NC, Croteau DL, Bohr VA. The mitochondrial transcription factor A functions in mitochondrial base excision repair. DNA Repair (Amst) 2010;9:1080–1089. doi: 10.1016/j.dnarep.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Escrivá H, Rodríguez-Peña A, Vallejo CG. Expression of mitochondrial genes and of the transcription factors involved in the biogenesis of mitochondria Tfam, NRF-1 and NRF-2, in rat liver, testis and brain. Biochimie. 1999;81:965–971. doi: 10.1016/s0300-9084(99)00223-0. [DOI] [PubMed] [Google Scholar]
- 11.Dinardo MM, Musicco C, Fracasso F, Milella F, Gadaleta MN, Gadaleta G, Cantatore P. Acetylation and level of mitochondrial transcription factor A in several organs of young and old rats. Biochem. Biophys. Res. Commun. 2003;301:187–191. doi: 10.1016/s0006-291x(02)03008-5. [DOI] [PubMed] [Google Scholar]
- 12.Picca A, Pesce V, Fracasso F, Joseph AM, Leeuwenburgh C, Lezza AMS. Aging and calorie restriction oppositely affect mitochondrial biogenesis through TFAM binding at both origins of mitochondrial DNA replication in rat liver. PLoS One. 2013;8:e74644. doi: 10.1371/journal.pone.0074644. http://dx.doi.org/10.1371/journal.pone 0074644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pesce V, Cormio A, Fracasso F, Lezza AM, Cantatore P, Gadaleta MN. Age-related changes of mitochondrial DNA content and mitochondrial genotypic and phenotypic alterations in rat hind-limb skeletal muscles. J. Gerontol. A Biol. Sci. Med. Sci. 2005;60:715–723. doi: 10.1093/gerona/60.6.715. [DOI] [PubMed] [Google Scholar]
- 14.Picca A, Fracasso F, Pesce V, Cantatore P, Joseph AM, Leeuwenburgh C, Gadaleta MN, Lezza AMS. Age- and calorie restriction-related changes in rat brain mitochondrial DNA and TFAM binding. AGE. 2013;35:1607–1620. doi: 10.1007/s11357-012-9465-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Navarro A, Boveris A. Rat brain and liver mitochondria develop oxidative stress and lose enzymatic activities on aging. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2004;287:R1244–R1249. doi: 10.1152/ajpregu.00226.2004. [DOI] [PubMed] [Google Scholar]
- 16.Anderson RM, Weindruch R. Metabolic reprogramming, caloric restriction and aging. Trends Endocrinol. Metab. 2010;21:134–141. doi: 10.1016/j.tem.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wallace DC. Mitochondrial genetics: a paradigm for aging and degenerative diseases? Science. 1992;256:628–632. doi: 10.1126/science.1533953. [DOI] [PubMed] [Google Scholar]
- 18.Wang CH, Wu SB, Wu YT, Wei YH. Oxidative stress response elicited by mitochondrial dysfunction: implication in the pathophysiology of aging. Exp. Biol. Med. (Maywood) 2013;238:450–460. doi: 10.1177/1535370213493069. [DOI] [PubMed] [Google Scholar]
- 19.Barazzoni R, Short KR, Nair KS. Effects of aging on mitochondrial DNA copy number and cytochrome c oxidase gene expression in rat skeletal muscle, liver, and heart. J. Biol. Chem. 2000;275:3343–3347. doi: 10.1074/jbc.275.5.3343. [DOI] [PubMed] [Google Scholar]
- 20.Pesce V, Cormio A, Fracasso F, Vecchiet J, Felzani G, Lezza AM, Cantatore P, Gadaleta MN. Age-related mitochondrial genotypic and phenotypic alterations in human skeletal muscle. Free Radic. Biol. Med. 2001;30:1223–1233. doi: 10.1016/s0891-5849(01)00517-2. [DOI] [PubMed] [Google Scholar]
- 21.Cassano P, Sciancalepore AG, Lezza AM, Leeuwenburgh C, Cantatore P, Gadaleta MN. Tissue-specific effect of age and caloric restriction diet on mitochondrial DNA content. Rejuvenation Res. 2006;9:211–214. doi: 10.1089/rej.2006.9.211. [DOI] [PubMed] [Google Scholar]
- 22.McInerny SC, Brown AL, Smith DW. Region-specific changes in mitochondrial D-loop in aged rat CNS. Mech. Ageing Dev. 2009;130:343–349. doi: 10.1016/j.mad.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 23.Pesce V, Nicassio L, Fracasso F, Musicco C, Cantatore P, Gadaleta MN. Acetyl-L-carnitine activates the peroxisome proliferator-activated receptor-γ coactivators PGC-1α/PGC-1β-dependent signaling cascade of mitochondrial biogenesis and decreases the oxidized peroxiredoxins content in old rat liver. Rejuvenation Res. 2012;15:136–139. doi: 10.1089/rej.2011.1255. [DOI] [PubMed] [Google Scholar]
- 24.Masoro EJ. Overview of caloric restriction and ageing: an update. Mech. Ageing Dev. 2005;126:913–922. doi: 10.1016/j.mad.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 25.Guarente L. Mitochondria—a nexus for aging, calorie restriction, and sirtuins? Cell. 2008;132:171–176. doi: 10.1016/j.cell.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fontana L. The scientific basis of caloric restriction leading to longer life. Curr. Opin. Gastroenterol. 2009;25:144–150. doi: 10.1097/MOG.0b013e32831ef1ba. [DOI] [PubMed] [Google Scholar]
- 27.Barja G. Mitochondrial oxygen consumption and reactive oxygen species production are independently modulated: implications for aging studies. Rejuvenation Res. 2007;10:215–224. doi: 10.1089/rej.2006.0516. [DOI] [PubMed] [Google Scholar]
- 28.Anderson R, Prolla T. PGC-1α in aging and anti-aging interventions. Biochim. Biophys. Acta. 2009;1790:1059–1066. doi: 10.1016/j.bbagen.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aspnes LE, Lee CM, Weindruch R, Chung SS, Roecker EB, Aiken JM. Caloric restriction reduces fiber loss and mitochondrial abnormalities in aged rat muscle. FASEB J. 1997;11:573–581. doi: 10.1096/fasebj.11.7.9212081. [DOI] [PubMed] [Google Scholar]
- 30.Kang CM, Kristal BS, Yu BP. Age-related mitochondrial DNA deletions: effect of dietary restriction. Free Radic. Biol. Med. 1998;24:148–154. doi: 10.1016/s0891-5849(97)00204-9. [DOI] [PubMed] [Google Scholar]
- 31.Stuart JA, Karahalil B, Hogue BA, Souza-Pinto NC, Bohr VA. Mitochondrial and nuclear DNA base excision repair are affected differently by caloric restriction. FASEB J. 2004;18:595–597. doi: 10.1096/fj.03-0890fje. [DOI] [PubMed] [Google Scholar]
- 32.Bua E, McKiernan SH, Aiken JM. Calorie restriction limits the generation but not the progression of mitochondrial abnormalities in aging skeletal muscle. FASEB J. 2004;18:582–584. doi: 10.1096/fj.03-0668fje. [DOI] [PubMed] [Google Scholar]
- 33.Cassano P, Lezza AM, Leeuwenburgh C, Cantatore P, Gadaleta MN. Measurement of the 4,834-bp mitochondrial DNA deletion level in aging rat liver and brain subjected or not to caloric restriction diet. Ann. N. Y. Acad. Sci. 2004;1019:269–273. doi: 10.1196/annals.1297.045. [DOI] [PubMed] [Google Scholar]
- 34.Pfaffl MW. A new mathematical model for relative quantification in real-time PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. http://dx.doi.org/10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fukui H, Moraes CT. Mechanisms of formation and accumulation of mitochondrial DNA deletions in aging neurons. Hum. Mol. Genet. 2009;18:1028–1036. doi: 10.1093/hmg/ddn437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krishnan KJ, Reeve AK, Samuels DC, Chinnery PF, Blackwood JK, Taylor RW, Wanrooij S, Spelbrink JN, Lightowlers RN, Turnbull DM. What causes mitochondrial DNA deletions in human cells? Nat. Genet. 2008;40:275–279. doi: 10.1038/ng.f.94. [DOI] [PubMed] [Google Scholar]
- 37.López-Torres M, Gredilla R, Sanz A, Barja G. Influence of aging and long-term caloric restriction on oxygen radical generation and oxidative DNA damage in rat liver mitochondria. Free Radic. Biol. Med. 2002;32:882–889. doi: 10.1016/s0891-5849(02)00773-6. [DOI] [PubMed] [Google Scholar]
- 38.Bestwick ML, Shadel GS. Accessorizing the human mitochondrial transcription machinery. Trends Biochem. Sci. 2013;38:283–291. doi: 10.1016/j.tibs.2013.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Falkenberg M, Gaspari M, Rantanen A, Trifunovic A, Larsson N-G, Gustafsson CM. Mitochondrial transcription factors B1 and B2 activate transcription of human mtDNA. Nat. Genet. 2002;31:289–294. doi: 10.1038/ng909. [DOI] [PubMed] [Google Scholar]
- 40.Gadaleta MN, Petruzzella V, Renis M, Fracasso F, Cantatore P. Reduced transcription of mitochondrial DNA in the senescent rat. Tissue dependence and effect of L-carnitine. Eur. J. Biochem. 1990;187:501–506. doi: 10.1111/j.1432-1033.1990.tb15331.x. [DOI] [PubMed] [Google Scholar]