Abstract
Telomere malfunction and other types of DNA damage induce an activin A-dependent stress response in mortal non-tumorigenic human mammary epithelial cells that subsequently induces desmoplastic-like phenotypes in neighboring fibroblasts. Some characteristics of this fibroblast/stromal response, such as reduced adipocytes and increased extracellular matrix content, are observed not only in tumor tissues but also in disease-free breast tissues at high risk for developing cancer, especially high mammographic density tissues. We found that these phenotypes are induced by repression of the fatty acid translocase CD36, which is seen in desmoplastic and disease-free high mammographic density tissues. In this study, we show that epithelial cells from high mammographic density tissues have more DNA damage signaling, shorter telomeres, increased activin A secretion and an altered DNA damage response compared to epithelial cells from low mammographic density tissues. Strikingly, both telomere malfunction and activin A expression in epithelial cells can repress CD36 expression in adjacent fibroblasts. These results provide new insights into how high mammographic density arises and why it is associated with breast cancer risk, with implications for the definition of novel invention targets (e.g. activin A, CD36) to prevent breast cancer.
Keywords: breast cancer, cell-cell interactions, CD36, activin A, mammographic density, stroma
Introduction
Histologic examination demonstrates that tumor stroma is morphologically distinct from disease-free stroma. It is characterized by increases in extracellular matrix (ECM), fibroblasts, immune and endothelial cells, cytokines and growth factors levels (1) and by fewer and smaller adipocytes (2, 3). Collectively, these changes are termed desmoplasia.
Seminal studies demonstrated that the stroma contributes to tumor initiation, progression and outcome. Tumor stromal cells (fibroblasts, adipocytes endothelial and immune cells) promote epithelial cell proliferation, mutagenesis, angiogenesis and migration and impair apoptosis and immunosurveillance (1, 4). Irradiation of mouse mammary gland stroma drives tumorigenesis of nonirradiated epithelial cells (5) and involuting mammary stroma facilitates progression of premalignant cells in xenografts (6, 7). Gene expression profiles of breast tumor-associated stroma are strongly associated with clinical outcome (8) and are remarkably similar to those of stroma surrounding ductal carcinomas in situ (DCIS) (9), suggesting that tumor stroma develops early in tumorigenesis.
Selected desmoplastic features are also observed in non-malignant breast tissues of women with high mammographic density (1-3, 10, 11). Mammographic density (MD) is determined by the relative amounts of radiolucent material (fat) and radiodense material (epithelial cells, fibroblasts and ECM) within the breast upon imaging. Radiodense areas, referred to as MD, exhibit histological characteristics of malignant stroma including low adipocyte content, high ECM and stromal cell content (1-3, 10, 11). Women with MD>75% have a 4- to 6-fold increase in invasive breast cancer compared to women with negligible MD (12, 13). It is estimated that almost 1/3 of breast cancers may be attributable to high MD (13).
Previously, we showed that CD36 is necessary and sufficient to coordinately control adipocyte and matrix accumulation, two phenotypes that histologically define MD (3). CD36, a widely expressed transmembrane receptor, modulates cell type- and ligand-specific phenotypes, including adipocyte differentiation, angiogenesis, apoptosis, TGFβ activation, cell–ECM interactions, ECM deposition and immune signaling (3, 14). CD36 expression is negligible in tumor stroma, in contrast to surrounding histologically disease-free tissue, and is inversely correlated with tumor size and grade (3). Strikingly, disease-free high mammographic density (HD) tissues (MD>70-75%) have reduced CD36 levels in multiple stromal components (adipocytes, endothelial cells, macrophages, and fibroblasts) compared with low mammographic density (LD) tissues (25%<MD<50%), suggesting that CD36 repression is a multicellular coordinated stromal program active in tissues at high risk for tumorigenesis (3).
In addition to desmoplasia and HD, increased ECM is seen in other pathologic conditions associated with a DNA damage response (DDR) and/or telomere malfunction: fibrosis after γ-irradiation (15), dyskeratosis congenita (16) and pulmonary (17) and liver fibrosis (18). Previously, we demonstrated that DNA damaging agents and/or telomere malfunction in mortal, non-tumorigenic variant human mammary epithelial cells (vHMEC) induce a DDR and activin A-dependent COX-2 expression (19). Fibroblasts from reduction mammoplasties (RMF) co-cultured with DNA-damaged vHMEC induce many genes consistent with a desmoplastic phenotype (e.g. ECM and inflammatory cytokines) and activin A secretion by damaged vHMEC is necessary and sufficient for this induction (20). In vivo, DCIS lesions whose epithelial cells exhibit shorter telomeres, increased activin A and COX-2, are surrounded by activated fibroblasts and increased immune infiltrate (20), suggesting that microenvironmental alterations, mimicking aspects of desmoplasia, occur even in the absence of an invasive tumor.
This manuscript presents in vitro and in vivo data supporting our hypothesis that HD is generated by factors(s) secreted by DNA-damaged epithelial cells (including damage caused by shortened telomeres) that repress CD36 causing induction of desmoplastic-like phenotypes in adjacent fibroblasts.
Materials and Methods
Human Subjects
Human tissues were accrued after informed consent and studied under institutionally approved protocols 10-02471 and 10-03756.
Human Tissue Analysis
Paraffin-embedded serial tissue sections, from 13 LD (25%<MD<50%) and 14 HD (MD>70%) disease-free women (Supplementary Table 1), were assessed for γH2AX levels by immunohistochemistry (3) and for telomere content by telomere-specific FISH (20). For immunohistochemistry, antigen retrieval was performed in citrate buffer (pH=6.0) for 30 minutes at 93°C prior to incubation with a γH2AX antibody (1:800, Millipore, #05-636) for 60 minutes at room temperature. For FISH, telomere-specific (Cy3-labeled) and centromere-specific (FITC-labeled) PNA probes were hybridized to tissue sections and the telomere to centromere intensity ratio calculated.
Isolation and propagation of human mammary epithelial cells (HMEC) and fibroblasts (HMF) from disease-free tissues
HMEC and vHMEC, which have silenced p16 (21), isolated from 12 biopsies of known MD (LD-HMEC/vHMEC or HD-HMEC/vHMEC) (Supplementary Table 2) and 4 reduction mammoplasties (RM) (Supplementary Table 3), and HMF, isolated from 14 biopsies of known MD (LD-HMF or HD-HMF) (Supplementary Table 2) and 10 reduction mammoplasties (RMF) (Supplementary Table 3), were propagated in MEGM and RPMI-1640+10%FBS, respectively (3, 4, 22, 23).
Etoposide treatment
LD-vHMEC and HD-vHMEC exposed to 50 μM etoposide or vehicle control (DMSO) for 24 hours were assessed for γH2AX intensity and foci number, cell viability, apoptosis or proliferation 0, 1, 3, 6, 12 and 24 hours after drug removal. Immunofluorescence detection of γH2AX was performed using the γH2AX antibody above (1:500) (20). Cell viability (fraction of live/dead cells) and apoptosis (caspase 3/7 activity) were measured using an ApoTox-Glo Triplex Assay (Promega) and proliferation (BrDU incorporation) using the Cell Proliferation Assay Kit (Cell Signaling). LD-vHMEC and HD-vHMEC exposed to 20 μM etoposide or vehicle for 3 hours were assessed for long-term survival in a colony formation assay 9 days after drug removal, and the surviving fractions (plating efficiency of cells exposed to etoposide/plating efficiency of cells exposed to vehicle) calculated. (24).
Telomere-Content Assays
Telomere content was assessed with genomic DNA isolated from HMEC, vHMEC and HMF using quantitative polymerase chain reaction (QPCR) as described (25). Telomeric DNA was expressed as the ratio of telomere cycle threshold (Ct) to the Human β globin gene Ct. Each sample was run in triplicate wells on each plate and averaged. Each target was analyzed in triplicate plates.
QPCR
QPCR was performed on a CFX-96 thermocycler using SsoFast Master Mix (Bio-Rad) and TaqMan primer-probe sets for each gene (Applied Biosystems) and the data analyzed using the standard curve method. Expression of beta-D-glucuronidase (GUSB) was used to normalize for variances in input cDNA. For all experiments, each sample was run in triplicate wells on each plate and averaged, and each gene was analyzed in triplicate plates.
Treatment of RMF with conditioned-media
LD-vHMEC and HD-vHMEC were plated in MEGM and conditioned their media for 48 hours. RMF were plated in RPMI-1640+10%FBS and grown for 24 hours in RPMI-1640+1%FBS before the media were replaced by a 1:1 mix of RPMI-1640+2%FBS and conditioned-media (or control unconditioned-media). RNA was isolated from RMF 48 hours after treatment with conditioned-media.
ELISA
vHMEC were plated in MEGM and conditioned their media for 26 hours. Activin A and TGF-β protein levels were measured in the conditioned-media using Duo-Set ELISA kits #DY338 and DY420, respectively (R&D Systems).
Treatment of RMF with Activin A
RMF, plated in RPMI-1640+10%FBS, were grown for 24 hours in RPMI-1640 without serum prior to the addition of: activin A (Sigma-Aldrich); prostaglandin E2 (Cayman Chemicals); COX-2 inhibitor, NS398 (Cayman Chemicals); Protein Kinase A (PKA) inhibitor, H89 (Sigma-Aldrich); Phosphatidylinositol 3-kinase (PI3K) inhibitor, LY294002 (Sigma-Aldrich); TGF-β Receptor 1 (TGFβR1) inhibitors, LY364947 and SB431542 (Sigma-Aldrich); p38 Mitogen Activated Protein Kinase (p38 MAPK) inhibitor SB203580 (Sigma-Aldrich); and MAPK Kinase (MAPKK) inhibitor, UO126 (Sigma-Aldrich).
Co-culture experiments
RMF were plated in 0.4μm pore Transwell dishes (Costar) and co-cultured with vHMEC expressing either control luciferase short hairpin (sh-Luciferase), activin A short hairpin (sh-activin A), vector control, TRF2 or hTERT (19, 20). RNA was isolated from RMF 48 hours after initiation of co-culture.
Immunocytochemistry
Immunocytochemistry was performed in RMF using primary antibodies against CD36 (1:20, #9154, Santa Cruz Biotechnology), fibronectin (1:100, #610077, BD Transduction) and α-smooth muscle actin (αSMA) (1:50, #M0851, Dako) (3).
Adipocyte differentiation experiment
RMF, plated in RPMI-1640+10%FBS, were grown for 24 hours in RPMI-1640 without serum prior to exposure to 80 ng/ml activin A for 48 hours. The media were then replaced with RPMI-1640+10%FBS, +/− 80 ng/ml Activin A and +/− 10 μM 15-deoxy-D12,14-Prostaglandin J2 (PJ2, Cayman Chemical) to induce adipocyte differentiation. Lipid accumulation was assayed after 1 week by Oil Red O staining (3).
Image acquisition, statistical analysis and image analysis are described in Supplementary Methods.
Results
HD Breast Tissues have Increased Basal γH2AX and Shortened Telomeres Compared to LD Breast Tissues
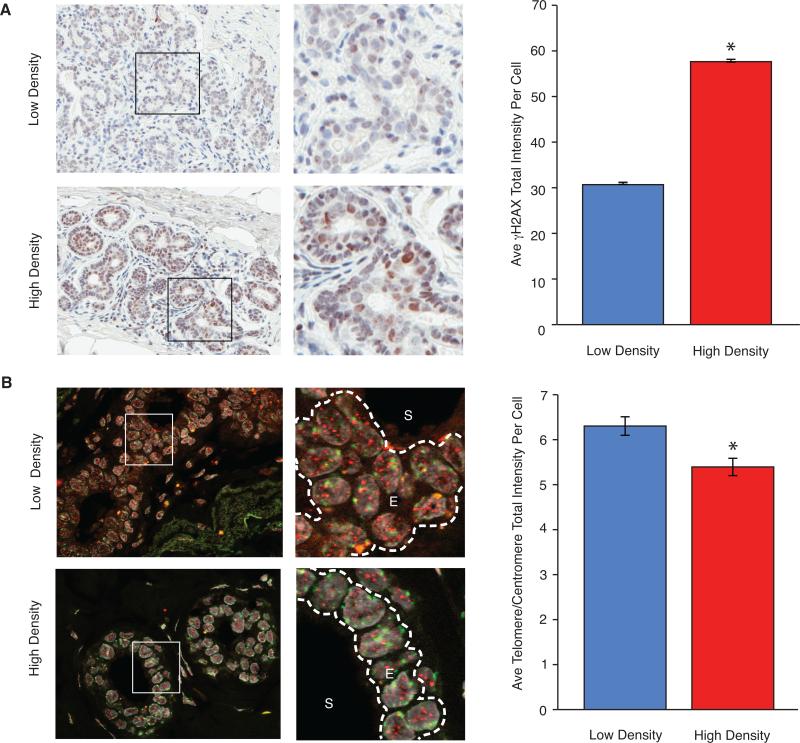
We hypothesized that HD epithelial cells have an elevated DDR and secrete factor(s) that repress CD36 expression and reprogram adjacent fibroblasts. To test this hypothesis, levels of γH2AX, a DNA damage marker, were assessed by immunohistochemistry (Figure 1A, left) in biopsies obtained from disease-free women with measured MD (Supplementary Table 1). HD tissues had higher γH2AX levels (1.9-fold, p<0.0001) than LD tissues (Figure 1A, right and Supplementary Figure 1A). To determine if the increased DDR observed in HD tissues was associated with telomere malfunction, telomere DNA content was assessed by FISH (Figure 1B, left). Telomere DNA content was reduced (1.2-fold, p<0.0001) in HD epithelial cells compared to LD epithelial cells (Figure 1B, right and Supplementary Figure 1B). CD36 levels, measured by immunohistochemistry in this cohort, were lower (4.5-fold, p<0.0001) in HD tissues than LD tissues (3). These in vivo data are consistent with our hypothesis that factors secreted by DNA-damaged HD epithelial cells repress CD36 in adjacent fibroblasts.
Figure 1. HD breast tissues have increased basal γH2AX and shortened telomeres compared to LD breast tissues.
γH2AX protein levels and telomere content assessed by immunohistochemistry and FISH, respectively, in serial paraffin breast tissue sections from 13 LD and 14 HD disease-free women. (A) Left: Representative bright field images (20X original magnification) of paraffin sections from 12 LD and 14 HD tissues stained for γH2AX (brown). Right: Average and SEM of γH2AX intensity per cell. (B) Left: Representative fluorescent images (63X original magnification) of paraffin sections from 12 LD and 11 HD tissues stained for telomere (red), centromere (green) and DNA (DAPI, grey). S: stromal cells, E: epithelial cells. Right: Average and SEM of telomere signal/centromere signal per epithelial cell nucleus.
HD Epithelial Cells Exhibit Increased γH2AX, Decreased Proliferation and Shortened Telomeres Compared to LD Epithelial Cells
To further assess the differences between LD and HD tissues in vitro, we isolated and propagated three cell types (3, 4, 22, 23) from biopsies of disease-free women with measured MD (Supplementary Table 2): (i) human mammary epithelial cells (HMEC) with an intact p16/Rb pathway; (ii) variant human mammary epithelial cells (vHMEC) with a compromised p16/Rb pathway and (iii) human mammary fibroblasts (HMF). We primarily used vHMEC for these experiments because: i) HMEC have limited proliferative capability (~10 population doublings) compared to vHMEC (~30-65 population doublings); ii) HMEC, which have intact cell cycle checkpoints (p16/Rb), undergo growth arrest upon telomere malfunction, whereas vHMEC, which lack p16, still proliferate (19, 22) and iii) vHMEC pre-exist in vivo in disease-free breast tissue (21).
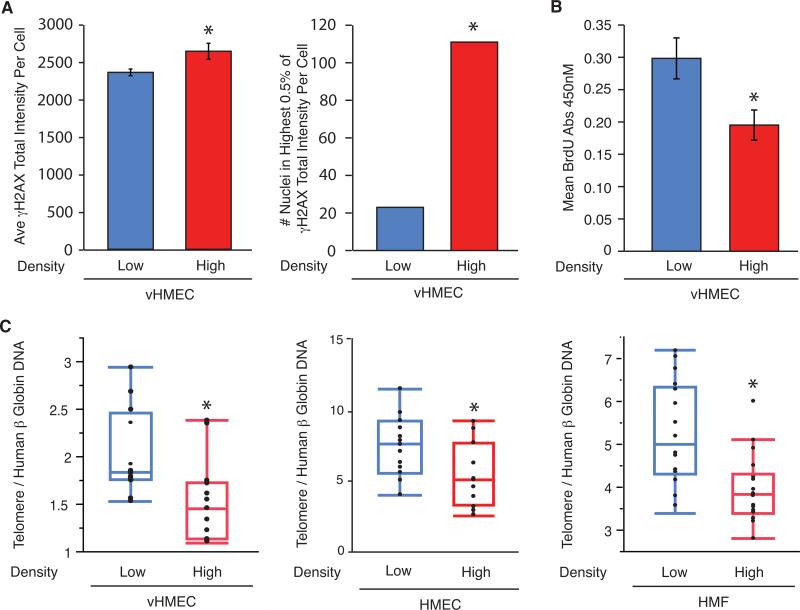
To determine if HD-vHMEC had a heightened basal DDR, as observed in vivo, basal γH2AX levels were measured. γH2AX levels were slightly higher (1.1-fold, p=0.05) in HD-vHMEC than LD-vHMEC (Figure 2A, left and Supplementary Figure 2A). Cell propagation in culture leads to selective expansion of cells capable of surviving and proliferating. Therefore, cells with the most pronounced DDR and the highest levels of γH2AX (“jackpots”) are likely lost from the population, potentially explaining why the difference in γH2AX levels between LD and HD cells is greater in vivo than in vitro. One might predict that “jackpots” would exist transiently in culture and in greater numbers in HD-vHMEC. To assess this possibility, cells with the 0.5% highest γH2AX levels (“jackpots”) were counted. ”Jackpots” were more frequent (4.8-fold, p<0.0001) in HD-vHMEC, as predicted (Figure 2A, right).
Figure 2. HD-vHMEC exhibit increased γH2AX, decreased proliferation and shortened telomeres compared to LD-vHMEC.
(A) Average and SEM of basal γH2AX intensity per nucleus (Left) and number of nuclei with the highest 0.5% γH2AX intensity (Right) in 3 LD-vHMEC and 3 HD-vHMEC. (B) Average and SEM of basal BrdU incorporation, measured by absorbance at 450nM, in 6 LD-vHMEC and 6 HD-vHMEC. (C) Quantitation of telomere QPCR data for vHMEC (4 LD and 5 HD, left), HMEC (5 LD and 4 HD, middle) and HMF (6 LD and 4 HD, right).
Since the induction of a DDR typically leads to growth arrest or apoptosis, we asked if HD-vHMEC showed reduced proliferation and/or increased apoptosis when compared to LD-vHMEC. HD-vHMEC showed decreased proliferation (1.5-fold, p=0.01) when assessed by BrDU incorporation (Figure 2B) but no significant difference in apoptosis when assayed for caspase 3/7 activity (Supplementary Figure 3B).
To determine if HD epithelial cells had reduced telomere content, as observed in vivo, telomere DNA was measured by QPCR in LD and HD vHMEC, HMEC and HMF (all at comparable population doublings: see Supplementary Figure 2B and Supplementary Methods). Telomere DNA was reduced 1.4-fold in both HD-vHMEC and HD-HMEC compared to LD-vHMEC and LD-HMEC (p=0.007 and 0.03, respectively; Figure 2C, left and middle, respectively) and 1.3-fold in HD-HMF compared to LD-HMF (p=0.0009; Figure 2C, right), demonstrating that this phenotype is not restricted to the epithelial compartment. These in vitro data demonstrate that HD-vHMEC have increased basal γH2AX levels, reduced proliferation, and reduced telomere content compared to LD-vHMEC, recapitulating our observations in vivo and indicating that LD and HD epithelial cells are intrinsically distinct.
HD-vHMEC Exposed to Exogenous DNA Damage Exhibit Enhanced γH2AX Levels and Viability Compared to LD-vHMEC
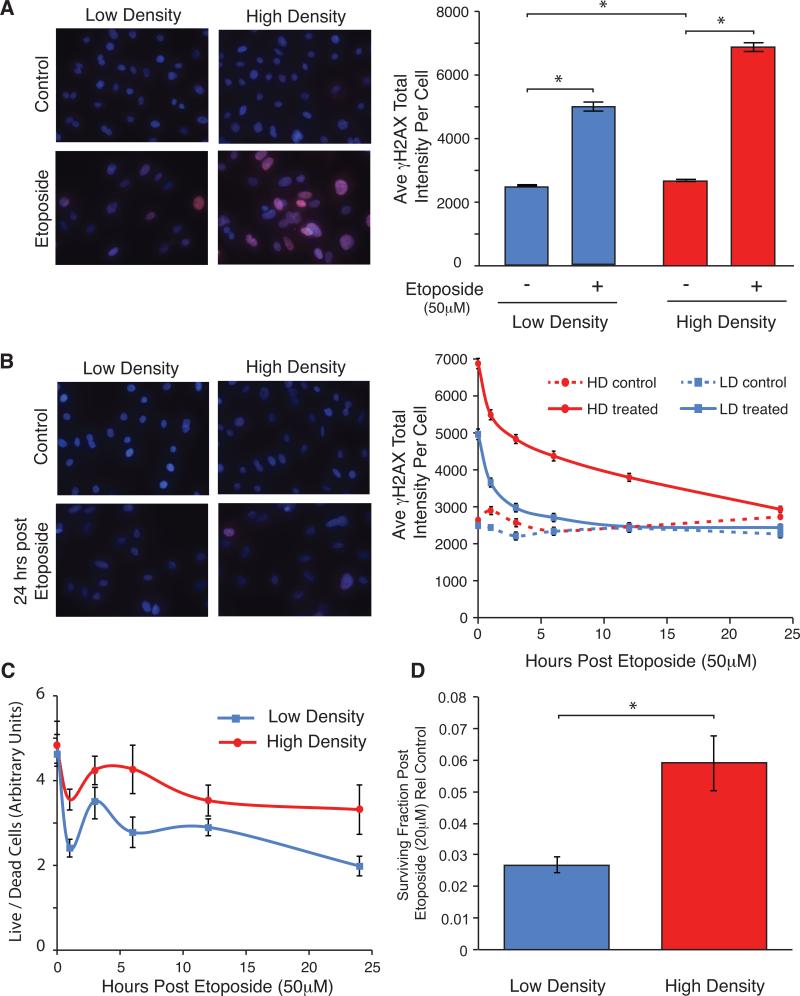
To assess if HD-vHMEC have a differential response to exogenous DNA damage, LD-vHMEC and HD-vHMEC were exposed to etoposide for 24 hours to induce double-strand DNA breaks, and γH2AX intensity quantitated (Figure 3A). LD-vHMEC and HD-vHMEC exhibited increased γH2AX intensity after etoposide exposure (2.0-fold, p<0.0001; 2.6-fold, p<0.0001; respectively). However, γH2AX intensity was higher (1.4-fold, p<0.0001) in HD-vHMEC.
Figure 3. HD-vHMEC exposed to exogenous DNA damage exhibit increased γH2AX and increased viability/survival compared to LD-vHMEC.
Three LD-vHMEC and 3 HD-vHMEC (A,B&D) or 6 LD-vHMEC and 6 HD-vHMEC (C) exposed to 50 μM etoposide for 24 hours (A,B&C) or 20 μM etoposide for 3 hours (D) were assessed for γH2AX intensity (A&B), cell viability (C) or long-term survival (D). (A&B, left) Representative fluorescent images (10X original magnification) of γH2AX staining (red) 0 hours (A) or 24 hours (B) after etoposide removal. (A&B, right) Average and SEM of γH2AX intensity per nucleus at 0 hours (A) or 1, 3, 6, 12 and 24 hours (B) after etoposide removal. (C) Average and SEM of cell viability (live/dead cells) 0, 1, 3, 6, 12 and 24 hours after etoposide removal. (D) Average and SEM of surviving fraction (plating efficiency of cells exposed to etoposide/plating efficiency of control cells) 9 days after etoposide removal.
To assess the ability of LD-vHMEC and HD-vHMEC to recover from etoposide-induced DNA damage, γH2AX levels were measured 0, 1, 3, 6, 12 and 24 hours after drug removal (Figure 3B). HD-vHMEC consistently exhibited significantly higher γH2AX intensity than LD-vHMEC. Importantly, while γH2AX intensity in LD-vHMEC returned to basal levels by 12 hours, γH2AX intensity in HD-vHMEC remained elevated for 24 hours, suggesting that DNA repair is less efficient in HD-vHMEC.
Although γH2AX intensity is frequently used as a readout of DDR, and correlates well with the amount of DNA damage (26) and γH2AX foci (27), we nonetheless counted the number of γH2AX foci in a subset of LD-vHMEC and HD-vHMEC (Supplementary Figure 3A). Consistent with γH2AX intensity measurements, both LD-vHMEC and HD-vHMEC exhibited increased γH2AX foci number (6.3-fold, p<0.0001; 9.8-fold, p<0.0001; respectively) after etoposide exposure. Furthermore, although not statistically significant, γH2AX foci number was higher in HD-vHMEC 0 and 24 hours after etoposide removal (1.1-fold and 1.4-fold, respectively).
Cell viability (Figure 3C) and apoptosis (Supplementary Figure 3B) were assessed 0, 1, 3, 6, 12 and 24 hours after etoposide removal. In spite of their increased and sustained DDR, HD-vHMEC exhibited increased cell viability compared to LD-vHMEC for the entire time course. Both LD-vHMEC and HD-vHMEC showed increased levels of apoptosis upon etoposide exposure. However, HD-vHMEC underwent less apoptosis than LD-vHMEC (although not statistically significant) for the entire time course, consistent with their increased viability. We also assessed the long-term survival of these cells after exposure to etoposide using a colony formation assay (Figure 3D), and found that HD-vHMEC formed more colonies than LD-vHMEC 9 days after etoposide removal (2.2-fold, p=0.006).
These in vitro data demonstrate that HD-vHMEC have elevated and persistent γH2AX levels, increased viability/survival and decreased apoptosis following exogenous DNA damage, compared to LD-vHMEC. Therefore, LD-vHMEC and HD-vHMEC have intrinsic differences in both their basal and induced DDR.
HD-vHMEC Secrete Factors that Repress CD36 in RMF to a Greater Extent than LD-vHMEC
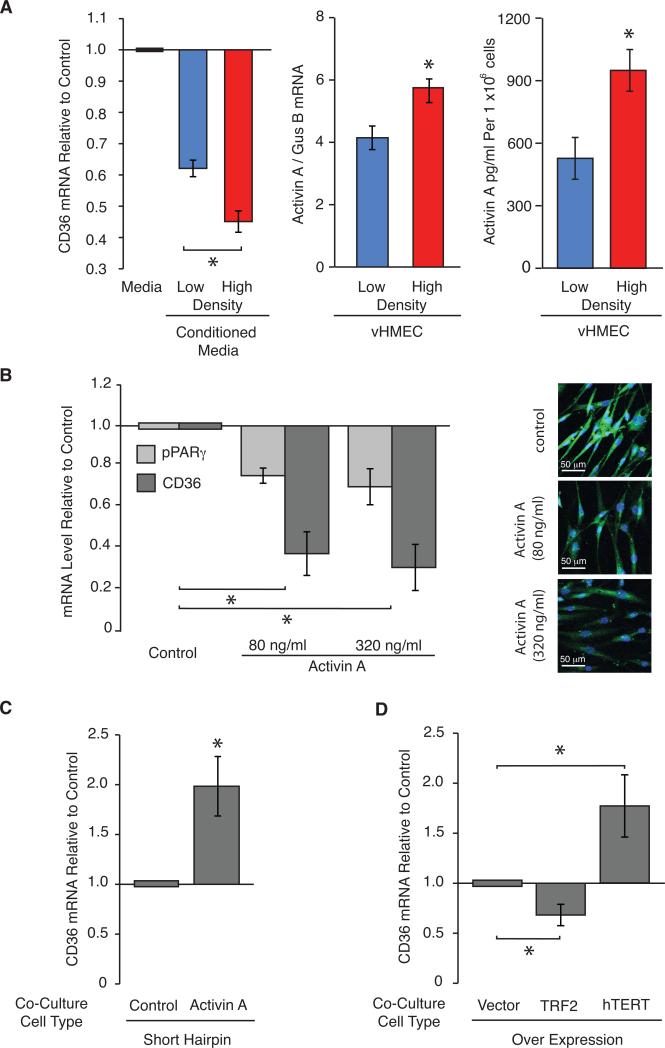
Having demonstrated that LD-vHMEC and HD-vHMEC are intrinsically different, we tested our hypothesis that secreted factor(s) from HD epithelial cells repress CD36 expression in adjacent stromal cells. CD36 mRNA levels were assessed in RMF (Supplementary Table 3) after exposure to control unconditioned-media or conditioned-media from LD-vHMEC or HD-vHMEC (Figure 4A, left). Conditioned-media from both LD-vHMEC and HD-vHMEC repressed CD36 in RMF compared to unconditioned-media (1.6-fold, p<0.0001; 2.2-fold, p<0.0001; respectively). However, consistent with our hypothesis, conditioned-media from HD-vHMEC repressed CD36 in RMF more than conditioned-media from LD-vHMEC (1.4-fold, p=0.0005).
Figure 4. Shortened telomeres and increased activin A secretion in HD-vHMEC compared to LD-vHMEC is necessary and sufficient for CD36 repression in RMF.
(A) Left: Two RMF exposed for 48 hours to control unconditioned-media, or media conditioned for 48 hours by 5 LD-vHMEC or 5 HD-vHMEC, were assayed for CD36 mRNA by QPCR. Average and SEM of CD36 mRNA fold-change relative to RMF exposed to control medium in 1 RMF. Middle: Average and SEM of activin A mRNA levels measured by QPCR in 5 LD-vHMEC and 5 HD-vHMEC. Right: Average and SEM of activin A protein levels measured by ELISA in 6 LD-vHMEC and 4 HD-vHMEC. (B) Left: CD36 and PPARγ mRNA levels were measured by QPCR in 2 RMF exposed to 80 and 320 ng/ml of activin A for 48 hours. Average and SEM of mRNA fold-change relative to untreated RMF. Right: Representative fluorescent images (20X original magnification) of CD36 protein staining (green) in 1 RMF exposed to 80 and 320 ng/ml of activin A for 48 hours. (C&D) Two RMF were co-cultured with 2 vHMEC for 24 hours and assayed for CD36 mRNA by QPCR. vHMEC expressed a short hairpin for luciferase (control) or for activin A (activin A) (C) or over-expressed mock (vector), TRF2 or hTERT (D). Average and SEM of CD36 mRNA fold-change relative to RMF co-cultured with control short hairpin (C) or vector control (D).
Activin A is Upregulated in HD-vHMEC Compared to LD-vHMEC
Induction of a DDR in vHMEC is associated with increased activin A (19). To assess if the heightened DDR observed in HD-vHMEC was associated with higher activin A, activin A mRNA and protein levels were measured in LD-vHMEC and HD-vHMEC. HD-vHMEC had higher levels of activin A mRNA (1.4-fold, p=0.03) and protein (1.8-fold, p=0.04) than LD-vHMEC (Figure 4A, middle and right, respectively).
Activin A is a TGF-β family member and TGF-β is known to induce ECM accumulation/fibrosis (28). Therefore, we evaluated TGF-β protein levels in LD-vHMEC and HD-vHMEC and in vHMEC with increased telomere malfunction, i.e. over-expressing TRF2 (TRF2-vHMEC), and vHMEC with reduced telomere malfunction, i.e. over-expressing hTERT (hTERT-vHMEC) (19). TGF-β protein levels were not significantly different between LD-vHMEC and HD-vHMEC nor between TRF2-vHMEC and hTERT-vHMEC (Supplementary Figure 4A&B, respectively), demonstrating that TGF-β does not contribute to the DDR in vHMEC in our experimental conditions.
We previously showed that activin A, induced by DDR, is necessary and sufficient for COX-2 induction in vHMEC (19). As predicted, COX-2 mRNA levels were higher (1.5-fold, p=0.09) in HD-vHMEC than LD-vHMEC (Supplementary Figure 4C). These data further support that the stress response previously identified by us in DNA-damaged vHMEC (19) is heightened in HD-vHMEC compared to LD-vHMEC.
Activin A and Telomere Malfunction in vHMEC Are Necessary and Sufficient for CD36 Repression in RMF
To determine if activin A was sufficient to repress CD36 in RMF, RMF were exposed to 80 and 320 ng/ml activin A. Exposure to 80 ng/ml activin A repressed CD36 mRNA (2.6-fold, p=0.01) and protein levels (Figure 4B, left and right, respectively). A similar repression was observed at 320 ng/ml. Since CD36 expression is primarily controlled by the transcription factor PPARγ (29), we asked if activin A-dependent CD36 repression could be mediated by PPARγ. RMF exposed to activin A repressed PPARγ mRNA at both doses (1.3-fold, p=0.007; 1.4-fold, p=0.015; respectively) (Figure 4B, left). To assess if activin A secretion by vHMEC was necessary for CD36 repression in RMF, RMF were co-cultured with vHMEC expressing short hairpin RNA to either a control (sh-Luciferase-vHMEC) or activin A (sh-activin A-vHMEC). RMF co-cultured with shactivin A-vHMEC had higher levels of CD36 mRNA (2.0-fold, p=0.007) than RMF co-cultured with sh-Luciferase-vHMEC (Figure 4C). Thus, activin A is sufficient to repress CD36 in RMF and activin A secretion by vHMEC is necessary for CD36 repression in RMF.
Activin A induces COX-2 and secretion of its product, Prostaglandin E2 (PGE2), in vHMEC (19). To ascertain if COX-2 expression and PGE2 secretion by vHMEC was necessary and/or sufficient for CD36 repression in RMF, RMF were exposed to activin A, PGE2 or a COX-2 inhibitor (NS398), or activin A and NS398 together and CD36 mRNA levels measured (Supplementary Figure 4D). CD36 expression was repressed in RMF exposed to activin A; but exposure to PGE2 did not repress CD36. In addition, exposure of RMF to NS398 did not affect activin A-mediated CD36 repression demonstrating that COX-2 induction, and the subsequent secretion of PGE2, by vHMEC is neither necessary nor sufficient for CD36 repression in RMF.
To evaluate if telomere malfunction in vHMEC was necessary and/or sufficient for CD36 repression in RMF, RMF were co-cultured with vector-vHMEC (control), TRF2-vHMEC with increased telomere malfunction or hTERT-vHMEC with reduced telomere malfunction (19). CD36 mRNA levels were repressed in RMF co-cultured with TRF2-vHMEC (1.5-fold, p=0.01) but elevated in RMF co-cultured with hTERT-vHMEC (1.8-fold, p=0.03) compared to RMF co-cultured with vector-vHMEC (Figure 4D). These data demonstrate that telomere malfunction in vHMEC is necessary and sufficient for CD36 repression in RMF and expand our previous report about cell-extrinsic consequences of telomere malfunction (20).
Transient Exposure to Activin A Persistently Represses CD36 in RMF
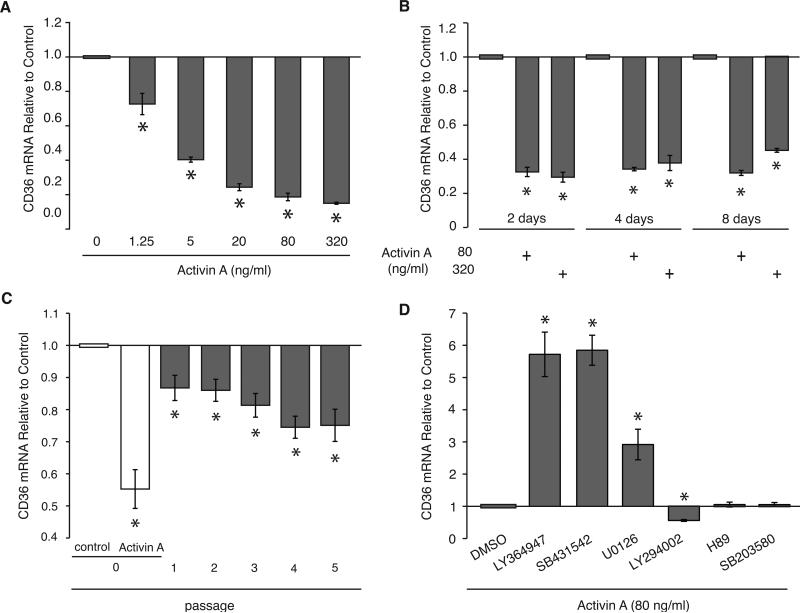
To gain further insights into the biological relevance of CD36 regulation by activin A, we analyzed the sensitivity and durability of this regulation in RMF in vitro. RMF exposed to physiological levels (~1.2 ng/ml) of activin A (30) for 48 hours showed CD36 repression in a dose-dependent manner, repression being observed (1.4-fold, p=0.05) with as little as 1.25 ng/ml of activin A (Figure 5A). Exposure of RMF to two doses of activin A for 2, 4 or 8 days repressed CD36 to a similar extent (2.6-fold to 3.4-fold, p<0.05) under all conditions demonstrating that a 2 day exposure to activin A is sufficient for maximum CD36 repression in RMF (Figure 5B). Importantly, RMF exposed to activin A for 48 hours then propagated in the absence of activin A for 5 passages exhibited sustained (and even increasing) CD36 mRNA repression (1.3-fold, p=0.0006 at P5) for several weeks after activin A removal (Figure 5C and Supplementary Figure 5). In summary, CD36 expression in RMF is exquisitely sensitive to physiological levels of activin A and even a brief exposure to activin A can result in prolonged CD36 repression.
Figure 5. Activin A represses CD36 in RMF, at low doses and persistently, in a TGFβR and MAPK-dependent manner.
(A-D) Average and SEM of CD36 mRNA fold-change relative to control cells, measured by QPCR. (A) One RMF exposed to the indicated doses of activin A for 48 hours. (B) Two RMF exposed to 80 and 320 ng/ml of activin A for 2, 4 or 8 days. (C) Four RMF exposed to activin A (80 ng/ml) for 48 hours and CD36 mRNA levels determined immediately after exposure (P0) and for 5 passages (P1-P5) after activin A removal. (D) Three RMF exposed for 48 hours to activin A (80 ng/ml) alone or in the presence of DMSO vehicle control or 10 μM various pathway inhibitors: TGFβR1 inhibitors (LY364947 and SB431542), MAPKK inhibitor (UO126), PI3K inhibitor (LY294002), PKA inhibitor (H89) or p38 MAPK inhibitor (SB203580).
Activin A-Dependent Repression of CD36 Requires Activin A/TGF-β Family Type I Receptor and MAPK Pathways
To elucidate the mechanism(s) by which activin A represses CD36 in RMF, RMF were exposed to activin A alone or in the presence of various pathway inhibitors or vehicle control (control RMF) (Figure 5D). Activin A, like other TGF-β family members, signals through the TGF-β family type I and type II receptors. We found that RMF exposed to activin A plus TGF-β family type I receptor (TGFβR1) inhibitors (LY364947 and SB431542) exhibited higher levels of CD36 (5.7-fold, p=0.0001; 5.8-fold, p<0.0001; respectively) than control RMF, demonstrating that activin A/TGFβR1 signaling is required for activin A-dependent CD36 repression. Previous studies demonstrated that MAPK represses CD36 and that activin A utilizes the MAPK pathway (19, 31). RMF exposed to activin A plus MAPK Kinase (MAPKK) inhibitor (UO126) had higher CD36 levels (2.9-fold, p=0.004) than controls. Thus, the MAPK pathway is necessary for activin A-dependent CD36 repression. In contrast, RMF exposed to activin A plus PI3K inhibitor (LY294002) exhibited lower CD36 levels (1.8-fold, p<0.0001) than controls, suggesting that the PI3K pathway is required for the induction of CD36 expression. Finally, RMF exposed to activin A plus PKA inhibitor (H89) or activin A plus p38 MAPK inhibitor (SB203580) had similar levels of CD36 to controls, demonstrating that neither of these pathways are involved in regulating CD36 expression in this context.
Activin A Modulates CD36-Dependent Phenotypes
If a DDR and increased activin A secretion in HD epithelial cells was responsible for the CD36-modulated desmoplastic-like phenotypes (low adipocyte and high ECM content) observed in HD tissues in vivo (3), activin A should modulate these phenotypes in RMF in vitro.
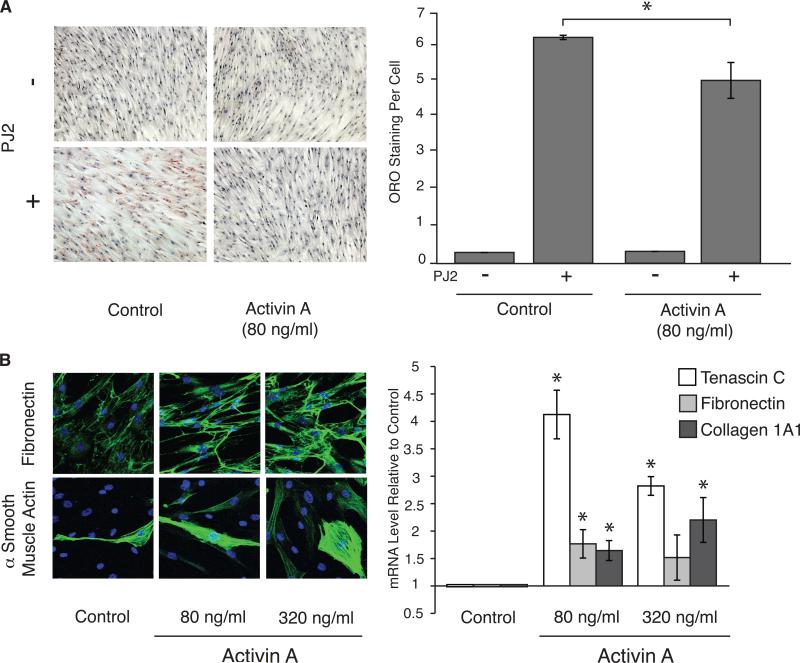
RMF were placed under proliferative or adipocyte differentiation conditions, in the absence or presence of activin A, and Oil Red O staining, an indicator of intracellular fat accumulation, was measured (Figure 6A) (32). RMF grown in the absence or presence of activin A both accumulated fat under differentiation conditions compared to proliferative conditions (15.7-fold and 25.4-fold, respectively, p<0.0001). However, RMF exposed to activin A accumulated significantly less fat under differentiation conditions than RMF grown without activin A (1.2-fold, p<0.0001).
Figure 6. Activin A modulates CD36-dependent phenotypes in vitro.
(A) Left: Representative bright field images (10X original magnification) of 2 RMF placed under proliferative (-PJ2) or adipocyte differentiation (+PJ2) conditions, in the absence (control) or presence of activin A (80 ng/ml), for 7 days and assessed for adipocyte formation by Oil Red O staining (red). Right: Average and SEM of Oil Red O staining per cell in 1 RMF. (B) 2 RMF were exposed to 80 and 320 ng/ml of activin A for 48 hours. (Left) Representative fluorescent images (20X original magnification) of Fibronectin (top) and αSMA (bottom) protein staining (green). (Right) Average and SEM of Tenascin C, Fibronectin, and Collagen 1A1 mRNA fold-change relative to untreated RMF, measured by QPCR.
RMF were exposed to two doses of activin A and protein and mRNA levels for selected ECM genes were assessed. Exposure of RMF to activin A induced fibronectin and α-SMA protein accumulation (Figure 6B, left) and Tenascin C, Fibronectin and Collagen 1A1 mRNAs (4.1-fold, p=0.0004; 1.8-fold, p=0.025; 1.7-fold, p=0.012; respectively, for 80 ng/ml) (Figure 6B, right) similarly at both doses. Thus, activin A can decrease fat accumulation and increase matrix accumulation in RMF in vitro, two prominent phenotypes of HD tissue modulated by CD36 expression.
Discussion
We previously showed, in vitro and in vivo, that CD36 is dramatically repressed in multiple stromal cell types within disease-free tissues from women with HD compared to women with LD (3). To define the mechanisms that account for CD36 repression, we tested the hypothesis that HD results from stress signaling in epithelial cells that induces the secretion of factors that repress CD36 and reprogram adjacent fibroblasts. Using cohorts previously used by us to study CD36-dependent phenotypes (3), we show that HD epithelial cells have more basal DNA damage (γH2AX intensity) than LD epithelial cells. Additionally, HD-vHMEC have more γH2AX foci after exogenous DNA damage, and take longer to resolve these foci, than LD-vHMEC. Paradoxically, HD-vHMEC also have increased viability/survival and decreased apoptosis after exogenous DNA damage, a property that could facilitate the escape of a mutated clone, leading to cancer. HD epithelial cells also have slightly shorter telomeres, both in vitro and in vivo, than LD epithelial cells. This relatively small difference is not surprising since telomere length is a tightly regulated phenotype.
We previously reported that DNA damage or telomere malfunction in epithelial cells results in increased secretion of activin A, which can act in a paracrine or autocrine fashion to induce its own expression, and the expression of many protumorigenic genes, in adjacent cells (19, 20). Similarly, we find that HD-vHMEC, with increased DNA damage, have higher activin A levels than LD-vHMEC. We describe a novel phenotype associated with this pathway by showing that CD36 expression in RMF is exquisitely sensitive to physiological levels of activin A (30), and that even a transient exposure to activin A can persistently repress CD36. Consistent with our hypothesis, conditioned-media from HD-vHMEC are more potent in repressing CD36 in RMF than conditioned-media from LD-vHMEC. Activin A and telomere malfunction in vHMEC are both necessary and sufficient for this repression. Importantly, activin A also modulates CD36-dependent desmoplastic-like phenotypes in vitro.
Our data suggest that CD36 repression may result from epigenetic modification, in addition to repression of its key regulator PPARγ. First, HD-HMFs maintain CD36 repression in culture over several passages in the absence of exogenous activin A or interaction with HD epithelial cells (3). Additionally, exposure to trichostatin A, a histone deacetylase inhibitor, increases CD36 expression more extensively in HD-HMF than LD-HMF (unpublished data). Finally, transient exposure to activin A persistently represses CD36 expression in RMF. Hence, even transient DNA damage in epithelial cells, and the subsequent secretion of activin A, could result in prolonged CD36 repression in adjacent fibroblasts.
We demonstrate that the activin A-dependent repression of CD36 in RMF requires both the activin A receptor (TGFβRI) and MAPK pathways, consistent with reports demonstrating that MAPK can regulate PPARγ function and repress CD36 (31) and with activin A's ability to repress PPARγ expression in RMF. Although TGF-β has been implicated in CD36 repression in macrophages (31), TGF-β levels are not elevated in HD-vHMEC, nor in vHMEC with telomere malfunction, suggesting that TGF-β does not participate in CD36 repression in this context. Finally, we show that, unlike many other pro-tumorigenic phenotypes previously described by us (19, 20), CD36 repression in RMF is not COX-2 dependent.
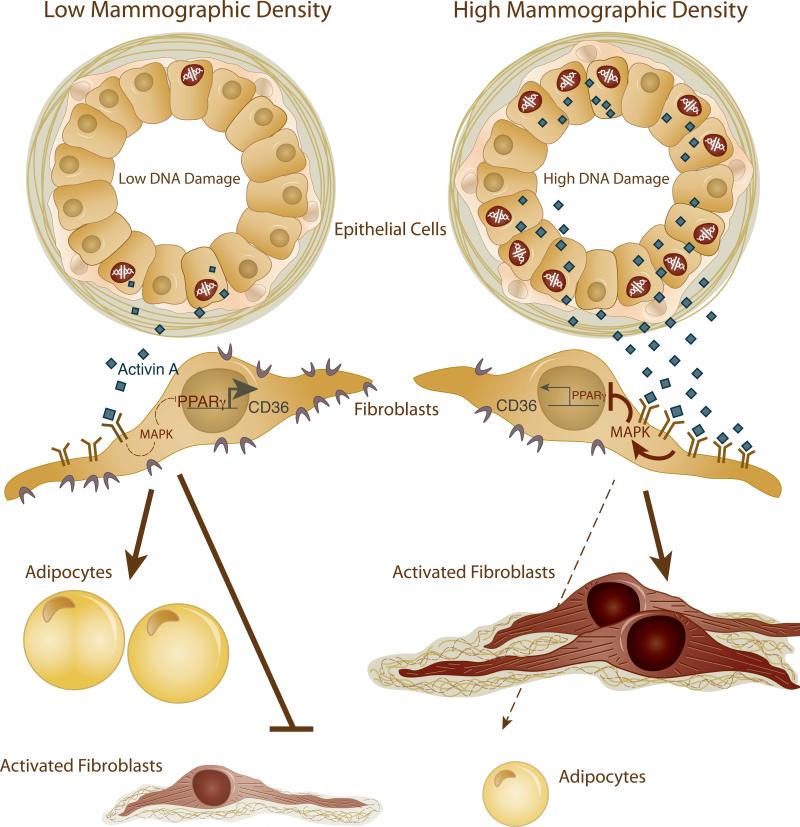
We envision a scenario in which: i) elevated basal DNA damage in HD epithelial cells results in increased activin A secretion; ii) activin A binds to its receptor on adjacent fibroblasts and activates the MAPK pathway; iii) MAPK pathway activation results in PPARγ phosphorylation and inhibition; iv) PPARγ inhibition leads to decreased CD36 transcription and subsequently, the induction of the desmoplastic-like phenotypes observed in HD tissues (Figure 7). In response, and as an extension, these desmoplastic-like fibroblasts can in turn promote motility of neighboring epithelial cells (20) and induce DNA damage of neighboring epithelial cells through the release of reactive oxygen species (ROS) (33). Damaged epithelial cells would further exacerbate pro-tumorigenic fibroblast phenotypes via the induction of activin A-dependent signaling pathways, thus propagating the DNA damage signal and the associated HD phenotypes throughout the tissue. Therefore, a breast with more DNA-damaged epithelial cells would exhibit more mammographically dense areas, leading to overall high MD. Our study highlights the reciprocal interactions between epithelial and stromal cells and their potential to induce carcinogenic processes and shows, for the first time, that there is a differential level of this cellular cross-talk in LD and HD tissues. The model described above suggests that these reciprocal interactions are initiated by a DNA damage event in the epithelial compartment. However, we cannot rule out that the initiating event occurs in the stromal compartment or, alternatively, simultaneously in both epithelial and stromal compartments. Regardless of the source of the initiating event, our data suggest that the alterations in LD and HD epithelial cells and fibroblasts are “intrinsic” and subsequently maintained in cells purified from tissues and propagated in vitro.
Figure 7.
Proposed model depicting the cross-talk between epithelial cells and fibroblasts in LD and HD tissues. Arrows and bars indicate induction or repression, respectively, of gene expression, protein activity or phenotypes. Solid and dotted lines indicate a strong or blunted/weak effect, respectively. Activin A and CD36 proteins are represented by diamonds or crescents, respectively.
Our data support one potential mechanism that contributes to MD (Figure 7). However, MD is a complex phenotype likely regulated by multiple pathways. Studies have implicated IGF-1 in the acquisition of HD (10). Interestingly, IGF-1 mRNA levels are higher (2-fold, p=0.09) in HD-HMF than LD-HMF and activin A induces IGF-1 expression (5.7-6.5-fold, p=0.0002) in RMF (unpublished data). In addition, although we ruled out the involvement of TGF-β in the DDR pathway, TGF-β could be involved in another context since it promotes fibrosis (28), a phenotype exhibited by HD.
MD is a heritable trait that can be modified by environmental factors (34). The same can be said of telomere length; for example chronic stress is correlated with shortened telomeres (35, 36). We, and others, have demonstrated that loss of telomere DNA can have cell-extrinsic consequences that may facilitate the development of a pro-tumorigenic stroma (20, 37). This may partially explain why loss of telomere DNA is associated with poor clinical outcome for women with breast cancer and increased risk of cancer (38, 39). This study is the first to suggest that telomere malfunction also contributes to HD.
In the breast, sex hormones drive expansion and involution of epithelial cells during menstrual cycling and lactation. Provocatively, expansion of epithelial cells during the luteal phase of the menstrual cycle is accompanied by modest but significant increases in MD (40) and coincides with the highest serum activin A levels (41). Successive expansion and involution may leave epithelial cells particularly vulnerable to DNA damage or telomere malfunction and select for genetically unstable cells able to resist apoptosis, like HD-vHMEC.
One might anticipate that as women age and their telomeres shorten, increased stress signaling would drive an increase in MD. However, MD remains the same or even decreases with age (42). This apparent contradiction could be explained by age-related lobular involution. This age-dependent loss of mammary epithelial cells would translate into decreased signaling to adjacent fibroblasts and a concomitant decrease in production of HD phenotypes. In fact, there is an inverse relationship between age-related involution and both MD and breast cancer risk (43, 44).
The demonstration that decreasing MD reduces breast cancer risk (45) provides tremendous opportunities for cancer prevention. Several drugs in clinical trials or already approved by the Federal Drug Administration modulate potential therapeutic targets identified in this study. Activin A is inhibited by competitive inhibitors of its receptor (e.g. bimagrumab), soluble receptor traps (e.g. dalantercept and sotatercept) and receptor kinase inhibitors (e.g. LY-2157299) (46) where as CD36 expression is increased by aspirin, dexamethasone, statins and adalimumab (47-50). The effects of these drugs on MD remain to be investigated.
Supplementary Material
Precis.
Findings provide new insights into how high mammographic density arises in the breast and why this condition is associated with breast cancer risk, with implications for the definition of novel invention targets to prevent breast cancer.
Acknowledgments
We thank Dr. Blackburn (UCSF) for thoughtful discussions and advice, Tlsty laboratory members, particularly Dr. Gascard, for editorial assistance and the Nikon Imaging Center (UCSF) for imaging support. We also thank Dr. Au (UCSF), Drs. Hornik and Kim (Kaiser Foundation Research Institute, Oakland, CA), Dr. Sukumar (Johns Hopkins Sidney Kimmel Comprehensive Cancer Center, Baltimore, MD) and Ms. Wiles (Cooperative Human Tissue Network, Nashville, TN) for providing breast tissue samples.
Grant Support
Support was provided by NIH/NCI PO1 CA107584 to T.D.T, K.K., and B.P. (under LBNL contract No. DE-AC02-05CH11231), NIH/NCI RO1 CA097214 to T.D.T, CBCRP grant 14OB-0165 to T.D.T, NIH/NCI U54 CA143803 to T.D.T. and B.P. (UC Riverside) and R01 Research Supplement for Underrepresented Minorities to C.A.F.
Footnotes
No conflicts of interest
References
- 1.Tlsty TD, Coussens LM. Tumor stroma and regulation of cancer development. Annu Rev Pathol. 2006;1:119–150. doi: 10.1146/annurev.pathol.1.110304.100224. [DOI] [PubMed] [Google Scholar]
- 2.Bing C, Trayhurn P. New insights into adipose tissue atrophy in cancer cachexia. Proc Nutr Soc. 2009;68:385–392. doi: 10.1017/S0029665109990267. [DOI] [PubMed] [Google Scholar]
- 3.DeFilippis RA, Chang H, Dumont N, Rabban JT, Chen YY, Fontenay GV, et al. CD36 Repression Activates a Multicellular Stromal Program Shared by High Mammographic Density and Tumor Tissues. Cancer Discov. 2012;2:826–839. doi: 10.1158/2159-8290.CD-12-0107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Olumi AF, Grossfeld GD, Hayward SW, Carroll PR, Tlsty TD, Cunha GR. Carcinoma-associated fibroblasts direct tumor progression of initiated human prostatic epithelium. Cancer Res. 1999;59:5002–5011. doi: 10.1186/bcr138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barcellos-Hoff MH, Ravani SA. Irradiated mammary gland stroma promotes the expression of tumorigenic potential by unirradiated epithelial cells. Cancer Res. 2000;60:1254–1260. [PubMed] [Google Scholar]
- 6.McDaniel SM, Rumer KK, Biroc SL, Metz RP, Singh M, Porter W, et al. Remodeling of the mammary microenvironment after lactation promotes breast tumor cell metastasis. Am J Pathol. 2006;168:608–620. doi: 10.2353/ajpath.2006.050677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lyons TR, O'Brien J, Borges VF, Conklin MW, Keely PJ, Eliceiri KW, et al. Postpartum mammary gland involution drives progression of ductal carcinoma in situ through collagen and COX-2. Nat Med. 2011;17:1109–1115. doi: 10.1038/nm.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Finak G, Bertos N, Pepin F, Sadekova S, Souleimanova M, Zhao H, et al. Stromal gene expression predicts clinical outcome in breast cancer. Nat Med. 2008;14:518–527. doi: 10.1038/nm1764. [DOI] [PubMed] [Google Scholar]
- 9.Sharma M, Beck AH, Webster JA, Espinosa I, Montgomery K, Varma S, et al. Analysis of stromal signatures in the tumor microenvironment of ductal carcinoma in situ. Breast Cancer Res Treat. 2010;123:397–404. doi: 10.1007/s10549-009-0654-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guo YP, Martin LJ, Hanna W, Banerjee D, Miller N, Fishell E, et al. Growth factors and stromal matrix proteins associated with mammographic densities. Cancer Epidemiol Biomarkers Prev. 2001;10:243–248. [PubMed] [Google Scholar]
- 11.Li T, Sun L, Miller N, Nicklee T, Woo J, Hulse-Smith L, et al. The association of measured breast tissue characteristics with mammographic density and other risk factors for breast cancer. Cancer Epidemiol Biomarkers Prev. 2005;14:343–349. doi: 10.1158/1055-9965.EPI-04-0490. [DOI] [PubMed] [Google Scholar]
- 12.Boyd NF, Byng JW, Jong RA, Fishell EK, Little LE, Miller AB, et al. Quantitative classification of mammographic densities and breast cancer risk: results from the Canadian National Breast Screening Study. J Natl Cancer Inst. 1995;87:670–675. doi: 10.1093/jnci/87.9.670. [DOI] [PubMed] [Google Scholar]
- 13.Byrne C, Schairer C, Wolfe J, Parekh N, Salane M, Brinton LA, et al. Mammographic features and breast cancer risk: effects with time, age, and menopause status. J Natl Cancer Inst. 1995;87:1622–1629. doi: 10.1093/jnci/87.21.1622. [DOI] [PubMed] [Google Scholar]
- 14.Silverstein RL, Febbraio M. CD36, a scavenger receptor involved in immunity, metabolism, angiogenesis, and behavior. Sci Signal. 2009;2:re3. doi: 10.1126/scisignal.272re3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stone HB, Coleman CN, Anscher MS, McBride WH. Effects of radiation on normal tissue: consequences and mechanisms. Lancet Oncol. 2003;4:529–536. doi: 10.1016/s1470-2045(03)01191-4. [DOI] [PubMed] [Google Scholar]
- 16.Mitchell JR, Wood E, Collins K. A telomerase component is defective in the human disease dyskeratosis congenita. Nature. 1999;402:551–555. doi: 10.1038/990141. [DOI] [PubMed] [Google Scholar]
- 17.Alder JK, Chen JJ, Lancaster L, Danoff S, Su SC, Cogan JD, et al. Short telomeres are a risk factor for idiopathic pulmonary fibrosis. Proc Natl Acad Sci U S A. 2008;105:13051–13056. doi: 10.1073/pnas.0804280105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wiemann SU, Satyanarayana A, Tsahuridu M, Tillmann HL, Zender L, Klempnauer J, et al. Hepatocyte telomere shortening and senescence are general markers of human liver cirrhosis. FASEB J. 2002;16:935–942. doi: 10.1096/fj.01-0977com. [DOI] [PubMed] [Google Scholar]
- 19.Fordyce C, Fessenden T, Pickering C, Jung J, Singla V, Berman H, et al. DNA damage drives an activin a-dependent induction of cyclooxygenase-2 in premalignant cells and lesions. Cancer Prev Res (Phila) 2010;3:190–201. doi: 10.1158/1940-6207.CAPR-09-0229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fordyce CA, Patten KT, Fessenden TB, DeFilippis RA, Hwang ES, Zhao J, et al. Cell-extrinsic consequences of epithelial stress: activation of protumorigenic tissue phenotypes. Breast Cancer Res. 2012;14:R155. doi: 10.1186/bcr3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holst CR, Nuovo GJ, Esteller M, Chew K, Baylin SB, Herman JG, et al. Methylation of p16(INK4a) promoters occurs in vivo in histologically normal human mammary epithelia. Cancer Res. 2003;63:1596–1601. [PubMed] [Google Scholar]
- 22.Romanov SR, Kozakiewicz BK, Holst CR, Stampfer MR, Haupt LM, Tlsty TD. Normal human mammary epithelial cells spontaneously escape senescence and acquire genomic changes. Nature. 2001;409:633–637. doi: 10.1038/35054579. [DOI] [PubMed] [Google Scholar]
- 23.Stampfer M, Hallowes RC, Hackett AJ. Growth of normal human mammary cells in culture. In Vitro. 1980;16:415–425. doi: 10.1007/BF02618365. [DOI] [PubMed] [Google Scholar]
- 24.Treszezamsky AD, Kachnic LA, Feng Z, Zhang J, Tokadjian C, Powell SN. BRCA1- and BRCA2-deficient cells are sensitive to etoposide-induced DNA double-strand breaks via topoisomerase II. Cancer Res. 2007;67:7078–81. doi: 10.1158/0008-5472.CAN-07-0601. [DOI] [PubMed] [Google Scholar]
- 25.Lin KW, Yan J. The telomere length dynamic and methods of its assessment. J Cell Mol Med. 2005;9:977–989. doi: 10.1111/j.1582-4934.2005.tb00395.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ismail IH, Wadhra TI, Hammarsten O. An optimized method for detecting gamma-H2AX in blood cells reveals a significant interindividual variation in the gamma-H2AX response among humans. Nucleic Acids Res. 2007;35:e36. doi: 10.1093/nar/gkl1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kataoka Y, Bindokas VP, Duggan RC, Murley JS, Grdina DJ. Flow cytometric analysis of phosphorylated histone H2AX following exposure to ionizing radiation in human microvascular endothelial cells. J Radiat Res. 2006;47:245–57. doi: 10.1269/jrr.0628. [DOI] [PubMed] [Google Scholar]
- 28.Branton MH, Kopp JB. TGF-beta and fibrosis. Microbes Infect. 1999;1:1349–1365. doi: 10.1016/s1286-4579(99)00250-6. [DOI] [PubMed] [Google Scholar]
- 29.Nagy L, Tontonoz P, Alvarez JG, Chen H, Evans RM. Oxidized LDL regulates macrophage gene expression through ligand activation of PPARgamma. Cell. 1998;93:229–240. doi: 10.1016/s0092-8674(00)81574-3. [DOI] [PubMed] [Google Scholar]
- 30.Harada K, Shintani Y, Sakamoto Y, Wakatsuki M, Shitsukawa K, Saito S. Serum immunoreactive activin A levels in normal subjects and patients with various diseases. J Clin Endocrinol Metab. 1996;81:2125–2130. doi: 10.1210/jcem.81.6.8964839. [DOI] [PubMed] [Google Scholar]
- 31.Han J, Hajjar DP, Tauras JM, Feng J, Gotto AM, Jr, Nicholson AC. Transforming growth factor-beta1 (TGF-beta1) and TGF-beta2 decrease expression of CD36, the type B scavenger receptor, through mitogen-activated protein kinase phosphorylation of peroxisome proliferator-activated receptor-gamma. J Biol Chem. 2000;275:1241–1246. doi: 10.1074/jbc.275.2.1241. [DOI] [PubMed] [Google Scholar]
- 32.Chang H, DeFilippis RA, Tlsty TD, Parvin B. Graphical methods for quantifying macromolecules through bright field imaging. Bioinformatics. 2009;25:1070–1075. doi: 10.1093/bioinformatics/btn426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cirri P, Chiarugi P. Cancer associated fibroblasts: the dark side of the coin. Am J Cancer Res. 2011;1:482–497. [PMC free article] [PubMed] [Google Scholar]
- 34.Boyd NF, Dite GS, Stone J, Gunasekara A, English DR, McCredie MR, et al. Heritability of mammographic density, a risk factor for breast cancer. N Engl J Med. 2002;347:886–894. doi: 10.1056/NEJMoa013390. [DOI] [PubMed] [Google Scholar]
- 35.Broer L, Codd V, Nyholt DR, Deelen J, Mangino M, Willemsen G, et al. Meta-analysis of telomere length in 19 713 subjects reveals high heritability, stronger maternal inheritance and a paternal age effect. Eur J Hum Genet. 2013;21:1163–1168. doi: 10.1038/ejhg.2012.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Epel ES, Blackburn EH, Lin J, Dhabhar FS, Adler NE, Morrow JD, et al. Accelerated telomere shortening in response to life stress. Proc Natl Acad Sci U S A. 2004;101:17312–17315. doi: 10.1073/pnas.0407162101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Coppe JP, Desprez PY, Krtolica A, Campisi J. The senescence-associated secretory phenotype: the dark side of tumor suppression. Annu Rev Pathol. 2010;5:99–118. doi: 10.1146/annurev-pathol-121808-102144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fordyce CA, Heaphy CM, Bisoffi M, Wyaco JL, Joste NE, Mangalik A, et al. Telomere content correlates with stage and prognosis in breast cancer. Breast Cancer Res Treat. 2006;99:193–202. doi: 10.1007/s10549-006-9204-1. [DOI] [PubMed] [Google Scholar]
- 39.Willeit P, Willeit J, Mayr A, Weger S, Oberhollenzer F, Brandstatter A, et al. Telomere length and risk of incident cancer and cancer mortality. JAMA. 2010;304:69–75. doi: 10.1001/jama.2010.897. [DOI] [PubMed] [Google Scholar]
- 40.Ursin G, Parisky YR, Pike MC, Spicer DV. Mammographic density changes during the menstrual cycle. Cancer Epidemiol Biomarkers Prev. 2001;10:141–142. [PubMed] [Google Scholar]
- 41.Muttukrishna S, Fowler PA, George L, Groome NP, Knight PG. Changes in peripheral serum levels of total activin A during the human menstrual cycle and pregnancy. J Clin Endocrinol Metab. 1996;81:3328–3334. doi: 10.1210/jcem.81.9.8784092. [DOI] [PubMed] [Google Scholar]
- 42.Checka CM, Chun JE, Schnabel FR, Lee J, Toth H. The relationship of mammographic density and age: implications for breast cancer screening. AJR Am J Roentgenol. 2012;98:W292–295. doi: 10.2214/AJR.10.6049. [DOI] [PubMed] [Google Scholar]
- 43.Ghosh K, Hartmann LC, Reynolds C, Visscher DW, Brandt KR, Vierkant RA, et al. Association between mammographic density and age-related lobular involution of the breast. J Clin Oncol. 2010;28:2207–2212. doi: 10.1200/JCO.2009.23.4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Milanese TR, Hartmann LC, Sellers TA, Frost MH, Vierkant RA, Maloney SD, et al. Age-related lobular involution and risk of breast cancer. J Natl Cancer Inst. 2006;98:1600–1607. doi: 10.1093/jnci/djj439. [DOI] [PubMed] [Google Scholar]
- 45.Cuzick J, Warwick J, Pinney E, Duffy SW, Cawthorn S, Howell A, et al. Tamoxifen-induced reduction in mammographic density and breast cancer risk reduction: a nested case-control study. J Natl Cancer Inst. 2011;103:744–752. doi: 10.1093/jnci/djr079. [DOI] [PubMed] [Google Scholar]
- 46.Fields SZ, Parshad S, Anne M, Raftopoulos H, Alexander MJ, Sherman ML, et al. Activin receptor antagonists for cancer-related anemia and bone disease. Expert Opin Investig Drugs. 2013;22:87–101. doi: 10.1517/13543784.2013.738666. [DOI] [PubMed] [Google Scholar]
- 47.Vinals M, Bermudez I, Llaverias G, Alegret M, Sanchez RM, Vazquez-Carrera M, et al. Aspirin increases CD36, SR-BI, and ABCA1 expression in human THP-1 macrophages. Cardiovasc Res. 2005;66:141–149. doi: 10.1016/j.cardiores.2004.12.024. [DOI] [PubMed] [Google Scholar]
- 48.Matasic R, Dietz AB, Vuk-Pavlovic S. Dexamethasone inhibits dendritic cell maturation by redirecting differentiation of a subset of cells. J Leukoc Biol. 1999;66:909–914. doi: 10.1002/jlb.66.6.909. [DOI] [PubMed] [Google Scholar]
- 49.Ruiz-Velasco N, Dominguez A, Vega MA. Statins upregulate CD36 expression in human monocytes, an effect strengthened when combined with PPAR-gamma ligands Putative contribution of Rho GTPases in statin-induced CD36 expression. Biochem Pharmacol. 2004;67:303–313. doi: 10.1016/j.bcp.2003.09.006. [DOI] [PubMed] [Google Scholar]
- 50.Boyer JF, Balard P, Authier H, Faucon B, Bernad J, Mazieres B, et al. Tumor necrosis factor alpha and adalimumab differentially regulate CD36 expression in human monocytes. Arthritis Res Ther. 2007;9:R22. doi: 10.1186/ar2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.