Abstract
Background
Excessive and inappropriate immune responses are the hallmark of several autoimmune disorders, including the inflammatory bowel diseases (IBD): Crohn’s disease (CD) and ulcerative colitis (UC). A complex etiology involving both environmental and genetic factors influences IBD pathogenesis. The role of microRNAs (miRNAs), noncoding RNAs involved in regulating numerous biological processes, to IBD pathology, in terms of initiation and progression, remains ill-defined. In the present study, we evaluated the relationship between colon, peripheral blood, and saliva whole miRNome expression in IBD patients and non-inflammatory bowel disease (non-IBD) controls to identify miRNAs that could discriminate CD from UC. Quantitative real-time PCR (qRT-PCR) was used to validate and assess miRNA expression.
Results
Microarray analysis demonstrated that upwards of twenty six miRNAs were changed in CD and UC colon biopsies relative to the non-IBD controls. CD was associated with the differential expression of 10 miRNAs while UC was associated with 6 miRNAs in matched colon tissues. CD was associated with altered expression of 6 miRNAs while UC was associated with 9 miRNAs in whole blood. Expression of miR-101 in CD patients and miR-21, miR-31, miR-142-3p, and miR-142-5p in UC patients were altered in saliva.
Conclusions
Our results suggest that there is specific miRNA expression patterns associated with UC versus CD in three separate tissue/body fluids (colon, blood, and saliva). Further, the aberrant miRNA expression profiles indicate that miRNAs may be contributory to IBD pathogenesis, or at least reflect the underlying inflammation. Scrutinizing miRNA expression in saliva and blood samples may be beneficial in monitoring or diagnosing disease in IBD patients. A panel of miRNAs (miR-19a, miR-21, miR-31, miR-101, miR-146a, and miR-375) may be used as markers to identify and discriminate between CD and UC.
Electronic supplementary material
The online version of this article (doi:10.1186/s12865-015-0069-0) contains supplementary material, which is available to authorized users.
Keywords: microRNAs, IBD, Blood, Colon, Saliva
Background
Crohn’s disease (CD) and ulcerative colitis (UC) are the two primary and most prevalent forms of inflammatory bowel disease (IBD). Characterized by inappropriate and exacerbated immune responses within the gastrointestinal tract, the nature and location of the inflammatory changes defines each respective disease. Despite years of research, the exact cause of IBD is still relatively poorly understood. Nevertheless, it is clear that many factors, including genetic and epigenetic predispositions, the gut microbiota content, and environmental injuries or exposures, contribute significantly to the disease process. Linkage analysis and genome wide association studies (GWAS) have uncovered over 100 loci that have significant association with IBD [1-3]. Cytokines (IL-17), cytokine receptors (IL-23 receptor), and bacterial response elements (CARD15/NOD2, ATG16L1) are just a few of the pathways that have been found to be either mutated or otherwise altered in IBD patients and mouse models of chronic intestinal inflammation [2-5].
MicroRNAs (miRNAs), single stranded RNA molecules of 19–25 nucleotides, are poised to make significant contributions to defining the multifactorial etiology and pathobiology of IBD. Initially discovered in the early 1990s, this novel class of noncoding RNAs regulates gene expression post-transcriptionally to repress translation and/or promote mRNA degradation [6-9]. The biological footprint of miRNAs is widespread; over 30% of the genome is predicted to be actively regulated by miRNAs and studies have shown that miRNAs are involved in the control of a variety of normal cellular events including differentiation, organogenesis, and metabolism [10,11]. Further, aberrant expression of miRNAs has been associated with a growing number of disease states, including cancer and autoimmune diseases [12-17].
However, the consequence of how the alterations in miRNA expression occur and contribute to disease pathobiology remains mostly intangible at the moment. Understanding the role of miRNAs in the regulation of inflammation is an area of importance that may have broad significance to understanding the pathogenesis of IBD as well as a number of other diseases. Several studies have identified miRNAs that are associated with IBD in intestinal tissues and peripheral blood specimens [18-24]. In a previous study involving the interleukin-10 knockout (IL-10−/−) mice, a mouse model of chronic intestinal inflammation, we demonstrated selective dysregulation of miRNAs in colon tissue and peripheral blood leukocytes [25]. We therefore postulated that miRNA expression would be similarly disrupted in IBD patients with the possibility that some of the same miRNAs might have altered expression. The purpose of this study was to identify differentially expressed miRNAs that could selectively discriminate CD from UC and healthy controls using colon, blood, and saliva specimens. A further goal was to determine how the tissue microenvironment affected miRNA expression.
Results
Whole miRNome expression analysis in human IBD colon biopsies
To systematically analyze miRNA expression in IBD patients and identify candidate miRNAs for subsequent validation, expression analysis using oligonucleotide microarrays was initially performed on a small number of colon tissue biopsies from CD, UC, and non-IBD (Normal, NL) patients. Among the over 600 miRNA probes, the microarray detected 89 miRNAs with fold changes > 0.5 (Figure 1A) and 26 miRNAs with fold changes > 3.0 over the mean (Figure 1B).
Figure 1.
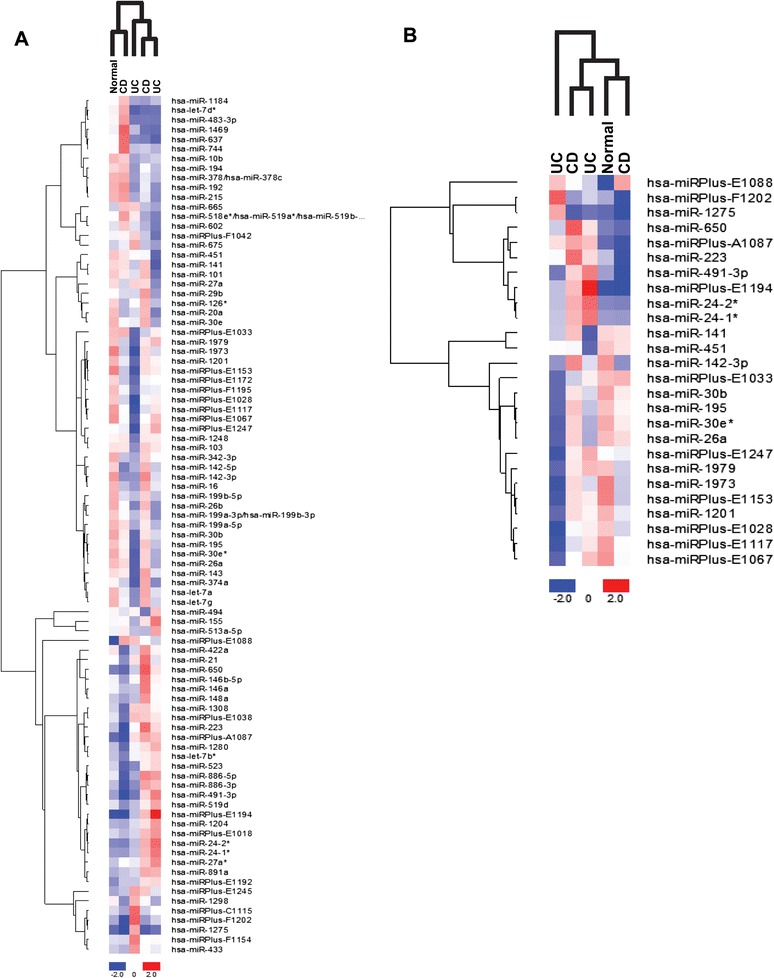
Heat map and unsupervised hierarchical clustering of miRNAs in IBD colon biopsies indicate dysregulation of miRNA expression. The miRCURY LNA microarray microRNA profiling service was used to examine miRNA expression in colon biopsies from Normal, CD, and UC subjects. (A) Heat map of fold changes > 0.5. (B) Heat map of fold changes > 3.0. Red color represents an expression level above mean, blue color represents expression lower than the mean. A Delta Log Median ratio of +/−1.0 is equal to a fold change of +/− 2.0.
miRNAs are differentially expressed in CD and UC intestinal biopsies
From the initial microarray screen, we selected 12 miRNAs for further validation. Among the qualities that factored into the inclusion of select miRNAs for validation was overall expression pattern, magnitude of expression, and previous associations with inflammation or disease. In terms of overall expression, we looked to include miRNAs that had consistent elevated or reduced expression in either the two CD or two UC biopsies relative to the non-IBD biopsy. miR-21, miR-26a, miR-101, miR-142-3p, miR-142-5p, miR-146a, miR-155, miR-223, and miR-494 were identified as candidate miRNAs in this manner. As an example miR-142-5p expression was decreased in both UC biopsies relative to the non-IBD biopsy. miR-26, miR-142-3p, and miR-223 were chosen as they were among the few miRNAs that had fold changes > 3.0 over the mean (Figure 1B). miR-19a, miR-31, and miR-375 were included as these miRNAs were elevated in our previous study with IL-10−/− mice or are associated with disease [25].
To validate miRNA expression, three separate condition-specific groups (NL, CD, or UC ) were each made by pooling total RNA from the corresponding NL, CD, or UC patient groups (n ≥ 20 biopsies per group) and analyzed via qRT-PCR for the selected miRNAs. In these pooled colon biopsies miR-31, miR-101, and miR-146a were significantly elevated in the CD samples while miR-375 was significantly reduced relative to the NL samples (Figure 2, column 1 & 4; p < 0.05). miR-19a, miR-21, miR-31, and miR-101 levels were significantly elevated in UC colon biopsies relative to the NL samples (Figure 2, column 1 & 4; p < 0.05). Secondary analysis using a Bonferroni correction (α = 0.05; n = 12) revealed that miR-146a was statistically elevated in CD biopsies (p < 0.004).
Figure 2.
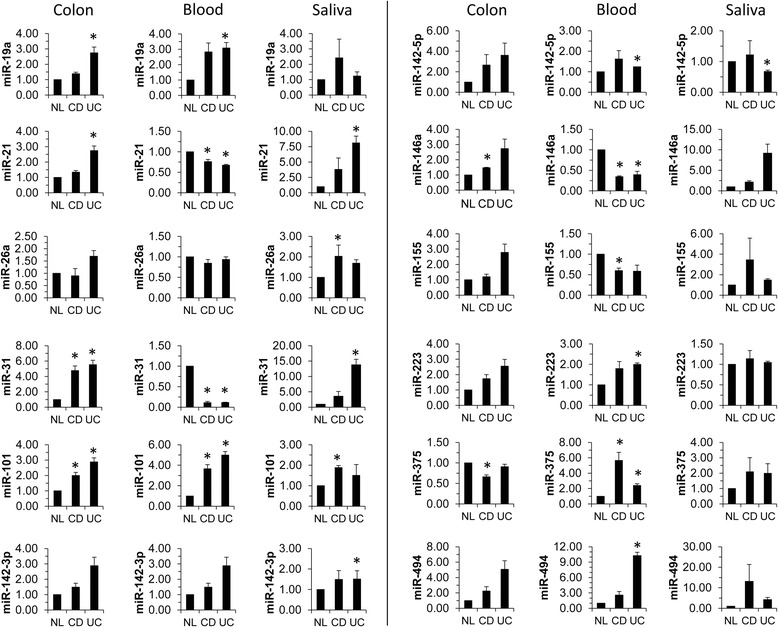
miRNA expression is altered in colon, blood, and saliva from Crohn’s disease and ulcerative colitis subjects. Total RNA from NL, CD, and UC colon biopsies was pooled and used for TaqMan qRT-PCR analysis for the indicated miRNAs. miRNA expression was normalized to U6 expression. Statistical significance was calculated using the Student’s t-test relative to the healthy NL control. At least 23 distinct specimens were included in each pooled group of colon biopsies. 30 and 5 distinct specimens were included in each pooled group of blood and saliva samples, respectively.
Circulating miRNAs are differentially expressed in CD and UC blood
To advance our knowledge in using less-invasive methods to screen for IBD, we characterized the extra-intestinal expression of miRNAs in peripheral blood. Peripheral blood is frequently used as a diagnostic tool for a varied number of diseases and conditions from simple cholesterol screenings to advanced genetic screens. Currently, there is not a single blood test that can make a diagnosis of IBD on its own merits; rather, multiple tests are required to come to a diagnosis of IBD. Thus, we explored the possibility of using whole blood miRNA expression profiling as an IBD diagnostic tool.
In order to determine if the miRNA profile uncovered in the colon biopsies could be replicated in a circulating extra-intestinal body fluid, whole blood was collected to examine miRNA expression levels. As above, total RNA from the blood specimens were pooled into three separate condition-specific groups (NL, CD, or UC) and analyzed for miRNA expression via qRT-PCR. A total of ten miRNAs (6 with CD and 9 with UC) had statistically significant altered expression in IBD peripheral blood pools versus non-IBD blood pools. In CD blood samples, miR-21, miR-31, miR-146a, and miR-155 were significantly reduced while miR-101 and miR-375 were significantly elevated relative to the NL samples (Figure 2, column 2 & 5; p < 0.05). miR-21, miR-31, and miR-146a were statistically significantly reduced and miR-19a, miR-101, miR142-5p, miR-223, miR-375, and miR-494 were statistically significantly elevated in UC blood samples relative to the NL samples (Figure 2, column 2 & 5; p < 0.05). Of these, miR-19a, miR-31, miR-101, miR-142-5p, miR-146a, miR-155, miR-223, miR-375, and miR-494 had not previously been associated with IBD to our knowledge. However, miR-21 has previously been shown to be differentially expressed in IBD [26,27]. In our previous study with IL-10−/− mice, miR-19a, miR-31, miR-101, miR-142-5p, miR-146a, miR-155, miR-223, and miR-375 were selectively dysregulated in whole blood of mice with mild intestinal pathology [25]. Secondary analysis using a Bonferroni correction (α = 0.05; n = 12) revealed that miR-21, miR-31, miR-142-5p, and miR-146a expression was significantly altered in CD and UC blood specimens (p < 0.004).
Evaluation of miRNA expression in oral fluid
Saliva is an important body fluid containing enzymatic proteins to begin digestion and antimicrobial proteins for immune protection. Similar to the peripheral blood, during pathophysiological states, such as cancer and inflammation, the composition of saliva can become altered as a reflection of these conditions [28,29]. Moreover, saliva provides a more accurate ‘real-time read-out’ than serum to screen and monitor the health status of patients. In addition, evidence suggests that saliva contains a transcriptome of thousands of mRNAs including miRNA [30-34]. Furthermore, due to the non-invasive nature of saliva collection for analytical purposes, saliva collection does not compromise a patient’s skin barrier nor does it require sedation, saliva is the ideal fluid for procuring multiple isolates to monitor disease progression. In our study, 5 miRNAs (1 CD and 4 UC) had statistically significant altered expression in pooled saliva samples from IBD patients relative to the non-IBD controls. In pooled saliva samples, miR-101 was significantly elevated in CD relative to the NL samples while miR-21, miR-31, and miR-142-3p were significantly elevated in UC relative to the NL samples (Figure 2, column 3; p < 0.05). miR-142-5p expression was significantly lowered in UC saliva samples relative to the NL samples (Figure 2, column 6; p < 0.05). No saliva specimens were statistically significant following a secondary analysis using a Bonferroni correction (α = 0.05; n = 12; p < 0.004).
To our knowledge this represents the first report of salivary miRNA diagnostics in IBD patients. Thus, examining the expression of miR-101, miR-21, miR-31, miR-142-3p, and miR-142-5p in saliva may be able to assist in diagnosing IBD.
The pooled colon, pooled blood, and pooled saliva specimen results are summarized in Additional file 1: Table S1 and Table S2 for CD and UC, respectively. miRNA expression profiles were altered in the extra-intestinal body fluids suggesting that these sources may effectively reflect the intestinal inflammation in IBD. miR-101, in particular, may be a key miRNA regulator in IBD, as it was statistically elevated in all three tissues from CD patients and two of the three UC tissues relative to the NL tissues.
miRNA expression is differentially expressed in matched colon biopsies from CD and UC subjects
As CD in particular is characterized by patchy areas of involved, inflamed tissues, we next sought to determine if a spatial relationship exists in miRNA expression patterns between these pockets of intestinal pathology and non-involved tissue in CD and UC. Paired endoscopically uninvolved (EU) and endoscopically involved (EI) colon biopsies from the same CD (Figure 3) or UC patients (Figure 4) were drawn from the previously analyzed pooled groups to examine their miRNA profiles. In the CD paired colon biopsies, nine miRNAs (miR-21, miR-31, miR-101, miR-142-3p, miR-142-5p, miR-155, miR-223, miR-375, and miR-494) were elevated at a statistically significant level in the endoscopically involved colonic tissue in comparison to the endoscopically uninvolved colonic tissue (Figure 3, Table 1). miR-21, miR-101, miR-142-5p, miR-146a, miR-155, and miR-223 were elevated at a statistically significant level in the UC endoscopically involved colonic tissue in comparison to the endoscopically uninvolved colonic tissue (Figure 4, Table 1). Although miR-31 was the most highly expressed miRNA in both the CD and UC matched pairs, it was statistically significant in only the CD pairs.
Figure 3.
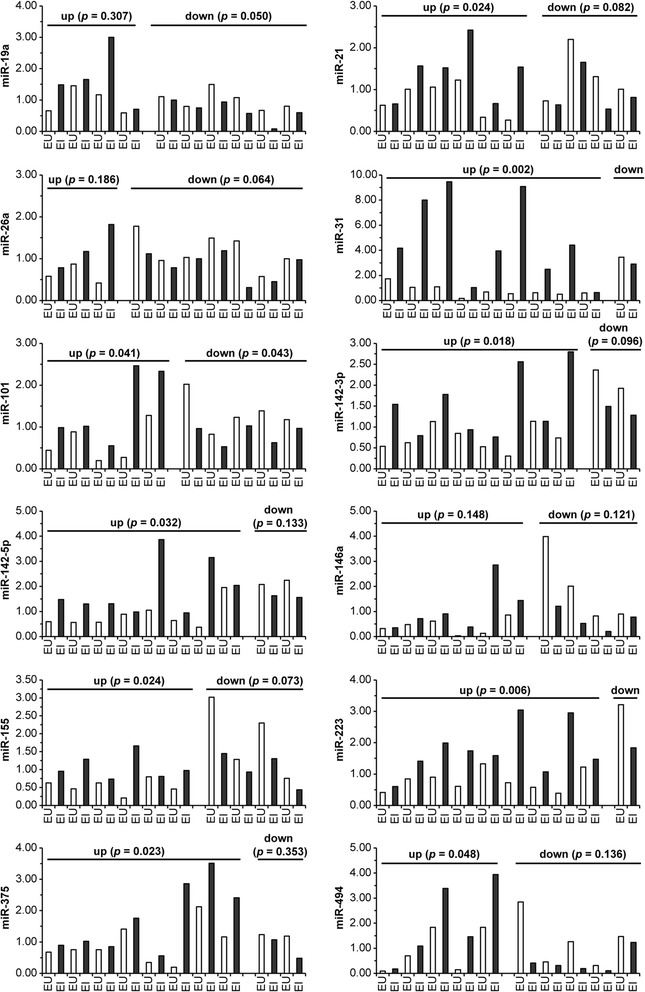
miRNA expression is differentially expressed in matched colon biopsies from Crohn’s disease subjects. Total RNA was isolated from matched endoscopically uninvolved (EU) and endoscopically involved (EI) colon biopsies from CD subjects. The RNA samples were used for TaqMan qRT-PCR analysis for the indicated miRNAs. miRNA expression was normalized to U6 expression. Pairings were subdivided according to miRNA expression trends. Statistical significance was calculated using the Student’s paired t-test relative to the endoscopically uninvolved CD tissue.
Figure 4.
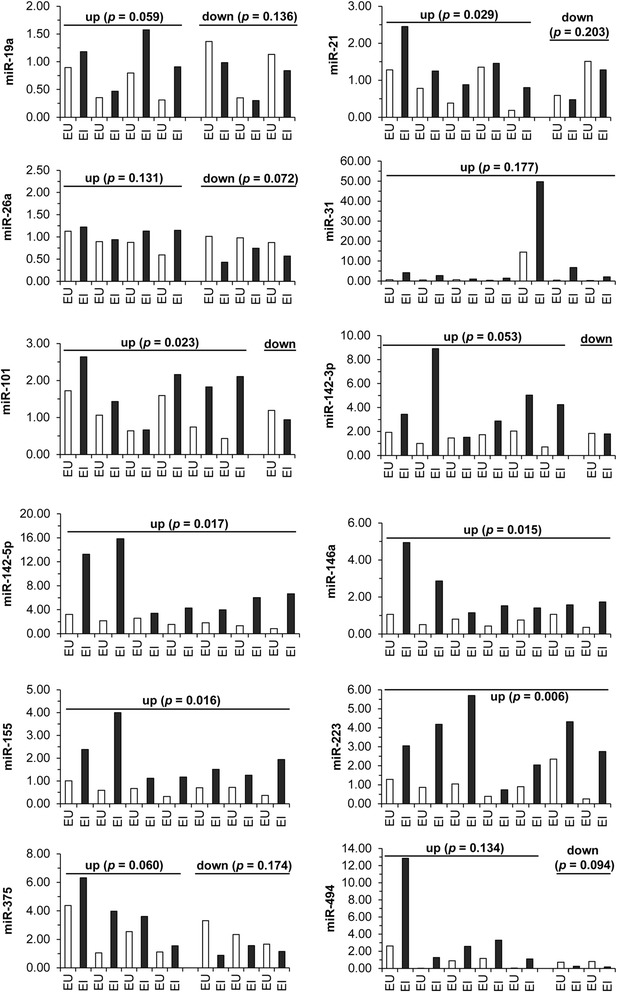
miRNA expression is differentially expressed in matched colon biopsies from ulcerative colitis subjects. Total RNA was isolated from matched endoscopically uninvolved (EU) and endoscopically involved (EI) colon biopsies from UC subjects. The RNA samples were used for TaqMan qRT-PCR analysis for the indicated miRNAs. miRNA expression was normalized to U6 expression. Pairings were subdivided according to miRNA expression trends. Statistical significance was calculated using the Student’s paired t-test relative to the endoscopically uninvolved UC tissue.
Table 1.
Summary of microRNA changes in paired colon biopsies from Crohn’s disease and ulcerative colitis subjects
| Crohn’s disease | Ulcerative colitis | |||||
|---|---|---|---|---|---|---|
| miRNA | N (Pairs) | Reduced ( p -value) | Elevated ( p -value) | N (Pairs) | Reduced ( p -value) | Elevated ( p -value) |
| miR-19a | 10 | 6 (0.050)* | 4 (0.307) | 7 | 3 (0.136) | 4 (0.059) |
| miR-21 | 10 | 4 (0.082) | 6 (0.024)* | 7 | 2 (0.203) | 5 (0.029)* |
| miR-26a | 10 | 7 (0.064) | 3 (0.186) | 7 | 3 (0.072) | 4 (0.131) |
| miR-31 | 10 | 1 (ND) | 9 (0.002)* | 7 | 0 (ND) | 7 (0.177) |
| miR-101 | 10 | 5 (0.043) | 5 (0.041)* | 7 | 1 (ND) | 6 (0.023)* |
| miR-142-3p | 10 | 2 (0.096) | 8 (0.018)* | 7 | 1 (ND) | 6 (0.053) |
| miR-142-5p | 10 | 2 (0.133) | 8 (0.032)* | 7 | 0 (ND) | 7 (0.017)* |
| miR-146a | 10 | 4 ( 0.121) | 6 (0.148) | 7 | 0 (ND) | 7 (0.015)* |
| miR-155 | 10 | 4 (0.073) | 6 (0.024)* | 7 | 0 (ND) | 7 (0.016)* |
| miR-223 | 10 | 1 (ND) | 9 (0.006)* | 7 | 0 (ND) | 7 (0.006)* |
| miR-375 | 10 | 2 (0.353) | 8 (0.023)* | 7 | 3 (0.174) | 4 (0.060) |
| miR-494 | 10 | 5 (0.136) | 5 (0.048)* | 7 | 2 (0.094) | 5 (0.134) |
ND, not determined due to insufficient number of samples in group.
*statistically significant values, p < 0.05.
In the miRNA microarray analysis, several of the non-annotated, human miRPlus miRNA candidates had altered expression (Figure 1A-B). miRPlus miRNA candidates are predicted miRNA sequences derived from Exiqon’s database of proprietary material, database mining, and publications. We investigated the expression of the top seven of these miRPlus miRNAs that were altered in the microarray analysis. In the matched CD biopsies, miRPlus-E1067 and miRPlus-E1117 were statistically significantly reduced while miRPlus-E1028 and miRPlus-F1202 were statistically significantly elevated in the endoscopically involved colonic tissue in comparison to the endoscopically uninvolved colonic tissue (Additional file 2: Figure S1). In the matched UC biopsies, miRPlus-E1067, miRPlus-E1088, and miRPlus-E1117 were statistically significantly reduced in the endoscopically involved colonic tissue in comparison to the endoscopically uninvolved colonic tissue (Additional file 3: Figure S2).
To confirm the reproducibility of the qRT-PCR analysis, biopsies were run in replicate; results for individual runs for 9 of the 12 miRNAs analyzed are shown (Additional file 4: Figure S3A). This confirms that there is little intra-sample variation between qRT-PCR runs.
We next decided to examine the expression of three miRNAs (miR-31, miR-146a, and miR-375) in pooled endoscopically uninvolved versus endoscopically involved IBD patient biopsies to determine if this panel of miRNAs could be used to separate CD from UC. A general pattern (Table 1) emerged for these 3 miRNAs in the paired analysis; miR-31 and miR-375 expression was increased at a statistically significant level in CD but not UC, while miR-146a is increased at a statistically significant level in UC but not CD. The CD and UC pairs from Figures 3 and 4 were pooled into endoscopically uninvolved versus endoscopically involved groups of CD or UC, respectively (CD EI, CD EU, UC EU, UC EI). Elevated expression of miR-31, miR-146a, and miR-375 was present in the CD pool while miR-31 and miR-146a were elevated in UC (Additional file 4: Figure S3B).
Roquin-1 and ATG16L1 expression is reduced in matched colon biopsies from CD but not UC subjects
To better understand the relationship that exists between miRNA expression and potential gene targets of interest, we used web-based target prediction programs (miRDB and TargetScan) to identify potential miRNA target genes (Additional file 1: Table S3). Of the predicted potential miRNA target genes, Roquin-1 (RC3H1), a RING finger E3 ligase with immunoregulatory properties in mice, was a common target for seven of the miRNAs with elevated expression. Autophagy related 16-like 1 (ATG16L1), one of the most commonly detected genetic variations in CD patients, was predicted to be a target of miR-142-3p [35,36]. Our unpublished observations and others have experimentally confirmed ATG16L1 as a regulatory target of miR-142-3p and miR-93-5p [37,38].
Analysis of Roquin-1 mRNA expression in the paired endoscopically uninvolved (EU) versus the endoscopically involved (EI) CD colon biopsies revealed statistically significant decreased expression of Roquin-1 in a subset of CD patients (Figure 5A). There was no appreciable difference in UC (Figure 5B). ATG16L1 expression was statistically decreased in a subset of CD patients (Figure 5C). Conversely, ATG16L1 expression was statistically increased in a subset of UC (Figure 5D).
Figure 5.
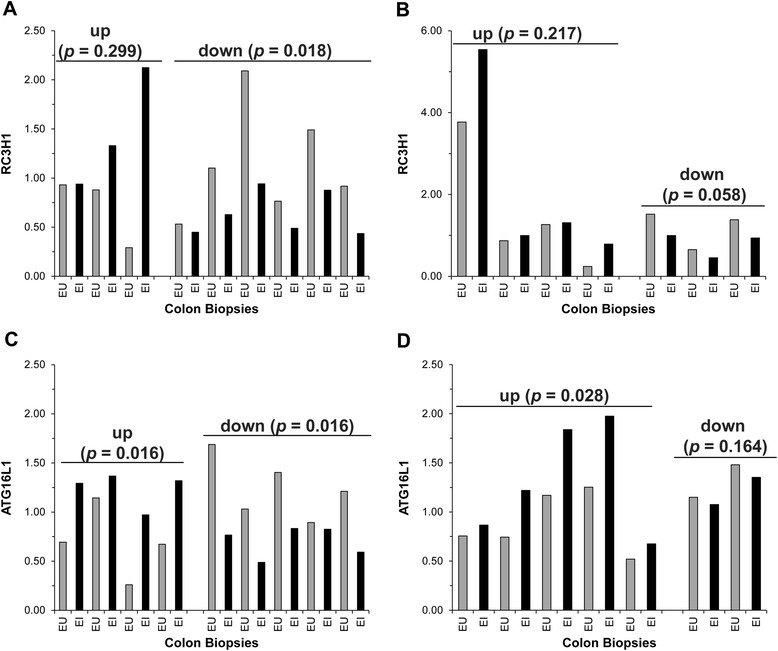
Roquin-1 and ATG16L1 expression is differentially expressed in a subset of CD and UC subjects. Total RNA from matched endoscopically uninvolved (EU) and endoscopically involved (EI) colon biopsies from (A, C) CD and (B, D) UC subjects was used to analyze Roquin-1 or ATG16L1 expression via qRT-PCR. Gene expression was normalized to GAPDH. Pairings were subdivided according to Roquin-1 or ATG16L1 expression trends. Statistical significance was calculated using the Student’s paired t-test relative to the endoscopically uninvolved IBD tissue.
Discussion
In the present study into the miRNA regulatory network of IBD, we were able to identify miRNAs (miR-19a, miR-21, miR-31, miR-101, miR-146a, and miR-375) that had statistically significant altered expression in pooled colon biopsy samples with several other miRNAs just outside the significance threshold (miR-26a, miR-142-3p, miR-155, miR-223, and miR-494; p ≤ 0.1). Of these miRNAs, miR-21, miR-31, miR-146a, and miR-375 have been identified in previous studies examining miRNA expression in CD and UC colon biopsies [18-21]. Further, these miRNAs are implicated in inflammation and cancer with miR-21 and miR-375 functioning as oncomiRs while miR-146a has been reported to be a tumor suppressor [39-41]. miR-31 is more complicated in that it has been reported to be both an oncomiR in lung cancer, and a tumor suppressor in breast cancer [42,43]. The increased expression of miR-146a and possibly miR-31 seem to be attempts to rein in the chronic inflammatory response in IBD. Previous studies that established miR-146a functionally opposing the effects of miR-155 via regulation of inflammatory signaling proteins, particularly NF-κB, support this [44-47]. miR-155 overexpression in mice results in a myeloproliferative disorder [44]. Conversely, miR-146a knockout mice develop hyperinflammatory and immunoproliferative disorders [45-47]. This suggests that, in IBD, the balance between these miRNAs with seemingly opposing functions is tilted and begs the question as to why the pro-inflammatory miRNAs win.
We additionally identified two miRNAs, miR-19a and miR-101, that have not previously been associated with IBD. miR-19a is a member of the miR-17-92 cluster and has been shown to be overexpressed in T-cell acute lymphoblastic leukemia and multiple myeloma where it was revealed that miR-19a negatively regulates the expression of CYLD and SOCS-1 respectively to promote cell survival and pathogenesis [48,49]. miR-101 has been associated with inflammation as a negative regulator of inducible costimulator (ICOS) and cancer cell stemness as a negative regulator of the corepressor C-terminal binding protein-2 (CtBP2) [50,51]. Once again, two miRNAs with seemingly opposing targets, miR-19a is pro-inflammatory while miR-101 is anti-proliferative, are both elevated in IBD.
Further, we were able to demonstrate that CD was associated with the differential expression of 10 miRNAs in a statistically significant manner (9 elevated, 1 decreased) while 6 miRNAs were elevated in a statistically significant manner in UC when comparing matched endoscopically uninvolved colon biopsies with endoscopically involved colon biopsies. Of these miRNAs, miR-21, miR-101, miR-142-5p, miR-155, and miR-223 had overlapping expression patterns between CD and UC. miR-146a was elevated in a statistically significant manner only in UC in the paired analysis while miR-19a, miR-31, miR-142-3p, miR-375, and miR-494 were altered in a statistically significant manner in CD.
Taken together, our analysis of the pooled colon samples versus the matched colon biopsies provides crucial but distinct information regarding the role of miRNAs in IBD. The former analysis suggests that a panel of miRNAs (miR-19a, miR-21, miR-31, miR-101, miR-146a, and miR-375) may be used as markers to identify and discriminate between CD and UC. The latter analysis provides more mechanistic information regarding miRNA involvement in disease pathology. Namely, it points to the underlying pathways that the miRNAs are altering to cause or promote disease. As an example, the overexpression of miR-31 and miR-146a, both of which target CD40L (predicted or experimentally confirmed), in the matched samples suggests that the CD40:CD40L costimulatory pathway plays an important role in promoting localized inflammation; CD40 and CD40L are elevated in IBD [52]. With further studies, we may even be able to distinguish patients at higher risk of colon cancer based on miRNA analysis.
As a proof of concept, we analyzed miRNA expression in the saliva of IBD patients. This new frontier of RNA diagnostics in oral fluids is being pioneered in the detection of cancer [33,53-56]. Although salivary miRNAs are emerging as a novel class of biomarkers in cancer, they have yet to be examined in other non-cancer diseases. In our study, we were able to detect differences in miRNA expression in the oral fluid from IBD patients. miR-21 in particular is a ubiquitous miRNA associated with many diseases and is an oncomiR [57,58]. Likewise, miR-31 and miR-101 have been implicated in cancer as well with the former being described as an oncomiR while the latter behaves as a tumor suppressor [43,59,60]. Finally, our results identified a potentially new IBD associated gene. In a subset of CD patients, RC3H1 expression was reduced in statistically significant manner in a subset of the endoscopically involved versus endoscopically uninvolved matched colony biopsy pairs. In mouse models deficient for Rc3h1, either the sanroque missense mutation (Rc3h1san/san) mouse or a genetrap knockout (Rc3h1gt/gt) mouse, an acute small intestinal inflammation develops among other phenotypes [50,61,62]. Roquin-1 is involved in the post-transcriptional regulation of mRNA. Roquin-1 localizes to P bodies and stress granules and plays a role in regulating mRNA turnover, and has been shown to regulate the expression of T cell coactivators, specifically ICOS and OX40 as well as the cytokines TNFα and IL-17. Given this role and the number of altered IBD-related miRNAs predicted or confirmed to target Roquin-1 (Additional file 1: Table S3), the importance of Roquin-1 in intestinal inflammation and human IBD necessitates further study.
This study also affirms the use of the IL-10−/− mouse as an effective model of IBD as miRNAs identified as differentially expressed in the CD and UC human samples were shared with the IL-10−/− mice [25]. Although there are frequently difficulties in going from bench to bedside, at least in this instance it seems that the IL-10−/− mouse model faithfully recreates some aspects of IBD.
Conclusions
In summary, selective sets of miRNA expression profiles may serve to separate CD and UC diagnostically. In this study, differentially expressed miRNAs in endoscopically involved versus endoscopically uninvolved CD and UC colon biopsies were discovered that indicate that a distinct miRNA profile exists within the intestinal microenvironment of IBD patients. Further, these study results provide support for the feasibility of using saliva and blood as diagnostic tools and advances the search for viable early detection methods for IBD. Importantly, this is the first report of altered miRNA expression in saliva samples from CD and UC patients.
Methods
Patients and controls
Fresh-frozen colonic mucosa was sampled via endoscopic pinch biopsies from the following groups: 1) CD, 2) UC, and 3) normal, healthy individuals scheduled for clinically indicated colonoscopies unrelated to IBD. For the CD and UC patients, one or two pinch biopsies were taken from endoscopically uninvolved appearing mucosa (EU) and a similar number from endoscopically involved (EI) tissue near the other site when possible. The pinch biopsies were immediately placed in RNALater for subsequent RNA isolation and analysis of miRNA expression. Blood and saliva samples were collected from the same groupings above albeit not the same individuals as from which the pinch biopsies were collected. Whole blood was collected in PAXgene Blood RNA Tubes (PreAnalytiX, Switzerland) while unstimulated saliva was collected in vessels containing the RNAprotect Saliva reagent (Qiagen, Valencia, CA). In total, 195 patient samples have been assessed from 35 controls, 42 CD, and 41 UC patients and broken down as: 23 UC pinch biopsies (7 matched pairs), 33 CD pinch biopsies (10 matched pairs), 34 normal pinch biopsies; 30 blood samples per group; and 5 saliva samples per group. All patient samples were collected under the auspices of protocols approved by The University of Texas Health Science Center at Houston Committee for the Protection of Human Subjects (the Institutional Review Board at UTHealth) and the Institutional Review Board for Baylor College of Medicine and Affiliated Hospitals. Written informed consent and informed assent (to level of age-related understanding) was obtained after the attending physician introduced the opportunity for study participation to the participants, or where appropriate, to the legal representative/parent(s) of the participants. Patient characteristics are detailed in Table 2.
Table 2.
Characteristics of IBD patients and controls
| CD | UC | Controls | |||
|---|---|---|---|---|---|
| No. of patients (total) | 42 | 41 | 35 | ||
| Patient specimens | |||||
| Pinch biopsies | 33 | 23 | 34 | ||
| Blood | 30 | 30 | 30 | ||
| Saliva | 5 | 5 | 5 | ||
| Sex | |||||
| Male, n (%) | 20 (47.6) | 28 (68.3) | 13 (37.1) | ||
| Female, n (%) | 22 (52.4) | 13 (31.7) | 22 (62.9) | ||
| Age (y) | |||||
| Mean | 42.7 | 47.6 | 56.6 | ||
| Male | 46.7 | 50.3 | 55.4 | ||
| Female | 39 | 41.8 | 57.4 | ||
| Range | 18-71 | 18-88 | 26-82 | ||
| Male | 18-71 | 27-88 | 26-82 | ||
| Female | 19-65 | 18-62 | 34-75 | ||
Isolation of RNA and real-time quantitative PCR (qRT-PCR)
Total RNA was isolated from colon biopsies using the miRNeasy Minikit (Qiagen) according to the manufacturer’s instructions. The RNeasy Protect Saliva Minikit (Qiagen) was used to isolate total RNA from the saliva samples with a minor modification recommended by the manufacturer. The PAXgene Blood miRNA Kit (PreAnalytiX) was used to isolate total RNA from the blood samples. cDNA was synthesized using either the High-Capacity cDNA Reverse Transcription Kit or the Taqman microRNA Reverse Transcription Kit (Applied Biosystems, Grand Island, NY). miRNA expression was quantified using TaqMan microRNA assays with the TaqMan Universal Master Mix II, No UNG reagent. To measure Roquin-1 and GAPDH transcript levels, either the Power SYBR Green PCR Master Mix (Applied Biosystems) or the TaqMan Universal Master Mix II, No UNG (Applied Biosystems) was used according to the manufacturer’s instructions. Samples were analyzed using the StepOnePlus real-time thermal cycler (Applied Biosystems) and software. Relative gene expression was calculated using the 2-ΔΔCt method and normalized to either GAPDH or U6 snRNA [63]. Roquin-1 and GAPDH gene specific primers were designed and purchased from Integrated DNA Technologies (Coralville, IA): Roquin-1 Forward, 5′-ACCAACCTTGCCTCCTACCT-3′, Roquin-1 Reverse, 5′-TAATCGCTGGTCCCTCATTC -3′; GAPDH Forward, 5′-TGCACCACCAACTGCTTAGC-3′, GAPDH Reverse, 5′-GGCATGGACTGTGGTCATGAG-3′. Taqman assays for mature miRNAs were purchased from Applied Biosystems.
miRNA Microarray
Total RNA (~1,000 ng each) from 1 healthy normal, 2 CD, and 2 UC colon biopsies were submitted to Exiqon (Woburn, MA) for miRNA expression analysis via the miRCURY LNA microarray microRNA profiling service. The miRNA array data has been deposited in the NCBI Gene Expression Omnibus (GEO) under the entry series GSE53867.
Statistical analysis
The statistical significance of data from these studies was determined using either an unpaired, two-sided Student’s t test (pooled samples) or a paired, two-sided Student’s t test (matched samples) in Excel. Data are presented as mean + SEM. Differences were considered statistically significant if p ≤ 0.05. A secondary analysis using the Bonferroni correction was included; differences were considered statistically significant if p ≤ 0.004.
Acknowledgments
This work has been supported by NIH grants DK035566, AI100159, Center Grant P30 DK56338, a Career Development Award from the Crohn’s & Colitis Foundation of America (CCFA), and an Institution-funded CCTS/KL2 career development grant. Dr. Graham and Dr. Hou are supported in part by the Office of Research and Development Medical Research Service Department of Veterans Affairs and VA HSR&D Center for Innovations in Quality, Effectiveness and Safety (#CIN 13–413), at the Michael E. DeBakey VA Medical Center, Houston, TX. Sample collection was supported in part by Public Health Service grant DK56338 which funds the Texas Medical Center Digestive Diseases Center. The contents are solely the responsibility of the authors and do not necessarily represent the official views of the VA or NIH.
Abbreviations
- IBD
Inflammatory bowel disease
- CD
Crohn’s disease
- UC
Ulcerative colitis
- miRNA
microRNA
- GWAS
Genome wide association studies
- IL-10
Interleukin 10
- qRT-PCR
Quantitative real-time PCR
- EU
Endoscopically uninvolved
- EI
Endoscopically involved
- NL
Normal
Additional files
MicroRNAs with statistically significant altered expression in Crohn’s disease by tissue site from pooled samples. Table S2. MicroRNAs with statistically significant altered expression in ulcerative colitis by tissue site from pooled samples. Table S3. Potential Targets for miRNAs with Elevated Expression in IBD.
miRPlus miRNA expression is altered in matched colon biopsies from Crohn’s disease subjects. Total RNA was isolated from matched endoscopically uninvolved (EU) and endoscopically involved (EI) colon biopsies from CD subjects. The RNA samples were used for SYBR Green qRT-PCR analysis for the indicated miRNAs. miRNA expression was normalized to U6 expression. Pairings were subdivided according to miRNA expression trends. Statistical significance was calculated using the Student’s paired t-test relative to the endoscopically uninvolved CD tissue.
miRPlus miRNA expression is altered in matched colon biopsies from ulcerative colitis subjects. Total RNA was isolated from matched endoscopically uninvolved (EU) and endoscopically involved (EI) colon biopsies from UC subjects. The RNA samples were used for SYBR Green qRT-PCR analysis for the indicated miRNAs. miRNA expression was normalized to U6 expression. Pairings were subdivided according to miRNA expression trends. Statistical significance was calculated using the Student’s paired t-test relative to the endoscopically uninvolved UC tissue.
Specificity and reproducibility of miRNA expression colon biopsies. (A) Replicate runs of RNA samples from matched endoscopically uninvolved versus endoscopically involved CD patient biopsies. Samples were analyzed via TaqMan qRT-PCR for the indicated miRNAs. (B) qRT-PCR analysis of miR-31, miR-146a, and miR-375 in pooled endoscopically uninvolved versus endoscopically involved CD and UC patient biopsies.
Footnotes
Competing interests
Dr. Graham is a consultant for RedHill Biopharma regarding novel therapies for Crohn’s disease.
Authors’ contributions
JSS, JRK, study concept and design; TA, BA, JH, HS, ARO, DYG, acquisition of samples and assessment of clinical disease; JSS acquisition of data; JSS, JRK, CS, analysis and interpretation of data; JSS, JRK, CS, statistical analysis; JSS, JRK, drafting of the manuscript; JSS, JRK, TA, BA, JH, HS, ARO, DYG, critical revision of the manuscript. All authors read and approved the final manuscript.
Contributor Information
Jeremy S Schaefer, Email: Jeremy.Schaefer@uth.tmc.edu.
Taraq Attumi, Email: attumi2002@yahoo.com.
Antone R Opekun, Email: aopekun@bcm.edu.
Bincy Abraham, Email: bincya@bcm.edu.
Jason Hou, Email: jkhou@bcm.edu.
Harold Shelby, Email: htshelby@bcm.edu.
David Y Graham, Email: dgraham@bcm.edu.
Charles Streckfus, Email: Charles.Streckfus@uth.tmc.edu.
John R Klein, Email: John.R.Klein@uth.tmc.edu.
References
- 1.Baptista ML, Amarante H, Picheth G, Sdepanian VL, Peterson N, Babasukumar U, et al. CARD15 and IL23R influences Crohn’s disease susceptibility but not disease phenotype in a Brazilian population. Inflamm Bowel Dis. 2008;14(5):674–9. doi: 10.1002/ibd.20372. [DOI] [PubMed] [Google Scholar]
- 2.Fujino S, Andoh A, Bamba S, Ogawa A, Hata K, Araki Y, et al. Increased expression of interleukin 17 in inflammatory bowel disease. Gut. 2003;52(1):65–70. doi: 10.1136/gut.52.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cho JH, Brant SR. Recent insights into the genetics of inflammatory bowel disease. Gastroenterology. 2011;140(6):1704–12. doi: 10.1053/j.gastro.2011.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lauriola M, Ugolini G, Rivetti S, Nani S, Rosati G, Zanotti S, et al. IL23R, NOD2/CARD15, ATG16L1 and PHOX2B polymorphisms in a group of patients with Crohn’s disease and correlation with sub-phenotypes. Int J Mol Med. 2011;27(3):469–77. doi: 10.3892/ijmm.2010.591. [DOI] [PubMed] [Google Scholar]
- 5.Montufar-Solis D, Schaefer J, Hicks MJ, Klein JR. Massive but selective cytokine dysregulation in the colon of IL-10−/− mice revealed by multiplex analysis. Int Immunol. 2008;20(1):141–54. doi: 10.1093/intimm/dxm126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403(6772):901–6. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 7.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–54. doi: 10.1016/0092-8674(93)90529-Y. [DOI] [PubMed] [Google Scholar]
- 8.Olsen PH, Ambros V. The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev Biol. 1999;216(2):671–80. doi: 10.1006/dbio.1999.9523. [DOI] [PubMed] [Google Scholar]
- 9.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75(5):855–62. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 10.Papagiannakopoulos T, Kosik KS. MicroRNAs: regulators of oncogenesis and stemness. BMC Med. 2008;6:15. doi: 10.1186/1741-7015-6-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liang Y, Ridzon D, Wong L, Chen C. Characterization of microRNA expression profiles in normal human tissues. BMC Genomics. 2007;8:166. doi: 10.1186/1471-2164-8-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Worley LA, Long MD, Onken MD, Harbour JW. Micro-RNAs associated with metastasis in uveal melanoma identified by multiplexed microarray profiling. Melanoma Res. 2008;18(3):184–90. doi: 10.1097/CMR.0b013e3282feeac6. [DOI] [PubMed] [Google Scholar]
- 13.Corsten MF, Miranda R, Kasmieh R, Krichevsky AM, Weissleder R, Shah K. MicroRNA-21 knockdown disrupts glioma growth in vivo and displays synergistic cytotoxicity with neural precursor cell delivered S-TRAIL in human gliomas. Cancer Res. 2007;67(19):8994–9000. doi: 10.1158/0008-5472.CAN-07-1045. [DOI] [PubMed] [Google Scholar]
- 14.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6(11):857–66. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 15.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105(30):10513–8. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pauley KM, Satoh M, Chan AL, Bubb MR, Reeves WH, Chan EK. Upregulated miR-146a expression in peripheral blood mononuclear cells from rheumatoid arthritis patients. Arthritis Res Ther. 2008;10(4):R101. doi: 10.1186/ar2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dai Y, Huang YS, Tang M, Lv TY, Hu CX, Tan YH, et al. Microarray analysis of microRNA expression in peripheral blood cells of systemic lupus erythematosus patients. Lupus. 2007;16(12):939–46. doi: 10.1177/0961203307084158. [DOI] [PubMed] [Google Scholar]
- 18.Wu F, Guo NJ, Tian H, Marohn M, Gearhart S, Bayless TM, et al. Peripheral blood microRNAs distinguish active ulcerative colitis and Crohn’s disease. Inflamm Bowel Dis. 2011;17(1):241–50. doi: 10.1002/ibd.21450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu F, Zikusoka M, Trindade A, Dassopoulos T, Harris ML, Bayless TM, et al. MicroRNAs are differentially expressed in ulcerative colitis and alter expression of macrophage inflammatory peptide-2 alpha. Gastroenterology. 2008;135(5):1624–35. doi: 10.1053/j.gastro.2008.07.068. [DOI] [PubMed] [Google Scholar]
- 20.Wu F, Zhang S, Dassopoulos T, Harris ML, Bayless TM, Meltzer SJ, et al. Identification of microRNAs associated with ileal and colonic Crohn’s disease. Inflamm Bowel Dis. 2010;16(10):1729–38. doi: 10.1002/ibd.21267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coskun M, Bjerrum JT, Seidelin JB, Nielsen OH. MicroRNAs in inflammatory bowel disease–pathogenesis, diagnostics and therapeutics. World J Gastroenterol. 2012;18(34):4629–34. doi: 10.3748/wjg.v18.i34.4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin J, Welker NC, Zhao Z, Li Y, Zhang J, Reuss SA, et al. Novel specific microRNA biomarkers in idiopathic inflammatory bowel disease unrelated to disease activity. Mod Pathol. 2014;27(4):602–8. doi: 10.1038/modpathol.2013.152. [DOI] [PubMed] [Google Scholar]
- 23.Lin J, Cao Q, Zhang J, Li Y, Shen B, Zhao Z, et al. MicroRNA expression patterns in indeterminate inflammatory bowel disease. Mod Pathol. 2013;26(1):148–54. doi: 10.1038/modpathol.2012.131. [DOI] [PubMed] [Google Scholar]
- 24.Duttagupta R, DiRienzo S, Jiang R, Bowers J, Gollub J, Kao J, et al. Genome-wide maps of circulating miRNA biomarkers for ulcerative colitis. PLoS One. 2012;7(2):e31241. doi: 10.1371/journal.pone.0031241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schaefer JS, Montufar-Solis D, Vigneswaran N, Klein JR. Selective upregulation of microRNA expression in peripheral blood leukocytes in IL-10−/− mice precedes expression in the colon. J Immunol. 2011;187(11):5834–41. doi: 10.4049/jimmunol.1100922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paraskevi A, Theodoropoulos G, Papaconstantinou I, Mantzaris G, Nikiteas N, Gazouli M. Circulating MicroRNA in inflammatory bowel disease. J Crohns Colitis. 2012;6(9):900–4. doi: 10.1016/j.crohns.2012.02.006. [DOI] [PubMed] [Google Scholar]
- 27.Zahm AM, Thayu M, Hand NJ, Horner A, Leonard MB, Friedman JR. Circulating microRNA is a biomarker of pediatric Crohn disease. J Pediatr Gastroenterol Nutr. 2011;53(1):26–33. doi: 10.1097/MPG.0b013e31822200cc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Streckfus CF, Mayorga-Wark O, Arreola D, Edwards C, Bigler L, Dubinsky WP. Breast cancer related proteins are present in saliva and are modulated secondary to ductal carcinoma in situ of the breast. Cancer Invest. 2008;26(2):159–67. doi: 10.1080/07357900701783883. [DOI] [PubMed] [Google Scholar]
- 29.Nguyen CQ, Hu MH, Li Y, Stewart C, Peck AB. Salivary gland tissue expression of interleukin-23 and interleukin-17 in Sjogren’s syndrome: findings in humans and mice. Arthritis Rheum. 2008;58(3):734–43. doi: 10.1002/art.23214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Patel RS, Jakymiw A, Yao B, Pauley BA, Carcamo WC, Katz J, et al. High resolution of microRNA signatures in human whole saliva. Arch Oral Biol. 2011;56(12):1506–13. doi: 10.1016/j.archoralbio.2011.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alevizos I, Alexander S, Turner RJ, Illei GG. MicroRNA expression profiles as biomarkers of minor salivary gland inflammation and dysfunction in Sjogren’s syndrome. Arthritis Rheum. 2011;63(2):535–44. doi: 10.1002/art.30131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weber JA, Baxter DH, Zhang S, Huang DY, Huang KH, Lee MJ, et al. The microRNA spectrum in 12 body fluids. Clin Chem. 2010;56(11):1733–41. doi: 10.1373/clinchem.2010.147405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park NJ, Zhou H, Elashoff D, Henson BS, Kastratovic DA, Abemayor E, et al. Salivary microRNA: discovery, characterization, and clinical utility for oral cancer detection. Clin Cancer Res. 2009;15(17):5473–7. doi: 10.1158/1078-0432.CCR-09-0736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Michael A, Bajracharya SD, Yuen PS, Zhou H, Star RA, Illei GG, et al. Exosomes from human saliva as a source of microRNA biomarkers. Oral Dis. 2010;16(1):34–8. doi: 10.1111/j.1601-0825.2009.01604.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cummings JR, Cooney R, Pathan S, Anderson CA, Barrett JC, Beckly J, et al. Confirmation of the role of ATG16L1 as a Crohn’s disease susceptibility gene. Inflamm Bowel Dis. 2007;13(8):941–6. doi: 10.1002/ibd.20162. [DOI] [PubMed] [Google Scholar]
- 36.Zhang HF, Qiu LX, Chen Y, Zhu WL, Mao C, Zhu LG, et al. ATG16L1 T300A polymorphism and Crohn’s disease susceptibility: evidence from 13,022 cases and 17,532 controls. Hum Genet. 2009;125(5–6):627–31. doi: 10.1007/s00439-009-0660-7. [DOI] [PubMed] [Google Scholar]
- 37.Zhai Z, Wu F, Dong F, Chuang AY, Messer JS, Boone DL, et al. Human autophagy gene ATG16L1 is post-transcriptionally regulated by MIR142-3p. Autophagy. 2014;10(3):468–79. doi: 10.4161/auto.27553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu C, Chen J, Xu HG, Zhou X, He Q, Li YL, et al. MIR106B and MIR93 prevent removal of bacteria from epithelial cells by disrupting ATG16L1-mediated autophagy. Gastroenterology. 2014;146(1):188–99. doi: 10.1053/j.gastro.2013.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.de Souza Rocha Simonini P, Breiling A, Gupta N, Malekpour M, Youns M, Omranipour R, et al. Epigenetically deregulated microRNA-375 is involved in a positive feedback loop with estrogen receptor alpha in breast cancer cells. Cancer Res. 2010;70(22):9175–84. doi: 10.1158/0008-5472.CAN-10-1318. [DOI] [PubMed] [Google Scholar]
- 40.Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133(2):647–58. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen G, Umelo IA, Lv S, Teugels E, Fostier K, Kronenberger P, et al. miR-146a inhibits cell growth, cell migration and induces apoptosis in non-small cell lung cancer cells. PLoS One. 2013;8(3):e60317. doi: 10.1371/journal.pone.0060317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Valastyan S, Reinhardt F, Benaich N, Calogrias D, Szasz AM, Wang ZC, et al. A pleiotropically acting microRNA, miR-31, inhibits breast cancer metastasis. Cell. 2009;137(6):1032–46. doi: 10.1016/j.cell.2009.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 43.Liu X, Sempere LF, Ouyang H, Memoli VA, Andrew AS, Luo Y, et al. MicroRNA-31 functions as an oncogenic microRNA in mouse and human lung cancer cells by repressing specific tumor suppressors. J Clin Invest. 2010;120(4):1298–309. doi: 10.1172/JCI39566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.O’Connell RM, Rao DS, Chaudhuri AA, Boldin MP, Taganov KD, Nicoll J, et al. Sustained expression of microRNA-155 in hematopoietic stem cells causes a myeloproliferative disorder. J Exp Med. 2008;205(3):585–94. doi: 10.1084/jem.20072108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao JL, Rao DS, Boldin MP, Taganov KD, O’Connell RM, Baltimore D. NF-kappaB dysregulation in microRNA-146a-deficient mice drives the development of myeloid malignancies. Proc Natl Acad Sci U S A. 2011;108(22):9184–9. doi: 10.1073/pnas.1105398108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boldin MP, Taganov KD, Rao DS, Yang L, Zhao JL, Kalwani M, et al. miR-146a is a significant brake on autoimmunity, myeloproliferation, and cancer in mice. J Exp Med. 2011;208(6):1189–201. doi: 10.1084/jem.20101823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.O’Connell RM, Rao DS, Baltimore D. microRNA regulation of inflammatory responses. Annu Rev Immunol. 2012;30:295–312. doi: 10.1146/annurev-immunol-020711-075013. [DOI] [PubMed] [Google Scholar]
- 48.Pichiorri F, Suh SS, Ladetto M, Kuehl M, Palumbo T, Drandi D, et al. MicroRNAs regulate critical genes associated with multiple myeloma pathogenesis. Proc Natl Acad Sci U S A. 2008;105(35):12885–90. doi: 10.1073/pnas.0806202105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ye H, Liu X, Lv M, Wu Y, Kuang S, Gong J, et al. MicroRNA and transcription factor co-regulatory network analysis reveals miR-19 inhibits CYLD in T-cell acute lymphoblastic leukemia. Nucleic Acids Res. 2012;40(12):5201–14. doi: 10.1093/nar/gks175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu D, Tan AH, Hu X, Athanasopoulos V, Simpson N, Silva DG, et al. Roquin represses autoimmunity by limiting inducible T-cell co-stimulator messenger RNA. Nature. 2007;450(7167):299–303. doi: 10.1038/nature06253. [DOI] [PubMed] [Google Scholar]
- 51.Cui TX, Kryczek I, Zhao L, Zhao E, Kuick R, Roh MH, et al. Myeloid-derived suppressor cells enhance stemness of cancer cells by inducing MicroRNA101 and suppressing the corepressor CtBP2. Immunity. 2013;39(3):611–21. doi: 10.1016/j.immuni.2013.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Danese S, Sans M, Fiocchi C. The CD40/CD40L costimulatory pathway in inflammatory bowel disease. Gut. 2004;53(7):1035–43. doi: 10.1136/gut.2003.026278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Alajez NM, Shi W, Wong D, Lenarduzzi M, Waldron J, Weinreb I, et al. Lin28b promotes head and neck cancer progression via modulation of the insulin-like growth factor survival pathway. Oncotarget. 2012;3(12):1641–52. doi: 10.18632/oncotarget.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brinkmann O, Wong DT. Salivary transcriptome biomarkers in oral squamous cell cancer detection. Adv Clin Chem. 2011;55:21–34. doi: 10.1016/B978-0-12-387042-1.00002-2. [DOI] [PubMed] [Google Scholar]
- 55.Matse JH, Yoshizawa J, Wang X, Elashoff D, Bolscher JG, Veerman EC, et al. Discovery and prevalidation of salivary extracellular microRNA biomarkers panel for the noninvasive detection of benign and malignant parotid gland tumors. Clin Cancer Res. 2013;19(11):3032–8. doi: 10.1158/1078-0432.CCR-12-3505. [DOI] [PubMed] [Google Scholar]
- 56.Yoshizawa JM, Wong DT. Salivary microRNAs and oral cancer detection. Methods Mol Biol. 2013;936:313–24. doi: 10.1007/978-1-62703-083-0_24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Folini M, Gandellini P, Longoni N, Profumo V, Callari M, Pennati M, et al. miR-21: an oncomir on strike in prostate cancer. Mol Cancer. 2010;9:12. doi: 10.1186/1476-4598-9-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Namwat N, Chusorn P, Loilome W, Techasen A, Puetkasichonpasutha J, Pairojkul C, et al. Expression profiles of oncomir miR-21 and tumor suppressor let-7a in the progression of opisthorchiasis-associated cholangiocarcinoma. Asian Pac J Cancer Prev. 2012;13(Suppl):65–9. [PubMed] [Google Scholar]
- 59.Hao Y, Gu X, Zhao Y, Greene S, Sha W, Smoot DT, et al. Enforced expression of miR-101 inhibits prostate cancer cell growth by modulating the COX-2 pathway in vivo. Cancer Prev Res. 2011;4(7):1073–83. doi: 10.1158/1940-6207.CAPR-10-0333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Strillacci A, Valerii MC, Sansone P, Caggiano C, Sgromo A, Vittori L, et al. Loss of miR-101 expression promotes Wnt/beta-catenin signalling pathway activation and malignancy in colon cancer cells. J Pathol. 2013;229(3):379–89. doi: 10.1002/path.4097. [DOI] [PubMed] [Google Scholar]
- 61.Schaefer JS, Montufar-Solis D, Nakra N, Vigneswaran N, Klein JR. Small intestine inflammation in Roquin-mutant and Roquin-deficient mice. PLoS One. 2013;8(2):e56436. doi: 10.1371/journal.pone.0056436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Montufar-Solis D, Vigneswaran N, Nakra N, Schaefer JS, Klein JR. Hematopoietic not systemic impairment of Roquin expression accounts for intestinal inflammation in Roquin-deficient mice. Sci Rep. 2014;4:4920. doi: 10.1038/srep04920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25(4):402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]


