Abstract
Background
Ezetimibe lowers plasma levels of low-density lipoprotein (LDL) cholesterol by inhibiting the activity of the Niemann–Pick C1-like 1 (NPC1L1) protein. However, whether such inhibition reduces the risk of coronary heart disease is not known. Human mutations that inactivate a gene encoding a drug target can mimic the action of an inhibitory drug and thus can be used to infer potential effects of that drug.
Methods
We sequenced the exons of NPC1L1 in 7364 patients with coronary heart disease and in 14,728 controls without such disease who were of European, African, or South Asian ancestry. We identified carriers of inactivating mutations (nonsense, splice-site, or frameshift mutations). In addition, we genotyped a specific inactivating mutation (p.Arg406X) in 22,590 patients with coronary heart disease and in 68,412 controls. We tested the association between the presence of an inactivating mutation and both plasma lipid levels and the risk of coronary heart disease.
Results
With sequencing, we identified 15 distinct NPC1L1 inactivating mutations; approximately 1 in every 650 persons was a heterozygous carrier for 1 of these mutations. Heterozygous carriers of NPC1L1 inactivating mutations had a mean LDL cholesterol level that was 12 mg per deciliter (0.31 mmol per liter) lower than that in noncarriers (P = 0.04). Carrier status was associated with a relative reduction of 53% in the risk of coronary heart disease (odds ratio for carriers, 0.47; 95% confidence interval, 0.25 to 0.87; P = 0.008). In total, only 11 of 29,954 patients with coronary heart disease had an inactivating mutation (carrier frequency, 0.04%) in contrast to 71 of 83,140 controls (carrier frequency, 0.09%).
Conclusions
Naturally occurring mutations that disrupt NPC1L1 function were found to be associated with reduced plasma LDL cholesterol levels and a reduced risk of coronary heart disease. (Funded by the National Institutes of Health and others.)
Ezetimibe, a drug that is commonly prescribed to reduce plasma levels of low-density lipoprotein (LDL) cholesterol, inhibits the function of the protein encoded by the Niemann–Pick C1-like 1 gene (NPC1L1).1 NPC1L1 protein, which is expressed in the small intestine and liver, functions as a transporter of dietary cholesterol from the gut lumen into intestinal enterocytes.2,3 Because of its ability to block sterol absorption by about 50%,4 ezetimibe lowers plasma LDL cholesterol levels by 15 to 20%.5 However, it is uncertain whether inhibiting NPC1L1 — either through ezetimibe treatment or by other means — reduces the risk of clinical coronary heart disease.6
Naturally occurring DNA sequence variants in humans that affect the activity of one or more protein targets can be used to estimate the potential efficacy and toxicity of a drug targeting such proteins.7,8 Genomewide association studies have identified common DNA sequence variants in NPC1L1 associated with modest alterations in plasma LDL cholesterol levels.9 However, it is difficult to discern precisely how variants that are discovered through genomewide association studies affect the activity of a gene.
In contrast, some DNA mutations that arise in the protein-coding sequence can completely inactivate a gene. Inactivating mutations can be single-base changes that introduce a stop codon and that lead to premature truncation of a protein (nonsense mutations), insertions or deletions (indels) of DNA that scramble the protein translation beyond the variant site (frameshift mutations), or point mutations at modification sites of the nascent pre–messenger RNA transcript that alter the splicing process10 (splice-site mutations). Because such mutations — which are variously termed protein-disruptive, protein-inactivating, loss-of-function, or null — profoundly affect protein function, they are typically very rare in the population as a consequence of natural selection.
We tested the hypothesis that protein-inactivating mutations in NPC1L1 reduce both the LDL cholesterol level and the risk of coronary heart disease. We sequenced the coding regions of NPC1L1 in a large number of persons, identified carriers of mutations that inactivate this gene, and determined whether persons who carry a heterozygous inactivating mutation had a lower LDL cholesterol level and a lower risk of coronary heart disease than noncarriers of these mutations.
Methods
Study Design
We conducted this study using data and DNA samples from 16 case–control studies and cohort studies. All study participants provided written informed consent for genetic studies. The first and last authors designed the study. The institutional review boards at the Broad Institute and each participating site approved the study protocols. The first and last authors vouch for the accuracy and completeness of the data and all analyses.
Study Participants
During the first phase of the study, we sequenced the 20 protein-coding exons in NPC1L1 in samples obtained from 22,092 participants from seven case–control studies and two prospective cohort studies (see Table S1 in the Supplementary Appendix, available with the full text of this article at NEJM.org). The case–control studies included the Exome Sequencing Project Early-Onset Myocardial Infarction (ESP-EOMI) study conducted by the National Heart, Lung, and Blood Institute,11 the Italian Atherosclerosis Thrombosis and Vascular Biology (ATVB) study,12 the Ottawa Heart Study (OHS),13 the Precocious Coronary Artery Disease (PROCARDIS) study,14 the Pakistan Risk of Myocardial Infarction Study (PROMIS),15 the Registre Gironi del COR (Gerona Heart Registry or REGICOR) study,16 and the Munich Myocardial Infarction (Munich-MI) study.17 The prospective cohort studies included the Atherosclerosis Risk in Communities (ARIC) study18 and the Jackson Heart Study (JHS).19
During the second phase of the study, we genotyped the most common inactivating mutation in NPC1L1 on the basis of data obtained during the sequencing phase (p.Arg406X) in nine independent sample sets from a total of 91,002 participants (Table S2 in the Supplementary Appendix). These nine sample sets were from participants in the ARIC study (participants who did not undergo sequencing), the Vanderbilt University Medical Center Biorepository (BioVU),20 the Genetics of Diabetes Audit and Research Tayside (GoDARTS) study,21 the German North and German South Coronary Artery Disease studies,22 the Mayo Vascular Diseases Biorepository (Mayo),23 PROCARDIS (participants who did not undergo sequencing), the Women’s Genome Health Study (WGHS),24 and the Women’s Health Initiative (WHI).25
Clinical Data
Data obtained for all the participants from both the sequencing and genotyping phases of the study included a medical history and laboratory assessment for cardiovascular risk factors, as described previously for each study. The participants were of African ancestry (2836 participants from ARIC, 2251 from JHS, and 455 from ESP-EOMI), South Asian ancestry (1951 participants from PROMIS), or European ancestry (all the other participants).
For each study cohort, available clinical data were used to define coronary heart disease. The definitions, which therefore varied from cohort to cohort, are provided in Tables S1 and S2 in the Supplementary Appendix.
Sequencing and Genotyping
Sequence data for NPC1L1 were extracted from exome sequences generated at the Broad Institute, the Human Genome Sequencing Center at Baylor College of Medicine, or the University of Washington with the use of protocols that are described in the Supplementary Appendix. Briefly, sequence reads were aligned to the human reference genome (build HG19), and the basic alignment files for sequenced samples were combined for the purpose of identifying variant positions. Single-nucleotide variants (SNVs) and indels were identified, and quality control procedures were applied to remove outlier samples and outlier variants, as described in the Supplementary Appendix.
For the purposes of this study, we defined inactivating mutations as any one of the following: SNVs leading to a stop codon substitution (nonsense mutations), SNVs occurring within two base pairs of an exon–intron boundary (splice-site mutations), or DNA insertions or deletions leading to a change in the reading frame and the introduction of a premature stop codon (frameshift mutations). The positions of nonsense, splice-site, and frameshift mutations were based on the complementary DNA reference sequence for NPC1L1 (NM_013389.2) with the ATG initiation codon, encoding methionine, numbered as residue 1 or p.Met1.
To obtain additional data for a particular nonsense mutation (p.Arg406X) observed from sequencing NPC1L1, we genotyped the variant site in additional samples using the HumanExome Bead-Chip Kit (Illumina), according to the manufacturer’s recommended protocol. (See the Methods section in the Supplementary Appendix for details.)
Technical Validation of Sequencing and Genotyping
To assess the accuracy of next-generation sequencing methods, we performed Sanger sequencing on samples obtained from all participants who carried inactivating mutations in the ATVB study. To assess the accuracy of the genotyping of NPC1L1 p.Arg406X with the HumanExome BeadChip kit, we compared these genotypes with those derived from next-generation sequencing for a subset of samples.
Statistical Analysis
We first tested the association between NPC1L1 protein-inactivating mutations and plasma lipid levels. For participants who were receiving lipid-lowering therapy, we accounted for an average reduction in total cholesterol and LDL cholesterol levels of 20% and 30%, respectively,26 by adjusting the measured values accordingly. We did not adjust levels of high-density lipoprotein (HDL) cholesterol or triglycerides in these participants. Status with respect to the use of lipid-lowering medication was available for participants in ARIC, JHS, Munich-MI, PROCARDIS, REGICOR, and WGHS. When possible, we combined primary data for studies that included only one participant with an inactivating mutation with data for other studies involving participants of the same ancestry in order to create a larger data set. We performed regression analysis with a linear model that was adjusted for age and sex, along with an indicator variable for the study if applicable, to test for an association between the presence of inactivating mutations in NPC1L1 and levels of total cholesterol, LDL cholesterol, HDL cholesterol, and log-transformed triglyceride levels in each sample set. We combined results first within ancestry groups and then across ancestry groups, using fixed-effects meta-analyses.
We next tested for an association between protein-inactivating mutations in NPC1L1 and the risk of coronary heart disease. In each study, we estimated the odds ratio for disease among carriers of any NPC1L1 inactivating mutation, as compared with noncarriers. We then calculated the summary odds ratios and 95% confidence intervals for coronary heart disease among carriers, using a Mantel–Haenszel fixed-effects meta-analysis without continuity correction, a method that is robust with low (and even zero) counts and resultant odds ratios. A P value of less than 0.05 was considered to indicate statistical significance. The R software program (R Project for Statistical Computing) was used for all analyses.
Results
Rare Inactivating Mutations in NPC1L1
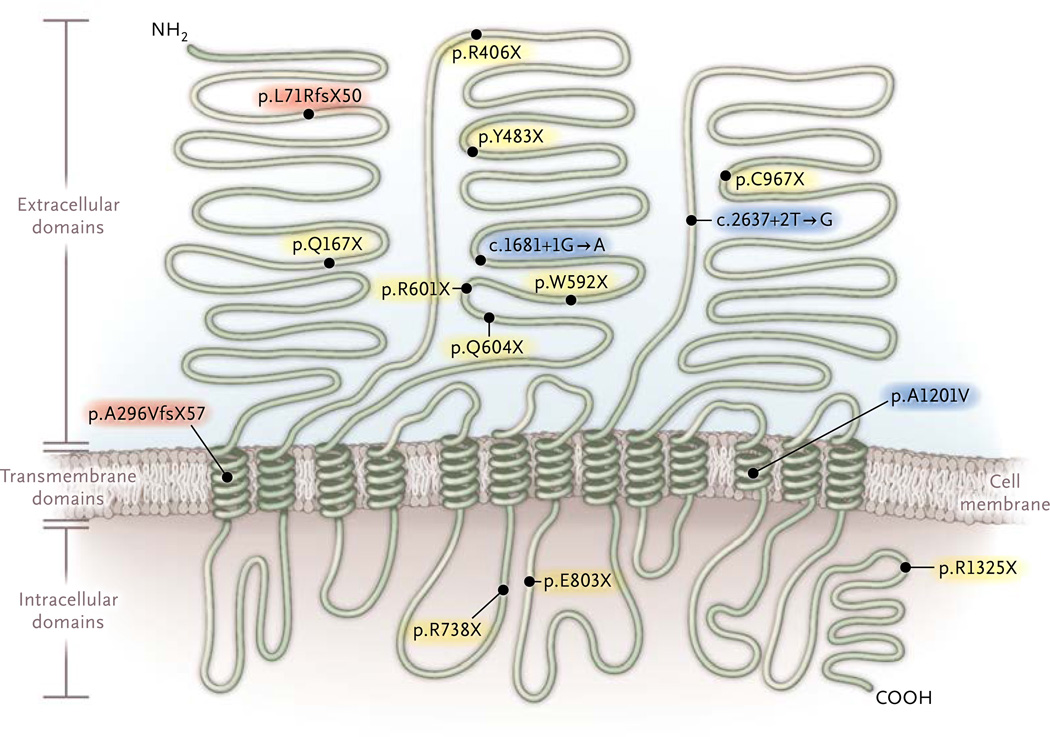
After sequencing NPC1L1 in 7364 patients with coronary heart disease and in 14,728 controls without such disease, we identified 15 mutations that were expected to inactivate NPC1L1 (Table 1). These mutations included 10 nonsense single-nucleotide substitutions, 3 single-nucleotide substitutions that were predicted to disrupt splicing, and 2 frame-shift indels (Fig. 1). In aggregate, these 15 mutations were seen in a total of 34 participants with heterozygous mutations; no homozygotes or compound heterozygotes were identified. NPC1L1 inactivating mutations were rare, with such variants found in approximately 1 in 650 participants.
Table 1.
Inactivating Mutations in NPC1L1 in Participants from 16 Studies. *
| Cohort and Mutation | Type of Mutation |
Study (No. of Participants/ No. of Carriers) |
Ancestry |
|---|---|---|---|
| Sequencing and genotyping cohorts |
All studies (113,094/82) | ||
| Sequencing cohort | |||
| All participants† | All studies in sequencing cohort (22,092/34) |
||
| p.L71RfsX50 | Frameshift | ARIC (2836/1), JHS (2251/1) | African |
| p.Q167X | Nonsense | OHS (1953/1) | European |
| p.A296VfsX57 | Frameshift | ATVB (3539/3) | European |
| p.R406X | Nonsense | ATVB (3539/4), PROCARDIS (1902/1), ARIC (5718/2) |
European |
| p.Y483X | Nonsense | PROMIS (1951/1) | South Asian |
| c.1681+1G→A | Splice-site | ARIC (2836/2), JHS (2251/1) | African |
| p.W592X | Nonsense | ARIC (5718/1) | European |
| p.R601X | Nonsense | ARIC (2836/1) | African |
| p.Q604X | Nonsense | ESP-EOMI (455/1), ARIC (2836/2) | African |
| p.R738X | Nonsense | REGICOR (783/2) | European |
| p.E803X | Nonsense | ARIC (5718/1) | European |
| c.2637+2T→G | Splice-site | ARIC (5718/1), Munich-MI (704/1) | European |
| p.C967X | Nonsense | ARIC (2836/1) | African |
| p.A1201V | Splice-site | JHS (2251/2) | African |
| p.R1325X | Nonsense | ARIC (2836 of African ancestry/1), JHS (2251 of African ancestry/2), ARIC (5718 of European ancestry/1) |
African and European |
| Genotyping cohort | |||
| All participants‡ | All studies in genotyping cohort (91,002/48) |
||
| p.R406X | Nonsense | ARIC (5237/4), BioVU (21,143/12), German North (7350/1), German South (8176/3), GoDARTS (3765/4), Mayo (2669/2), PROCARDIS (2227/1), WGHS (22,617/11), WHI (17,818/10) |
European |
ARIC denotes Atherosclerosis Risk in Communities, ATVB Atherosclerosis Thrombosis and Vascular Biology, BioVU Vanderbilt University Medical Center Biorepository, ESP-EOMI Exome Sequencing Project Early-Onset Myocardial Infarction, German North German North Coronary Artery Disease Study, German South German South Coronary Artery Disease Study, GoDARTS Genetics of Diabetes Audit and Research Tayside, JHS Jackson Heart Study, Mayo Mayo Vascular Diseases Biorepository, Munich-MI Munich Myocardial Infarction, OHS Ottawa Heart Study, PROCARDIS Precocious Coronary Artery Disease, PROMIS Pakistan Risk of Myocardial Infarction Study, REGICOR Registre Gironi del COR, WGHS Women’s Genome Health Study, and WHI Women’s Health Initiative.
The number of participants in the sequencing cohort includes 7364 patients with coronary heart disease and 14,728 controls without such disease.
The number of participants in the genotyping cohort includes 22,590 patients with coronary heart disease and 68,412 controls without such disease.
Figure 1. Inactivating Mutations in NPC1L1 Identified in the Study.
Black circles indicate individual mutations along with the effect expected to lead to NPC1L1 inactivation. Mutations p.L71RfsX50 and p.A296VfsX57 (red shading) are indels that shift the open reading frame and induce a premature termination codon after an additional 50 and 57 amino acids, respectively. Mutations c.1681+1G→A, c.2637+2T→G, and p.A1201V (c.3602C→T) (blue shading) alter the splicing process at sites of modification of the nascent pre-messenger RNA transcript (splice-site mutations). All other mutations (yellow shading) are single-nucleotide variants that introduce a termination codon. The locations of the three main extracellular domains, 13 transmembrane domains, and intracellular domains are based on data from Betters and Yu.27 NH2 denotes the N-terminal at which protein translation is initiated, and COOH the C-terminal at which translation terminates.
The most frequently observed individual mutation was p.Arg406X, which had a minor allele frequency of 0.02% among participants of European ancestry (seven alleles observed in 29,198 chromosomes) and was not observed in participants of African or South Asian ancestry. We genotyped this single variant in an additional 22,590 participants with coronary heart disease and in 68,412 controls. Among these 91,002 participants, we identified 48 additional heterozygous carriers (Table 1). The baseline characteristics of participants carrying NPC1L1 inactivating mutations and those without such mutations were similar across all 16 studies (Table S3 in the Supplementary Appendix).
As a quality-control measure to assess the accuracy of next-generation sequencing, we performed Sanger sequencing and independently confirmed the presence of inactivating mutations in all carriers who were identified in the ATVB study. (See the Supplementary Appendix for details.) In a similar effort to assess the quality of genotyping, we compared genotype calls for p.Arg406X across 4092 samples that had undergone both genotyping and sequencing. On the basis of these data, we observed 100% specificity and sensitivity in identifying p.Arg406X carriers with the use of genotyping.
NPC1L1 Mutations and Plasma Lipid Levels
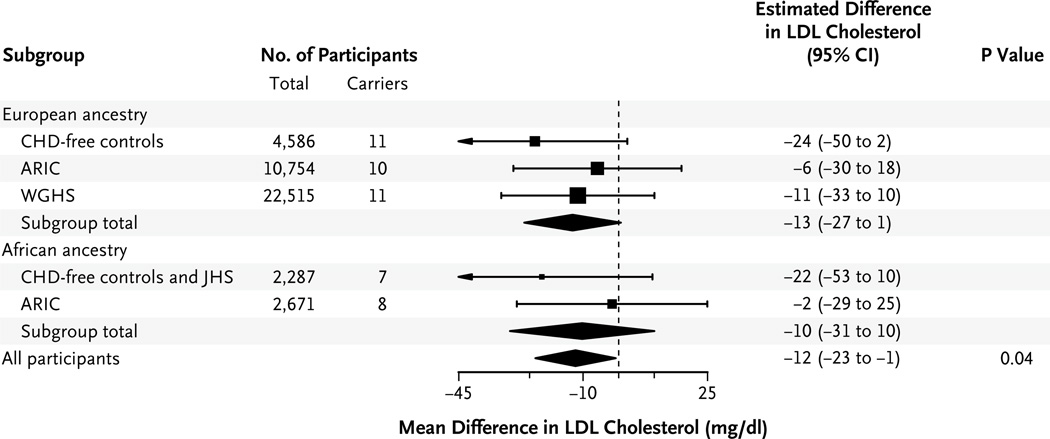
Plasma lipid measurements were available for 13,626 participants in the ARIC study, 2082 in the JHS, and 22,515 in the WGHS. In addition, plasma lipid levels were available for 5405 controls without coronary heart disease from the ATVB, ESP-EOMI, Munich-MI, OHS, PROCARDIS, and REGICOR studies. LDL cholesterol levels were available for 42,813 of these 43,628 study participants. To minimize the effect of ascertainment bias, we excluded patients with coronary heart disease from case–control studies in the lipids analysis. As compared with noncarriers, carriers of inactivating NPC1L1 mutations had significantly lower levels of total cholesterol (mean adjusted difference, −13 mg per deciliter [0.34 mmol per liter]; P = 0.03) (Table 2) and LDL cholesterol (mean adjusted difference, −12 mg per deciliter [0.31 mmol per liter]; P = 0.04) (Table 2 and Fig. 2). Triglyceride levels were also reduced among carriers, although the difference was not significant (mean change, −12%; P = 0.11). We did not observe any significant difference in HDL cholesterol levels between carriers and non-carriers, with an increase of 2 mg per deciliter (0.05 mmol per liter) among carriers (P = 0.29). Participants of European and African ancestry had a similar magnitude of LDL reduction (−13 mg per deciliter and −10 mg per deciliter [0.26 mmol per liter], respectively) (Fig. 2).
Table 2.
Association between the Presence of Inactivating Mutations in NPC1L1 and Plasma Lipid Levels.*
| Variable | Mean Difference between Carriers and Noncarriers* |
P Value |
|---|---|---|
| Cholesterol (mg/dl) | ||
| Total | −13 | 0.03 |
| Low-density lipoprotein | −12 | 0.04 |
| High-density lipoprotein | 2 | 0.29 |
| Triglycerides (% change) | −12 | 0.11† |
The mean difference is the summary effect estimate for carriers of inactivating mutations in NPC1L1, as compared with noncarriers, after adjustment for age, sex, and study. Participants from population-based studies (ARIC, JHS, and WGHS) and controls without coronary heart disease from case–control studies were included in this analysis. To convert the values for cholesterol to millimoles per liter, multiply by 0.02586.
This P value was calculated with the use of natural log transformation of the values.
Figure 2. Association between the Presence of Inactivating Mutations in NPC1L1 and LDL Cholesterol Levels, According to Genetic Ancestry.
In each group of participants, we tested the association between the presence of inactivating mutations in NPC1L1 and plasma levels of low-density lipoprotein (LDL) cholesterol, after adjustment for age, sex, and study. The squares indicate the estimated adjusted difference in the LDL cholesterol level for carriers, as compared with noncarriers, in each ancestry group. The sizes of the squares are inversely proportional to the variance of the estimates. The diamonds indicate the combined results, based on a fixed-effects meta-analysis performed first within and then across ancestry groups. Participants from population-based studies — the Atherosclerosis Risk in Communities (ARIC) study, the Jackson Heart Study (JHS), and the Women’s Genome Health Study (WGHS) — and controls without coronary heart disease (CHD) from case–control studies were included in this analysis. To convert the values for cholesterol to millimoles per liter, multiply by 0.02586.
Association between NPC1L1 Mutations and coronary Risk
Carriers of the 15 inactivating mutations that we identified in NPC1L1 were underrepresented among patients with coronary heart disease, as compared with controls (Table 3). In total, only 11 participants among 29,954 patients with coronary heart disease had an inactivating mutation (carrier frequency, 0.04%) in contrast to 71 of 83,140 controls (carrier frequency, 0.09%). This represented a 53% reduction in the risk of coronary heart disease among carriers of inactivating NPC1L1 mutations (odds ratio for disease among carriers, 0.47; 95% confidence interval [CI], 0.25 to 0.87; P = 0.008) (Table 3, and Table S4 in the Supplementary Appendix).
Table 3.
Association between the Presence of Inactivating Mutations in NPC1L1 and the Risk of Coronary Heart Disease (CHD).
| Inactivating Mutation | Mutation Carriers | Total Participants | Carrier Frequency | |||
|---|---|---|---|---|---|---|
| With CHD |
Without CHD |
With CHD |
Without CHD |
Participants with CHD |
Participants without CHD |
|
| number | percent | |||||
| All mutations* | 11 | 71 | 29,954 | 83,140 | 0.04 | 0.09 |
| p.L71RfsX50 | 0 | 2 | 709 | 4,378 | 0 | 0.05 |
| p.Q167X | 0 | 1 | 966 | 987 | 0 | 0.10 |
| p.A296VfsX57 | 0 | 3 | 1,794 | 1,745 | 0 | 0.17 |
| p.R406X | 6 | 49 | 26,507 | 75,654 | 0.02 | 0.06 |
| p.Y483X | 0 | 1 | 844 | 1,107 | 0 | 0.09 |
| c.1681+1G→A† | 0 | 3 | 709 | 4,378 | 0 | 0.07 |
| p.W592X | 1 | 0 | 1,157 | 4,561 | 0.09 | 0 |
| p.R601X | 1 | 0 | 474 | 2,362 | 0.21 | 0 |
| p.Q604X | 0 | 3 | 652 | 2,639 | 0 | 0.11 |
| p.R738X | 0 | 2 | 382 | 401 | 0 | 0.50 |
| p.E803X | 1 | 0 | 1,157 | 4,561 | 0.09 | 0 |
| c.2637+2T→G† | 1 | 1 | 1,525 | 4,897 | 0.07 | 0.02 |
| p.C967X | 0 | 1 | 474 | 2,362 | 0 | 0.04 |
| p.A1201V† | 0 | 2 | 235 | 2,016 | 0 | 0.10 |
| p.R1325X | 1 | 3 | 1,866 | 8,939 | 0.05 | 0.03 |
The overall odds ratio for coronary heart disease in mutation carriers, as compared with noncarriers, was 0.47 (95% confidence interval, 0.25 to 0.87; P = 0.008) on the basis of a meta-analysis of independent samples.
This mutation was predicted to disrupt messenger RNA splicing.
We observed a reduced risk of coronary heart disease among both participants of African descent and those of European descent. In the African ancestry subgroup, only 2 of 887 patients with coronary heart disease carried an NPC1L1 inactivating mutation (carrier frequency, 0.23%), as compared with 13 of 4655 controls (carrier frequency, 0.28%), representing a 17% reduction in the risk of coronary heart disease among carriers (cohort-based meta-analysis odds ratio, 0.83). In participants of European ancestry, 9 of 28,223 patients with coronary heart disease carried the mutation (carrier frequency, 0.03%), as compared with 57 of 77,378 controls (carrier frequency, 0.07%), representing a 57% reduction in the risk of coronary heart disease among carriers (cohort-based meta-analysis odds ratio, 0.43).
Discussion
We sequenced the protein-coding regions of NPC1L1 in 22,092 participants and identified 15 rare mutations that were expected to disrupt the protein. We also genotyped the most frequently observed of these inactivating mutations (p.Arg406X) in an additional 91,002 participants. Carriers of any NPC1L1 inactivating mutation had a mean LDL cholesterol level that was 12 mg per deciliter lower than the level in noncarriers, along with a 53% lower risk of coronary heart disease. These results show that lifelong inactivation of one copy of NPC1L1 is protective against coronary heart disease. The observation that genetic inhibition of NPC1L1 reduces the risk of coronary heart disease increases the prior probability that pharmacologic inhibition of NPC1L1 will also reduce the risk of disease. In 2002, ezetimibe was initially approved as a therapeutic agent in the United States on the basis of the capacity of the drug to lower LDL cholesterol levels. Although it has been assumed that any pharmacologic means of lowering LDL cholesterol levels will reduce the risk of coronary heart disease, the findings from the Ezetimibe and Simvastatin in Hypercholesterolemia Enhances Atherosclerosis Regression (ENHANCE) trial have led some observers to question this assumption.28 In ENHANCE, the addition of ezetimibe to background statin therapy in patients with familial hypercholesterolemia did not reduce the progression of carotid intima–media thickness, a surrogate measure for atherosclerosis.6 In the ongoing phase 3, randomized Improved Reduction of Outcomes: Vytorin Efficacy International Trial (IMPROVE-IT; ClinicalTrials .gov number, NCT00202878), investigators are evaluating whether the addition of ezetimibe to background simvastatin therapy will reduce the risk of recurrent cardiovascular events in patients with a recent acute coronary syndrome.29
Our findings do not predict with certainty that ezetimibe will be found to reduce cardiovascular risk in the IMPROVE-IT trial or other clinical studies, for several reasons. First, lifelong genetic inhibition, as tested in our study, has important differences from pharmacologic inhibition that is initiated in adulthood and lasts for several years. Second, our genetic study focuses on a first cardiovascular event, whereas IMPROVE-IT is evaluating recurrent events. Finally, the net clinical benefit of a pharmacologic therapy is a complex interplay among multiple factors, including many that are specific to the drug (e.g., toxic effects) and that would not be tested in a genetic model such as the one used in our study.
The reduction in the risk of coronary heart disease (53%) that we observed among carriers exceeds the reduction that would be expected for a decrease of 12 mg per deciliter in LDL cholesterol on the basis of results from statin trials.26 Several factors may explain this difference. Modest reductions in plasma lipid levels over a lifetime, as achieved in carriers of an inactivating mutation, appear to lead to a larger modification of the risk of coronary heart disease than pharmacologic treatment that is initiated later in life. Such an effect has been observed in persons with genetic loss of function in several lipid genes.30 In addition to affecting LDL cholesterol levels, genetic loss of NPC1L1 function is associated with reduced plant sterol absorption.31,32 Levels of plant sterols are markedly elevated in patients with autosomal recessive sitosterolemia, a disease that is associated with accelerated atherosclerotic vascular disease even among patients without significantly elevated plasma LDL cholesterol levels.33 These observations raise the possibility that genetic inhibition of NPC1L1 may also lower the risk of coronary heart disease by reducing the absorption of noncholesterol sterols. Also, the effect of NPC1L1 inhibition on cardiovascular risk in our study may be overestimated owing to the “winner’s curse”34 phenomenon, in which the effects of newly discovered associations are inflated as compared with the true effect sizes. Our results suggest a broad range of plausible risk estimates associated with these mutations.
Several limitations of the study deserve mention. The combined statistical evidence supporting a protective association with coronary heart disease (P = 0.008) is significant for a test of a single hypothesis but falls short of the exomewide significance threshold that would be used to account for multiple hypothesis testing across all genes (P = 1.7×10−6 on the basis of a Bonferroni correction for 21,000 protein-coding and 9000 long noncoding RNA genes).35 This stringent threshold is used to limit false positive results of genetic association studies35 involving many hypotheses, in which the prior probability of a true association is low. Here, however, we have evaluated a gene that is known to alter LDL cholesterol levels, a proven causal factor for coronary heart disease. Therefore, the prior probability that this gene alters the risk of coronary heart disease is considerably higher than that for a random gene drawn from the genome.
In addition, we focused only on the classes of genetic variation — nonsense, splice-site, and frameshift — that are clearly expected to lead to a loss in NPC1L1 function and did not include missense variants. Although some missense mutations in NPC1L1 clearly inhibit function, many others have no effect on the protein.31,32,36,37 On average, the inclusion of neutral missense variants has been shown to dilute association signals and decrease statistical power.38 As a result of our focus on rare inactivating mutations, the associations that we discovered are based on a relatively modest number of observations. Finally, we were unable to evaluate whether NPC1L1 inactivating mutations lead to other phenotypic consequences.
In conclusion, on the basis of sequencing and genotyping in 113,094 study participants, we found that inactivating mutations in NPC1L1 were associated with both reduced LDL cholesterol levels and a reduced risk of coronary heart disease. Whether pharmacologic therapies that are focused on inhibiting NPC1L1 function reduce the risk of coronary heart disease remains to be determined.
Supplementary Material
Acknowledgments
The views expressed in this article are solely those of the authors and do not necessarily represent the official views of the National Human Genome Research Institute (NHGRI), the National Heart, Lung, and Blood Institute (NHLBI), or the National Institutes of Health (NIH).
Supported by grants from the NHLBI (K08HL114642, to Dr. Stitziel; T32HL007208, to Dr. Peloso; R01HL107816, to Dr. Kathiresan; RC2HL102926, to Dr. Nickerson; and RC2HL102925, to Dr. Gabriel), a grant from the NHGRI (5U54HG003067-11, to Drs. Gabriel and Lander), a grant from the Foundation for Barnes–Jewish Hospital (to Dr. Stitziel), a Banting Fellowship from the Canadian Institutes of Health Research (to Dr. Do), and a grant from the Donovan Family Foundation, an investigator-initiated research grant from Merck, and a grant from Fondation Leducq (all to Dr. Kathiresan). Funders for the various studies that are discussed in this article are listed in the Supplementary Appendix.
Appendix
The authors are as follows: Nathan O. Stitziel, M.D., Ph.D., Hong-Hee Won, Ph.D., Alanna C. Morrison, Ph.D., Gina M. Peloso, Ph.D., Ron Do, Ph.D., Leslie A. Lange, Ph.D., Pierre Fontanillas, Ph.D., Namrata Gupta, Ph.D., Stefano Duga, Ph.D., Anuj Goel, M.Sc., Martin Farrall, F.R.C.Path., Danish Saleheen, M.B., B.S., Ph.D., Paola Ferrario, Ph.D., Inke König, Ph.D., Rosanna Asselta, Ph.D., Piera A. Merlini, M.D., Nicola Marziliano, Ph.D., Maria Francesca Notarangelo, M.D., Ursula Schick, M.S., Paul Auer, Ph.D., Themistocles L. Assimes, M.D., Ph.D., Muredach Reilly, M.D., Robert Wilensky, M.D., Daniel J. Rader, M.D., G. Kees Hovingh, M.D., Ph.D., Thomas Meitinger, M.D., Thorsten Kessler, M.D., Adnan Kastrati, M.D., Karl-Ludwig Laugwitz, M.D., David Siscovick, M.D., M.P.H., Jerome I. Rotter, M.D., Stanley L. Hazen, M.D., Ph.D., Russell Tracy, Ph.D., Sharon Cresci, M.D., John Spertus, M.D., M.P.H., Rebecca Jackson, M.D., Stephen M. Schwartz, Ph.D., Pradeep Natarajan, M.D., Jacy Crosby, Ph.D., Donna Muzny, M.S., Christie Ballantyne, M.D., Stephen S. Rich, Ph.D., Christopher J. O’Donnell, M.D., Goncalo Abecasis, Ph.D., Shamil Sunyaev, Ph.D., Deborah A. Nickerson, Ph.D., Julie E. Buring, Sc.D., Paul M. Ridker, M.D., Daniel I. Chasman, Ph.D., Erin Austin, Ph.D., Zi Ye, M.D., Ph.D., Iftikhar J. Kullo, M.D., Peter E. Weeke, M.D., Christian M. Shaffer, B.S., Lisa A. Bastarache, M.S., Joshua C. Denny, M.D., Dan M. Roden, M.D., Colin Palmer, Ph.D., Panos Deloukas, Ph.D., Dan-Yu Lin, Ph.D., Zheng-zheng Tang, Ph.D., Jeanette Erdmann, Ph.D., Heribert Schunkert, M.D., John Danesh, M.B., Ch.B., D.Phil., Jaume Marrugat, M.D., Ph.D., Roberto Elosua, M.D., Ph.D., Diego Ardissino, M.D., Ruth McPherson, M.D., Hugh Watkins, M.D., Ph.D., Alex P. Reiner, M.D., James G. Wilson, M.D., David Altshuler, M.D., Ph.D., Richard A. Gibbs, Ph.D., Eric S. Lander, Ph.D., Eric Boerwinkle, Ph.D., Stacey Gabriel, Ph.D., and Sekar Kathiresan, M.D.
Drs. Stitziel and Won contributed equally to this article.
The authors’ affiliations are as follows: the Cardiovascular Division, Department of Medicine (N.O.S., S.C.), Division of Statistical Genomics (N.O.S.), and Department of Genetics (S.C.), Washington University School of Medicine, St. Louis; Center for Human Genetic Research (H.-H.W., G.M.P., R.D., P.N., D. Altshuler, S.K.), Cardiovascular Research Center (H.-H.W., G.M.P., R.D., P.N., S.K.), and Cardiology Division (C.J.O., S.K.), Massachusetts General Hospital, the Department of Medicine, Harvard Medical School (H.-H.W., G.M.P., R.D., P.N., C.J.O., D. Altshuler, S.K.), and the Divisions of Genetics (S.S.) and Preventive Medicine (J.E.B., P.M.R., D.I.C.), Brigham and Women’s Hospital and Harvard Medical School, Boston, the Program in Medical and Population Genetics, Broad Institute, Cambridge (H.-H.W., G.M.P., R.D., P. Fontanillas, N.G., P.N., S.S., D. Altshuler, E.S.L., S.G., S.K.), and the National Heart, Lung, and Blood Institute Framingham Heart Study, Framingham (C.J.O.) — all in Massachusetts; the Human Genetics Center, University of Texas Health Science Center at Houston (A.C.M., J.C., E.B.), and the Human Genome Sequencing Center (D.M., R.A.G., E.B.), and Section of Atherosclerosis and Vascular Medicine (C.B.), Baylor College of Medicine — both in Houston; the Departments of Genetics (L.A.L.), and Biostatistics (D.-Y.L.), University of North Carolina, Chapel Hill; Dipartimento di Biotecnologie Mediche e Medicina Traslazionale, Università degli Studi di Milano, Milan (S.D., R.A.), Azienda Ospedaliera-Universitaria di Parma, Parma (P.A.M., N.M., M.F.N., D. Ardissino), and Associazione per lo Studio della Trombosi in Cardiologia, Pavia (P.A.M., D. Ardissino) — all in Italy; the Division of Cardiovascular Medicine, Radcliffe Department of Medicine and the Wellcome Trust Centre for Human Genetics, University of Oxford, Oxford (A.G., M.F., H.W.), Public Health and Primary Care, University of Cambridge (J.D.), and Wellcome Trust Sanger Institute (P.D.), Cambridge, Medical Research Institute, University of Dundee, Dundee (C.P.), and William Harvey Research Institute, Barts and the London School of Medicine and Dentistry, Queen Mary University of London, London (P.D.) — all in the United Kingdom; the Departments of Genetics (D.J.R.) and Biostatistics and Epidemiology (D. Saleheen) and the Cardiovascular Institute (M.R., R.W.), Perelman School of Medicine, University of Pennsylvania, Philadelphia; Center for Noncommunicable Diseases, Karachi, Pakistan (D. Saleheen); Institut für Medizinische Biometrie und Statistik (P. Ferrario, I.K.) and Institut für Integrative und Experimentelle Genomik (J.E.), Universität zu Lübeck, and DZHK (German Research Center for Cardiovascular Research) partner site Hamburg–Lübeck–Kiel (J.E.), Lübeck, Institut für Humangenetik, Helmholtz Zentrum, Neuherberg (T.M.), and Institut für Humangenetik (T.M.) and Medizinische Klinik (K.-L.L.), Klinikum rechts der Isar, and Deutsches Herzzentrum München (T.K., A.K., H.S.),Technische Universität München, and German Center for Cardiovascular Research (DZHK), partner site Munich Heart Alliance (T.M., A.K., K.-L.L., H.S.), Munich — all in Germany; the Division of Public Health Sciences, Fred Hutchinson Cancer Research Center (U.S., S.M.S., A.P.R.), and the Cardiovascular Health Research Unit, Departments of Medicine and Epidemiology (D. Siscovick), Department of Epidemiology (S.M.S., A.P.R.), and Department of Genome Sciences (D.A.N.), University of Washington — both in Seattle; School of Public Health, University of Wisconsin–Milwaukee, Milwaukee (P.A.); Stanford Cardiovascular Institute and the Division of Cardiovascular Medicine, Stanford University, Stanford (T.L.A.), and Institute for Translational Genomics and Population Sciences, Los Angeles Biomedical Research Institute at Harbor–UCLA Medical Center, Torrance (J.I.R.) — both in California; the Department of Vascular Medicine, Academic Medical Center, Amsterdam (G.K.H.); the Department of Cardiovascular Medicine, Cleveland Clinic, Cleveland (S.L.H.), and the Division of Endocrinology, Diabetes and Metabolism, Department of Medicine, Ohio State University, Columbus (R.J.) — both in Ohio; the Departments of Pathology and Biochemistry, University of Vermont College of Medicine, Burlington (R.T.); St. Luke’s Mid America Heart Institute, University of Missouri–Kansas City, Kansas City (J.S.); Center for Public Health Genomics, University of Virginia, Charlottesville (S.S.R.); Center for Statistical Genetics, Department of Biostatistics, University of Michigan, Ann Arbor (G.A.); the Division of Cardiovascular Diseases, Mayo Clinic, Rochester, MN (E.A., Z.Y., I.J.K.); the Departments of Medicine (P.E.W., C.M.S., J.C.D., D.M.R.), Biomedical Informatics (L.A.B., J.C.D.), Pharmacology (D.M.R.), and Biostatistics (Z.T.), Vanderbilt University, Nashville; the Department of Cardiology, Laboratory of Molecular Cardiology, Copenhagen University Hospital Rigshospitalet, Copenhagen (P.E.W.); King Abdulaziz University, Jeddah, Saudi Arabia (P.D.); Grupo de Epidemiología y Genética Cardiovascular, Institut Hospital del Mar d’Investigacions Mèdiques (IMIM), Barcelona (J.M., R.E.); the Division of Cardiology, University of Ottawa Heart Institute, Ottawa (R.M.); and the Department of Physiology and Biophysics, University of Mississippi Medical Center, Jackson (J.G.W.).
Footnotes
Disclosure forms provided by the authors are available with the full text of this article at NEJM.org.
References
- 1.Garcia-Calvo M, Lisnock J, Bull HG, et al. The target of ezetimibe is Niemann-Pick C1-Like 1 (NPC1L1) Proc Natl Acad Sci U S A. 2005;102:8132–8137. doi: 10.1073/pnas.0500269102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davis HR, Jr., Zhu LJ, Hoos LM, et al. Niemann-Pick C1 Like 1 (NPC1L1) is the intestinal phytosterol and cholesterol transporter and a key modulator of whole-body cholesterol homeostasis. J Biol Chem. 2004;279:33586–33592. doi: 10.1074/jbc.M405817200. [DOI] [PubMed] [Google Scholar]
- 3.Altmann SW, Davis HR, Jr., Zhu LJ, et al. Niemann-Pick C1 Like 1 protein is critical for intestinal cholesterol absorption. Science. 2004;303:1201–1204. doi: 10.1126/science.1093131. [DOI] [PubMed] [Google Scholar]
- 4.Sudhop T, Lütjohann D, Kodal A, et al. Inhibition of intestinal cholesterol absorption by ezetimibe in humans. Circulation. 2002;106:1943–1948. doi: 10.1161/01.cir.0000034044.95911.dc. [DOI] [PubMed] [Google Scholar]
- 5.Ezzet F, Wexler D, Statkevich P, et al. The plasma concentration and LDL-C relationship in patients receiving ezetimibe. J Clin Pharmacol. 2001;41:943–949. doi: 10.1177/00912700122010915. [DOI] [PubMed] [Google Scholar]
- 6.Kastelein JJ, Akdim F, Stroes ES, et al. Simvastatin with or without ezetimibe in familial hypercholesterolemia. N Engl J Med. 2008;358:1431–1443. doi: 10.1056/NEJMoa0800742. [Erratum, N Engl J Med 2008;358:1977.] [DOI] [PubMed] [Google Scholar]
- 7.Cohen JC, Boerwinkle E, Mosley TH, Jr., Hobbs HH. Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N Engl J Med. 2006;354:1264–1272. doi: 10.1056/NEJMoa054013. [DOI] [PubMed] [Google Scholar]
- 8.Plenge RM, Scolnick EM, Altshuler D. Validating therapeutic targets through human genetics. Nat Rev Drug Discov. 2013;12:581–594. doi: 10.1038/nrd4051. [DOI] [PubMed] [Google Scholar]
- 9.Teslovich TM, Musunuru K, Smith AV, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krawczak M, Reiss J, Cooper DN. The mutational spectrum of single base-pair substitutions in mRNA splice junctions of human genes: causes and consequences. Hum Genet. 1992;90:41–54. doi: 10.1007/BF00210743. [DOI] [PubMed] [Google Scholar]
- 11.Do R, Stitziel NO, Won H-H, et al. Multiple rare alleles at LDLR and APOA5 confer risk for early-onset myocardial infarction. Nature. doi: 10.1038/nature13917. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Atherosclerosis, Thrombosis, and Vascular Biology Italian Study Group. No evidence of association between prothrombotic gene polymorphisms and the development of acute myocardial infarction at a young age. Circulation. 2003;107:1117–1122. doi: 10.1161/01.cir.0000051465.94572.d0. [DOI] [PubMed] [Google Scholar]
- 13.McPherson R, Pertsemlidis A, Kavaslar N, et al. A common allele on chromosome 9 associated with coronary heart disease. Science. 2007;316:1488–1491. doi: 10.1126/science.1142447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clarke R, Peden JF, Hopewell JC, et al. Genetic variants associated with Lp(a) lipoprotein level and coronary disease. N Engl J Med. 2009;361:2518–2528. doi: 10.1056/NEJMoa0902604. [DOI] [PubMed] [Google Scholar]
- 15.Saleheen D, Zaidi M, Rasheed A, et al. The Pakistan Risk of Myocardial Infarction Study: a resource for the study of genetic, lifestyle and other determinants of myocardial infarction in South Asia. Eur J Epidemiol. 2009;24:329–338. doi: 10.1007/s10654-009-9334-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sentí M, Tomás M, Marrugat J, Elosua R. Paraoxonase1–192 polymorphism modulates the nonfatal myocardial infarction risk associated with decreased HDLs. Arterioscler Thromb Vasc Biol. 2001;21:415–420. doi: 10.1161/01.atv.21.3.415. [DOI] [PubMed] [Google Scholar]
- 17.Crosby J, Peloso GM, Auer PL, et al. Loss-of-function mutations in APOC3, triglycerides, and coronary disease. N Engl J Med. 2014;371:22–31. doi: 10.1056/NEJMoa1307095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.The ARIC investigators. The Atherosclerosis Risk in Communities (ARIC) Study: design and objectives. Am J Epidemiol. 1989;129:687–702. [PubMed] [Google Scholar]
- 19.Taylor HA., Jr. The Jackson Heart Study: an overview. Ethn Dis. 2005;15(Suppl 6):S6–1–S6–3. [PubMed] [Google Scholar]
- 20.Roden DM, Pulley JM, Basford MA, et al. Development of a large-scale de-identified DNA biobank to enable personalized medicine. Clin Pharmacol Ther. 2008;84:362–369. doi: 10.1038/clpt.2008.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Doney AS, Fischer B, Leese G, Morris AD, Palmer CN. Cardiovascular risk in type 2 diabetes is associated with variation at the PPARG locus: a Go-DARTS study. Arterioscler Thromb Vasc Biol. 2004;24:2403–2407. doi: 10.1161/01.ATV.0000147897.57527.e4. [DOI] [PubMed] [Google Scholar]
- 22.Erdmann J, Stark K, Esslinger UB, et al. Dysfunctional nitric oxide signalling increases risk of myocardial infarction. Nature. 2013;504:432–436. doi: 10.1038/nature12722. [DOI] [PubMed] [Google Scholar]
- 23.Ye Z, Kalloo FS, Dalenberg AK, Kullo IJ. An electronic medical record-linked biorepository to identify novel biomarkers for atherosclerotic cardiovascular disease. Glob Cardiol Sci Pract. 2013;2013:82–90. doi: 10.5339/gcsp.2013.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ridker PM, Chasman DI, Zee RY, et al. Rationale, design, and methodology of the Women’s Genome Health Study: a genome-wide association study of more than 25,000 initially healthy American women. Clin Chem. 2008;54:249–255. doi: 10.1373/clinchem.2007.099366. [DOI] [PubMed] [Google Scholar]
- 25.The Women’s Health Initiative Study Group. Design of the Women’s Health Initiative clinical trial and observational study. Control Clin Trials. 1998;19:61–109. doi: 10.1016/s0197-2456(97)00078-0. [DOI] [PubMed] [Google Scholar]
- 26.Baigent C, Blackwell L, Emberson J, et al. Efficacy and safety of more intensive lowering of LDL cholesterol: a meta-analysis of data from 170,000 participants in 26 randomised trials. Lancet. 2010;376:1670–1681. doi: 10.1016/S0140-6736(10)61350-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Betters JL, Yu L. NPC1L1 and cholesterol transport. FEBS Lett. 2010;584:2740–2747. doi: 10.1016/j.febslet.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krumholz HM. Emphasizing the burden of proof: the American College of Cardiology 2008 Expert Panel Comments on the ENHANCE Trial. Circ Cardiovasc Qual Outcomes. 2010;3:565–567. doi: 10.1161/CIRCOUTCOMES.110.959577. [DOI] [PubMed] [Google Scholar]
- 29.Cannon CP, Giugliano RP, Blazing MA, et al. Rationale and design of IMPROVE-IT (IMProved Reduction of Outcomes: Vytorin Efficacy International Trial): comparison of ezetimbe/simvastatin versus simvastatin monotherapy on cardiovascular outcomes in patients with acute coronary syndromes. Am Heart J. 2008;156:826–832. doi: 10.1016/j.ahj.2008.07.023. [DOI] [PubMed] [Google Scholar]
- 30.Cohen JC, Stender S, Hobbs HH. APOC3, coronary disease, and complexities of Mendelian randomization. Cell Metab. 2014;20:387–389. doi: 10.1016/j.cmet.2014.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cohen JC, Pertsemlidis A, Fahmi S, et al. Multiple rare variants in NPC1L1 associated with reduced sterol absorption and plasma low-density lipoprotein levels. Proc Natl Acad Sci U S A. 2006;103:1810–1815. doi: 10.1073/pnas.0508483103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fahmi S, Yang C, Esmail S, Hobbs HH, Cohen JC. Functional characterization of genetic variants in NPC1L1 supports the sequencing extremes strategy to identify complex trait genes. Hum Mol Genet. 2008;17:2101–2107. doi: 10.1093/hmg/ddn108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Berge KE, Tian H, Graf GA, et al. Accumulation of dietary cholesterol in sitosterolemia caused by mutations in adjacent ABC transporters. Science. 2000;290:1771–1775. doi: 10.1126/science.290.5497.1771. [DOI] [PubMed] [Google Scholar]
- 34.Kraft P. Curses — winner’s and otherwise — in genetic epidemiology. Epidemiology. 2008;19:649–651. doi: 10.1097/EDE.0b013e318181b865. [DOI] [PubMed] [Google Scholar]
- 35.MacArthur DG, Manolio TA, Dim-mock DP, et al. Guidelines for investigating causality of sequence variants in human disease. Nature. 2014;508:469–476. doi: 10.1038/nature13127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hegele RA, Guy J, Ban MR, Wang J. NPC1L1 haplotype is associated with inter-individual variation in plasma low-density lipoprotein response to ezetimibe. Lipids Health Dis. 2005;4:16. doi: 10.1186/1476-511X-4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang J, Williams CM, Hegele RA. Compound heterozygosity for two non-synonymous polymorphisms in NPC1L1 in a non-responder to ezetimibe. Clin Genet. 2005;67:175–177. doi: 10.1111/j.1399-0004.2004.00388.x. [DOI] [PubMed] [Google Scholar]
- 38.Zuk O, Schaffner SF, Samocha K, et al. Searching for missing heritability: designing rare variant association studies. Proc Natl Acad Sci U S A. 2014;111:E455–E464. doi: 10.1073/pnas.1322563111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.