Abstract
Direct reprogramming provides a fundamentally new approach for the generation of patient-specific cells. Here, by screening a pool of candidate transcription factors, we identify that a combination of three factors, MITF, SOX10 and PAX3, directly converts mouse and human fibroblasts to functional melanocytes. Induced melanocytes (iMels) activate melanocyte-specific networks, express components of pigment production and delivery system, and produce melanosomes. Human iMels properly integrate into the dermal-epidermal junction, and produce and deliver melanin pigment to surrounding keratinocytes in a 3D organotypic skin reconstruct. Human iMels generate pigmented epidermis and hair follicles in skin reconstitution assays in vivo. The generation of iMels has important implications for studies of melanocyte lineage commitment, pigmentation disorders and cell replacement therapies.
Introduction
Melanocytes are neural crest derived pigment-producing cells localized at the basal layer of epidermis and hair follicles. Melanocytes synthesize melanin pigment within specialized lysosome-related structures known as melanosomes. Melanin pigment is then transferred to adjacent keratinocytes to form pigmented skin or hairs, protecting skin from UV irradiation1. Loss of melanocytes in the skin (vitiligo) affects 0.1% to greater than 8% population worldwide and depigmentation may be the source of severe psychological distress, diminished quality of life, and increased risk of psychiatric morbidity2. Autologous melanocyte transplant is effective in treating stable vitiligo. However, it is not widely clinically used due to issues such as donor site infection, scar or irregular pigmentation3. Therefore, there is an unmet clinical need to efficiently generate scalable amount of autologous melanocytes.
The discovery that fibroblasts can be directly reprogrammed into muscle cells and neurons by combinations of lineage-enriched transcription factors4–11 leads us to hypothesize that melanocytes can be generated directly from fibroblasts without going through a pluripotent state. Previous studies have demonstrated a critical role of microphthalmia-associated transcription factor (MITF) in melanocyte linage determination from neural crest cells12 and forced expression of MITF in NIH3T3 fibroblasts converted them into melanocyte-like cells13. However, such induced cells expressed only some of the melanocyte specific markers and lacked functional characteristics of melanocytes13. Here, by screening a pool of candidate transcription factors, we discover that three factors, SOX10, MITF and PAX3, are sufficient to directly reprogram human and mouse fibroblasts to melanocytes. iMels acquire phenotypical and functional characteristics of normal melanocytes. Generation of functional melanocytes by direct reprogramming methods provides a potential source for autologous melanocytes to treat skin pigmentation disorders.
Results
Transcription factor screening to discover melanocyte direct reprogramming factors
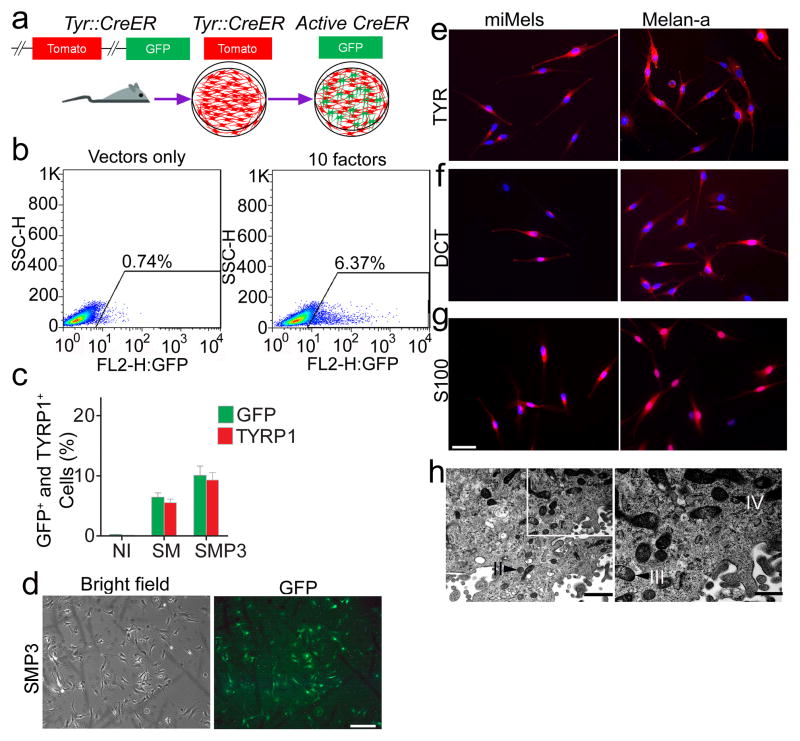
Reasoning that multiple transcription factors would probably be required to reprogram fibroblasts into functional melanocytes, we selected 10 candidate transcription factors that are related to neural crest lineage determination and melanocyte differentiation (Supplementary Table S1)12, 14–20. To efficiently monitor melanocyte differentiation by flow cytometric analysis, we developed a transcription factor screening assay using tail fibroblasts (TTFs) from Tyrosinase-CreER; Gt(ROSA)26Sor tm4(ACTB-tdTomato,-EGFP)Luo/J reporter mice21. Tyrosinase (TYR)-driven CreER-expression in melanocytes converts these mouse cells from expressing red fluorescent protein (RFP) to green fluorescent protein (GFP) upon treatment with 4-hydroxytamoxifen (4-HT). This makes it possible to monitor the emergence of TYR-positive (TYR+) melanocytes by detecting GFP expression (Fig. 1a). Retroviruses carrying 10 candidate mouse transcription factors were prepared and a mixture of all the factors was used to infect the TTFs. Expression of transgenes was confirmed by qRT-PCR (Supplementary Fig. S1) and virus packaged with vector only was used as a negative control. Twelve days after infection with all factors, GFP-positive (GFP+) cells were observed in the presence of 4-HT (Fig. 1b), indicating activation of the TYR promoter in cells. Significantly fewer GFP+ cells were detected in the control, vector only cells (Fig. 1b). Next, we sought to determine the minimal set of genes required for melanocyte induction from fibroblasts. Given the known dominant role of SOX10 during neural crest lineage differentiation, SOX10 was introduced into TTFs combined with every other single factor firstly. The greatest number of GFP+ cells was produced when SOX10 was combined with MITF (Supplementary Fig. S2a). However, the SOX10/MITF combination elicited modest reprogramming efficiencies with GFP+ cells comprising 6.44 % of all cells. Therefore, we added a third transcription factor (from the 8 remaining) and analyzed the percentage of GFP+ cells using each combination. SOX10/MITF/PAX3 and SOX10/MITF/SOX9 combinations increased the generation of GFP+ cells, compared to other combinations (Supplementary Fig. S2b). The addition of a fourth factor to the SOX10/MITF/PAX3 or SOX10/MITF/SOX9 combinations failed to further increase the percentage of GFP+ cells (Supplementary Fig. S4b), including SOX10/MITF/PAX3/SOX9 combination (Supplementary Fig. S2c). To further confirm melanocytic reprogramming, we examined the percentage of TYRP1-positive (TYRP1+) cells using flow cytometric analysis after reprogramming with different combinations of transcription factors. The results demonstrated that the SOX10/MITF/PAX3 combination induced the highest percentage of TYRP1+ cells (Supplementary Fig. S3). Statistical analysis showed that the SOX10/MITF/PAX3 combination activated higher GFP and TYRP1 expression, compared to other combinations (Fig. 1c and Supplementary Fig. S4a). Therefore, melanocytes induced by SOX10/MITF/PAX3 (SMP3) were characterized in additional studies.
Figure 1. Screening for melanocyte direct reprogramming factors.
a. Scheme for melanocyte direct reprogramming transcription factor (TF) screening. Screening was performed using fibroblasts from the Tyrosinase-CreER/Gt(ROSA)26Sor tm4(ACTB-tdTomato,-EGFP)Luo/J mice. When tyrosinase (TYR) was activated, Cre activation resulted in excision of the tomato cassette and expression of GFP in the presence of 4-HT. b. Flow cytometric analysis of the GFP+ cells after cells were infected with viruses packaged with 10 candidate factors (right panel) or vector only (left panel). c. Flow cytometric analysis of GFP and TYRP1 expression after cells were infected with control vectors (NI), SOX10 and MITF (SM) or SOX10, MITF and PAX3 (SMP3). Representative data are from three independent experiments. d. Morphology of FACS sorted GFP+ cells. Mouse fibroblasts were infected with SMP3 and sorted based on GFP expression at Day 14. Scale bar indicates 50 μm. e–g. Immunostaining analysis of mouse induced mouse melanocytes (miMels) using antibodies specific for TYR (e), DCT (f) and S100 (g). Secondary antibody was labeled with Alexa Fluro 594. DAPI was used to stain the nuclei. Melan-a mouse melanocytes were used as a positive control. Scale bar indicates 30 μm. h. Electron microscopy analyses showed that miMels contained many mature melanosomes in the cytoplasm. Arrow heads point to the different stages of melanosomes, including stage II, III and IV. Scale bar indicates 400 nm (left panel) and 200 nm (right panel).
Characterization of mouse induced melanocytes
We monitored the GFP+ cell population daily under a fluorescence microscope after TTFs derived from Tyrosinase(TYR)-CreER/Gt(ROSA)26Sor tm4(ACTB-tdTomato,-EGFP)Luo/J mice were infected with virus carrying the SMP3 combination. GFP+ cells with melanocytic morphology emerged 14 days after induction (Fig. 1d, and Supplementary Fig. S5a and S5b). Most of the sorted GFP+ cells showed TYR expression by FACS (Supplementary Fig. S5c) and expressed the protein components of pigment production and delivery machineries including TYR, DCT, Melan-A and SILV (Fig. 1e and 1f, and Supplementary Fig. S6a) by immunocytochemistry. We also observed that S100, a calcium binding protein, is highly expressed in the induced cells (Fig. 1g). Transcriptional analysis by reverse transcription–polymerase chain reaction (RT–PCR) revealed the expression of multiple melanocyte-specific genes, including TYR, TYRP1, DCT, SILV, P as well as endogenous MITF, SOX10 and PAX3 (Supplementary Fig. S6b). Meanwhile, transgenic SOX10, MITF and PAX3 were still expressed in the GFP+ cells (Supplementary Fig. S6c). Electron microscopy (EM) showed that GFP+ induced cells produced melanosomes at different developmental stages (Fig. 1h), including mature melanin-containing (types III and IV) melanosomes.
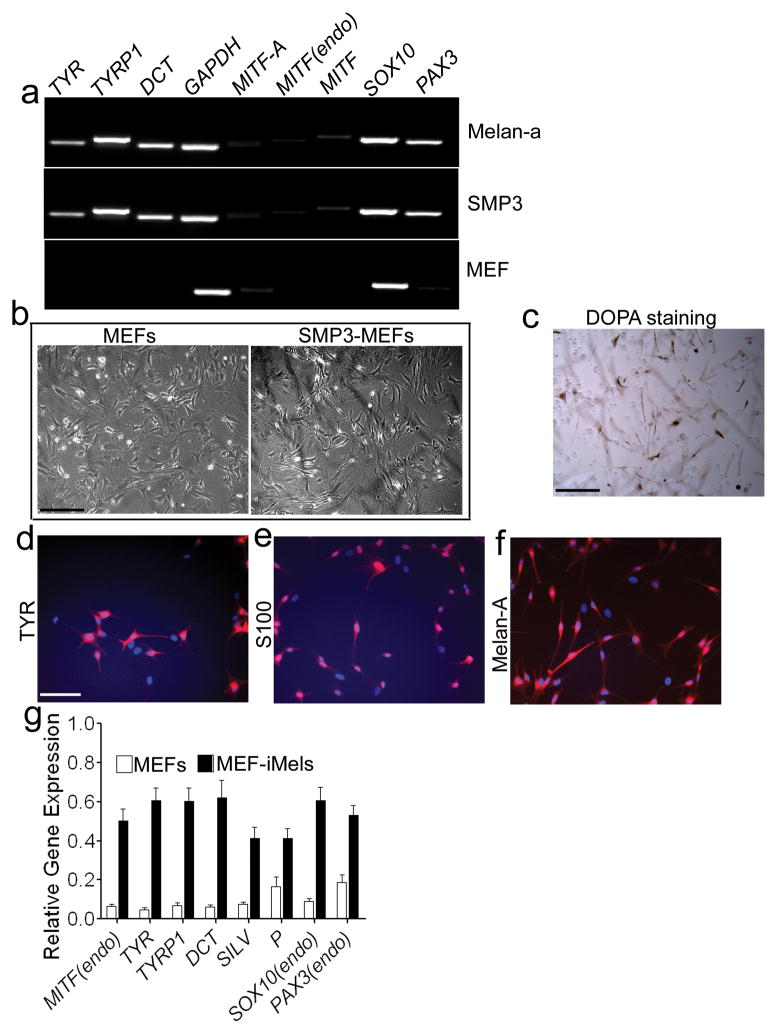
We then tested the SMP3 combination in mouse embryonic fibroblasts (MEFs) and TTFs derived from adult C57BL/6 mice. We found that melanocyte-specific markers, including TYR, TYRP1 and DCT were expressed as early as 5 days after MEFs were infected with the SMP3 combination (Fig. 2a). Since melanocytes are more resistant to G418 than fibroblasts22, we cultured the SMP3-infeced MEFs on layers of XB2 keratinocyte feeder cells for 14 days with G418. G418-resistent cells with typical melanocyte morphology showed strong Dopa activity (Fig. 2b and 2c). The majority of the G418-resistant cells expressed TYR, Melan-A and S100 (Fig. 2d-2f) and displayed melanocyte-specific gene expression patterns (Fig. 2g). Similar results were obtained when adult TTFs were infected with the SOX10, MITF-GFP and PAX3 (SMGFPP3) combination and these adult TTFs expressed melanocytic markers after infection with SMGFPP3 (Supplementary Fig. S7a). GFP+ cells (Supplementary Fig. S7b) were sorted out using FACS and cultured in melanocyte inducing medium. As expected, the reprogrammed GFP+ cells showing typical melanocyte morphologies (Supplementary Fig. S7c) displayed higher expression levels of melanocytic markers, compared to adult TTFs infected with vector only (Supplementary Fig. S7d).
Figure 2. Direct reprogramming of MEFs to iMels.
a. RT-PCR analysis of melanocytic markers in MEFs which were reprogrammed by SMP3 combination. MEFs infected with SMP3 were collected for RT-PCR analysis at Day 5 after infection. MEFs and melan-a mouse melanocytes were used as negative and positive controls, respectively. The melanocytic markers included TYR, TYRP1, DCT, MITF (endo), SOX10 and PAX3. GAPDH here was used as an internal control. b. Morphologies of parental MEFs and SMP3-induced MEFs (SMP3-MEFs). MEFs (left panel) were infected with viruses containing SMP3, cultured for 19 days and then selected under G418 and photographed (right panel). Scale bar, 50 μm. c. DOPA activity in mouse iMels. Reprogrammed cells were stained by Dopamine and showed Dopa activity. Scale bar, 50 μm. d–f. Immunocytochemical staining of iMels derived from MEFs (MEF-iMels) using antibodies specific for TYR, S100 and Melan-A. Scale bar, 25 μm. g. qRT-PCR analysis of melanocytic markers, including MITF (endo), TYR, TYRP1, DCT, P, SOX10 (endo) and PAX3 (endo) in MEF-iMels and MEFs. Data shown are mean ± SD of the expression from three independent experiments.
Direct reprogramming of human fetal dermal fibroblasts to iMels
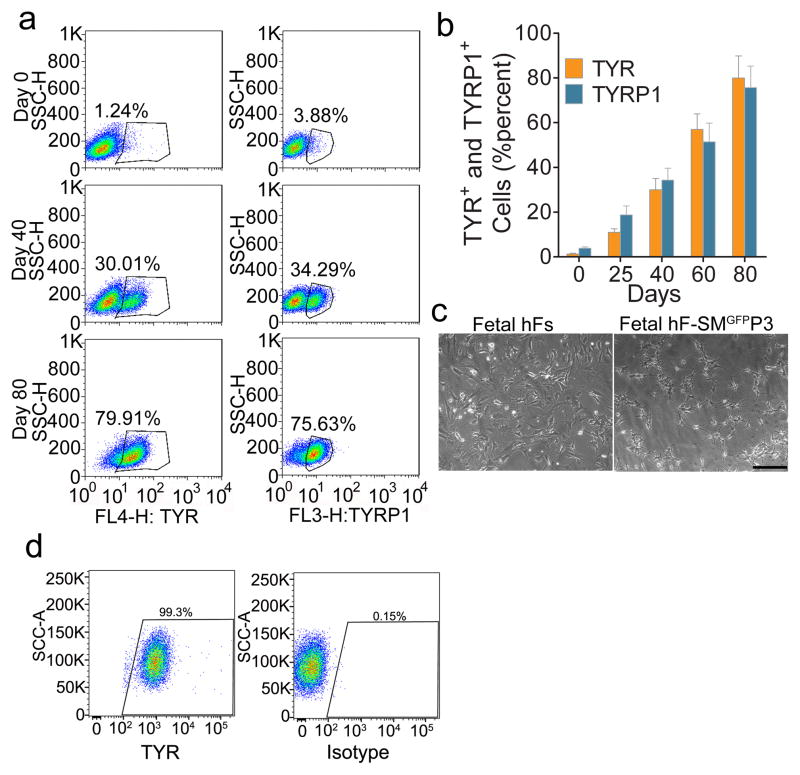
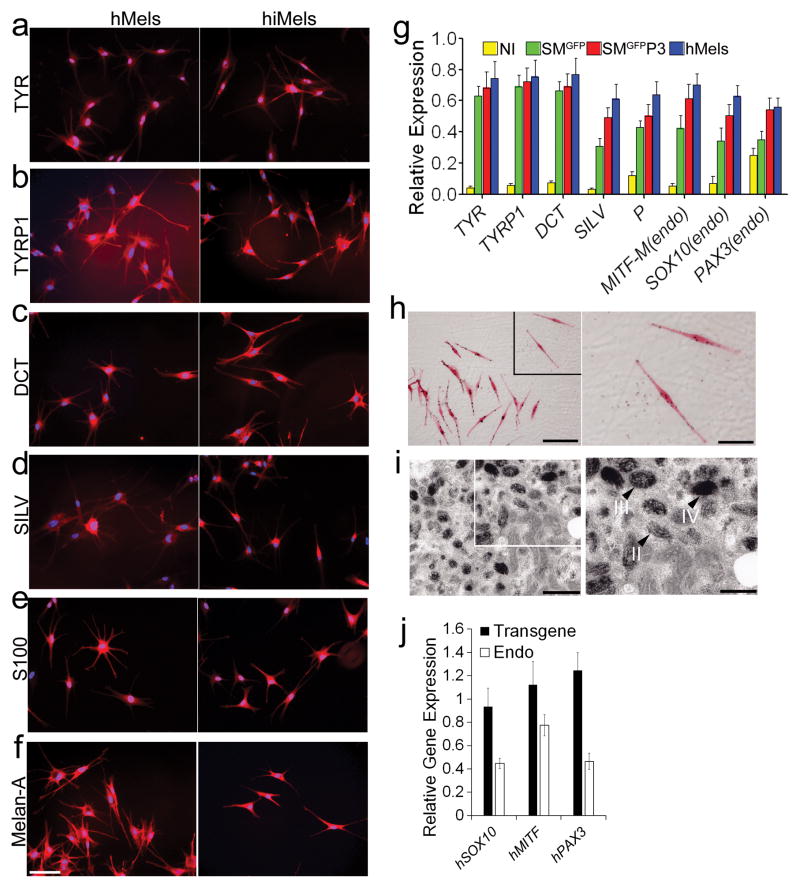
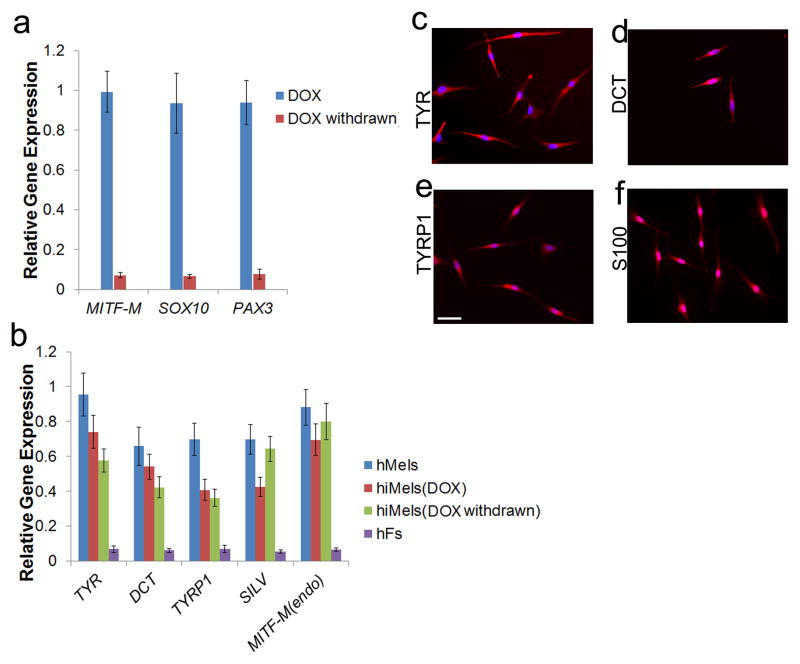
To test whether human fibroblasts can be directly reprogrammed into melanocytes, we infected passaged primary human fetal dermal fibroblasts (fetal hFs) with the human SMGFPP3 combination. SMGFPP3-infected cells were cultured under G418 selection until the cell population with typical melanocyte morphology overwhelmed other cell populations. During this process, we analyzed the percentage of TYR+ and TYRP1+ cells in the culture using flow cytometric analysis. About 40% of cells with typical melanocyte morphology were observed by Day 40 (Fig. 3a–3c), and the majority of the cells showed typical melanocyte immuno-phenotypes by Day 80 (Fig 3a and 3b). We continued to culture these melanocytic cells and found that 99.3% of the enriched cells were TYR+ cells at Day 100 (Fig. 3d). Except TYR (Fig 4a), hiMels expressed TYRP1, DCT, SILV, S100 and Melan-A (Fig 4b–4f). qRT-PCR analysis further confirmed that the melanocyte-specific gene network was activated (Fig. 4g). Endogenous expression of PAX3, SOX10 and MITF was also induced (Fig. 4g). Meanwhile, hiMels were capable of producing melanin pigment, as revealed by Fontana-Mason staining (Fig. 4h). To further confirm the melanocyte identity, we found that hiMels contained authentic melanosomes from early stage (type II) to mature melanin-containing (types III and IV) melanosomes (Fig. 4i). As expected, we found that transgenic PAX3, SOX10 and MITF were still expressed in hiMels (Fig. 4j). Concerned that the melanocyte phenotype and function might be dependent on continued transgene expression, we introduced the viruses that express doxycycline inducible PAX3, SOX10 and MITF into fetal hFs. Transgenic expression of PAX3, SOX10 and MITF was induced for 2 weeks and then silenced by withdrawing doxycycline from the culture medium (Fig. 5a). The silenced cells were cultured for another 80 days and analyzed by qRT-PCR and immunostaining assays. The melanocytic markers continued to be expressed without exogenous PAX3, SOX10 and MITF expression (Fig. 5b and 5c-5f). These data indicate that the induced melanocytic phenotype is stable and independent of transgene expression.
Figure 3. Direct reprogramming of human fibroblasts to iMels.
a. Representative flow cytometry plots for analyses of TYR+ and TYRP1+ cells in SMGFPP3 induced cells at indicated time points. Fetal hFs were infected with SMGFPP3, sorted and selected in the medium containing G418. Cells were collected and flow cytometrically analyzed at Day 0, 40 and 80. b. Percentage of TYR+ and TYRP1+ cells in SMGFPP3 induced cells at indicated time points. Human fetal fibroblasts (fetal hFs) were infected with SMGFPP3, sorted and selected in media containing G418. Cells were analyzed by flow cytometry at Day 0, 25, 40 and 80. Representative data are from three independent experiments. c. Cell morphology of fetal hFs and iMels derived from fetal hFs. Representative images of fetal hFs and iMels derived from fetal hFs (fetal hF-SMGFPP3) were photographed at Day 40. Scale bar indicates 50 μm. d. Representative flow cytometry plots for analyses of TYR+ cells after cells were reprogramming by SMGFPP3 by Day 100.
Figure 4. Characterization of directly reprogrammed human iMels from fetal dermal fibroblasts.
a–f. Immunostaining analysis of human iMels (hiMels) derived from fetal hFs and normal human skin melanocytes (hMels) using antibodies specific for TYR(a), TYRP1 (b), DCT (c), SILV (d), S100 (e) and Melan-A (f). The secondary antibody was labeled with Alexa Fluro 594. DAPI was used to stain the nuclear DNA. Scale bar indicates 20 μm. g. qRT-PCR analysis of melanocytic specific markers in fetal hFs infected with vector virus only (NI), SMGFP and SMGFPP3 and hMels. Melanocytic markers included TYR, TYRP1, DCT and SILV, and endogenous SOX10, MITF and PAX3. Data shown here are mean ± SD of the expression from three independent experiments. h. Fontana-Masson staining showed melanin pigments in hiMels. Scale bars indicate 25 μm in left panel and 10 μm in right panel. Arrow heads point to the melanin pigment. i. Electron microscopy images of hiMels with many mature melanosomes in the cytoplasm. II: stage II melanosome; III: stage III melanosome; IV: stage IV melanosome. Scale bars indicate 800 nm in left panel and 400 nm in right panel. Arrow heads point to melanosomes. j. qRT-PCR analysis of transgenic and endogenous (Endo) expression of human PAX3 (hPAX3), human SOX10 (hSXO10) and human MITF (hMITF) in hiMels. Data shown are mean ± SD of the expression from three independent experiments.
Figure 5. Generation of hiMels using inducible system.
Human MITF-M, SOX10 and PAX3 were subcloned into Doxycycline inducible viral vectors. Fetal hFs were infected with viruses carrying these three transgenes. a. qRT-PCR analysis of the transgenic expression of SOX10, MITF and PAX3 in the presence or absence of DOX. b. qRT-PCR analysis of melanocytic markers in hiMels which were stopped treating by DOX (DOX withdrawn), hiMels with DOX persistent induction (DOX), hMels and fetal hFs. hMels and fetal hFs here were used as positive and negative controls, respectively. Data shown are mean ± SD of the expression from three independent experiments. c–f. Immunostaining analysis of melanocytic markers including TYR (c), DCT (d), TYRP1 (e) and S100 (f) in hiMels which showed no transgenic SOX10, MITF and PAX3 expression without treating by DOX. Scale bar, 30 μm.
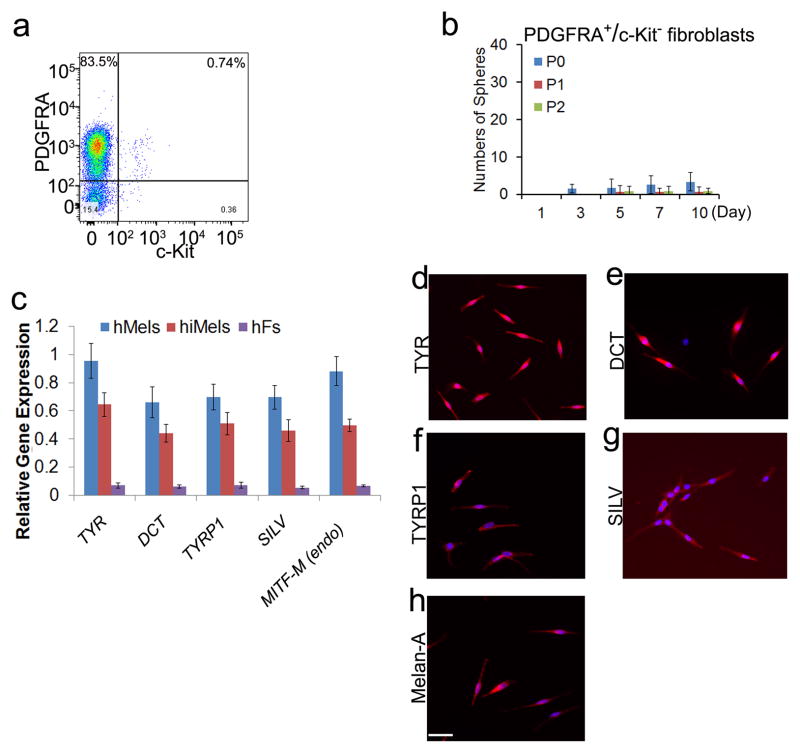
Next, we performed sphere formation assays to test presence of adult stem cells in primary fetal fibroblasts in the current melanocyte culture condition. We found that Passage 0 (P0) primary fetal dermal fibroblasts can form some spheres; however these sphere forming cells decreased dramatically with passaging and by Passage 2 (P2) fetal fibroblasts which were used in the melanocyte induction experiments formed few spheres (Supplementary Fig. S8). To exclude the possibility that contaminating dermal adult stem cells or melanocyte stem cells in the fetal fibroblast cultures contributed to melanocyte induction, we cultured P2 fetal fibroblasts in the induction medium for 40 days. After this incubation period, we did not detect any cells that expressed TYR, TYRP1, DCT or Melan-A (Supplementary Fig. S9a). Similarly, flow cytometric analysis showed few TYR+ or TYRP1+ cell populations in the culture (Supplementary Fig. S9b).
To further clarify the purity and identity of the starting fibroblast population, we used a fibroblast marker, PDGFRA23, to purify fibroblasts and a melanocyte marker, c-Kit, to gate out melanocytes using the MACS MicroBead technology (Fig. 6a). Firstly, we found that the PDGFRA+/c-KIT− fibroblasts formed few spheres (Fig. 6b). The P2 PDGFRA+/c-KIT--purified fibroblasts were then reprogrammed by the human SMGFPP3 factors group. Expression of melanocytic markers was confirmed by qRT-PCR and immunostaining (Fig. 6c and 6d). These results further support that iMels are reprogrammed from dermal fibroblasts.
Figure 6. Characterization of hiMels induced from purified human fetal fibroblasts.
Human fetal PDGFRA+/c-Kit− fibroblasts were reprogrammed using SMGFPP3. a. Flow cytometric analysis of the percentage of PDGFRA+ and c-Kit+ cells in primary human fetal fibroblasts. Representative data are from three independent experiments. b. Sphere formation capacity of PDGFRA+/c-Kit− fibroblasts from primary fetal fibroblasts. P0 fibroblasts were MACS microbeads purified using an antibody against PDGFRA (positive selection) and c-Kit (negative selection). PDGFRA+/c-Kit− fibroblasts were experimented in the sphere formation assays. P0 indicates Passage 0, P1 indicates Passage 1 and P2 indicates Passage 2. Representative data are from three independent experiments. c. qRT-PCR analysis of melanocytic markers in hMels, hiMels derived from human fetal PDGFRA+/c-Kit− fibroblasts (hiMels) and human fetal PDGFRA+/c-Kit− fibroblasts (hFs). Data shown here are mean ± SD of gene expression from three independent experiments. d-h. Immunostaining analysis of melanocytic markers in hiMels including TYR (d), DCT (e), TYRP1 (f), SILV (g) and Melan-A (h). Scale bar, 30 μm.
Molecular characterization of hiMels
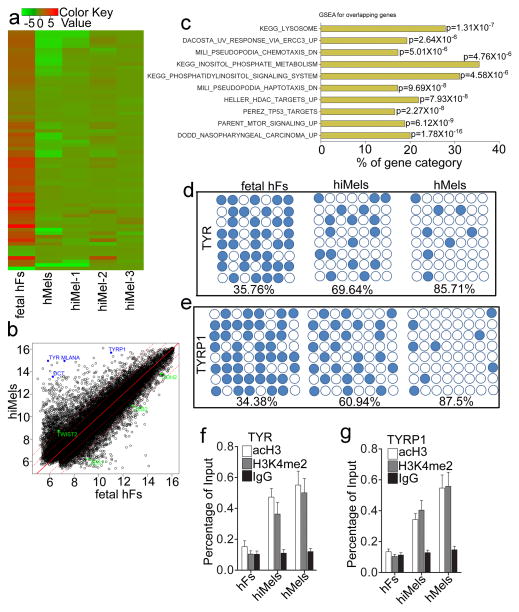
Global expression analysis showed that hiMels clustered with human adult melanocytes rather than with the parental fibroblasts, as illustrated by unsupervised hierarchical clustering (Fig. 7a). Of note, many representative genes encoding rate limiting enzymes for pigmented melanin production (such as TYR, TYRP1 and DCT) were upregulated in hiMels (Fig. 7b). In addition, Melan-A was highly expressed in hiMels (Fig. 7b). Moreover, we analyzed the MSigDB gene set collection for its enrichment in both hiMels and hMels. As shown in Fig. 7c, hiMels derived from human fibroblasts gained the characteristic of melanocytes (KEGG_LYSOSOME gene set and DACOSTA_UV_RESPONSE _VIA_ ERCC3 pathways) and lost the expression of fibroblast specific gene network (MILI_PSEUDOPODIA_HAPTOTAXIS_DN and MILI_PSEUDOPODIA_ CHEMOTAXIS _DN pathways24).
Figure 7. Molecular characterization of induced human melanocytes.
a. Heat-map of genes differentially expressed in RNA-microarray analysis performed on human fetal fibroblasts (fetal hFs), induced melanocytes derived from human fetal fibroblast (hiMels) and normal skin melanocytea (hMels). b. Scatter plots show that melanocytic markers are expressed in hiMels, but not in fetal hFs. c. Gene Set Enrichment Analysis (GSEA) for the overlapping genes between hMels and hiMels. Many gene sets including KEGG_LYSOSOME, DACOSTA_UV_RESPONSE_VIA_ERCC3, PARENT_MTOR_SIGNALING_UP, MILI_PSEUDOPODIA_HAPTOTAXIS_DN and MILI_PSEUDOPODIA_CHEMOTAXIS_DN were enriched in hiMels and hMels. d and e. DNA methylation analysis of the promoters of TYR (d) and TYRP1 (e) in fetal hFs, hiMels and hMels. Open circles indicate unmethylated CpG dinucleotides, while closed circles indicate methylated CpGs. f and g. Histone modification analysis of promoters of TYR and TYRP1 in fetal hFs, hiMels and hMels. Chromatin immunoprecipitation was performed using antibodies against dimethylated histone H3K4 (H3K4me2) and H3 acetylation (acH3). TYR and TYRP1 promoters showed enrichment for the active states (H3K4 me2 and acH3) in hiMels and hMels. In fetal hFs, TYR and TYRP1 promoters appeared in the inactive state. Representative data are from three independent experiments.
We next analyzed the DNA methylation status of TYR and TYRP1 promoters, as indicators of gene activation. As expected, TYR and TYRP1 promoters were highly demethylated in hiMels (percentage of demethylation: TYR, 69.64%; TYRP1, 60.94%) and human melanocytes (percentage of demethylation: TYR, 85.71%; TYRP1, 87.5%), whereas these same regions were highly methylated in the parental fibroblasts (percentage of demethylation: TYR, 35.67%; TYRP1, 34.38%) (Fig. 7d and 7e). We then performed chromatin immunoprecipitation (ChIP) assays to analyze histone modifications in TYR and TYRP1 promoter regions. We found that hiMels and hMels showed high levels of H3K4me2 methylation and acH3 acetylation, compared to hiMels’ parental fibroblasts (Fig. 7f and 7g).
Functional characterization of hiMels in vitro and in vivo
To study the biological functions of hiMels, we generated 3D organotypic skin equivalents using hiMels, parental fetal fibroblasts and foreskin keratinocytes as previously described25. Melan-A and S100 positive melanocytes were found to locate near the dermal-epidermal junction (Supplementary Fig. S10a). Fontana-Masson staining showed melanin pigment in the basal and supra-basal layer of keratinocytes, indicating that the iMels produce pigment and also transfer pigment to surrounding keratinocytes (Supplementary Fig. S10a). In addition, we found that melanin production in hiMels-constituted 3D skin reconstruct increased upon α-MSH stimulation (Supplementary Fig. S10b).
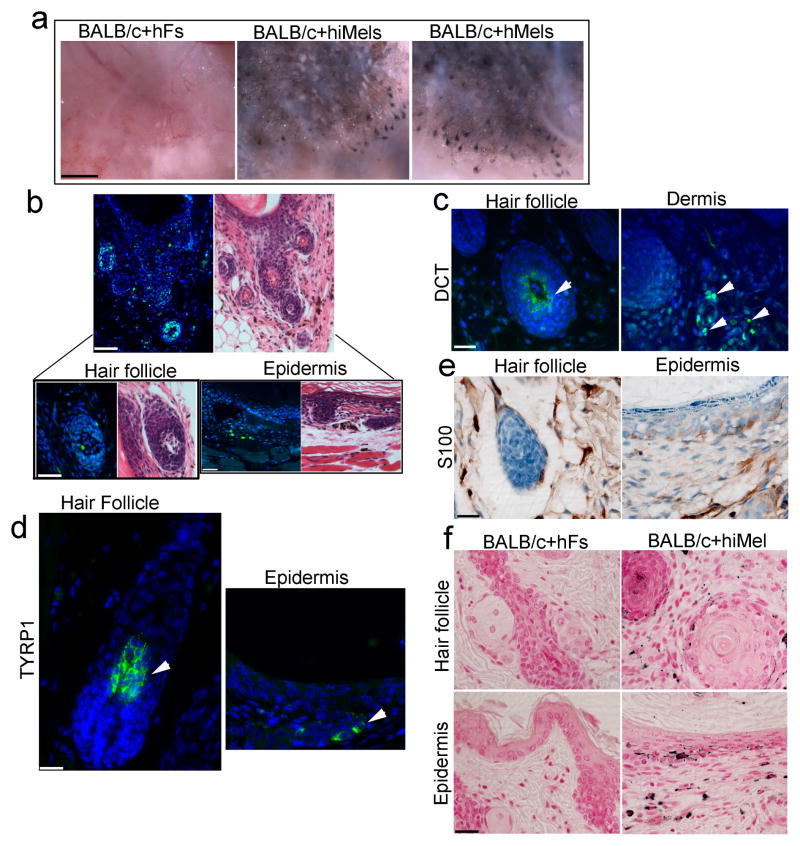
To investigate the in vivo function of hiMels, we developed a novel assay to assess skin and hair pigmentation using a modified skin reconstitution assay26–28. In this assay, hiMels were mixed with epithelial cells and dermal fibroblasts derived from neonatal BALB/c albino mouse skin and then transplanted into the back skin of nude mice. Human normal skin melanocytes were used as positive controls. When human normal skin melanocytes or hiMels were mixed with cells prepared from BALB/c albino mouse skin, pigmented hair follicles formed (Fig. 8a, middle and right panels). To avoid the possibility of contaminating melanocytes or melanocyte stem cells in the parental fibroblasts, we mixed parental fibroblasts with BALB/c-derived epithelial cells and dermal fibroblasts and then transplanted the cell mixture into the back skin of nude mice. Only white hair was produced under these conditions (Fig. 8a, left panel). The grafts were harvested and histologic examination revealed epidermal lined cysts with many hair follicles (Fig. 8b). We found that hiMel specifically recognized by Alu-probe labeling with FITC were localized near the dermal-epidermal junction, inside the hair follicles and in the dermis (Fig. 8b). DCT+ cells can be seen in the interfollicular dermis and bulb region of hair follicles (Fig. 8c). Meanwhile, TYRP1+ cells were observed in hair follicles and the epidermis (Fig. 8d). Similarly, S100+ cells were seen near the dermal-epidermal junction in the epidermis and in the hair follicles (Fig. 8e). We then performed Fontana-Masson staining to study the distribution of melanin pigment in the regenerated epidermis and hair follicles. Melanin pigment was detected in the epidermis, hair follicle epithelium and hair shafts (Fig. 8f), indicating that the hiMels were able to transfer pigment to the surrounding keratinocytes in vivo (Fig. 8f). The pigmentation patterns formed in the skin reconstitution assays using hiMels were identical to those of normal skin melanocytes. We did not find any melanin pigment in the skin reconstitution assays using parental fibroblasts (Fig. 8f). These data indicate that hiMels are functionally identical to normal skin melanocytes.
Figure 8. Functional analysis of induced human melanocytes in vivo.
a. Skin reconstitution assays showed pigmented hair follicles using hiMels. hiMels, hMels or fetal hFs combined with neonatal mouse dermal fibroblasts and epithelial cells derived from BALB/c (albino) mouse skin. Cells were injected into the back skin of an immunodeficient mouse. After 3 weeks, grafts were photographed from the underside of the skin. Pigmented hair follicles were observed in hiMels or hMels groups in the skin reconstitution assays, whereas pigmented hair follicles were not observed in the group containing fetal hFs. Scale bar, 5 mm. n=5. b. Human specific Alu staining (green nuclei) confirmed human origin of iMels (left, upper panel) and H&E staining of consecutive section showed a cyst with hair follicle formation (right, upper panel); Scale bar, 200 μm. Human cells located in the bulb region of a hair follicle (left, lower panel) and basal layer of epidermis (right, lower panel); Scale bar, 50 μm. c and d. Immunostaining of the xenografts from the patch assays using antibodies against human DCT (c) and TYRP1 (d). DCT+ cells were present in the interfollicular dermis and bulb region of hair follicles. TYRP1+ cells were observed in both hair follicles and the epidermis. Scale bar, 20 μm. e. Immunohistochemical staining of the xenografts using the antibody against human S100. S100+ positive cells were located in epidermis, hair follicle and the dermis. Scale bar, 20 μm. f. Fontana-Masson staining of the xenografts showed no pigment in xenografts formed by fetal hFs and mouse cells (left); whereas abundant melanin pigment was evident in the epidermis and follicular epithelium when hiMels were included in the assays (right). Scale bar, 20 μm.
To test whether MITF alone is sufficient to generate functional melanocytes in vivo, we cultured MITF-infected fibroblasts for 50 days in the melanocyte inducing medium. We then transplanted MITF-infected fibroblasts with BALB/c-derived epithelial cells and dermal fibroblasts mixture to mouse back skin. We did not find any pigment in the hair follicles and epidermis in the skin reconstruct (Supplementary Fig. S11a and S11b), indicating that MITF alone is insufficient to convert fibroblast to melanocyte.
Direct reprogramming of human adult dermal fibroblasts to iMels
We then used the same procedure to human adult fibroblasts (adult hFs) and infected them with the SMGFPP3 virus combination. The SMGFPP3-infected cells were cultured for 15 days, and GFP+ cells were then sorted and cultured in melanocyte inducing medium for 40 days. qRT-PCR analysis of the GFP+ cells showed the activated melanocytic network for melanin production and transfer (Supplementary Fig. S12a). We also detected that key melanocytic markers including TYR, DCT, S100 and Melan-A were expressed in cells with typical melanocyte morphology (Supplementary Fig. S12b). Notably, only 6% of the adult hFs expressed melanocytic markers, indicating that the adult fibroblast reprogramming efficiency was much lower than that for fetal fibroblasts (Supplementary Fig. 12c). We performed flow cytometric analysis to measure the percentage of the TYR+ and TYRP1+ cells in the adult reprogrammed cells and found only 3.27% and 2.16%, respectively (Supplementary Fig. S12d).
Discussion
Our results demonstrate that SOX10, MITF and PAX3 are sufficient to reprogram mouse and human fibroblasts to functional melanocytes. iMels generated using our method acquire all the biochemical and functional characteristics of normal melanocytes.
Substantial progress has been achieved with the discovery of transdifferentiation of somatic cells to other lineage specific cell types4, 5, 7–11, 29, suggesting that somatic cell fates can be interconverted without transiting through distinct hierarchies. We previously showed that a multipotent cell population in the foreskin dermis can be differentiated into melanocytes30. However, this approach is limited by the small percentage (0.085%) of dermal stem cells present in foreskin and the relatively low differentiation efficiency of these cells to melanocytes. In addition, the foreskin dermal stem cell population gradually disappears with serial passage. To exclude the possibility that these dermal stem cells contributed to melanocyte direct reprogramming, we used a fibroblast cell surface marker, PDGFRA23, to sort out PDGFRA+/c-Kit− fibroblasts. We showed that these purified fibroblasts can also be reprogrammed to melanocytes, supporting that iMels are directly reprogrammed from dermal fibroblasts. Nevertheless, despite our extensive efforts to characterize the starting dermal fibroblasts, we cannot completely exclude the possibility that some of the iMels might be derived from multipotent cells in the dermis.
In translational medicine, generation of functional melanocytes by direct reprogramming establishes a means for obtaining a scalable source of autologous melanocytes, which then could be used for developing cell-based treatments for pigmentary disorders such as vitiligo and hypopigmentation associated with congenital disorders. Reprogramming fibroblasts to melanocytes specifically from patients with melanoma also serves as a powerful strategy for studying the etiology of melanoma.
Methods
Cell culture
TTFs were isolated from the Tyrosinase-CreER/Gt (ROSA)26Sor tm4(ACTB-tdTomato,-EGFP)Luo/J transgenic and C57BL/6 mice. Tails were peeled, minced into 1cm pieces, placed on culture dishes, and incubated in MEF media (Dulbecco’s modified Eagle medium (DMEM; Invitrogen) containing 10% fetal bovine serum (FBS; Hyclone), non-essential amino acids (Invitrogen), sodium pyruvate and penicillin/streptomycin (Invitrogen)) for 5 days. MEFs were isolated from Day 14.5 mouse embryos. Cells were split no more than three times in all experiments. Human fetal fibroblasts were isolated from 20-week old fetal skin (Advanced Bioscience Resources, Inc; Alameda, CA). Human adult fibroblasts were obtained from discarded normal skin after surgery following a protocol approved by the University of Pennsylvania Institutional Review Board. Human skin samples were mechanically dissociated, plated on gelatin-coated dishes and cultured in MEF media. XB2, an immortal line of mouse keratinocytes, was culture in MEF media. HEK 293T cells and human fibroblasts were cultured in DMEM supplemented with 10% fetal bovine serum (FBS), 2mM L-glutamine, 100 unit/ml penicillin and 100μg/ml of streptomycin (all from Invitrogen).
Viral infection
For mouse cell infection, viruses were packaged by transfecting retroviruses and pECO into 293T cells. For human cell infection, pantropic viruses were packaged by transfecting the retrovirus vectors, pUMVC and pCMV-VSVG into 293T cells. To improve the pantropic retrovirus infection efficiency, we concentrated the virus using the Retro-X™ Concentrator (Clontech) according to manufacturer’s instruction. After 48h, growth medium was replaced with mouse melanocyte inducing medium containing RPMI 1640 (Invitrogen), 10% FBS, 10ng/ml bFGF (Invitrogen), 100ng/ml SCF (R&D), 100 nM ET-3 (American Peptide Company), 20pM cholera toxin (CT) (Sigma-Aldrich) and 200nM 12-O-tetradecanoyl-phorbel 13-acetate (TPA) (Sigma-Aldrich). Human melanocyte inducing medium contained medium 254, 10ng/ml bFGF, 100ng/ml SCF, 100 nM ET-3. For G418 selection, 60 ug/mL G418 was added in melanocyte induction media.
Flow cytometry and cell sorting
Cells (2.5 × 106 cells/ml) were stained with antibodies against TYR (Abcam), TYRP1 (Sigma), c-Kit (eBioscience) and PDGFRA (BioLegend). To detect intracellular proteins, staining was carried out on cells fixed with 4% paraformaldehyde (Electron Microscopy Sciences) in PBS. Staining was performed in PBS with 2% FBS. Stained cells and GFP+ cells were analyzed using an LSRII flow cytometer (BD). For FACS sorting, cells were sorted at a concentration of 106 cells/ml in PBS/2% FBS using a FACSAriaTMII (BD) cell sorter (Upenn Flow Cytometry Facility). PDGFRA+/c-Kit− fibroblasts were sorted from P1 fetal hFs and cultured as P0. For magnetic bead sorting, the Miltenyi MACS bead sorting system was used according to the manufacturer’s guidelines. Data were analyzed using FlowJo software (Treestar). The working solutions of antibodies are shown in Supplementary Table S3.
Immunofluorescence and immunochemistry
Monolayer cells were fixed with 4% paraformaldehyde and stained with primary antibodies specific for TYR, TYRP1, DCT (polyclonal; a gift from Dr. V.J. Hearing, Bethesda, MD), Melan-A, SILV and S100 (polyclonal; Dako). After washing, cells were incubated with the appropriate Alexa Fluor® 594-labeled secondary antibodies (Invitrogen). Paraffin-embedded slides were deparaffinized, followed by antigen retrieval and staining as described above. Immunofluorescence for DCT was performed as described above. Immunohistochemical staining for S100, melan-A and Fontana-Masson was performed on paraffin-embedded slides using standard immunoperoxidase techniques. The working solutions of antibodies are shown in Supplementary Table S3.
qRT-PCR
RNA was extracted from single cultures, using RNA mini kits (Qiagen) according to manufacturer’s instructions. We performed reverse transcription reactions using SuperScript™ III First-Strand Synthesis Kit (Invitrogen). qPCR was performed using SYBR Green Supermix (Bio-Rad) and reactions were analyzed using the Bio-Rad qPCR detection system. The primers used are listed in Supplementary Table S2.
Global gene profiling and array analysis
Micro-array raw data generated from Illumina Chips were normalized, background-corrected, and summarized using the R package “lumi”31. To reduce false positives, unexpressed probes were removed from analysis and 21,758 probes were examined in all experiments described herein. The R package “limma”32, 33 was used to evaluate differential gene expression analysis, followed by multiple test correction by the Benjamini and Hochberg procedure 3734. The genes with the adjusted p values < 0.05 and fold changes > 4 were subjected to the two-way clustering analysis to generatethe heat maps. GSEA analysis was performed as described previously 35. Briefly, the top bound genes in the iMel and parental Fibroblast data sets were selected, and the overlapping genes were subjected to analysis by using the MSigDB gene set collection.
Bisulfite genomic sequencing
Bisulfite treatment was performed using the CpGenome modification Kit (Millipore), according to the manufacturer’s recommendations. The PCR primers used are listed in the Supplementary Table S2. Amplified products were cloned into pCR2.1-TOPO (Invitrogen). Ten randomly selected clones were sequenced with the M13 forward and M13 reverse primers for each gene. Sequencing was performed at the University of Pennsylvania sequencing facility. CpG methylation of the sequence was analyzed by BiQ Analyzer software.
Chromatin immunoprecipitation
107 hFFs, hiMels or human melanocytes were fixed with 1% formaldehyde at room temperature for 10 min and then lysed in 1 ml lysis buffer (50 mM Tris-HCl, pH 8.0, 10 mM EDTA, 1% SDS, and protease inhibitors) on ice for 20 min. The lysate was split into three tubes and sonicated. After 10 min centrifugation, the supernatant was pre-cleared by incubating at 4 °C for 4 h with agarose beads pre-blocked with BSA (1 mg BSA for 10 ml beads) in IP buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 2 mM EDTA, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, protease inhibitors). A total of 100 μl of pre-cleared chromatin diluted in 1 ml IP buffer in the presence of 20 μg antibody was used for each immunoprecipitation reaction, according to the manufacturer’s protocol. The antibodies used for this study were: anti-acH3 (Millipore), anti-dimethyl K4 of H3 (H3K4me2, Millipore), and normal rabbit IgG (Sigma). The precipitate was purified and analyzed by qPCR. PCR primers are listed in Supplementary Table S2.
Sphere formation assay
Sphere formation assay was performed as described previously36. Briefly, dissociated cells were plated in serum-free media in low-cell-density cultures with exogenous growth factors used at final concentrations of 10 ng/ml bFGF and 100 ng/ml SCF. Sphere colonies were generated and counted under the microscopy.
Human 3D skin reconstructs
Human 3D skin reconstructs were generated as described previously25. Briefly, inserts of tissue culture trays (Organogenesis, Canton, MA) were coated with 1 ml bovine collagen I (Organogenesis) and layered with 3 ml collagen I containing 7.5×104 fibroblasts. After 4 to 7 days at 37°C, hiMels were mixed with keratinocytes and seeded on top of the dermal reconstructs at a ratio of 1:5 (hiMels to keratinocytes). After 4 days, human keratinocytes were added and the cells were cultured in skin reconstruct media: keratinocyte serum-free medium (Invitrogen) with 60 μg/ml bovine pituitary extract, 2% dialyzed fetal bovine serum (FBS, Invitrogen), 4.5 ng/ml bFGF (Invitrogen), 100 nM ET-3 (sigma), and 10 ng/ml SCF (R&D system). Cultures were submerged in media containing 1 ng/ml epidermal growth factor (EGF) (Invitrogen) for 2 days, 0.2 ng/ml EGF for another 2 days, then maintained at the air—liquid interface and incubated in high-calcium (2.4 mM) media. Two weeks later, skin reconstructs were harvested, fixed in 10% neutral buffered formalin for 3 hours, and processed by routine histological methods. For α-MSH treatment assay, 50 nM α-MSH was added to induce the melanin production in the cultured reconstructed skin.
Pigmented skin reconstitution assays with melanocytes
The protocol has been approved by the University of Pennsylvania Institutional Animal Care and Use Committee. Skin reconstitution assays were performed as described previously27, 28. Briefly, truncal skin from one day postnatal BALB/c mice, a mouse strain with non-pigmented white hair coat, was removed and rinsed in free PBS. The skin was laid flat in PBS containing Dispase (2.5 mg/ml, Invitrogen) at 4 °C overnight. Inductive dermal cells and epidermal cells were isolated as previously described37–39. In brief, epidermis and dermis were separated firstly. Epidermal and dermal cells were then isolated for skin reconstitution. Trichogenic cells were assayed in male nude (nu/nu) mice (Charles River) at 7–9 weeks. For each intracutaneous injection, 1 × 106 BALB/c neonatal dermal cells and 10,000 epidermal aggregates were used. In order to test the ability of hiMels to participate in the formation of hair follicles and produce pigmented shafts in the reconstitution assay, 0.5X106 hiMels were added to the neonatal BALB/c dermal and epidermal cell mixture for each injection. As positive controls, 0.5X106 cultured human melanocytes were added in separate injections with the mouse cells. The cell mixtures were resuspended in 50–70 μl of DMEM-F12 medium (Invitrogen) and injected (25-gauge needle) into the hypodermis of the mouse skin, forming a bleb. The injection site was marked by a black tattoo puncture (242 Permanent Black Pigment, Aims, Hornell, New York). The skin reconstruct was harvested at two weeks after injection and the newly formed hair follicles were examined under a dissection microscope. Two independent experiments with different batches of hiMels were performed, with 2 duplicate sets each time.
In situ hybridization
Briefly, paraffin slides were dehydrated, antigen retrieved, and hybridized with Alu DNA probe (BioGenex PR-1001-01, ready-to-use): heat slide at 85°C for 10 min, and then incubate at 37°C overnight. The slides were then incubated with antibody specific for fluorescein, biotin-labeled (BioGenex AS2505-16, ready to use), and finally incubated with secondary antibody labeled with Streptavidin-Alexa Fluo 488. DAPI was used to label nuclear DNA.
Statistical analysis
Student’s t-test or ANOVA was used to analyze gene expression and flow cytometric data. qPCR data were analyzed after being normalized for β-actin loading control. Error bars indicate standard deviation. Statistical significance was determined if two-sided P<0.05.
Supplementary Material
Acknowledgments
We would like to thank Dr. M. Boserberg for providing the Tyr:cre; BRAFca mice; the histology laboratory at the Department of Pathology and Laboratory Medicine for assistance in histological studies, Anatomic pathology test development center for immunohistochemical stains, Penn Flow Cytometry Facility and the Cooperative Human Tissue Network. This research was supported by the US National Institutes of Health grant R01-AR054593, P30-AR057217 and P01-CA 25874
Footnotes
Author contributions
R.Y. and X.X. designed the studies, conducted the experiments, interpreted the data and wrote the manuscript. Y.Z., L.L., S.L., M.B., A.N., R.C. and W.G. conducted biochemical and biological studies. M.H. and G.C. interpreted the data and wrote the manuscript. Z.W. analysed the microarray data.
Accession Codes: Microarray data has been deposited in Gene Expression Omnibus under accession number GSE51028.
References
- 1.Slominski A, Tobin DJ, Shibahara S, Wortsman J. Melanin pigmentation in mammalian skin and its hormonal regulation. Physiol Rev. 2004;84:1155–1228. doi: 10.1152/physrev.00044.2003. [DOI] [PubMed] [Google Scholar]
- 2.Alikhan A, Felsten LM, Daly M, Petronic-Rosic V. Vitiligo: a comprehensive overview Part I. Introduction, epidemiology, quality of life, diagnosis, differential diagnosis, associations, histopathology, etiology, and work-up. J Am Acad Dermatol. 2011;65:473–491. doi: 10.1016/j.jaad.2010.11.061. [DOI] [PubMed] [Google Scholar]
- 3.Hann SK, Im S, Bong HW, Park YK. Treatment of stable vitiligo with autologous epidermal grafting and PUVA. J Am Acad Dermatol. 1995;32:943–948. doi: 10.1016/0190-9622(95)91329-7. [DOI] [PubMed] [Google Scholar]
- 4.Caiazzo M, et al. Direct generation of functional dopaminergic neurons from mouse and human fibroblasts. Nature. 2011;476:224–227. doi: 10.1038/nature10284. [DOI] [PubMed] [Google Scholar]
- 5.Huang P, et al. Induction of functional hepatocyte-like cells from mouse fibroblasts by defined factors. Nature. 2011;475:386–389. doi: 10.1038/nature10116. [DOI] [PubMed] [Google Scholar]
- 6.Ieda M, et al. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142:375–386. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pang ZP, et al. Induction of human neuronal cells by defined transcription factors. Nature. 2011;476:220–223. doi: 10.1038/nature10202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qian L, et al. In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature. 2012;485:593–598. doi: 10.1038/nature11044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sekiya S, Suzuki A. Direct conversion of mouse fibroblasts to hepatocyte-like cells by defined factors. Nature. 2011;475:390–393. doi: 10.1038/nature10263. [DOI] [PubMed] [Google Scholar]
- 10.Song K, et al. Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature. 2012;485:599–604. doi: 10.1038/nature11139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vierbuchen T, et al. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Adameyko I, et al. Sox2 and Mitf cross-regulatory interactions consolidate progenitor and melanocyte lineages in the cranial neural crest. Development. 2012;139:397–410. doi: 10.1242/dev.065581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tachibana M, et al. Ectopic expression of MITF, a gene for Waardenburg syndrome type 2, converts fibroblasts to cells with melanocyte characteristics. Nat Genet. 1996;14:50–54. doi: 10.1038/ng0996-50. [DOI] [PubMed] [Google Scholar]
- 14.Southard-Smith EM, Kos L, Pavan WJ. Sox10 mutation disrupts neural crest development in Dom Hirschsprung mouse model. Nat Genet. 1998;18:60–64. doi: 10.1038/ng0198-60. [DOI] [PubMed] [Google Scholar]
- 15.Hornyak TJ, Hayes DJ, Chiu LY, Ziff EB. Transcription factors in melanocyte development: distinct roles for Pax-3 and Mitf. Mech Dev. 2001;101:47–59. doi: 10.1016/s0925-4773(00)00569-4. [DOI] [PubMed] [Google Scholar]
- 16.Galibert MD, Yavuzer U, Dexter TJ, Goding CR. Pax3 and regulation of the melanocyte-specific tyrosinase-related protein-1 promoter. J Biol Chem. 1999;274:26894–26900. doi: 10.1074/jbc.274.38.26894. [DOI] [PubMed] [Google Scholar]
- 17.Baumer N, et al. Retinal pigmented epithelium determination requires the redundant activities of Pax2 and Pax6. Development. 2003;130:2903–2915. doi: 10.1242/dev.00450. [DOI] [PubMed] [Google Scholar]
- 18.Saito H, et al. Melanocyte-specific microphthalmia-associated transcription factor isoform activates its own gene promoter through physical interaction with lymphoid-enhancing factor 1. J Biol Chem. 2002;277:28787–28794. doi: 10.1074/jbc.M203719200. [DOI] [PubMed] [Google Scholar]
- 19.Prince S, Wiggins T, Hulley PA, Kidson SH. Stimulation of melanogenesis by tetradecanoylphorbol 13-acetate (TPA) in mouse melanocytes and neural crest cells. Pigment Cell Res. 2003;16:26–34. doi: 10.1034/j.1600-0749.2003.00008.x. [DOI] [PubMed] [Google Scholar]
- 20.Curran K, et al. Interplay between Foxd3 and Mitf regulates cell fate plasticity in the zebrafish neural crest. Dev Biol. 2010;344:107–118. doi: 10.1016/j.ydbio.2010.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dankort D, et al. Braf(V600E) cooperates with Pten loss to induce metastatic melanoma. Nat Genet. 2009;41:544–552. doi: 10.1038/ng.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sviderskaya EV, et al. Complementation of hypopigmentation in p-mutant (pink-eyed dilution) mouse melanocytes by normal human P cDNA, and defective complementation by OCA2 mutant sequences. J Invest Dermatol. 1997;108:30–34. doi: 10.1111/1523-1747.ep12285621. [DOI] [PubMed] [Google Scholar]
- 23.Collins CA, Kretzschmar K, Watt FM. Reprogramming adult dermis to a neonatal state through epidermal activation of beta-catenin. Development. 2011;138:5189–5199. doi: 10.1242/dev.064592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mili S, Moissoglu K, Macara IG. Genome-wide screen reveals APC-associated RNAs enriched in cell protrusions. Nature. 2008;453:115–119. doi: 10.1038/nature06888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li L, Fukunaga-Kalabis M, Herlyn M. The three-dimensional human skin reconstruct model: a tool to study normal skin and melanoma progression. J Vis Exp. 2011 doi: 10.3791/2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morris RJ, et al. Capturing and profiling adult hair follicle stem cells. Nat Biotechnol. 2004;22:411–417. doi: 10.1038/nbt950. [DOI] [PubMed] [Google Scholar]
- 27.Zheng Y, et al. Organogenesis from dissociated cells: generation of mature cycling hair follicles from skin-derived cells. J Invest Dermatol. 2005;124:867–876. doi: 10.1111/j.0022-202X.2005.23716.x. [DOI] [PubMed] [Google Scholar]
- 28.Zheng Y, et al. Mature hair follicles generated from dissociated cells: a universal mechanism of folliculoneogenesis. Dev Dyn. 2010;239:2619–2626. doi: 10.1002/dvdy.22398. [DOI] [PubMed] [Google Scholar]
- 29.Pfisterer U, et al. Direct conversion of human fibroblasts to dopaminergic neurons. Proc Natl Acad Sci U S A. 2011;108:10343–10348. doi: 10.1073/pnas.1105135108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li L, et al. Human dermal stem cells differentiate into functional epidermal melanocytes. J Cell Sci. 2010;123:853–860. doi: 10.1242/jcs.061598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Du P, Kibbe WA, Lin SM. lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24:1547–1548. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- 32.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 33.Smyth GK, editor. Bioinformatics and Computational Biology Solutions using R and Bioconductor. Springer; New York: 2005. [Google Scholar]
- 34.Benjamini YH. Y Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society. Series B. 1995;57 [Google Scholar]
- 35.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tropepe V, et al. Retinal stem cells in the adult mammalian eye. Science. 2000;287:2032–2036. doi: 10.1126/science.287.5460.2032. [DOI] [PubMed] [Google Scholar]
- 37.Lichti U, et al. In vivo regulation of murine hair growth: insights from grafting defined cell populations onto nude mice. J Invest Dermatol. 1993;101:124S–129S. doi: 10.1111/1523-1747.ep12363165. [DOI] [PubMed] [Google Scholar]
- 38.Weinberg WC, et al. Reconstitution of hair follicle development in vivo: determination of follicle formation, hair growth, and hair quality by dermal cells. J Invest Dermatol. 1993;100:229–236. doi: 10.1111/1523-1747.ep12468971. [DOI] [PubMed] [Google Scholar]
- 39.Prouty SM, Lawrence L, Stenn KS. Fibroblast-dependent induction of a murine skin lesion with similarity to human common blue nevus. Am J Pathol. 1996;148:1871–1885. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.