Abstract
The sequence type 11 Klebsiella pneumoniae strain Kpn-3002cz was confirmed to harbor two NDM-1-encoding plasmids, pB-3002cz and pS-3002cz. pB-3002cz (97,649 bp) displayed extensive sequence similarity with the blaNDM-1-carrying plasmid pKPX-1. pS-3002cz (73,581 bp) was found to consist of an IncR-related sequence (13,535 bp) and a mosaic region (60,046 bp). A 40,233-bp sequence of pS-3002cz was identical to the mosaic region of pB-3002cz, indicating the en bloc acquisition of the NDM-1-encoding region from one plasmid by the other.
TEXT
The emergence and spread of carbapenemase-producing Enterobacteriaceae (CPE) have caused a public health crisis of global dimensions (1). One of the carbapenemase groups observed in CPE is metallo-β-lactamases (MβLs), mainly of the VIM, IMP, and NDM types. An NDM-1 enzyme was first described in Klebsiella pneumoniae and Escherichia coli isolated in Sweden in 2008 from an Indian patient transferred from a New Delhi hospital (2). Since then, NDM-1-producing bacteria, including clinical isolates of Enterobacteriaceae and Acinetobacter baumannii, have been reported from the Indian subcontinent but also worldwide (1). The aim of the present study was to describe the first case of an NDM-1-producing K. pneumoniae isolate (Kpn-3002cz) identified in the Czech Republic. We also describe the sequences of two blaNDM-1-carrying plasmids harbored by Kpn-3002cz.
In March 2013, a 61-year-old female was admitted to the Central Military Hospital in Prague, Czech Republic. Since the patient was previously hospitalized (November to December 2012) in Kosice's hospital (Slovak Republic), routine screening for CPE was performed according to the recommendation published by the Ministry of Health of the Czech Republic. K. pneumoniae Kpn-3002cz, which was nonsusceptible to carbapenems, as determined by the broth dilution method (3) and interpreted according to the EUCAST criteria (http://www.eucast.org/), was isolated from a urine sample. Of all the drugs tested, Kpn-3002cz was susceptible to gentamicin only (Table 1).
TABLE 1.
Antibiotic susceptibilities of the K. pneumoniae Kpn-3002cz isolate producing NDM-1 and CMY-4 and its E. coli DH5α transformants producing NDM-1
| Isolate (plasmid) | MIC (mg/liter) fora: |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pip | Tzp | Ctx | Caz | Fep | Atm | Imp | Mem | Ert | Gen | An | Cml | Sxt | Col | Cip | |
| K. pneumoniae Kpn-3002cz | >64 | >64 | >8 | >32 | >16 | 8 | 32 | 8 | 32 | 1 | 32 | 32 | >32 | 4 | >8 |
| E. coli DH5α (pB-3002cz) | 64 | 64 | >8 | >32 | >16 | 0.094 | 2 | 0.75 | 1 | 0.5 | 8 | 2 | 1 | 0.25 | ≤0.06 |
| E. coli DH5α (pS-3002cz) | 64 | 64 | >8 | >32 | 8 | 0.032 | 1.5 | 0.19 | 1 | ≤0.12 | 4 | 2 | 1 | 0.25 | ≤0.06 |
| E. coli DH5α (recipient) | ≤0.5 | 1 | ≤0.06 | ≤0.25 | ≤0.125 | 0.047 | 0.19 | 0.032 | 0.008 | ≤0.12 | ≤0.5 | ≤1 | 1 | ≤0.5 | ≤0.06 |
Pip, piperacillin; Tzp, piperacillin-tazobactam (inhibitor fixed at 4 μg/ml); Ctx, cefotaxime; Caz, ceftazidime; Fep, cefepime; Atm, aztreonam; Imp, imipenem; Mem, meropenem; Ert, ertapenem; Gen, gentamicin; An, amikacin; Cml, chloramphenicol; Sxt, trimethoprim-sulfamethoxazole; Col, colistin; Cip, ciprofloxacin.
Carbapenemase production was hypothesized to exist because of a positive result in the matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) meropenem hydrolysis assay (4). Kpn-3002cz was MβL positive with the meropenem-EDTA combined-disk test (5). PCR screening for various MβL genes (2, 6) and sequencing revealed the presence of blaNDM-1. Isoelectric focusing of β-lactamase extracts showed that Kpn-3002cz also produces β-lactamases, with pIs of 7.6 (most likely the species-specific penicillinase) and 9.0 (a highly basic β-lactamase, as is common among AmpC enzymes). PCR screening for various ampC genes (7) and sequencing revealed the presence of blaCMY-4 that was in accordance with production of the β-lactamase, with a pI of 9.0. Multilocus sequence typing (MLST) (8) assigned Kpn-3002cz to sequence type 11 (ST11), which belongs to clonal complex 258 (CC258) (9).
Attempts to transfer β-lactam resistance to the E. coli A15 laboratory strain by conjugation in mixed broth cultures were unsuccessful. Next, plasmid DNA of Kpn-3002cz was extracted using a Nucleobond AX midi kit (Macherey-Nagel, Düren, Germany) and used to transform E. coli strain DH5α cells. β-Lactam-resistant transformants were obtained on Luria-Bertani agar plates with ampicillin (50 mg/liter) (Table 1) and confirmed to be NDM producers by PCR (2) and MALDI-TOF MS hydrolysis assay (4). Plasmid content analysis was carried out by pulsed-field gel electrophoresis of the total DNA digested with S1 nuclease (10). Then, the DNA was transferred to Hybond-N+ nylon membrane (Amersham Biosciences, Buckingham, United Kingdom) and hybridized with a digoxigenin-labeled blaNDM-specific probe. The results revealed the presence of multiple plasmids in Kpn-3002cz, including molecules of ∼80 kb, ∼100 kb, ∼160 kb, and ∼200 kb. Moreover, plasmid analysis showed two types of transformants carrying either the ∼80-kb plasmid pS-3002cz or the ∼100-kb plasmid pB-3002cz. Both plasmids hybridized with the blaNDM probe (not shown).
Plasmid DNAs extracted from E. coli DH5α transformants were sequenced using the 454 Genome Sequencer Junior system (Roche, Prague, Czech Republic) on a standard DNA fragment library. The reads were assembled using the GS de novo Assembler software. The sequence gaps on the plasmids were filled by sequencing of the PCR-produced fragments. The BLAST algorithm (www.ncbi.nlm.nih.gov/BLAST), IS Finder (www-is.biotoul.fr/), and ORF Finder (www.bioinformatics.org/sms/) were used for data analysis.
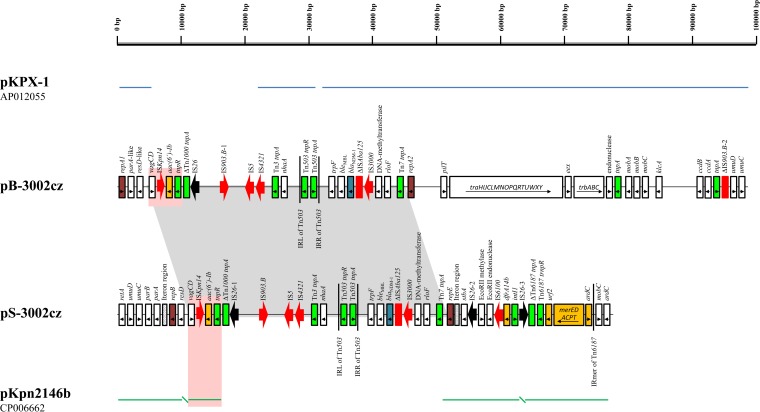
Plasmid pB-3002cz was found to be 97,649 bp in size, with an average G+C content of 53.2%. The sequencing data indicate that pB-3002cz is a derivative of the NDM-1-encoding plasmid pKPX-1 (90% coverage, 99% identity). pKPX-1 was characterized from an ST11 K. pneumoniae strain isolated in Taiwan from a patient previously hospitalized in a New Delhi (India) medical center (11). The similarities between pB-3002cz and pKPX-1 were localized in two main segments (Fig. 1). The first segment is composed of a contiguous 70,412-bp sequence (nucleotides [nt] 1 to 6065 and 33303 to 97649) containing a part of the plasmidic backbone and the NDM-1-encoding region. The plasmidic scaffold of pB-3002cz possesses a conjugative transfer region (tra-trb), the mobABC, ccdAB, vagCD, and umuDC operons, and two repA genes. Similarly to pKPX-1, none of the repA genes were assigned to any of the known incompatibility (Inc) groups by the PCR-based replicon typing (PBRT) method (12). The NDM-1-encoding region is composed of bleMBL, trpF, blaNDM-1, and a 259-bp fragment of ISAba125 (ΔISAba125). ISAba125 is disrupted by an IS3000 element, as has also been found in the IncH plasmid pNDM-MAR (13). The remaining sequence of pB-3002cz, including the second pKPX-1-like segment (nt 21402 to 31550), comprises a mosaic structure containing intact insertion sequences (n = 5) and fragments of various transposons (n = 4). Furthermore, pB-3002cz showed a deletion of two big fragments of pKPX-1 being responsible for resistance to several antibiotics and for plasmid maintenance.
FIG 1.
Linear maps of two NDM-encoding plasmids, pB-3002cz and pS-3002cz. Open reading frames (ORFs) are shown as rectangles (arrows within the rectangles indicate the direction of transcription). Intact insertion sequences (ISs) are represented by arrows, while truncated ISs appear as rectangles. The replicons of the plasmids are indicated by purple rectangles. blaNDM-1 genes, resistance genes, and transposases are shown in blue, orange, and green, respectively. The black arrows indicate IS26 elements; the other IS elements are shown in red. The remaining genes, including plasmidic scaffold regions, are indicated by white rectangles. The blue lines above the map of pB-3002cz correspond to highly similar sequences from plasmid pKPX-1, while the green line below the map of pS-3002cz corresponds to highly similar sequences from plasmid pKpn2146b. The gray shading shows the presence of the same NDM-1-encoding mosaic region on both pB-3002cz and pS-3002cz. Homologous regions, which might have been involved in a recombination event leading to the shaping of pS-3002cz, are indicated by pink shading. IRL, left inverted repeat; IRR, right inverted repeat.
The second plasmid, pS-3002cz, is 73,581 bp in size, with an average G+C content of 53.9%. Sequence analysis showed that pS-3002cz is a fusion derivative of pB-3002cz and pKpn2146b. pKpn2146b is one of the four plasmids harbored by K. pneumoniae strain ATCC BAA-2146 (Kpn2146), the first NDM-1-producing isolate from the United States (14). Kpn2146, which also belongs to ST11, was recovered from a patient who had received medical care in India. Of note is that pKpn2146b was not found to harbor blaNDM-1, which was carried by the IncA/C plasmid pNDM-US (14). pS-3002cz contains a 40,233-bp sequence (nt 10402 to 50634) encoding the NDM-1, which was identical to the entire mosaic region of pB-3002cz (100% coverage, 99% identity) (Fig. 1). The remaining 33,348-bp sequence of pS-3002cz consists of two segments sharing extensive similarities with sequences carried by pKpn2146b (100% coverage, 99% identity). The first segment, a contiguous 20,721-bp sequence (nt 1 to 10401 and 63262 to 73581), is composed of an IncR-derived fragment, which includes a repB gene (nt 6909 to 7775) being 100% identical to that found in the IncR plasmid pKP1780 (15), and a truncated Tn6187 transposon (ΔTn6187) (14). The second segment is an 11,807-bp sequence (nt 50635 to 62441) containing the repE gene (nt 51498 to 52253). repE showed limited nucleotide similarity (86%) with the replication region of the IncFIA plasmid pmk115 (12), which explains the failure of the PBRT method to classify pS-3002cz. However, pS-3002cz was found to be positive in an IncR-specific PCR assay (16).
In conclusion, this study presents the first NDM-1-producing K. pneumoniae strain in the Czech Republic. However, the clinical data of the patient indicate that the isolate was probably imported from Slovakia, a country with a currently unknown epidemiological situation regarding CPE (17). The production of NDM-1 occurs in ST11, which has been associated with NDM-1-producing isolates having an epidemiological link to the Indian subcontinent (18) and which has recently caused outbreaks in Greece (19).
Of note is that in Kpn-3002cz, blaNDM-1 is carried on two plasmids, pB-3002cz and pS-3002cz, sharing the same NDM-1-encoding mosaic region (Fig. 1). Sequencing data suggest that the progenitor of pB-3002cz is a pKPX-1-like plasmid. Considering that pKPX-1-like plasmids are not common, the presence of pB-3002cz and pKPX-1 in two K. pneumoniae strains of the same ST underlines the importance of pKPX-1-like plasmids in the dissemination of the blaNDM-1 gene in different parts of the world. Furthermore, sequencing data indicate that en bloc acquisition of the NDM-1-encoding mosaic region from pB-3002cz by an IncR-type plasmid is a plausible hypothesis regarding the formation of pS-3002cz. At a certain step of the evolution of pS-3002cz, pKPX-1 and an IncR pKpn2146b-like plasmid may have formed a fusion structure via a recombination event between a homologous region (nt 6033 to 10111 in pB-3002cz; Fig. 1) upstream of the resD gene. After the resolution of the fusion structure, pS-3002cz evolved further through rearrangements facilitated by ISs. These findings underscore the spreading potential of large segments carrying resistance determinants, such as blaNDM-1, through the reshuffling of enterobacterial plasmids.
Nucleotide sequence accession numbers.
The nucleotide sequences of the plasmids pB-3002cz and pS-3002cz have been assigned the GenBank accession numbers KJ958926 and KJ958927, respectively.
ACKNOWLEDGMENTS
This work was supported by research project grant NT11032-6/2010 from the Ministry of Health of the Czech Republic and by the Charles University Research Fund (project no. P36) and CEITEC (grant CZ.1.05/1.1.00/02.0068). C. C. Papagiannitsis was supported by the project “Support of establishment, development, and mobility of quality research teams at the Charles University” (registration no. CZ.1.07/2.3.00/30.0022), and M. Dolejska was supported by the project “Molecular biology as a tool for integrating biomedical disciplines at VFU Brno” (registration no. CZ.1.07/2.3.00/30.0014), financed by The Education for Competitiveness Operational Programme (ECOP), funded by the European Social Fund (ESF) and the government budget of the Czech Republic.
REFERENCES
- 1.Nordmann P, Poirel L, Walsh TR, Livermore DM. 2011. The emerging NDM carbapenemases. Trends Microbiol 19:588–595. doi: 10.1016/j.tim.2011.09.005. [DOI] [PubMed] [Google Scholar]
- 2.Yong D, Toleman MA, Giske CG, Cho HS, Sundman K, Lee K, Walsh TR. 2009. Characterization of a new metallo-β-lactamase gene, blaNDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother 53:5046–5054. doi: 10.1128/AAC.00774-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.European Committee on Antimicrobial Susceptibility Testing. 2003. Determination of minimum inhibitory concentrations (MICs) of antibacterial agents by broth dilution. Clin Microbiol Infect 9:1–7. doi: 10.1046/j.1469-0691.2003.00790.x.12691538 [DOI] [Google Scholar]
- 4.Hrabák J, Studentová V, Walková R, Zemlicková H, Jakubu V, Chudácková E, Gniadkowski M, Pfeifer Y, Perry JD, Wilkison K, Bergerová T. 2012. Detection of NDM-1, VIM-1, KPC, OXA-48, and OXA-162 carbapnemases by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J Clin Microbiol 50:2441–2443. doi: 10.1128/JCM.01002-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hrabák J, Chudáčková E, Papagiannitsis CC. 2014. Detection of carbapenems in Enterobacteriaceae: a challenge for diagnostic microbiological laboratories. Clin Microbiol Infect, 20:839–853. doi: 10.1111/1469-0691.12678. [DOI] [PubMed] [Google Scholar]
- 6.Ellington MJ, Kistler J, Livermore DM, Woodford N. 2007. Multiplex PCR for rapid detection of genes encoding acquired metallo-β-lactamases. J Antimicrob Chemother 59:321–322. doi: 10.1093/jac/dkl481. [DOI] [PubMed] [Google Scholar]
- 7.Pérez-Pérez FJ, Hanson ND. 2002. Detection of plasmid-mediated AmpC beta-lactamase genes in clinical isolates by using multiplex PCR. J Clin Microbiol 40:2153–2162. doi: 10.1128/JCM.40.6.2153-2162.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diancourt L, Passet V, Verhoef J, Grimont PAD, Brisse S. 2005. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolate. J Clin Microbiol 43:4178–4182. doi: 10.1128/JCM.43.8.4178-4182.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Woodford N, Turton JF, Livermore DM. 2011. Multiresistant Gram-negative bacteria: the role of high-risk clones in the dissemination of antibiotic resistance. FEMS Microbiol Rev 35:736–755. doi: 10.1111/j.1574-6976.2011.00268.x. [DOI] [PubMed] [Google Scholar]
- 10.Barton BM, Harding GP, Zuccarelli AJ. 1995. A general method for the detecting and sizing large plasmids. Anal Biochem 226:235–240. doi: 10.1006/abio.1995.1220. [DOI] [PubMed] [Google Scholar]
- 11.Huang TW, Chen TL, Chen YT, Lauderdale TL, Liao TL, Lee YT, Chen CP, Liu YM, Lin AC, Chang YH, Wu KM, Kirby R, Lai JF, Tan MC, Siu LK, Chang CM, Fung CP, Tsai SF. 2013. Copy number change of the NDM-1 sequence in a multidrug-resistant Klebsiella pneumoniae clinical isolate. PLoS One 8:e62774. doi: 10.1371/journal.pone.0062774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threfall EJ. 2005. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods 63:219–228. doi: 10.1016/j.mimet.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 13.Hudson CM, Bent ZW, Meagher RJ, Williams KP. 2014. Resistance determinants and mobile genetic elements of an NDM-1-encoding Klebsiella pneumoniae strain. PLoS One 9:e99209. doi: 10.1371/journal.pone.0099209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Villa L, Poirel L, Nordmann P, Carta C, Carattoli A. 2012. Complete sequencing of an IncH plasmid carrying the blaNDM-1, blaCTX-M-15, and qnrB1 genes. J Antimicrob Chemother 67:1645–1650. doi: 10.1093/jac/dks114. [DOI] [PubMed] [Google Scholar]
- 15.Papagiannitsis CC, Miriagou V, Giakkoupi P, Tzouvelekis LS, Vatopoulos AC. 2013. Characterization of pKP1780, a novel IncR plasmid from the emerging Klebsiella pneumoniae ST147, encoding the VIM-1 metallo-β-lactamsase. J Antimicrob Chemother 68:2259–2262. doi: 10.1093/jac/dkt196. [DOI] [PubMed] [Google Scholar]
- 16.García-Fernández A, Fortini D, Veldman K, Mevius D, Carattoli A. 2009. Characterization of plasmids harbouring qnrS1, qnrB2 and qnrB19 genes in Salmonella. J Antimicrob Chemother 63:274–281. doi: 10.1093/jac/dkn470. [DOI] [PubMed] [Google Scholar]
- 17.Glasner C, Albiger B, Buist G, Tambić Andrašević A, Canton R, Carmeli Y, Friedrich AW, Giske CG, Glupczynski Y, Gniadkowski M, Livermore DM, Nordmann P, Poirel L, Rossolini GM, Seifert H, Vatopoulos AC, Walsh T, Woodford N, Donker T, Monnet DL, Grundmann H, European Survey on Carbapenemase-Producing Enterobacteriaceae (EuSCAPE) Working Group . 2013. Carbapenemase-producing Enterobacteriaceae in Europe: a survey among national experts from 39 countries, February 2013. Euro Surveill 18:pii=20525 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20525. [DOI] [PubMed] [Google Scholar]
- 18.Giske CG, Fröding I, Hasan CM, Turlej-Rogacka A, Toleman M, Livermore D, Woodford N, Walsh TR. 2012. Diverse sequence types of Klebsiella pneumoniae contribute to the dissemination of blaNDM-1 in India, Sweden, and the United Kingdom. Antimicrob Agents Chemother 56:2735–2738. doi: 10.1128/AAC.06142-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Voulgari E, Gartzonika C, Vrioni G, Politi L, Priavali E, Levidiotou-Stefanou S, Tsakris A. 2014. The Balkan region: NDM-1-producing Klebsiella pneumoniae ST11 clonal strain causing outbreaks in Greece. J Antimicrob Chemother 69:2091–2099. doi: 10.1093/jac/dku105. [DOI] [PubMed] [Google Scholar]